Abstract
Purpose of review
Recent developments have generated renewed interest in the possibility of curing HIV-1 infection. This review describes some of the practical challenges that will need to be overcome if curative strategies are to be successful.
Recent findings
The latent reservoir for HIV-1 in resting memory CD4+ T cells is the major barrier to curing the infection. The most widely discussed approach to curing the infection involves finding agents that reverse latency in resting CD4+ T cells, with the assumption that the cells will then die from viral cytopathic effects or be lysed by host cytolytic T lymphocytes (CTL). A major challenge is the development of in vitro models that can be used to explore mechanisms and identify latency reversing agents (LRA). Although several models have been developed, including primary cell models, none of them may fully capture the quiescent state of the cells that harbor latent HIV-1 in vivo. An additional problem is that LRA that do not cause T cell activation may not lead to the death of infected cells. Finally, measuring the effects of LRAs in vivo is complicated by the lack of correlation between different assays for the latent reservoir.
Summary
Progress on these practical issues is essential to finding a cure.
Keywords: HIV latency, eradication, reservoir
Introduction
The recent cure of a single patient with HIV-1 infection using a bone marrow transplantation strategy [1] has renewed hopes that eradication may eventually become possible on a large scale. Finding ways to cure the infection without the risks inherent in bone marrow transplantation has become a major research priority [2,3]. The major barrier to HIV-1 eradication is a small pool of resting memory CD4+ T cells that carry stably integrated, replication-competent HIV-1 genomes [4–11]. There is great current interest in developing novel strategies for targeting this reservoir. However, the practical challenges inherent in doing this remain daunting. The purpose of this article is to describe some of these practical challenges in a quantitative way.
Viral dynamics
HIV-1 eradication strategies can be best understood through a consideration of fundamental models of viral dynamics. When patients start on a potent antiretroviral drug, plasma virus levels undergo rapid exponential decay [12,13] which can be described by a simple mathematical model, originally developed by Nowak [12,14,15] and Perelson [13,16,17], in which free virus interacts with uninfected cells to generate infected cells, which go on to produce virus at a characteristic rate. Most of productively infected cells are activated CD4+ T cells (Figure 1A). Antiretroviral drugs block new infection of susceptible cells without blocking virus production by cells that are already infected, and although the free virus in the plasma decays rapidly, cells that were infected before therapy was started can continue to produce virus. Therefore, the decay of viremia is really determined by the decay rate of the productively infected cells, symbolized here as a. From a. we can determine the half life of the cells that produce most of the plasma virus. It is simply ln2 / a, and it is very short, only one day [12,13].
Figure 1.
Viral dynamics in HIV-1 infection. (A) Basic model of viral dynamics based. Free virus (v) interacts with uninfected cells to generate productively infected cells at a rate that depends on the concentrations of each and a rate constant β. These cells produce virus at rate k . Productively infected cells and free virus undergo exponential decay at rates a and u, respectively. The production and clearance rates for uninfected cells are not shown. The block imposed by ART is indicated by a black box. (B) Expanded model to account for a second phase of viral decay. This phase is due to another population of infected cells with a slower decay rate (m). (C) Futher expansion of the model to account for latently infected cells. Productively infected CD4+ T lymphoblasts can transition into a latent state at rate l, and these cells can be reactivated at rate r. The decay rate of the latent population (a′) is much slower than the decay rate of productively infected cells (a′≪a). (D) The fundamental dynamic in patients on ART. ART blocks new infections, and labile populations decay. The decay rate of the latent reservoir (a′) is so slow as to be negligible.
If the initial therapy is monotherapy, preexisting resistant variants can grow out. However, if the initial treatment is an appropriate combination of antiretroviral drugs, then viremia falls to below the limit of detection of clinical assays, as was shown in pioneering studies in 1997 [17–19]. The decay is biphasic, due to the presence of another population of cells which become infected and produce virus, but which decay at a slower rate (t1/2 =14 days, Figure 1B). Combination antiretroviral therapy (ART) blocks infection of these cells, and after most of the activated CD4+ T cells have died, viremia falls at the second slower decay rate. Initial predictions in 1997 that ART might be curative were based on this second slower decay rate [17]. However, at about this same time, it was hypothesized that a third population of infected cells with an even slower decay rate might prevent eradication. These are latently infected, resting memory CD4+ T cells [4,5].
The existence of a latent reservoir for HIV-1 in resting memory cells can be considered as a consequence of the normal way in which immunologic memory is established. When a resting CD4+ T cell encounters antigen, it undergoes blast transformation and divides, ultimately generated many activated effector cells of the same specificity. At the conclusion of the immune response, many of these activated cells die, but some survive and return to a resting state as long lived memory cells that allow future responses to the same antigen. HIV-1 replicates mainly in the activated cells, resulting in their death at rate a. The virus does not replicate well in resting T cells as a result of low dNTP pools and other factors [20,21]. However, on rare occasions activated T cells can become infected as they are returning back to a resting state. This results in a stably integrated viral genome in a long lived memory T cell.
Interestingly, as the cell returns to a resting memory state, HIV-1 gene expression is turned off. One reason is that HIV-1 gene expression is heavily dependent upon the host transcription factor NFκB, which is excluded from the nucleus in resting cells [22–25]. The end result is a stably integrated but transcriptionally silent form of the viral genome in a long lived memory T cell. This is a perfect mechanism for viral persistence; it allows the virus to persist essentially as pure information, unaffected by immune responses or antiretroviral drugs. If the cell becomes activated in the future, it can begin to produce virus again.
The scenario described above was simply a hypothesis until it was demonstrated that replication-competent virus could be released following the activation of resting memory CD4+ T cells from infected individuals. A quantitative viral outgrowth assay was used to demonstrate the presence of latently infected cells in infected individuals [4,5] and persistence of these cells in patients on suppressive ART [4–11]. The viral outgrowth assay is based on the model. To detect latently infected cells, it is necessary to reverse latency by inducing global T cell activation [26]. Resting CD4+ T cells from patients on ART are plated in limiting dilution and subjected to maximum activation with the mitogen phytohemaglutinnin (PHA), which induces 100% of the cells to undergo blast transformation (Figure 2). Latently infected cells can then produce virus which is expanded through coculture with two additions of CD4+ lymphoblasts from normal donors. After two weeks, free virus is measured in the supernatant by Elisa assay for p24 antigen. The frequency of cells that were induced to release replication-competent virus is determined by Poisson statistics and is generally around 1/106 resting CD4+ T cells. Although the frequency is low, it does not decrease significantly even after years of suppressive HAART [9,10]. The low frequency of latently infected cells may reflect the fact that latency is only established if cells are infected in a narrow time window after activation when levels of the HIV-1 coreceptor CCR5 are high but conditions for viral gene expression are suboptimal, allowing the cells to escape viral cytopathic effects and return to a resting memory state.
Figure 2.
Quantitative viral outgrowth assay used to define the latent reservoir [5,6,26]. See text for details.
This model emphasizes the close connection between immunologic memory and HIV-1 latency. Interestingly, antigen exposure and memory cell generation begin at birth. This provides an explanation for the remarkable case of an infant who acquired HIV-1 infection in utero infection but was treated with suppressive ART within 30 hours of birth (Persaud et al., 2013, submitted). This child appears to have been cured, probably because viral replication was suppressed before a stable reservoir in memory T cells was generated.
Given the existence of this latent pool, the model described above needs to be expanded because infected CD4+ T cells can transition into and out of a latent state (Figure 1C). ART blocks new infection, and labile populations decay. The decay rate of the pool of latently infected cells is so slow as to be negligible [9,10]. This leaves the dynamic situation shown in Figure 1D. Every day, a small number of latently infected cells become activated and produce virus. Thus viremia does not continue to decay but rather levels off at values that are below the limit of detection of current clinical assays. In most patients, this residual viremia is around 1 copy/ml [27,28]. It appears to reflect the activation of latently infected cells and possibly release of virus from other stable reservoirs [29].
Eliminating latently infected cells
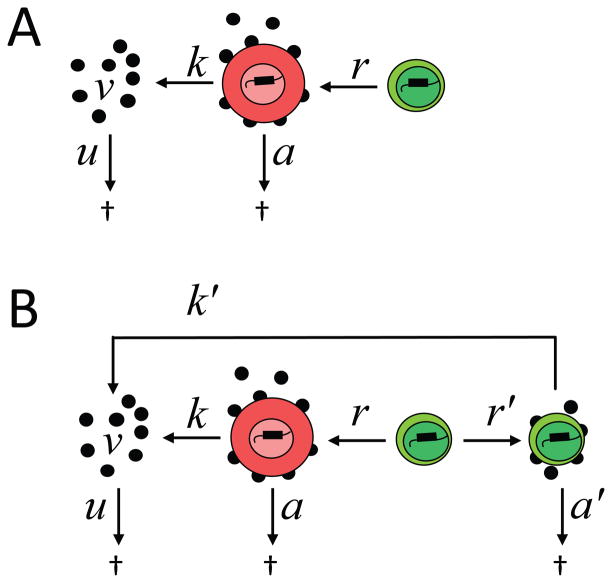
Initial approaches to eliminating latently infected cells made use of non-specific T cell activation with IL-2 [30] or anti-CD3 antibodies [31] to accelerate the normally slow rate at which latently infected cells become activated, with the assumption that the cells would then decay at rate a (Figure 3A). However global T cell activation is associated with cytokine storm and unacceptable toxicity. Therefore there has been interest in finding a way to turn on virus gene expression without inducing T cell activation (Figure 3B). However, this approach raises important questions. First, will the cells actually die? It cannot be assumed that the cells will decay at rate a because these cells are in a very different state than the activated CD4+ T cells that produce most of the plasma virus. This issue is discussed below. The first question is whether it is possible to induce HIV-1 gene expression without inducing global T cell activation.
Figure 3.
Strategies for targeting latently infected cells. (A) Strategies involving T cell activation. Initial strategies focused on inducing global T cell activation to accelerate the normally slow rate (r) at which latently infected cells become activated, with the assumption that the cells will then die rapidly at rate a. New infections are largely blocked by ART and are not shown. The decay rate of the latent reservoir is extremely slow and is not shown. (B) Current strategies. (B) Alternative strategies. Because of the toxicity associated with inducing global T cell activation, alternative strategies seeks to reactivate latent HIV-1 without inducing T cell activation. The rate of reactivation of HIV-1 gene expression (r′) must be greater than the normal rate r. Because the cells are not activated, the rate of virus production (k′) may be lower than the rate of virus production by activated CD4+ T cells (k′). However, the cells also survive longer (a′≪a). The relationship between these rate constants will determine whether latency reversing agents cause a transient increase in residual viremia and a long term decrease in the size of the latent reservoir.
In vitro models of HIV-1 latency have proven useful in the analysis of the molecular mechanisms involved in HIV-1 latency and in the identification of agents that reverse latency. The simplest models are transformed epithelial cell lines transfect with HIV-1 LTR reporter constructs. These are being used in ongoing chemical library screens by pharmaceutical companies. Transformed T cell lines have been used extensively, and through elegant selection protocols, it is possible to obtain clonal populations in which all the cells carry an integrated provirus [32]. However the cells that harbor latent HIV-1 in vivo are in G0, and therefore recent efforts have focused on non-transformed, primary T cells models (reviewed in [33]). Ultimately latency reversing agents should be tested in cells from patients, but increased in vivo relevance comes at a cost in screening throughput. Most of what we know about the mechanism of latency comes from studies in transformed cells.
Multiple mechanisms have been identified in studies by Anthony Fauci, Warner Greene, Jon Karn, Matija Peterlin, David Margolis, Eric Verdin and many others. These mechanisms will not be discussed in detail here but are summarized in several recent reviews [34–36]. The mechanisms include the sequestration of critical host factors needed for initiation and elongation of transcription as well as repressive chromatin modifications such as DNA methylation and histone deactylation. These mechanisms suggest potential latency reversing agents, but it is currently unclear which of these mechanisms is most important in vivo. To date, histone deacetylase (HDAC) inhibitors have received the most attention as potential latency reversing agents, although there is controversy over the precise mechanism involved [37–40]. In a recent clinical study, a single dose of the the HDAC inhibitor vorinostat (suberoylanilide hydroxamic acid or SAHA) was shown to cause a measurable increase in cell associated HIV-1 RNA in CD4+ T cells from peripheral blood [40].
Because transformed cell lines do not precisely mimick the quiescent state of the cells have harbor latent HIV-1 in vivo, we developed on a primary T cell model which essentially recapitulates the way in which latency is established in vivo [41]. Primary CD4+ T cells are first transduced with the bcl-2 which allows them to survive in vitro but otherwise behave normally. They can be activated, infected an HIV-1 reporter virus carrying GFP, and then cultured for long enough to allow some of these cells to revert to a quiescent state and turn off HIV-1 gene expression. In this model, latency can be readily reversed with T cell activation. This model can be used to screen for compounds that reverse latency without inducing global T cell activation. To date, several classes of latency reversing agents have been identified using this model [41–43] One of these classes includes the drug disulfiram, which has been used clinically for over 50 years in the treatment of alcoholism. A clinical trial of disulfiram in patients on HAART is underway.
A major concern in all studies of latency reversing agents is that although the agents may reactivate latent HIV-1 in cell line and primary cell models, they may not work as well in vivo. There may be some additional aspect of HIV-1 latency in vivo that is not captured by any of these models. Thus although vorinostat works well in various in vitro models and induces some increase in cell associated HIV-1 RNA in vivo, it does not induce virus production by cells from patients on ART [44]. It is clearly best to test putative latency reversing agents with cells from patients. However, this is difficult because the frequency of latently infected cells is very low, averaging only 1/106 resting CD4+ T cells. Thus for example from a large 200 ml blood sample, it may be possible to purify 20 x 106 resting CD4+ T cells, which means that there are only 20 latently infected cells to work with. Of course the sample must be split so that positive and negative controls can be done. Thus it is extremely difficult to evaluate latency reversing agents using cells from patients on ART. Because of this frequency problem, few of the latency reversing agents being considered today have actually been shown to reverse latency in cells from patient on ART.
Assuming that it will be possible to find agents that reverse latency in vivo, the next major issue is whether the cells will die following reversal of latency. It cannot be assumed that they will die rapidly at decay at rate a because they are not in an activated state. This issue can be addressed in primary cell models of latency. When latency is reversed through T cell activation, infected cells die quickly. However, when latency is reversed with a HDAC inhibitor that does not induce T cell activation, the cells do not die [45]. Of course in vivo it is possible that they would be lysed by HIV-1-specific cytolytic T lymphocytes (CTL). To examine this possibility, we isolated resting CD4+ T cells from selected donors, transduced them with bcl-2, infected them with an HIV-1 reporter virus, and allowed them to return to a quiescent state. We then obtained a second blood sample from the same donor and isolated CD8+ T cells. These cells are then cocultured with autologous CD4+ T cells in which latency has been reversed with an HDAC inhibitor. Then we simply followed the disappearance of productively infected cells. If the experiment is performed with cells from normal donors, infected cells are not cleared because there is no HIV-1-specific CTL response. If the experiment is performed with cells from elite suppressors, who generally have strong HIV-1-specific CTL responses, productively infected cells are readily cleared. However, the critical issue is what happens with cells from patients on ART because latency reversing strategies will be implemented in the setting of ART. We found that for most patients on ART, infected cells were not cleared because of qualitative and quantitative defects in the HIV-1-specific CTL response [45]. Importantly, these defects could be reversed by in vitro stimulation of the CD8+ T cells with Gag peptides, suggesting that it may be necessary to combine latency reversing strategies with therapeutic vaccination.
Measuring changes in the latent reservoir
Even if promising strategies for eliminating latently infected cells can be developed, there remains the problem of how measuring reductions in the reservoir in patients participating in eradication trials. There is no clinical assay for the reservoir. The viral outgrowth assay used to define the reservoir is difficult, expensive, and time-consuming, requiring 2–3 week of tissue culture in a BLS3 facility [26]. PCR assays would be much simpler, but it has been unclear how well PCR assays correlate with the viral outgrowth assay. Therefore a collaborative study was undertaken to compare 11 different approaches for measure persistent HIV-1 using a set of samples from two well characterized cohorts of patients who started HAART either during acute or chronic infection [46]. The viral outgrowth assay was used a standard of comparison. With this assay, infected cell frequencies varied over a two log range centered around 1/106, with lower values in patients starting HAART during acute infection The simplest alternative approach would be to measure proviral DNA in unfractionated peripheral blood mononuclear cells (PBMC by PCR). This was done using a very accurate droplet digital PCR [47] which gave infected cell frequencies about two logs higher than those obtained with the viral outgrowth assay . The results of the two assays were not well correlated. One potential reason is that the viral outgrowth assay is done on purified resting CD4+ T cells and we thought the correlation might be better if the PCR assay were also done on resting cells, not PBMC. However, even when the droplet digital PCR assay is done on DNA from purified resting CD4+ T cells, the correlation with viral outgrowth remains poor. This reflects the fact that the ratio between infected cells frequencies determined by viral outgrowth and by PCR varies dramatically from patient to patient, by thousands of fold. If this ratio was constant from patient to patient, then DNA PCR would make a convenient surrogate measure in cross sectional analysis. Unfortunately, it is not constant.
The stable reservoir for HIV-1 is present in the form of integrated HIV-1DNA [4,5]. Some PCR assays such as Alu PCR specifically detect integrated HIV-1 DNA [48,49] and not the unintegrated form that is the dominant species in untreated patients [50]. When Alu PCR was used, infected cells frequencies were similar to those observed with droplet digital PCR, indicating that most of the HIV-1 DNA in patients on ART is integrated [46]. A modest correlation with viral outgrowth was observed with Alu PCR assays on PBMC, but the correlation was weaker when the PCR was done on purified resting CD4+ T cells. HIV-1 DNA levels were also measured by qPCR in CD4+ T cells from rectal biopsies. Here the infected cell frequencies were even higher than these observed in blood, but again not well correlated with viral outgrowth. 2 LTR circles, a non-functional circular form of the viral genome, were below the detection limit in most patients. Residual viremia, which is an indication of ongoing virion production in patients on ART [27,28,51,52], was below the limit of detection in many patients even with an extremely sensitive assay with a limit of detection below 1 copy/ml.
Together these results raise serious issues regarding the choice of an assay to monitor the size of the reservoir in patients participating in eradication studies. The very large (>100 fold) discrepancy between infected cells frequencies measured by viral outgrowth vs. PCR suggests that there are many proviruses that are not detected in the culture assay. We have termed these non-induced proviruses. This term was chosen carefully; non-induced does not mean that they are non-inducible. All we know is that after a single round of T cell activation, they were not induced to produce virus replication competent virus that could grow out in a two week in vitro expansion. Some of these proviruses may have been lethally hypermutated by APOBEC3G which deaminates cytidines on the minus strand of viral cDNA during reverse transcription [53–56]. This results in G→A hypermutation which converts many tryptophan codons to stop codons, which are found in every open reading frame. Some non-induced proviruses may contain large internal deletions, as was suggested by Ivan Hirsch several years ago [57]. The presence of defective proviruses raises the possibility that most of what is detected by PCR is irrelevant. In addition, PCR assays may not be appropriate for longitudinal monitoring of patients in eradication trials because eradication strategies depend on the production of viral proteins, which can be recognized by an augmented CTL response. However, cells with defective proviruses may not make viral proteins and may not be eliminated even by strategies that effectively eliminate cells with replication-competent virus.
Even more disturbing is the possibility that some of the non-induced proviruses may be completely intact at the primary sequence level. Although they were not induced to produce replication-competent virus after a single round of T cell activation, it remains possible that they could be induced to do so in vivo under some conditions. The frequency of the cells carrying these potentially replication-competent non-induced proviruses could be substantially higher than the frequency of cells that score in the viral outgrowth assay. If these intact non-induced proviruses can be induced in vivo, then the latent reservoir is substantially larger than previously thought. Until the potential threat posed by intact, non-induced proviruses is fully understood, the viral outgrowth assay may represent the best available measure of the reservoir. Fortunately, simpler versions of the assay have recently been developed (Laird et al, submitted).
These results highlight the problems that are likely to be encountered in monitoring patients participating in eradication trials. The only widely available virologic assay is the RT-PCR assay for HIV-1 RNA in plasma. For most patients on ART, levels of viremia are already below the limit of detection of this assay. It is possible that latency reversing agents will cause a transient increase in residual viremia. This is an important because it would mean that an available clinical assay could be used to determine whether these agents are having an effect in a given patient. Latency reversing agents need to reactivate latent HIV-1 at a rate that is greater than the normal rate at which latently infected cells become activated. Because the resulting cells remain in a resting state, they may produce virus at a rate that is lower than the rate of virus production by activated cells [Shan et al., in press]. However, they also decay at a much slower [45]. Therefore it is possible that these agents will cause a transient increase in residual viremia. In initial clinical studies with vorinostat, no increase in residual viremia was seen.
Conclusions
Significant practical challenges must be overcome before HIV-1 eradication can become a reality on wide scale. These include the development of model systems that can be used to understand mechanisms of latency and to identify agents that reverse latency in vivo. In addition, we need a better understanding of the fate of cells following reversal of latency. Unless the cells die or are lysed by host cytolytic effector cells, reversing latency will have little benefit. Finally, we need reliable, scalable assays for the latent reservoir to use in monitoring progress in eradication efforts.
Key Points.
The stable latent reservoir for HIV-1 in resting memory CD4+ T cells is a major barrier to HIV-1 eradication.
A better understanding of models systems to study HIV-1 latency is urgently needed.
Reactivating latent HIV-1 may not be sufficient to induce the elimination of latently infected cells.
Improved assays that distinguish replication-competent vs. defective proviruses are needed so that the effectiveness of eradication strategies can be monitored clinically.
Acknowledgments
This work was supported by an ARCHE Collaborative Research Grant from the Foundation for AIDS Research (amFAR 108165-50-RGRL), by the Martin Delaney CARE and DARE Collaboratories (NIH grants AI096113 and 1U19AI096109), by the Johns Hopkins Center for AIDS Research, and by NIH grant 43222 (RFS), and by the Howard Hughes Medical Institute.
Footnotes
Conflict of Interest
None.
References
- 1.Hutter G, Nowak D, Mossner M, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 2.Richman DD, Margolis DM, Delaney M, et al. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 3•.Deeks SG, Autran B, et al. The International AIDS Society Scientific Working Group on HIV Cure. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol. 2012;12:607–614. doi: 10.1038/nri3262. An excellent review and summary of the research agenda for eradication studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chun TW, Finzi D, Margolick J, et al. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. 1995;1:1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 5.Chun TW, Carruth L, Finzi D, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 6.Finzi D, Hermankova M, Pierson T, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 7.Wong JK, Hezareh M, Gunthard HF, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 8.Chun TW, Stuyver L, Mizell SB, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finzi D, Blankson J, Siliciano JD, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 10.Siliciano JD, Kajdas J, Finzi D, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 11.Strain MC, Gunthard HF, Havlir DV, et al. Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: intrinsic stability predicts lifelong persistence. Proc Natl Acad Sci U S A. 2003;100:4819–4824. doi: 10.1073/pnas.0736332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei X, Ghosh SK, Taylor ME, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 13.Ho DD, Neumann AU, Perelson AS, et al. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 14.Nowak MA, Lloyd AL, Vasquez GM, et al. Viral dynamics of primary viremia and antiretroviral therapy in simian immunodeficiency virus infection. J Virol. 1997;71:7518–7525. doi: 10.1128/jvi.71.10.7518-7525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wodarz D, Nowak MA. Mathematical models of HIV pathogenesis and treatment. Bioessays. 2002;24:1178–1187. doi: 10.1002/bies.10196. [DOI] [PubMed] [Google Scholar]
- 16.Perelson AS, Neumann AU, Markowitz M, et al. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 17.Perelson AS, Essunger P, Cao Y, et al. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 18.Hammer SM, Squires KE, Hughes MD, et al. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N Engl J Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 19.Gulick RM, Mellors JW, Havlir D, et al. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 20••.Berger A, Sommer AF, Zwarg J, et al. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 2011;7:e1002425. doi: 10.1371/journal.ppat.1002425. Important description of a new restriction factor that affects HIV-1 replication by modulating nucleotide pools. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Laguette N, Sobhian B, Casartelli N, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. Important description of a new restriction factor that affects HIV-1 replication by modulating nucleotide pools. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siekevitz M, Josephs SF, Dukovich M, et al. Activation of the HIV-1 LTR by T cell mitogens and the trans-activator protein of HTLV-I. Science. 1987;238:1575–1578. doi: 10.1126/science.2825351. [DOI] [PubMed] [Google Scholar]
- 23.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 24.Bohnlein E, Lowenthal JW, Siekevitz M, et al. The same inducible nuclear proteins regulates mitogen activation of both the interleukin-2 receptor-alpha gene and type 1 HIV. Cell. 1988;53:827–836. doi: 10.1016/0092-8674(88)90099-2. [DOI] [PubMed] [Google Scholar]
- 25.Duh EJ, Maury WJ, Folks TM, et al. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proc Natl Acad Sci U S A. 1989;86:5974–5978. doi: 10.1073/pnas.86.15.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siliciano JD, Siliciano RF. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol. 2005;304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- 27.Palmer S, Wiegand AP, Maldarelli F, et al. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 2003;41:4531–4536. doi: 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmer S, Maldarelli F, Wiegand A, et al. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A. 2008;105:3879–3884. doi: 10.1073/pnas.0800050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey JR, Sedaghat AR, Kieffer T, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80:6441–6457. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davey RT, Jr, Bhat N, Yoder C, et al. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci U S A. 1999;96:15109–15114. doi: 10.1073/pnas.96.26.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulkosky J, Nunnari G, Otero M, et al. Intensification and stimulation therapy for human immunodeficiency virus type 1 reservoirs in infected persons receiving virally suppressive highly active antiretroviral therapy. J Infect Dis. 2002;186:1403–1411. doi: 10.1086/344357. [DOI] [PubMed] [Google Scholar]
- 32.Jordan A, Bisgrove D, Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003;22:1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Yang HC. Primary cell models of HIV latency. Curr Opin HIV AIDS. 2011;6:62–67. doi: 10.1097/COH.0b013e3283412568. A recent review comparing the various primary cell models of HIV-1 latency. [DOI] [PubMed] [Google Scholar]
- 34.Williams SA, Greene WC. Regulation of HIV-1 latency by T-cell activation. Cytokine. 2007;39:63–74. doi: 10.1016/j.cyto.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Siliciano RF, Greene WC. HIV Latency. Cold Spring Harb Perspect Med. 2011;1:a007096. doi: 10.1101/cshperspect.a007096. A recent review of the entire field of HIV-1 latency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36••.Karn J. The molecular biology of HIV latency: breaking and restoring the Tat-dependent transcriptional circuit. Curr Opin HIV AIDS. 2011;6:4–11. doi: 10.1097/COH.0b013e328340ffbb. An excellent review of the molecular aspects of HIV-1 latency, focusing on Tat. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ylisastigui L, Archin NM, Lehrman G, et al. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS. 2004;18:1101–1108. doi: 10.1097/00002030-200405210-00003. [DOI] [PubMed] [Google Scholar]
- 38.Archin NM, Espeseth A, Parker D, et al. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses. 2009;25:207–212. doi: 10.1089/aid.2008.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Contreras X, Schweneker M, Chen CS, et al. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J Biol Chem. 2009;284:6782–6789. doi: 10.1074/jbc.M807898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Archin NM, Liberty AL, Kashuba AD, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. An important paper showing that administration of a single dose of an HDAC inhibitor can increase HIV-1 RNA levels in the resting CD4+ T cells of patients on ART. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang HC, Xing S, Shan L, et al. Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J Clin Invest. 2009;119:3473–3486. doi: 10.1172/JCI39199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Xing S, Bullen CK, Shroff NS, et al. Disulfiram reactivates latent HIV-1 in a Bcl-2-transduced primary CD4+ T cell model without inducing global T cell activation. J Virol. 2011;85:6060–6064. doi: 10.1128/JVI.02033-10. Example of the use of a primary cell model to identify latency reversing agents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Xing S, Bhat S, Shroff NS, et al. Novel structurally-related compounds reactivate latent HIV-1 in a Bcl-2 transduced primary CD4+ T cell model without inducing global T cell activation. J Antimicrob Chemother. 2012;67:398–403. doi: 10.1093/jac/dkr496. Example of the use of a primary cell model to identify latency reversing agents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44•.Blazkova J, Chun TW, Belay BW, et al. Effect of histone deacetylase inhibitors on HIV production in latently infected, resting CD4(+) T cells from infected individuals receiving effective antiretroviral therapy. J Infect Dis. 2012;206:765–769. doi: 10.1093/infdis/jis412. A study challenging the notion that HDAC inhibitors can increase HIV-1 production by resting CD4+ T cells from patients on ART. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45••.Shan L, Deng K, Shroff NS, et al. Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity. 2012;36:491–501. doi: 10.1016/j.immuni.2012.01.014. A paper showing that reversal of latency alone may be insufficient for eradication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46••.Eriksson S, Graf EH, Dahl V, et al. Comparative Analysis of Measures of Viral Reservoirs in HIV-1 Eradication Studies. PLoS Pathog. 2013;9:e1003174. doi: 10.1371/journal.ppat.1003174. A collaborative study comparing several different approaches for measuring the latent reservoir. The relatively weak correlation between results of the viral outgrowth and PCR-based assays raises important questions about how the reservoir should be monitored in eradication studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47•.Strain MC, Richman DD. New assays for monitoring residual HIV burden in effectively treated individuals. Curr Opin HIV AIDS. 2013;8:106–110. doi: 10.1097/COH.0b013e32835d811b. A thoughtful review of approaches for measuring persistent HIV-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Doherty U, Swiggard WJ, Jeyakumar D, et al. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J Virol. 2002;76:10942–10950. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49•.Graf EH, Mexas AM, Yu JJ, et al. Elite suppressors harbor low levels of integrated HIV DNA and high levels of 2-LTR circular HIV DNA compared to HIV+ patients on and off HAART. PLoS Pathog. 2011;7:e1001300. doi: 10.1371/journal.ppat.1001300. This study illustrates the value of elegant PCR methods that specifically detect the integrate form of HIV-1 DNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bukrinsky MI, Stanwick TL, Dempsey MP, Stevenson M. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science. 1991;254:423–427. doi: 10.1126/science.1925601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dornadula G, Zhang H, VanUitert B, et al. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA. 1999;282:1627–1632. doi: 10.1001/jama.282.17.1627. [DOI] [PubMed] [Google Scholar]
- 52.Maldarelli F, Palmer S, King MS, et al. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS Pathog. 2007;3:e46. doi: 10.1371/journal.ppat.0030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 54.Harris RS, Bishop KN, Sheehy AM, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 55.Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 56.Kieffer TL, Kwon P, Nettles RE, et al. G-->A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J Virol. 2005;79:1975–1980. doi: 10.1128/JVI.79.3.1975-1980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanchez G, Xu X, Chermann JC, Hirsch I. Accumulation of defective viral genomes in peripheral blood mononuclear cells of human immunodeficiency virus type 1-infected individuals. J Virol. 1997;71:2233–2240. doi: 10.1128/jvi.71.3.2233-2240.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]