Abstract
The ability of cells to sense oxygen is a highly evolved process that facilitates adaptations to the local oxygen environment and is critical to energy homeostasis. In vertebrates, this process is largely controlled by three intracellular prolyl-4-hydroxylases (PHD) 1–3. These related enzymes share the ability to hydroxylate the hypoxia-inducible transcription factor (HIF), and therefore control the transcription of genes involved in metabolism and vascular recruitment. However, it is becoming increasingly apparent that PHD controls much more than HIF signaling, with PHD3 emerging as an exceptionally unique and functionally diverse PHD isoform. In fact, PHD3-mediated hydroxylation has recently been purported to function in such diverse roles as sympathetic neuronal and muscle development, sepsis, glycolytic metabolism, and cell fate. PHD3 expression is also highly distinct from that of the other PHD enzymes, and varies considerably between different cell types and oxygen concentrations. This review will examine the evolution of oxygen sensing by the HIF family of PHD enzymes, with a specific focus on the complex nature of PHD3 expression and function in mammalian cells.
Keywords: PHD3, EGLN3, HIF–PHD, hypoxia-inducible factor, hypoxia, oxygen sensing
Introduction
Molecular oxygen (O2): nothing has shaped our planet more than this simple molecule. Its first appearance in significant quantities occurred nearly 2.5 billion years ago as a product of cyanobacteria photosynthesis in Earth’s expansive seas.1–3 Over the next 2 billion years, a “Great Oxygenation Event” occurred. This accumulation of O2 in the atmosphere stimulated the evolution of more highly efficient aerobic respiration. It also necessitated the evolution of antioxidant systems, as O2 is a highly reactive and potentially damaging molecule. Ultimately, the result was the appearance of a complex signaling system for sensing and controlling O2 usage within the cell, allowing organisms to balance the life-giving yet poisonous properties of O2.
The foundation for this evolutionary feat began with the origin of a prolyl-4-hydroxylase (PHD) enzyme, which was likely spurred by the initial appearance of O2. This happened sometime before the divergence of amoebozoa (Dictyostelium discoideum) from other eukaryotes around 2 billion years ago (Figure 1).4–6 By utilizing O2 as a substrate for posttranslational modification of signaling proteins, this primitive PHD allowed the integration of environmental signals (for example, fluctuating O2 concentrations) with cell signaling pathways that control critical events in the lifecycle of these organisms.5,6 Around 800 million years ago, atmospheric O2 levels rose even more considerably, and organisms began to utilize and transport O2 in more complex ways. In the animal kingdom (metazoans), a hypoxia-inducible transcription factor (HIF) evolved, which integrated O2-sensing (PHD) ability with control of blood cell production, vascular recruitment, and metabolism.1 It was later during the evolution of vertebrate metazoans that the single PHD gene split into three independent cytoplasmic PHD enzymes (PHD 1–3).7 Overall, these events enabled the evolution of large, complex organisms with exquisite integration of O2 sensing, O2 delivery, metabolism, and other critical cellular processes.1
Figure 1.
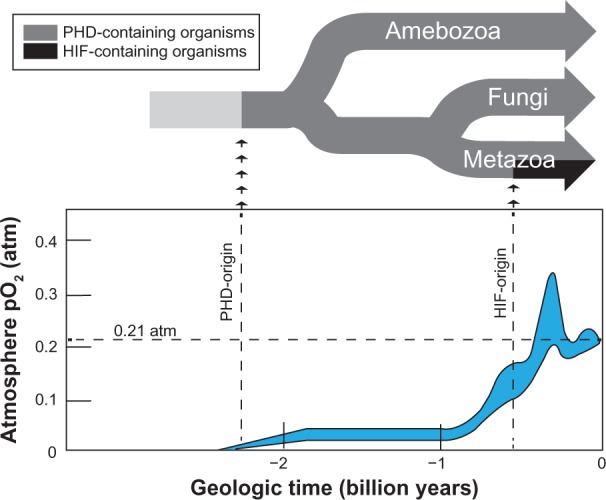
The evolution of PHD and HIF in relation to the oxygenation of Earth.
Notes: At the top of the figure, the phylogenetic tree of the PHD-containing groups amebozoa and opisthokonta (metazoan and fungi)5,6 is depicted with approximate branch points relative to the Earth’s geological age. The arrows indicate the approximate origins of the PHD and HIF genes.4,7,86 At the bottom of the figure, the partial pressure of atmospheric O2 is graphed in relation to time, as adapted from Holland et al.3 The gray area represents the range of the estimate.3 In addition, 0.21 atm (the current pO2) is marked as a dotted line. Holland HD. The oxygenation of the atmosphere and oceans. Philos Trans R Soc Lond B Biol Sci. 2006;361(1470):903–9153, by permission of the Royal Society.
Abbreviations: PHD, prolyl-4-hydroxylase; HIF, hypoxia-inducible transcription factor; pO2, partial pressure of oxygen.
The temporal relationship between PHD and HIF evolution is often overlooked, as the literature commonly focuses on HIF pathway regulation when discussing PHD function. Nonetheless, PHD enzymes predate metazoans and, therefore, HIF evolution. This fact strongly implies that the roots of the PHD-mediated O2 sensing system run much more deeply within the cell signaling machinery than simple control of the HIF pathway. A recent increase in the identification of non-HIF PHD targets seems to validate this assumption, with PHD3 leading the way with its broad involvement in cell survival, differentiation, metabolism, and cell migration. The following review will provide a brief overview of the expression and function of the HIF–PHD enzymes, and will then focus on PHD3 and evidence for its broad functionality.
The metazoan HIF–PHD family
PHD3 (PHD3/EGLN3/SM-20) is one of three cytoplasmic Fe-2-dependent PHDs (PHD1/EGLN2; PHD2/EGLN1; and PHD3/EGLN3/SM-20) that hydroxylate HIF-α subunits at two conserved proline residues. Along with their requirement for Fe-2, these enzymes consume molecular O2 and 2-oxolglutarate (2-OG), yielding succinate, CO2, and a 4-hydroxylated proline residue (Figure 2). The mammalian genome also encodes for two multi-subunit collagen PHD enzymes and an endoplasmic reticulum membrane-associated PHD (P4H-TM) that is capable of HIF hydroxylation.8,9 Although these other PHDs are related to the HIF–PHDs in their requirement for O2, 2-OG, and Fe-2, they are localized to the endoplasmic reticulum and share little sequence homology with the HIF–PHDs.9 Therefore, further discussion of PHD enzymes will be in reference only to HIF–PHD 1, 2, and 3.
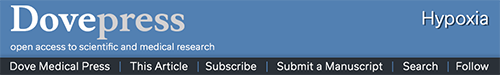
Figure 2.
The interaction between metabolism and PHD enzymatic activity.
Notes: It is apparent that 2-oxoglutarate from the TCA cycle is utilized, along with molecular oxygen, to hydroxylate a proline residue on carbon 4. The reaction produces CO2 and succinate (left). Hydroxylation is thought to involve a transient OOH molecule, which is stabilized by Fe-2 in the PHD3 catalytic domain. One oxygen atom subsequently becomes incorporated into succinate, and the other in the proline–OH group (right). During oxidative stress, Fe-2 is susceptible to oxidation by reactive oxygen species, which inhibits hydroxylase activity. Fe 3 is reduced back to Fe-2 by ascorbate/Vit C.
Abbreviations: TCA, tricarboxylic; PHD, prolyl-4-hydroxylase; Vit, vitamin; ROS, reactive oxygen species; OH, hydroxyl group.
Effectors of PHD enzymatic activity
Examination of the enzymatic mechanism of the PHDs uncovers an elegant design that is simultaneously sensitive to O2 concentration, tricarboxylic (TCA) cycle disturbances, and redox status within the cell. The sensitivity of the PHDs to these parameters is notable in the context of severe disturbances of metabolism and oxidative stress, which often present as abnormal accumulation of HIF proteins.
TCA cycle/metabolism
Errors in metabolism are well known in activating the O2-sensing pathways of the cell. This is due to the unique sensitivity of the PHD enzymatic mechanism to components of cellular metabolism. Molecular O2 itself is consumed directly in the hydroxylation reaction, with one O2 atom becoming incorporated into the hydroxyl group on the receptive proline residue.10 Concurrently, 2-OG from the TCA cycle undergoes oxidative decarboxylation to succinate (see Figure 2).10
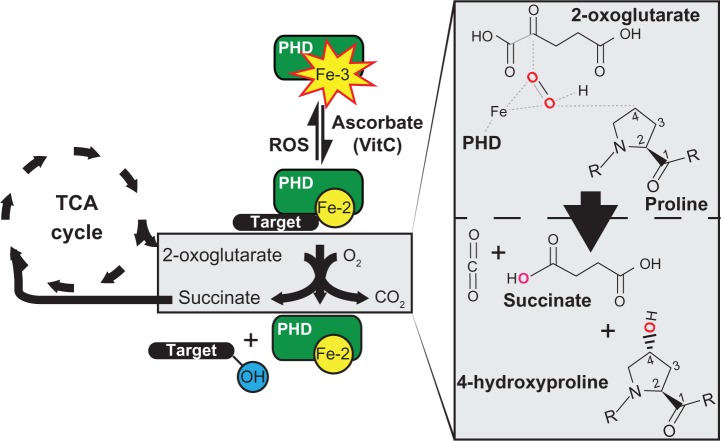
This exquisite sensitivity of the PHDs to cellular metabolism is observable in the context of numerous benign and malignant tumors containing mutations in TCA cycle enzymes. These include inactivating mutations in succinate dehydrogenase in paragangliomas and pheochromocytomas, and fumarate hydratase leiomyoma and renal cancer.11 Furthermore, gain-of-function mutations in isocitrate dehydrogenase (IDH) 1/2 in acute myeloid leukemia and gliomas have also been shown to result in activation of the HIF pathway.12
The mechanism for HIF activation in the presence of these mutations is thought to be due to the accumulation of metabolic intermediates. For example, mutations in fumarate hydratase and succinate dehydrogenase lead to the accumulation of fumarate and succinate, respectively, which likely act through product inhibition of the active site of PHD.11,13 Mutations in IDH, on the other hand, promote the abnormal production of (R)-2-hydroxyglutarate (2-HG). It should be noted that 2-HG is similar in structure to 2-OG, but is not an appropriate substrate for oxidative decarboxylation. Similarly, dimethyloxalylglycine is a synthetic 2-OG analog used in cell culture to inhibit PHD activity, and likely works through a similar mechanism of action (see Figure 3).
Figure 3.
Effectors of PHD3 enzymatic activity.
Notes: Proline hydroxylation requires 2-oxoglutarate from the TCA cycle, and O2 for enzymatic function (blue boxes). The TCA cycle and electron transport chain are the source of many inhibitors of PHD enzymatic activity (yellow boxes). Build-up of fumarate occurs upstream of FH-mut and succinate buildup occurs upstream of SDH-mut. Gain-of-function mutations in the IDH1-mut produce (R)-2-hydroxyglutarate, another PHD inhibitor. Electron leak from complex 3 of the electron transport chain results in superoxide production (O2−) in the presence of O2, which is converted to H2O2 by catalase. ROS, such as H2O2 can oxidize Fe-2 to Fe-3. PHD inhibitors commonly used in research include dimethyloxalylglycine, desferrioxamine, and cobalt-chloride (Co-2) (upper right yellow box).
Abbreviations: MnSOD, manganese superoxide dismutase; FH, fumarate hydratase; mut, mutations; SDH, succinate dehydrogenase; Co-2, cobalt-chloride; IDH, isocitrate dehydrogenase; IDH1, cytoplasmic isoform if isocitrate dehydrogenase; PHD, prolyl-4-hydroxylase; TCA, tricarboxylic; ROS, reactive oxygen species; OH, hydroxyl group.
Redox state
Another layer of PHD enzymatic regulation occurs due to PHD inactivation by reactive oxygen (ROS) species (see Figure 2).14 This is due to the sensitivity of Fe-2 to oxidation by ROS.14 Oxidized iron (Fe-3) is not compatible with PHD enzymatic activity, resulting in increased stability of HIF.15 The effect of ROS on HIF stability is well documented in many conditions where mitochondrial function is compromised. This may occur, in part, to a premature leak of electrons from complexes 1 and 3 of the electron transport chain (ETC), which results in superoxide production (O2−).16 This O2− is dismutated to H2O2 by manganese superoxide dismutase, and ultimately results in PHD inactivation by the mechanism described above (see Figure 3).17,18
This reliance of PHDs on Fe-2 explains the role of ascorbate/vitamin C in proline hydroxylation. Ascorbate promotes the reduction of Fe-3 to Fe-2, and is therefore able to recover PHD enzymatic activity in the event of Fe-2 oxidation.19 This is the mechanism for the phenotype observed in physiologic ascorbate deficiency (scurvy), where collagen PHDs become dysfunctional due to the inability to replenish Fe-2.20 Although all PHDs apparently share the same mechanism of action,21 it is unclear if the HIF–PHDs are affected in this particular pathologic state. Nonetheless, a lack of ascorbate in cell culture media has been attributed to aberrancies in the HIF pathway in vitro.22,23
Other conditions where intracellular iron pools are compromised or rendered unavailable to PHDs also inhibit PHD activity. This is the basis for the usage of the iron chelating agent desferrioxamine in experiments involving O2-sensing pathways. Cobalt (Co-2) is also used as an in vitro PHD inhibitor, presumably through competition with Fe-2 in the active site (see Figure 3).24
PHD structure
Amino acid residues within the human PHD3 catalytic domain are highly conserved with that of PHD1 and 2; however, N-terminal sequences are highly divergent. In fact, the PHD3 N-terminus is over 150+ amino acids shorter in length than both PHD1 and PHD2 (Figure 4). This suggests that N-terminal residues dictate the functional diversity of the PHD enzymes. Among other vertebrates, sequence variation within individual PHD isoforms is generally minimal. However, there is a notable exception with rat PHD3, which differs significantly from other vertebrate PHD3 sequences by the presence of a large N-terminal region containing a mitochondrial localization sequence (Figure 3).25 This point becomes important when interpreting PHD3 data from rat studies, which make up a large portion of the PHD3 literature.
Figure 4.
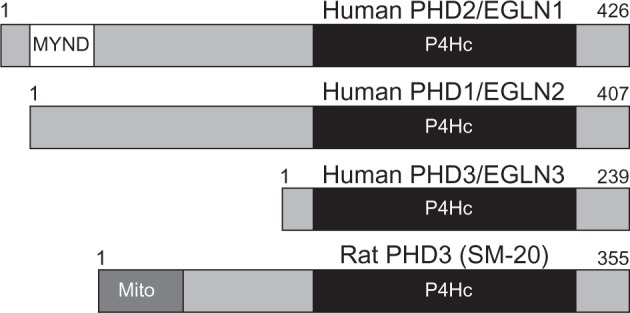
Models of human PHD1–3 and rat PHD3 (SM-20) proteins.
Notes: PHD1, PHD2, and three proteins all share highly homologous P4Hc-containing sequences in their N-terminus. PHD2 is the longest PHD and contains a MYND. Rat PHD3 (SM-20) is unique in that it contains a Mito.
Abbreviations: PHD, prolyl-4-hydroxylase; EGLN, egg-laying-defective nine; MYND, C-terminal MYND-type zinc finger domain; P4Hc, prolyl-4-hydroxylase; SM-20, smooth muscle-20; Mito, N-terminal mitochondrial localization signal.
PHD3 target proteins
The PHD family’s conserved PHD activity targets HIF-α, and a growing list of other intracellular proteins that contain a conserved Leu-X-X-Leu-Ala-Pro (LXXLAP) motif, where X represents any amino acid. Early studies identified the LXXLAP motif as consensus for PHD-mediated hydroxylation.26 However, studies have shown that substitutions within the LXXLAP motif can still allow for PHD-mediated hydroxylation.27,28 Recent studies involving PHD3 have supported this by identifying putative PHD3 target proteins that are hydroxylated on prolines outside of LXXLAP motifs.29–31
Identification of these targets is a difficult hurdle to overcome when studying PHD biology. This can be attributed, in part, to the small-sized, noncharged nature of the hydroxyl modification. Nonetheless, several strategies have been published, including the use of 14C-labeled 2-OG in in vitro hydroxylation assays, with the production of 14C-labeled CO2 as a surrogate indicator for hydroxylation.32 A hydroxyproline antibody also exists and has been utilized with limited success in cases where the hydroxylated protein is first immunoprecipitated.30 Other investigators have used liquid chromatography followed by tandem mass spectroscopy analysis of tryptic fragments from isolated proteins.29–31
Although the latter type of analysis is quite popular, it has several intrinsic problems, which may raise questions regarding some of the currently published data. First, metal present during protein processing can theoretically result in proline oxidation, leading to cis/trans-4-hydroxyproline.33 Also, it should be noted that leucine and isoleucine have nearly identical molecular weights to hydroxyl-proline (POH), which could lead to misinterpretation of LXXLAL or LXXLAI as a peptide containing a LXXLAPOH. These obstacles necessitate careful consideration when planning and interpreting hydroxyproline data. The use of isotope-labeled synthetic peptides containing 4-hydroxyproline as positive controls, may allow for more reliable identification of true hydroxyproline-containing peptides as the mass spectrometry ionization patterns of these synthetic peptides can be used for comparison against experimental data. Furthermore, the use of hydroxylase inhibitors should be used as controls to demonstrate the reduction of hydroxylated prolines within putative target proteins.
Upon the identification of hydroxylated prolines on target proteins, the assignment of a specific PHD as the hydroxylase responsible for this hydroxylation should also be made with care. The PHD family has previously been demonstrated to assemble into higher-order complexes composed of heteromultimeric constituents of the various PHD enzymes.34 Coimmunoprecipitated complexes may contain several different PHDs that may be responsible for target hydroxylation. Therefore, in vitro experiments identifying direct protein–protein interactions and/or hydroxylation reactions should be performed when identifying novel PHD target proteins.
Other PHD3-interacting proteins
Aside from hydroxylation targets, PHD3 appears to have many effector proteins that have not been reported as targets for hydroxylation. This list far outweighs the list of identified PHD3 targets (for references, see Table 1). Some of these studies ruled out hydroxylation due to the absence of an effect when PHD3 was mutated at an active site histidine, or by use of a hydroxylase inhibitor. This suggests that PHD3 has hydroxylase-independent effects, and supports previous studies that suggest protein-protein complexes involving PHD3 may play significant functional roles within the cell.34,35
Table 1.
List of PHD3-targets and interacting proteins along with the methods used
| PHD3 target/interacting protein | Method for determining interaction | Method for determining hydroxylation |
|---|---|---|
| hCLK2 (P374, P419, P422)29 | Co-IP (E + OE); GST-PD | LC-MS/MS with peptide |
| PKM2 (P403, P408)30 | Co-IP (OE); GST-PD | Orbitrap LC-MS; HP-Ab |
| PKM253 | Co-IP (E); GST-PD | N/A |
| β2-AR (P382, P395)31 | Co-IP (OE); GST-PD | LC-MS/MS |
| hPRP1987 | Co-IP (E + OE); GST-PD | N/A |
| IKKβ88 | Co-IP (E + OE); GST-PD | N/A |
| Morg190 | Co-IP (IVTT proteins); Co-IP (OE) | N/A |
| Siah234 | Co-IP (OE); GST-PD | N/A |
| Bcl-270 | Co-IP (OE) | N/A |
| Proteosome (20S core)35 | Colocalization by IF | N/A |
| ATF-491 | Y2HB; Co-IP (OE in insect cells) | N/A |
| OS-992 | Co-IP (OE) | N/A |
| MAGE-1193 | Co-IP (OE) | N/A |
| TRiC94 | Co-IP (OE) | N/A |
| PHD334 | Co-IP (OE) | N/A |
| PHD234 | Co-IP (OE) | N/A |
| PHD134 | Co-IP (OE) | N/A |
Notes: Column 1 lists all non-HIF proteins known to be targets of and/or that interact with PHD3. The specific prolines targeted for hydroxylation are listed (if applicable). Column 2 lists the methods used to determine the interaction of each protein with PHD3. GST-PD use purified proteins (unless otherwise noted). For the peptide, isotope-labeled synthetic hydroxylated peptide was used as a control. For N/A, some publications demonstrated that the hydroxylase mutant PHD3 had same effect as PHD3-wild type, and therefore did not assay for hydroxylation of the interacting protein.
Abbreviations: PHD, prolyl-4-hydroxylase; Co-IP, coimmunoprecipitation; E, endogenous proteins; OE, overexpressed proteins; GST-PD, GST-pulldown experiments using purified proteins; LC, liquid chromatography; MS, mass spectrometry; PKM2, pyruvate kinase isoenzyme; HP-Ab, hydroxyproline antibody; N/A, not attempted; IKKβ, IkappaB kinase beta; IVTT, in vitro translated and transcribed proteins; IF, immunofluorescence; ATF-4, AMP-dependent transcription factor 4; Y2HB, yeast 2 hybrid; MAGE-11, melanoma antigen gene protein-A11; HIF, hypoxia-inducible transcription factor; S, subunit; hCLK2, TEL2/telmomere maintenance 2; hPRP19, pre-RNA processing factor 19 homolog (of S. cerevisiae); Bcl, B-Cell lymphoma; OS-9, osteosarcoma amplified 9.
HIF – the classic PHD target
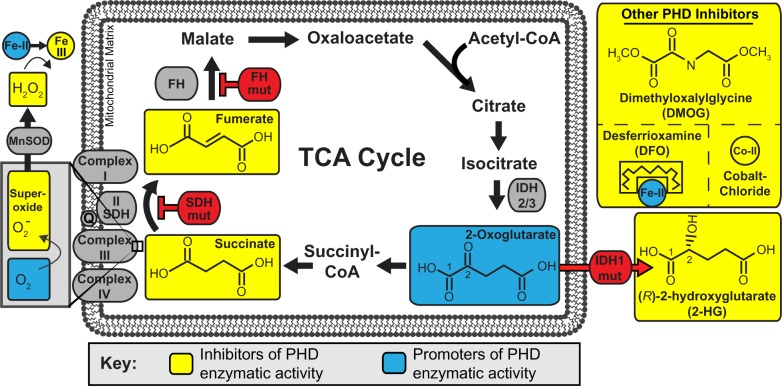
The first PHD targets to be recognized were HIF-1α and HIF-2α (collectively called HIF-α).26 HIF-α proteins are unique basic-helix–loop-helix transcription factors that function to regulate the transcription of genes that control critical processes like metabolism, cell cycle, and vascular recruitment (there are several excellent reviews36–38). The key to the regulation of the HIF-α protein is their posttranslational regulation by PHD enzymes. In the presence of O2 (normoxia), PHD enzymes quickly hydroxylate HIF-α subunits on two highly conserved proline residues that reside in conserved LXXLAP hydroxylation motifs in the HIF-α O2-dependent degradation domain (ODD). It is interesting to note here that PHD 1–3 have different affinities for either the N-terminal ODD (NODD) proline or the C-terminal ODD (CODD) proline of HIF-α.39,40 PHD3 almost exclusively hydroxylates the CODD, whereas PHD1 and PHD2 are able to hydroxylate both CODD and NODD prolines, but have higher affinity for the NODD.39,40 The implications of this remain enigmatic. When hydroxylated, these prolines on HIF-α serve as binding sites for the von Hipple–Lindau ubiquitin ligase, leading to ubiquitination and degradation of HIF-α (Figure 5).
Figure 5.
HIF-1α regulation.
Notes: In the presence of O2, HIF-1α is hydroxylated by PHD1, PHD2, or PHD3 on two proline residues in the O2-dependent domain. These hydroxylated proline residues serve as binding sites for VHL, which binds and Ub HIF-1α. Hydroxylation on asparagine 803 in the C-TAD by FIH blocks p300/CPB recruitment, which further inhibits HIF transcriptional transactivation. In the absence of O2, HIF is not hydroxylated and VHL does not bind (bottom of figure). HIF-1α protein levels are subsequently stabilized and HIF-1α becomes localized to the nucleus where it dimerizes with HIF-1β via the PAS domain and interacts with DNA through the bHLH domain. P300/CBP is recruited through binding to the C-TAD.
Abbreviations: HIF, hypoxia-inducible transcription factor; bHLH, basic helix–loop–helix; DNA, deoxyribonucleic acid; PAS, Per-aryl hydrocarbon receptor nuclear translocator-Sim; VHL, Von Hippel–Lindau tumor suppressor; NODD, nonoxygen-dependent domain; CODD, C-terminal oxygen-dependent degradation domain Ub, ubiquitinates; FIH, factor-inhibiting hypoxia-inducible transcription factor; OH, hydroxide; PHD, prolyl-4-hydroxylase, C-TAD, C-terminal activation domain; CPB, creb-binding protein.
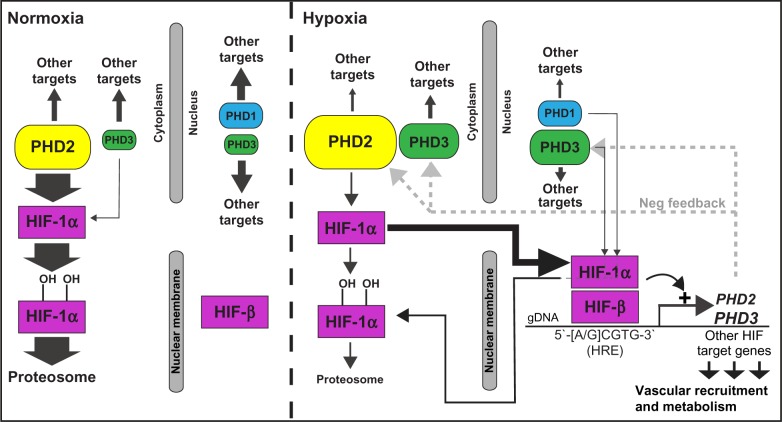
Conversely, when O2 is limited (hypoxia) PHD enzymes are less active, leading to increased HIF-α stability and nuclear localization.41 In the nucleus, HIF-α dimerizes with the constitutively expressed HIF-β/aryl hydrocarbon receptor nuclear translocator nuclear subunit.42,43 The HIF-α/HIFβ heterodimer (collectively termed HIF) binds to deoxyribonucleic acid (DNA) on 5′(A/G)CGTG-3′ sequences called hypoxia response elements (HRE), and recruits p300/CBP to its C-terminal transactivation domain, leading to transcriptional transactivation of HIF target genes.43,44 HIF target genes are numerous, and include enzymes of the glycolytic pathway, vascular endothelial growth factor, as well as PHD2 and PHD3.37,45,46 Collectively, upregulated HIF target genes act synergistically to increase energy production in the absence of O2 and to restore vascular supply by stimulating vascular recruitment and vessel growth.37 For an overview of the HIF pathway and its control by PHD enzymes, see Figure 6.
Figure 6.
HIF pathway regulation by the PHD enzymes.
Notes: HIF-1 can be a substrate for any of the three PHD proteins; however, PHD3, which is the most inducible PHD under hypoxic conditions, has broader substrate specificity and can hydroxylate a number of non-HIF-1α targets.
Abbreviations: PHD, prolyl-4-hydroxylase; HIF, hypoxia-inducible transcription factor; OH, hydroxyl group; gDNA, genomic deoxyribonucleic acid; HRE, hypoxia response elements; Neg, negative.
Regulation of PHD3 expression and localization
Of the three PHD enzymes, PHD3 is the most dynamic with regard to its expression, localization, and stability. Interestingly, O2 levels control each of these parameters. The following will discuss these unique regulatory properties of PHD3.
Control of PHD3 transcription
PHD3 induction by hypoxia
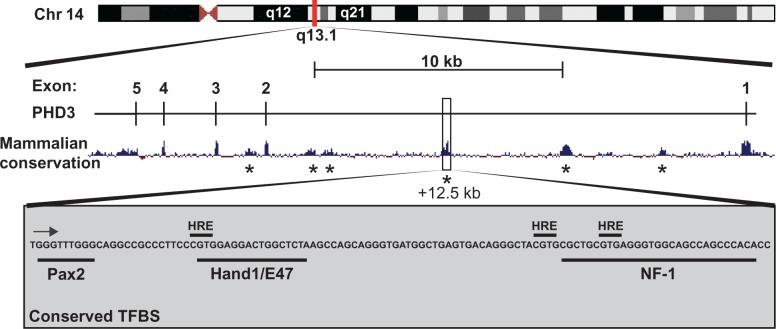
The induction of PHD3 expression by hypoxia is exquisite. A study performed by Appelhoff et al40 nicely demonstrates this property. Here, relative expression levels of the three PHD enzymes were analyzed over ten human cancer cell lines. While PHD3 expression during normoxia is the lowest of the three PHDs, it is induced by an average of 36.2-fold after 16 hours in 1% O2. This is compared to only a threefold induction of PHD2, and no hypoxic induction of PHD1 (relative expression data from Appelhoff et al40 has been summarized, including statistical analysis, and are displayed in Figure 7). At the messenger ribonucleic acid level, PHD3 upregulation by hypoxia occurs by binding of HIF to an HRE, resulting in transcriptional transactivation.46,47 Although there are numerous HRE consensus sequences throughout the PHD3 gene, HIF binding under hypoxic conditions has been demonstrated to occur at a specific site in intron 1 approximately +12.5 kb downstream of the transcription start site.46–48 When analyzed at the nucleotide base level using the University of California Santa Cruz Genome Browser (GRCH37/hg19), three HRE elements are present in this region, along with conserved binding sites for Pax2, Hand1/E47, and nuclear factor-1 (Figure 8).49,50 Due to the close proximity of these three HREs, it is unclear which is utilized by HIF-1α. However, these HREs overlap the Hand1/E47 and nuclear factor-1 sites, suggesting that occupancy of these sites could affect HIF binding to the region. Indeed, evidence suggests that stress response transcription factors (including Hand1/E47) are overrepresented in regions containing functional HREs, and may “fine tune” the hypoxia-induced expression at the gene level.48
Figure 7.
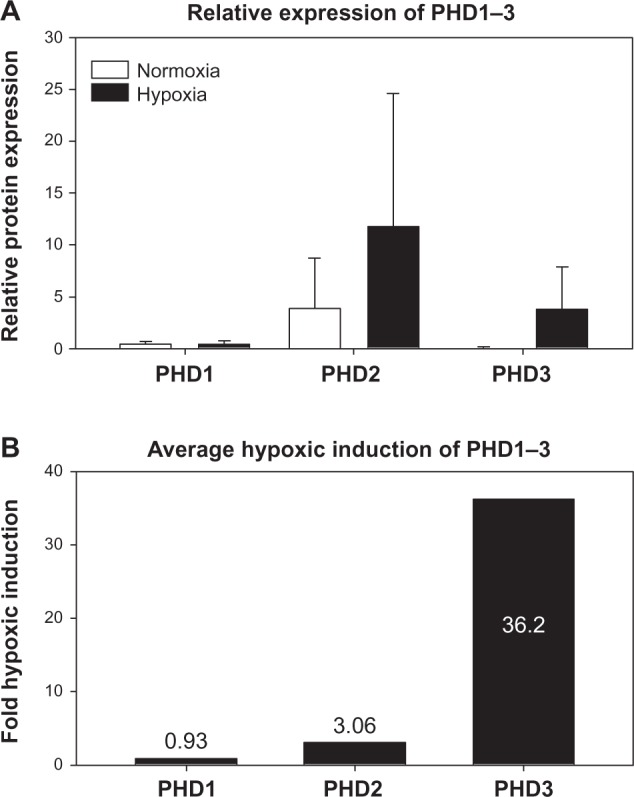
Relative expression of PHD1, PHD2, and PHD3 in ten human cancer cell lines.
Notes: The relative expression of PHD enzymes was determined by Appelhoff et al40 in ten human cancer cell lines: U2-OS, BxPC3, MCF7, ZR751, MDA-435, BT-474, HEP3B, HeLa, 833K, and SuSa cell lines under normoxic and hypoxic (1% O2) conditions for 16 hours. (A) Appelhoff et al’s40 data were recompiled and averaged to demonstrate general trends in the relative expression of PHD1, PHD2, and PHD3. Error bars represent one standard deviation. (B) The average fold hypoxic-induction of each PHD is re-graphed from the data above ^40.
Abbreviation: PHD, prolyl-4-hydroxylase.
Figure 8.
PHD3 regulatory elements.
Notes: The PHD3 gene with mammalian conservation is depicted in relation to its position on chromosome 14 q13.1 (modified from the UCSC Genome Browser hg19 [http://genome.ucsc.edu/]).49 The highly conserved region at ∼+12 kb upstream of exon 1 is expanded (bottom), and putative HRE are indicated (gray box), along with conserved TF binding sites, as predicted by the UCSC hg19 “TFBS conserved” track. The black arrow indicates the direction of 3′–5′ sequence orientation. *Represents highly conserved sequences located in intron 1 and 2 that may represent important regulatory regions.
Abbreviations: Chr, chromosome; PHD, prolyl-4-hydroxylase; HRE, hypoxia response elements; NF-1, nuclear factor-1; TF, transcription factor; UCSC, University of California Santa Cruz; TFBS, transcription factor binding sites.
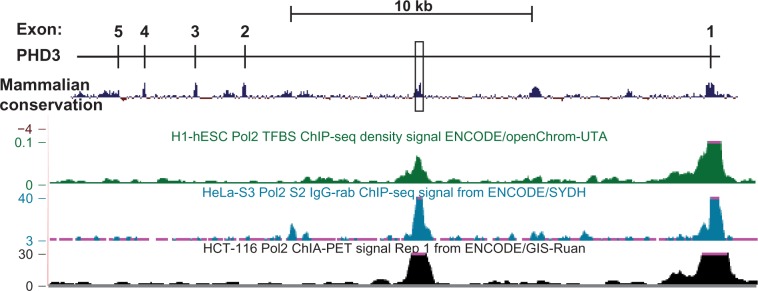
The importance of the +12.5 kb region of PHD3 in transcriptional control can be seen in polymerase 2 (Pol 2) ChIP-seq data mining analysis. Several Pol 2 ChIP-seq tracks in the University of California Santa Cruz Genome Browser (GRCH37/hg19) indicate a biphasic Pol 2 signal at both the PHD3 promoter and +12 kb region in intron 1 (Figure 9).49,50 This does not appear to occur in PHD1 and PHD2, and to our knowledge, the significance of this has not yet been demonstrated. However, it does not appear to be associated with an alternative transcriptional start site, as no alternative transcripts have been published to originate in this region. This may represent a “poised” normoxic state where chromosomal looping creates a physical association of the promoter and HRE containing +12 kb region. Upon hypoxic stimulation, HIF binding to this region could stimulate activation of Pol 2 transcription. Experiments that combine Pol 2 and HIF-1α ChIP-seq under both normoxic and hypoxic conditions would provide significant insight into the mechanisms surrounding the induction of PHD3 transcription by hypoxia.
Figure 9.
Polymerase 2 interaction with +12 kb region of the PHD3 gene.
Notes: The PHD3 locus is represented with mammalian conservation (modified from the UCSC Genome Browser hg19 [http://genome.ucsc.edu/]).49 ChIP-Seq signal data for polymerase 2 in H1-hESC, HeLa-S3, and HCT-116 cells were mined from several ENCODE data tracks (as indicated) using the UCSC Genome Browser hg19 (http://genome.ucsc.edu/).50
Abbreviations: PHD, prolyl-4-hydroxylase; hESC, human embryonic stem cells; TFBS, transcription factor binding sites; ENCODE, Encyclopedia of DNA Elements; ChIA-PET, chromatin interaction analysis with paired-end tag sequencing; ChIP-Seq, chromatin immunoprecipitation sequencing.
The precise role(s) for PHD3 upregulation under hypoxic conditions is not clear. However, in the context of the HIF pathway, evidence suggests that it plays a predominant role in regulating HIF-2α.40 Here, it may act similarly to the PHD2–HIF-1α feedback loop that has been nicely demonstrated in experiments by Millonig et al.51 These authors demonstrate that acute induction of hypoxia leads to a transient rise in HIF-1α protein expression. Following a brief time period, these levels subsequently return back to baseline despite continued hypoxia. This can be attributed to hypoxia-induced PHD2 protein expression, which enables cells to effectively increase HIF-1α hydroxylation under lower O2 concentrations. When PHD2 activity is eliminated by colbalt chloride treatment, HIF-1α levels remain high rather than returning to baseline.51
Another function of PHD3 upregulation during hypoxia may be to control metabolism. Remarkably, PHD3 has been demonstrated to interact with the M2-isoform of pyruvate kinase, altering both the flux of carbons through glycolysis and the regulation of metabolic gene expression via interaction with HIF.30,52,53
Hypoxia-independent regulation of PHD3
Aside from the pronounced regulatory control of PHD3 by hypoxia and HIF-1α, PHD3 transcriptional regulation by other factors has been extensively described. However, most of these experiments were performed in the rat, where PHD3 was first discovered.54 This initial characterization placed PHD3 in the realm of immediate-early genes. These immediate-early genes are responsive to rapid transcriptional upregulation by the growth agonists platelet-derived growth factor angiotensin 2 and serum, and are not inhibited at the transcriptional level by cyclohexamide (protein synthesis inhibitor).54 Here, the induction of PHD3 by serum appears dependent on an SP1/SP3 binding site in the proximal PHD3 promoter.55 In murine myeloid 32D cells, PHD3 is also induced by c-Myc, which is known to synergize with SP1/SP3 to promote transcription.56 In endothelial cells, PHD3 can be induced downstream of interferon-γ in a JAK/STAT1-dependent manner.57 Other, larger studies focusing on global transcriptional changes downstream of specific stimuli have indicated that PHD3 may be induced by mechanism involving p53 in rat fibroblasts, and via signaling downstream of nerve growth factor (NGF) withdrawal in rat neurons, where PHD3 appears to mediate apoptosis.25,58,59
Another regulatory layer controlling the expression of PHD3 is DNA methylation. We have previously demonstrated that several human cancer cell lines of diverse tissue origin silence PHD3 expression in a DNA methylation-dependent manner.60 PHD3 promoter methylation has also been demonstrated in primary human B-cell malignancies.61 The fact that PHD3 is silenced in such a wide variety of tumor types suggests that PHD3 affects important signaling pathways that are common to the progression of many malignancies.
Regulation of PHD3 expression via alternative splicing
A final, and less well-addressed mechanism by which PHD3 expression can be controlled by the cell is alternative splicing. Following transcription of PHD3, at least three transcripts of varying size can be detected in many tissues and cell lines.62 These represent full-length PHD3, a form missing half of exon 1 (PHD3Δ1), and a form in which exon 4 is missing (PHD3Δ4).62 To our knowledge, no one has yet detected endogenous alternative transcripts that are translated to proteins; however, forced expression of these isoforms by expression vectors does result in detectable protein of the appropriate reduced molecular weight. Furthermore, it appears that the PHD3Δ4 isoform retains hydroxylase activity, whereas PHD3Δ1 is hydroxylase deficient.62 It will be interesting to see what, if any, role these alternative transcripts play in cell signaling.
Control of PHD3 protein stability
In cell line studies under normoxic conditions, PHD3 protein levels are maintained at levels significantly below that of PHD1 and PHD2 (Figure 7).40 However, during periods of hypoxia, PHD3 protein levels are remarkably increased. Although some of this regulation occurs via transcriptional control, a significant portion is a result of increased PHD3 protein stability under hypoxic conditions. The mechanism for this appears to be mediated by the E3 ubiquitin ligase Siah2.34,63 Under normoxia, the PHD3 protein is present in small molecular weight fractions that represent homomultimers of the PHD3 protein. These small complexes of PHD3 appear to be enzymatically active, but also accessible to Siah2-mediated ubiquitination, followed by degradation. In hypoxia, PHD3 forms larger molecular weight complexes consisting of heteromultimers of PHD3 with other PHD proteins. In this form, PHD3 is not accessible to Siah2 for ubiquitination.34
Control of PHD3 localization
Another unique aspect of PHD3 regulation is its subcellular localization. Whereas PHD1 and PHD2 are expressed almost exclusively in the nucleus and cytoplasm respectively, PHD3 is expressed both in the nucleus and cytoplasm.27,64 Furthermore, in the presence of plentiful O2, PHD3 appears to be largely sequestered in aggregates that colocalize with the proteasome.35 The mechanism of this aggregation is unknown, but may affect the hydroxylation potential, proapoptotic activity, as well as the Siah2-mediated PHD3 degradation, as discussed above.34,35
Interestingly, the localization of rat PHD3 (smooth muscle-20) differs from that of other mammals. This is likely due to the evolution of a large N-terminal sequence containing a mitochondrial localization sequence.59 Evidence suggests that this mitochondrial targeting sequence becomes cleaved in a fraction of rat PHD3, allowing cytoplasmic/nuclear localization similar to that of other mammals.59 The exact function of the rat PHD3 mitochondrial localization sequence, or if there is a functional difference between rat PHD3 and PHD3 of other mammals, is not known. Nevertheless, because of this additional protein localization motif in rats, caution should be used when interpreting PHD3 data from rat cells, as these data may be qualitatively different from the human PHD3 phenotype.
PHD3 function
Although the PHDs were originally characterized for their control of the HIF pathway, PHD3 does not appear to be a major player in HIF regulation under nonstressed conditions. This is evident in mouse studies, where embryos of PHD3−/− mice do not show abnormalities in the placenta, an organ critically dependent on the HIF pathway for normal development.65,66 In fact, PHD3−/− mice appear grossly normal in the absence of any insults and survive normally into adulthood.65 In contrast, PHD2−/− embryos die between day 12.5 and day 14.5 of embryonic development, and display an abnormal placenta, with significantly increased levels of HIF-1α, HIF-2α, and vascular endothelial growth factor protein expression upon immunohistochemical examination.65
Even though these data do not show a major role for PHD3 in normal developmental processes that involve the HIF pathway, there is evidence that PHD3 does regulate HIF-2α during hypoxia followed by reoxygenation in cancer cell lines.40 This may signify that PHD3 plays more of a role in regulating the HIF pathway during pathological processes that involve hypoxia and/or hypoxia-reoxygenation (ischemia/reperfusion). Although evidence supporting this hypothesis is limited, a recent study has shown that PHD3−/− mice have an aberrant response to infection, which can produce regions of tissue hypoxia due to inadequate perfusion.67 The extent to which this PHD3-related phenotype is related to PHD3’s effects on the hypoxia-response pathway, as opposed to its effects on other non-HIF signaling pathways, remains to be determined.
As suggested above, there are numerous publications that describe a role for PHD3 in non-HIF signaling pathways (see following sections for specific citations). These include, but are not limited to control of sympathetic neuronal development, myogenic signaling, cell migration, metabolism, and cell survival. The following will discuss the evidence for PHD3 in cell signaling mechanisms that fall outside of HIF pathway regulation.
Muscle development, differentiation, and survival
PHD3 appears to have an important relationship to muscle. In fact, upon PHD3’s initial discovery by Wax et al in 1994,54 it was named smooth muscle-20. Wax proceeded to report high expression of PHD3 in smooth, skeletal, and cardiac muscle (as well as in brain and nerve cells), with no detectable transcript in fibroblasts.54 Interestingly, PHD3 expression in these cells was upregulated in response to the vasoactive growth factors angiotensin 2 and platelet-derived growth factor, and this expression was not inhibited by pretreatment by cyclohexamide. Therefore, PHD3 was categorized as an immediate early gene in rat vascular smooth muscle.54 Thirteen years later, the role of PHD3 expression in muscle cells was elaborated; Fu et al68,69 demonstrated that PHD3 promotes myogenic differentiation in part through its stabilization of myogenin and inhibition of nuclear factor-kappa beta signaling during skeletal muscle myogenesis. Others have described a proapoptotic role for PHD3 in rat embryonic heart-derived H9c2 cells and neonatal rat ventricular cardiomyocytes.70,71 Here, PHD3 was found to induce apoptosis in response to stress in the form of doxorubicin or hypoxia-reoxygenation. Knockdown of PHD3 (by small interfering ribonucleic acid or by endogenous miR-20a), or inhibition of PHD hydroxylase activity appeared to mitigate the PHD3 proapoptotic response.70,71
Immune system function
PHD3−/− mice display marked defects in immune system function. Specifically, knockout mice produce a hyperimmune response to septic stimuli that includes increased recruitment of macrophages to internal organs, and increased proinflammatory cytokine production.67 This phenotype is specific for the loss of PHD3, as PHD1−/− and PHD2+/− mice are normal in this regard. Furthermore, this effect is specific to the innate immune system, as sublethal irradiation of wild type mice, followed by bone marrow transplant from PHD3−/− mice results in a similar hyperimmune response to sepsis.67 In vivo experiments indicate that PHD3 expression is a predominant feature of proinflammatory macrophages, and PHD3 loss appears to result in an increased macrophage migratory and phagocytic capacity.67,72 Thus, PHD3 expression appears, in part, to limit the proinflammatory response of macrophages to pathogens.
PHD3 also appears to play a key role in neutrophil survival.73 PHD3−/− mice have reduced neutrophilic inflammation in models of lipopolysaccharide-induced acute lung injury and dextran sodium sulfate-induced colitis. This was attributed to the decreased survival of PHD3−/− neutrophils due to hypoxic conditions that are present within these areas of inflammation.73 Furthermore, the effects of PHD knockout on hypoxic survival were confirmed in in vitro experiments, where PHD3−/− neutrophils underwent an increase in apoptosis under hypoxic laboratory conditions.73
Cell motility/migration
A possible role for proline hydroxylation in cell motility can be found in studies of the slime mold, D. discoideum, which use O2 gradients to direct cell motility into more favorable environments.74 This function could likely be downstream of O2 sensing by a PHD that is evolutionarily related to mammalian PHD1–3 enzymes.5,7 In mammals, the literature also supports a more specific role for PHD3 in the regulation of cell motility and cytoskeletal dynamics. PHD3−/− neurons have an apparent defect in axon guidance, where PHD3 loss resulted in increased axonal growth and branching.75 In another report, knockout of PHD3 was shown to result in an exacerbated macrophage-mediated immune response to abdominal sepsis.67 This was characterized by an increased migratory capacity of PHD3−/− macrophages.67 A similar effect of PHD3 loss can be found in cancer cells, as knockdown of PHD3 was shown to result in an increase in cell invasion through Matrigel™ (BD Biosciences, San Jose, CA, USA) in human pancreatic adenocarcinoma cell lines.76
Regulation of survival and apoptotic pathways
Many studies demonstrate a clear role for PHD3 in the regulation of both cell survival and apoptotic pathways. Much of this research, however, has been done in the context of rat PC12 (pheochromocytoma) cells and primary rat sympathetic neurons. As discussed in this review, rat PHD3 contains a large N-terminal sequence and mitochondrial localization signal that is not present in other mammals. Due to this property, it is difficult to translate some of the results of these studies into humans. (Table 2 provides a comprehensive list of studies involving experiments on the prosurvival and proapoptotic effects of PHD3). Aside from the structural differences between rat and human PHD3, some of the apparent disparities of PHD3’s effects on cell survival appear to be due, in part, to its functional dependency on O2. In fact, many of these studies report a requirement for PHD3-enzymatic activity for its proapoptotic effects. Therefore, it seems likely that PHD3’s role in apoptosis or survival is at least partially dependent on the presence or absence of O2, respectively.
Table 2.
List of publications relevant to PHD3 and cell survival
| Effector | Hydroxy-dependent? | Major conclusion | Cell type | YearRef |
|---|---|---|---|---|
| hCLK2Ψ | Yes | PHD3 promotes activation of the DNA damage response through hydroxylation of hCLK2 following ionizing radiation | MEFs and mouse thymus | 201229 |
| PKM2Ψ | Yes | PHD3 promotes metabolic reprogramming under hypoxia (1% O2) through its interaction with PKM2 and HIF | HeLa (cervical cancer) | 201130 |
| Unknown | No | PHD3 expression enhances cell cycle progression and overall cell-survival during prolonged hypoxia (1% O2) | UT-SCC 2,7,8,9,33 (head and neck squamous cell carcinoma) | 201177 |
| Unknown | Unknown | PHD3 expression is negatively correlated with Bcl-2 expression, and positively correlated with advanced stage and poor differentiation | Nonsmall cell lung cancer | 201178 |
| Unknown | Yes | PHD3−/− neutrophils show reduced hypoxic survival compared to neutrophils from PHD3 wild type mice | Primary mouse neutrophils | 201173 |
| hPRP19 | Unknown | hPRP19 interacts with PHD3 and inhibits PHD3-mediated apoptosis under prolonged hypoxia (1% O2) | Rat PC12 (pheochromocytoma) | 201087 |
| IKKβ | No | PHD3 inhibits NFκB activity through association with IKKβ – preventing IKKβ phosphorylation; PHD3 expression negatively correlates with colorectal tumor grade and metastasis | HCT-116, SW480 (colorectal cancer), human colorectal cancer specimens | 201088 |
| Unknown | Yes* | Esterified 2-OG induces apoptosis in xenografts – dependent on PHD3 expression | HCT-116 (colon carcinoma), A431 (SSC), A375 (melanoma) | 201089 |
| Unknown | Yes* | PHD3 overexpression induces caspase 3 cleavage in the presence of O2. DMOG inhibits this effect. | MiaPaca2, Panc1 (pancreatic adenocarcinoma) | 201076 |
| Bcl-2 | Yes | PHD3 expression promotes apoptosis by interacting with Bcl-2 to prevent Bcl-2/Bax complex on mitochondria | H9c2 (rat cardiomyoblast) | 201070 |
| KIF1Bβ | Unknown | KIF1Bβ acts downstream of PHD3 to induce apoptosis in neural-crest derived cells but not cells derived from kidney or fibroblasts | PC12 (rat pheochromocytoma), SK-Mel28 (melanoma) | 200884 |
| Unknown | Yes | PHD3 expression induces 20S proteosomal aggregation and apoptosis in an O2 -dependentmanner | HeLa (cervical cancer) | 200835 |
| Unknown | Unknown | NGF withdrawal from sympathetic neurons results in c-jun-mediated PHD3 upregulation and PHD3-induced apoptosis | PC12 (rat pheochromocytoma) | 200582 |
| Unknown | Unknown | PHD3 expression stimulates cytochrome C release in neuronal cells but not HEK293 or CV-1 cells | Rat PC12 (pheochromocytoma) and neonatal sympathetic neuronal cells | 200383 |
| Unknown | Unknown | PHD3 expression induces caspase-3 dependent apoptosis downstream of cytochrome c release | Rat primary embryonic sympathetic neurons | 200159 |
| Unknown | Unknown | PHD3 induces neuronal cell death following NGF withdrawal | Rat primary embryonic sympathetic neurons | 199925 |
Notes:
Studies utilized PHD inhibitors to establish enzymatic dependency;
effector proteins were demonstrated to be directly hydroxylated by PHD3.
Abbreviations: PHD3, prolyl-4-hydroxylase; Ref, reference; DNA, deoxyribonucleic acid; MEF, mouse embryonic firbroblasts; PKM2, pyruvate kinase isoenzyme; HIF, hypoxia-inducible transcription factor; UT-SCC, ; PC12, pheochromocytoma 12; IKKβ, IkappaB kinase beta; NFκB, nuclear factor-kappa beta; 2-OG, 2-oxolglutarate; SSC, spermatogonial stem cells; DMOG, dimethyloxalylglycine; NGF, nerve growth factor; S, subunit; hCLK2, TEL2/telmomere maintenance 2; hPRP19, pre-RNA processing factor 19 homolog (of S. cerevisiae); Bcl, B-Cell lymphoma; KIF1B, kinesin family member 1B.
Prosurvival studies
Although demonstrations of PHD3’s proapoptotic effects are found in a vast majority of PHD3 studies, these effects have been shown to play a prosurvival role under hypoxic conditions in several cell types. In head and neck carcinoma cells, PHD3 appears to promote cell cycle progression from the G1-S phase.77 One mechanism for this increased survival under hypoxia may be due to PHD3’s effects on cell metabolism. Both PHD3 and pyruvate kinase M2 isoform have been shown to complex with nuclear HIF-1α, promoting the transcription of genes involved in glycolysis under hypoxic conditions.30 This may allow for increased adenosine triphosphate production through the glycolytic pathway when O2 is limiting. Another study similarly showed an interaction between pyruvate kinase M2 isoform and PHD3 in the cytosol, where the complex appears to regulate the flux of carbons through glycolysis.53 Thus, PHD3 may act to promote survival under hypoxic conditions both indirectly (promoting transcription of metabolic genes) and directly (by modulating metabolic processes themselves).
The role of PHD3 in hypoxic cell survival is also evident from studies of PHD3 expression from tumor samples. Here, PHD3 is often upregulated compared to its expression in normal tissue.76,78–81 The high expression of PHD3 in these samples suggests that it is not acting to induce apoptosis. In fact, PHD3 expression was correlated with increased aggressiveness in pancreatic exocrine and endocrine cancers. However, it was not determined whether PHD3 expression was the reason for this increase in aggressiveness, or if the correlation was simply due to the presence of tumor hypoxia, which upregulates PHD3 expression. Regardless, these studies suggest that upregulation of PHD3 under hypoxic conditions in some cancers does not result in apoptosis, but rather may promote tumor progression and/or survival.
Proapoptotic studies
Despite the evidence of PHD3’s prosurvival attributes, evidence for proapoptotic functions dominates the PHD3 literature. In fact, one of the major phenotypes discovered in PHD3−/− mice was due to a decrease of PHD3-mediated apoptosis in neurons during sympathoadrenal development.75 Overall, this resulted in enlarged sympathetic ganglia and reduced target tissue innervation.75 Indeed, several studies have demonstrated a significant increase in PHD3 transcript levels following the withdrawal of NGF from cultures of neuronally-derived cell lines.25,82,83 In particular, NGF withdrawal from the rat pheochromocytoma cell line, PC12, results in a c-jun-dependent upregulation of PHD3, followed by cytochrome c release and caspase-dependent cell death.83 It has also been proposed that the kinesin KIF1Bβ may act downstream of PHD3 to mediate these proapoptotic effects.84 KIF1Bβ acts as a molecular motor, and is responsible for synaptic vesicle transport. It is difficult to hypothesize precisely how KIF1Bβ might modulate cell survival in this regard. Clearly more work must be done in this area.
The clear role for PHD3 in neuronal apoptosis predicts that PHD3 mutation may be involved in the progression of neuronal malignancies. To date, however, PHD3 mutations have not been reported in any type of malignancy. On the other hand, inhibition of PHD3 activity through other mechanisms may contribute to survival of cancer cells in this disease. For example, SDH mutations are common in pheochromocytoma.82 These mutations lead to a buildup of succinate, which can inhibit PHD3 activity through product inhibition of the hydroxylation reaction.13,82 Similarly, gain-of-function mutations in IDH result in the abnormal production of 2-HG, which also inhibits PHD activity in some gliomas.85 The idea that PHD3 inhibition due to mutations in TCA enzymes acts as a driving force for these types of malignancies is an exciting proposition; however, the precise mechanism remains fairly obscure and warrants further research.
Outside of neurons, PHD3 also has proapoptotic effects. It is purported to interact with Bcl-2, inhibiting the formation of the antiapoptotic Bax–Bcl-2 complex in muscle cells.70 However, PHD3 has also been demonstrated to promote apoptosis through hydroxylation of the human biological clock protein 2 (hCLK2), which in turn promotes apoptosis through activation of the ATR–Chk1–P53 DNA damage response pathway following ionizing radiation.29 In the HeLa cervical cancer cell line, transfection of a green fluorescent protein–PHD3 vector in normoxic conditions induced PHD3 localization into large punctate cytoplasmic aggregates followed by apoptosis.35 These “aggresomes” colocalized with the 20S proteasome and were associated with caspase-3 cleavage and apoptosis.35 This process appeared to require O2 (and thus PHD3-enzymatic activity) as inhibition of PHD3 with the 2-OG analog dimethyloxalylglycine, and mutation of PHD3, caused a more diffuse cytoplasmic localization, which protected cells from the proapoptotic effects of PHD3.35 Whether PHD3’s interaction with “aggresomes”, Bcl-2, and hCLK2 are related, or are cell-type/stimulus dependent remains to be elucidated. Nonetheless, PHD3-mediated apoptosis appears to require molecular O2 and hydroxylase activity.
Conclusion
Over the last 2 billion years, molecular O2 has accumulated and persisted on Earth. Its presence has promoted many functions of life, including the evolution of aerobic respiration. However, it has also antagonized life through its capacity to oxidize and inactivate biological molecules. The evolution of the prolyl-hydroxylase was an evolutionary answer to the antagonistic effects of O2, providing the cell with a tool to sense and adapt to changes in O2 concentration.
The proline-hydroxylation reaction itself requires O2 and 2-OG, and the PHD catalytic core contains a Fe-2 atom that is sensitive to oxidation by ROS. This allows tight integration of cell signaling with O2 concentration, TCA cycle function, and oxidative stress in the cell. Predictably, an enzyme with this sensitivity to react simultaneously to an array of metabolic cues should be involved in a broad array of cell signaling processes.
In the case of PHD3, a plethora of recent literature is beginning to support such roles. In fact, data argue that PHD3 is involved in cell differentiation, death, survival, and metabolism. Furthermore, PHD3 expression may modulate complex processes such as cell migration and immune system function, providing a direct link between O2 and these important biological processes. There is no doubt that continued research into the functions of PHD3 and other proline hydroxylases will continue to evolve our understanding of the intimate link between O2 sensing and cell signaling.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Taylor CT, McElwain JC. Ancient atmospheres and the evolution of oxygen sensing via the hypoxia-inducible factor in metazoans. Physiology (Bethesda) 2010;25(5):272–279. doi: 10.1152/physiol.00029.2010. [DOI] [PubMed] [Google Scholar]
- 2.Catling DC, Claire MW. How Earth’s atmosphere evolved to an oxic state: a status report. Earth Planet Sci Lett. 2006;237(1–2):1–20. [Google Scholar]
- 3.Holland HD. The oxygenation of the atmosphere and oceans. Philos Trans R Soc Lond B Biol Sci. 2006;361(1470):903–915. doi: 10.1098/rstb.2006.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parfrey LW, Lahr DJ, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A. 2011;108(33):13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.West CM, van der Wel H, Wang ZA. Prolyl 4-hydroxylase-1 mediates O2 signaling during development of Dictyostelium. Development. 2007;134(18):3349–3358. doi: 10.1242/dev.000893. [DOI] [PubMed] [Google Scholar]
- 6.Hughes BT, Espenshade PJ. Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. EMBO J. 2008;27(10):1491–1501. doi: 10.1038/emboj.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rytkönen KT, Williams TA, Renshaw GM, Primmer CR, Nikinmaa M. Molecular evolution of the metazoan PHD-HIF oxygen-sensing system. Mol Biol Evol. 2011;28(6):1913–1926. doi: 10.1093/molbev/msr012. [DOI] [PubMed] [Google Scholar]
- 8.Myllyharju J. Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol. 2003;22(1):15–24. doi: 10.1016/s0945-053x(03)00006-4. [DOI] [PubMed] [Google Scholar]
- 9.Koivunen P, Tiainen P, Hyvärinen J, et al. An endoplasmic reticulum transmembrane prolyl 4-hydroxylase is induced by hypoxia and acts on hypoxia-inducible factor alpha. J Biol Chem. 2007;282(42):30544–30552. doi: 10.1074/jbc.M704988200. [DOI] [PubMed] [Google Scholar]
- 10.Cardinale GJ, Rhoads RE, Udenfriend S. Simultaneous incorporation of 18 O into succinate and hydroxyproline catalyzed by collagen proline hydroxylase. Biochem Biophys Res Commun. 1971;43(3):537–543. doi: 10.1016/0006-291x(71)90647-4. [DOI] [PubMed] [Google Scholar]
- 11.Pollard PJ, Brière JJ, Alam NA, et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum Mol Genet. 2005;14(15):2231–2239. doi: 10.1093/hmg/ddi227. [DOI] [PubMed] [Google Scholar]
- 12.Dang L, Jin S, Su SM. IDH mutations in glioma and acute myeloid leukemia. Trends Mol Med. 2010;16(9):387–397. doi: 10.1016/j.molmed.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Maxwell PH. A common pathway for genetic events leading to pheochromocytoma. Cancer Cell. 2005;8(2):91–93. doi: 10.1016/j.ccr.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 14.Tuderman L, Myllylä R, Kivirikko KI. Mechanism of the prolyl hydroxylase reaction. 1. Role of co-substrates. Eur J Biochem. 1977;80(2):341–348. doi: 10.1111/j.1432-1033.1977.tb11888.x. [DOI] [PubMed] [Google Scholar]
- 15.Schofield CJ, Ratcliffe PJ. Signalling hypoxia by HIF hydroxylases. Biochem Biophys Res Commun. 2005;338(1):617–626. doi: 10.1016/j.bbrc.2005.08.111. [DOI] [PubMed] [Google Scholar]
- 16.Spitz DR, Azzam EI, Li JJ, Gius D. Metabolic oxidation/reduction reactions and cellular responses to ionizing radiation: a unifying concept in stress response biology. Cancer Metastasis Rev. 2004;23(3–4):311–322. doi: 10.1023/B:CANC.0000031769.14728.bc. [DOI] [PubMed] [Google Scholar]
- 17.Guzy RD, Hoyos B, Robin E, et al. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1(6):401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Kaewpila S, Venkataraman S, Buettner GR, Oberley LW. Manganese superoxide dismutase modulates hypoxia-inducible factor-1 alpha induction via superoxide. Cancer Res. 2008;68(8):2781–2788. doi: 10.1158/0008-5472.CAN-07-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myllylä R, Kuutti-Savolainen ER, Kivirikko KI. The role of ascorbate in the prolyl hydroxylase reaction. Biochem Biophys Res Commun. 1978;83(2):441–448. doi: 10.1016/0006-291x(78)91010-0. [DOI] [PubMed] [Google Scholar]
- 20.Peterkofsky B. Ascorbate requirement for hydroxylation and secretion of procollagen: relationship to inhibition of collagen synthesis in scurvy. Am J Clin Nutr. 1991;54(Suppl 6):1135S–1140S. doi: 10.1093/ajcn/54.6.1135s. [DOI] [PubMed] [Google Scholar]
- 21.Hoffart LM, Barr EW, Guyer RB, Bollinger JM, Krebs C. Direct spectroscopic detection of a C-H-cleaving high-spin Fe(IV) complex in a prolyl-4-hydroxylase. Proc Natl Acad Sci U S A. 2006;103(40):14738–14743. doi: 10.1073/pnas.0604005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knowles HJ, Raval RR, Harris AL, Ratcliffe PJ. Effect of ascorbate on the activity of hypoxia-inducible factor in cancer cells. Cancer Res. 2003;63(8):1764–1768. [PubMed] [Google Scholar]
- 23.Pagé EL, Chan DA, Giaccia AJ, Levine M, Richard DE. Hypoxia-inducible factor-1alpha stabilization in nonhypoxic conditions: role of oxidation and intracellular ascorbate depletion. Mol Biol Cell. 2008;19(1):86–94. doi: 10.1091/mbc.E07-06-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho VT, Bunn HF. Effects of transition metals on the expression of the erythropoietin gene: further evidence that the oxygen sensor is a heme protein. Biochem Biophys Res Commun. 1996;223(1):175–180. doi: 10.1006/bbrc.1996.0865. [DOI] [PubMed] [Google Scholar]
- 25.Lipscomb EA, Sarmiere PD, Crowder RJ, Freeman RS. Expression of the SM-20 gene promotes death in nerve growth factor-dependent sympathetic neurons. J Neurochem. 1999;73(1):429–432. doi: 10.1046/j.1471-4159.1999.0730429.x. [DOI] [PubMed] [Google Scholar]
- 26.Epstein AC, Gleadle JM, McNeill LA, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107(1):43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 27.Huang J, Zhao Q, Mooney SM, Lee FS. Sequence determinants in hypoxia-inducible factor-1alpha for hydroxylation by the prolyl hydroxylases PHD1, PHD2, and PHD3. J Biol Chem. 2002;277(42):39792–39800. doi: 10.1074/jbc.M206955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D, Hirsilä M, Koivunen P, et al. Many amino acid substitutions in a hypoxia-inducible transcription factor (HIF)-1alpha-like peptide cause only minor changes in its hydroxylation by the HIF prolyl 4-hydroxylases: substitution of 3,4-dehydroproline or azetidine-2-carboxylic acid for the proline leads to a high rate of uncoupled 2-oxoglutarate decarboxylation. J Biol Chem. 2004;279(53):55051–55059. doi: 10.1074/jbc.M410287200. [DOI] [PubMed] [Google Scholar]
- 29.Xie L, Pi X, Mishra A, Fong G, Peng J, Patterson C. PHD3-dependent hydroxylation of HCLK2 promotes the DNA damage response. J Clin Invest. 2012;122(8):2827–2836. doi: 10.1172/JCI62374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo W, Hu H, Chang R, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145(5):732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie L, Xiao K, Whalen EJ, et al. Oxygen-regulated beta(2)-adrenergic receptor hydroxylation by EGLN3 and ubiquitylation by pVHL. Sci Signal. 2009;2(78):ra33. doi: 10.1126/scisignal.2000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kivirikko KI, Myllylä R. Posttranslational enzymes in the biosynthesis of collagen: intracellular enzymes. Meth Enzymol. 1982;82(Pt A):245–304. doi: 10.1016/0076-6879(82)82067-3. [DOI] [PubMed] [Google Scholar]
- 33.Stadtman ER. Oxidation of free amino acids and amino acid residues in proteins by radiolysis and by metal-catalyzed reactions. Annu Rev Biochem. 1993;62:797–821. doi: 10.1146/annurev.bi.62.070193.004053. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama K, Gazdoiu S, Abraham R, Pan ZQ, Ronai Z. Hypoxia-induced assembly of prolyl hydroxylase PHD3 into complexes: implications for its activity and susceptibility for degradation by the E3 ligase Siah2. Biochem J. 2007;401(1):217–226. doi: 10.1042/BJ20061135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rantanen K, Pursiheimo J, Högel H, Himanen V, Metzen E, Jaakkola PM. Prolyl hydroxylase PHD3 activates oxygen-dependent protein aggregation. Mol Biol Cell. 2008;19(5):2231–2240. doi: 10.1091/mbc.E07-11-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20(1):51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Semenza GL. Oxygen homeostasis. Wiley Interdiscip Rev Syst Biol Med. 2010;2(3):336–361. doi: 10.1002/wsbm.69. [DOI] [PubMed] [Google Scholar]
- 38.Semenza GL. Regulation of oxygen homeostasis by hypoxia-inducible factor 1. Physiology (Bethesda) 2009;24:97–106. doi: 10.1152/physiol.00045.2008. [DOI] [PubMed] [Google Scholar]
- 39.Villar D, Vara-Vega A, Landázuri MO, Del Peso L. Identification of a region on hypoxia-inducible-factor prolyl 4-hydroxylases that determines their specificity for the oxygen degradation domains. Biochem J. 2007;408(2):231–240. doi: 10.1042/BJ20071052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Appelhoff RJ, Tian YM, Raval RR, et al. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem. 2004;279(37):38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- 41.Kallio PJ, Okamoto K, O’Brien S, et al. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1alpha. EMBO J. 1998;17(22):6573–6586. doi: 10.1093/emboj/17.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92(12):5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J Biol Chem. 1995;270(3):1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- 44.Arany Z, Huang LE, Eckner R, et al. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci U S A. 1996;93(23):12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Metzen E, Stiehl DP, Doege K, Marxsen JH, Hellwig-Bürgel T, Jelkmann W. Regulation of the prolyl hydroxylase domain protein 2 (phd2/egln-1) gene: identification of a functional hypoxia-responsive element. Biochem J. 2005;387(Pt 3):711–717. doi: 10.1042/BJ20041736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pescador N, Cuevas Y, Naranjo S, et al. Identification of a functional hypoxia-responsive element that regulates the expression of the egl nine homologue 3 (egln3/phd3) gene. Biochem J. 2005;390(Pt 1):189–197. doi: 10.1042/BJ20042121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schödel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117(23):e207–e217. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Villar D, Ortiz-Barahona A, Gómez-Maldonado L, et al. Cooperativity of stress-responsive transcription factors in core hypoxia-inducible factor binding regions. PLoS ONE. 2012;7(9):e45708. doi: 10.1371/journal.pone.0045708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyer LR, Zweig AS, Hinrichs AS, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41(Database issue):D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenbloom KR, Sloan CA, Malladi VS, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41(Database issue):D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Millonig G, Hegedüsch S, Becker L, Seitz HK, Schuppan D, Mueller S. Hypoxia-inducible factor 1 alpha under rapid enzymatic hypoxia: cells sense decrements of oxygen but not hypoxia per se. Free Radic Biol Med. 2009;46(2):182–191. doi: 10.1016/j.freeradbiomed.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 52.Luo W, Semenza GL. Pyruvate kinase M2 regulates glucose metabolism by functioning as a coactivator for hypoxia-inducible factor 1 in cancer cells. Oncotarget. 2011;2(7):551–556. doi: 10.18632/oncotarget.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen N, Rinner O, Czernik D, et al. The oxygen sensor PHD3 limits glycolysis under hypoxia via direct binding to pyruvate kinase. Cell Res. 2011;21(6):983–986. doi: 10.1038/cr.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wax SD, Rosenfield CL, Taubman MB. Identification of a novel growth factor-responsive gene in vascular smooth muscle cells. J Biol Chem. 1994;269(17):13041–13047. [PubMed] [Google Scholar]
- 55.Menzies K, Liu B, Kim WJ, Moschella MC, Taubman MB. Regulation of the SM-20 prolyl hydroxylase gene in smooth muscle cells. Biochem Biophys Res Commun. 2004;317(3):801–810. doi: 10.1016/j.bbrc.2004.03.115. [DOI] [PubMed] [Google Scholar]
- 56.Kyo S, Takakura M, Taira T, et al. Sp1 cooperates with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (hTERT) Nucleic Acids Res. 2000;28(3):669–677. doi: 10.1093/nar/28.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gerber SA, Yatsula B, Maier CL, Sadler TJ, Whittaker LW, Pober JS. Interferon-gamma induces prolyl hydroxylase (PHD)3 through a STAT1-dependent mechanism in human endothelial cells. Arterioscler Thromb Vasc Biol. 2009;29(9):1363–1369. doi: 10.1161/ATVBAHA.109.192542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Madden SL, Galella EA, Riley D, Bertelsen AH, Beaudry GA. Induction of cell growth regulatory genes by p53. Cancer Res. 1996;56(23):5384–5390. [PubMed] [Google Scholar]
- 59.Lipscomb EA, Sarmiere PD, Freeman RS. SM-20 is a novel mitochondrial protein that causes caspase-dependent cell death in nerve growth factor-dependent neurons. J Biol Chem. 2001;276(7):5085–5092. doi: 10.1074/jbc.M008407200. [DOI] [PubMed] [Google Scholar]
- 60.Place TL, Fitzgerald MP, Venkataraman S, et al. Aberrant promoter CpG methylation is a mechanism for impaired PHD3 expression in a diverse set of malignant cells. PLoS ONE. 2011;6(1):e14617. doi: 10.1371/journal.pone.0014617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hatzimichael E, Dasoula A, Shah R, et al. The prolyl-hydroxylase EGLN3 and not EGLN1 is inactivated by methylation in plasma cell neoplasia. Eur J Haematol. 2010;84(1):47–51. doi: 10.1111/j.1600-0609.2009.01344.x. [DOI] [PubMed] [Google Scholar]
- 62.Cervera AM, Apostolova N, Luna-Crespo F, Sanjuan-Pla A, Garcia-Bou R, McCreath KJ. An alternatively spliced transcript of the PHD3 gene retains prolyl hydroxylase activity. Cancer Lett. 2006;233(1):131–138. doi: 10.1016/j.canlet.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 63.Nakayama K, Frew IJ, Hagensen M, et al. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell. 2004;117(7):941–952. doi: 10.1016/j.cell.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 64.Metzen E, Berchner-Pfannschmidt U, Stengel P, et al. Intracellular localisation of human HIF-1 alpha hydroxylases: implications for oxygen sensing. J Cell Sci. 2003;116(Pt 7):1319–1326. doi: 10.1242/jcs.00318. [DOI] [PubMed] [Google Scholar]
- 65.Takeda K, Ho VC, Takeda H, Duan LJ, Nagy A, Fong GH. Placental but not heart defects are associated with elevated hypoxia-inducible factor alpha levels in mice lacking prolyl hydroxylase domain protein 2. Mol Cell Biol. 2006;26(22):8336–8346. doi: 10.1128/MCB.00425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dunwoodie SL. The role of hypoxia in development of the Mammalian embryo. Dev Cell. 2009;17(6):755–773. doi: 10.1016/j.devcel.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 67.Kiss J, Mollenhauer M, Walmsley SR, et al. Loss of the oxygen sensor PHD3 enhances the innate immune response to abdominal sepsis. J Immunol. 2012;189(4):1955–1965. doi: 10.4049/jimmunol.1103471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fu J, Menzies K, Freeman RS, Taubman MB. EGLN3 prolyl hydroxylase regulates skeletal muscle differentiation and myogenin protein stability. J Biol Chem. 2007;282(17):12410–12418. doi: 10.1074/jbc.M608748200. [DOI] [PubMed] [Google Scholar]
- 69.Fu J, Taubman MB. Prolyl hydroxylase EGLN3 regulates skeletal myoblast differentiation through an NF-kappaB-dependent pathway. J Biol Chem. 2010;285(12):8927–8935. doi: 10.1074/jbc.M109.078600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Y, Huo Z, Yan B, et al. Prolyl hydroxylase 3 interacts with Bcl-2 to regulate doxorubicin-induced apoptosis in H9c2 cells. Biochem Biophys Res Commun. 2010;401(2):231–237. doi: 10.1016/j.bbrc.2010.09.037. [DOI] [PubMed] [Google Scholar]
- 71.Frank D, Gantenberg J, Boomgaarden I, et al. MicroRNA-20a inhibits stress-induced cardiomyocyte apoptosis involving its novel target Egln3/PHD3. J Mol Cell Cardiol. 2012;52(3):711–717. doi: 10.1016/j.yjmcc.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 72.Escribese MM, Sierra-Filardi E, Nieto C, et al. The prolyl hydroxylase PHD3 identifies proinflammatory macrophages and its expression is regulated by activin A. J Immunol. 2012;189(4):1946–1954. doi: 10.4049/jimmunol.1201064. [DOI] [PubMed] [Google Scholar]
- 73.Walmsley SR, Chilvers ER, Thompson AA, et al. Prolyl hydroxylase 3 (PHD3) is essential for hypoxic regulation of neutrophilic inflammation in humans and mice. J Clin Invest. 2011;121(3):1053–1063. doi: 10.1172/JCI43273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sternfeld J, David CN. Oxygen gradients cause pattern orientation in Dictyostelium cell clumps. J Cell Sci. 1981;50:9–17. doi: 10.1242/jcs.50.1.9. [DOI] [PubMed] [Google Scholar]
- 75.Bishop T, Gallagher D, Pascual A, et al. Abnormal sympathoadrenal development and systemic hypotension in PHD3−/− mice. Mol Cell Biol. 2008;28(10):3386–3400. doi: 10.1128/MCB.02041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Su Y, Loos M, Giese N, et al. PHD3 regulates differentiation, tumour growth and angiogenesis in pancreatic cancer. Br J Cancer. 2010;103(10):1571–1579. doi: 10.1038/sj.bjc.6605936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Högel H, Rantanen K, Jokilehto T, Grenman R, Jaakkola PM. Prolyl hydroxylase PHD3 enhances the hypoxic survival and G1 to S transition of carcinoma cells. PLoS ONE. 2011;6(11):e27112. doi: 10.1371/journal.pone.0027112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen S, Zhang J, Li X, Luo X, Fang J, Chen H. The expression of prolyl hydroxylase domain enzymes are up-regulated and negatively correlated with Bcl-2 in non-small cell lung cancer. Mol Cell Biochem. 2011;358(1–2):257–263. doi: 10.1007/s11010-011-0976-1. [DOI] [PubMed] [Google Scholar]
- 79.Su C, Huang K, Sun L, et al. Overexpression of the HIF hydroxylase PHD3 is a favorable prognosticator for gastric cancer. Med Oncol. 2012;29(4):2710–2715. doi: 10.1007/s12032-012-0171-6. [DOI] [PubMed] [Google Scholar]
- 80.Couvelard A, Deschamps L, Rebours V, et al. Overexpression of the oxygen sensors PHD-1, PHD-2, PHD-3, and FIH Is associated with tumor aggressiveness in pancreatic endocrine tumors. Clin Cancer Res. 2008;14(20):6634–6639. doi: 10.1158/1078-0432.CCR-07-5258. [DOI] [PubMed] [Google Scholar]
- 81.Gossage L, Zaitoun A, Fareed KR, et al. Expression of key hypoxia sensing prolyl-hydroxylases PHD1, -2 and -3 in pancreaticobiliary cancer. Histopathology. 2010;56(7):908–920. doi: 10.1111/j.1365-2559.2010.03566.x. [DOI] [PubMed] [Google Scholar]
- 82.Lee S, Nakamura E, Yang H, et al. Neuronal apoptosis linked to EglN3 prolyl hydroxylase and familial pheochromocytoma genes: developmental culling and cancer. Cancer Cell. 2005;8(2):155–167. doi: 10.1016/j.ccr.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 83.Straub JA, Lipscomb EA, Yoshida ES, Freeman RS. Induction of SM-20 in PC12 cells leads to increased cytochrome c levels, accumulation of cytochrome c in the cytosol, and caspase-dependent cell death. J Neurochem. 2003;85(2):318–328. doi: 10.1046/j.1471-4159.2003.01688.x. [DOI] [PubMed] [Google Scholar]
- 84.Schlisio S, Kenchappa RS, Vredeveld LC, et al. The kinesin KIF1Bbeta acts downstream from EglN3 to induce apoptosis and is a potential 1p36 tumor suppressor. Genes Dev. 2008;22(7):884–893. doi: 10.1101/gad.1648608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294(5545):1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 87.Sato M, Sakota M, Nakayama K. Human PRP19 interacts with prolyl-hydroxylase PHD3 and inhibits cell death in hypoxia. Exp Cell Res. 2010;316(17):2871–2882. doi: 10.1016/j.yexcr.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 88.Xue J, Li X, Jiao S, Wei Y, Wu G, Fang J. Prolyl hydroxylase-3 is down-regulated in colorectal cancer cells and inhibits IKKbeta independent of hydroxylase activity. Gastroenterology. 2010;138(2):606–615. doi: 10.1053/j.gastro.2009.09.049. [DOI] [PubMed] [Google Scholar]
- 89.Tennant DA, Gottlieb E. HIF prolyl hydroxylase-3 mediates alpha-ketoglutarate-induced apoptosis and tumor suppression. J Mol Med. 2010;88(8):839–849. doi: 10.1007/s00109-010-0627-0. [DOI] [PubMed] [Google Scholar]
- 90.Hopfer U, Hopfer H, Jablonski K, Stahl RA, Wolf G. The novel WD-repeat protein Morg1 acts as a molecular scaffold for hypoxia-inducible factor prolyl hydroxylase 3 (PHD3) J Biol Chem. 2006;281(13):8645–8655. doi: 10.1074/jbc.M513751200. [DOI] [PubMed] [Google Scholar]
- 91.Köditz J, Nesper J, Wottawa M, et al. Oxygen-dependent ATF-4 stability is mediated by the PHD3 oxygen sensor. Blood. 2007;110(10):3610–3617. doi: 10.1182/blood-2007-06-094441. [DOI] [PubMed] [Google Scholar]
- 92.Baek JH, Mahon PC, Oh J, et al. OS-9 interacts with hypoxia-inducible factor 1alpha and prolyl hydroxylases to promote oxygen-dependent degradation of HIF-1alpha. Mol Cell. 2005;17(4):503–512. doi: 10.1016/j.molcel.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 93.Aprelikova O, Pandolfi S, Tackett S, et al. Melanoma antigen-11 inhibits the hypoxia-inducible factor prolyl hydroxylase 2 and activates hypoxic response. Cancer Res. 2009;69(2):616–624. doi: 10.1158/0008-5472.CAN-08-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Masson N, Appelhoff RJ, Tuckerman JR, et al. The HIF prolyl hydroxylase PHD3 is a potential substrate of the TRiC chaperonin. FEBS Lett. 2004;570(1–3):166–170. doi: 10.1016/j.febslet.2004.06.040. [DOI] [PubMed] [Google Scholar]