Abstract
Background
Deletion 13q14.3 is the most common cytogenetic abnormality in chronic lymphocytic leukemia (CLL). Previously it was reported that miR-15/16 is the target of 13q14 deletions and plays a tumor suppressor role by suppressing Bcl-2. Therefore, Bcl-2 expression was examined more closely to determine whether it would predict 13q14 deletion status.
Methods
A multi-color flow panel consisting of anti-Bcl-2/anti-lambda/anti-kappa/CD19/CD5/CD3/CD20 was performed. The ability of Bcl-2 to predict 13q14 deletion was tested using the conventional Bcl-2 index (c-index): mean fluorescence intensity (MFI) of CLL clone/MFI of residual T-cells. Fifty-four untreated CLL/MBL patients were studied. Bimodal Bcl-2 expression was evaluated to test the ability of Bcl-2 to detect intra-clonal heterogeneity. Other CLL prognostic markers including CD38, CD49d, CD26, and CD69 were evaluated. FISH was performed on selected sorted populations.
Results
The Bcl-2 c-index strongly predicts del13q14 p<0.0001. A statistically significant association was observed between the percentage of cells carrying the deletion and the level of Bcl-2 expression p<0.05. Cells sorted based on Bcl-2 expression showed enrichment of both hemi-and homozygous del 13q14 cells. Also we observed that an alteration in Bcl-2 level over time predicts changes in 13q14 deletion status. And a statistically significant correlation between the bimodal pattern of CD69 expression and the presence of 13q14 deletion was found p<0.0001.
Conclusion
Bcl-2 expression using the c-index strongly predicts 13q14 deletion and can be used to distinguish homozygous, heterozygous, and diploid CLL clonal cells. Further systematic studies of this biomarker are needed for confirmation and expansion of these findings.
Keywords: chronic lymphocytic leukemia, Bcl-2, Flow cytometry, del 13q14, hemizygous/homozygous deletion
Introduction
Chronic lymphocytic leukemia (CLL) is the most common adult leukemia in western countries. CLL is characterized by accumulation of CD5 positive malignant B-cells in peripheral lymphoid organs, bone marrow (BM) and peripheral blood (PB) [1]. Fluorescence in situ hybridization (FISH) analysis has shown that chromosomal abnormalities can be found in more than 80% of CLL cases. Hemizygous and/or homozygous deletions of chromosome band 13q14 constitute the most frequent cytogenetic abnormalities in CLL and are seen in approximately half of the cases [2, 3]. Trisomy 12 (15%), 11q22–23 deletion (12%), and 17p13 deletion (8%) were less frequent [3, 4].
MicroRNAs (miRNAs) are small non-coding RNA genes that regulate gene expression and can be involved in human tumorigenesis [5–12]. The single-stranded miRNA binds specific mRNA through sequences that are imperfectly complementary to the target mRNA, mainly to the 3′UTR. The bound mRNA remains un-translated, resulting in reduced levels of the corresponding protein [13, 14]. In humans, miR-15a and miR-16 are clustered intronicaly within 0.5-kilo bases in band 13q14 [15]. Deletions involving miR-15a and miR-16-1, located in that cluster or their down-regulation, were found in approximately 65% of CLL patients [5]. Also the homologous 13q14 deletion has been described in an NZB mouse model for CLL [16]. A germ-line mutation in the primary precursor of miR-16-1/miR-15a located 7 bp after the 3′end of miR-16-1 was also reported to cause low levels of miRNA expression in vitro and in vivo in CLL patients [17]. Moreover, it has been experimentally shown that miR-15a and miR-16 expression inversely correlates with Bcl-2 expression and that both microRNAs negatively regulate Bcl-2 at a post-transcriptional level. Therefore, the lack of miR-15a and miR-16-1 in the majority of CLL cases does result in up-regulation of the Bcl-2 protein [18]. The earliest report for Bcl-2 elevation in CLL was reported by Honanda et al., Hoffbrand et al., and McConkey et al., all noted that this increased expression of Bcl-2 was independent of (14; 18) translocation [19–21]. Although Bcl-2 expression is characteristic of B-cells in CLL [19–27], high levels are not seen in all patients [23, 24, 27, and 28].
These observations suggested that down regulation of miR-15/16 as part of 13q14 deletion contributes to an increase in Bcl-2 expression. Therefore, we explored the relationship between the Bcl-2 expression and 13q14 deletion. A new flow cytometric gating strategy was used to evaluate Bcl-2 expression. The c-index (mean fluorescence intensity (MFI) of the CLL clone/MFI of residual T-cells) was used to quantitate this important biomarker and its relationship to 13q14 deletion status. Optimal c-index cutoff values were determined to differentiate 13q14 deletion from non-13q14 deletion cases. The group negative for the 13q14 deletion was used to study the basal level of Bcl-2 expression in CLL. Intra-clonal Bcl-2 heterogeneity was carefully examined as it may relate to overall complexity of the tumor and might signify and separate two different populations with different 13q14 deletion status. For example, intra-clonal Bcl-2 bimodality in CLL cases may represent two different populations that had either a mixture of hemizygous and homozygous 13q14 deletion, or a mixture of hemizygous and diploid CLL clonal cells.
Material & Methods
Patients and Samples
This study used heparinized peripheral blood samples obtained from 54 newly diagnosed, untreated CLL patients. Thirty-three (61.1%) had del 13q14, and 21 (40.7%) cases were designated as non-13q14 deletion and used as a control to study the basal Bcl-2 level in CLL cases. Rai stage and lymphocyte doubling time (LDT) were obtained from the clinical record. The diagnosis of CLL was made on the basis of clinical examination, as well as morphological and immunological criteria according to the international workshop on CLL iwCLL and Hallek et al. [29]. No long-term follow-up is available. These patients were enrolled on an NHLBI IRB approved clinical study, registered with clinicaltrials.gov under identifier (NCT00923507), and under NCI study 97-C-0178 (clinicaltrial.gov, identifier: NCT00019370).
Intracellular Bcl-2 Staining
For each sample, 100 μl of washed whole blood cells (Phosphate Buffer Saline × 2) were stained with the following panel for 30 min in the dark at room temperature (surface staining): anti-lambda PE, CD19 PerCp Cy5.5, CD5 PE Cy7, anti-kappa APC, CD3 APC Cy 7 (these reagents obtained from BD Biosciences, San Diego, USA), and CD20 eFluor 450 (eBiosciences, San Diego, CA). After surface staining, fixation was performed using 100 μl fixation medium (Reagent A, Caltag, Burlingame, CA) for 15 minutes. The cells were then washed once with 3 ml of 5% bovine serum albumin (BSA) in PBS. Permeabilization was performed using 100 μl of permeabilization medium (Reagent B, Caltag, Burlingame, CA), and 10 μl of FITC-conjugated anti-Bcl-2 (clone 124, DAKO Cytomation, Glostrup, Denmark) or the corresponding isotype matched control from Caltage (Burlingame, CA). The cells were vortexed 1–2 times and incubated in the dark for 20 min at room temperature. Cells were then washed once in 3 ml of 5% BSA/PBS, re-suspended in 300–500 ml of the same buffer, and immediately run on a FACS Canto II flow cytometer (Becton Dickinson, CA, USA). Isotype-matched controls were used to define the threshold line separating positive and negative cell populations such that less than 1% of isotype-positive cells were present to the right of the line. Initially, the cases were identified as CLL based on the presence of a clonal light chain restricted B-cell population that was positive for CD19, CD5, and dim for CD20. Bcl-2 expression was evaluated over CLL clonal cells.
Surface Staining for CLL Prognostic Markers
Surface staining for CD38, CD49d, CD26, and CD69 were performed as follows: 100 μl of washed whole blood [Phosphate Buffer Saline (PBS) × 2] was added to each of three pre-wetted tubes containing CD19-peridinin chlorophyll Cy5.5 (PerCP Cy5.5); CD69- allophycocyanin (APC); CD5 phycoerythrin Cy-7 (PE Cy 7); CD3- allophycocyanin-Cy7 (APC Cy-7); CD20 eFluor450, and CD45 V500. In each of the three tubes, one of the following reagents was added CD38/CD49d/CD26-phycoerythrin (PE). The cells were stained for 30 minute in the dark at room temperature. The cells were lysed with 2 ml of 1x BD FACS ™ Lysing Solution for 10 minutes, washed once with PBS/ Azide, and re-suspended in 0.5 ml of 1% paraformaldehyde. The CLL clone (CD19+ve, CD20 dim, and CD5+ve cells) was analyzed for the prognostic surface biomarkers.
Flowcytometry
FACS Canto II with FACS Diva software was used for acquisition. Cytometry setup and tracking beads (CST, BD) were used to initialize PMT settings and daily QC/QA performance characteristics. Unstained control cells as well as single stained tubes for FITC, PE, Per CPCy5.5, PE Cy7, APC, APC Cy7, and eFluor 450 were prepared and used to set the flow cytometric compensation. In some experiments, rat anti-mouse kappa light chain Comp Beads (BD) were used to set the compensation and were stained according to the manufacturer’s instructions. The number of events collected in these experiments varied from 500,000 to 1 million events. FlowJo software (Tree Star, Ashland, OR) was used for data analysis and display. The expression was reported positive if the percentage of positive CLL cells was greater than or equal to the following cutoff values: ≥20%, ≥30%, ≥10%, and ≥30% for CD 38, CD49d, CD26, and CD69 respectively.
Bcl-2 Gating Strategy and Analysis of Expression
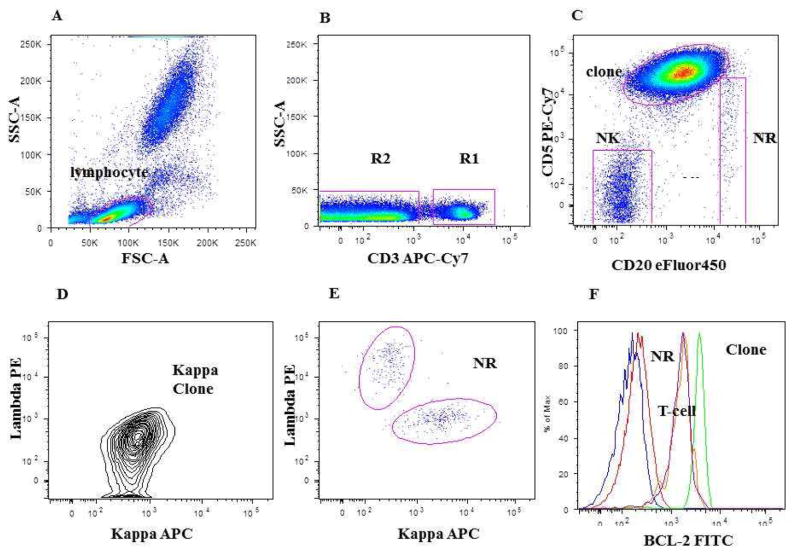
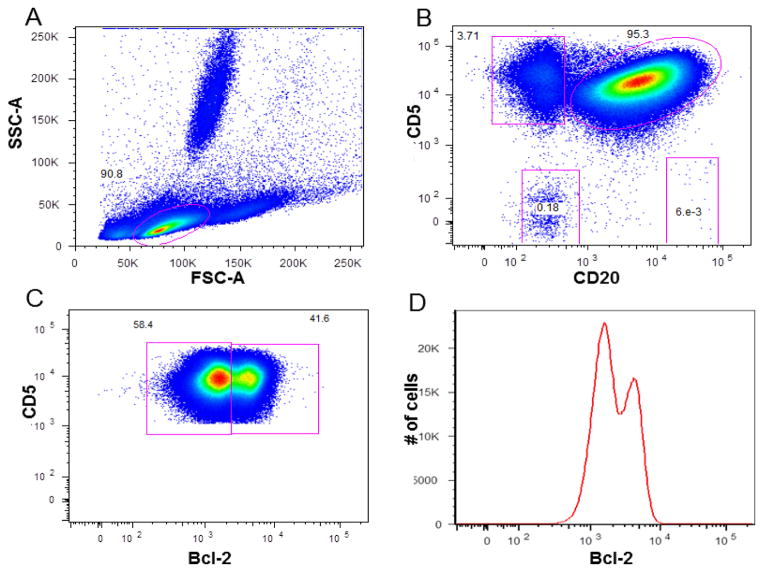
A doublet exclusion gate based upon (FSC-A vs. FSC-W) was utilized to gate on singlet cells, and then within this gate a lymphocyte gate was drawn using FSC-A vs. SSC-A characteristics. Using a dot plot of CD3 vs. SSC-A within singlet gate and lymphocyte gate, (R1) gate was drawn around the CD3 positive cells (patient’s residual T-cell) and (R2) gate on the CD3 negative cells. Within the (R2 gate) the CLL clone (CD19 positive, CD20 dim, CD5 positive cells), normal remaining (NR) polyclonal B cells (CD19 positive, CD20 bright, CD5 negative cells), and patient NK cells (CD19 negative, CD20 negative, CD5 negative cells) were identified using a dual-color histogram CD20 vs. CD5. A subsequent dot plot using lambda in x-axis, and kappa in y-axis was performed gated on the clonal B-cell population to identify light chain restriction and on polyclonal normal remaining B-cell population to confirm polyclonality. (See Fig. 1 for gating strategy).
Figure 1. Gating strategy for Bcl-2 expression analysis using whole blood.
A: is the FSC-A vs. SSC-A gating on lymphocytes. B: CD3 vs. SSC-A gating on CD3+ve patient residual T-cell (R1 gate); and on CD3-ve cells (R2). C: Dot plot CD20 vs. CD5 gated on R2: CLL clone, NR (normal remaining polyclonal B cells), and NK cell gates shown. D: shows monoclonal kappa clone expression gated on CLL clone. E: shows the polyclonal kappa and lambda expression gated on NR B-cells. F: single-color histogram showing the overlay of Bcl-2 expression on the Clonal B-cell, patient residual T-cells, and normal remaining polyclonal B-cells together with the Bcl-2 isotype matched control and unstained cells.
Bcl-2 mean fluorescence intensity (MFI) was evaluated on the clonal CLL cells, residual patient T-cells, and normal remaining polyclonal B-cells. The conventional Bcl-2 index (c-index) = MFI of CLL cells/MFI of T-cells was used to evaluate Bcl-2 expression. In this study several cutoff values have been studied for the c-index. The sensitivity and specificity for each cutoff value was evaluated. Also Bcl-2 was analyzed for bimodal expression. Bimodality was defined as two well-defined and discrete populations of cells within the malignant clone. Other CLL prognostic markers were also examined for bimodal expression.
Interphase FISH on Patient Samples
Interphase FISH was performed as previously described [30]. Briefly, buffy coat cells from fresh, heparinized peripheral blood were cultured in duplicate overnight without mitogens and for 96 hours with B-cell mitogens. Cells were harvested and fixed in 3:1 methanol: glacial acetic acid. Fresh slides were made from the fixed cell pellets and hybridized with commercially available probes (Abbott Molecular, Inc., formerly Vysis, Downers Grove, IL) to detect deletions in 13q14.3 (D13S319, D13S25), 11q23 (MLL) or 11q22.3 (ATM), and 17p13.1 (TP53); and to detect trisomy 12 (CEP 12 DNA). A minimum of 200 interphase nuclei were scored for hybridization signals for each probe.
FISH Staining on Sorted Cells
Sorting of the cell populations was performed based on the apparent difference in Bcl-2 MFI (intra- or inter-clonal) bimodality. Interphase FISH was performed on sorted cells using a Vysis D13S319 spectrum orange/ LSI 13q34 spectrum green probe set (Vysis Inc., No. 32–191018), which includes the probe and the hybridization buffer for defined del 13q14 x1, del 13q14x2, and diploid cells. Sorting was performed using a BD FACS Aria II machine (Bioinformatics, San Jose, CA)
FISH on sorted cells was carried out according to the manufacturer’s instructions and as described by Degheidy et al., 2007 [31]. Briefly, the slides were fixed in methanol: acetic acid (MAA) for at least 30 minutes then incubated in 2 X standard saline citrate (SSC) at 37°C for at least 1 hour. Following dehydration through graded alcohols, the probe was suspended in hybridization buffer and added to the slide. A cover slip was applied and sealed on using rubber cement. Using a thermo Brite Hybrite machine (Abbott Molecular), denaturation was performed at 75°C for 5 minutes, followed by hybridization at 37°C for 16 to 24 hours. Post-hybridization washes consisted of 2 washes (2 minutes each) in pre-warmed 0.4 X SSC/0.3XNP-40 (70°C); followed by 5 minutes in 2X SSC/0.1X NP-40 at room temperature. 4-6-diamino-2-phenylindole (DAPI) was added to the slide and used as a counter stain.
Statistical Analysis
The associations between patients’13q14 deletion status (yes/no) and markers/variables of interest were assessed using the Student t-test for continuous variables, and the chi-square test for categorical variables. Multivariable regression models were constructed to adjust for patients’ ages. Pearson’s correlation test was used to evaluate the strength of association between %13q14 deletion and the biomarkers of interest. To assess the power of Bcl-2-expression as a diagnostic biomarker for 13q14 deletion, we constructed receiver operating characteristic curves (ROC), and calculated the sensitivity, specificity, positive (PPV) and negative (NPV) predictive values for different Bcl-2 c-index expression cut off points [32]. All statistical tests were performed using SPSS version 17 for Windows (SPSS Inc., Chicago, IL).
Results
Patient demographic data
A total of 54 untreated CLL patients (mean age +/− S.D. 61.81+/−8.44) were studied. There were 32 males (59.3%) and 22 females (40.7%) with 1.45:1 male: female ratio. In all, 37 cases were Binet stage A (69%), and 17 cases were Binet stage B & C (31%). Also, 36 cases (67%) were Rai stage 0–1, and 18 cases (33%) Rai stage 2–4. Thirty-three patients (61.1%) had del 13q14 (age 63.4+/− 9.03), and 21 (40.7%) cases were designated as non-13q14 deletion (age 59.5+/−7.06). Based on interphase FISH analysis, 33 patients had a 13q14 deletion; 25 of these patients had 13q14 deletion as the sole abnormality, and 8 patients had a second abnormality (5 patients had 11q deletion, 2 patients had 17p deletion, and 1 patient had trisomy 12). Of the twenty-one patients designated as non-13q14 deletion, 2 had 11q deletion and 8 had trisomy 12 as the sole abnormality. The remaining 11 patients had normal FISH results.
Bcl-2 expression and its relation to 13q14 deletion
Bcl-2 c-index (See Bcl-2 gating strategy and expression analysis in the Material & Methods section and Figure.1) was used to quantify Bcl-2 expression in this study. Among the studied population, the majority (17/21, 80.9%) of the non13q14 deletion cases had a low Bcl-2 c-index (c- index =1.4). On the other hand the majority of the del 13q14 cases (26/33, 78.7%) had a high Bcl-2 c-index (c- index = 2.0). (See Figure 2 and Tables 1A and 1B).
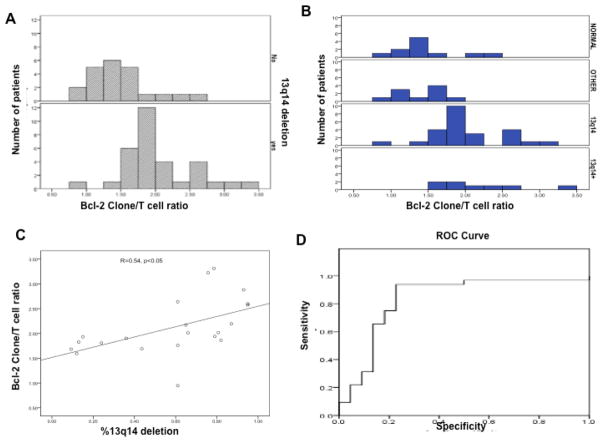
Figure 2.
A. Distribution of Bcl-2 c-index (Clone /T cell) in CLL patients with and without 13q14 deletion. B. Distribution of Bcl-2 expression using Bcl-2 c-index among four different CLL subpopulations based on cytogenetic abnormality (Normal, Other, del13q14, del 13q14+) See the material and method section. C. The correlation between the percentage of 13q14 deletion cells within the CLL clone and the Bcl-2 c-index (p< 0.05). D. The Receiver Operating Characteristic (ROC) curve for Bcl-2 c-index to predict 13q14 deletion in CLL patients.
Table 1.
Mean, SD, and p value for Bcl-2 c-index for cases with normal FISH, del 13q14 and cases with other cytogenetic abnormality.
| Table 1A
| |||||
|---|---|---|---|---|---|
| N | Mean | Std. Deviation | P-value | ||
|
| |||||
| c-index | Normal | 11 | 1.4735 | .41588 | Ref* |
| Deletion | 33 | 2.0441 | .51933 | 0.006 | |
| Others | 10 | 1.3690 | .29723 | 0.46 | |
| Table 1B
| |||||
|---|---|---|---|---|---|
| DELETION | N | Mean | Std. Deviation | p-value | |
|
| |||||
| c-index | non-deletion | 22 | 1.4796 | .43774 | <0.0001 |
| deletion | 33 | 2.0251 | .51583 | ||
There is no statistically significant difference between the groups with other cytogenetic abnormality compared to the group with normal FISH (p value= 0.46 using the c-index). So we combined both groups and referred to them as non-deletion 13q14 in table 1(B). C-index, conventional index; SD, standard deviation; Normal: CLL cases with normal FISH, others: CLL cases with other cytogenetic abnormality, deletion: CLL cases with deletion 13q14.
There is no statistically significant difference between the CLL cases that had other cytogenetic abnormalities compared to those cases with normal cytogenetics (p value= 0.46) using the Bcl-2 c-index (See Table 1A). Therefore, we combined both groups (cases with other abnormalities and cases with normal cytogenetics) and referred to them as non-13q14 deletion group and this designation was used for all the subsequent statistical analysis. The non-13q14 deletion cases were used to define the basal level of Bcl-2 expression. Cases with normal cytogenetics had a mean Bcl-2 c-index of 1.47 +/− 0.415. Cases with other cytogenetic abnormalities (non-13q14 deletion) had a mean Bcl-2 c-index of 1.36 +/− 0.297. On the other hand, del 13q14 cases had a mean value of 2.04 +/− 0.519 and Bcl-2 c-index strongly correlated with 13q14 deletion status with p <0.0001 (Table 1A and 1B).
Several cutoff values were evaluated for their ability to predict 13q14 deletion (1.25, 1.5, 1.6, 1.75, and 2.0). We found that a cutoff value of 1.6 led to the best correlation for the following parameters: sensitivity, specificity, PPV, and NPV (87%, 77.1%, 84.4%, and 77.3%, respectively). The sensitivities, specificities, PPVs, and NPVs for all of the above cutoff values are summarized in Table 2.
Table 2.
Correlation of Sensitivity, Specificity, PPV, and NPV for choosing optimal Bcl-2 c-index cutoff values.
| Values | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|
| C-index (MFI of Clone/MFI of T-cells) | ||||
| 1.25 | 96.9% | 31.8% | 67.4% | 87.5% |
| 1.50 | 93.8% | 59.1% | 76.9% | 86.7% |
| 1.60 | 87.0% | 77.1% | 84.4% | 77.3% |
| 1.75 | 75.0% | 81.8% | 85.7% | 69.2% |
| 2.0 | 37.5% | 86.4% | 80.0% | 48.7% |
PPV, positive predictive value; NPV, negative predictive value; MFI, mean fluorescent intensity.
Also a statistically significant correlation was observed between the percentage of cells carrying 13q14 deletion and the Bcl-2 c-index (R=0.54, p< 0.05, see Figure 2C).
Deletion 13q14 and surface co-expression of CD38, CD49d, CD26, and CD69
We studied the expression of the prognostic biomarkers CD38, CD49d, CD26 and CD69 on clonal CLL cells and correlation with the presence of 13q14 deletion. None of these markers demonstrated a statistically significant association with the presence of 13q14 deletion (p values = 0.07, 0.79, 0.61, and 0.46 for CD38, CD49d, CD26 and CD69, respectively).
Bimodal Antigen Expression is frequent in CLL
Bimodal CD69 expression was observed in 33/54 (61.1 %), bimodal CD38 in 7/54 (12.9%), bimodal CD5 in 2/54 (3.7%), and bimodal CD20 in 5/54 (9.2%) patients.
CD69 Expression pattern correlates with 13q14 deletion
All 54 CLL cases were analyzed for CD69 expression. Positivity verses negativity for CD69 did not correlate with the presence of 13q14 deletion (p=0.46). These cases were further classified based on the pattern of CD69 expression as follows: homogenously negative, homogenously positive, bimodal expression with more than 50% of cells showing bright CD69, and bimodal expression with less than 50% of cells showing bright CD69 expression. Figure 3 shows different patterns of CD69 expression that ranged from negative to positive and bimodal expression.
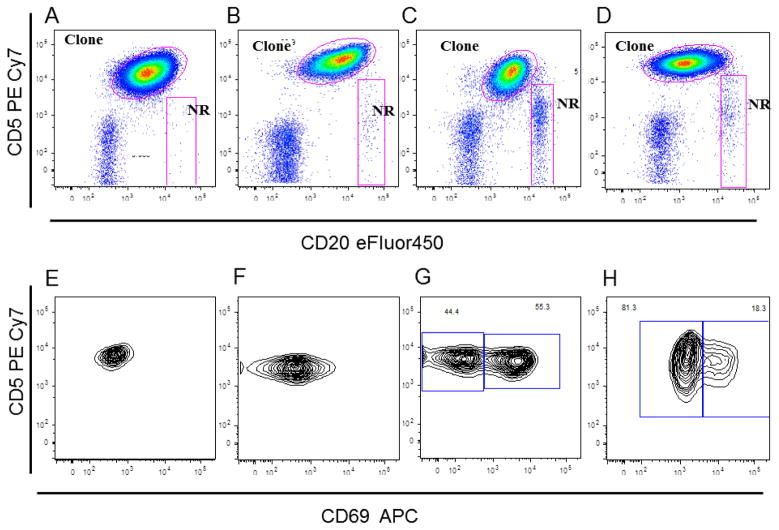
Figure 3. Pattern of CD69 expression over the CLL clonal cells.
The sequential analysis gates are based on R2 gate as seen before in figure 1. A, B, C, and D: Dot plot shows CD20 vs. CD5 gated on R2 gate for 4 different CLL cases that showed different CD69 APC expression pattern. The CLL clonal cells and normal remaining polyclonal B-cells (NR) gates were also shown. E, F, G, and H: Dot plot shows CD69 in x-axis vs. CD5 in y-axis gated on CLL clonal cells gate in A, B, C, and D. Homogenously negative (E), homogenously positive (F), bimodal expression with more than 50% of cells showed CD69 bright (G), and bimodal expression with less than 50% of cells showed bright CD69 expression. Note: the different pattern of CD69 expression ranged from negative, positive, and bimodal CD69 expression.
Cases were designated as positive when CD69 expression was uni-modal and expressed in more than 30% of CLL cells; as negative when CD69 expression was uni-modal and present on 30% or less of the CLL cells; and as bimodal, when there were two clearly distinct CD69 positive populations. Of interest, bimodal CD69 expression showed a strong statistically significant correlation with 13q14 deletion status with p value <0.0001 and 0.0005 before and after age adjustment, respectively
Bcl-2 Bimodality and FISH analysis on sorted cells
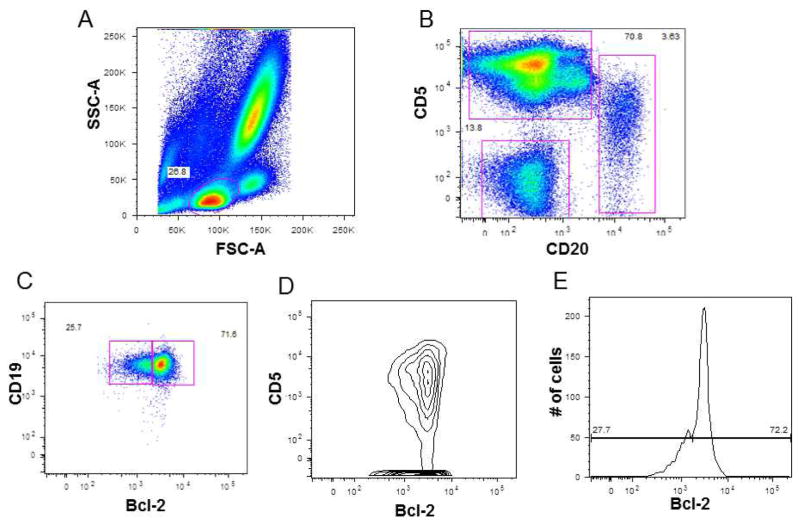
During the course of the study, intra-clonal bimodal Bcl-2 expression was found in 3/54 (5.5%) cases, including one MBL case and two CLL cases. Intra-clonal Bcl-2 bimodality is defined as two discrete cell subsets that showed two different Bcl-2 expression intensities within the CLL clonal cell (CD19 positive, CD20 dim, CD5 positive cells). Figure 4 demonstrates an MBL case with ≈ 72% of all B cells showing bright Bcl-2 expression (which represents about 12.7% of total lymphocytes). This case had 13% hemizygous del 13q14 (corresponds with the 12.7% bright Bcl-2 population) and 87% diploid cells performed on Ficoll-Hypaque lymphocytes. Figure 5 shows one of the cases with intra-clonal Bcl-2 bimodality. Flow cytometric analysis of this case revealed that approximately 42% of the CLL cells have bright Bcl-2 expression while the remaining CLL cells have dim Bcl-2 expression. This case had 53% homozygous 13q14 deletion and 40% hemizygous 13q14 deletion. FISH on the sorted cells showed enrichment of 13q14 X1 in the dim Bcl-2 population and enrichment of 13q14 X2 in the Bcl-2 bright population. These cases are example of how Bcl-2 can separate intra-clonal homozygous from hemizygous 13q14 deletion based on bright and dim Bcl-2 expression. This phenomenon was also seen in CLL cases that had a mixture of hemizygous and diploid cells in their CLL clone, and in CLL cases that showed inter-clonal Bcl-2 bimodality (defined as two different Bcl-2 expression intensities based on differences in CLL surface expression of CD19, CD20, and CD5; e.g. CD5 bright and CD5 dim cells) n=4 (data not shown).
Figure 4. Bimodal Bcl-2 expression in MBL.
A: is the FSC-A vs. SSC-A gating on lymphocytes. B: Dot plot CD20 vs. CD5 gated on Lymphocyte (T-cells, B-cells, and NK-cells gates are shown). C, D, and E: showing bimodal Bcl-2 expression in B-cells. C: Dot plot Bcl-2 vs. CD19 gated on B-cells. D: Dot plot Bcl-2 vs. CD5 gated on B-cells. E: Single color histogram showing bimodal Bcl-2 expression with about 72% of B-cells showing bright Bcl-2 expression and about 28% showing dim Bcl-2 expression.
Figure 5. Bimodal Bcl-2 expression in CLL.
A: is the FSC-A vs. SSC-A gating on lymphocytes. B: Dot plot CD20 vs. CD5 gated on Lymphocyte (T-cells, Clone, NK-cells, and normal remaining polyclonal B-cells (NB) gates are shown). C: Dot plot Bcl-2 vs. CD5 gated on CLL clonal cells showing the bimodal Bcl-2 expression whereas, about 40% showing bright Bcl-2 expression and about 60 % is showing dim expression. D: Single color histogram showing bimodal Bcl-2 expression in CLL clonal cells.
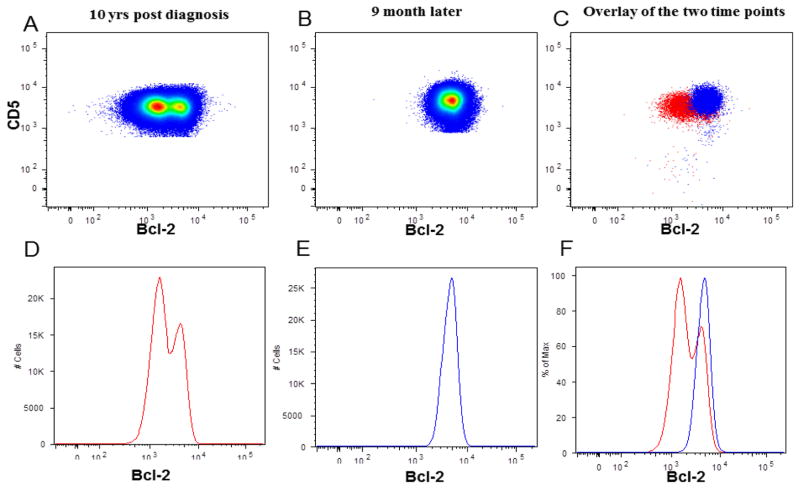
Another CLL case with bimodal Bcl-2 involved a patient with known FISH analysis at presentation (Bcl-2 analysis not performed) and subsequent FISH and Bcl-2 analyses performed at two time points 10 years after diagnosis (Figure. 6). At diagnosis, FISH showed that 96% of the CLL cells had a hemizygous 13q14 deletion. When the patient was studied 10 years later, FISH analysis revealed a mixture of 53% of cells with homozygous 13q14 deletions and 40% with hemizygous 13q14 deletion and flow cytometry showed bimodal Bcl-2 expression. Nine months later, prior to treatment, Bcl-2 expression and FISH were repeated. At this time, Bcl-2 expression intensity was uni-modal with more than 90% bright Bcl-2 cells compared to the previous results. FISH revealed that 96% of the clonal cells at that time had homozygous 13q14 deletions.
Figure 6. Correlation between Bcl-2 expression and 13q14 status in two different time points.
A, B, and C: Dot plots of BCl-2 vs. CD5 showing Bcl-2 expression in two different time point in the same case. A: Dot plot showing bimodal Bcl-2 expression in the 1st time point (10 years from diagnosis). B: Dot plot showing the uni-modal Bcl-2 expression in the second time point 9 month later. C: is the overlay of Bcl-2 expression for the two time points. D, E, and F: single color histogram showing the bimodal expression of Bcl-2 in A, uni-modal Bcl-2 expression in B, and the overlay in C.
Discussion
Deletion 13q14 is the most common cytogenetic abnormality in CLL. This deletion is associated with down regulation of miR-15a and miR-16-1 and a consequent increase in Bcl-2 expression [18, 33, and 34]. Based on this observation, we studied the association between Bcl-2 expression and deletion 13q14 in untreated CLL/MBL cases. In this study 54 CLL/MBL cases were studied: 33 cases had 13q14 deletions, and 21 cases were designated as non-13q14 deletion. Bcl-2 level was evaluated using a new gating strategy and expressed as the c-index (MFI of CLL cells/MFI of T-cells). The non-deletion 13q14 cases were used to define the basal level of Bcl-2 expression. Intra- and inter-clonal Bcl-2 heterogeneity was also evaluated. We found that a high Bcl-2 c-index (value above 1.6) strongly predicts the presence of 13q14 deletion, p<0.0001. Also a statistically significant correlation was observed between the percentage of cells carrying the 13q14 deletion and a high Bcl-2 c-index, p <0.05.
The prognostic indicators CD38, CD49d, CD26, and CD69 were also studied. There was no statistically significant correlation observed between the presence of 13q14 deletion and CD38, CD49d, CD69 and CD26 positive expressions. However, a strong statistically significant correlation was observed between the presence of a bimodal pattern of CD69 expression and the presence of 13q14 deletion p<0.0001.
Intra-clonal Bcl-2 heterogeneity was observed in two CLL cases (3.7%) and in one individual with MBL (1.85%). On the other hand, inter-clonal Bcl-2 bimodality was observed in five CLL cases (9.2%, data not shown). Also we observed an increase in the level of Bcl-2 expression over time in a single patient, and this was associated with clonal evolution.
Croce and colleagues cloned the bcl-2 gene from a 14;18 chromosome translocation in 1984 [35]. Its constitutive expression in follicular lymphoma is well known. Bcl-2 expression is normally thought to be on the nuclear membrane, mitochondrial membrane and endoplasmic reticulum membrane. Also it may be sequestered in the nucleus. There is one report of its surface expression by McCarthy [36]. Although most CLL cells express levels of Bcl-2 protein comparable to those seen in follicular lymphoma with t(14;18)(q32; q21.3), several studies have shown that some express lower levels [37, and 38]. The causes of this variation are unknown. In addition to the pro-survival signaling, dimer formation with Bax was often noted. In fact, Goolsby et al. showed flow cytometric evidence that the normal Bcl-2 apoptotic regulation pathway is intact [39].
Different mechanisms have been proposed to explain increased Bcl-2 expression in CLL cells, including promoter hypo-methylation [19], loss of microRNA (miRNA) expression (specifically, loss of miRNA-15a and miRNA-16-1 that are frequently deleted in CLL) [5]. Two major promoters control Bcl-2 transcription, P1 and P2. Also Majid et al. reported lack of an association between Bcl-2 level of expression and promotor single nucleotide polymorphisms [40]. The up-regulation of Bcl-2 is associated with down regulation of the mir 15-a/16-1 complex subsequent to 13q14 deletion. These two micro RNAs are contained in a minimally deleted region that also contains DLEU-7. Also, previous studies showed the cooperation of miR-15/16 and DLEU-7 in the pathogenesis of CLL. In this study, we observed that there is an increase in Bcl-2 level of expression using Bcl-2 c-index in the 13q14 deletion group compared to the non-deletion group p<0.0001. Furthermore, we observed that the level of Bcl-2 expression varies based on the percentage of cells harboring 13q14 deletion p<0.05 (Figure 2C). These results support the previous reported data that the level of Bcl-2 expression is inversely related to miR-15/16 deletion [16] and related to disease prognosis and progression [27, and 28].
Owing to the heterogeneity of Bcl-2 expression in CLL and the variation in 13q14 deletion status, we questioned whether bimodality in Bcl-2 expression equated to heterozygous or homozygous 13q14 deletion status. We also wondered if increases in Bcl-2 expression over time predict clonal evolution. To answer these questions, Bcl-2 bright and dim populations were sorted from bimodal samples. FISH on sorted Bcl-2 bright and dim populations was performed. Our data shows that the distinct bimodal Bcl-2 sub-populations differ in their 13q14 deletion status with either a mixture of hemizygous del 13q14 x1 and diploid CLL clonal cells or homozygous (del 13q14 x2) and hemizygous (del 13q14 x1).
Bright Bcl-2 sorted cells were enriched for the hemizygous 13q14 deletion cells. On the other hand, dim Bcl-2 sorted cells were enriched for the diploid clonal cells (one MBL, and one CLL case). Also there was enrichment of the Bcl-2 bright population with homozygous 13q14 deletion cells versus enrichment of the Bcl-2 dim CLL population with hemizygous deleted cells in one CLL case. It would seem that the level of Bcl-2 expression is relative with diploid cells having the least, heterozygous deletions with intermediate values and homozygous deletions with the highest values (personal observation).
We also observed that increases in Bcl-2 level and/or change in modality pattern over the time corresponded conversion from a mono-allelic deletion to a bi-allelic deletion in a CLL case (Figure 6). At diagnosis 96% hemizygous 13q14 deletion was noted. Ten years post-diagnosis a flow cytometric bi-modal Bcl-2 expression pattern was observed and a mixture of 53% of cells with homozygous 13q14 deletion and 40% with hemizygous 13q14 deletions was found by FISH. Eight months later, prior to treatment, Bcl-2 and FISH studies were repeated. Bcl-2 expression was uni-modal (greater than 90% bright Bcl-2 cells). Interphase FISH revealed that 96% of the nuclei scored cells at that time had homozygous 13q14 deletion. This single case suggests that monitoring Bcl-2 expression intensity can predict clonal evolution and it might be a useful biomarker to direct the repeat of FISH studies to evaluate further clonal evolution.
Bimodal distribution of a particular antigen within the CLL tumor cells suggests two discrete cell subsets based on the degree to which the antigen is expressed. Previously Ghia et al., [1] reported bimodal CD38 expression in CLL and showed that CD38 bimodal expression in CLL cases had similar survival outcome to the uni-modal CD38 positive cases. Also Cocco et al. reported bimodal expression of CD38, CD13, CD20, CD11c, CD5, FMC7, and surface immunoglobulin [41]. In our study, we had similar findings as we noticed seven cases with bimodal CD38 expression, five cases with bimodal CD20 expression, two cases with bimodal CD5 expression, and thirty-three cases with bimodal CD69 expression. Although our study did not show any significant association between the levels of expression of CD38, CD69, CD49d, and CD26, and 13q14 deletion, there was a significant association between CD69 bimodal expression and 13q14 deletion p<0.0001. Further studies to elucidate the significance of the bimodal CD69 activation pattern are needed.
In conclusion, 13q14 deletion is correlated with increased Bcl-2 expression. A high intensity of expression is related to the number of cells with the deletion versus the diploid cells and the ratio of hemizygous to homozygous deletions. These data combined with serial observation of 13q14 clonal evolution in a single patient suggests that further studies in this area would be of interest.
Acknowledgments
Research support: This project was supported in part by Heba Degheidy’s appointment to the Research Participation Program at CBER administered by the Oak Ridge Institute for Science and Education through US DOE and US FDA. And by the Intramural Research Program of the National, Heart, Lung and Blood Institute, National Cancer Institute, National Institutes of Health.
The mention of commercial products, their sources, or their use in connection with material reported here is not to be construed as either an actual or implied endorsement of such products by the Department of Health and Human Services.
References
- 1.Ghia P, Ferreri AM, Caligaris-Cappio F. Chronic lymphocytic leukemia. Crit Rev Oncol Hematol. 2007 Dec;64(3):234–46. doi: 10.1016/j.critrevonc.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Ouillette P, Erba H, Kujawski L, Kaminski M, Shedden K, Malek SN. Integrated genomic profiling of chronic lymphocytic leukemia identifies subtypes of deletion 13q14. Cancer Res. 2008;68(4):1012–1021. doi: 10.1158/0008-5472.CAN-07-3105. [DOI] [PubMed] [Google Scholar]
- 3.Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, Döhner K, Bentz M, Lichter P. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000 Dec 28;343(26):1910–6. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 4.Rowntree C, Duke V, Panayiotidis P, Kotsi P, Palmisano GL, Hoffbrand AV, Foroni L. Deletion analysis of chromosome 13q14.3 and characterisation of an alternative splice form of LEU1 in B cell chronic lymphocytic leukemia. Leukemia. 2002 Jul;16:1267–1275. doi: 10.1038/sj.leu.2402551. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sc USA. 2002 Nov 26;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michael MZ, O’Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced Accumulation of Specific MicroRNAs in Colorectal Neoplasia 1. Mol Cancer Res. 2003 Oct;1(12):882–891. [PubMed] [Google Scholar]
- 7.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004 Mar 2;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004 Feb;39(2):167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 10.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced Expression of the let-7 MicroRNAs in Human Lung Cancers in Association with Shortened Postoperative Survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 11.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 MicroRNA Family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 12.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs down regulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 15.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 16.Raveche ES, Salerno E, Scaglione BJ, Manohar V, Abbasi F, Lin YC, Fredrickson T, Landgraf P, Ramachandra S, Huppi K, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109:5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri MA, et al. MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 18.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102(39):13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanada M, Delia D, Aiello A, Stadtmauer E, Reed JC. Bcl-2 gene hypomethylation and high-level expression in B-cell chronic lymphocytic leukemia. Blood. 1993;82:1820–1828. [PubMed] [Google Scholar]
- 20.Hoffbrand AV, Panayiotidis P, Reittie J, Ganeshaguru K. Autocrine and paracrine growth loops in chronic lymphocytic leukemia. Semin Hematol. 1993;30:306–317. [PubMed] [Google Scholar]
- 21.McConkey DJ, Chandra J, Wright S, Plunkett W. Apoptosis sensitivity in chronic lymphocytic leukemia is determined by endogenous endonuclease content and relative expression of bcl-2 and bax. J Immunol. 1996;156:2624–2630. [PubMed] [Google Scholar]
- 22.Pepper C, Bentley P, Hoy T. Regulation of clinical chemoresistance by bcl-2 and bax oncoproteins in B-cell chronic lymphocytic leukaemia. Br J Haematol. 1996;95:513–517. doi: 10.1046/j.1365-2141.1996.d01-1927.x. [DOI] [PubMed] [Google Scholar]
- 23.Robertson LE, Plunkett W, McConnell K, Keating MJ, McDonnell TJ. Bcl-2 expression in chronic lymphocytic leukemia and its correlation with the induction of apoptosis and clinical outcome. Leukemia. 1996;10:456–459. [PubMed] [Google Scholar]
- 24.Thomas A, El Rouby S, Reed JC, Krajewski S, Silber R, Potmesil M, Newcomb EW. Drug-induced apoptosis in B-cell chronic lymphocytic leukemia: relationship between p53 gene mutation and bcl-2/bax proteins in drug resistance. Oncogene. 1996;12:1055–1062. [PubMed] [Google Scholar]
- 25.Gottardi D, Alfarano A, De Leo AM, Stacchini A, Aragno M, Rigo A, Veneri D, Zanotti R, Pizzolo G, Caligaris-Cappio F. In leukaemic CD5+ B cells the expression of BCL-2 gene family is shifted toward protection from apoptosis. Br J Haematol. 1996;94:612–618. doi: 10.1046/j.1365-2141.1996.d01-1856.x. [DOI] [PubMed] [Google Scholar]
- 26.Schena M, Larsson LG, Gottardi D, Gaidano G, Carlsson M, Nilsson K, Cappio FC. Growth- and differentiation-associated expression of bcl-2 in B-chronic lymphocytic leukemia cells. Blood. 1992;79:2981–2989. [PubMed] [Google Scholar]
- 27.Quijano S, López A, Rasillo A, Sayagués JM, Barrena S, Sánchez ML, Teodosio C, Giraldo P, Giralt M, Pérez MC, et al. Impact of Trisomy 12, del(13q), del(17p), and del(11q) on the Immunophenotype, DNA Ploidy Status, and Proliferative Rate of Leukemic B-Cells in Chronic Lymphocytic Leukemia. Cytometry B Clin Cytom. 2008;74B:139–149. doi: 10.1002/cyto.b.20390. [DOI] [PubMed] [Google Scholar]
- 28.Nückel H, Frey UH, Bau M, Sellmann L, Stanelle J, Dürig J, Jöckel KH, Dührsen U, Siffert W. Association of a novel regulatory polymorphism (−938C>A) in the BCL2 gene promoter with disease progression and survival in chronic lymphocytic leukemia. Blood. 2007;109 (1):290–297. doi: 10.1182/blood-2006-03-007567. [DOI] [PubMed] [Google Scholar]
- 29.Hallek M, Cheson BD, Catovsky D, Cappio FC, Dighiero G, Döhner H, Hillmen P, Keating MJ, Montserrat E, Rai KR, Kipps TJ. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–56. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenwald A, Chuang EY, Davis RE, Wiestner A, Alizadeh AA, Arthur DC, Mitchell JB, Marti GE, Fowler DH, Wilson WH, Staudt LM. Fludarabine treatment of patients with chronic lymphocytic leukemia induces a p53-dependent gene expression response. Blood. 2004;104:1428–1434. doi: 10.1182/blood-2003-09-3236. [DOI] [PubMed] [Google Scholar]
- 31.Degheidy H, Fouda M, Shahin D, Shamaa S, El-Bedewy A, Abd El-Ghaffar H. Diagnostic and Prognostic Utility of t(14;18) in Follicular Lymphoma. Acta Haematol. 2007;118:231–236. doi: 10.1159/000112474. [DOI] [PubMed] [Google Scholar]
- 32.Grzybowski M, Younger JG. Statistical methodology: III. Receiver operating characteristic (ROC) curves. Acad Emerg Med. 1997;4:818–826. doi: 10.1111/j.1553-2712.1997.tb03793.x. [DOI] [PubMed] [Google Scholar]
- 33.Mertens D, Philippen A, Ruppel M, Allegra D, Bhattacharya N, Tschuch C, Wolf S, Idler I, Zenz T, Stilgenbauer S. Chronic lymphocytic leukemia and 13q14: miRs and more. Leuk Lymphoma. 2009;50(3):502–505. doi: 10.1080/10428190902763509. [DOI] [PubMed] [Google Scholar]
- 34.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsujimoto Y, Finger LR, Yunis J, Nowell PC, Croce CM. Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. 1984 Nov 30;226(4678):1097–9. doi: 10.1126/science.6093263. [DOI] [PubMed] [Google Scholar]
- 36.McCarthy BA, Boyle E, Wang XP, Guzowski D, Paul S, Catera R, Trott J, Yan XJ, Croce CM, Damle R, Yancopoulos S, Messmer BT, Lesser M, Allen SL, Rai KR, Chiorazzi N. Surface Expression of Bcl-2 in Chronic LymphocyticLeukemia and Other B-Cell Leukemias and Lymphomas Without a Breakpoint t(14;18) Mol Med. 2008;14 (9 – 10):618– 627. doi: 10.2119/2008.00061.McCarthy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson LE, Plunkett W, McConnell K, Keating MJ, McDonnell TJ. Bcl-2 expression in chronic lymphocytic leukemia and its correlation with the induction of apoptosis and clinical outcome. Leukemia. 1996;10:456–459. [PubMed] [Google Scholar]
- 38.Kitada S, Andersen J, Akar S, Zapata JM, Takayama S, Krajewski S, Wang HG, Zhang X, Bullrich F, Croce CM, Rai K, Hines J, Reed JC. Expression of apoptosis-regulating proteins in chronic lymphocytic leukemia: correlations with in vitro and in vivo chemoresponses. Blood. 1998;91:3379–3389. [PubMed] [Google Scholar]
- 39.Goolsby C, Paniagua M, Tallman M, Gartenhaus RB. Bcl-2 regulatory pathway is functional in chronic lymphocytic leukemia. Cytometry B Clin Cytom. 2005 Jan;63B(1):36–46. doi: 10.1002/cyto.b.20034. [DOI] [PubMed] [Google Scholar]
- 40.Majid A, Tsoulakis O, Walewska R, Gesk S, Siebert R, Kennedy DBJ, Dyer MJSD. BCL2 expression in chronic lymphocytic leukemia: lack of association with the BCL2 938A>C promoter single nucleotide polymorphism. 2008;111:874–877. doi: 10.1182/blood-2007-07-098681. [DOI] [PubMed] [Google Scholar]
- 41.Cocco AE, Osei ES, Thut DM, Edinger AK, Powers JJ, Fu P, Meyerson HJ. Bimodal cell populations are common in chronic lymphocytic leukemia but do not impact overall survival. Am J Clin Pathol. 2005 Jun;123(6):818–825. doi: 10.1309/14XK-ERAY-LUL3-H2HT. [DOI] [PubMed] [Google Scholar]