Abstract
The cytoplasmic C-terminus of connexin43 (Cx43) interacts with numerous signaling complexes. We hypothesize that signal complex docking to the Cx43 C-terminus (CT) is required to propagate the molecules being shared by gap junctions. We have previously shown that Cx43 impacts the responsiveness of osteoblasts to FGF2 in a PKCδ- and ERK-dependent manner, converging on Runx2 activity. Here, we mapped the interaction domain of Cx43 and PKCδ to amino acids 243–302 of the Cx43 CT by GST pulldown assay. Using Runx2-responsive luciferase reporter assays, a Cx43 deletion construct (Cx43 S244Stop), which lacks the C-terminus (amino acids 244 to 382), failed to support the Cx43-dependent potentiation of transcription following FGF2 treatment in MC3T3 osteoblast-like cells. Similarly, overexpression of Cx43 S244Stop could not mimic the ability of the full length Cx43 to stimulate expression of osteoblast genes. In contrast to full length Cx43, overexpression of just the Cx43 CT (amino acids 236 to 382) inhibited both transcription from a Runx2 reporter and signaling via PKCδ and ERK. Inhibition of signaling by the CT did not occur in HeLa cells, which lack endogenous Cx43. In summary, the data support a model in which an intact Cx43 is required for both signal propagation/permeability (i.e., channel function) and local recruitment of signaling complexes to the CT (i.e., docking function) in order to mediate its cellular effects. Further, while the CT alone has channel independent activity, it is opposing to the effect of overexpression of the full length Cx43 channel.
Keywords: Gap Junction, Bone, Fibroblast Growth Factor 2, Protein Kinase C delta, Signal Transduction, Cell-to-Cell Communication
INTRODUCTION
Gap junctions are intercellular channels formed by hexamers of connexins (called a hemichannel or connexon) on one cell that dock with a hemichannel on an adjacent cell. The resultant gap junction forms an aqueous pore between the cytoplasm of two cells that permits the direct intercellular exchange of ions, small molecules and second messengers. In addition, gap junctions can function as unopposed hemichannels, serving as a direct conduit between the cytosol and extracellular fluid. Each connexin monomer contains four transmembrane domains, two extracellular loops, a cytoplasmic loop, and cytosolic N-terminus and C-terminus (Fig. 1A).
Fig. 1.
Binding of PKCδ to the C-terminus of Cx43. (A) Left, this model shows the structure of Cx43 with its four transmembrane domains, two extracellular loops, a cytoplasmic loop, and intracellular N-terminus and C-terminus. Right, the amino acid composition of the Cx43 CT (amino acids 227–382) is shown. Putative interaction domains for complex binding with PKCδ are indicated in bold. (B) Top, a schematic of the Cx43 CT GST fusion constructs used for the GST-pulldown assays is shown. The full length Cx43 is shown as a reference with amino acid number indicated above. The CX43 CT domain is indicated in black. The deleted domains are indicated by a white box containing an X. Bottom, GST-pulldown assay of the input and bead fractions of whole cell extracts from MC3T3 cells, following SDS-PAGE and western blotting with anti-PKCδ antibodies. “GST beads” refers to beads conjugated to GST alone, rather than a GST-coupled Cx43 CT construct.
In bone, osteoblasts and osteocytes are highly interconnected via gap junctions composed primarily of connexin43 (Cx43). In these cells, Cx43 has been shown to play an important role in transmitting hormone-, mechanical load- and growth factor-induced signals, ultimately affecting bone quality and geometry via both classic cell-to-cell communication through gap junctions or via hemichannel activity [Batra et al., 2012b; Civitelli, 2008]. Mouse models of Cx43 gene deletion (both globally and osteoblast-specific conditional knockout models) have underscored the fundamental importance of Cx43 in skeletal function, bone mass acquisition and bone quality [Bivi et al., 2011a; Chung et al., 2006; Lecanda et al., 2000; Watkins et al., 2011; Zhang et al., 2011]. Despite the importance of Cx43 in bone, the molecular mechanisms by which this gap junction protein exerts its influence on osteoblast and osteocyte function are only beginning to emerge.
Work from our group has shown that overexpression of Cx43 in MC3T3 osteoblasts can enhance the activity of the essential osteogenic transcription factor, Runx2, in response to the fibroblast growth factor-2 (FGF2) [Lima et al., 2009; Niger et al., 2012]. This is consistent with the work of others showing that overexpression of Cx43 can enhance osteogenic capacity and responsiveness to extracellular cues in vitro [Gramsch et al., 2001; Lecanda et al., 1998; Rossello et al., 2009] and in vivo [Rossello et al., 2009]. The ability of Cx43 to potentiate the osteoblast response to FGF2 requires the activity of PKCδ and ERK cascades [Lima et al., 2009; Niger et al., 2012]. Also, PKCδ is transiently recruited to the Cx43 channel, interacting with the C-terminus (CT) of Cx43, prior to nuclear translocation following FGF2 treatment [Niger et al., 2010a].
Among the ~21 connexin proteins, the CT-domain is the least conserved [Palatinus et al., 2012]. The Cx43 CT has been implicated in the regulation of Cx43 function and as a target of kinases and other signaling machinery [Herve et al., 2012]. The Cx43 CT binds numerous signaling complexes, such as β-catenin, src, and PKCs [Ai et al., 2000; Bowling et al., 2001; Doble et al., 2000; Loo et al., 1995; Toyofuku et al., 2001; Xu et al., 2001], and is a target for phosphorylation by numerous kinases, like ERK, PKA and PI3K [Cruciani and Mikalsen, 2002; Giepmans, 2004]. Intriguingly, it is possible that the CT is not only a target of phosphorylation but may also act as a docking platform for signaling complexes and that this docking function is required to propagate signals that have passed through the gap junction channel. In addition, a growing number of studies have described novel, channel-independent functions for the Cx43 CT [Kameritsch et al., 2012; Vinken et al., 2012].
In order to gain insight into how Cx43 regulates osteoblast function, the goal of this study was to determine the functional relevance of the Cx43 and PKCδ complex. Specifically, we asked if the Cx43 CT is a docking platform for signaling complexes, if the Cx43 CT is required for the ability of Cx43 to amplify signaling and Runx2 activity in response to FGF2 and whether the Cx43 CT could function independent of channel activity.
MATERIALS AND METHODS
Cell culture
MC3T3 clone 4 cells (ATCC, Manassas, VA) were cultured as previously described [Gupta et al., 2010]. HeLa cells (ATCC) were cultured in αMEM containing 10% fetal bovine serum (Hyclone, Logan, UT), penicillin (50IU/ml) and streptomycin (50μg/ml). Cells were used at passage < 15. For experiments involving FGF2 treatment, cells were serum starved in media containing 0.1% fetal bovine serum and 0.3% bovine serum albumin for 24 hours. FGF2 (Millipore, Billerica, MA) was added to the starvation media at 10ng/ml. The vehicle diluent for FGF2 (phosphate buffered saline containing 0.1% bovine serum albumin) was used as a negative control for FGF2. Cell viability was routinely monitored by a CCK-8 assay (Dojindo Molecular Technologies, Bethesda, MD) according to manufacturer's directions.
Plasmid constructs
The pSFFV-Cx43 construct, which contains the full-length rat Cx43 open reading frame cloned into the EcoR1 site of the pSFFV-neo plasmid [Beyer et al., 1987], was provided by Dr. Thomas Steinberg (Washington University, St Louis, MO). The pSFFV-neo empty vector [Fuhlbrigge et al., 1988] was provided by Dr. Gabriel Nunez (University of Michigan, Ann Arbor, MI). The Cx43 S244Stop construct contains the coding sequence for amino acids 1–243 of the rat Cx43 gene cloned into the EcoRI site of pSFFV-neo. The p6xOSE2 luciferase reporter construct, which contains 6 tandem copies of the Runx2-binding OSE2 element from the mouse osteocalcin promoter cloned upstream of a minimal promoter [Ducy and Karsenty, 1995], and the mutated p6xmutOSE2 luciferase plasmid, which contains non-functional Runx2-binding elements, were provided by Dr. Gerard Karsenty (Columbia University, New York, NY). The pCMV3XFLAGm43CT plasmid, which contains the sequence encoding amino acids 236–382 (C-terminus) of the murine Cx43 gene, was provided by Dr. Cecilia Lo via Addgene (Addgene plasmid 17664, Cambridge, MA). The bacterial expression plasmid (pGEX-3X-Cx43 CT) encoding the GST-fused rat Cx43 C-terminus (amino acids, 227–382) and the eight deletion mutants, described in Akiyama et al. [Akiyama et al., 2005], were provided by Dr. Takuya Ogawa (Nara Institute of Science and Technology, Nara, Japan).
GST-pulldown assay
The GST-pulldown assay was performed as described previously [Niger et al., 2010a]. Briefly, GST-tagged Cx43 CT constructs were expressed in E.coli prior to crosslinking to glutathione-sepharose beads (GE Healthcare, Piscataway, NJ). Immobilized beads were incubated with whole cell extracts from MC3T3 cells, washed and the bound material eluted. Subsequently, the eluted material was subjected to SDS-PAGE and examined by Western blotting with anti-PKCδ antibodies (Cell Signaling Technologies, Beverly, MA). This experiment was repeated a minimum of two times. Data from a representative experiments is shown.
Western blotting
Western blots were performed on whole cell extracts, as described previously [Niger et al., 2010a]. The anti-GAPDH antibodies were purchased from Millipore. The anti-phospho-ERK1/2, anti-ERK1/2, anti-phospho-PKCδ Thr 505, anti-total PKCδ and anti-FLAG M2 and antibodies were purchased from Cell Signaling Technologies (Beverly, MA). All experiments were repeated a minimum of three times. Data from a representative experiment is shown. Blots were quantified using the Image Lab (v4.1) software (Biorad, Hercules, CA).
Transient transfections and luciferase reporter assays
Transient transfections were carried out as described previously [Niger et al., 2012] with the exception that X-tremegene 9 (Roche, Indianapolis, IN) was used as the transfection reagent. In addition, transfection efficiency was monitored by co-transfection with a pRL-TK renilla luciferase reporter (Promega) and luciferase activity assessed using a dual luciferase assay reagent as reported by Dyer et al [Dyer et al., 2000]. Luciferase reporter assays were performed as described previously [Niger et al., 2011]. All experiments were repeated a minimum of three times in triplicate wells. Data from representative experiments are shown. Luciferase data shown on a single graph were from the same experiment so that all other variables are kept constant.
RNA isolation and Real Time PCR
Total RNA was extracted from cells in culture using Trizol reagent (Invitrogen) and used to prepare cDNA using RevertAid First Strand cDNA Synthesis kit (Fermentas, Glen Burnie, MD) according to the manufacturer's recommendations. The primer sets used for these reactions are: GAPDH, CGT GTT CCT ACC CCC AAT GT (For), TGT CAT CAT ACT TGG CAG GTT TCT (Rev); RPL13, CGA AAC AAG TCC ACG GAG TCA (For), GAG CTT GGA GCG GTA CTC CTT (Rev); HPRT, AGC AGT ACA GCC CCA AAA TGG (For), AAC AAA GTC TGG CCT GTA TCC AA (Rev); Cx43, GCT TCT GGG TCC TTC AGA TCA T (For), CAT AGA ACA CAT GGG CCA AGT ACA (Rev); Bglap, CAC AGA TGC CAC GCC CA (For), TGC CCT CCT GCT TGG ACA (Rev); Col1a1, CTT CAC CTA CAG CAC CCT TGT G (For), GAT GAC TGT CTT GCC CCA AGT T (Rev); Sp7/Osterix, CCC TAT GGC TCG TGG TAC AAG (For), CAT GTC CCA CCA AGG AGT AGG T (Rev); Runx2, CCG TGG CCT TCA AGG TTG T (For), CGG CCA TGA CGG TAA CCA (Rev); Tnfsf11/RANKL, ACC AGC ATC AAA ATC CCA AGT T (For), TCA GAA TTG CCC GAC CAG TT (Rev); Tnfrsf11b/osteoprotegerin GCG TGC AGC GGC ATC T (For), AGG CTC TCC ATC AAG GCA AGA (Rev). Gene expression was normalized to the expression of GAPDH, HPRT and RPL13 using geNorm v3.5 software (Ghent University Hospital Ghent, Belgium), as described previously [Niger et al., 2013]. Experiments were repeated a minimum of three times in triplicate wells. Data from representative experiments are shown.
Scrape loading dye coupling assay
Assessment of gap junctional coupling by scrape loading dye transfer assays were performed in confluent cultures of transiently transfected MC3T3 cells, as described previously [Niger et al., 2010b]. Experiments were repeated a minimum of two times. Data from a representative experiment is shown.
Immunofluorescence microscopy
Immunofluorescence labeling of MC3T3 cells cultured on glass coverslips was performed as described previously [Niger et al., 2010b]. The anti-Cx43 antibodies were purchased from Sigma (St. Louis, MO). Actin was labeled by incubation of the fixed cells with 1μM TRITC-labeled phalloidin for 30 minutes prior to mounting. Once collected images were processed (contrast, brightness, and merged) in Adobe Photoshop CS4 (Adobe Systems, Inc, San Jose, CA). Experiments were repeated three times. Data from a representative experiment is shown.
Statistical Analysis
Graphs depict means, and error bars indicate standard deviation. Samples were compared by an ANOVA for unpaired samples. Where a statistical difference was indicated (p-value <0.05), we performed a Dunnett's post-hoc test.
RESULTS
Previously, we have shown that PKCδ is in a complex with Cx43 by co-immunoprecipitation and GST-pulldown with the Cx43 CT [Niger et al., 2010a]. In order to refine the responsible interaction domain(s), we performed GST pulldown assays with a series of Cx43 CT constructs cloned in frame with an N-terminal GST tag. The CT (amino acids 227–382) was able to form a complex with PKCδ in whole cell extracts from MC3T3 osteoblast-like cells (Fig. 1). In contrast, the complex of PKCδ with Cx43 CT was nearly eliminated in Cx43 CT constructs lacking amino acids 243–262, 263–282, and 283–302. These data suggest that the complex of PKCδ and Cx43 CT requires a domain within amino acids 243–302 of Cx43. In addition, there was a reduction in the ability to pulldown PKCδ with a construct lacking the final 20 amino acids of the Cx43 CT (amino acids 363–382). Accordingly, we generated a truncated Cx43 construct (Cx43 S244Stop) that lacked most of the CT, to test its ability to function in osteoblastic cells.
Increasing Cx43 expression in MC3T3 osteoblasts can amplify ERK and PKCδ signal pathway activation and the transcriptional activity of Runx2, the master regulator of osteoblastogenesis, in response to FGF2 [Lima et al., 2009; Niger et al., 2012]. To test if the Cx43 CT is required for the enhancement of Runx2 activation, we examined the ability of a full-length Cx43 (amino acids 1–382) and the Cx43 S244Stop (amino acids 1–242) construct to activate transcription from a Runx2 luciferase reporter (p6xOSE2-Luc). Consistent with our previous work [Lima et al., 2009; Niger et al., 2012] and the work of others [Kim et al., 2003; Willis et al., 2002; Xiao et al., 2002], FGF2 stimulated Runx2 activity (Fig. 2A). Also as expected, overexpression of a full-length Cx43 (amino acids 1–382) enhanced the transcriptional activity of the Runx2 reporter in response to FGF2. Importantly, this was an effect that could not be recapitulated by the Cx43 S244Stop construct, suggesting that the CT domain is required to amplify FGF2-responses in osteoblasts. Qualitative real time PCR confirmed similar levels of Cx43 overexpression between the full-length and truncated constructs (Fig. 2B). Likewise, scrape loading dye transfer assays showed a similar increase in gap junctional intercellular communication between the full-length and truncated constructs relative to empty vector transfected MC3T3 cells (Fig 2C), indicating that the Cx43 S244Stop construct was as generally as effective as the full-length Cx43 construct at supporting gap junctional communication. Similar to the Runx2 transcriptional data, FGF2-stimulated phosphorylation of both PKCδ and ERK was enhanced to a greater degree when Cx43 was overexpressed than when Cx43 S244Stop was overexpressed (Fig 2D). In total, these data reveal that deletion of the Cx43 CT prevents the potentiation of Runx2 activity and signal transduction despite supporting gap junctional intercellular communication.
Fig. 2.
A C-terminus truncated Cx43 mutant prevents the potentiation of Runx2 despite supporting gap junctional intercellular communication. (A) Luciferase reporter assays were performed on MC3T3 cells co-transfected with a Runx2-reponsive luciferase reporter (p6xOSE2-Luc) and pSFFV-neo (empty vector), a full-length pSFFV-Cx43 (Cx43), or pSFFV-Cx43 S244Stop (Cx43 S244Stop) expression vector. Subsequently, the cells were treated with or without FGF2 (10ng/ml, 4h). Data are shown as means ± s.d. *, P<0.05, relative to the corresponding FGF2-treated empty vector control. (B) Quantitative real time RT-PCR of RNA isolated from transfected cells demonstrated a similar increase in Cx43 expression in cells transfected with pSFFV-Cx43 and pSFFV-Cx43 S244Stop vectors relative to the empty vector control. *, P<0.05, relative to the empty vector control. (C) Overexpression of Cx43 or Cx43 S244Stop increased the diffusion of the low molecular weight Lucifer yellow tracer between cells relative to the empty vector control, following scrape loading. (D) MC3T3 cells were transiently transfected with Cx43 or Cx43 S244Stop. Serum starved cells were stimulated with FGF2 for 5 minutes (ERK) or 15 minutes (PKCδ) prior to lysis. Whole cell extracts were subjected to western blotting with anti-phospho-ERK, anti-total ERK, anti-phospho-PKCδ Thr 505 or anti-total PKCδ antibodies. Because the ERK and PKCδ blots were from separate lysates, the blots probed with anti-GAPDH antibodies are shown below the blot for which they serve as the load control. Densitometric quantitation of the bands is shown to the right. Data from a representative experiment are shown.
In order to extend these findings into osteoblast function, we performed quantitative real time PCR on samples from MC3T3 osteoblasts that were transiently transfected with full length Cx43 or the Cx43 S244Stop construct. The osteoblast genes examined are inhibited by loss of Cx43 in murine models of Cx43 ablation [Chung et al., 2006; Lecanda et al., 2000; Watkins et al., 2011; Zhang et al., 2011]. Notably, overexpression of Cx43 enhanced the expression of Bglap (osteocalcin), Col1a1, Sp7 (Osterix), and Tnfrs11b (osteoprotegerin) (Fig. 3). Conversely, overexpression of the Cx43 S244Stop construct failed to amplify expression of these genes, supporting the notion that the Cx43 CT is needed for Cx43 to affect osteoblast function. The expression of Runx2 and Tnfs11 (RANKL) were unaffected by Cx43 or Cx43 S244Stop, in line with results from loss of Cx43 studies [Bivi et al., 2011a; Watkins et al., 2011].
Fig 3.
Overexpression of Cx43 but not Cx43 S244Stop stimulates the expression of osteoblast genes. (A) Quantitative real time PCR was performed on MC3T3 cells transfected with and pSFFV-neo (empty vector), a full-length pSFFV-Cx43 (Cx43), or pSFFV-Cx43 S244Stop (Cx43 S244Stop) expression vector. Relative gene expression from triplicate samples is shown for (A) Bglap, (B) Col1a1, (C) Runx2, Sp7/Osterix, (D) Tnfsf11/RANKL and (E) Tnfrsf11b/osteoprotegerin. Data are shown as means ± s.d. *, P<0.05, relative to the empty vector control. n.s., no significant difference relative to the empty vector control. Data from a representative experiment are shown.
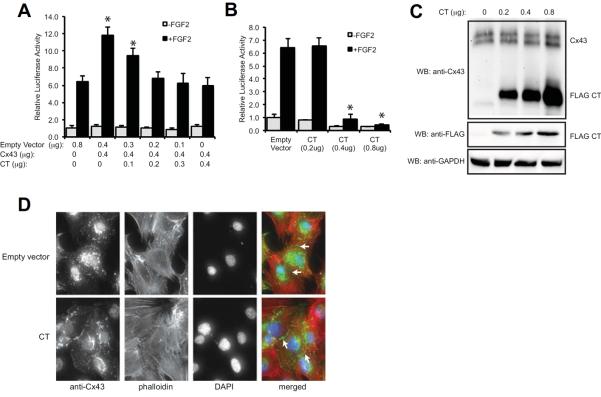
Next we examined if overexpression of just the Cx43 CT alone could mimic the effects of full-length Cx43 expression, which would indicate that the CT domain is necessary and sufficient to carry out the influence of Cx43 on Runx2, gene expression and signal transduction. Indeed, channel independent action of the Cx43 CT has been previously reported [Kameritsch et al., 2012; Vinken et al., 2012]. Using a luciferase reporter assay, overexpression of just the Cx43 CT (amino acids 236–382) dose-dependently inhibited the ability of full-length Cx43 overexpression to stimulate Runx2 activity in FGF2 treated MC3T3 cells (Fig. 4A). Likewise, overexpression of the Cx43 CT dose-dependently inhibited Runx2 activity in MC3T3 cells expressing only endogenous levels of Cx43 (Fig. 4B). Thus Cx43 CT overexpression produced precisely the opposite effect of overexpression of the full-length Cx43. FGF2, full-length Cx43 and Cx43 CT overexpression had no effect on a mutated p6xmutOSE2-Luc reporter (data not shown). Western blotting revealed a dose-dependent increase in the expression of the Cx43 CT in these cells with no effect on endogenous Cx43 expression (Fig. 4C).
Fig. 4.
The Cx43 CT antagonizes the ability of Cx43 to stimulate Runx2 activity in MC3T3 cells. (A) Luciferase reporter assays were performed on MC3T3 cells co-transfected with a Runx2-reponsive luciferase reporter (p6xOSE2-Luc) and pSFFV-neo (empty vector) or a full-length pSFFV-Cx43 (Cx43) and increasing concentrations of pCMV3XFLAGm43CT plasmid, which encodes amino acids 236–382 of the Cx43 CT, at the indicated concentrations. Subsequently the cells were treated with or without FGF2 (10ng/ml, 4h). Data are shown as means ± s.d. *, P<0.05, relative to the corresponding FGF2-treated empty vector control. (B) Luciferase reporter assays were performed on MC3T3 cells co-transfected with a Runx2-reponsive luciferase reporter (p6xOSE2-Luc) and increasing concentrations of pCMV3XFLAGm43CT plasmid without the transfection of exogenous full-length Cx43. Subsequently the cells were treated with or without FGF2 (10ng/ml, 4h). Data are shown as means ± s.d. *, P<0.05, relative to the corresponding FGF2-treated empty vector control. (C) Western blots from transfected MC3T3 cells transfected with increasing concentrations of pCMV3XFLAGm43CT plasmid. The blots were probed with anti-Cx43, anti-FLAG and anti-GAPDH (load control) antibodies. (D) MC3T3 cells were transiently transfected with 0.8μg pCMV3XFLAGm43CT plasmid followed by immunofluorescent labeling with anti-Cx43 antibodies (green). Phalloidin (red) and DAPI (blue) staining were also performed. Arrows show labeling at cell-to-cell junctions. A representative image is shown.
To ensure that the Cx43 CT did not alter the subcellular distribution of full-length Cx43, we examined Cx43 localization by immunofluorescence microscopy. In empty vector transfected MC3T3 cells, Cx43 is abundantly expressed in a perinuclear compartment, as well as in a defined punctate pattern at areas of cell-to-cell contact (Fig. 4D). There is no obvious change in the pattern of Cx43 expression when cells are transiently transfected with the Cx43 CT.
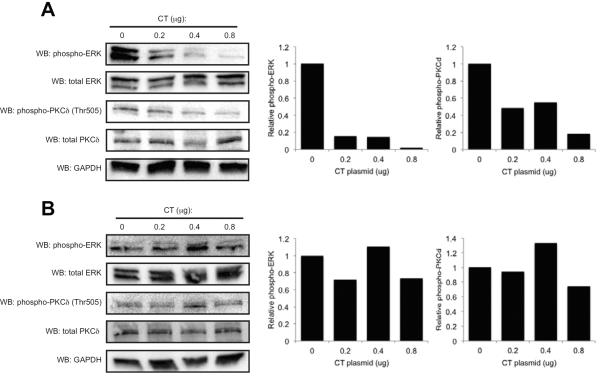
We then examined if overexpression of the Cx43 CT could affect signaling downstream of FGF2. We have previously demonstrated that overexpression of Cx43 can enhance ERK and PKCδ phosphorylation in response to FGF2 in these cells [Lima et al., 2009; Niger et al., 2012] and that the Cx43 S244Stop construct was less effective at stimulating activation of these pathways (Fig 2D). Consistent with the effects of the Cx43 CT on Runx2 transcriptional activity, western blotting revealed that the Cx43 CT dose-dependently inhibited phospho-ERK and phospho-PKCδ (Fig. 5A) levels in transfected MC3T3 cells. In contrast, the levels of phospho-ERK and phospho-PKCδ in HeLa cells, which lack endogenous gap junctions, were unaffected by overexpression of the CT (Fig. 5B).
Fig 5.
Overexpression of the Cx43 CT inhibits the activation of ERK and PKCδ in MC3T3, but not HeLa cells. MC3T3 cells (A) or HeLa cells (B) were transiently transfected with increasing concentrations of pCMV3XFLAGm43CT plasmid. The cells were lysed 72 hours post-transfection. Whole cell extracts were subjected to western blotting with anti-phospho-ERK, anti-total ERK, anti-phospho-PKCδ Thr 505 or anti-total PKCδ antibodies. Anti-GAPDH antibodies were used as load controls. Densitometric quantitation of the bands is shown to the right. Representative data are shown.
DISCUSSION
In addition to the ability of Cx43 to serve as conduit for the intercellular exchange of ions, metabolites and second messengers, a number of signal complexes bind to the C-terminus of Cx43 [Herve et al., 2012]. In this study, we showed in MC3T3 osteoblast-like cells that the intact Cx43 channel and CT domain of Cx43 are required for the ability of Cx43 to amplify the transcriptional activity of Runx2, increase signaling through ERK and PKCδ and stimulate osteoblast gene expression. Indeed, a CT-deficient Cx43 truncation could not recapitulate the effect the full length Cx43 on Runx2 activity, signal transduction or the activation of osteoblast genes. These finding are consistent with a recent study in osteoblasts in which the Cx43 CT domain is required for the anti-apoptotic action of parathyroid hormone [Bivi et al., 2011b]. Enhancement of osteoblast gene expression by overexpression of the full length channel has never been shown but is perhaps expected, as loss of Cx43 can impact osteoblast gene expression, including down regulation of Bglap, Col1a1, Sp7/Osterix, and Tnfrsf11b/Osteoprotegerin [Bivi et al., 2012; Chung et al., 2006; Lecanda et al., 1998; Lecanda et al., 2000; Li et al., 2006; Watkins et al., 2011; Zhang et al., 2011].
We also show that the Cx43 CT domain alone cannot substitute for the action of the full-length Cx43. Rather, we found the opposite, as CT overexpression in MC3T3 cells inhibited Runx2 transcriptional activity and attenuated the signaling cascades that are stimulated by full-length Cx43 overexpression [Lima et al., 2009; Niger et al., 2012]. These finding are consistent with the observation that a Cx43 CT construct has an antagonistic effect on Cx43 hemichannel opening in osteocyte-like cells exposed to fluid flow [Batra et al., 2012a]. These data are relevant because several studies have indicated gap junction/channel-independent effects of the Cx43 CT [Crespin et al., 2010; Dang et al., 2003; Dang et al., 2006; Moorby and Patel, 2001]. Unlike these studies, our data revealed that the Cx43 full-length channel and Cx43 CT had opposing effects.
Overexpression of the Cx43 CT did not alter the expression or cellular distribution of endogenous Cx43, and deletion of the Cx43 CT (Cx43 S244Stop) did not affect cell-to-cell communication as assessed by scrape loading. However, it is worth mentioning that we do not rule out that finer level details of cell-to-cell permeability are impacted by the loss of the Cx43 CT. Some Cx43 CT deletions, while functional, reduce gap junction permeability [Behrens et al., 2010]. However, our findings are consistent with a study using a similar CT-truncated Cx43 construct (lacking amino acids 242–382), which forms functional gap junctions albeit with longer plaque size at the plasma membrane [Wayakanon et al., 2012]. Previously, we showed that cell-to-cell communication is required for the Cx43-dependent potentiation of Runx2 signaling in osteoblasts [Niger et al., 2012]. The current data indicate that, even if the communication is present, the entire Cx43 protein must be expressed for the Cx43-dependent effects on osteoblasts, and that the Cx43 CT alone is necessary (in the context of the intact Cx43 channel) but insufficient to enhance Runx2 activity and ERK and PKCδ signaling. These data suggest that the CT domain, when present in the context of an intact Cx43 protein, serves one purpose, while free CT has an opposing, channel-independent function. However, the failure of overexpressed Cx43 CT to inhibit ERK and PKCδ signaling in cells that lack endogenous Cx43 expression suggests that the mode of action of the overexpressed Cx43 CT may be by antagonizing the function of the full-length Cx43. For example, the inhibitory effect of Cx43 CT overexpression on signaling may involve competition for factors that are recruited to the intact Cx43 channel, thereby preventing efficient downstream signaling.
We hypothesize a model in which Cx43 serves not only as a conduit for the exchange of second messengers and other small molecules, but also serves as a docking platform for signaling complexes. As such, signal complex docking with Cx43 and intercellular communication of second messengers are intertwined. In this model, it is not only the charge and size selective permeability of a gap junction that determines its biologic activity, but also the unique profile of signal molecules that are recruited to the gap junction channel. In fact, such a model may help to explain the vast biodiversity of connexins. These connexins often have overlapping permeability. The least conserved domain of connexins tends to be the C-terminus, which is largely where the signaling complexes interact. Thus, the connexin, and specifically the C-terminus, may specifically recruit signaling complexes to the gap junction plaque, forming a signaling microdomain in the cell that can appropriately respond to the communicated signals and second messengers that are propagated by the gap junction channel. Further each connexin (e.g., Cx43 versus Cx37) may recruit its own unique subset of signaling complexes, lending unique functionality to each connexin. In the three dimensional context of the cell, the local recruitment of signaling complexes to the area of the cell-to-cell exchange of signal molecules seems the most efficient way to sense and respond to shared molecules between gap junction coupled cells.
The data presented here are consistent with this model. The failure of CT deficient Cx43 channel to support osteoblast signaling and gene expression, and the opposing actions of full-length Cx43 and the Cx43 CT on osteoblast signaling and transcription suggests that the channel function and docking function of Cx43 are both required for Cx43 to enhance osteoblast function.
It is likely that these effects are not unique to bone cells, as numerous studies in many cell systems have shown that altering gap junctional communication or alteration of the Cx43 CT influences signal transduction, gene expression, cell migration and survival. Likewise, the convergence of Cx43 on Runx2, PKCδ and ERK is probably not the only mechanism by which Cx43 influences bone cell function. Indeed, we and others have shown that multiple signaling pathways downstream of Cx43 influence diverse osteoblast functions including gene expression and apoptosis [Batra et al., 2012a; Bivi et al., 2011b; Niger et al., 2011; Plotkin et al., 2005; Taylor et al., 2007].
In conclusion, these data demonstrate the requirement for the full-length, intact Cx43 protein to amplify the response of osteoblasts to FGF2 treatment. Further, the antagonistic action of the Cx43 CT to this response in cells expressing endogenous Cx43 indicates that the Cx43 CT is critically important for downstream signal pathway activation and the regulation of cellular responses. In total, these data provide insight into how Cx43 regulate bone cell function, which is important not only to treating genetic disease of Cx43 function, such as oculodenotdigital dysplasia, but also to finding effective ways to regulate bone formation.
ACKNOWLEDGEMENTS
This study was supported by grants, AR052719 and AR063631, from the National Institutes of Health/National Institute for Arthritis, Musculoskeletal and Skin Diseases.
Footnotes
DISCLOSURES No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS CH performed the experiments, collected data, assisted in data interpretation and writing of the manuscript. JPS conceived the experiments, assisted in data interpretation and writing of the manuscript.
REFERENCES
- Ai Z, Fischer A, Spray DC, Brown AM, Fishman GI. Wnt-1 regulation of connexin43 in cardiac myocytes. J Clin Invest. 2000;105:161–71. doi: 10.1172/JCI7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama M, Ishida N, Ogawa T, Yogo K, Takeya T. Molecular cloning and functional analysis of a novel Cx43 partner protein CIP150. Biochem Biophys Res Commun. 2005;335:1264–71. doi: 10.1016/j.bbrc.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Batra N, Burra S, Siller-Jackson AJ, Gu S, Xia X, Weber GF, DeSimone D, Bonewald LF, Lafer EM, Sprague E, Schwartz MA, Jiang JX. Mechanical stress-activated integrin alpha5beta1 induces opening of connexin 43 hemichannels. Proc Natl Acad Sci U S A. 2012a;109:3359–64. doi: 10.1073/pnas.1115967109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra N, Kar R, Jiang JX. Gap junctions and hemichannels in signal transmission, function and development of bone. Biochim Biophys Acta. 2012b;1818:1909–18. doi: 10.1016/j.bbamem.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J, Kameritsch P, Wallner S, Pohl U, Pogoda K. The carboxyl tail of Cx43 augments p38 mediated cell migration in a gap junction-independent manner. Eur J Cell Biol. 2010;89:828–38. doi: 10.1016/j.ejcb.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Beyer EC, Paul DL, Goodenough DA. Connexin43: a protein from rat heart homologous to a gap junction protein from liver. J Cell Biol. 1987;105:2621–9. doi: 10.1083/jcb.105.6.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivi N, Condon KW, Allen MR, Farlow N, Passeri G, Brun LR, Rhee Y, Bellido T, Plotkin LI. Cell autonomous requirement of connexin 43 for osteocyte survival: consequences for endocortical resorption and periosteal bone formation. J Bone Miner Res. 2011a doi: 10.1002/jbmr.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivi N, Lezcano V, Romanello M, Bellido T, Plotkin LI. Connexin43 interacts with betaarrestin: A pre-requisite for osteoblast survival induced by parathyroid hormone. J Cell Biochem. 2011b;112:2920–30. doi: 10.1002/jcb.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivi N, Nelson MT, Faillace ME, Li J, Miller LM, Plotkin LI. Deletion of Cx43 from osteocytes results in defective bone material properties but does not decrease extrinsic strength in cortical bone. Calcif Tissue Int. 2012;91:215–24. doi: 10.1007/s00223-012-9628-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling N, Huang X, Sandusky GE, Fouts RL, Mintze K, Esterman M, Allen PD, Maddi R, McCall E, Vlahos CJ. Protein kinase C-alpha and -epsilon modulate connexin-43 phosphorylation in human heart. J Mol Cell Cardiol. 2001;33:789–98. doi: 10.1006/jmcc.2000.1349. [DOI] [PubMed] [Google Scholar]
- Chung DJ, Castro CH, Watkins M, Stains JP, Chung MY, Szejnfeld VL, Willecke K, Theis M, Civitelli R. Low peak bone mass and attenuated anabolic response to parathyroid hormone in mice with an osteoblast-specific deletion of connexin43. J Cell Sci. 2006;119:4187–98. doi: 10.1242/jcs.03162. [DOI] [PubMed] [Google Scholar]
- Civitelli R. Cell-cell communication in the osteoblast/osteocyte lineage. Arch Biochem Biophys. 2008;473:188–92. doi: 10.1016/j.abb.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespin S, Bechberger J, Mesnil M, Naus CC, Sin WC. The carboxy-terminal tail of connexin43 gap junction protein is sufficient to mediate cytoskeleton changes in human glioma cells. J Cell Biochem. 2010;110:589–97. doi: 10.1002/jcb.22554. [DOI] [PubMed] [Google Scholar]
- Cruciani V, Mikalsen SO. Connexins, gap junctional intercellular communication and kinases. Biol Cell. 2002;94:433–43. doi: 10.1016/s0248-4900(02)00014-x. [DOI] [PubMed] [Google Scholar]
- Dang X, Doble BW, Kardami E. The carboxy-tail of connexin-43 localizes to the nucleus and inhibits cell growth. Mol Cell Biochem. 2003;242:35–8. [PubMed] [Google Scholar]
- Dang X, Jeyaraman M, Kardami E. Regulation of connexin-43-mediated growth inhibition by a phosphorylatable amino-acid is independent of gap junction-forming ability. Mol Cell Biochem. 2006;289:201–7. doi: 10.1007/s11010-006-9162-2. [DOI] [PubMed] [Google Scholar]
- Doble BW, Ping P, Kardami E. The epsilon subtype of protein kinase C is required for cardiomyocyte connexin-43 phosphorylation. Circ Res. 2000;86:293–301. doi: 10.1161/01.res.86.3.293. [DOI] [PubMed] [Google Scholar]
- Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol. 1995;15:1858–69. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer BW, Ferrer FA, Klinedinst DK, Rodriguez R. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal Biochem. 2000;282:158–61. doi: 10.1006/abio.2000.4605. [DOI] [PubMed] [Google Scholar]
- Fuhlbrigge RC, Fine SM, Unanue ER, Chaplin DD. Expression of membrane interleukin 1 by fibroblasts transfected with murine pro-interleukin 1 alpha cDNA. Proc Natl Acad Sci U S A. 1988;85:5649–53. doi: 10.1073/pnas.85.15.5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giepmans BN. Gap junctions and connexin-interacting proteins. Cardiovasc Res. 2004;62:233–45. doi: 10.1016/j.cardiores.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Gramsch B, Gabriel HD, Wiemann M, Grummer R, Winterhager E, Bingmann D, Schirrmacher K. Enhancement of connexin 43 expression increases proliferation and differentiation of an osteoblast-like cell line. Exp Cell Res. 2001;264:397–407. doi: 10.1006/excr.2000.5145. [DOI] [PubMed] [Google Scholar]
- Gupta RR, Yoo DJ, Hebert C, Niger C, Stains JP. Induction of an osteocyte-like phenotype by fibroblast growth factor-2. Biochem Biophys Res Commun. 2010;402:258–64. doi: 10.1016/j.bbrc.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herve JC, Derangeon M, Sarrouilhe D, Giepmans BN, Bourmeyster N. Gap junctional channels are parts of multiprotein complexes. Biochim Biophys Acta. 2012;1818:1844–65. doi: 10.1016/j.bbamem.2011.12.009. [DOI] [PubMed] [Google Scholar]
- Kameritsch P, Pogoda K, Pohl U. Channel-independent influence of connexin 43 on cell migration. Biochim Biophys Acta. 2012;1818:1993–2001. doi: 10.1016/j.bbamem.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim JH, Bae SC, Choi JY, Kim HJ, Ryoo HM. The protein kinase C pathway plays a central role in the fibroblast growth factor-stimulated expression and transactivation activity of Runx2. J Biol Chem. 2003;278:319–26. doi: 10.1074/jbc.M203750200. [DOI] [PubMed] [Google Scholar]
- Lecanda F, Towler DA, Ziambaras K, Cheng SL, Koval M, Steinberg TH, Civitelli R. Gap junctional communication modulates gene expression in osteoblastic cells. Mol Biol Cell. 1998;9:2249–58. doi: 10.1091/mbc.9.8.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecanda F, Warlow PM, Sheikh S, Furlan F, Steinberg TH, Civitelli R. Connexin43 deficiency causes delayed ossification, craniofacial abnormalities, and osteoblast dysfunction. J Cell Biol. 2000;151:931–44. doi: 10.1083/jcb.151.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhou Z, Saunders MM, Donahue HJ. Modulation of connexin43 alters expression of osteoblastic differentiation markers. Am J Physiol Cell Physiol. 2006;290:C1248–55. doi: 10.1152/ajpcell.00428.2005. [DOI] [PubMed] [Google Scholar]
- Lima F, Niger C, Hebert C, Stains JP. Connexin43 potentiates osteoblast responsiveness to fibroblast growth factor 2 via a protein kinase C-delta/Runx2-dependent mechanism. Mol Biol Cell. 2009;20:2697–708. doi: 10.1091/mbc.E08-10-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo LW, Berestecky JM, Kanemitsu MY, Lau AF. pp60src-mediated phosphorylation of connexin 43, a gap junction protein. J Biol Chem. 1995;270:12751–61. doi: 10.1074/jbc.270.21.12751. [DOI] [PubMed] [Google Scholar]
- Moorby C, Patel M. Dual functions for connexins: Cx43 regulates growth independently of gap junction formation. Exp Cell Res. 2001;271:238–48. doi: 10.1006/excr.2001.5357. [DOI] [PubMed] [Google Scholar]
- Niger C, Buo AM, Hebert C, Duggan BT, Williams MS, Stains JP. ERK acts in parallel to PKCdelta to mediate the connexin43-dependent potentiation of Runx2 activity by FGF2 in MC3T3 osteoblasts. Am J Physiol Cell Physiol. 2012;302:C1035–44. doi: 10.1152/ajpcell.00262.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niger C, Hebert C, Stains JP. Interaction of connexin43 and protein kinase C-delta during FGF2 signaling. BMC Biochem. 2010a;11:14. doi: 10.1186/1471-2091-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niger C, Howell FD, Stains JP. Interleukin-1beta increases gap junctional communication among synovial fibroblasts via the extracellular-signal-regulated kinase pathway. Biol Cell. 2010b;102:37–49. doi: 10.1042/BC20090056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niger C, Lima F, Yoo DJ, Gupta RR, Buo AM, Hebert C, Stains JP. The transcriptional activity of osterix requires the recruitment of Sp1 to the osteocalcin proximal promoter. Bone. 2011;49:683–92. doi: 10.1016/j.bone.2011.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niger C, Luciotti MA, Buo AM, Hebert C, Ma V, Stains JP. The regulation of Runx2 by FGF2 and connexin43 requires the inositol polyphosphate/protein kinase Cdelta cascade. J Bone Miner Res. 2013 doi: 10.1002/jbmr.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatinus JA, Rhett JM, Gourdie RG. The connexin43 carboxyl terminus and cardiac gap junction organization. Biochim Biophys Acta. 2012;1818:1831–43. doi: 10.1016/j.bbamem.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin LI, Aguirre JI, Kousteni S, Manolagas SC, Bellido T. Bisphosphonates and estrogens inhibit osteocyte apoptosis via distinct molecular mechanisms downstream of extracellular signal-regulated kinase activation. J Biol Chem. 2005;280:7317–25. doi: 10.1074/jbc.M412817200. [DOI] [PubMed] [Google Scholar]
- Rossello RA, Wang Z, Kizana E, Krebsbach PH, Kohn DH. Connexin 43 as a signaling platform for increasing the volume and spatial distribution of regenerated tissue. Proc Natl Acad Sci U S A. 2009;106:13219–24. doi: 10.1073/pnas.0902622106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AF, Saunders MM, Shingle DL, Cimbala JM, Zhou Z, Donahue HJ. Mechanically stimulated osteocytes regulate osteoblastic activity via gap junctions. Am J Physiol Cell Physiol. 2007;292:C545–52. doi: 10.1152/ajpcell.00611.2005. [DOI] [PubMed] [Google Scholar]
- Toyofuku T, Akamatsu Y, Zhang H, Kuzuya T, Tada M, Hori M. c-Src regulates the interaction between connexin-43 and ZO-1 in cardiac myocytes. J Biol Chem. 2001;276:1780–8. doi: 10.1074/jbc.M005826200. [DOI] [PubMed] [Google Scholar]
- Vinken M, Decrock E, Leybaert L, Bultynck G, Himpens B, Vanhaecke T, Rogiers V. Non-channel functions of connexins in cell growth and cell death. Biochim Biophys Acta. 2012;1818:2002–8. doi: 10.1016/j.bbamem.2011.06.011. [DOI] [PubMed] [Google Scholar]
- Watkins M, Grimston SK, Norris JY, Guillotin B, Shaw A, Beniash E, Civitelli R. Osteoblast connexin43 modulates skeletal architecture by regulating both arms of bone remodeling. Mol Biol Cell. 2011;22:1240–51. doi: 10.1091/mbc.E10-07-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayakanon P, Bhattacharjee R, Nakahama K, Morita I. The role of the Cx43 C-terminus in GJ plaque formation and internalization. Biochem Biophys Res Commun. 2012;420:456–61. doi: 10.1016/j.bbrc.2012.03.018. [DOI] [PubMed] [Google Scholar]
- Willis DM, Loewy AP, Charlton-Kachigian N, Shao JS, Ornitz DM, Towler DA. Regulation of osteocalcin gene expression by a novel Ku antigen transcription factor complex. J Biol Chem. 2002;277:37280–91. doi: 10.1074/jbc.M206482200. [DOI] [PubMed] [Google Scholar]
- Xiao G, Jiang D, Gopalakrishnan R, Franceschi RT. Fibroblast growth factor 2 induction of the osteocalcin gene requires MAPK activity and phosphorylation of the osteoblast transcription factor, Cbfa1/Runx2. J Biol Chem. 2002;277:36181–7. doi: 10.1074/jbc.M206057200. [DOI] [PubMed] [Google Scholar]
- Xu X, Li WE, Huang GY, Meyer R, Chen T, Luo Y, Thomas MP, Radice GL, Lo CW. Modulation of mouse neural crest cell motility by N-cadherin and connexin 43 gap junctions. J Cell Biol. 2001;154:217–30. doi: 10.1083/jcb.200105047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Paul EM, Sathyendra V, Davison A, Sharkey N, Bronson S, Srinivasan S, Gross TS, Donahue HJ. Enhanced osteoclastic resorption and responsiveness to mechanical load in gap junction deficient bone. PLoS One. 2011;6:e23516. doi: 10.1371/journal.pone.0023516. [DOI] [PMC free article] [PubMed] [Google Scholar]