Abstract
The Brilliant Cresyl Blue (BCB) test relies on G6PDH activity and a simple protocol for the selection of higher quality oocytes. Although the BCB+ oocytes of all the species that have been investigated are characterized by superior quality when compared to BCB- counterparts, application of the test for embryo production still remains an open issue. The aim of our study was to compare BCB+ and the control oocytes (not subjected to the BCB test) in terms of selected aspects of cytoplasmic maturation (mtDNA copy number, mitochondria distribution, relative transcript abundance of six marker genes). The results of our study revealed more relevant differences within the BCB+ and the control oocytes (before and after IVM) than between the two categories of oocytes. There was no difference in the transcript abundance of the BCB+ and the control oocytes in 5 out of 6 analyzed genes (BMP15, GDF9, ATP5A1, EEF1A, ZAR1) and in mtDNA content (pre-IVM 179609 vs. 176595 and post-IVM 187243 vs. 246984, respectively). With regard to mitochondria distribution in pre- and post-IVM oocytes, there was nonsignificant tendency for a more frequent occurrence of the expected patterns in the BCB+ group. The results of the present study do not support the application of BCB staining in a routine IVM protocol due to relatively high similarity in selected parameters characterizing cytoplasmic maturation of BCB+ and control oocytes. This high similarity may results from the limited amount of less competent BCB- oocytes (10%) still present among nonselected oocytes of proper morphology.
Keywords: Cytoplasmic maturation, Marker genes, mtDNA, Mitochondria, Oocyte quality
In the 1990s, Ericsson et al. [1] suggested a simple test for the selection of porcine oocytes having increased developmental competence. The test relies on measurement of the glucose-6-phosphate (G6PDH) enzyme activity. It was shown that this enzyme’s activity decreases, and yields the lowest oocyte level having a completed growth phase. The test involves staining immature cumulus-oocyte complexes (COCs) with Brilliant Cresyl Blue dye (BCB). The G6PDH enzyme converts the dye into a colorless form. Oocytes stained blue (BCB+, low G6PDH activity) are characterized by higher developmental competence (quality) when compared with colorless oocytes of reduced quality (BCB-, high activity of G6PDH). According to Wongsrikeao et al. [2], to select oocytes of superior quality, the BCB test should be applied only once, before IVM, due to the toxic effects of double exposure (before and after IVM). Up till now, the BCB test appeared to be a very useful tool for the selection of higher quality oocytes in, goats, cattle, mice and buffalo [3,4,5,6,7,8,9]. These studies carried out in vitro demonstrated that BCB+ oocytes are of superior quality compared with BCB- oocytes in terms of nuclear maturation, cleavage rate and blastocyst yield. The higher quality of the BCB+ oocytes is mainly due to better synchronized cytoplasmic maturation reflected by a significantly higher mtDNA copy number and transcript abundance (RA) of marker genes [9,10,11]. Moreover, BCB- bovine oocytes have been characterized by significantly lower RA of genes involved in replication and transcription of mitochondrial DNA. Similar observations have been made in pig oocytes. The nuclear genome-encoded proteins that regulate transcription and replication of mtDNA underwent significantly lower expression in BCB- oocytes [10, 12]. Moreover, Castaneda et al. [13] demonstrated higher lipid content in BCB+ oocytes and correlated this trait with higher quality.
Without a doubt, the BCB+ oocytes of all the investigated species were of superior quality compared with BCB- counterparts. Application of the BCB test to routine embryo production, however, still remains an open issue. According to published data, the BCB test is mainly used as a tool to distinguish two distinct categories of oocytes as experimental objects. However, in the majority of labs, the IVF protocols are based on COCs of proper morphology. These COCs are referred to in the present manuscript as the control group (not subjected to BCB staining). The comparison of these groups showed no significant differences in nuclear maturation efficiency, which is probably due to the limited proportion of BCB- oocytes, which varies from 10 to 30% in pigs and cattle [14]. Depending on the study and species, some differences have been observed in the blastocyst yield between BCB+ and control oocytes. With regard to pigs and horses, no significant differences have been noticed in the developmental potential of embryos derived from BCB+ and control oocytes [2, 3, 15]. On the other hand, a significantly higher blastocyst rate was reported for BCB+ oocytes of mice, goats and sheep [7, 16, 17]. Experiments done on bovine oocytes provided ambiguous results, as significant differences were observed in four out of six published experiments (Table 1) [4, 6, 9, 18,19,20]. It is worth investigating whether the slight differences between the control and BCB+ oocytes reflect the amount of BCB- oocytes in the control group or better synchronized cytoplasmic maturation of the BCB+ counterparts. It should also be noted that BCB staining may cause some negative effects. Pawlak et al. [21] observed a higher rate of chromosomal aberrations in porcine oocytes after BCB staining. A higher susceptibility to apoptosis of BCB-stained bovine oocytes as revealed by the relative transcript abundance of BCL-2 and BAX genes was noticed by Opiela et al. [9, 22]. Wongsrikeao et al. [2] reported that BCB staining after IVM had a negative impact on the cleavage and development of porcine embryos. However, there are no molecular studies demonstrating a negative impact of the BCB test on oocytes. The studies showing some negative effects point at increased sensitivity of a population of oocytes to BCB incubation rather than the toxicity of the BCB test per se.
Table 1. Comparison of BCB+ and control oocytes in selected animal species with respect to blastocyst rate .
| Species | Donor/treatment | CONTROL | BCB+ | Reference | ||
| % of blastocyst per | % of blastocyst per | |||||
| cleaved | oocytes | cleaved | oocytes | |||
| Cattle | Cow/heifer/IVF | 19.1 | 13.2 | 19.3 | 14.2 | [9] |
| Cattle | Cows/IVF | 25.0 | 16.3 | 28.7 | 18.5 | [20] |
| Cattle | Heifers/IVF | 5.2 | 3.4 | 12.3 | 9.2 | [18] |
| Cattle | Cows/IVF | 18.3 | 12.3 | 34.1 | 24.8 | [6] |
| Cattle | –/SCNT | 21.0 | 15.5 | 39.0 | 30.7 | [4] |
| Cattle | Cows/IVF | 25.7 | – | 35.7 | – | [19] |
| Pig | –/IVF ICSI | 20.6 | – | 15.1 | – | [3] |
| Pig | Prepubertal/IVF | 19.7 | 11.3 | 13.0 | 11.9 | [2] |
| Sheep | –/IVF | 30.8 | 20.8 | 48.1 | 34.5 | [17] |
| Horse | –/IVF ICSI | 13.6 | 7.0 | 20.0 | 9.2 | [15] |
| Goat | –/IVF | 0 | 0 | 7.9 | 3.6 | [7] |
The aim of our study was to compare BCB+ (oocytes subjected to BCB staining) and control oocytes (gametes of proper morphology not subjected to the BCB test) in terms of selected aspects of cytoplasmic maturation. We also aimed to demonstrate the relevance of BCB staining of porcine oocytes in a routine IVM procedure. We analyzed several markers of oocyte quality, to characterize maturing oocytes at the molecular level. These included mtDNA copy number, mitochondria distribution, and RA of six genes related to oocyte quality.
Materials and Methods
Unless stated otherwise, all chemicals and reagents used in this study were purchased from Sigma-Aldrich (Germany). The ovaries utilized in our experiment were collected post mortem from slaughtered gilts.
Recovery of cumulus-oocyte complexes (COCs)
COCs were collected from the ovaries of commercially slaughtered crossbred gilts when the gilts were 5–6 months old and weighed approximately 100–110 kg. The ovaries were excised and placed in a thermo insulated flask. Ovaries were transported to the laboratory within 2 h after the animal had been slaughtered. COCs were aspirated from 2–6 mm follicles with a syringe, placed in Hepes-TALP medium, and morphologically evaluated. Only COCs with evenly granulated cytoplasm and at least three layers of compact cumulus cells were selected for the experiment. COCs were divided in two groups – BCB and control (not subjected to BCB staining). The BCB group included oocytes incubated in BCB medium (PBS supplemented with 4 mg/ml BSA and 13 μM of BCB dye) for 90 min at 39 C in a humidified 5% CO2 atmosphere. Following exposure to BCB, COCs were transferred to PBS supplemented with 0.4% BSA and washed twice. COCs with the ooplasm stained blue were classified as more competent (BCB+), whereas COCs with a colorless ooplasm were classified as less competent (BCB-, Fig. 1). BCB- oocytes were excluded from further analysis. The control group included oocytes that had not been incubated in BCB dye and directly analyzed of matured in vitro. Both groups (BCB and control) were subjected to analysis before and after in vitro maturation. Consequently, four groups of porcine COCs were distinguished: 1) C (control pre-IVM), 2) C IVM (control post-IVM), 3) BCB (BCB-stained oocytes pre-IVM) and 4) BCB IVM (BCB+ oocytes post-IVM).
Fig. 1.
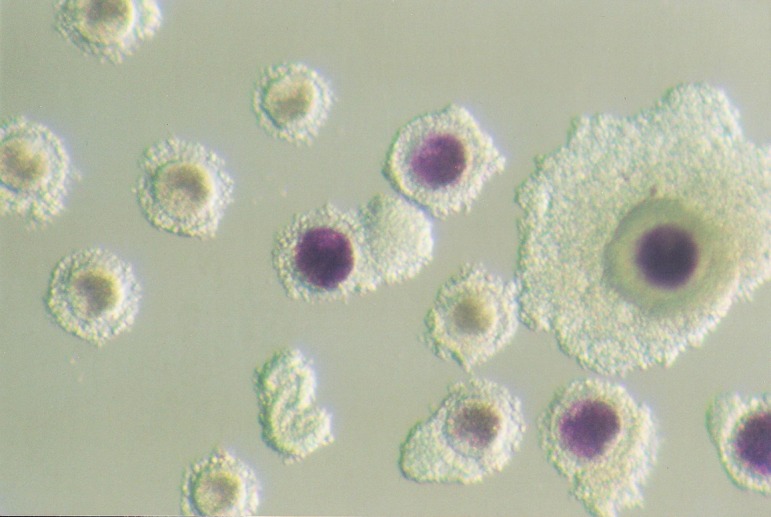
Porcine oocytes after BCB staining. Blue ooplasm characterizes more competent, BCB+ oocytes, whereas colorless ooplasm indicates BCB– oocytes with reduced developmental competence.
In vitro maturation (IVM)
In vitro maturation was carried out in basic NCSU23 medium supplemented with 10% (v/v) porcine follicular fluid, 10 U/ml hCG (Chorulon, Intervet), 10 U/ml eCG (Folligon, Intervet), 0.1 mg/ml cysteine, and 50 μg/μl gentamicin sulfate as previously described by Pawlak et al. [21]. The maturation protocol involved two steps: 1) an initial 20 h culture in hormone-supplemented NCSU23 medium and 2) an additional 24 h incubation in freshly made, hormone-free medium. Incubation was performed in 500 μl droplets of IVM medium (about 70 COCs per droplet) in four-well plates (NUNC, Thermo Scientific, Waltham, Ma,, USA), at 39 C in a 5% CO2 humidified atmosphere. Preparation of the follicular fluid (FF) included aspiration of the FF from the ovarian follicles (3–6 mm) and transfer of FF to conical tubes. The collected FF was centrifuged at 3000 rpm for 30 min, and aliquots of the resulting supernatant were frozen in liquid nitrogen in 1.5 ml tubes (Eppendorf Poland). Follicular fluid was stored for one month and used to supplement the IVM medium. Maturation efficiency (the rate of oocytes at the MII stage) was estimated by DAPI (Vector Laboratories) staining. After in vitro maturation, denuded oocytes were fixed in 4% paraformaldehyde for 30 min. at room temperature and placed on slides under coverslips in 1 μg/mL DAPI solution and further examined using a Zeiss Axiovert 200 inverted microscope. Maturation efficiency was calculated by dividing the number of matured oocytes [with visible chromatin of the first polar body (I pb) and oocyte chromosomes] by the number of all fixed oocytes with stained chromatin.
Mitochondrial DNA (mtDNA)
Sample preparation and total DNA extraction: Before analysis, all cumulus cells surrounding the oocytes were removed by vigorous pipetting. Denuded oocytes were washed 3× in a PBS supplemented with 0.2% (w/v) polyvinylpyrrolidone to ensure that there was no cumulus cell contamination of the samples. Individual oocytes were placed in 1.5 ml tubes with 200 μl PBS and immediately frozen in liquid nitrogen. Only oocytes exhibiting the first polar body after in vitro maturation were frozen and used for DNA isolation. All samples were stored at –80 C. Extraction of the genomic DNA and mtDNA was conducted using a High Pure PCR Template Preparation Kit (Roche) according to the manufacturer’s protocol. Briefly, samples were incubated in binding buffer and proteinase K for 10 min at 70 C. The next steps involved the use of an inhibitor removal buffer and a double washing. Finally, the total DNA from individual oocytes was eluted into a fresh 1.5 ml tube in 200 μl of Elution Buffer. DNA samples were stored at –20 C prior to analysis.
Standard curve preparation and real-time PCR analysis: Real-time absolute quantification analysis was conducted using the standard curve method. For this purpose, a desired sequence of the CYTB gene was amplified by PCR and visualized on 1.5% agarose gel. The PCR product was cut out from the gel, isolated and purified using a GeneJet Gel Extraction Kit (Fermentas, Burlington, ON, Canada). Although the primers for the CYTB gene did not yield nonspecific products as revealed by BLAST, standard gel electrophoresis and melting curve analysis, the amplicon was sequenced to confirm the specificity of mtDNA gene amplification. The procedure for sequencing was previously described by Cieslak et al. [23]. Briefly, prior to sequencing, amplicons were treated with exonuclease and alkaline phosphatase (Fermentas, Burlington, ON Canada) and further amplified by using a BigDye Terminator Sequencing Kit with labeled dideoxynucleotides according to the manufacturer’s protocol (Applied Biosystems, Warsaw, Poland). The DNA sequencing products were subsequently purified by gel filtration on Sephadex G-50 (Sigma Aldrich Chemie GmbH) and analyzed by capillary electrophoresis on an ABI 3130 Genetic Analyzer (Applied Biosystems). Based on the DNA concentration measured with a Nanodrop 2000c spectrophotometer (Thermo Scientific, Waltham, MA, USA), 10-fold serial dilutions of known concentrations (standards) were made. Each standard was used as a template for real-time PCR reaction in order to produce the appropriate standard curve. Finally, each real-time PCR run, contained one DNA standard of known concentration. All reactions were carried out using a LightCycler 2.0 system with a set of supplied reagents (Roche, Switzerland). The 20 μl reaction mixture consisted of 4 μl of LightCycler® FastStart DNA MasterPLUS SYBR Green I, 0.5 μM primers, 3 μl of isolated DNA and water. The set of primers for the CYTB gene published by El Shourbagy et al. was applied [10]. The reaction conditions were: denaturation for 10 min at 95 C, amplification for 40 cycles at 95 C for 15 sec, 55 C for 10 sec, 72 C for 15 sec and cooling at 40 C. Every reaction was followed by melting curve analysis to ensure the specificity of the amplification. Three steps were involved: 94 C, 55 C for 30 sec, and 95 C while the fluorescence was continuously measured. The temperature slope was set at 20 C/sec during amplification and at 0.1 C/sec during melting. Each DNA sample was analyzed in two independent PCR runs, and the mean value was taken for the calculations. All the presented values for mtDNA analysis are absolute and represent copy numbers. A category of mtDNA-poor oocytes (< 100 000 copies) was distinguished based on the results of El Shourbagy et al. [10] and Wai et al. [24] to highlight the rate of oocytes unsuitable for further fertilization or activation.
Relative transcript abundance
The relative (RA) was assessed in samples containing 25 oocytes. There were six independent replicates in each of the four experimental groups. Two reference genes were used in this study, ACTB and GAPDH, and 6 marker genes were described as developmentally important: BMP15, GDF9, ZAR1, GSTA2, ATP5A1 and EEF1A1. Total RNA was extracted using a mirVANA Paris Kit (Ambion) according to the protocol provided by the manufacturer. RNA was eluted with 100 μl Elution Buffer and precipitated using Pellet Paint NF Co-precipitant (Novagen, Merck) to 12 μl. The concentration was measured using a Nanodrop 2000c spectrofotomether. For reverse transcription (RT), 1 μg of each RNA sample was taken. The RT reaction was performed using a First Strand cDNA Synthesis Kit (Roche, Switzerland). Briefly, RNA with random hexamers were denatured at 65 C for 10 min. Then, a mixture consisting of reaction buffer, reverse transcriptase, RNase protector and dTT was added to each denatured sample and incubated at 55 C for 1 h. A FastStart HybProbe MasterPlus Kit was used for real-time PCR as described by the manufacturer (Roche, Switzerland). The reaction mix consisted of: 0.5 μM primers, 0.3 μM probes, 2 μl HybProbe Master mix, 1 μl cDNA and water. The reaction conditions were denaturation at 95 C for 10 min; 40 cycles of amplification at 95 C for 15 sec, 60 C for 8 sec, 72 C for 10 sec; and cooling at 40 C. The temperature slope was set at 20 C/sec. The calculation method was based on the standard curve as described in section 2.3.2. HybProbe probes for each gene were designed and synthesized by Tib Molbiol (Germany; Table 2). The fluorescence of the probes was detected by a LightCycler 2.0 on channel 640 or 610 after every annealing step. Each DNA sample was analyzed in two independent PCR runs. The mean value of each run was taken for calculations. The RA value was calculated in relation to the mean RA of the two reference genes.
Table 2. Sequences of starters (S) and probes (P) for real time PCR (5’-3’). For each gene, PCR elongation temperature, channel and length of amplicon (bp) are indicated (C).
| Gene | 5’-3’ | |
| ATP5A1 | S | AGTTGCTGAAGCAAGGACAGTAT; GTGTTGGCTGATAACGTGAGAC |
| P | TCCAGCTTATCAAGATAGCCCCTG; CACCCGCATAGATAACAGCTACTTGCT | |
| C | 610 nm, 60 C /147 bp | |
| BMP15 | S | CCTGATTAGGGAGCTGCTAGA; ATAAACCACAGTGGCTCTGAC |
| P | CTGGCATATACAGACCTTGGACTTTCC; CTGAGACCAAACCGGGTAGCCT | |
| C | 640 nm, 60 C / 262 bp | |
| EEF1A | S | TGGCTTTACAGCTCAGGTGATTA; CTTTTTCCCAGAACGACGA |
| P | AATGTGAGCTGTGTGGCAATCC; CACAGGTGCGTAACCAGCACTG | |
| C | 640 nm, 60 C / 144 bp | |
| GDF9 | S | GGGACCAGGTGACAGGA; GCCGTCACATCAATCTCAATC |
| P | ACAAGACTCTGCCTAAAGCTCCATACTC; TTTACCTTGAACTCACCATTCACATTTCAA | |
| C | 640 nm, 60 C / 250 bp | |
| GSTA2 | S | GGAGAGAGCCCTGATTGATATGTATA; CCCACAAGGTAGTCTTGTCCA |
| P | GCCACCTTGGCATCTTTTTCATTGG; TGGGCACAGTGGCAACAGCAAG | |
| C | 640 nm, 60 C / 271 bp | |
| ZAR1 | S | CTCAAGCAGTTTTGCAGAAC; GCATGGAAAAAGGCTCA |
| P | CTGTGACAGCACTTTCAGTTTCAAA; ATATTATTTAAGCGAAAAGCAACAAACCA | |
| C | 640 nm, 60 C / 292 bp | |
| ACTB | S | CAAAGCCAACCGTGAGAAGA; GTACCCCTCGTAGATGGGCA |
| P | TGTCCCTGTACGCCTCTGGCC; CACCACTGGCATCGTGATGGACTCCG | |
| C | 640 nm, 60 C / 121 bp | |
| GAPDH | S | CCCACGAGCACACCTCAGAA; TGCAGCCTGTACTCCCGCT |
| P | GGCAGCAGGTGGGACAGCAG; GGGTGCAGGAGAGGGGCGAC | |
| C | 640 nm, 60 C / 141 bp | |
Distribution of mitochondria
The mitochondria analysis was performed as previously described [25]. Briefly, mitochondria in oocytes were stained with 250 nM MitoTracker® CMTMRos (Life Technologies, Paisley, UK) diluted in PBS supplemented with 3% (w/v) BSA. Cumulus-oocyte complexes were washed 2 times in prewarmed and equilibrated 3% PBS/BSA, followed by incubation for 30 min with fluorescent probes. After COC staining, cumulus cells were removed by pipetting in a 0.025% hyaluronidase/PBS solution, and denuded oocytes were fixed in 2% paraformaldehyde for 30 min at 39 C. The oocytes were then stained with 0.5 μg/ml DAPI (Vector Laboratories, Burlingame, CA, USA) and placed in groups on slides with a single concave indentation under the coverslip.
Mitochondria patterns were analyzed using a confocal microscope equipped with Ar/Kr, and He/Ne lasers and the z-stack method (Zeiss, Germany) was used. The filters were 560 nm for MitoTracker® CMTMRos (mitochondria) and 420 nm for DAPI (chromatin). Active mitochondria were assessed through 4–5 optical sections. Objective (Plan Neofluar 40×/1.3 Oil DIC; Zeiss, Germany), pinhole, and filters were kept constant throughout the experiment. Classification of the mitochondria distribution patterns has been previously described in detail by Pawlak et al. [25]. Briefly, three patterns of mitochondria distribution were observed: a) peripheral - fluorescent signals homogeneously distributed in the peripheral ooplasm; b) diffused - mitochondria distributed homogeneously throughout the ooplasm and c) cortical - fluorescent signals covering most of the cytoplasmic volume without its central part.
Statistical analysis
Comparisons of the analyzed parameters characterizing cytoplasmic maturation of BCB+ and control oocytes before and after IVM, were performed with the Statistica Software (StatSoft, Krakow Poland). All the data, before computing, were subjected to testing for normal distribution with the KS test (Kolmogorov-Smirnov). The following tests were also applied: 1) the chi-square test for comparing the rates of oocytes with particular distribution patterns (mitochondria); 2) the Student’s t-test (mean mtDNA copy number) and Fisher’s exact test (the frequency of mtDNA-poor oocytes) and 3) the Student’s t-test and Mann-Whitney tests to compare the RAs of investigated genes. Correlation of the two independent real-time PCR runs for each DNA sample was calculated with the Spearman’s rank test.
Results
Maturation efficiency and mtDNA copy number
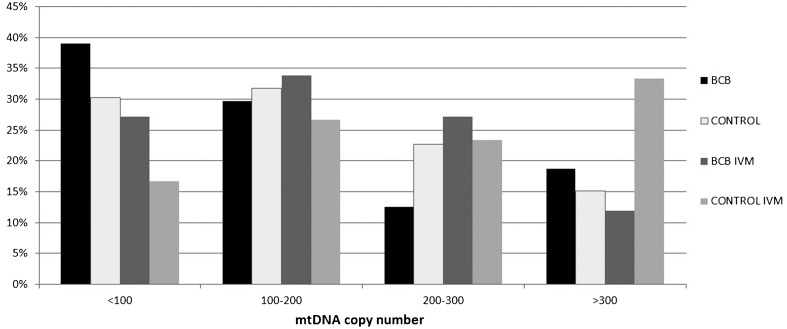
The BCB+ and control oocytes reached the metaphase II stage at similar rates (88.8 and 91.4% respectively; P>0.05) as revealed by DAPI staining. No significant differences were found between the control (n=66; 179 609; 10 502-610 000) and BCB+ (n=64; 176 595; 19 960-590 000) pre-IVM oocytes. After IVM, the mean mtDNA copy number increased significantly only in the control group (n=60; 246 984; 20 360-506 000; P<0.01), and the mean mtDNA copy number was significantly higher than in BCB+ post-IVM oocytes (n=59; 187 243; 9 720-510 000; P<0.05). The category of mtDNA-poor oocytes (<100 000 mtDNA copies) was represented in comparable frequencies in the pre-IVM (30 vs. 39%) and post-IVM (17 vs. 27%) control and BCB+ oocytes, respectively (P>0.05). After IVM, the rates of BCB+ and control oocytes in three mtDNA copy number categories (<100 000, 100 000-200 000, 200 000-300 000) were similar. However, the category with the highest mtDNA copy number (> 300 000) was significantly more represented in the group of control oocytes (12 vs. 33%, respectively; Fig. 2).
Fig. 2.
Distribution of porcine oocytes within the selected categories according to mtDNA copy number.
Mitochondria distribution
Similar frequencies of the three patterns of mitochondria distribution were observed in the control and BCB+ pre- and post-IVM oocytes. After IVM, an expected increase in the rate of oocytes with the diffused pattern, and a decrease in the rate of oocytes with the peripheral pattern, were found in oocytes – regardless of the experimental group. The most explicit differences were noticed in BCB+ oocytes before and after IVM (pre-IVM vs. post-IVM; Table 3). Post-IVM BCB+ oocytes were characterized by a higher frequency of the diffused pattern (38.5%) and a lower rate of oocytes with the peripheral pattern (23%) when compared with pre-IVM oocytes (15.9 and 50%, respectively; P<0.05).
Table 3. The percentages of oocytes with particular patterns of mitochondria distribution including maturation status.
| Group | Peripheral (%) | Cortical (%) | Diffused (%) |
| Control pre-IVM | 26 (54.2) | 15 (31.3) | 7 (14.6) |
| BCB+ pre-IVM | 22 (50.0) a | 15 (34.1) | 7 (15.9) b |
| Control post-IVM | 24 (36.9) | 24 (36.9) | 17 (26.2) |
| BCB+ post- IVM | 12 (23.0) a | 20 (38.5) | 20 (38.5) b |
The values within columns labeled with the same letter are significantly different (a P<0.01; b P<0.05).
Relative transcript abundance
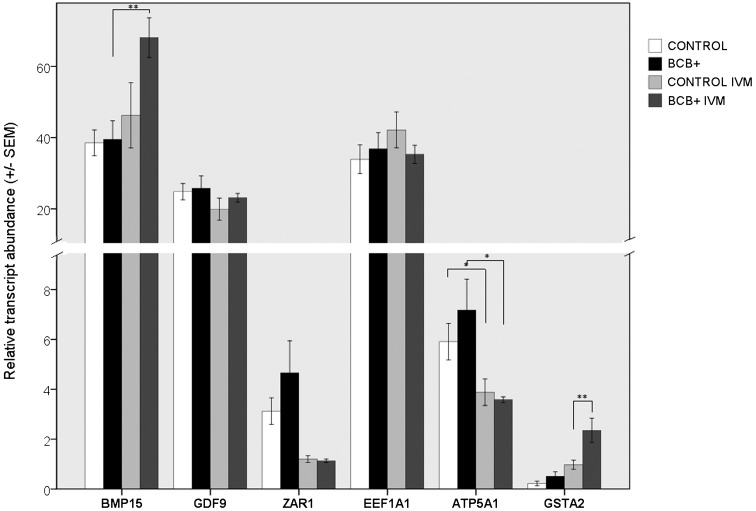
Pre-IVM oocytes of the control and BCB+ groups did not differ significantly in terms of the RAs of all the analyzed genes. After IVM, the RA of GSTA2 increased in both groups, and BCB+ oocytes had over a 2-fold higher RA than the control (P<0.01; Fig. 3). The RAs of GDF9 and EEF1A1 genes were constant in pre- and post-IVM oocytes and did not differ between the BCB+ and the control groups. On the other hand, the RA of the BMP15 gene increased significantly only in post-IVM BCB+ compared with pre-IVM BCB+ oocytes (P<0.01). Although the BCB+ and control post-IVM oocytes did not differ with regard to the RA of BMP15 (P=0.085), the BCB+ oocytes were characterized by over a 1.5 × higher RA of this gene. The RA of ATP5A1 decreased significantly in both groups after IVM (P<0.05). No differences were found in the RA of the ZAR1 gene in pre- and post-IVM oocytes of the BCB+ and control groups (Fig. 3).
Fig. 3.
Relative transcript abundance of analyzed genes in porcine oocytes.
Discussion
Although the BCB test has been successfully used to select oocytes of superior quality, applying the test to routine procedures of embryo production still remains questionable. Our results demonstrate that BCB+ and control porcine oocytes differed in some aspects of cytoplasmic maturation, however, the differences reflected the meiotic status of the oocytes (before or after IVM). The most interesting finding of the present study concerns the post-IVM oocytes, which differed in two parameters: the BCB+ oocytes were characterized by a higher RA for the GSTA2 gene and a higher incidence of the expected (diffused) pattern of mitochondria distribution. We have previously shown that cytoplasmic maturation is a complex phenomenon and should be investigated by multiple approaches. Only some processes accompanying oocyte maturation mark the quality of oocytes [25]. In this study, we focused on selected aspects of cytoplasmic maturation to search for parameters that would differentiate the BCB+ and control porcine oocytes. It should be noted that the control oocytes in several previous studies were not subjected to detailed analysis and usually served as a source of data on embryonic development. Moreover, the composition of the basic medium used for the BCB test may hamper comparison of the results of different studies and final interpretation of data. Although the majority of studies rely on PBS/BSA medium, some experiments involve additional energy substrates like pyruvate or glucose. However, the aspect of basic media composition has never been tested.
Mitochondria are crucial for oocyte maturation mainly as energy suppliers. It has been suggested by Krisher et al. that mitochondrial migration within a maturing oocyte is related to changes in the local energy requirements [26]. The peripheral distribution of mitochondria in pre-IVM oocytes indicates contact of the gametes with cumulus cells, whereas migration to the central ooplasm leading to a uniform redistribution reflects changes in energy requirements accompanying cytoplasmic maturation. Sun et al. postulated that under in vitro conditions, mitochondrial distribution can be disturbed, which impairs the cytoplasmic maturation of porcine oocytes [27]. Nevertheless, in this study, we showed that the frequency of the diffused pattern significantly increased in the BCB+ oocytes after IVM, whereas in the control oocytes, only a nonsignificant shift in the rate of oocytes with particular expected patterns was noticed. Our findings complement the results of Egerszegi et al., who found that pos-IVM BCB+ oocytes showed higher mitochondrial activity and a higher rate of oocytes with the expected homogenous distribution when compared with BCB– oocytes [28]. In the pig, the rate of BCB– oocytes does not usually exceed 30%. The similar rates of oocytes with particular distribution patterns in the BCB+ and control groups observed in our study, may reflect a limited share of the BCB– oocytes within the control group.
Another attribute of mitochondrion strongly related to oocyte quality, is the number of mtDNA copies [12, 26]. Until the embryo reaches the blastocyst stage, it relies on mtDNA accumulated during oocyte growth and maturation [12, 29]. Porcine oocytes containing less than 100 000 mtDNA copies (mtDNA-poor gametes) fail to fertilize, cleave, and develop as shown by El Shourbagy et al. [10]. It was also noted, that porcine post-IVM BCB+ oocytes contain significantly more mtDNA copies (222 000) than their BCB- counterparts (115 000) [10]. The mean mtDNA content in the control oocytes analyzed by El Shourbagy et al., was 138 000, which represents an intermediate value between BCB+ and BCB– gametes [10]. In our study, the mtDNA contents of the BCB+ and control oocytes were similar (176 595 and 179 609, respectively). Although the number of mtDNA copies fell into the range of the data presented by El Shourbagy et al., BCB+ oocytes were not superior in terms of the mtDNA copy number [10]. No significant increase in the mtDNA copy number in the BCB+ oocytes after IVM may seem surprising. However, this finding corroborates that of Spikings et al. (2005) who performed mtDNA analysis at three definite time points of the IVM process. Under the above conditions, the mtDNA content of BCB+ porcine oocytes decreased from 400 000 to about 320 000. On the other hand, in the BCB– oocytes, a huge (almost 7-fold) increase in mtDNA copy number was observed (from 66 000 to 500 000). Thus, the data described in our manuscript and that presented by Spikings et al. (2005) suggest an opposing phenomenon related to mtDNA synthesis during IVM in BCB+, control and BCB– oocytes. According to this hypothesis, the increase in mtDNA content in control oocytes observed in our study may result from dynamic mtDNA synthesis probably taking place in potential BCB– oocytes, which account for 10% of control oocytes. On the other hand, a large standard deviation of the data reflecting the known oocyte heterogeneity should be always considered.
In addition to the above, the rate of mtDNA-poor gametes (<100 000 copies) was similar in the control and BCB+ group, both before (30 vs. 39%) and after IVM (17 vs. 27%). Therefore, our findings on mtDNA content in oocytes do not support the applicability of the BCB test for routine IVF protocol in pigs. The upper limit of the mtDNA copy number, which would significantly benefit the oocyte or the future embryo, is not really known. One can only speculate that higher mtDNA content is more desirable, as the subsequent blastomeres receive a greater supply of mitochondria, and thus better access to ATP factories during development to the blastocyst stage.
Transcript abundance of genes regulating oocyte growth and maturation has been shown to mark the quality of porcine and bovine oocytes [30,31,32]. Although large amounts of data have been published on RA in oocytes and embryos of domestic animals, appropriate evidence concerning oocytes selected by the BCB test is still scarce. The only data published to date on pig oocytes, concern the RA of genes related to the properties of the zona pellucida [33, 34]. According to these studies, BCB+ oocytes contained more transcripts of genes regulating sperm-oocyte interaction compared with control oocytes. Torner et al. did excellent work on BCB+ and BCB– bovine oocytes using a microarray platform [11]. Several developmentally important genes were found to be differentially expressed between BCB+ and BCB- bovine oocytes. These results inspired us to perform an analysis of the transcript abundance of six recognized markers of oocyte quality (BMP15, GDF9, ZAR1, GSTA2, ATP5A1, EEF1A1) in BCB+ and control porcine oocytes. The most interesting finding of our study was the significant increase in RA of the BMP15 gene in post-IVM BCB+ oocytes, the level of which was over 1.5 × higher than that in the control post-IVM group – in which significant change was noticed. Due to the important role in folliculogenesis, BMP15 may have a significant impact on oocyte quality. Our result corroborates that of Paczkowski et al., who observed higher RA of the BMP15 gene in oocytes collected from cyclic gilts when compared to oocytes from prepubertal females [30]. It must be stressed, that oocytes from cyclic gilts are of higher quality than those from prepubertal gilts. Before in vitro maturation, Torner et al. did not show differences in the RA of the BMP15 gene between bovine BCB+ and BCB– oocytes [11]. Since the highest RA of this gene accompanied the onset of cumulus cell expansion, we may assume that the transcript level of the BMP15 gene is more indicative as a quality marker in post-IVM porcine oocytes.
Another marker gene, GSTA2 (glutathione transferase), was characterized by a more than two twofold higher RA in BCB+ post-IVM oocytes compared to the control post-IVM oocytes. Paczkowski et al. [30] and Romar et al. [35] reported significantly more transcripts of genes engaged in cell protection against oxidative stress (reductase and glutathione peroxidase) in more competent porcine oocytes. On the other hand, Puglisi [36] postulated that increased RA of the GSTA2 gene may be a response to cellular stress. Whether oocytes are being providently protected or subjected to stress conditions needs to be elucidated. Considering the results of Romar [35] and Puglisi [36], increased GSTA2 expression in BCB+ oocytes may suggest a response to BCB test conditions. This hypothesis complements previous results showing that oocyte incubation for an additional 90 minutes in BCB solution exerted a negative effect on chromosome segregation [21]. Therefore, increased RA of the GSTA2 gene during BCB incubation caused by stress conditions, should not be overlooked.
With regard to the transcript abundance of the GDF9 gene, the BCB+ and the control oocytes did not differ. There is, however, contradictory evidence concerning the RA of this gene published for porcine oocytes. We did not find a significant change in RA during IVM, which is consistent with data reported by Prochazka et al. [37]. On the other hand, in the study of Li et al., the RA of this gene decreased during IVM [38]. Studies on various species have revealed some differences in the onset and duration of GDF9 expression depending on the cell type (oocyte, cumulus cells, granulosa). The function and expression of GDF9 still remains a matter of debate. Additionally, the BCB+ and control oocytes displayed similar transcript levels of the ZAR1 gene. This finding confirmed the previous observations in bovine oocytes, in which no differences between BCB+, BCB–, and control oocytes were noticed [20]. Considering the above results, it may be postulated that the reduced quality of the control oocytes are probably not the result of the differential expression of three (BMP15, GDF9, ZAR1) maternal-effect genes.
In addition to the maternal effect genes, we investigated two genes regulating cell metabolism (EEF1A1, ATP5A1). The gene, ATP5A1, encodes a subunit of ATP synthase, a key protein for energy production, and this gene was found to be more abundant in less competent, BCB– bovine oocytes [11]. The product of the EEF1A1 gene is a subunit of the eukaryotic elongation factor responsible for biosynthesis of proteins. An increased RA of this gene was demonstrated in more competent bovine oocytes (BCB+) [11]. Although we did not find significant differences in transcript abundance of ATP5A1 and EEF1A1 genes between BCB + and control oocytes, the presented data is crucial. Torner et al. hypothesized that the higher RA of ATP5A1 may reflect intensive oocyte metabolism at the onset of cytoplasmic maturation [11]. After IVM, we observed a significant decrease in RA of the ATP5A1 gene in both groups of oocytes. Therefore, according to Torner’s hypothesis, we can assume that BCB+ and control porcine oocytes have similar maturation potentials and a sufficient energy stores for maturation and fertilization. Investigation of individual genes, though, may turn out to be insufficient to draw far-reaching conclusions.
In conclusion, the BCB+ and control porcine oocytes showed a relatively high similarity in selected parameters characterizing cytoplasmic maturation. In our opinion, this high similarity may result mainly from a limited (10%) share of the less competent BCB– oocytes still present among nonselected oocytes of proper morphology. Therefore, our results do not justify the application of the BCB test for routine IVM protocol of porcine oocytes.
Acknowledgments
This work was supported by an international grant from the Ministry of Science and Higher Education in Poland (no 451/N-COST/2009/0) operating within the European COST Action (FA0702, GEMINI - Maternal Interaction with Gametes and Embryos. Piotr Pawlak is a grant holder of the 2012 European Social Fund (Human Capital Program) and the recipient of a Stipend for Young Researchers from the 2013 START program of the Foundation for Polish Science. We would like to acknowledge Dr Mariusz Mackowski for help in sequencing and interpretation of results.
References
- 1.Ericsson SA, Boice ML, Funahashi H, Day BN. Assessment of porcine oocytes using brilliant cresyl blue. Theriogenology 1993; 39: 214 [abstract] [Google Scholar]
- 2.Wongsrikeao P, Otoi T, Yamasaki H, Agung B, Taniguchi M, Naoi H, Shimizu R, Nagai T. Effects of single and double exposure to brilliant cresyl blue on the selection of porcine oocytes for in vitro production of embryos. Theriogenology 2006; 66: 366–372 [DOI] [PubMed] [Google Scholar]
- 3.Ishizaki C, Watanabe H, Bhuiyan MM, Fukui Y. Developmental competence of porcine oocytes selected by brilliant cresyl blue and matured individually in a chemically defined culture medium. Theriogenology 2009; 72: 72–80 [DOI] [PubMed] [Google Scholar]
- 4.Bhojwani S, Alm H, Torner H, Kanitz W, Poehland R. Selection of developmentally competent oocytes through brilliant cresyl blue stain enhances blastocyst development rate after bovine nuclear transfer. Theriogenology 2007; 67: 341–345 [DOI] [PubMed] [Google Scholar]
- 5.Manjunatha BM, Gupta PS, Devaraj M, Ravindra JP, Nandi S. Selection of developmentally competent buffalo oocytes by brilliant cresyl blue staining before IVM. Theriogenology 2007; 68: 1299–1304 [DOI] [PubMed] [Google Scholar]
- 6.Alm H, Torner H, Lohrke B, Viergutz T, Ghoneim IM, Kanitz W. Bovine blastocyst development rate in vitro is influenced by selection of oocytes by brilliant cresyl blue staining before IVM as indicator for glucose-6-phosphate dehydrogenase activity. Theriogenology 2005; 63: 2194–2205 [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-González E, Lopez-Bejar M, Izquierdo D, Paramio MT. Developmental competence of prepubertal goat oocytes selected with brilliant cresyl blue and matured with cysteamine supplementation. Reprod Nutr Dev 2003; 43: 179–187 [DOI] [PubMed] [Google Scholar]
- 8.Roca J, Martinez E, Vazquez JM, Lucas X. Selection of immature pig oocytes for homologous in vitro penetration assays with the brilliant cresyl blue test. Reprod Fertil Dev 1998; 10: 479–485 [DOI] [PubMed] [Google Scholar]
- 9.Opiela J, Katska-Ksiazkiewicz L, Lipiński D, Słomski R, Bzowska M, Ryńska B. Interactions among activity of glucose-6-phosphate dehydrogenase in immature oocytes, expression of apoptosis-related genes BC-2 and Bax, and developmental competence following IVP in cattle. Theriogenology 2008; 69: 546–555 [DOI] [PubMed] [Google Scholar]
- 10.El Shourbagy SH, Spikings EC, Freitas M, St John JC. Mitochondria directly influence fertilisation outcome in the pig. Reproduction 2006; 131: 233–245 [DOI] [PubMed] [Google Scholar]
- 11.Torner H, Ghanem N, Ambros C, Hölker M, Tomek W, Phatsara C, Alm H, Sirard MA, Kanitz W, Schellander K, Tesfaye D. Molecular and subcellular characterisation of oocytes screened for their developmental competence based on glucose-6-phosphate dehydrogenase activity. Reproduction 2008; 135: 197–212 [DOI] [PubMed] [Google Scholar]
- 12.Spikings EC, Alderson J, St John JC. Regulated mitochondrial DNA replication during oocyte maturation is essential for successful porcine embryonic development. Biol Reprod 2007; 76: 327–335 [DOI] [PubMed] [Google Scholar]
- 13.Castaneda CA, Kaye P, Pantaleon M, Phillips N, Norman S, Fry R, D’Occhio MJ. Lipid content, active mitochondria and brilliant cresyl blue staining in bovine oocytes. Theriogenology 2013; 79: 417–422 [DOI] [PubMed] [Google Scholar]
- 14.Pawlak P, Renska N, Pers-Kamczyc E, Warzych E, Lechniak D. The quality of porcine oocytes is affected by sexual maturity of the donor gilt. Reprod Biol. 2011; 11: 1–18 [DOI] [PubMed] [Google Scholar]
- 15.Mohammadi-Sangcheshmeh A, Held E, Ghanem N, Rings F, Salilew-Wondim D, Tesfaye D, Sieme H, Schellander K, Hoelker M. G6PDH-activity in equine oocytes correlates with morphology, expression of candidate genes for viability, and preimplantative in vitro development. Theriogenology 2011; 76: 1215–1226 [DOI] [PubMed] [Google Scholar]
- 16.Wu YG, Liu Y, Zhou P, Lan GC, Han D, Miao DQ, Tan JH. Selection of oocytes for in vitro maturation by brilliant cresyl blue staining: a study using the mouse model. Cell Res 2007; 17: 722–731 [DOI] [PubMed] [Google Scholar]
- 17.Mohammadi-Sangcheshmeh A, Soleimani M, Deldar H, Salehi M, Soudi S, Hashemi SM, Schellander K, Hoelker M. Prediction of oocyte developmental competence in ovine using glucose-6-phosphate dehydrogenase (G6PDH) activity determined at retrieval time. J Assist Reprod Genet 2012; 29: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pujol M, López-Béjar M, Paramio MT. Developmental competence of heifer oocytes selected using the brilliant cresyl blue (BCB) test. Theriogenology 2004; 61: 735–744 [DOI] [PubMed] [Google Scholar]
- 19.Mirshamsi SM, Karamishabankareh H, Ahmadi-Hamedani M, Soltani L, Hajarian H, Abdolmohammadi AR. Combination of oocyte and zygote selection by brilliant cresyl blue (BCB) test enhanced prediction of developmental potential to the blastocyst in cattle. Anim Reprod Sci 2013; 136: 245–251 [DOI] [PubMed] [Google Scholar]
- 20.Mota GB, Batista RI, Serapião RV, Boité MC, Viana JH, Torres CA, de Almeida Camargo LS. Developmental competence and expression of the MATER and ZAR1 genes in immature bovine oocytes selected by brilliant cresyl blue. Zygote 2010; 18: 209–216 [DOI] [PubMed] [Google Scholar]
- 21.Pawlak P, Pers-Kamczyc E, Renska N, Kubickova S, Lechniak D. Disturbances of nuclear maturation in BCB positive oocytes collected from peri-pubertal gilts. Theriogenology 2011; 75: 832–840 [DOI] [PubMed] [Google Scholar]
- 22.Opiela J, Kątska-Książkiewicz L. The utility of Brilliant Cresyl Blue (BCB) staining of mammalian oocytes used for in vitro embryo production (IVP). Reprod Biol 2013; 13: 177–183 [DOI] [PubMed] [Google Scholar]
- 23.Cieslak J, Flisikowska T, Schnieke A, Kind A, Szydlowski M, Switonski M, Flisikowski K. Polymorphisms in the promoter region of the adiponectin (ADIPOQ) gene are presumably associated with transcription level and carcass traits in pigs. Anim Genet. 2013; 44: 340–343 [DOI] [PubMed] [Google Scholar]
- 24.Wai T, Ao A, Zhang X, Cyr D, Dufort D, Shoubridge EA. The role of mitochondrial DNA copy number in mammalian fertility. Biol Reprod 2010; 83: 52–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pawlak P, Cieslak A, Warzych E, Zejden Z, Szumacher-Strabel M, Molinska-Glura M, Lechniak D. No single way to explain cytoplasmic maturation of oocytes from prepubertal and cyclic gilts. Theriogenology 2012; 78: 2020–2030 [DOI] [PubMed] [Google Scholar]
- 26.Krisher RL, Brad AM, Herrick JR, Sparman ML, Swain JE. A comparative analysis of metabolism and viability in porcine oocytes during in vitro maturation. Anim Reprod Sci 2007; 98: 72–96 [DOI] [PubMed] [Google Scholar]
- 27.Sun QY, Wu GM, Lai L, Park KW, Cabot R, Cheong HT, Day BN, Prather RS, Schatten H. Translocation of active mitochondria during pig oocyte maturation, fertilization and early embryo development in vitro. Reproduction 2001; 122: 155–163 [PubMed] [Google Scholar]
- 28.Egerszegi I, Alm H, Rátky J, Heleil B, Brüssow KP, Torner H. Meiotic progression, mitochondrial features and fertilisation characteristics of porcine oocytes with different G6PDH activities. Reprod Fertil Dev 2010; 22: 830–838 [DOI] [PubMed] [Google Scholar]
- 29.Ramalho-Santos J, Varum S, Amaral S, Mota PC, Sousa AP, Amaral A. Mitochondrial functionality in reproduction: from gonads and gametes to embryos and embryonic stem cells. Hum Reprod Update 2009; 15: 553–572 [DOI] [PubMed] [Google Scholar]
- 30.Paczkowski M, Yuan Y, Fleming-Waddell J, Bidwell CA, Spurlock D, Krisher RL. Alterations in the transcriptome of porcine oocytes derived from prepubertal and cyclic females is associated with developmental potential. J Anim Sci 2011; 89: 3561–3571 [DOI] [PubMed] [Google Scholar]
- 31.Yuan Y, Ida JM, Paczkowski M, Krisher RL. Identification of developmental competence-related genes in mature porcine oocytes. Mol Reprod Dev 2011; 78: 565–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wrenzycki C, Herrmann D, Lucas-Hahn A, Korsawe K, Lemme E, Niemann H. Messenger RNA expression patterns in bovine embryos derived from in vitro procedures and their implications for development. Reprod Fertil Dev 2005; 17: 23–35 [DOI] [PubMed] [Google Scholar]
- 33.Kempisty B, Jackowska M, Piotrowska H, Antosik P, Woźna M, Bukowska D, Brüssow KP, Jaśkowski JM. Zona pellucida glycoprotein 3 (pZP3) and integrin β2 (ITGB2) mRNA and protein expression in porcine oocytes after single and double exposure to brilliant cresyl blue test. Theriogenology 2011; 75: 1525–1535 [DOI] [PubMed] [Google Scholar]
- 34.Antosik P, Kempisty B, Bukowska D, Jackowska M, Włodarczyk R, Budna J, Brüssow KP, Lianeri M, Jagodziński PP, Jaśkowski JM. Follicular size is associated with the levels of transcripts and proteins of selected molecules responsible for the fertilization ability of oocytes of puberal gilts. J Reprod Dev 2009; 55: 588–593 [DOI] [PubMed] [Google Scholar]
- 35.Romar R, De Santis T, Papillier P, Perreau C, Thélie A, Dell’Aquila ME, Mermillod P, Dalbiès-Tran R. Expression of maternal transcripts during bovine oocyte in vitro maturation is affected by donor age. Reprod Domest Anim 2011; 46: e23–e30 [DOI] [PubMed] [Google Scholar]
- 36.Puglisi R, Cambuli C, Capoferri R, Giannino L, Lukaj A, Duchi R, Lazzari G, Galli C, Feligini M, Galli A, Bongioni G. Differential gene expression in cumulus oocyte complexes collected by ovum pick up from repeat breeder and normally fertile Holstein Friesian heifers. Anim Reprod Sci. 2013; 141: 26–33 [DOI] [PubMed] [Google Scholar]
- 37.Prochazka R, Nemcova L, Nagyova E, Kanka J. Expression of growth differentiation factor 9 messenger RNA in porcine growing and preovulatory ovarian follicles. Biol Reprod 2004; 71: 1290–1295 [DOI] [PubMed] [Google Scholar]
- 38.Li HK, Kuo TY, Yang HS, Chen LR, Li SS, Huang HW. Differential gene expression of bone morphogenetic protein 15 and growth differentiation factor 9 during in vitro maturation of porcine oocytes and early embryos. Anim Reprod Sci 2008; 103: 312–322 [DOI] [PubMed] [Google Scholar]