Abstract
A primary goal of metabonomics research is biomarker discovery for human diseases based on differences in metabolic profiles between healthy and diseased patient populations. One of the most significant challenges in biomarker discovery is validation, which implicitly depends on the coefficient of variation (CV) associated with the measurement technique. This paper investigates how the CV of metabolite resonances measured by nuclear magnetic resonance spectroscopy (NMR) depends on signal-to-noise ratio (SNR) and normalization method. CVs were calculated for NMR resonance peaks in a series of NMR spectra of five synthetic urine samples collected over an eight-month period. An inverse correlation was detected between SNR and CV for all normalization methods. Small peaks with SNR<15 tended to have larger CVs (15–30%) compared to peaks with the highest SNR>150, which typically had smaller CVs (5–10%). The inverse relationship between CV and SNR roughly obeyed a log10 dependence. Quotient normalization (QN) tended to produce smaller CVs for smaller peaks, but larger CVs for the strongest peaks in the data, compared to no normalization, normalization to total intensity (NTI) or normalization to an internal standard (NIS). Consequently, quotient normalization appears optimal for validating low concentration metabolites. NTI or NIS appear superior to QN for samples that have very small variation in total signal intensity. While the inverse relationship between CV and log10(SNR) did not strictly hold for all metabolites, weaker concentration metabolites will likely require more rigorous validation as potential biomarkers since they tend to have poorer reproducibility.
Keywords: Nuclear Magnetic Resonance, Metabonomics, Metabolomics, Coefficient of Variation, Biomarker Validation
1. Introduction
Nuclear magnetic resonance spectroscopy (NMR)-based metabonomics research aimed at biomarker discovery for human diseases has increased significantly over the last decade [1–3]. The increase in popularity is due to the promise that metabonomics research could contribute to personalized health care, non-invasive diagnosis of diseases, earlier diagnosis of diseases that have high fatality rates, and potential biomarker discovery for diseases that currently are difficult to diagnose [4, 5]. The potential importance of biomarkers discovered using a metabonomics approach are discussed in a recent review [6].
Metabonomics has shown great promise in identifying potential biomarkers for human diseases, specifically in human cancers [7], and metabonomics in oncology has recently been reviewed [8, 9]. One area of metabonomics research that could benefit from further study is biomarker validation. Currently there are very few papers available that address this issue. Serkova et al. has written the most comprehensive review on biomarker validation relating to metabonomics research [10]. Most of the work with biomarker validation has been studied in epidimeology. Reliability models have been used to help with biomarker validation [11]. Puntmann has recently written a review that outlines biomarker terminology and validation as well as giving examples related to cardiovascular disease [12]. These examples do not pertain to metabonomics studies but the principles involved should be considered when developing a biomarker that has been discovered through metabonomics research.
One of the first things that must be considered for validation of NMR-based biomarkers is analytical reproducibility. Keun et al. has determined that the analytical reproducibility of NMR measurements appeared to be very good when data gathered on common samples at different sites were compared [13]. Ebbels et al. analyzed spectral variation between rat strains using a range of statistics including standard deviation, skewness, and kurtosis [14]. A few papers have used coefficient of variation (CV), also referred to as relative standard deviation (RSD), to look at the reproducibility of various NMR-based metabonomics data sets. Teahan et al. showed mean unedited CPMG spectra colored with CVs at each point after 10 repeated measurements over three hours. Based on these spectra, it was determined that the unedited spectrum contained no more than 1% deviation and most points in the CPMG spectra contained < 5% variation [15]. Another paper determined the reproducibility of NMR data by looking at urinary spectra over a period of seven months and the CV of each point was plotted vs. the chemical shift to obtain a “spectrum” that showed where the highest and lowest CV was present in the original spectra. In this case the authors determined that the areas with a high CV contained the noise region of the spectra and that the CV of relevant metabolite signals had a CV in the range of 0 – 10% [16]. Sysi-Aho et al. looked at the CV distribution as it related to the type of spectral normalization applied in a UPLC/MS application of metabolomics. They tested three different types of normalization, NOMIS, 3STD, and L2N, and compared it to the raw data. It was determined that the NOMIS normalization method, which stands for normalization using optimal selection of multiple internal standards, produced the lowest median CV [17]. In 3STD, three internal standards with different retention times were used and the internal standard used for normalization depended on the peak retention time [18]. L2N is based on normalization to sum of squares of peak intensities [19] with some adjustments [17]. Finally, Parsons et al. looked at the CV focusing on bucketed NMR spectra by normalizing the data to unity, also known as total intensity. They looked at various NMR data sets and used boxplots to show the inter- and intra-individual metabolic variation between the different types of samples. They concluded that it was important to use CVs to assess the quality of metabonomics datasets and recommended creating a database of CV values for other researchers to consult [20].
All of the previous studies that have looked at the CV of NMR data for metabonomic studies have used either human or animal samples. Even though several papers have shown that freeze-thaw and long-term storage do not contribute to sample degradation when evaluated by principal component analysis (PCA), some degradation can still take place over time [21–24]. Such degradation could cause an increase in CVs due to the possibility of changing peak intensities. In this paper we looked at five synthetic urine samples starting with Surine™ (Dyna-Tek Industries, Lenexa, KS, USA) and adding various small molecules that could be present in urine. These samples allowed CV analysis strictly focused on instrumental and analytical reproducibility with minimal expected sample changes over time. CV analysis was performed after using quotient normalization (QN) [25], normalization to total intensity (NTI), normalization to an internal standard (NIS), and no normalization (NN). Peaks were grouped into distinct SNR groups and CV analysis was evaluated to determine how the CV depended on the metabolite concentrations.
2. Experimental
2.1 Materials
Synthetic Urine Samples. Five different synthetic urine samples were prepared by adding specified amounts of a number of compounds to 50 mL of Surine™ (Dyna-Tek Industries, Lenexa, KS, USA). The five resulting synthetic urine samples contained anywhere from 9 to 17 added components (Table 1). The components in synthetic urines were chosen by their NMR spectroscopic characteristics which included range of resonant frequencies, relaxation times, peaks overlap and splittings. 500 µL of each mixture was transferred to an eppendorf tube and 200 µL of buffer (0.3 mM KH2PO4, pH 7.2) added to the mixture. The samples were then centrifuged for 5 min. 540 µL of each sample was then transferred into a 5 mm NMR tube and 60 µL of a 10% trimethylsilyl propionate (TSP)/D2O solution was added. The solution was then mixed and sealed in a glass-sealed NMR tube to prevent degradation. Five different synthetic urine samples were prepared in this manner. Some samples shared common components, but the five samples had unique overall compositions. The concentrations of the components, which ranged from 63 µM to 1.1 mM, were chosen to span a typical concentration range of majority human urine metabolites [26].
Table 1.
Composition of five synthetic urine samples.
| Sample #1 | Sample #2 | Sample #3 | Sample #4 | Sample #5 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Compon ents |
[X] in µM |
Compon ents |
[X] in µM |
Compon ents |
[X] in µM |
Compon ents |
[X] in µM |
Compon ents |
[X] in µM |
| 3 | 164 | 22 | 404 | 1 | 141 | 5 | 440 | 2 | 330 |
| 5 | 317 | 7 | 674 | 2 | 214 | 6 | * | 3 | 257 |
| 7 | 547 | 8 | 84 | 4 | 1088 | 7 | 390 | 5 | 669 |
| 10 | 443 | 9 | 405 | 12 | 286 | 16 | 565 | 8 | 184 |
| 35 | 266 | 11 | 241 | 15 | 343 | 14 | 287 | 35 | 510 |
| 11 | 261 | 15 | 121 | 17 | 360 | 24 | 411 | 11 | 375 |
| 13 | 485 | 20 | 548 | 20 | 582 | 21 | 1095 | 17 | 411 |
| 16 | 437 | 28 | 346 | 28 | 436 | 26 | 783 | 18 | 110 |
| 18 | 61 | 29 | 323 | 31 | 211 | 30 | 503 | 20 | 1062 |
| 19 | 138 | 33 | 165 | 36 | 650 | 23 | 416 | ||
| 23 | 256 | 37 | 116 | 25 | 157 | ||||
| 25 | 143 | 27 | 754 | ||||||
| 27 | 507 | 28 | 296 | ||||||
| 29 | 331 | 29 | 380 | ||||||
| 32 | 63 | 30 | 512 | ||||||
| 33 | 225 | 31 | 390 | ||||||
| 34 | 221 | 33 | 203 | ||||||
1) 2-hydroxycinnamic acid; 2) 4,4’-dimethylbenzophenone; 3) 4-androstene-3,17-dione; 4) acetamide; 5) adipic acid; 6) agarose; 7) benzoic acid; 8) biotin; 9) camphor; 10) cis-1,2-cyclohexanediol; 11) citric acid; 12) d-3-phenyllactic acid; 13) d-galactose; 14) DL-4-fluorophenylalanice; 15) d-mannose; 16) d-ribose; 17) d-xylose; 18) erythromycin; 19) fumaric acid monoethyl ester; 20) glycine; 21) imidazole; 22) l-alanine; 23) l-ascorbic acid; 24)l-glutamic acid; 25) lithocholic acid; 26) l-serine; 27) malonic acid; 28) menthol; 29) n-acetyl-l-valine; 30) p-dimethoxybenzene; 31) phenylphosphonic acid; 32) quinine; 33) sucrose; 34) trans-1,2-cyclohexanediol; 35) trans-aconitic acid; 36) trimethylamine-N-oxide; 37) uridine
Concentration for agarose could not be computed due to the varying sizes of agarose present in the stock compound
2.2 Data Collection
NMR Data Collection. 1H NMR spectra were acquired at 298K on a Bruker US2 Avance™ III 850 MHz spectrometer operating at 850.10 MHz equipped with a 5 mm TXI triple resonance probe with inverse detection and controlled by TopSpin 2.1.4 (Bruker, Germany). All data were collected using a spectral width of 20.0 ppm. Two 1H NMR experiments optimized by Bruker (Bruker BioSpin, Billerica, MA) for metabonomics studies were run on all samples: a standard 1D pulse and acquire pulse sequence employing water-suppression using pre-saturation (zgpr) and the one-dimensional first increment of a nuclear Overhauser effect spectroscopy (NOESY) experiment with pre-saturation (noesygppr1d). All experiments included on-resonance presaturation of water achieved by irradiation during a recycle delay of 4.5 s with pulse power levels of 49.02 dB and 54.89 dB for the zgpr, and 1D NOESY, respectively. The 90° pulse width was determined for every sample using the automatic pulse calculation feature in TopSpin [27]. All pulse widths were 15 ± 0.5 µs. The zgpr experiment was used to screen samples and assure that presaturation and shimming were sufficient to collect reliable data. Quality of shimming was determined by measuring the full width at half height (FWHH) of the TSP peak, which was deemed acceptable when the FWHH was 1.0 ±0.5 Hz. One-dimensional zgpr 1H spectra were acquired using 2 transients and 2 dummy scans, with 65 K points per spectrum. This resulted in an acquisition time of 1.92 s. Once the spectrum was determined to be of acceptable quality the 1D NOESY experiment was collected. The one-dimensional first increment of the NOESY experiment was collected using 4 transients with 4 dummy scans. 65K points per spectrum were collected which resulted in an acquisition time of 1.92 s, and the mixing time was 0.01 s. All NMR spectra were processed using the AU program apk0.noe. This AU automatically phase corrected, baseline corrected, and corrected for chemical shift registration relative to TSP, which was set to 0 ppm in TopSpin 2.1.4. Manual phase correction was applied to achieve optimal phase correction after automatic phase correction. This process was repeated on the same samples over an eight-month period to mimic a longitudinal NMR-based metabonomic study of animal or human biofluid samples.
2.3 Data Analysis
After processing, spectra were imported into AMIX 3.9.11 (Bruker Biospin, Billerica, MA, USA) and in this software, bucket tables were created using a manual pattern for each synthetic urine sample. Each bucket width was matched to a peak in the overlaid spectra. Bucket tables were exported into Excel (Microsoft Office 2010) to enable further calculations. The different types of normalization, including normalization to total intensity (NTI), normalization to an internal 0.1 mM TSP standard (NIS), quotient normalization (QN) and no normalization (NN), were performed in Excel. NTI was calculated by dividing each bucket intensity by the sum of all bucket intensities in the spectrum, and NIS was calculated by dividing each bucket by the intensity of the TSP peak. QN was calculated after NTI as reported in the literature using the spectrum of the first measurement as a reference [25]. CVs for each bucket were calculated in Excel by taking the standard deviation of the bucket computed over the 12 spectra for each mixture and dividing by the mean bucket intensity. Signal-to-noise ratios (SNR) were calculated for each bucket from the five synthetic urine mixtures. The un-normalized bucket table was used to calculate the average SNR using the standard deviation of the noise for each spectrum in the chemical shift range between 9.5 – 10 ppm. After the standard deviation was calculated, the bucket intensity was divided by the calculated noise to determine the SNR of that bucket for that spectrum. This resulted in 12 different SNR being calculated for each bucket for each synthetic urine sample. The average of these 12 SNRs was calculated to determine the average SNR of that bucket across the 12 different spectra. The CVs of all buckets were evaluated for the four different types of spectral normalization (including NN) and assessed with regard to averaged bucket SNR. The un-normalized bucket table was used in SNR calculations since this ratio was independent of the normalization technique applied to the data.
3. Results And Discussion
3.1 Sample degradation test
For each sample, the first and last measured spectra were examined to study the sample aging. In this test, NTI was applied since it is commonly used in urine data analysis. First, the ratio of the last spectra and first spectra was calculated, and all buckets were included in the analysis. To study the variation of the ratios, standard deviations and CVs were both calculated and listed in Table S1. The results indicated that all the samples had very low standard deviations and CVs except Sample 3, which had slightly higher values. The correlation coefficients of the two measurements were also studied for all five samples. Similar to the CVs results, all the samples had very high R2 (>0.99) including Sample 3 which indicated the peaks were highly correlated (Table S1). These analyses indicated that sample degradation could be neglected with the possible exception Sample 3.
3.2 Internal Standard Variation Associated with Normalization Method
The performance of PCA, Partial Least Squares - Discriminant Analysis (PLS-DA) and statistical tests of significance, like a Student’s t-test, when applied to NMR data sets, can depend on the data normalization technique applied to the data [28]. This raises the question as to what degree the choice of spectral normalization technique affects the analytical reproducibility of the measured spectral intensities, i.e. the CVs? One way to address this question is to compute the CV for the internal standard, in our case, TSP, over multiple measurements. Since the concentration of the internal TSP standard was controlled to be identical in every sample, one can assess the peak intensity variability introduced by the normalization technique by comparing the TSP peak CVs. The optimal normalization technique should produce the smallest standard deviation in the peak intensity across the entire set of measurements. A normalization technique would be considered inferior if it introduced systematic error into the normalized peak intensities, as measured by a larger TSP peak CV.
Table 2 shows the CVs for the TSP peak for three different normalization techniques applied to the five different synthetic urine datasets. All three normalization methods produced small TSP CVs (<6%). However, NTI, by far, produces the smallest average CV (1.31) compared to NN (2.41), and QN (3.24). Interestingly, calculation of the CV of the CVs for each of the three different normalization methods further highlighted the differences between the normalization techniques, illustrating that NTI not only produced that smallest average CV, but also the smallest variation in CV across the three normalization techniques. For example, the CVs of the TSP peak in the five synthetic urines for NN ranged from 0.96 to 5.71, and the CV of these CVs was relatively high (79%). By contrast, after NTI or QN, the CV of CVs was much lower, 31% and 25% for QN and NTI, respectively, indicating that NTI performed better than QN or NN. Finally, all CVs for QN were larger than NTI. QN is intended to be used when the total intensity of each spectrum in a comparison group experiences significant variation. However, in this study, the higher CV of CVs, as well as the higher CVs indicated that NTI was superior to QN in cases where variation in the overall intensity of the spectra was not large.
Table 2.
The CVs of the internal standard, TSP, for three different normalization techniques. The last row is CV of the CVs for the five different samples computed for each of the three normalization techniques. The CV values are reported in %.
| Synthetic urine sample |
NN | NTI | QN |
|---|---|---|---|
| 1 | 2.13 | 1.47 | 3.49 |
| 2 | 2.09 | 1.04 | 2.46 |
| 3 | 5.71 | 1.79 | 4.86 |
| 4 | 0.96 | 1.00 | 2.97 |
| 5 | 1.19 | 1.24 | 2.42 |
| Average (Standard deviation) | 2.42 (1.91) | 1.31 (0.33) | 3.24 (1.01) |
| CV of CVs | 79.1 | 25.0 | 31.0 |
3.3 Coefficient of Variation Relationship to Signal-to-Noise Ratio
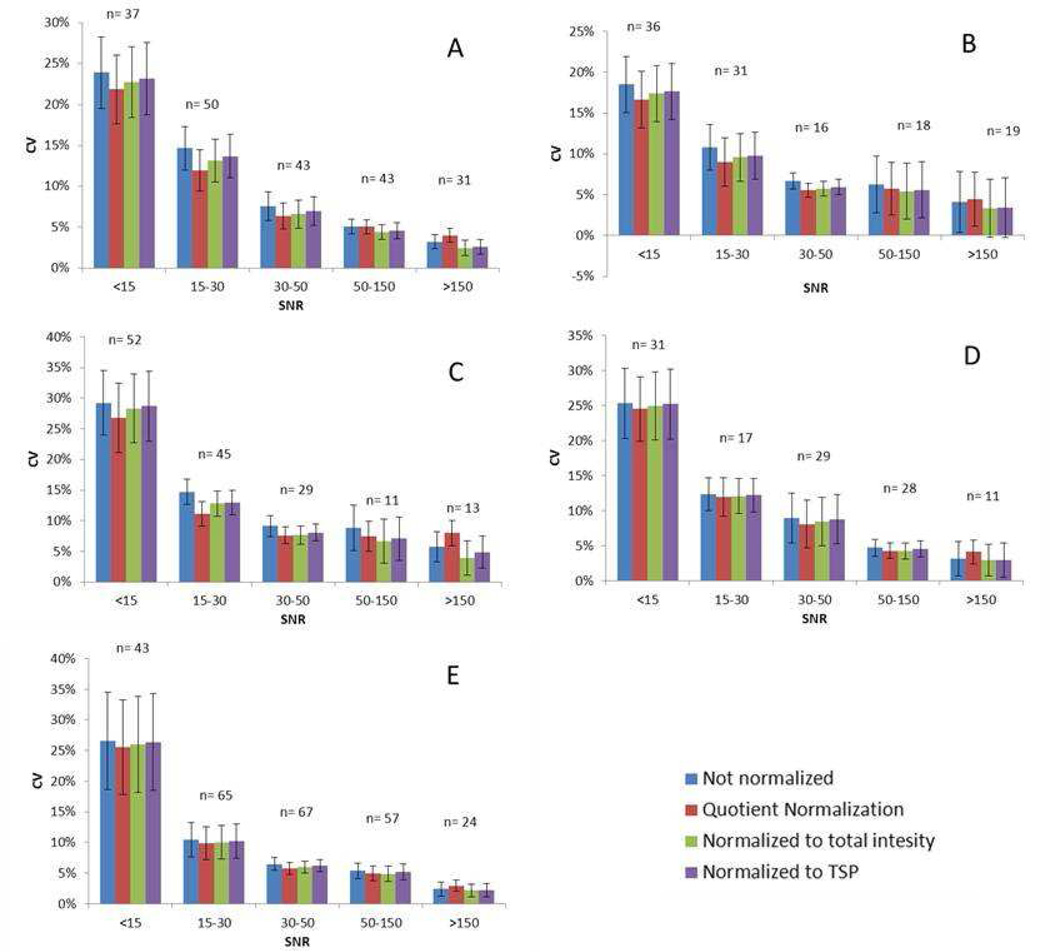
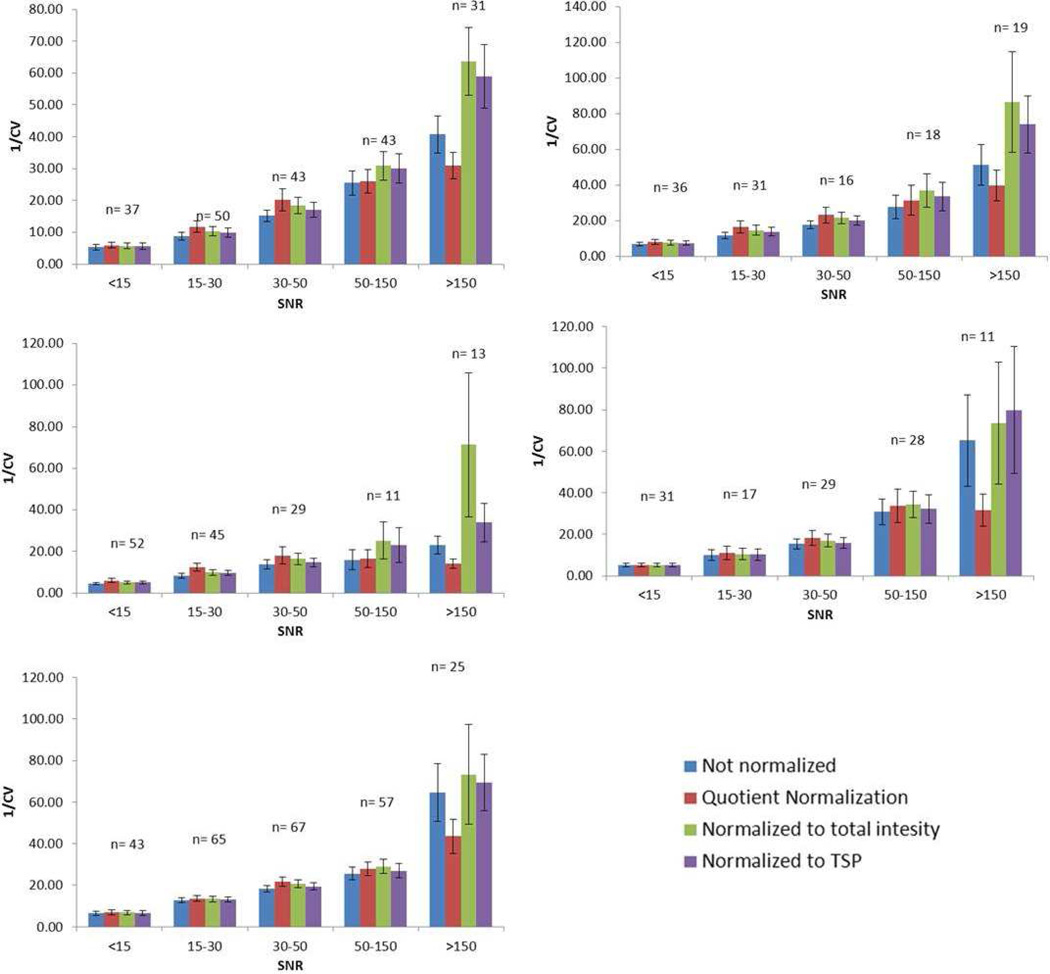
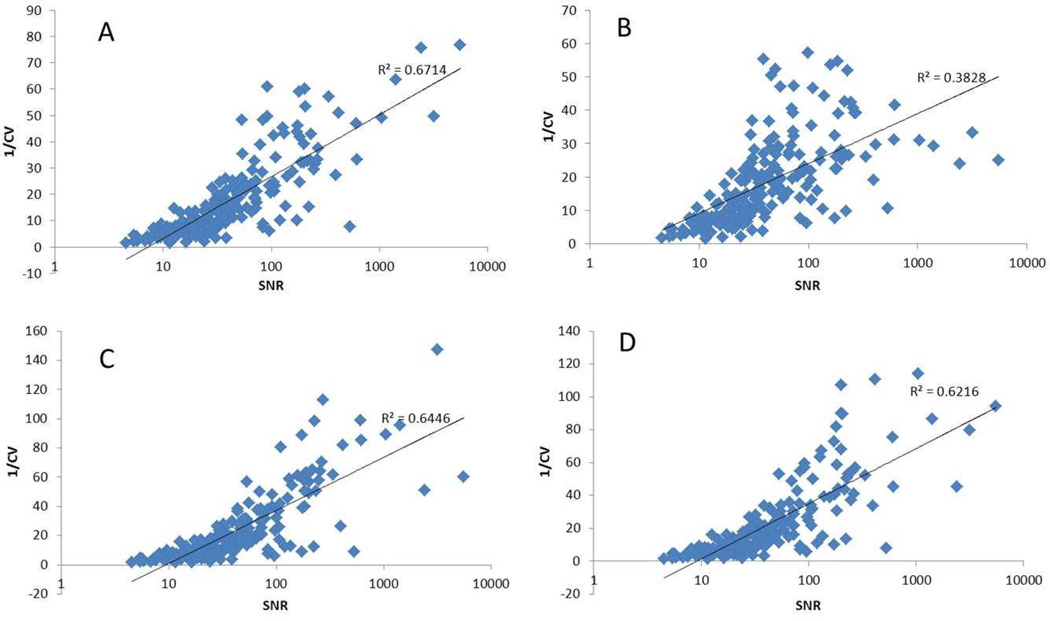
The influence of normalization method on the CV of all peaks was examined to determine whether or not a correlation existed between CV and SNR. Anywhere from 100 to 250 peaks were identified in the five different synthetic urine samples. The peak CVs were grouped into five categories based on SNR as follows: > 150, 150 – 50, 50 – 30, 30 – 15, and <15 (Table 3). Figure 1 shows the average CV in each SNR range for the four different normalization methods for each of the five synthetic urine samples. There were different numbers of buckets in each S/N category, but more than 15 peaks represented most groups. Figures 1A through 1E show the results for synthetic urine samples 1–5, respectively. The CV was below 15% for most peaks, except for peaks with a SNR<15, which had a significantly higher average CV compared to other categories, ranging from 15–30%. In general, an inverse relationship was observed between CV and SNR for all normalization methods (Figure 2). All peaks with SNR > 30 tended to have relatively small CVs (<10%). As a consequence, plots of CV versus SNR were relatively insensitive for illustrating an inverse relationship between CV and SNR for peaks in this category. A plot of 1/CV vs the log10(SNR) (Figure 3) for all peaks confirmed an inverse correlation between CV and SNR even for peaks in the highest SNR categories. Although there was some scatter about the best-fit straight line defined by the log10 relationship between CV and SNR, the trend was clear as supported by the correlation coefficients (R2) indicated in the plots. The CV was observed to decrease more slowly as the peaks became stronger. In other words, the CV of the strong peaks was influenced less by SNR than weak peaks.
Table 3.
The number of peaks observed in each of the five synthetic urine samples.
| Synthetic urine sample |
<15 | 15–30 | 30–50 | 50–150 | >150 | Total peaks |
|---|---|---|---|---|---|---|
| 1 | 37 | 50 | 43 | 43 | 31 | 204 |
| 2 | 36 | 31 | 16 | 18 | 19 | 120 |
| 3 | 52 | 45 | 29 | 11 | 13 | 150 |
| 4 | 31 | 17 | 29 | 28 | 11 | 116 |
| 5 | 43 | 65 | 67 | 57 | 25 | 257 |
Figure 1.
Graph showing the average CVs for the five SNR ranges for the five different synthetic urine samples that were investigated. Samples 1 – 5 are A – E, respectively. Error bars show the confidence intervals (CI) of the CV contained in that SNR range. CI was calculated by xm ± 1.96×sm, and xm is the mean of the data and sm is mean standard deviation.
Figure 2.
Graph showing the average reciprocal CVs for the five SNR ranges for the five synthetic urine samples. Samples 1 – 5 are A – E, respectively. Error bars show the confidence intervals (CI) of the CV contained in that SNR range. CI was calculated by xm ± 1.96×sm, and xm is the mean of the data and sm is mean standard deviation.
Figure 3.
Scatter plots of reciprocal of CV versus SNR of synthetic urine #1. The plots show A) no normalization, B) quotient normalization, C) Normalized to total intensity and D) normalized to TSP. The SNR was plotted using a log10 scale. All plots were fit using a log10 model.
The data also indicated small differences in CV depending on the normalization method applied. For small peaks (SNR<30), NN produced the largest CV, QN produced the smallest CV, and the other two methods had intermediate CV values. However, in majority cases, peaks with SNR > 50, and especially peaks with a SNR>150, the NTI and NIS normalization methods produced lower CVs than the other two methods. In other words, QN tended to produce smaller CVs for smaller peaks but larger CVs for larger peaks. Hence, QN tended to balance the relationship between CV and SNR, which was supported by the R2 values reported in Figure 3 and Table 4, where QN tended to produce smaller correlation coefficients compared to the other methods. Since large peaks tended to have smaller CVs, the slight increase in CV introduced by QN should not have a large effect on validation for peaks in this category, and therefore QN appears to be a good choice when low concentration peaks are of great interest.
Table 4.
R2 for the log10 fit of the five synthetic urine samples for four different data normalization methods.
| Synthetic urine sample |
NN | QN | NTI | NIS |
|---|---|---|---|---|
| 1 | 0.62 | 0.38 | 0.64 | 0.62 |
| 2 | 0.63 | 0.41 | 0.57 | 0.60 |
| 3 | 0.57 | 0.16 | 0.56 | 0.61 |
| 4 | 0.67 | 0.37 | 0.61 | 0.64 |
| 5 | 0.68 | 0.43 | 0.62 | 0.66 |
3.4 A Strict Inverse SNR and CV Relationship is Not Always Observed
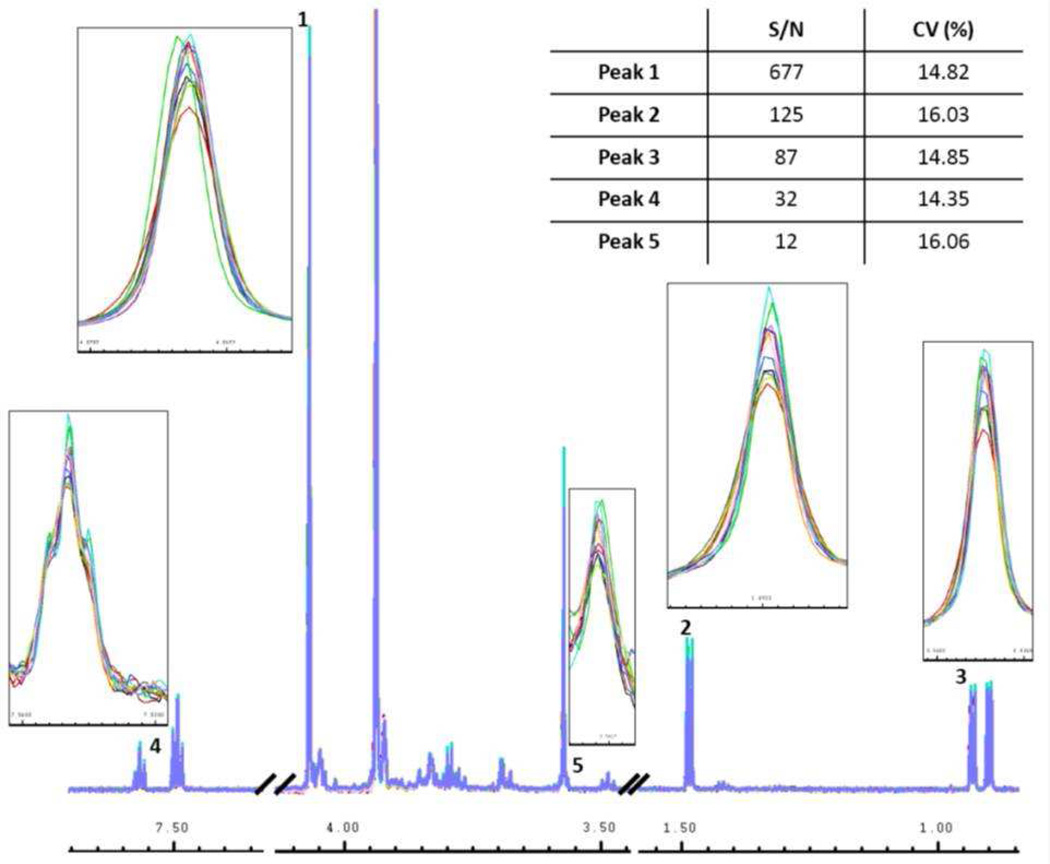
Though a log10 relationship between CV and SNR was supported by the R2 values, the fit of the CVs to log10 (SNR) indicate significant scatter. For example, if we consider one peak, or a small sample of peaks, in each SNR category, this inverse trend may not hold. To illustrate this point, the 12 spectra from synthetic urine sample #2 were overlaid and selected peaks with similar CVs in each SNR category shown in Figure 4. These peaks, labeled 1 – 5 in Figure 4, had SNRs ranging from 12/1 to 677/1. The CVs of the peaks, however, were quite similar, with values of 14.82%, 16.03%, 14.85%, 14.35%, and 16.06% for peaks 1 – 5, respectively. The insets in Figure 4 show that even though these peaks had significantly different SNRs, they all experienced about the same magnitude of intensity variance. These data illustrate that despite the general trend of an inverse relationship between CV and log10 (SNR), peaks corresponding to metabolites at high concentrations will not necessarily exhibit better analytical reproducibility, i.e. CV, compared to peaks associated with metabolites at weak concentrations.
Figure 4.
Overlay of all 12 spectra of synthetic urine sample #2 with the peaks marked 1–5 containing a peak from approximately each S/N range shown in Figure 1. The insets show a zoomed in look of each peak to illustrate magnitude of the variation occurring in each peak. The inset table shows the SNR of each peak and its corresponding CV.
4. Conclusions
Validation of potential disease biomarkers from NMR-based metabonomics studies is still far from routine. Currently, many potential metabolic biomarkers have been discovered from academic metabonomics research efforts for several cancers and other diseases, but at this time, virtually none are used for clinical diagnosis due to the difficulty associated with biomarker validation. In order for a biomarker to be validated, the analytical reproducibility, or CV, must be smaller than the effect size for diagnosis, i.e. the CV must be smaller than the smallest significant change in metabolite concentration associated with the detection of the disease. Otherwise, analytical reproducibility is a limiting factor in the ability to make a disease diagnosis. Several investigations have used CV analysis to determine the analytical reproducibility of NMR data used in metabonomic studies. Those papers have shown that good data typically contains CVs in the range of 0 – 10% and a high degree of reproducibility has been observed for several different types of samples.
In this paper we characterized the CVs of peaks from five synthetic urines that contained a variety of small molecules found in urine using replicated data collected over an 8-month period. Peak CVs ranged from less than 1% to greater than 60%. However, the majority of the CVs for the peaks fell in the range of 1 – 15%. Our data indicates that CV and SNR have a weak but clear inverse relationship and a log10 fit can be applied to represent the trend. The CV for peaks with small SNRs generally had larger CV values. Accordingly, metabolites at higher concentrations in solution tended to have smaller CVs, and generally should be easier to validate in terms of concentration changes associated with disease. On the other hand, small peaks (SNR<15) will generally require larger effect sizes in order for validation [30].
Finally, we explored how spectral normalization affected the magnitude and distribution of CVs. Our results indicated that QN produced higher average CVs in the large SNR range but produced smaller CVs in the small SNR range compared to NN. If lower concentration metabolites (small peaks) are putative biomarkers being considered for validation, QN is a good choice since it should have minimal influence on the CVs of strong peaks in the dataset but tends to increase the reproducibility of small peaks. CVs for large peaks were still in the 15% range after QN. In the study of the CVs of the TSP peaks, NTI had better performance than QN. Hence, NTI appears superior to QN for samples that have very small variation in total signal intensity, such as cell line study comparisons. For NTI and NIS, similar performance was observed, and they were both better than NN data in almost all categories. NTI or NIS is useful in reducing systematic errors during validation of potential disease biomarker. Urine concentrations can vary from sample to sample based on diet, water in-take, and diurnal variation among other things [29] which will make NTI less powerful. In such cases, QN will likely produce the best results for validation. While the various normalization techniques exhibit different performance depending on SNR, one can always tailor the acquisition conditions to obtain better SNR as required for the analysis. Collectively, these observations have important consequences for validation of NMR-based biomarkers of human disease, since the data suggests that it will be essential to characterize the CV of any given putative biomarker, notwithstanding its typical concentration in control patients.
Supplementary Material
Highlights.
We examine how signal-to-noise ratio is correlated with coefficient of variance for NMR resonances
We explore how data normalization affects coefficient of variance for NMR resonances
We assess how coefficient of variance should be considered for validation of biomarkers in NMR based metabonomics studies
Acknowledgments
MAK acknowledges support by a grant from the NIH/NCI (1R15CA152985). The instrumentation used in this work was obtained with the support of Miami University and the Ohio Board of Regents with funds used to establish the Ohio Eminent Scholar Laboratory where the work was performed. We would also like to acknowledge support from Bruker Biospin, Inc that enabled development of the statistical significance analysis software used in the analysis of the data reported in this paper. The data collection was conducted at the Ohio Biomedicine Center of Excellence in Structural Biology and Metabonomics at Miami University. The authors acknowledge Lindsey Romick-Rosendale for providing some of the raw NMR data used for the demonstration exercises. We acknowledge Dr. Donald Stec at Vanderbilt University for supplying the recipes for the various synthetic urine samples.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gowda GA, Zhang S, Gu H, Asiago V, Shanaiah N, Raftery D. Metabolomics-based methods for early disease diagnostics. Expert Rev Mol Diagn. 2008;8:617–633. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindon JC, Holmes E, Bollard ME, Stanley EG, Nicholson JK. Metabonomics technologies and their applications in physiological monitoring, drug safety assessment and disease diagnosis. Biomarkers. 2004;9:1–31. doi: 10.1080/13547500410001668379. [DOI] [PubMed] [Google Scholar]
- 3.Wilson ID. The practice of NMR spectroscopy in drug metabolism studies. Drugs and the Pharmaceutical Sciences. 2009;186:373. [Google Scholar]
- 4.NordströM A, Lewensohn R. Metabolomics: Moving to the clinic. J Neuroimmune Pharmacol. 2010;5:4–17. doi: 10.1007/s11481-009-9156-4. [DOI] [PubMed] [Google Scholar]
- 5.Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008;134:714–717. doi: 10.1016/j.cell.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 6.Kell DB. Metabolomic biomarkers: Search, discovery and validation. Expert Rev Mol Diagn. 2007;7:329–333. doi: 10.1586/14737159.7.4.329. [DOI] [PubMed] [Google Scholar]
- 7.Griffin JL, Kauppinen RA. Tumour metabolomics in animal models of human cancer. Journal of Proteome Research. 2007;6:498–505. doi: 10.1021/pr060464h. [DOI] [PubMed] [Google Scholar]
- 8.Serkova NJ, Glunde K. Metabolomics of cancer. Methods Mol Biol. 2009;520:273–295. doi: 10.1007/978-1-60327-811-9_20. [DOI] [PubMed] [Google Scholar]
- 9.Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: A review. Clin Cancer Res. 2009;15:431–440. doi: 10.1158/1078-0432.CCR-08-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serkova NJ, Niemann CU. Pattern recognition and biomarker validation using quantitative 1h-NMR-based metabolomics. Expert Rev Mol Diagn. 2006;6:717–731. doi: 10.1586/14737159.6.5.717. [DOI] [PubMed] [Google Scholar]
- 11.Taioli E, Kinney P, Zhitkovich A, Fulton H, Voitkun V, Cosma G, Frenkel K, Toniolo P, Garte S, Costa M. Application of reliability models to studies of biomarker validation. Environ Health Perspect. 1994;102:306–309. doi: 10.1289/ehp.94102306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puntmann VO. How-to guide on biomarkers: Biomarker definitions, validation and applications with examples from cardiovascular disease. Postgrad Med J. 2009;85:538–545. doi: 10.1136/pgmj.2008.073759. [DOI] [PubMed] [Google Scholar]
- 13.Keun HC, Ebbels TM, Antti H, Bollard ME, Beckonert O, Schlotterbeck G, Senn H, Niederhauser U, Holmes E, Lindon JC, Nicholson JK. Analytical reproducibility in (1)H NMR-based metabonomic urinalysis. Chem Res Toxicol. 2002;15:1380–1386. doi: 10.1021/tx0255774. [DOI] [PubMed] [Google Scholar]
- 14.Ebbels TM, Holmes E, Lindon JC, Nicholson JK. Evaluation of metabolic variation in normal rat strains from a statistical analysis of 1H NMR spectra of urine. J Pharm Biomed Anal. 2004;36:823–833. doi: 10.1016/j.jpba.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 15.Teahan O, Gamble S, Holmes E, Waxman J, Nicholson JK, Bevan C, Keun HC. Impact of analytical bias in metabonomic studies of human blood serum and plasma. Anal Chem. 2006;78:4307–4318. doi: 10.1021/ac051972y. [DOI] [PubMed] [Google Scholar]
- 16.Dumas ME, Maibaum EC, Teague C, Ueshima H, Zhou B, Lindon JC, Nicholson JK, Stamler J, Elliott P, Chan Q, Holmes E. Assessment of analytical reproducibility of 1h NMR spectroscopy based metabonomics for large-scale epidemiological research: The intermap study. Anal Chem. 2006;78:2199–2208. doi: 10.1021/ac0517085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sysi-Aho M, Katajamaa M, Yetukuri L, Oresic M. Normalization method for metabolomics data using optimal selection of multiple internal standards. BMC Bioinformatics. 2007;8:93. doi: 10.1186/1471-2105-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bijlsma S, Bobeldijk L, Verheij E, Ramaker R, Kochhar S, Macdonald I, Van Ommen B, Smilde A. Large-scale human metabolomics studies: A strategy for data (pre-) processing and validation. Analytical Chemistry. 2006;78:567–574. doi: 10.1021/ac051495j. [DOI] [PubMed] [Google Scholar]
- 19.Crawford L, Morrison J. Computer methods in analytical mass spectrometry - identification of an unknown compound in a catalog. Analytical Chemistry. 1968;40:1464. [Google Scholar]
- 20.Parsons HM, Ekman DR, Collette TW, Viant MR. Spectral relative standard deviation: A practical benchmark in metabolomics. Analyst. 2009;134:478–485. doi: 10.1039/b808986h. [DOI] [PubMed] [Google Scholar]
- 21.Lauridsen M, Hansen SH, Jaroszewski JW, Cornett C. Human urine as test material in 1H NMR-based metabonomics: Recommendations for sample preparation and storage. Anal Chem. 2007;79:1181–1186. doi: 10.1021/ac061354x. [DOI] [PubMed] [Google Scholar]
- 22.Saude E, Adamko D, Rowe B, Marrie T, Sykes B. Variation of metabolites in normal human urine. Metabolomics. 2007:439–451. [Google Scholar]
- 23.Gika HG, Theodoridis GA, Wilson ID. Liquid chromatography and ultra-performance liquid chromatography-mass spectrometry fingerprinting of human urine: Sample stability under different handling and storage conditions for metabonomics studies. J Chromatogr A. 2008;1189:314–322. doi: 10.1016/j.chroma.2007.10.066. [DOI] [PubMed] [Google Scholar]
- 24.Maher AD, Zirah SF, Holmes E, Nicholson JK. Experimental and analytical variation in human urine in 1H NMR spectroscopy-based metabolic phenotyping studies. Anal Chem. 2007;79:5204–5211. doi: 10.1021/ac070212f. [DOI] [PubMed] [Google Scholar]
- 25.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in H-1 NMR metabonomics. Analytical Chemistry. 2006;78:4281–4290. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- 26.Saude E, Adamko D, Rowe B, Marrie T, Sykes B. Variation of metabolites in normal human urine. Metabolomics. 2007;3:439–451. [Google Scholar]
- 27.Wu P, Otting G. Rapid pulse length determination in high-resolution NMR. Journal of Magnetic Resonance. 2005;176:115–119. doi: 10.1016/j.jmr.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 28.Warrack B, Hnatyshyn S, Ott K, Reily M, Sanders M, Zhang H, Drexler D. Normalization strategies for metabonomic analysis of urine samples. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 2009;877:547–552. doi: 10.1016/j.jchromb.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Slupsky CM, Rankin KN, Wagner J, Fu H, Chang D, Weljie AM, Saude EJ, Lix B, Adamko DJ, Shah S, Greiner R, Sykes BD, Marrie TJ. Investigations of the effects of gender, diurnal variation, and age in human urinary metabolomic profiles. Analytical Chemistry. 2007;79:6995–7004. doi: 10.1021/ac0708588. [DOI] [PubMed] [Google Scholar]
- 30.Goodpaster A, Romick-Rosendale L, Kennedy M. Statistical significance analysis of nuclear magnetic resonance-based metabonomics data. Analytical Biochemistry. 2010;401:134–143. doi: 10.1016/j.ab.2010.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.