Abstract
This unit describes a method for the separation of quadruplex species formed from the same sequence via size exclusion chromatography (SEC). Polymorphism is inherent to quadruplex formation, and even relatively simple quadruplex forming sequences, such as the human telomere sequence d(GGG(TTAGGG)3), can form a myriad of possible configurations. High Performance Liquid Chromatography (HPLC), especially reverse phase and anion exchange methods, has been a mainstay of nucleic acids research and purification for many decades. We have applied these methods for the separation of individual quadruplex species formed in a mixture from the same parent sequence.
Keywords: SEC, quadruplex, size exclusion chromatography, telomere
Introduction
This unit describes a facile method for the separation of quadruplex topologies, and possibly conformations, formed from the same sequence via size exclusion chromatography (SEC). This unit details a general method for the separation of quadruplex DNA (Basic Protocol: Size Exclusion Chromatography of G-Quadruplexes), outlines some useful procedures for handling and preparing quadruplex samples (Basic Protocol: Size Exclusion Chromatography of G-Quadruplexes, Support Protocol: Preparation of quadruplex sample), and presents and discusses the results for the separation of several quadruplex forming sequences.
Until recently there has been no effective method for accomplishing the isolation of a single quadruplex species from a mixture of configurations formed from the same sequence. This lack has led to a trend to modify quadruplex-forming sequences to achieve a single significantly enriched species for study with the goal that the product of this modification will be in some way embody the diverse range of species which may be actually be present in solution or in vivo. In reality, it is unlikely that the product of such modifications would be either the exact biologically relevant configuration or that the single “isolated” configuration and its properties would be sufficiently representative of the original diversity. This SEC method could be utilized for greater understanding of quadruplex formation and equilibrium. Separation may also afford the opportunity to separate more than one configuration from a single sequence and set of annealing conditions. With the possibility of separation comes the chance to study these complex systems in greater detail and to gain a better insight into quadruplex structure, stability, and formation as quadruplexes are becoming more and more attractive as drug targets and for biotechnology applications.
Basic Protocol: Size Exclusion Chromatography of G-Quadruplexes
This protocol describes a general method for the separation of quadruplex species formed from the same parent sequence. High Performance Liquid Chromatography (HPLC), especially reverse phase and anion exchange methods, has been a mainstay of nucleic acids research and purification for many decades and is now being applied to the separation of quadruplex DNA. This SEC method could be utilized for greater understanding of the polymorphism inherent to quadruplex formation and to enhance the study of quadruplex structure, the dynamics of quadruplex formation, the effects of small molecule quadruplex-interacting compounds, or simply produce a consistent configuration for nanotechnology or biotechnology applications. Each step of the method section is explained in detail and is then followed by a set of critical points. This method can be easily modified to accommodate individual sample requirements and procedures.
Strategic planning
It is important to plan the procedure thoroughly, especially if the user is relatively unfamiliar with high performance liquid chromatography. Materials must be gathered and prepared ahead of time. For example, putative quadruplex DNA must be annealed and HPLC mobile phase must be prepared and degassed well ahead of time. Several key steps such as equilibration and calibration of the SEC column cannot be accomplished on the fly and are very time consuming. The separation step often takes as long as 10 hours due to the slow flow rate and the relatively large total elution volume involved. Further, this method may need to be modified based on the constraints imposed by the handling and preparation protocols of individual samples. While this method attempts to be thorough, the common HPLC usage aspects should be considered during application. For example, each component of the buffers used in quadruplex sample preparation or chosen as the mobile phase should be checked for compatibility with the selected SEC column. Incompatible buffer components may damage the column and lead to unsatisfactory results. This method assumes that the user has access to the necessary equipment and the equipment and supplies available in any standard biochemistry laboratory.
Materials
Quadruplex sample (For sample specifications and preparations see SUPPORT PROTOCOL: Preparation of quadruplex sample)
HPLC
Superdex 75 10/300 size exclusion column (GE Healthcare 17-5174-01)
HPLC mobile phase (For buffer specification see Reagents and Solutions.)
Distilled, deionized water
Glycerol
Column Calibration Kit (GE Healthcare Catalog No. 28-4038-41)
1.5 ml microcentrifuge tubes for fraction collection (optional)
Set up HPLC and prepare column with mobile phase
-
Attach SEC column to HPLC pump output. Be sure to follow all manufacturer recommendations and procedures for individual column and HPLC equipment selection.
Size exclusion columns are generally shipped and stored with 20% ethanol/80% water as a preservative. The general method for setting up and equilibration of a SEC column includes washing out the preservative with water followed by equilibration with mobile phase. The manufacturer's recommendations for initial set up and equilibration should be followed. For the Superdex 75 10/300 column initial water wash of at least 2 column volumes, i.e. at least 50 ml, is recommended followed by similar equilibration with mobile phase. Flow rates of 0.1 to 0.2 ml per minute are appropriate.
Pump approximately 2 column volumes of distilled, deionized water through the column at 0.1 to 0.2 ml/min.
Switch to mobile phase reservoir and pump 2 column volumes through the column at 0.1 to 0.2 ml/min.
Column Calibration
-
4
Make sure that instrument is turned on and working properly.
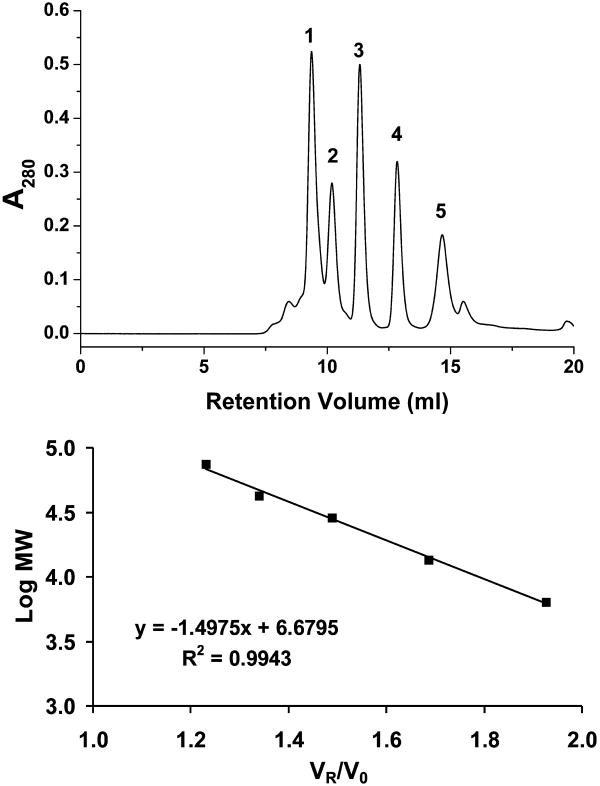
Column calibration serves to check not only the function of the column, but to ensure that the entire HPLC system, from sample injection to recording results, is working as it should. Size exclusion columns are typically calibrated with a set of proteins of known molecular weight. Each supplier will have their own recommendations for calibration and the manufacturer's specifications should be followed. For calibration purposes the Superdex 75 10/300 column was run at a flow rate of 0.10 ml/min and elution was monitored at A280.
-
5
Set up HPLC for use. Turn on typical UV-vis detectors about 20 minutes in advance for stable measurements. Set flow rate to 0.1 ml/min and set absorbance measurement to A280.
-
6
Prepare protein standards as described below or as individual kit requires.
The elution of proteins was calibrated using standards of known molecular weight from the Gel Filtration Calibration Kit LMW. (Catalog No. 28-4038-41 GE Healthcare) Protein standards, bovine lung aprotonin (6.5 kDa), ribonuclease A (13.7 kDa), carbonic anhydrase (29 kDa), ovalbumin (43 kDa), conalbumin (75 kDa), and blue dextran (1,000 kDa) were individually prepared in elution buffer with 5% glycerol at 10 mg/ml. An aliquot of blue dextran was then diluted to 1 mg/ml in elution buffer with 5% glycerol. The remaining protein standards were then combined such that each individual component had a concentration of 2.0 mg/ml. When using calibration kits with blue dextran do not mix protein standards with the blue dextran sample. Blue dextran can precipitate some protein samples.
-
7
Inject V0 sample. (Blue dextran)
The void volume (V0) is determined by injection of 100 μl of 1 mg/ml blue dextran in mobile phase with 5% glycerol.
-
8
Allow V0 sample to elute and record results.
-
9
Inject protein standards.
Protein standards, bovine lung aprotonin (6.5 kDa), ribonuclease A (13.7 kDa), carbonic anhydrase (29 kDa), ovalbumin (43 kDa), and conalbumin (75 kDa) were individually prepared in elution buffer with 5% glycerol at 10 mg/ml and combined such that each individual component had a concentration of 2.0 mg/ml and injected in 100 μl aliquots.
The entire set of protein standards chosen for calibration may be injected in one sample if they are well separated and easily identifiable. If not, each standard may be injected singly or in sets or two or more. If the protein samples selected are not well separated slowing the flow rate may better resolve them. In general, if the column, flow rate, and buffer selected cannot resolve a set of protein standards ranging from roughly 80kDa to 6kDa, then the condition will not be appropriate for quadruplex separation.
-
10
Allow VR samples to elute and record results.
The resulting retention volumes (VR) can be graphed as VR/V0 vs. Log MW to produce a linear calibration.
-
11
Graph the results as VR/V0 vs. Log MW to produce a linear calibration. Compare this result to the results expected for the column.
Periodic calibration is necessary to insure that column performance is at a maximum (Figure 1). Calibration with protein standards is convenient because it allows direct comparison to known performance data for the column. To date, no quadruplex calibration standards exist. It should also be kept in mind that commercially available protein standards can yield different results based on differences in buffer conditions.
Figure 1.
Calibration of the Superdex 75 10/300 column. TOP: A mixture of proteins of known molecular weights resolved on the column. 1. conalbumin (75 kDa) VR=9.36, 2. ovalbumin (43 kDa) VR=10.19, 3. carbonic anhydrase (29 kDa) VR=11.32, 4. ribonuclease A (13.7 kDa) VR=12.84, 5. bovine lung aprotonin (6.5 kDa) VR=14.66. BOTTOM: The linear calibration curve developed by graphing VR/V0 vs. Log MW. V0 was determined to be 7.61 ml by injection of blue dextran as described in the method.
Separate quadruplex sample
-
12
Make sure that instrument on turned on and working properly.
-
13
Set up HPLC for use. As in the calibration step, turn on typical UV-vis detectors about 20 minutes in advance for stable measurements. At this point the HPLC detector can be set to monitor A260 or A280.
-
14
Set flow rate to 0.05 ml/min.
Before injection of quadruplex sample, set the flow rate to 0.05 ml/minute and insure that the instrument is set to record the correct absorbance wavelength. For DNA samples this value can remain at A280 or be set to A260 as desired.
Careful attention to flow rate is, in general, critical for success when using SEC. For quadruplex DNA, separation efficiency is reduced drastically by increased flow rates. Care should also be taken that total run time allows for total elution of previous samples.
-
15
Inject quadruplex sample.
Sample injection can be accomplished manually or via the autosampler. Injection volumes can vary from 10 to 500 μl depending on the concentration of the DNA sample, column specifications, and detector specifications. Initial trial runs are useful for determining the appropriate DNA concentration and sample size.
-
16
Allow quadruplex sample to elute and record results. If desired, collect fractions as described below.
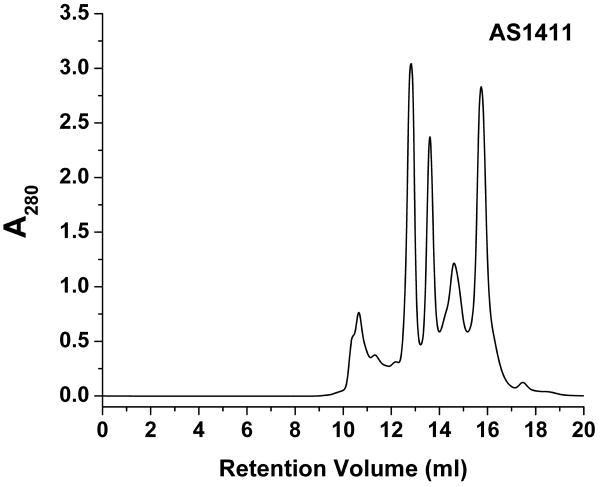
General total elution volumes are on the order of 20 to 30 ml, therefore total run times can be up to ten hours. Faster flow rates may be possible for quadruplex forming sequences which are well separated by the column. Initial trial runs are useful for determining appropriate flow rates. Elution of quadruplex DNA can vary drastically depending on sequence. The best results, such as those for AS1411 (Figure 2), will produce well isolated peaks.
Figure 2.
Typical elution profile of AS1411. At a glance, there are at least eight isolatable species with varying levels of separation.
Sample elution and collection of fractions (Optional)
-
17
During sample elution fractions can be collected with any commercial fraction collector or collected by hand as each fraction elutes.
-
18
If the fraction collection method does not adequately match fractions with chromatogram, then orientation of chromatogram to fraction can usually be accomplished via standard UV-vis analysis with low volume cuvettes or via low volume devices such as the Nanodrop (Thermo Scientific).
Since UV-vis detectors generally have much smaller path lengths than desktop UV-vis instruments, the actual optical density of the collected fraction will be much higher than indicated by the detector. For the Waters 2998 UV-vis Photodiode Array Detector the path length is 1 mm, thus an A260 of 1.0 will result in a fraction with an A260 of 10.0. If one assumes a typical optical density for quadruplex forming sequences of, say, 250,000 L/(M·cm), then a sample with an A260 of 10.0 has a concentration of 40 μM. In many cases the collected fractions will be concentrated enough so that further dilution may be necessary for UV-vis measurements or other downstream applications.
-
19
When the desired fractions have been isolated and collected they should be refrigerated or frozen immediately.
-
20
Once the individual fractions have been collected they may be combined with succeeding sets of identical fractions.
In the case of a downstream application requiring a fairly concentrated sample such as NMR or calorimetry it may be possible to use individual fractions as samples without further processing. If more concentrated samples are needed then concentration via a commercially available spin column, such as the Millipore Microcon YM-3, may be an option. Samples may also be lyophilized and re-hydrated at lower volumes to concentrate them. Fractions of samples which are prone to re-equilibration or degradation of the quadruplex species should be frozen at -80°C until use. Individual samples should be tested to insure that they are stable in the conditions they will be exposed to via a high-resolution technique such as NMR.
Support Protocol: Preparation of quadruplex sample
Quadruplex DNA samples should be prepared as individual lab protocols or experimental needs require. All oligonucleotides used to produce data in this protocol (Table 1) were obtained from IDT as purified, dry, de-salted pellets. The oligonucleotides were then dissolved in water and dialyzed into buffer and diluted to a final concentration ranging from 200 μM to 1 mM. Samples were then equilibrated for ten minutes at 100°C in a water bath, followed by gradual cooling to room temperature overnight. The DNA samples were then refrigerated until use.
Table 1.
Quadruplex forming sequences used in this method. All oligonucleotides were ordered from Integrated DNA Technologies of Coralville, Iowa.
| Sequence Name: | Sequence: |
|---|---|
| AS1411 | GGTGGTGGTGGTTGTGGTGGTGGTGG |
| Her2 | AGGAGAAGGAGGAGGTGGAGGAGGAGGGC |
| c-myc | TGGGGAGGGTGGGGAGGGTGGGGAAGG |
| G4T4G4T4G4T4 | GGGGTTTTGGGGTTTTGGGGTTTT |
| HuTel 22 | AGGGTTAGGGTTAGGGTTAGGG |
| HuTel Hybrid 1 | AAAGGGTTAGGGTTAGGGTTAGGGAA |
| HuTel Hybrid 2 | TTAGGGTTAGGGTTAGGGTTAGGGTT |
| TG4T | TGGGGT |
Materials
DNA sample
Annealing buffer
Distilled deionized water
Dialysis unit
1 to 2 liter beakers
Hotplate or water bath
Magnetic stir plate
Teflon stir bar
Aluminum foil
Dissolve DNA sample in distilled, deionized water. In general, it is better to dissolve the sample at a slightly higher concentration, say 1.5×, than the final desired concentration.
-
Dissolved DNA sample should be placed into the dialysis unit of choice, dialyzed against distilled, deionized water, and then dialyzed into the annealing buffer. For samples of up to 1 ml in volume, dialysis into 1000 ml volumes is recommended with at least 2 changes of buffer. Gentle stirring is recommended.
The dialysis unit should be of the appropriate size and MWCO for the sequence being prepared. Most quadruplex forming sequences are no more than 30 bases in length with an average molecular weight of about 7,000 to 8,000. The authors prefer Slide-A-Lyzer Dialysis Cassettes of 0.2-0.5 ml total volume and 2,000 MWCO (Thermo Sci. Cat. No. 66205.) for this step. Other systems can be used.
After dialysis the sample should then be transferred to an appropriately sized microcentrifuge tube, approximately 1.5 to 2 ml total volume, with a screw cap featuring a rubber gasket.
Turn on water bath. Allow time for water bath to come to 100°C. Alternatively, a laboratory beaker with boiling water may be used instead. If using a beaker of boiling water 1 to 2 liters of water volume is recommended.
Immerse quadruplex samples in water bath using a commercially available float. Do not allow the top of the tube and cap to be immersed to reduce the possibility for contamination.
After 10 minutes, turn off water bath and allow sample to cool in the water bath over night. If using a beaker of boiling water remove beaker from heat source and cover with aluminum foil to allow slower, more even cooling.
-
7
Samples should be checked for concentration and to confirm quadruplex formation.
Reagents and Solutions
Quadruplex forming oligonucleotide sample
Any quadruplex forming sequence is sufficient, but the best separation results have been achieved with sequences un-related to the human telomere sequence. Results for several sequences will be reported (Table 1.).
NOTE: Any oligonucleotide sample used for this procedure should be dissolved and dialyzed into water before dilution/dialysis into annealing buffer to insure purity. Do not depend on the quantities of DNA stated by the manufacturer. Measure DNA quantities and check purity directly whenever possible.
Quadruplex annealing buffer
Many different annealing buffers and profiles are acceptable. For this method the annealing buffer is 100 mM KCl, 25 mM K2HPO4, pH=7.0. The buffer is prepared with K2HPO4 and titrated from an approximate pH of 9.0 to pH=7.0 with HCl so that the total K+ concentration is known.
NOTE: Phosphate and cacodylate buffers are generally best for this application. Buffers such as Tris, Tricine, HEPES, MOPS, TAPS, MES, BES, TES and acetate are not recommended for use with this method. Phosphate buffers should be avoided if working with some divalent ions such as Mg2+ and Ca2+. Due to the sensitivity of quadruplex formation to ion type and concentration care should be taken not to introduce inappropriate ions as counter ions to buffer constituents.
CAUTION: Cacodylate buffer contains arsenic and should be handled and disposed of accordingly.
SEC column
All size exclusion columns are similar in construction and function. For this method the Superdex 75 10/300 column (Catalog No. 17-5174-01) from GE Healthcare was used.
NOTE: Other SEC columns may be used. Various column matrixes may have slightly different characteristics such as particle size, pore size, and hydrodynamic properties so results may vary. Experimentation to reveal the best column material and column configuration for your application is recommended.
HPLC system
The HPLC system should be capable of reliable flow rates of 0.1 - 0.05 ml/minute and be equipped with either a manual injection port or an autosampler system, and a UV/fluorescence or UV-vis detector. Computer control and fraction collection are optional. In this case a Waters 600 pump and controller equipped with a Waters 2998 UV-vis Photodiode Array Detector, a Waters 2707 Autosampler, and a Waters Fraction collector III was used. The autosampler was equipped with a 250 μl sample loop. The system was operated via computer equipped with Empower Software (Waters).
HPLC mobile phase
Typically has the same as the annealing buffer, but the composition may vary. Great care and consideration should be used when choosing mobile phase conditions for quadruplex separation. Quadruplex formation has been shown to be sensitive to variations in buffer composition and to the presence of organic solvents such as PEG or acetonitrile. Any variation between mobile phase and annealing buffer may cause variation in stability of the various quadruplex species in solution. Buffer components should have little or no absorbance in the far UV region. For this method the mobile phase was 100 mM KCl, 25 mM K2HPO4, pH=7.0.
NOTE: All buffers, solvents and samples used should be filtered to 0.22 μm. Alternatively, samples may be spun at max speed for 10 minutes.
Column calibration kit
GE Healthcare (Catalog No. 28-4038-41)
Mobile phase with 5% Glycerol
For hydration, mixing, and storage of protein samples. 5 ml glycerol added to mobile phase to a total volume of 100 ml.
Optional miscellaneous supplies
Additional reagents, equipment, and supplies for dissolving samples, dialyzing samples, collecting fractions, filtering buffers, UV-vis measurements, and other non-specified operations such as 1.5 ml and 2.0 ml microcentrifuge tubes, bottle top and syringe filters, storage bottles, ultra-pure water, and glassware as needed.
Commentary
Background Information
In the presence of certain monovalent cations, guanine-rich DNA or RNA with at least four runs of two guanines can form three-dimensional structures called G-quadruplexes. G-quadruplexes are made up of stacks of two or more square planer arrays of four guanines, called a G-quartet. Each G-quartet involves Hoogsten hydrogen bonds between the guanine N1 and N2 amino and the O6 and N7 imino group, respectively, and coordination of a cation such as Na+, K+, or NH4+ to the O6 of the guanines (Phan, Kuryavyi et al. 2007). Quadruplex-forming sequences have been found throughout the genome (Huppert and Balasubramanian 2005) but are most commonly associated with telomeric DNA. The telomere, a repeat of d(GGGTTA) in humans, caps the ends of chromosomes and participates in critical functions such as regulating cell cycle, replication, and cell senescence (Counter, Avilion et al. 1992; Wright, Tesmer et al. 1997). Activation of telomerase, an enzyme that is responsible for maintaining the length of telomeric DNA, has been found to be involved in 90% of all cancers (Kim, Piatyszek et al. 1994; Shay and Bacchetti 1997) while not expressed in normal somatic cells. Potential quadruplex-forming sequences have also been found in the promoter regions of many oncogenes, such as c-myc (Ambrus, Chen et al. 2005), c-kit (Fernando, Reszka et al. 2006; Phan, Kuryavyi et al. 2007; Hsu, Varnai et al. 2009), bcl-2 (Dai, Chen et al. 2006; Dai, Dexheimer et al. 2006), VEGF (Sun, Guo et al. 2005), and HIF-1α (De Armond, Wood et al. 2005), and in the 5′-untranslated regions of RNA oncogenic sequences (Huppert and Balasubramanian 2005; Patel, Phan et al. 2007). A search of the human genome has revealed in excess of 350,000 potential quadruplex-forming sequences (Huppert and Balasubramanian 2005; Huppert and Balasubramanian 2007). Therefore quadruplex-forming sequences have become attractive targets for therapeutic strategies. Specifically, there is strong interest in using small molecules to stabilize these complex structures and therefore regulate telomerase function or oncogene expression (Hahn, Stewart et al. 1999; Neidle and Parkinson 2002; Neidle 2010). The first successful application of this concept was the discovery of a bisamidoanthroquinone that was shown to inhibit human telomerase by binding to and stabilizing a G-quadruplex formed in the human telomere (Sun, Thompson et al. 1997). This work was followed by stabilization of the quadruplex formed by the sequence in the promoter region of the proto-oncogene c-myc by a cationic porphyrin, TMPyP4, which suppressed c-myc transcriptional activation (Simonsson, Pecinka et al. 1998; Siddiqui-Jain, Grand et al. 2002).
Development of quadruplex-based therapeutics is not limited to their role as potential therapeutic drug targets, quadruplexes themselves can act as therapeutic agents. Some examples include AS1411, which is currently in phase IIB clinical trials, T40214, an inhibitor of IL-6-stimulated Stat3 activation and suppressor of Stat3-mediated up regulation of bcl-x and mcl-1 gene expression, and T22AG a potent inhibitor of the nuclear factor necessary for Ki-ras expression (Jing, Li et al. 2003; Cogoi, Quadrifoglio et al. 2004; Ireson, Djeha et al. 2005; Jing and Tweardy 2005; Ireson and Kelland 2006). Quadruplexes have also been shown to have antiviral activity and have been demonstrated to be effective against HIV-1 in vivo (Bishop, Guy-Caffey et al. 1996; Suzuki, Miyano-Kurosaki et al. 2002). Quadruplex-forming sequences have also been found in many other species and researchers are now beginning to uncover putative G-quadruplex forming sequences in prokaryotes creating possibilities for a wide range of potential applications (Rawal, Kummarasetti et al. 2006). Quadruplex DNA is also of interest for other bio/nanotechnology applications such as biosensors, nano-switches, nano-wires, and as possible logic gates (Huppert 2007). Due to their ability to recognize proteins with a high degree of specificity, and because of their high degree of stability and nuclease resistance, quadruplexes are rapidly becoming attractive for development of novel therapeutics and other bio/nanotechnology applications (Cogoi, Quadrifoglio et al. 2004; Jing, Sha et al. 2005; Qi, Lin et al. 2006).
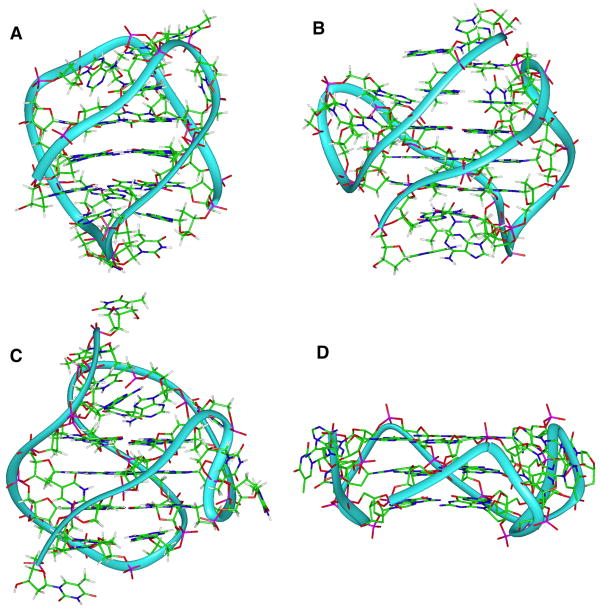
Polymorphism is an intrinsic feature of quadruplex formation for most quadruplex-forming DNA sequences (Figure 3). When factors such as strand orientation, loop type and arrangement, and glycosyl torsion angles are considered, even the relatively simple sequences, such as the human telomeric sequences d(GGGTTA)n, can theoretically fold into over 200 unimolecular conformations (Lane, Chaires et al. 2008). Formation of tetramolecular and bimolecular structures can lead to an even greater degree of polymorphism. For more complex sequences, such as those typically found in promoter sequences, the potential for polymorphism is multiplied by the possibility of the formation of alternative G-quartets due to different numbers of guanines in each run or by more than four runs of guanines in the sequence. Factors such as ion type, DNA concentration, the presence of organic solvents or biological molecules, and even annealing protocols can also greatly influence which quadruplex conformations are formed (Li, Correia et al. 2005; Webba da Silva 2007; Xue, Kan et al. 2007; Lane, Chaires et al. 2008).
Figure 3.
Examples of possible topologies of the human telomere sequence. A) Basket (Protein database code: 143D), B) hybrid1 (2HY9), C) Hybrid2 (2JPZ), and D) propeller (1KF1).
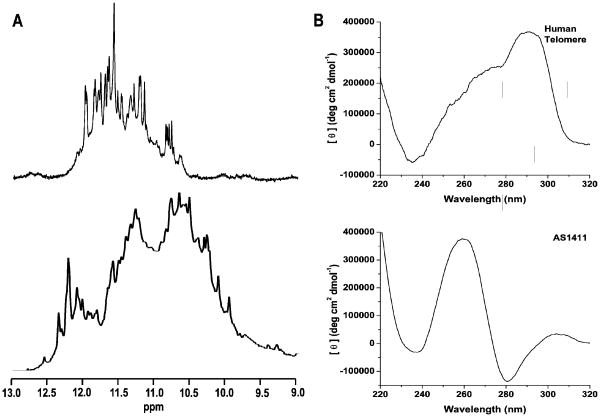
This innate polymorphism is an issue for those who wish to investigate quadruplex structure, the dynamics of quadruplex formation, the effects of small molecule quadruplex-interacting compounds, or simply produce a consistent configuration for nanotechnology or biotechnology applications. Structural and biophysical studies of quadruplex DNA have been severely hindered by the inability to isolate and study the individual quadruplex species that may occur in solution. Low resolution methods such as CD, UV-vis, AUC, gel electrophoresis, or calorimetry generally are not able to differentiate between quadruplex conformations with nearly identical physical properties. It has been shown that the CD spectra of quadruplex mixtures present as an average of the features of each individual component in the mixture (Dailey, Miller et al. 2010). Another example of this issue can be found in the use of the electromobility shift assay (EMSA) to measure interaction between DNA/RNA binding proteins such as nucleolin and quadruplex DNA. The goal in this assay is to measure interaction of two individual species, but when multiple quadruplex species are present then questions arise as to the specificity of each individual species and the possibly of perturbing the equilibrium of species in solution. High resolution techniques such as NMR are of limited use for examining the specific details of these complex mixtures (Dailey, Miller et al. 2010) (Figure 4).
Figure 4.
A. 1H NMR of the imino regions of the human telomere sequence (AG3(T2AG3)3) and of AS1411 ((G2T)4TGT(G2T)3G2). The human telomere exhibits 23-24 GN1H signals, indicative of the presence of two quadruplex species. AS1411 yields an irresolvable mixture of GN1H resonances known to be the result of more than 8 quadruplex species. B. CD of the human telomere sequence and of AS1411. The CD data provides no indication of the polymorphism evident from the NMR data. Conventional wisdom would associate both sets of CD data with a single, parallel stranded species in solution.
Polymorphism has been generally dealt with by altering the sequence of interest to produce an assumed enriched species for study. This usually means either adding bases at the 5′ or 3′ ends of the sequence or lengthening, shortening, or eliminating guanines from putative loop regions to ensure that a single fold is enriched (Dai, Carver et al. 2008; Yang and Okamoto 2010). Sequence modifications can also include chemical modification of the guanine base or sugar. Inclusion of O6-methyl guanine, inosine, or 6-thioguanine has been shown to destabilize quadruplex formation (Marathias, Sawicki et al. 1999; Spackova, Cubero et al. 2004; Mekmaysy, Petraccone et al. 2008; Petrovic and Polavarapu 2008). Substitution of 8-aminoguanine accelerates formation of tetramolecular parallel quadruplexes such as those formed by TG4T and TG5T (Gros, Avino et al. 2008). Incorporation of 8-methylguanine or 8-bromoguanine into a quadruplex forming sequence is known to stabilize quadruplex structures by assuming the syn glycosyl conformation (Esposito, Randazzo et al. 2004; Virgilio, Esposito et al. 2005; Virgilio, Esposito et al. 2005). RNA and LNA also force adoption of a syn glycosyl bond conformation and are useful for controlling the resulting quadruplex topology (Tang and Shafer 2006; Kumar and Maiti 2007; Qi and Shafer 2007; Bonifacio, Church et al. 2008). Replacement of guanine in telomeric sequences with 7,8-dihydro-8-oxoguanine, the product of oxidative damage to guanine, produces stabilization when occurring at the 5′ position of a G triplet and destabilization when occurring in the middle of a G triplet (Szalai, Singer et al. 2002). Modification of the sugar phosphate backbone by insertion of 5′-5′ or 3′-3′ polarity inversion has also been shown to have a large effect on quadruplex formation and stability (Esposito, Virgilio et al. 2005). Reduction of quadruplex polymorphism does not stop at sequence modification. Solvent conditions can greatly affect the range of configurations formed. Choice of monovalent cation, primarily K+ or Na+, or inclusion of divalent cations, such as Mn2+, Co2+, Ni2+, Mg2+, Pb2+, or Sr2+, can stabilize or destabilize G-quadruplex structures. For example, K+ concentration alone has been shown to determine the overall topology for some human telomere based sequences (Gray, Li et al. 2009; Gray, Petraccone et al. 2010), and inclusion of divalent cations has been shown to induce a transition from an antiparallel to a parallel G-quadruplex structure for the G4T4G4 sequence (Blume, Guarcello et al. 1997; Miyoshi, Nakao et al. 2001). Addition of organic or biological molecules such as PEG, acteonitrile, proteins, or polysaccharides can contribute greatly to quadruplex formation and stability (Xue, Kan et al. 2007; Miller, Buscaglia et al. 2010).
While these methods can yield a less complex mixture of species and have been used to produce many solution and crystal structures, and topologies deduced from low resolution methods, the relevance of these results is uncertain at best. The general concept seems to be that if a mixture of quadruplex topologies exists in equilibrium that the alterations simply perturb that equilibrium in favor of a single topology. Even if this is so, the results of such modifications and manipulations represent only a small number of the topologies possible in solution. These results may not reflect the diversity of species that may be actually present in solution for the parent sequence and the modified sequences represent a further removal from conditions found in vivo (Lane, Chaires et al. 2008). Further, in many cases the results of these modifications are evaluated with low resolution techniques such as CD. The incorrectly generally accepted assumption for interpreting CD data related to quadruplex structure states that parallel quadruplexes generally display a negative peak near 240 nm and a positive peak near 265 nm, and that antiparallel quadruplexes are characterized by a negative peak near 260 nm and a positive peak near 295 nm (Balagurumoorthy, Brahmachari et al. 1992; Giraldo, Suzuki et al. 1994; Hazel, Huppert et al. 2004; Kypr, Kejnovska et al. 2009). However, many exceptions to this assumption are known to exist (Jing, Rando et al. 1997; Dapic, Bates et al. 2002; Dapic, Abdomerovic et al. 2003). In reality, CD data is a result of base stacking interactions and does not provide any information on strand orientation so no inference of overall quadruplex structure or topology is possible without supporting structural data.
Thus far small molecule quadruplex-based therapeutics have yet to live up to their initial promise. While many attempts have been made to find small molecules that stabilize quadruplex DNA and thereby regulate cancer cell growth by either impeding telomerase function or via oncogene regulation, none of these potential drugs have yet made it to market. Only one small molecule, CX-3543, has been successful in clinical trials. CX-3543, a small molecule fluoroquinone derivative also known as Quarfloxin. It was designed to act via interaction with a quadruplex formed in the c-myc promoter region, but was later discovered not to function as designed, but to inhibit nucleolin/ribosomal DNA complexation, thus interfering with Pol I transcription and inducing apoptosis (Drygin, Siddiqui-Jain et al. 2009). There are several likely reasons for the disconnect between potential and reality for therapeutics based on quadruplex DNA. First, in any drug screening process it is difficult to isolate interaction with the desired target from secondary interactions that may cloud interpretation of results (Hopkins 2009; Keiser, Setola et al. 2009). This is especially true of quadruplex binding compounds as there may be a cornucopia of potential targets available in vivo. Second and more importantly to the topic being discussed here, the extreme polymorphism inherent in quadruplex formation and our lack of understanding of these complex systems and their possible structures and roles in vivo makes the screening of small molecules for any sort of specificity for these structures problematic at best. This likely contributes to the polypharmacology displayed by many “selective” quadruplex-binding compounds. Conversely, once quadruplex polymorphism is better understood then perhaps this feature may contribute to producing drugs that fully realize the initial promise of the quadruplex.
Ordinarily the solution to the dilemmas presented by the extreme polymorphism associated with quadruplex formation would be separation. However, until recently, separation of diverse quadruplexes formed by the same sequence was unreported. One of the simplest HPLC methods to employ is size exclusion chromatography. SEC is often used to estimate the molar mass of proteins, DNA, and other polymers. At first glance it seems counterintuitive to choose SEC for such an application as one would suppose that quadruplex configurations formed from the same sequence must have the same molecular weight and hydrodynamic properties. It is important to realize that the results of SEC are only an approximation of molecular weight, not an absolute value. Normally SEC separates according to particle size. While molar mass does roughly correlate with particle size, other factors such as folding compactness and hydrodynamic properties of the material in question also directly relate to the apparent particle size in solution. This is also true of other methods, such as analytical ultracentrifugation (AUC) and gel electrophoresis, which are used to generate similar data. AUC in particular can be used to estimate molecular weight and, through sedimentation velocity experiments, can provide information about hydrodynamic properties, thermodynamic properties, and particle shape (Garbett, Mekmaysy et al. 2009). Gel electrophoresis, which separates as a function of particle size vs. charge, has been used to estimate the differences in quadruplex configuration of the human telomere sequence based on changes in size and compactness of the quadruplex structure (Xue, Kan et al. 2007). Keeping these limitations in mind, SEC has many advantages over other methods. First, SEC is non-destructive. Samples subjected to SEC can be collected and reused for other applications. SEC is ideal for downstream applications such as ITC, DSC, CD, UV-vis, and NMR as it accomplishes a complete buffer exchange during the procedure while preserving a buffer reservoir that can be used to dilute samples, to prepare other samples, and for baseline measurements. This inherent mildness makes SEC ideal for preserving biological activity. Second, SEC is easily scalable. This method achieves significant separation with a Superdex 75 10/300 column. Should increased throughput be desired, larger sample sizes can be accommodated by wider diameter columns. For SEC increased separation is generally a function of column length. Therefore use of a longer column or simply running two or more columns in series can easily increase separation. Third, readily available automation makes HPLC an extremely time efficient method. The ease of separation, fraction collection, and versatility identifies SEC as a new tool for quadruplex research.
Critical Parameters and Troubleshooting
Size exclusion chromatography is a relatively straight forward technique and is generally very easy to execute. However, there are several key points to this method that must be adhered to. First, the flow rate of 0.05 ml/min should not be increased unless it can be clearly demonstrated that an increased flow rate does not impair resolution. This may be the case since many different SEC columns are available for use. Second, any buffer introduced to the SEC column should be investigated thoroughly for column compatibility. Third, the best results are achieved when the annealing buffer matches the mobile phase. In some cases, such as the addition or of an organic constituent like PEG or acetonitrile or a change in ion type or concentration, differences between buffer and mobile phase can result in complete rearrangement of quadruplex configuration resulting in a misleading chromatographic outcome. In some cases an annealing buffer/mobile phase mismatch can result in damage to the column. For example, if the annealing buffer includes divalent ions such as Ca2+ and Mg2+, as may be necessary for formation and stability of a specific quadruplex, then a mobile phase containing phosphate will not only result in precipitants, possibly damaging the column, but in inaccessibility of these ions in solution. Finally, while quadruplex structures are known for their stability and resistance to nucleases, some quadruplex samples, once isolated, are known to re-equilibrate. Therefore any fractions that are collected for downstream applications should be refrigerated or frozen at once to prevent degradation. If resolution is not sufficient for separation of chosen quadruplex forming sequence then a longer column may be used. Alternatively, two columns in series may provide the desired resolution. Any changes to the method as outlined should be investigated thoroughly before implementation. General laboratory safety is also a concern when hazardous materials, such as organic solvents and cacodylate buffers, are involved.
Anticipated Results
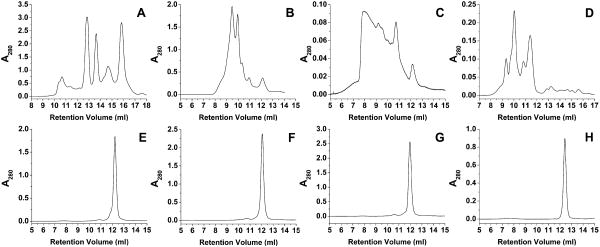
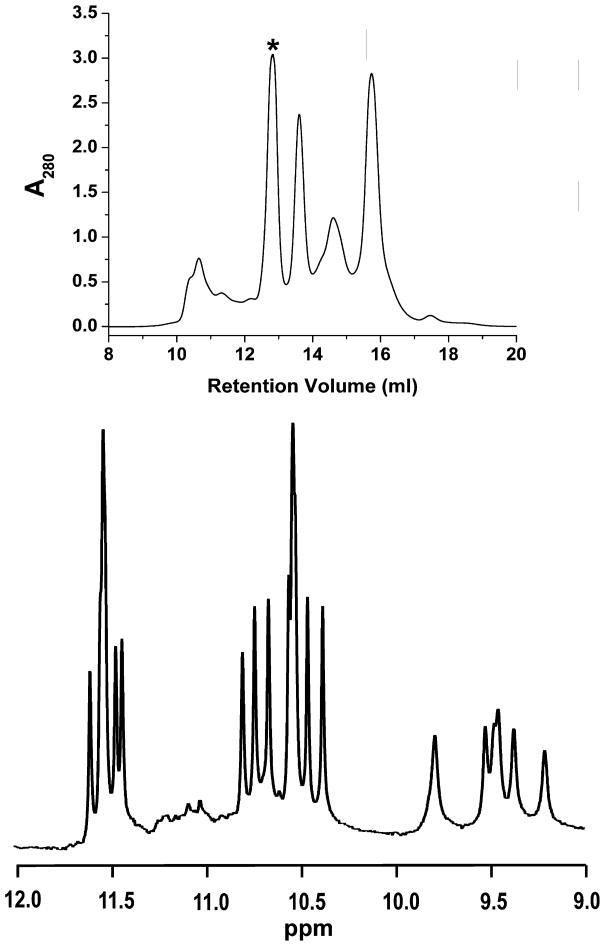
Several examples of the application of SEC to the separation of biologically relevant quadruplex-forming sequences are shown in Figure 5. An initial inspection of the results of several separations reveals that separation of promoter sequences and telomere sequences are very different. AS1411, the promoter sequences, and G4T4G4T4G4T4 show profound separation and resolution of species (Figure 5A – 5D). On the other hand, even telomere sequences that are known to produce at least two species in solution in roughly equal proportions, such as HuTel 22 (Dai, Punchihewa et al. 2007), do not demonstrate resolution (Figure 5E – 5H). When compared to the protein calibration curves results from the separation of AS1411, at 8,272.3 g/mole, results in species that elute with apparent molecular weights ranging from 45,000 g/mole to less than 6,000 g/mole (Figure 6). One possible answer to this observation would be the formation of duplex and tetramer species, yet when fractions eluting with an apparent molecular weight of 14,000 g/mole are collected and examined via 1H NMR the material is shown to be a single, monomeric species (Figure 6). The G4T4G4T4G4T4 sequence, with a molecular weight of 7538.9 g/mole, demonstrates a series of putative quadruplex species that range from apparent molecular weights of 23,000 to 75,000 g/mole. The results from the sequences based on the human telomere sequence are much less dramatic with elution corresponding to about 20,000 g/mole. Finally, TG4T elutes with an apparent molecular weight of about 16,000g/mole.
Figure 5.
HPLC results for each of the sequences listed. A. AS1411, B. Her2, C. c-myc, D. G4T4G4T4G4T4, E. HuTel 22, F. HuTel Hybrid 1, G. HuTel Hybrid 2, H. TG4T
Figure 6.
Resolution of AS1411 polymorphism. TOP: Typical chromatographic separation for AS1411 demonstrating multiple isolatable species with apparent molecular weights ranging from 45,000 g/mole to less than 6,000 g/mole. BOTTOM: 1HNMR of the fraction of AS1411 corresponding to roughly 14,000 g/mole. (Marked with a * in the chromatogram above.) The spectrum shows roughly 16 GN1H signals corresponding to a single, monomeric species.
Quadruplex systems based on the human telomere sequence and on more irregularly repeating sequences, such as those found in many promoter regions, can be expected to behave and separate differently. The differences in chromatographic resolution might be explained by the differences in how these sequences fold. Telomere sequences have very similar loop arrangements and G-quartet stacking. No matter the resulting quadruplex topology the human telomere sequence is limited to three different loop arrangements of equal length (Webba da Silva 2007). More irregular sequences can exhibit greater differences in loop length and arrangement, are capable of more diverse stacking of the G-quartets, and may even form alternate G-quartets. Yet, it seems implausible that these relatively small changes in loop topology can generate the huge changes in hydrodynamic properties necessary to explain the SEC results and still remain undetectable by other biophysical means.
It is also unlikely that associative interactions with the column matrix are responsible for such large differences elution volumes. Any such interaction should result in increased retention volumes, not retention volumes close to V0. The hallmark of such interactions is retention volumes greater than the total solvent permeable column volume (VT), roughly 24 ml for the Superdex 75 10/300, of the column (Frigon, Leypoldt et al. 1983). Thus far no quadruplex sample has yielded retention volumes of greater than 24 ml so associative interactions can probably be eliminated. It has been shown that elution of protein samples via SEC can depend greatly upon the ionic nature of the molecule and the ionic strength of the mobile phase. At low ionic strength, some negatively charged species can experience exclusion by repulsive interaction with the column matrix whereas cationic species may be selectively retained (Frigon, Leypoldt et al. 1983; Le Maire, Aggerbeck et al. 1986; le Maire, Ghazi et al. 1987). Based on this model, the results from separation of quadruplex DNA might be interpreted as differences in and accessibility of the charge on the molecule. This would also explain why quadruplex systems based on the human telomere sequence and on more irregular sequences, such as those found in many promoter regions, behave so differently. Telomere sequences form quadruplexes with very similar loop arrangements and G-quartet stacking and thus, identical ionic nature. More irregular sequences, typical of quadruplex forming promoter sequences, can exhibit greater differences in loop arrangement are capable of more diverse stacking of the G-quartets resulting in greater differences in ionic nature. This phenomenon may also explain the differences between separation of dsDNA and quadruplex DNA via SEC. If this is the case then careful manipulation of mobile phase conditions may be useful for isolation of specific configurations. In any case, it is likely that the separation results demonstrated here are not solely due to size exclusion, but to repulsive interactions as well. This putative repulsive interaction is unpredictable because it seems to depend upon the unique characteristics of each individual sequence. Therefore, while SEC may be used to effect a separation of quadruplex species, it is essentially useless to predict the molecular weight or stokes radius of quadruplex species in solution. On the other hand, SEC may be a useful bellwether for evaluating changes in equilibrium during formation due to alteration of sequence or annealing conditions.
This approach facilitates the structural determination of the actual species formed by a parent sequence without the need for extensive sequence modification and possible structural perturbation. This method could also be used in conjunction with sequence modification to validate that the actual enriched species is present in the parent mixture. SEC also opens the door to resolving structures for many quadruplex forming sequences, such as G4T4G4T4G4T4, that were once too complex and not amiable to sequence modification. An added benefit is that SEC purification may yield more than one isolatable species providing the opportunity for simultaneous study of several structures. In addition to separation, this method can be used for several useful applications. First, quadruplex DNA is of great interest as a drug target this method might be utilized for a variety of applications including identification of protein/quadruplex interactions or drug/quadruplex binding studies from the parent mixtures. Small molecule/quadruplex interactions can be evaluated by incubating a small molecule of interest with a quadruplex and then eluting via SEC and then comparing multiple absorbances or, if the small molecule contains a fluorophore, fluorescence intensity. Protein/quadruplex interactions could be evaluated using methods similar to those described for gel electrophoresis this method could easily provide information about the stoichiometry of such interactions and, potentially, information about small molecule interference with protein/DNA interactions. Second, on column footprinting experiments and digestion experiments could be accomplished. Again, methods similar to those developed for gel electrophoresis may be adapted to this technique.
Time Considerations
Separation of most quadruplex samples, from annealing to separation, can usually be accomplished over the course of three days or so. However, several steps of this procedure are time consuming and any strategic planning should take these steps into account. For example, due to low flow rates the separation step usually takes approximately 10 hours per run. Repetition of this method offers some economies of scale as, once performed, some steps of this method, such as calibration, need only be repeated periodically. Once the HPLC system is set up and calibrated, the HPLC time for each sample drops to about one day. Unless stability is proven by NMR or another high resolution method any fractions collected should be refrigerated or frozen at once to prevent degradation especially if the researcher is planning to combine fractions from several different HPLC runs for a single use.
Literature Cited
- Ambrus A, Chen D, et al. Solution structure of the biologically relevant g-quadruplex element in the human c-MYC promoter. Implications for g-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- Balagurumoorthy P, Brahmachari SK, et al. Hairpin and Parallel Quartet Structures for Telomeric Sequences. Nucleic Acids Res. 1992;20:4061–4067. doi: 10.1093/nar/20.15.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop JS, Guy-Caffey JK, et al. Intramolecular G-quartet motifs confer nuclease resistance to a potent anti-HIV oligonucleotide. J Biol Chem. 1996;271:5698–5703. doi: 10.1074/jbc.271.10.5698. [DOI] [PubMed] [Google Scholar]
- Blume SW, Guarcello V, et al. Divalent transition metal cations counteract potassium-induced quadruplex assembly of oligo(dG) sequences. Nucleic Acids Res. 1997;25:617–625. doi: 10.1093/nar/25.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacio L, Church FC, et al. Effect of locked-nucleic acid on a biologically active G-quadruplex. A structure-activity relationship of the thrombin aptamer. International Journal of Molecular Sciences. 2008;9:422–433. doi: 10.3390/ijms9030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoi S, Quadrifoglio F, et al. G-rich oligonucleotide inhibits the binding of a nuclear protein to the Ki-ras promoter and strongly reduces cell growth in human carcinoma pancreatic cells. Biochemistry. 2004;43:2512–2523. doi: 10.1021/bi035754f. [DOI] [PubMed] [Google Scholar]
- Counter CM, Avilion AA, et al. Telomere Shortening Associated with Chromosome Instability Is Arrested in Immortal Cells Which Express Telomerase Activity. Embo Journal. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Carver M, et al. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008;90:1172–1183. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Chen D, et al. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Dexheimer TS, et al. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J Am Chem Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Punchihewa C, et al. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation. Nucleic Acids Res. 2007;35:2440–2450. doi: 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey MM, Miller MC, et al. Resolution and characterization of the structural polymorphism of a single quadruplex-forming sequence. Nucleic Acids Res. 2010;38:4877–4888. doi: 10.1093/nar/gkq166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapic V, Abdomerovic V, et al. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapic V, Bates PJ, et al. Antiproliferative activity of G-quartet-forming oligonucleotides with backbone and sugar modifications. Biochemistry. 2002;41:3676–3685. doi: 10.1021/bi0119520. [DOI] [PubMed] [Google Scholar]
- De Armond R, Wood S, et al. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1 alpha promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- Drygin D, Siddiqui-Jain A, et al. Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 2009;69:7653–7661. doi: 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]
- Esposito V, Randazzo A, et al. Effects of an 8-bromodeoxyguanosine incorporation on the parallel quadruplex structure [d(TGGGT)](4) Organic & Biomolecular Chemistry. 2004;2:313–318. doi: 10.1039/b314672c. [DOI] [PubMed] [Google Scholar]
- Esposito V, Virgilio A, et al. A new class of DNA quadruplexes formed by oligodeoxyribonucleotides containing a 3 ′-3 ′ or 5 ′-5 ′ inversion of polarity site. Chemical Communications. 2005;31:3953–3955. doi: 10.1039/b504455c. [DOI] [PubMed] [Google Scholar]
- Fernando H, Reszka AP, et al. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigon RP, Leypoldt JK, et al. Disparity between Stokes Radii of Dextrans and Proteins as Determined by Retention Volume in Gel-Permeation Chromatography. Analytical Chemistry. 1983;55:1349–1354. doi: 10.1021/ac00259a037. [DOI] [PubMed] [Google Scholar]
- Garbett NC, Mekmaysy CS, Chaires JB. Sedimentation Velocity Ultracentrifugation Analysis for Hydrodynamic Characterization of G-Quadruplex Structures. In: Bauman P, Walker JM, editors. G-quadruplex DNA. Springer; New York: 2009. pp. 97–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo R, Suzuki M, et al. Promotion of Parallel DNA Quadruplexes by a Yeast Telomere Binding-Protein - a Circular-Dichroism Study. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:7658–7662. doi: 10.1073/pnas.91.16.7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray RD, Li J, et al. Energetics and Kinetics of a Conformational Switch in G-Quadruplex DNA. Journal of Physical Chemistry B. 2009;113:2676–2683. doi: 10.1021/jp809578f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray RD, Petraccone L, et al. Characterization of a K+-induced conformational switch in a human telomeric DNA oligonucleotide using 2-aminopurine fluorescence. Biochemistry. 2010;49:179–194. doi: 10.1021/bi901357r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros J, Avino A, et al. 8-amino guanine accelerates tetramolecular G-quadruplex formation. Chemical Communications. 2008;25:2926–2928. doi: 10.1039/b801221k. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Stewart SA, et al. Inhibition of telomerase limits the growth of human cancer cells. Nature Medicine. 1999;5:1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- Hazel P, Huppert J, et al. Loop-length-dependent folding of G-quadruplexes. J Am Chem Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- Hopkins AL. Drug discovery: Predicting promiscuity. Nature. 2009;462:167–168. doi: 10.1038/462167a. [DOI] [PubMed] [Google Scholar]
- Hsu STD, Varnai P, et al. A G-Rich Sequence within the c-kit Oncogene Promoter Forms a Parallel G-Quadruplex Having Asymmetric G-Tetrad Dynamics. J Am Chem Soc. 2009;131:13399–13409. doi: 10.1021/ja904007p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert JL. Four-stranded DNA: cancer, gene regulation and drug development. Philos Transact A Math Phys Eng Sci. 2007;365:2969–2984. doi: 10.1098/rsta.2007.0011. [DOI] [PubMed] [Google Scholar]
- Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireson C, Djeha H, et al. Preclinical anticancer properties of a G-rich oligonucleotide-based aptamer, AS1411. Clinical Cancer Research. 2005;11:9114s–9114s. [Google Scholar]
- Ireson CR, Kelland LR. Discovery and development of anticancer aptamers. Molecular Cancer Therapeutics. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- Jing N, Li YD, et al. Targeting Stat3 with G-quartet oligodeoxynucleotides in human cancer cells. DNA and Cell Biology. 2003;22:685–696. doi: 10.1089/104454903770946665. [DOI] [PubMed] [Google Scholar]
- Jing N, Sha W, et al. Rational drug design of G-quartet DNA as anti-cancer agents. Curr Pharm Des. 2005;11:2841–2854. doi: 10.2174/1381612054546761. [DOI] [PubMed] [Google Scholar]
- Jing N, Tweardy DJ. Targeting Stat3 in cancer therapy. Anticancer Drugs. 2005;16:601–607. doi: 10.1097/00001813-200507000-00002. [DOI] [PubMed] [Google Scholar]
- Jing NJ, Rando RF, et al. Ion selective folding of loop domains in a potent anti-HIV oligonucleotide. Biochemistry. 1997;36:12498–12505. doi: 10.1021/bi962798y. [DOI] [PubMed] [Google Scholar]
- Keiser MJ, Setola V, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Kumar N, Maiti S. Role of locked nucleic acid modified complementary strand in quadruplex/Watson-Crick duplex equilibrium. J Phys Chem B. 2007;111:12328–12337. doi: 10.1021/jp072705u. [DOI] [PubMed] [Google Scholar]
- Kypr J, Kejnovska I, et al. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane AN, Chaires JB, et al. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Maire M, Aggerbeck LP, et al. The use of high-performance liquid chromatography for the determination of size and molecular weight of proteins: a caution and a list of membrane proteins suitable as standards. Anal Biochem. 1986;154:525–535. doi: 10.1016/0003-2697(86)90025-4. [DOI] [PubMed] [Google Scholar]
- le Maire M, Ghazi A, et al. The use of gel chromatography for the determination of sizes and relative molecular masses of proteins. Interpretation of calibration curves in terms of gel-pore-size distribution. Biochem J. 1987;243:399–404. doi: 10.1042/bj2430399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Correia JJ, et al. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marathias VM, Sawicki MJ, et al. 6-Thioguanine alters the structure and stability of duplex DNA and inhibits quadruplex DNA formation. Nucleic Acids Res. 1999;27:2860–2867. doi: 10.1093/nar/27.14.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekmaysy CS, Petraccone L, et al. Effect of O6-methylguanine on the stability of G-quadruplex DNA. J Am Chem Soc. 2008;130:6710–6711. doi: 10.1021/ja801976h. [DOI] [PubMed] [Google Scholar]
- Miller MC, Buscaglia R, et al. Hydration Is a Major Determinant of the G-Quadruplex Stability and Conformation of the Human Telomere 3′ Sequence of d(AG(3)(TTAG(3))(3)) J Am Chem Soc. 2010;132:1705–1707. doi: 10.1021/ja105259m. [DOI] [PubMed] [Google Scholar]
- Miyoshi D, Nakao A, et al. Structural transition of d(G4T4G4) from antiparallel to parallel G-quartet induced by divalent cations. Nucleic Acids Res Suppl. 2001;1:259–260. doi: 10.1093/nass/1.1.259. [DOI] [PubMed] [Google Scholar]
- Neidle S. Human telomeric G-quadruplex: the current status of telomeric G-quadruplexes as therapeutic targets in human cancer. Febs Journal. 2010;277:1118–1125. doi: 10.1111/j.1742-4658.2009.07463.x. [DOI] [PubMed] [Google Scholar]
- Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nature Reviews Drug Discovery. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- Patel DJ, Phan AT, et al. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: Diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic AG, Polavarapu PL. Quadruplex structure of polyriboinosinic acid: Dependence on alkali metal ion concentration, pH and temperature. Journal of Physical Chemistry B. 2008;112:2255–2260. doi: 10.1021/jp075873v. [DOI] [PubMed] [Google Scholar]
- Phan AT, Kuryavyi V, et al. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J Am Chem Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan AT, Kuryavyi V, et al. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Lin CP, et al. G-quadruplexes induce apoptosis in tumor cells. Cancer Res. 2006;66:11808–11816. doi: 10.1158/0008-5472.CAN-06-1225. [DOI] [PubMed] [Google Scholar]
- Qi J, Shafer RH. Human telomere quadruplex: refolding and selection of individual conformers via RNA/DNA chimeric editing. Biochemistry. 2007;46:7599–7606. doi: 10.1021/bi602392u. [DOI] [PubMed] [Google Scholar]
- Rawal P, Kummarasetti VBR, et al. Genome-wide prediction of G4 DNA as regulatory motifs: Role in Escherichia coli global regulation. Genome Research. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. European Journal of Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- Siddiqui-Jain A, Grand CL, et al. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsson T, Pecinka P, et al. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackova N, Cubero E, et al. Theoretical study of the guanine -> 6-thioguanine substitution in duplexes, triplexes, and tetraplexes. J Am Chem Soc. 2004;126:14642–14650. doi: 10.1021/ja0468628. [DOI] [PubMed] [Google Scholar]
- Sun D, Thompson B, et al. Inhibition of human telomerase by a G-quadruplex-interactive compound. J Med Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- Sun DY, Guo KX, et al. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J, Miyano-Kurosaki N, et al. Inhibition of human immunodeficiency virus type 1 activity in vitro by a new self-stabilized oligonucleotide with guanosine-thymidine quadruplex motifs. Journal of Virology. 2002;76:3015–3022. doi: 10.1128/JVI.76.6.3015-3022.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalai VA, Singer MJ, et al. Site-specific probing of oxidative reactivity and telomerase function using 7,8-dihydro-8-oxoguanine in telomeric DNA. J Am Chem Soc. 2002;124:1625–1631. doi: 10.1021/ja0119651. [DOI] [PubMed] [Google Scholar]
- Tang CF, Shafer RH. Engineering the quadruplex fold: nucleoside conformation determines both folding topology and molecularity in guanine quadruplexes. J Am Chem Soc. 2006;128:5966–5973. doi: 10.1021/ja0603958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgilio A, Esposito V, et al. 8-Methyl-2′-deoxyguanosine incorporation into parallel DNA quadruplex structures. Nucleic Acids Res. 2005;33:6188–6195. doi: 10.1093/nar/gki924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgilio A, Esposito V, et al. Effects of 8-methyl-2 ′-deoxyadenosine incorporation into quadruplex forming oligodeoxyribonucleotides. Bioorganic & Medicinal Chemistry. 2005;13:1037–1044. doi: 10.1016/j.bmc.2004.11.037. [DOI] [PubMed] [Google Scholar]
- Webba da Silva M. Geometric formalism for DNA quadruplex folding. Chemistry. 2007;13:9738–9745. doi: 10.1002/chem.200701255. [DOI] [PubMed] [Google Scholar]
- Wright WE, Tesmer VM, et al. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes & Development. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Kan ZY, et al. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J Am Chem Soc. 2007;129:11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- Yang D, Okamoto K. Structural insights into G-quadruplexes: towards new anticancer drugs. Future Med Chem. 2010;2:619–646. doi: 10.4155/fmc.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]