Abstract
Background
Elevated levels of CD23, a natural regulator of IgE production, have been shown to decrease the signs of lung inflammation in mice. The aim of this study was to study the involvement of ADAM10, the primary CD23 sheddase, in experimental asthma.
Methods
ADAM10 was blocked either by using mice with a B cell specific deletion of the protease or pharmacologically by intranasal administration of selective ADAM10 inhibitors. Airway hypersensitivity (AHR) and bronchoaveolar lavage fluid (BALF) eosinophilia and select BALF cytokine/chemokine levels were then determined.
Results
Using an IgE and mast cell dependent mouse model, B cell specific ADAM10 -/- mice (C57B/6 background) exhibited decreased eosinophilia and AHR when compared to littermate controls. Treatment of C57B/6 mice with selective inhibitors of ADAM10 resulted in an even further decrease in BALF eosinophilia, as compared with the ADAM10-/- animals. Even in the Th2 selective strain, Balb/c, BALF eosinophilia was reduced from 60 to 23% respectively. In contrast when an IgE/mast cell independent model of lung inflammation was used, the B cell ADAM10-/- animals and ADAM10 inhibitor treated animals had lung inflammation levels that were similar to the controls.
Conclusions
These results thus show that ADAM10 is important in the progression of IgE dependent lung inflammation. The use of the inhibitor further suggested that ADAM10 was important for maintaining Th2 levels in the lung. These results thus suggest that decreasing ADAM10 activity could be beneficial in controlling asthma and possible other IgE dependent diseases.
Keywords: ADAM10, Asthma, CD23, IgE, Th2
Introduction
The contribution of IgE to allergy was first established in the late 1960s (1) and with the development of anti-IgE therapy its role in asthma has become better appreciated. In experimental asthma models in mice the therapy leads to decreases in lung eosinophil infiltration, reduced airway hypersensitivity (AHR) and decreased IL-5 and IL-13 production (2), which changes are also seen in human patients(3). Although clearly beneficial, its cost-effectiveness in severe asthmatics has been questioned (4). In addition, anti-IgE therapy is only approved for those whose IgE levels fall in the 30 and 700 IU/ml range (5). Thus, additional therapies, especially for patients with IgE levels in excess of this range are needed.
CD23, the low affinity-receptor of IgE, is a type II integral transmembrane protein. It is predominantly expressed as a trimer at the cell surface (6) and it is in this oligomerized state that CD23 bound to IgE can decrease the synthesis of IgE (7). CD23 is a member of the C-type lectin family, making it unique among Fc receptors. IgE is bound by the lectin head of CD23 and the trimer has an effective affinity for IgE in the nM range. In addition to being a membrane bound protein, CD23 after cleavage, is also found as a soluble factor (sCD23). In humans sCD23 has been shown to have the opposite role of membrane CD23, as it increases B cell proliferation and IgE synthesis(8).
When administered in conjunction with antigen, agonist antibodies specific for the IgE binding site of CD23 cause decreased levels of IgE while having no effects on other isotypes (9). Anti-CD23 also decreased signs of experimental asthma in both IgE dependent and independent mouse models (10). One study (10) additionally examined CD23 transgenics with respect to experimental asthma induction and found decreased eosinophils in the BALF as well as reduced AHR (by penh); however, no mechanism was presented. Finally, when anti-CD23 therapy was used in a clinical trial there was a 40% drop in free serum IgE (11). While not sufficient for clinical efficacy, this trial did show that CD23 is a useful target to decrease IgE levels in humans.
The identification of A Disintegrin and Metalloproteinase 10 (ADAM10) as the primary CD23 sheddase (12) suggests an alternative method to promote the CD23 regulatory activity of IgE, namely by modulating B cell ADAM10 levels and/or activity. The ADAM10 mediated cleavage of CD23 primarily takes place after CD23 is sorted into exosomes in an ADAM10 dependent manner (13). While global ADAM10-/- mice die at embryonic day 9.5, ADAM10 B cell conditional knock outs (called ADAM10-/- from this point) have a large increase in surface CD23 and a corresponding decrease in sCD23 (14). Additionally, we have recently shown that the inhibition of ADAM10 in human B cell in vitro cultures decreases the synthesis of IgE (15). In this paper we show that when surface levels of CD23 are increased, the features of IgE dependent experimental asthma are reduced. We show that CD23Tg mice and ADAM10 B cell specific knockouts, which both have high levels of surface CD23, are less susceptible to IgE dependent asthma. Furthermore, we show that treatment of mice intranasally with ADAM10 inhibitors show substantially reduced responses to OVA. The mechanism is potentially due to a reduction in IgE and/or in the Th2 response.
Material and Methods
Reagents
Chicken Ovalbumin (OVA) and Imject Alum Adjuvant were purchased from Sigma (St. Louis, MO) and Pierce (Rockford, IL), respectively. Cytokines were measured using multiplex kits from Biorad (Hercules, CA) according to the manufacturer’s instructions. The ADAM10 hydroxamate inhibitors, INC008765 and INC009588 (16), were synthesized by the Incyte Corporation. These inhibitors are very selective for ADAM10 as shown by both cell free as well as cell based assays requiring at least 5 fold higher concentrations to inhibit MMP12 and at least 20 fold to inhibit any other enzymes including ADAM17 (16).
Mice
CD23 transgenics were described previously (17) and have been backcrossed 12 generation onto a Balb/c background. Littermates that were negative for the transgene were used as controls. B cell selective ADAM10-/- mice were also described previously (14) and are on a C57B/6 background. Mice negative for the cre gene were used as littermate (LM) controls. Female C57BL/6J and Balb/c mice were purchased from Jackson laboratory (Bar Harbor, ME) and were used in the inhibitor studies. Female mice ages 8-12 weeks were used in the experiments. All mouse protocols were approved by the VCU Institutional Animal Care and Use Committee.
Asthma models
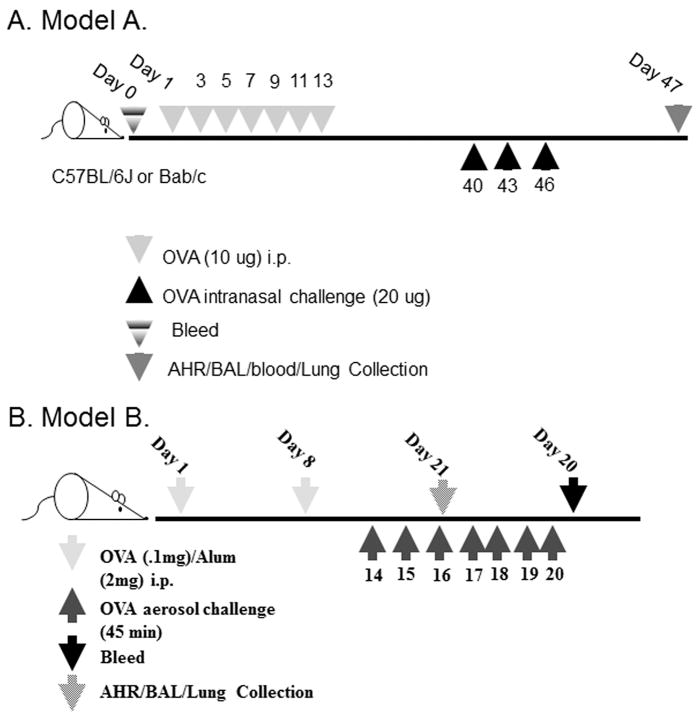
Two asthma models were used and are shown in Figure 1. Model A was developed by Williams et al. and was shown to be mast cell as well as IgE dependent (18). Model B was developed here at VCU and was shown to be independent of IgE and mast cells through the use of IgE-/- (19) and mast cell deficient mice (W/Wsh), in both cases the eosinophil levels in the BALF and lung tissue were similar to wild type mice (data not shown).
Figure 1. Models of asthma used.
(A) Mast cell/IgE dependent model. This model (18) was used in Figures 2(a, c), 3 and 4 and 5. Called Model A in text. (B) Mast cell/IgE independent model. Called Model B in text.
FlexiVent
In some experiments, 18 hours after the final OVA challenge, airway resistance was measured using the Flexivent system (Scireq, Montreal, Canada) (19). Mice were anesthetized by intraperitoneal (IP) injection of 206.7 mg/kg of ketamine and 41.7 mg/kg of xylazine. Following cannulation and paralyzation mice were ventilated and measurements of baseline lung function were taken, mice were exposed to aerosols containing increased doses (10, 25, 50 mg/ml for C57B/6 or 2.5, 5, 10, 25 for BALB/c) of Acetyl-β-methylcholine chloride (Methacholine) (Sigma). Newtonian resistance (Rn), tissue damping (G) and tissue elastance (H) were all measured at baseline (PBS) and 50 mg/ml of methacholine using the Flexivent software version 5.3.
BALF/lung collection
Following the Flexivent analysis, BALF was collected by lavaging the lungs with 1 ml PBS. The BALF was centrifuged and supernatant fluids were saved for further analysis. Pelleted cells were resuspended in 100 μl PBS, counted, cytospun onto slides and stained with Diff-Quik (Siemens Healthcare Diagnostics, Deerfield, IL). Percentages were determined by counting of at least 100 leukocyte cells per cytospin. Additionally, lung and heart tissue were fixed in 10% formulin (Fisher) and embedded in paraffin by the VCU pathology core. 5 μm sections were cut onto microscope slides and stained with hemotoxin and eosin (H&E). A Nikon eclipse with a SPOT Flex Shifting Pixel Color Mosaic (Diagnostic Instrumental inc., Sterling Heights, MI) camera was used to take pictures of the sections.
ELISAs
Total mouse IgE was measured as previously described (20). For OVA specific IgE, R1E4 (rat anti-mouse IgE) was used as the capture antibody and OVA-DNP-Biotin in combination with Streptavidin-alkaline phosphatase (AP) was used for detection. Mouse IgE anti-DNP was used as a standard. For OVA-specific IgG1, OVA was used to capture the antibody. Samples and standard (serum from a mouse hyper immunized with OVA was used as an internal control) were detected with an AP-goat anti mouse IgG1. The liver enzymes aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were measured as previously described (21).
Real time PCR
Total RNA was extracted from disrupted lung tissue and purified as explained previously(14). For analysis the following previously published primers were used: GATA3 (22), T bet(22), IL- 25 (23), and IL-33 (23).
Statistics
All statistics were done using the student two-tailed T test, with bonferroni correction when multiple comparisons were being made. All bars represent standard error.
Results
ADAM10-/- and CD23Tg mice have decreased signs of allergic lung inflammation in an IgE dependent model
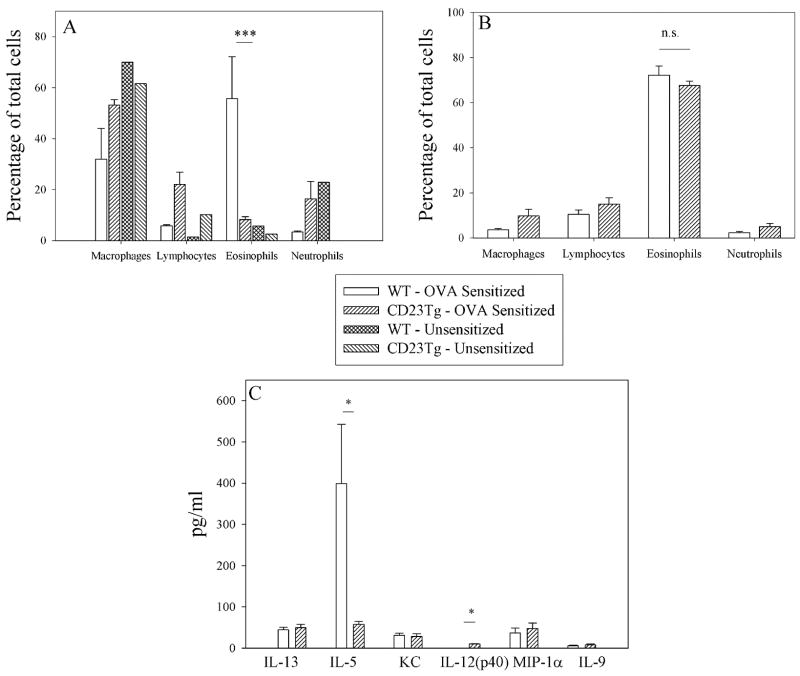
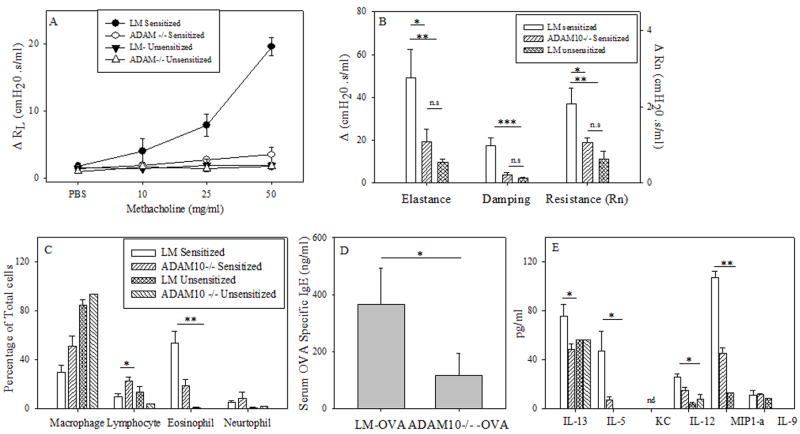
To test the importance of CD23 and ADAM10 in asthma induction we used the murine models of allergic lung inflammation shown in figure 1. In agreement with Haczku et al.(10), we found that CD23Tg mice (Balb/c background), which have suppressed IgE (17) and have high expression of CD23 on lymphocytes, exhibited decreased eosinophilia (Fig 2a) when tested in an IgE-dependent asthma model (model A). We also observed decreased levels of the chemokine IL-5 (Fig 2c) which may explain the decreased eosinophilia seen. In contrast to the IgE-dependent model (model A) no differences in BALF eosinophilia was seen (Fig. 2b) when model B was used. This indicates that the decrease in asthma severity observed in the CD23Tg occurs because of decreased IgE in these animals. Given that ADAM10 is the CD23 sheddase and that mice which lack B cell ADAM10 have greatly increased CD23 expression (14) we next tested whether these mice would have reduced inflammation with model A. Lung AHR in ADAM10-/- mice (C57B/6 background), as determined by total lung resistance (RL) (Fig 3a) and forced oscillation technique (Fig 3b) was significantly lower than in the LM sensitized mice. Fig 3c shows that the ADAM10-/- mice also had significantly reduced eosinophilia in the BALF. Total BALF cell numbers (data not shown) and percentages of other cell types in the ADAM10-/- mice closely resembled the unsensitized animals. ADAM10-/- mice also had reduced serum antigen-specific IgE levels (Fig 3d). In addition, significant decreases in both Th1 and Th2 cytokines were observed (Fig 3e). In contrast, when asthma was induced with model B, the ADAM10-/- (data not shown) like the CD23Tg (Fig 2b) was not significantly different from the LM sensitized animals with respect to AHR or BALF eosinophilia (data not shown). Additionally, the Th1 cytokines IL-12 and MIP1-α were still significantly decreased in the IgE independent model, when comparing the OVA sensitized ADAM10-/- mice to LM controls (data not shown). Over all these data show that increasing CD23 by inhibiting ADAM10 on B cells has the potential for strongly inhibiting the IgE dependent, but not the IgE independent lung inflammatory response by reducing IgE levels.
Figure 2. Overexpression of CD23 in CD23Tg can control eosinophilia in an IgE dependent model but not an IgE independent model.
(A) Asthma was induced in Balb/c LM and CD23Tg using model A. BALF were collected and the percentages of cells were determined as explained in the Material and Methods by counting at least 300 cells. Shown is the average of four mice. (B) Asthma was induced in Balb/c LM and CD23Tg using model B. The percentage of cells was determined as in A. Shown in the average of at least 4 mice per group. (C) The cytokines in the BALF of the mice used in A were determined using multiplex kits according to the manufactures’ instructions. * = P value <.05 and ** = P value <.01
Figure 3. ADAM10-/- decreases the severity of disease in a mast cell dependent model.
(A) Mice were sensitized and challenged using model A (mice are on a C57B/6 background). On day 47 lung function was assessed by measuring total lung resistance as explained in the Material and Methods; shown is the average of at least 4 mice per treated group. Significance is between ADAM10 -/- and LM. (B) Rn (Newton Resistance), G (Tissue Dampening) and H (Elastance) were determined at baseline (PBS) and after the addition of 50 mg/ml of methacholine, with the average of the three maximum points at each dose being used for each mouse. Shown is the average of at least 4 mice per group. (C) Cells from the BALF were counted and stained as explained in the materials and methods. Percentage of cells was then determined in the BALF by counting of at least one hundred cells and numbers presented is the average of at least four mice. (D) The levels of OVA specific IgE in the serum were determined by ELISA, shown is the average of at least five animals. (E) Cytokine levels in the BALF was determined by multiplex analysis, shown is the average of at least four animals (same key as in C). * = P value <.05, ** = P value <.01 and *** = P value < .001
ADAM10 inhibition blocks the induction of experimental asthma
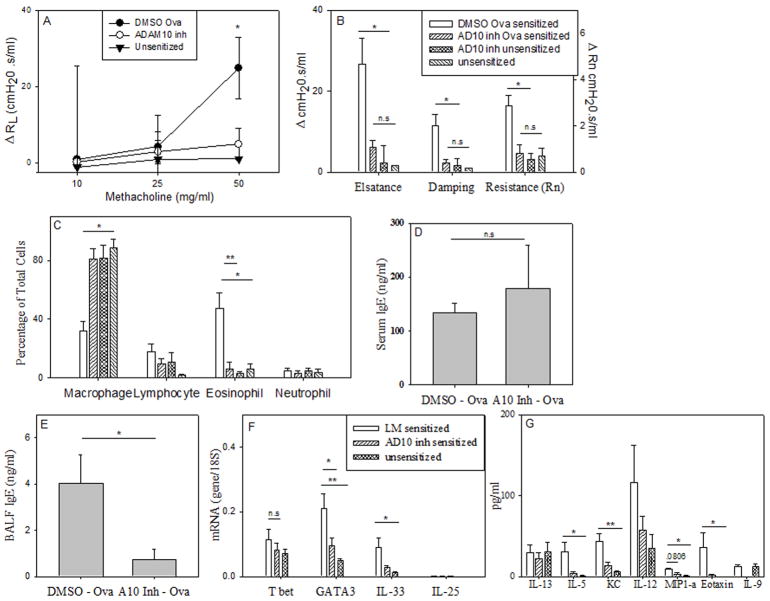
Given the above finding that mice lacking B cell ADAM10 have minimal pathology in IgE dependent lung inflammation, we sought to determine if inhibiting ADAM10 activity in WT mice would have the same affect. Experiments were conducted with two hydroxamate inhibitors of ADAM10 (both were found to act similar, thus the data from both were combined). They were dissolved in DMSO and administered intranasally during the induction of the asthma model. The inhibitors were administered intranasally at a concentration of 1 mg/kg (16), a dose where ADAM10 should selectively be inhibited, in DMSO/PBS every three days throughout the experiment. As a control, the ADAM10 inhibitors were also administered to unsensitized mice and found to have no effect (Fig 4 b, c). Using the IgE-dependent model (model A) we found that the inhibitors significantly decreased the signs of allergic airway inflammation, while having no toxic effects as measured by weight loss as well as the levels of liver enzymes (ALT, AST) in the serum (Table 1). The carrier (5% DMSO in PBS) was well tolerated; responses of the LM controls in Fig 3c were similar to Fig 4c. Thus, any reduction in asthma severity was due to inhibition of ADAM10 activity. We also found that similar to the ADAM10-/- (Fig 3), AHR as measured by forced oscillation technique (Fig 4b), as well as RL (Fig 4a) was decreased by the administration of the ADAM10 inhibitor. The eosinophilia in the BALF was also found to be significantly reduced (Fig 4c); this decrease was even more drastic than in the ADAM10-/-, from ~50% to 20% in the ADAM10-/- (Fig 3c) and from ~50% to 5% (Fig 4c) in WT mice treated with the inhibitor. Thus, this shows that ADAM10 expression by other cells, in addition to the B cell, is important in the progression of experimental asthma. The inhibitors are also possible blocking ADAM10 activity on epithelial cells as they have been shown to control the allergic response (reviewed in (24)) and have high expression of ADAM10 (25). Of note although the percentage of macrophages was increased, there were actually less macrophages in the BALF (data not shown) after inhibitor treatment. To determine if this effect was through an inhibition of IgE synthesis, as it was with the ADAM10-/-, we measured the levels of total IgE in the serum (Fig 4d) but found it was unchanged from the LM. However, local production was decreased, as total IgE in the BALF was significantly decreased (Fig 4e). Hydroxamate inhibitors are known to have a short half-life in serum (26), however as clearly shown by our data when administered intranasally the inhibitory activity is sufficient for at least a 3 day period. The effects on IgE suggest local activity, however, blocking only the local production of IgE is advantageous as it has been shown that local but not systemic, production of IgE (27) is important for disease progression.
Figure 4. ADAM10 inhibitor blocks the induction of an IgE/mast cell dependent model potentially through blocking the Th2 response.
Asthma was induced using the IgE-dependent model (Model A). 1 mg/kg in a 20:1 mixture of PBS/DMSO of an ADAM10 inhibitor was injected intranasal every three days throughout the experiment in wild type C57B/6. The control mice (wild type C57B/6) received just the PBS/DMSO mixture. (A) On day 47 AHR was measured using total lung resistance (Significance is between ADAM10 inhibitor treated and LM) and (B) forced oscillation measurements as explained in Fig 3b. Data represents the average of data from at least four sensitized mice and two unsensitized mice. (C) Percentage of BALF cells were determined as in Fig 2a. Shown is the average of at least three animals (same key as B). (D) Total IgE in the serum as determined by ELISA, average of at least four animals. (E) Total IgE in the BALF as determined by ELISA, average of at least three animals. (F) qPCR of total lung RNA was performed using the primers listed in material and methods. Shown is the average of at least five animals. The unsensitized groups were combined, as no difference where seen between them. (G) BALF cytokine levels as determined by multiplex ELISA. Shown is the average of at least five animals per group. Again unsensitized groups were combined. (same key as F). Number above comparison line equals P value, * = P value < .05 and ** = P value <.01
Table 1. Liver Enzyme analysis indicates lack of toxicity for ADAM10 inhibitors.
Just prior to measurement of AHR the mice where weighed. Shown is the average of at least 8 mice per group. The serum isolated after sacrifice of three mice of similar treatment were pooled together and then AST and ALT levels were measured in the serum as explained in the Material and Methods. Shown is the average of at least two different pools.
| Parameter | DMSO – OVA Sensitized | AD10 inh – OVA Sensitized |
|---|---|---|
| Weight (time of sacrifice) | 20.48889 ± 0.503721 | 20.42727 ± 0.282872 |
| AST (aspartate aminotransferase) | 13.666 ± 6.81 | 15.558 ± 6.87 |
| ALT (alanine aminotransferase) | 2.589382 ± 0.931133 | 3.636805 ± 0.668527 |
To test if ADAM10 inhibitors had further effects beyond that of inhibiting IgE, we isolated total lung mRNA from treated and control mice and using qPCR, measured the level of several components known to be important in asthma progression, including the Th2 response. In the ADAM10-/- mice we found that the decrease in serum OVA-specific IgE correlated with a decrease in Th2 cytokines (Fig 3e). To test what effect administration of the inhibitor was having on T cell responses, mRNA levels of the dominate T cell transcription factors were measured by qPCR. Analysis demonstrated that GATA3 message levels were significantly decreased, while Tbet was not (Fig 4f). Recently two cytokines, IL-25 and 33 (reviewed in (28)), have been implicated in the induction of a Th2 response. While minimal IL-25 levels were detected, message for IL-33 was significantly decreased in mice treated with the ADAM10 inhibitor. These results show that ADAM10 is also important in the initiation of the Th2 response in the lung. Interestingly, when cytokines were analyzed in the BALF, IL-13 levels were not decreased (Fig 4g) when given the inhibitor, in spite of the known association of IL-13 and AHR (29). Alternative cell sources, not affected by ADAM10 inhibition, of IL-13 production (30) could potentially explain this finding. It is interesting to speculate that blockade of ADAM10 decreases IL-13 ability (29) to act on epithelial cells in the lung. The decreased eosinophilia (Fig 3c) observed after ADAM10 inhibition can be explained by the strong inhibition of IL-5, the primary chemotactic factor in the recruitment of eosinophils, as well as by significant decreases in eotaxin and KC which are also involved in eosinophil recruitment (Fig 4g). Finally, IL-9 was also found to be decreased by the addition of the ADAM10 inhibitor. IL-9 is produced both by Th2 cells as well as the new subclass of T helper cells, Th9, and is known to enhance airway inflammation (31). Overall, the multiple changes help explain how the inhibition of ADAM10 suppresses IgE-dependent lung inflammation.
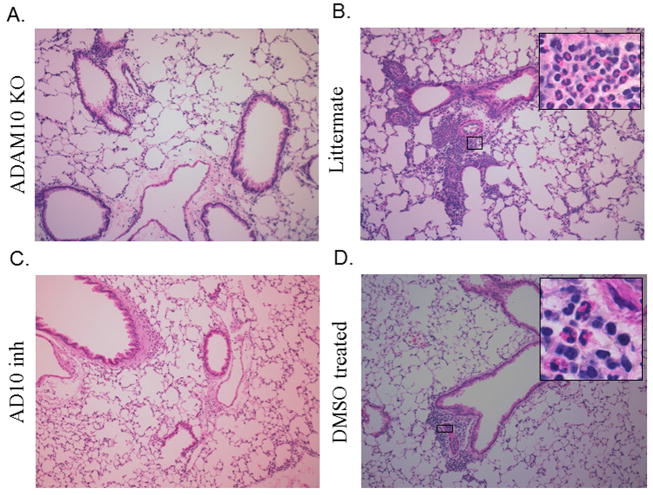
In order to confirm our findings in the BALF lung section from both the ADAM10-/- and inhibitor treated as well as their respective controls were stained with H&E. As is seen by the representative pictures in Fig 5(a, c) that when ADAM10 was blocked either genetically or with an inhibitor there was a decrease in inflammation compared to their controls (Fig 5b, d), including the infiltration of eosinophils. These results thus show that ADAM10 is important not only in the recruitment to the BALF of eosinophils but also to the lung tissue.
Figure 5. ADAM10 inhibition also blocks the inflammatory response in the lung tissue.
The lungs from mice where asthma had been induced using Model A, where fixed and sectioned as explained in the Material and Methods. The sections were then de-paraffinized and stained with H&E. Larger photomicrographs are 10x with select inserts from areas showing obvious inflammation at 100x. (A) Photomicrographs using lungs from ADAM10 -/- mice or (B) their littermate controls. (C) Photomicrographs using lungs from C57B/6 that received INCB008765 throughout the induction of asthma or (D) DMSO control lungs. Photos shown are representative from experiments using 4 mice per group.
When the ADAM10 inhibitor administration was repeated in BALB/c mice using model A, similar results were seen expect the eosinophilia decrease in the BALF was not as drastic (60% to 23% in the Balb/c compared to 50% to 5% in the C57B/6 after inhibitor treatment). Finally, when the ADAM10 inhibitor was administered every three days during the induction of model B (IgE independent), no effect was seen. Overall these results show that regardless of the mouse background, the administration of an ADAM10 inhibitor is able to block the induction of experimental asthma.
Discussion
Although, it has previously been shown that if the cleavage of CD23 is blocked with a stabilizing antibody or if CD23 is overexpressed that the signs of lung inflammation were decreased (10), it was not known what role ADAM10 played in this or how CD23 was able to control lung inflammation. Here in we showed that overexpression of CD23 or blocking ADAM10 decreases the Th2 response as well as the other signs of lung inflammation, including eosinophilia, AHR and IgE production.
The mechanism of the reduced signs of experimental asthma after ADAM10 inhibition is currently under investigation. Based on the findings reported we propose two possibilities. First, as we found that administration of the inhibitor blocked IgE production (Fig 3e), this decrease in IgE would be reflected in decreased mediators released from mast cells after antigen challenge, thus resulting in decreased signs of experimental asthma, including AHR. One of the reasons for decreased IgE production is reduced cleavage of CD23, as we have previously shown that inhibitors of ADAM10 (15) and loss of ADAM10 (14) reduce CD23 cleavage both in vitro and in vivo respectively. Additionally, using a hu-PBL model in SCID mice, the inhibition of CD23 cleaveage was previously shown to correlate with decreased IgE synthesis (32). Secondly, the stimulatory activity of IgE complexes that bind to CD23 and enhance antigen presentation has been well documented by the Heyman laboratory (reviewed in (33)). In a recent paper we showed that ADAM10 is not only the sheddase of CD23, but also sorts CD23 into exosomes (13). Once released from the cell, the CD23 containing exosomes could bind IgE complexes and cause increased antigen presentation and T cell responses. The ADAM10 B cell conditional knockouts do not have these CD23 containing exosomes (13), and the lack of these CD23 containing exosomes could possibly explain part of the inhibition of the Th1, as well as the Th2, responses. Such exosomes, containing bound IgE complexes, would be anticipated to enhance dendritic cell activation of T cells.
Overall, the combination of using B cell ADAM10 knockouts as well as hydroxamate inhibitors of ADAM10, clearly indicates an important role for ADAM10, and CD23, in Th2-induced asthmatic disease, and suggests that hydroxamate inhibitors of ADAM10, directly administered to the airway, may have utility to modulate this disease.
Acknowledgments
We thank John Tew and Keith Brooks for their review and comments on the manuscript. Also we thank Drew Jones for his help in developing the IgE/mast cell independent model and Jorge Almenara in his help in sectioning of the lungs. Microscopy was performed at the VCU Department of Anatomy and Neurobiology Microscopy Facility, supported, in part, with funding from NIH-NINDS Center core grand (5P30NSD4763-02). Support for this work came from the NIH grants RO1AI18697 and 1U19AI077435.
Footnotes
Author Contributions
JM, contributed to each of the figures, JF contributed to part of fig 1, SN contributed to the interpretation and understanding of the AHR. DG created the ADAM10-/- mice, AF and AK contributed to the creation of the asthma models and finally DC gave guidance for the studies in his laboratory. AD and CK measured the liver enzymes in the serum. HM gave guidance on interpretation of inflammation in the lungs.
The authors have no conflicting financial interests.
Reference List
- 1.Johansson SG. IgE in allergic diseases. Proc R Soc Med. 1969 Sep;62(9):975–976. doi: 10.1177/003591576906200947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kang JY, Kim JW, Kim JS, Kim SJ, Lee SH, Kwon SS, et al. Inhibitory effects of anti-immunoglobulin E antibodies on airway remodeling in a murine model of chronic asthma. J Asthma. 2010 May;47(4):374–380. doi: 10.3109/02770901003801972. [DOI] [PubMed] [Google Scholar]
- 3.Djukanovic R, Wilson SJ, Kraft M, Jarjour NN, Steel M, Chung KF, et al. Effects of treatment with anti-immunoglobulin E antibody omalizumab on airway inflammation in allergic asthma. Am J Respir Crit Care Med. 2004 Sep 15;170(6):583–593. doi: 10.1164/rccm.200312-1651OC. [DOI] [PubMed] [Google Scholar]
- 4.Wu AC, Paltiel AD, Kuntz KM, Weiss ST, Fuhlbrigge AL. Cost-effectiveness of omalizumab in adults with severe asthma: results from the Asthma Policy Model. J Allergy Clin Immunol. 2007 Nov;120(5):1146–1152. doi: 10.1016/j.jaci.2007.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avila PC. Does anti-IgE therapy help in asthma? Efficacy and controversies. Annu Rev Med. 2007;58:185–203. doi: 10.1146/annurev.med.58.061705.145252. [DOI] [PubMed] [Google Scholar]
- 6.Kilmon MA, Shelburne AE, Chan-Li Y, Holmes KL, Conrad DH. CD23 Trimers Are Pre-associated On The Cell Surface Even In The Absence Of Its Ligand, IgE. J Immunol. 2003 doi: 10.4049/jimmunol.172.2.1065. In press. [DOI] [PubMed] [Google Scholar]
- 7.Kilmon MA, Ghirlando R, Strub MP, Beavil RL, Gould HJ, Conrad DH. Regulation of IgE production requires oligomerization of CD23. J Immunol. 2001 Sep 15;167(6):3139–3145. doi: 10.4049/jimmunol.167.6.3139. [DOI] [PubMed] [Google Scholar]
- 8.Conrad DH, Ford JW, Sturgill JL, Gibb DR. CD23: an overlooked regulator of allergic disease. Curr Allergy Asthma Rep. 2007 Sep;7(5):331–337. doi: 10.1007/s11882-007-0050-y. [DOI] [PubMed] [Google Scholar]
- 9.Flores-Romo L, Shields J, Humbert Y, Graber P, Aubry JP, Gauchat JF, et al. Inhibition of an in vivo antigen-specific IgE response by antibodies to CD23. Science. 1993 Aug 20;261(5124):1038–1041. doi: 10.1126/science.8351517. [DOI] [PubMed] [Google Scholar]
- 10.Haczku A, Takeda K, Hamelmann E, Loader J, Joetham A, Redai I, et al. CD23 exhibits negative regulatory effects on allergic sensitization and airway hyperresponsiveness. Am J Respir Crit Care Med. 2000 Mar;161(3 Pt 1):952–960. doi: 10.1164/ajrccm.161.3.9905046. [DOI] [PubMed] [Google Scholar]
- 11.Rosenwasser LJ, Meng J. Anti-CD23. Clin Rev Allergy Immunol. 2005 Aug;29(1):61–72. doi: 10.1385/CRIAI:29:1:061. [DOI] [PubMed] [Google Scholar]
- 12.Weskamp G, Ford JW, Sturgill J, Martin S, Docherty AJ, Swendeman S, et al. ADAM10 is a principal ‘sheddase’ of the low-affinity immunoglobulin E receptor CD23. Nat Immunol. 2006 Dec;7(12):1293–1298. doi: 10.1038/ni1399. [DOI] [PubMed] [Google Scholar]
- 13.Mathews JA, Gibb DR, Chen BH, Scherle P, Conrad DH. CD23 Sheddase A Disintegrin and Metalloproteinase 10 (ADAM10) Is Also Required for CD23 Sorting into B Cell-derived Exosomes. J Biol Chem. 2010 Nov 26;285(48):37531–37541. doi: 10.1074/jbc.M110.141556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibb DR, El SM, Kang DJ, Rowe WJ, El SR, Cichy J, et al. ADAM10 is essential for Notch2-dependent marginal zone B cell development and CD23 cleavage in vivo. J Exp Med. 2010 Mar 15;207(3):623–635. doi: 10.1084/jem.20091990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sturgill JL, Mathews J, Scherle P, Conrad DH. Glutamate signaling through the kainate receptor enhances human immunoglobulin production. J Neuroimmunol. 2011 Jan 5; doi: 10.1016/j.jneuroim.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou BB, Peyton M, He B, Liu C, Girard L, Caudler E, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006 Jul;10(1):39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payet ME, Woodward EC, Conrad DH. Humoral response suppression observed with CD23 transgenics. J Immunol. 1999 Jul 1;163(1):217–223. [PubMed] [Google Scholar]
- 18.Williams CM, Galli SJ. Mast cells can amplify airway reactivity and features of chronic inflammation in an asthma model in mice. J Exp Med. 2000 Aug 7;192(3):455–462. doi: 10.1084/jem.192.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hantos Z, Collins RA, Turner DJ, Janosi TZ, Sly PD. Tracking of airway and tissue mechanics during TLC maneuvers in mice. J Appl Physiol. 2003 Oct;95(4):1695–1705. doi: 10.1152/japplphysiol.00104.2003. [DOI] [PubMed] [Google Scholar]
- 20.Caven TH, Shelburne A, Sato J, Chan-Li Y, Becker S, Conrad DH. IL-21 dependent IgE production in human and mouse in vitro culture systems is cell density and cell division dependent and is augmented by IL-10. Cell Immunol. 2005 Dec;238(2):123–134. doi: 10.1016/j.cellimm.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 21.REITMAN S, FRANKEL S. A colorimetric method for the determination of serum glutamic oxalacetic and glutamic pyruvic transaminases. Am J Clin Pathol. 1957 Jul;28(1):56–63. doi: 10.1093/ajcp/28.1.56. [DOI] [PubMed] [Google Scholar]
- 22.Wang SY, Yang M, Xu XP, Qiu GF, Ma J, Wang SJ, et al. Intranasal delivery of T-bet modulates the profile of helper T cell immune responses in experimental asthma. J Investig Allergol Clin Immunol. 2008;18(5):357–365. [PubMed] [Google Scholar]
- 23.Ishii A, Oboki K, Nambu A, Morita H, Ohno T, Kajiwara N, et al. Development of IL-17-mediated delayed-type hypersensitivity is not affected by down-regulation of IL-25 expression. Allergol Int. 2010 Dec;59(4):399–408. doi: 10.2332/allergolint.10-OA-0218. [DOI] [PubMed] [Google Scholar]
- 24.Lambrecht BN, Hammad H. The role of dendritic and epithelial cells as master regulators of allergic airway inflammation. Lancet. 2010 Sep 4;376(9743):835–843. doi: 10.1016/S0140-6736(10)61226-3. [DOI] [PubMed] [Google Scholar]
- 25.Dijkstra A, Postma DS, Noordhoek JA, Lodewijk ME, Kauffman HF, ten Hacken NH, et al. Expression of ADAMs (“a disintegrin and metalloprotease”) in the human lung. Virchows Arch. 2009 Apr;454(4):441–449. doi: 10.1007/s00428-009-0748-4. [DOI] [PubMed] [Google Scholar]
- 26.Fridman JS, Caulder E, Hansbury M, Liu X, Yang G, Wang Q, et al. Selective inhibition of ADAM metalloproteases as a novel approach for modulating ErbB pathways in cancer. Clin Cancer Res. 2007 Mar 15;13(6):1892–1902. doi: 10.1158/1078-0432.CCR-06-2116. [DOI] [PubMed] [Google Scholar]
- 27.Chvatchko Y, Kosco-Vilbois MH, Herren S, Lefort J, Bonnefoy JY. Germinal center formation and local immunoglobulin E (IgE) production in the lung after an airway antigenic challenge. J Exp Med. 1996 Dec 1;184(6):2353–2360. doi: 10.1084/jem.184.6.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett NA, Austen KF. Innate cells and T helper 2 cell immunity in airway inflammation. Immunity. 2009 Sep 18;31(3):425–437. doi: 10.1016/j.immuni.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walter DM, McIntire JJ, Berry G, McKenzie AN, Donaldson DD, DeKruyff RH, et al. Critical role for IL-13 in the development of allergen-induced airway hyperreactivity. J Immunol. 2001 Oct 15;167(8):4668–4675. doi: 10.4049/jimmunol.167.8.4668. [DOI] [PubMed] [Google Scholar]
- 30.Akbari O, Stock P, Meyer E, Kronenberg M, Sidobre S, Nakayama T, et al. Essential role of NKT cells producing IL-4 and IL-13 in the development of allergen-induced airway hyperreactivity. Nat Med. 2003 May;9(5):582–588. doi: 10.1038/nm851. [DOI] [PubMed] [Google Scholar]
- 31.Soroosh P, Doherty TA. Th9 and allergic disease. Immunology. 2009 Aug;127(4):450–458. doi: 10.1111/j.1365-2567.2009.03114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayer RJ, Bolognese BJ, Al-Mahdi N, Cook RM, Flamberg PL, Hansbury MJ, et al. Inhibition of CD23 processing correlates with inhibition of IL-4-stimulated IgE production in human PBL and hu-PBL-reconstituted SCID mice. Clin Exp Allergy. 2000 May;30(5):719–727. doi: 10.1046/j.1365-2222.2000.00812.x. [DOI] [PubMed] [Google Scholar]
- 33.Hjelm F, Carlsson F, Getahun A, Heyman B. Antibody-mediated regulation of the immune response. Scand J Immunol. 2006 Sep;64(3):177–184. doi: 10.1111/j.1365-3083.2006.01818.x. [DOI] [PubMed] [Google Scholar]