Abstract
The Wnt/β-catenin pathway is a critical stem cell regulator and plays important roles in neuroepithelial cells during early gestation. However, the role of Wnt/β-catenin signaling in radial glia, a major neural stem cell population expanded by midgestation, remains poorly understood. This study shows that genetic ablation of β-catenin with hGFAP-Cre mice inhibits neocortical formation by disrupting radial glial development. Reduced radial glia and intermediate progenitors are found in the β-catenin-deficient neocortex during late gestation. Increased apoptosis and divergent localization of radial glia in the subventricular zone are also observed in the mutant neocortex. In vivo and in vitro proliferation and neurogenesis as well as oligodendrogenesis by cortical radial glia or by dissociated neural stem cells are significantly defective in the mutants. Neocortical layer patterning is not apparently altered, while astrogliogenesis is ectopically increased in the mutants. At the molecular level, the expression of the transcription factor Pax6 is dramatically diminished in the cortical radial glia and the sphere-forming neural stem cells of β-catenin-deficient mutants. Chromatin immunoprecipitation and luciferase assays demonstrate that β-catenin/Tcf complex binds to Pax6 promoter and induces its transcriptional activities. The forced expression of Pax6 through lentiviral transduction partially rescues the defective proliferation and neurogenesis by β-catenin-deficient neural stem cells. Thus, Pax6 is a novel downstream target of the Wnt/β-catenin pathway, and β-catenin/Pax6 signaling plays critical roles in self-renewal and neurogenesis of radial glia/neural stem cells during neocortical development.
Keywords: Neural stem cells, Radial glia, Wnt/β-catenin, Pax6, Proliferation, Neurogenesis
Introduction
Neural stem cells (NSCs) are self-renewing and multipotent, generating all types of neurons and macroglia as well as ependymal cells [1–4]. The self-renewal and differentiation of NSCs are precisely regulated by the interplay of cell-intrinsic factors and extrinsic signals during neural development and adult neurogenesis [5–8]. Wnt/β-catenin signaling is a crucial stem cell regulator and plays various important roles in development and disease [9–12]. Wnt/β-catenin signaling agonists or Wnt3a proteins support self-renewal of murine and human embryonic stem cells (ESCs) by preventing their differentiation, and are required to maintain pluripotency of ESCs in cultures [13–17]. Wnt/β-catenin signaling also plays critical roles in adult hematopoietic, intestinal, skin, mammary gland, and muscle stem cells, and in somatic cell reprogramming [18–24].
Important roles of Wnt/β-catenin signaling have also been increasingly revealed in developing and adult NSCs and progenitors [7, 25–27]. In the cerebral cortex, Wnt/β-catenin signaling is required for hippocampal development and the maintenance of an adult neurogenic niche in the dentate gyrus [28–31]. Transgenic mice expressing a constitutively active β-catenin allele in neuroepithelial cells exhibit dramatically enlarged brains caused by a tangential expansion of the cortical precursors or NSCs [32]. Genetic inactivation of the Wnt coreceptor Lrp6 leads to defective cortical precursor proliferation and neuronal production [33, 34]. Wnt/β-catenin signaling or its upstream regulator Tlx promotes proliferation and in vitro expansion of NSCs and progenitors derived from the proliferating subventricular zone (SVZ) of either developing or adult mouse cortex [35, 36]. Wnt/β-catenin signaling also promotes proliferation and neurogenesis of peripheral NSCs during olfactory development and adult regeneration [37]. However, the downstream molecular mechanisms of Wnt/β-catenin signaling in NSCs, particularly in developing neocortex, remain poorly understood.
NSCs exist initially as neuroepithelial stem cells in the neural tube, then transform to radial glia, a recently recognized major NSC population in the developing CNS, which eventually give rise to adult NSCs [38–43]. Radial glial cells divide symmetrically or asymmetrically for self-renewal or differentiation and express long apical radial processes that guide migration of newborn neurons [41, 44, 45]. Pax6, a paired-box transcription factor with dominant expression in radial glia of the cortical SVZ, is a critical regulator of radial glial and ESC neurogenesis [46–49]. We hypothesize that Wnt/ β-catenin signaling controls radial glial proliferation and neurogenesis for cortical development through transcriptional activation of Pax6.
β-Catenin is the central player in the canonical Wnt signaling pathway. After Wnt binds to cell-surface receptors Fzd and Lrp, β-catenin is stabilized in the cytoplasm and translocated to the nucleus where it activates the Tcf/Lef1 transcription complex to regulate the promoter activity of Wnt signaling target genes [50, 51]. To address our hypothesis in this study, we conditionally ablated β-catenin in radial glia, which caused a dramatic inactivation of Pax6 expression in the cortical radial glia and hypoplastic cortical development. Through clonal NSC cultures combined with molecular biological and viral transduction rescue approaches, we demonstrated a novel β-catenin/Pax6 signaling cascade that may control NSC/radial glial self-renewal and neurogenesis during neocortical development.
Materials and Methods
Animals
The conditional loss-of-function of β-catenin(ex2–6)flox mice, the hGFAP (human glial fibrillary acidic protein)-driven Cre transgenic mice, and the Cre reporter Rosa26-lacZ mice were obtained through the Jackson Laboratory (Bar Harbor, ME, www.jax.org) and described by the original contributors [52–54]. Mutants were genotyped by PCR of genomic DNA prepared from tail or limb biopsies. Mice were housed in the vivarium of the UC Davis School of Medicine (Davis and Sacramento, CA). All research procedures using laboratory mice were approved by the UC Davis Animal Care and Use Committee and conform to NIH guidelines.
Neural Sphere Culture
The cortical tissues were dissected from the β-catenin-cKO (β-catenin(ex2–6)flox/flox;hGFAP-Cre) and the wild-type(β-catenin(ex2–6)flox/flox) control brains, with or without the Cre reporter Rosa26-lacZ at the postnatal day 3. Cells were maintained in the Neurobasal Medium (Gibco) with 2% B27, 1% N2, 20 ng/ml epidermal growth factor (EGF), 20 ng/ml basic fibroblast growth factor (bFGF), and 2 mM L-glutamine (all from Invitrogen) at 37°C in 5% CO2 chamber [37]. The medium was half refreshed and the growth factors were replenished every 2 days. The initial passage up to 5 days in vitro (DIV) was recorded as passage 0.
Neural Sphere Diameter, Growth Curve, and Sphere-Forming Assays
Neurosphere diameters were measured from pictured images at passage 3. Only spheres with a diameter >25 mm were counted. To measure the growth curve of the neurospheres, cells were dissociated from the primary neurospheres and seeded at 2 × 104 cells per milliliter (10,000 cells per 0.5 ml in triplicate) into the 24-well plates. Total cell numbers were counted at passages 2–8. For the sphere-forming assay, cells were seeded at 2 × 104 cells per milliliter and the sphere numbers were counted at 5 DIV at passages 1–4.
X-Gal Staining
X-gal staining was performed for Rosa26-lacZ;hGFAP-Cre genetic fate mapping of the sphere-forming cells at passage 3. Spheres were washed twice in phosphate-buffered saline (PBS), fixed for 5 minutes at room temperature in 1% paraformaldehyde (PFA). After washing in PBS, the spheres were transferred to a freshly prepared X-gal staining solution and incubated in a parafilm-sealed culture plate overnight at 37°C. The X-gal staining solution consisted with 1 mg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal), 40 mM citric acid/sodium phosphate, pH 6.0, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM NaCl, and 2 mM MgCl2.
BrdU Incorporation and TUNEL Assays
In vivo BrdU incorporation and TUNEL assays were performed as described previously [37]. For in vitro proliferation and apoptosis assays, dissociated neurosphere cells at passage 3 were seeded onto chamber slides (BD Biosciences) for attachment for 48 hours with the same culture condition. A final concentration of 10 μM BrdU was added and incubated for 24 hours prior to immunolabeling. TUNEL assay was conducted on the dissociated sphere cells at passage 3 according to the manual of Deadend Fluorometric TUNEL System (Promega, Madison, WI, www.promega.com).
Immunohistochemistry and Immunocytochemistry
These were performed as described previously [37]. Frozen coronal brain sections (10–14 μm) were prepared with a cryostat for immunohistochemistry. The following primary antibodies were used: mouse anti-bromodeoxyuridine (BrdU) (DAKO Cytomation), rabbit anti-GFAP (DAKO), goat anti-Nestin (Santa Cruz Biotech, Dallas, TX, www.scbt.com), mouse anti-NeuN (Millipore, Billerica, MA, www.millipore.com), mouse anti-O4 (R&D, Minneapolis, MN, www.rndsystems.com), rabbit anti-Sox2 (Epitomics, Burlingame, CA, www.epitomics.com), rabbit anti-Tbr2 (Millipore), and mouse anti-Tuj1 (Millipore). All images were collected by a laser scanning spectral confocal microscope system (Nikon Eclipse TE2000-E2, Tokyo, Japan, www.nikon.com) and processed with the Adobe Photoshop 7.0 software (Adobe Systems, San Jose, CA, www.adobe.com).
Neural Sphere Differentiation Assays
For the differentiation of NSCs, primary neurospheres were dissociated with trypsin (Gibco, Carlsbad, CA, www.lifetechnologies.com) for 15 minutes, immediately followed by addition of trypsin inhibitors (Gibco) and mechanical trituration. Dissociated cells were plated on glass coverslips pre-coated with poly(D-lysine) (Sigma, St. Louis, MO, www.sigmaaldrich.com) in neurobasal medium containing 1% fetal calf serum (Gibco), 1 μg/ml insulin, 20 μM dopamine, and 100 μM ascorbic acid (all from Sigma) without EGF and bFGF. After 5 days of differentiation, cells were fixed with 4% PFA for 15 minutes and then processed for immunocytochemistry.
Lentiviral Construction, Production, and Infection
Full length Pax6 cDNA was inserted into the pLentiviral vector just after the C-terminal of the FLAG-tag sequence (pLenti-Pax6). The recombinant lentiviruses were produced by cotransfecting 293T cells with 7.5 μg of pLenti-Pax6, 7.5 μg of psPAX2, and 3.5 μg of pMD2.G. The lentiviral titers were determined by limiting dilution with the pLenti-EGFP as a reference. For infections, 5 × 104 dissociated sphere cells were seeded in the six-well plates. The viruses were added to the cells in the presence of polybrene (Santa Cruz Biotech) on the second day. After 24-hour infection, the viruses were washed out, and the cells were returned to the culture for 48 hours prior to immunocytochemistry and differentiation assays.
Western Blot
Cultured NSCs were lysed in the radioimmunoprecipitation assay buffer (Santa Cruz Biotech) mixed with proteinase inhibitors (10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM phenyl-methylsulfonyl fluoride, 0.2 mM sodium orthovanadate, and 1 mM NaF), and homogenized with a sonicator. Protein concentration of each sample was determined by the BCA protein assay kit (Thermo Scientific, Waltham, MA, www.thermoscientific.com). Protein (50 μg) was electrophoresed on 15% SDS-polyacrylamide gel and transferred to the nitrocellulose membrane (Bio-Rad, Hercules, CA, www.bio-rad.com). The membrane was blocked by 5% non-fat dry milk in Tris Buffered Saline with Tween 20 (TBSF) (10 mM Tris-Cl, pH 7.5, 150 mM NaCl, and 0.05% Tween 20) and then incubated with the primary antibody diluted in the blocking buffer overnight at 4°C. Mouse anti-β-catenin (ab22656, Abcam, Cambridge, UK, www.abcam.com), goat anti-Actin (sc-1616, Santa Cruz Biotech), rabbit anti-FLAG (A00170, Genscript), and rabbit anti-Gapdh (PA1–16780, Pierce, Rockford, IL, www.piercenet.com) were used. After washing, the blots were incubated with the secondary antibodies conjugated with the fluorescence (LI-COR Biosciences, Lincoln, NE, www.licor.com) at 1:10,000 in TBST or with the horseradish peroxidase (GE Healthcare, Fairfield, CT, www.gehealthcare.com) at 1:40,000 for 1 hour at room temperature. Immunoblots were visualized with the Odyssey Imaging System (ODYSSEY CLx, LI-COR Biosciences) or with the Chemiluminescent HRP Substrate (Millipore) according to the manufacturer’s instruction.
RNA Isolation and Real-Time Quantitative PCR
Total RNA was isolated from the tissues or cultured cells by the RNeasy purification kit (QIAGEN, Venlo, Netherlands, www.qiagen.com). After denaturing the total RNA at 70°C for 10 minutes, cDNA was synthesized with the oligo-dT primer and the reverse transcriptase (Bio-Rad). Real-time quantitative PCR was performed using the SYBR Green Master Mix (Roche, Basel, Switzerland, www.roche.com). Accumulation of fluorescent products was monitored by real-time PCR using lightcycler 480 systems (Roche). A melting curve was generated to ensure that a single peak of the predicted Tm was produced and no primer-dimer complexes were present. The mRNA levels of β-catenin, Pax6, or Tbr2 were normalized to the mRNA levels of the housekeeping gene Gapdh to allow comparisons among different experimental groups using the delta Ct method [37]. The following primer pairs were used for qPCR:
Gapdh, 5′-TGCTGAGTATGTCGTGGAGTCT-3′, 5′-CATATTTCTC GTGGTTCACACC-3′; β-catenin, 5′-GTGCAATTCCTGAGCTGACA-3′, 5′-CTTAAAGATGGCCAGCAAGC-3′; Pax6, 5′-AGTTCTTCGCAACC TGGCTA-3′, 5′-GTGTTCTCTCCCCCTCCTTC-3′; Tbr2, 5′-CTCCTCT CACCCCAACAGAG-3′, 5′-GAAGGTCGGGTCAGGGTAAT-3′.
In Situ Hybridization and H&E Staining
Nonradioactive in situ hybridization was performed using digoxigenin-labeled riboprobes as described previously [37]. The in situ probes were amplified by RT-PCR from mouse brain cDNAs with primer pairs referred to Allen Brain Atlas (www.brain-map.org). After color development, the sections were dehydrated and mounted by coverslips. For H&E staining, mouse brains were fixed in 4% PFA at 4°C overnight and dehydrated through gradient ethanol. The fixed samples were embedded in paraffin, sectioned with a microtome (Microm), and stained with H&E solutions according to the standard protocol. Bright-field digital images were collected on an Olympus DSU Spinning Disk confocal microscope (Olympus BX61, Center Valley, PA, www.olympusamerica.com).
Luciferase Assay
The 497-bp promoter region of the mouse Pax6 gene, which contains a conserved Tcf/Lef-binding site, and the same promoter region with the binding site deleted were amplified by PCR and cloned into the pGL2 basic vector to acquire the pPax6-WT-Luc and pPax6-Mut-Luc constructs, respectively (Fig. 6A). Transient transfection was performed in L cells and primary cortical cells with Lipofectamine 2000 reagent following the manufacturer’s instructions (Invitrogen). Cells were transfected with pPax6-WT-Luc or pPax6-Mut-Luc in combination with a control expression vector pcDNA3 or the expression constructs of Lef1, dnLef1 (dominant negative Lef1), and/or aβ-catenin (constitutively active β-catenin). Renilla luciferase reporter plasmid pRL-CMV (2 ng) was also cotransfected into each sample as an internal control. Primary cortical cells were prepared from the wild-type embryos by dissecting the tissue in cold PBS, which was then digested at 37°C in Hank’s solution containing 0.025% trypsin (Gibco), centrifuged at 1,000 rpm for 10 minutes, cultured in 10% fetal bovine serum for 12 hours, and treated with 12 mM LiCl (Sigma). After 24 hours of transfection, luciferase activity was assessed with a dual luciferase reporter assay system (Promega) according to the manufacturer’s instructions. The enzyme activity was normalized for efficiency of the transfection on the basis of Renilla luciferase activity levels and reported as relative light units. All reporter assays were performed in triplicate and at least two individual experiments, and standard errors are denoted by bars in the figures.
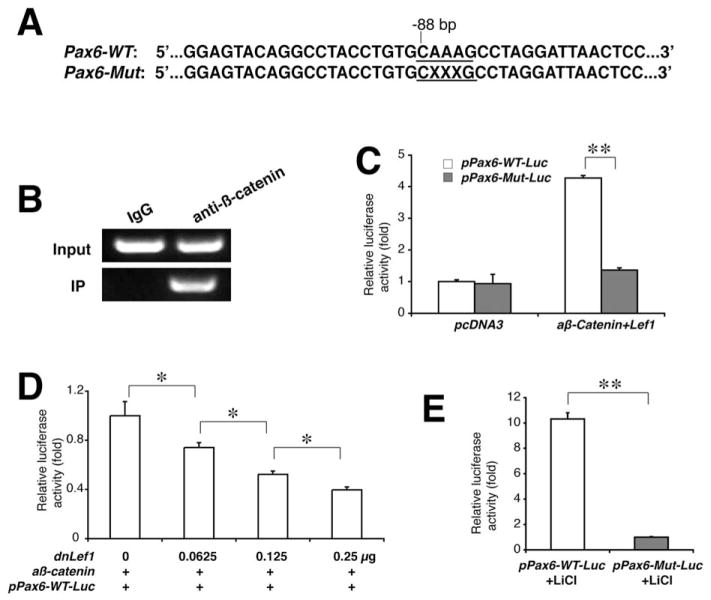
Figure 6.
Transcriptional activation of the Pax6 promoter by Wnt/β-catenin signaling. (A): The wild-type and mutated (×××, deleted base pairs) Tcf/Lef1-binding sites are shown in the mouse Pax6 promoter region. (B): Chromatin immunoprecipitation demonstrates the specific recruitment of the Tcf/Lef1-binding site in the Pax6 promoter region by β-catenin antibodies, not by the nonspecific IgG, from the wild-type neocortex of the P3 mouse. (C): Luciferase reporter assays demonstrate the specific activation of the wild-type promoter, not the mutated, after cotransfected with Lef1 and active β-catenin cDNAs. (D): Dose-dependent repression of the wild-type Pax6 promoter activity by dominant negative (dn) Lef1. (E): Luciferase assays on the primary neocortical cells. The cells were isolated from P3 mouse brains and transfected with either intact pPax6-WT-Luc or mutated pPax6-Mut-Luc plasmids, then stimulated by lithium chloride. *, p < .05; **, p < .01.
Chromatin Immunoprecipitation
Chromatin Immunoprecipitation (ChIP) was performed using the Chromatin Immunoprecipitation Assay Kit (Upstate, Billerica, MA, www.millipore.com) according to the manufacturer’s instructions. For each experimental condition, 30 mg neocortical tissue dissected from the wild-type mouse was used. Tissue was cross-linked with 1% formaldehyde for 15 minutes at room temperature. After sonication, the lysate was immunoprecipitated using an antibody against β-catenin (Santa Cruz Biotech). Rabbit IgG was used as a negative control. To amplify the β-catenin/Tcf binding site of the Pax6 promoter, the following sequences of the primers were used: 5′-CAGTTC AGGCACCAGGTTTT-3′, 5′-CTGCATGCTGGAGCTGGT-3′.
Statistical Analysis
At least three normal controls and three mutants were used for each statistical evaluation. Significances were assessed by Student’s t test or pairwise (one-way analysis of variance, ANOVA) when appropriate. In all cases, p ≤ .05 was considered significant.
Results
Conditional Ablation of β-Catenin in Radial Glia Disrupts Pax6 Expression, Radial Glia and Progenitor Productions, and Neocortical Development
To address the role and underlying mechanisms of β-catenin signaling in the radial glia/NSCs during neocortical development, we used the Cre-loxP conditional gene-targeting approach by crossing β-catenin(ex2–6)flox mice [52] with the hGFAP-Cre transgenic mice [53]. The Cre recombinase activity driven by hGFAP promoter has been demonstrated in the multipotent radial glia of the forebrain as early as embryonic day 13.5 (E13.5) [53, 55]. Real-time PCR demonstrated that the β-catenin mRNAs were markedly decreased in the embryonic and postnatal neocortex of the conditional knockout β-catenin(ex2–6)flox/flox;hGFAP-Cre (abbreviated as β-catenin-cKO) mice (Supporting Information Fig. S1A). In situ hybridization demonstrated that the expression of Pax6, one of the earliest neurogenic transcription factors in radial glia [47], was not apparently altered at E13.5 (Supporting Information Fig. S2A), but it was clearly diminished at E15.5 and drastically diminished at E16.5, in the ventricular (VZ)/SVZ of the mutant cortex (Fig. 1A).
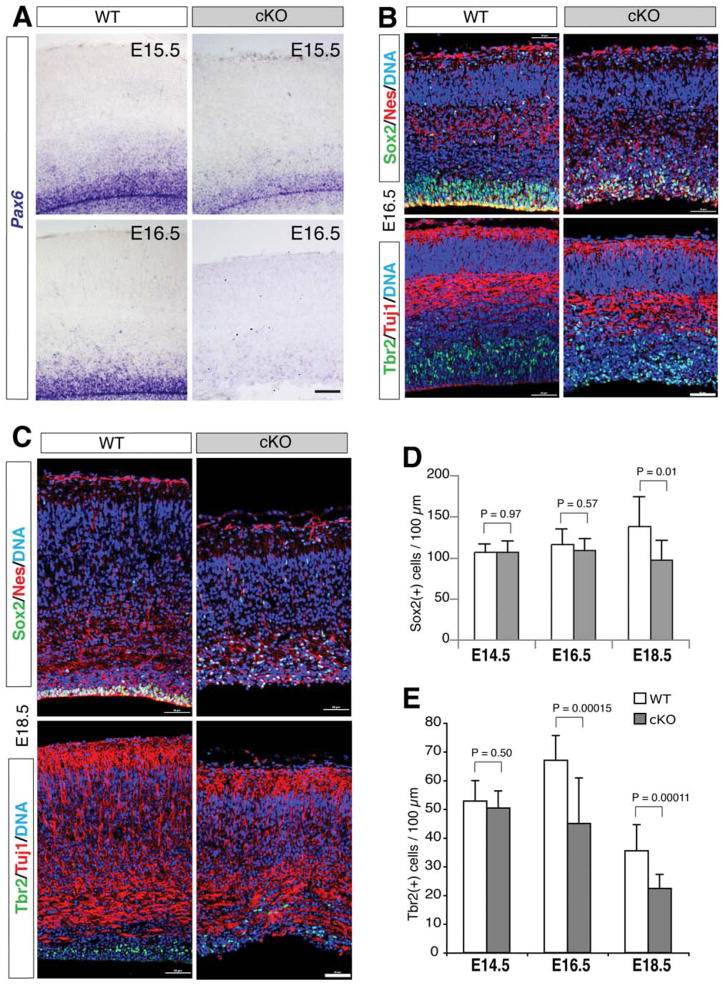
Figure 1.
Conditional ablation of β-catenin with hGFAP-Cre disrupts radial glia and intermediate progenitors in late embryonic neocortex. (A): In situ hybridization of Pax6 mRNAs. (B, C): Double immunofluorescence of Sox2/Nestin and Tbr2/Tuj1. (D, E): Histograms of the numbers of Sox2(+) and Tbr2(+) cells in 100-μm-wide neocortical columns. Scale bars =50 μm.
Double immunolabeling of Sox2 and Nestin for radial glia/ NSCs showed no significant changes of Sox2(+) cells at E14.5 and E16.5, but a 30% reduction of Sox2(+) cells was detected at E18.5 of the mutant neocortex (Fig. 1B–1D and Supporting Information Fig. S2B). Also, the compact localization of Sox2(+);Nestin(+) radial glia in the VZ/SVZ was disrupted in the mutants at both E16.5 and E18.5 (Fig. 1B, 1C). Moreover, approximately 33% and 37% reductions of Tbr2(+) intermediate progenitors were detected in the mutant neocortex of E16.5 and E18.5, respectively, while no apparent change of Tuj1 immunolabeling was found in these mutants (Fig. 1B, 1C, 1E). These results indicate a defective production of radial glia and intermediate progenitors in the mutant embryonic neocortex. As a consequence, although statistically insignificant at E16.5, the thickness of the mutant neocortex was significantly reduced from E18.5 onward (Supporting Information Fig. S1B, S1C). The mutant mice were postnatally viable, while many died prior to wean or adulthood.
Repressed Neurosphere Formation from β-Catenin-Deficient Radial Glia/NSCs
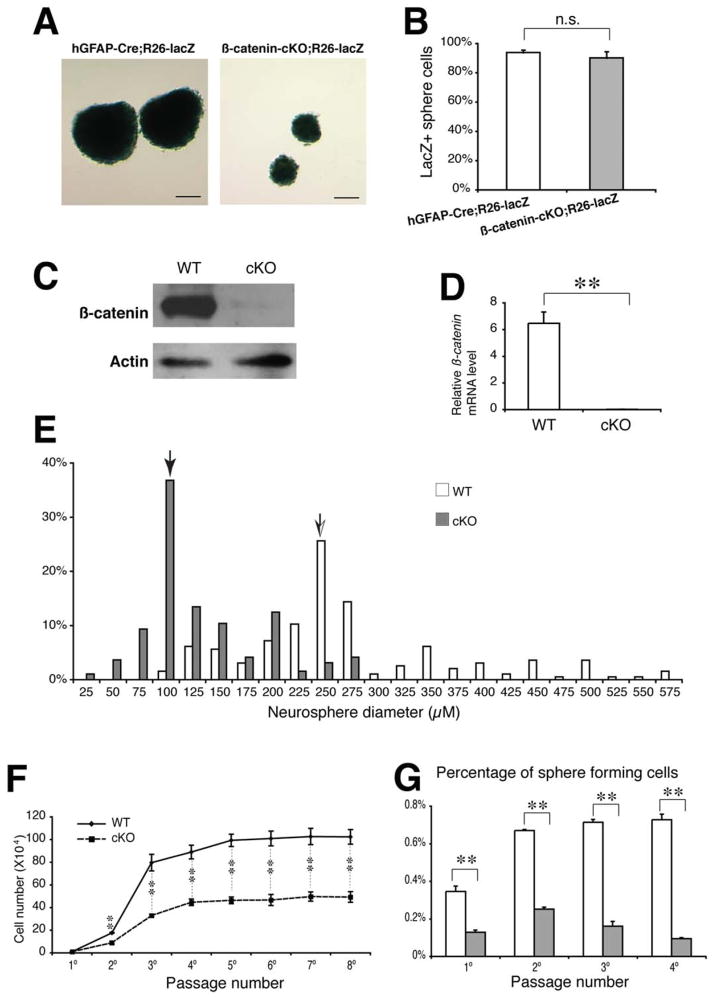
To address the role of β-catenin signaling in the self-renewal of the cortical radial glia/NSCs, we performed neurosphere assays for NSCs derived from the dissociated cerebral cortex of neonatal mice. Genetic fate mapping using the Cre reporter Rosa26-lacZ mouse demonstrated that most wild-type (93.8%) or mutant (90.2%) neurospheres are positive by X-gal staining, indicating they originated from the hGFAP-Cre activated radial glia/NSCs (Fig. 2A, 2B). Accordingly, β-catenin mRNAs or proteins had been effectively ablated in the mutant neurospheres (Fig. 2C, 2D). Neurosphere size is a rough parameter for self-renewal potential of NSCs. Mutant neurospheres were significantly smaller than the wild-type. We measured diameters of 193 mutant and 195 wild-type neurospheres at passage 3. The median diameter of the mutant neurospheres (36.8%) was 100 μm, while the wild-type (25.6%) was 250 μm (Fig. 2E). The average diameter of the mutant was 129 μm, and the wild-type neurosphere was 264 μm. The difference was statistically significant.
Figure 2.
Repressed neurosphere formation and self-renewal potentials of the neocortical radial glia/neural stem cells cultured from the β-catenin-cKO mutants. (A, B): X-gal staining and the percentages of the hGFAP-Cre;Rosa26-lacZ-fate-mapped neurospheres derived from the controls and mutants. (C, D): Immunoblot and real-time PCR results for the β-catenin proteins or mRNAs in the normal and mutant neurospheres. (E): Percentage distributions of the control and mutant neurosphere diameters. Arrows indicate the median sizes of the control or mutant neurospheres. (F): Growth curves for expanded cell numbers from the primary up to eighth passages of the neurospheres. (G): Percentages of the sphere-forming cells at the primary, secondary, third, and fourth passages of the neurosphere cultures. **, p < .01. Scale bars =150 μm. Abbreviation: hGFAP, human glial fibrillary acidic protein.
The growth curve of the neurospheres from multiple generations is a more accurate parameter for the self-renewal capabilities of the NSCs. We cultured the neurospheres for seven consecutive passages starting with equal cell numbers (2 × 104) of the wild-type and the mutant neurospheres at passage 1, and counted the total cell numbers in each sample at each passage from passages 2–8. The average cell numbers (from triplicate samples) of the wild-type neurospheres increased sharply by 8.8-fold at passage 2 and by 40-fold at passage 3 (Fig. 2F). The increased cell numbers reached the peak around passage 4 and have little changes from passage 5 onward. The average cell numbers of the mutant neurospheres increased by much lower fold, 4.5 at passage 2 and 16.5 at passage 3, and reached the peak by passage 4, which shows only 50% or less of the total cell numbers when compared with that in the wild-type at the same passage (Fig. 2F).
Moreover, we performed sphere-forming assays by analyzing the percentages of the cells that formed neurospheres at each of the first four consecutive passages. The percentages of the sphere-forming cells were 2.7-fold decreased in the mutant cultures compared with the wild-type cultures at passages 1 and 2, respectively (Fig. 2G). Remarkably, the percentages of the mutant sphere-forming cells decreased steadily at each successive passage after passage 2, while the percentages of the wild-type sphere-forming cells maintained at a similar level from passages 2–4 (Fig. 2G). These findings indicate that loss of β-catenin signaling prevents self-renewal of the neocortical NSCs.
Diminished Proliferation in β-Catenin-Deficient SVZ and NSCs
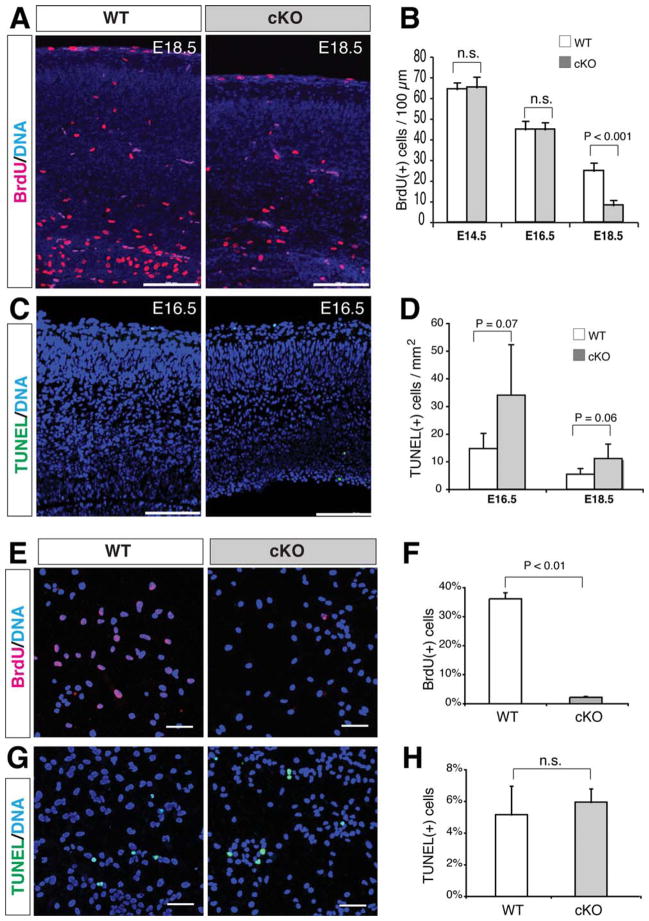
To illuminate the cellular mechanisms responsible for repressed neocortical NSC populations in the mutants, we performed BrdU incorporation and TUNEL assays in vivo and in vitro. The percentage of BrdU(+) proliferating cells in the mutant neocortex was significantly lower than that in the wild-type controls at E18.5, but not at E14.5 and E16.5 (Fig. 3A, 3B and Supporting Information Fig. S3A). The percentages of the TUNEL(+) apoptotic cells were increased (while just out the border of statistical significance) in the mutant neocortex at both E16.5 and E18.5 (Fig. 3C, 3D and Supporting Information Fig. S3B). Consistently, the percentage of BrdU(+) proliferating cells in the dissociated sphere cells of the mutants was significantly lower than that in the wild-type controls (Fig. 3E, 3F). However, the percentages of the TUNEL(+) apoptotic cells were not significantly different between the mutant and wild-type sphere cells (Fig. 3G, 3H). In agreement with the in vivo results, we also detected that the mRNAs of the cortical transcription factor Pax6 and its potential downstream factor Tbr2 were significantly diminished in β-catenin-deficient neurospheres compared with that in the wild-type spheres (Supporting Information Fig. S4A, S4B). These results suggest that β-catenin regulates Pax6 expression and proliferation in neocortical radial glia/NSCs.
Figure 3.
In vivo and in vitro changes of proliferation and apoptosis in the β-catenin-cKO neocortex or dissociated neurosphere cells. (A, B): Significantly reduced BrdU(+) cells (per 100-μm-wide neocortical column) in E18.5 mutants. (C, D): Increased TUNEL(+) cells (per square mm neocortex) in E16.5 and E18.5 mutants. (E–H): Significantly reduced BrdU(+) percentage and no difference of TUNEL(+) percentage in the dissociated mutant sphere cells compared with those in the wild-type. n.s., no statistical difference (p > .05). Scale bars in A and C, 100 μm; scale bars in E and G, 50 μm.
Impaired Neuronal and Glial Differentiations of β-Catenin-Deficient Radial Glia/NSCs
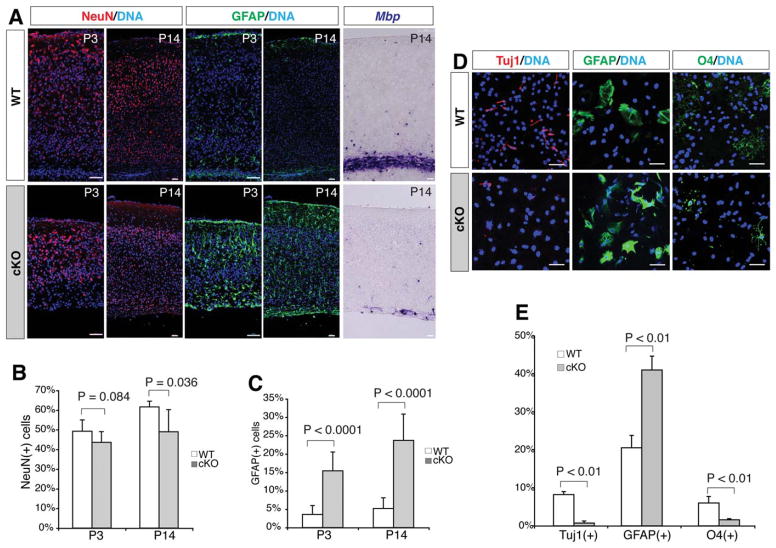
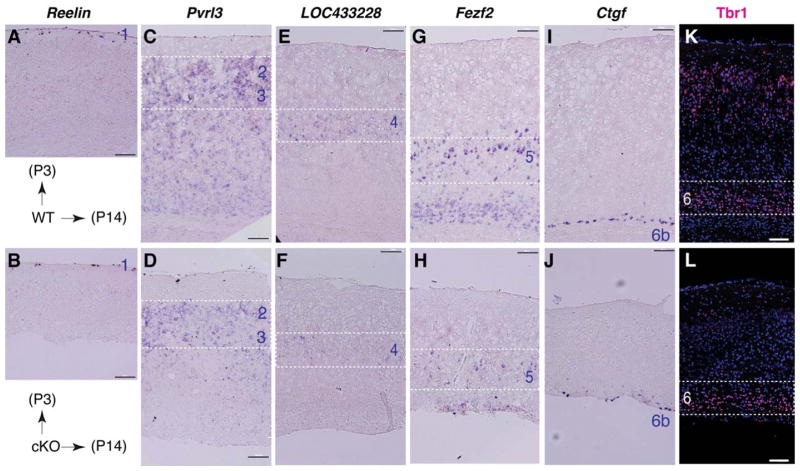
To analyze the effects of the β-catenin signaling on differentiation of the cortical radial glia/NSCs, we examined lineage-specific markers in vivo and in vitro. Immunohistochemistry showed that the percentages of the NeuN(+) neurons declined from 49.2% in the control to 43.6% in the mutant neocortex at P3 (p =.084), and from 61.6% in the control to 49.0% in the mutant neocortex at P14 (p =.036) (Fig. 4A, 4B). However, the percentages of the GFAP(+) radial glia and astrocyte lineage cells in the mutant cortical plate increased significantly from that in the wild-type control, by 4.3-fold at P3 and by 4.6-fold at P14 (Fig. 4A, 4C). In contrast, the in situ signals of Mbp mRNAs expressed in the mature oligodendrocytes were dramatically diminished in the cortical white matter region of the P14 mutants (Fig. 4A). In vitro assays showed that production of the Tuj1(+) neurons and O4(+) oligodendrocytes differentiated from β-catenin-deficient NSCs was decreased by 11-fold and 4-fold, respectively, while production of the GFAP(+) astrocytes from the mutants was increased by twofold, from those in the wild-type controls (Fig. 4D, 4E). These consistent in vivo and in vitro results suggest that β-catenin signaling promotes neurogenesis and oligodendrogenesis, but represses astrogliogenesis from radial glia/NSCs during neocortical development. In addition, in situ hybridization and immunohistochemistry for representative neocortical layer markers revealed that cortical layer-specific neuronal patterning and migration were not significantly affected, but respective neuronal numbers were decreased in the layers 2–6b of the mutants at P14 (Fig. 5).
Figure 4.
Repressed neurogenesis and oligodendrogenesis, and increased astrogliogenesis by β-catenin-cKO neural stem cells in vivo and in vitro. (A): Immunohistochemistry of the neuronal nuclei NeuN and astrocyte marker GFAP at P3 and P14, and in situ hybridization of the mature oligodendrocyte marker Mbp in P14 neocortex. Note the overall thinner neocortex in the mutants. (B, C): Reduced NeuN(+) and increased GFAP(+) cell percentages in P3 and P14 mutant neocortex. (D): Immunocytochemistry of the differentiated cortical sphere cells with antibodies against neuronal class III β-tubulin (Tuj1), GFAP, and oligodendrocyte marker O4. (E): Percentages of Tuj1(+), O4(+), and GFAP(+) cells differentiated from the wild-type or mutant neurospheres. Scale bars =50 μm. Abbreviation: GFAP, glial fibrillary acidic protein.
Figure 5.
Representative neocortical layer markers in postnatal wild-type and β-catenin-cKO neocortex. (A-J): In situ hybridization of Reelin (layer 1) at P3, and Pvrl3 (layers 2/3), LOC433228 (layer 4), Fezf2 (layer 5), and Ctgf (layer 6b) at P14 normal and mutant neocortex. (K, L): Tbr1 immunolabeled layer 6 neurons in normal control and mutant neocortex at P14. Note the overall thinner neocortex and reduced marker signals in layers 2–6b of the mutants. Scale bars =100 μm.
Pax6 Is a Downstream Target of the Wnt/β-Catenin Signaling
To test whether Pax6 is a direct transcriptional target of the Wnt/β-catenin signaling, we carried out ChIP and promoter luciferase assays. A Tcf/Lef1-binding site, the Wnt-responsive element (WRE), was found in the presumptive promoter region located at −88 bp proximal upstream of the mouse Pax6 gene (Fig. 6A). ChIP results from the cortical tissue samples of neonatal mice demonstrated that the β-catenin antibody, but not the nonspecific IgG, could pull-down the WRE of the Pax6 promoter (Fig. 6B). Luciferase assay results demonstrated that the luciferase activities driven by Pax6 promoter with the wild-type, but not the mutated WRE, were specifically upregulated by constitutively active β-catenin and Lef1 (Fig. 6C), which also showed a dose-dependent repression competed by the dominant negative Lef1 (Fig. 6D). In primary cortical cell cultures, the luciferase activity of the Pax6 promoter with the wild-type WRE was highly responsive to lithium ion, a Wnt signaling agonist, while the promoter with mutated WRE showed little or no response under the same condition (Fig. 6E). These results suggest that Pax6 is transcriptionally regulated by Wnt/β-catenin signaling.
Lentiviral Transduction of Pax6 Rescues Defective Proliferation and Neurogenesis by β-Catenin-Deficient Neurosphere Cells
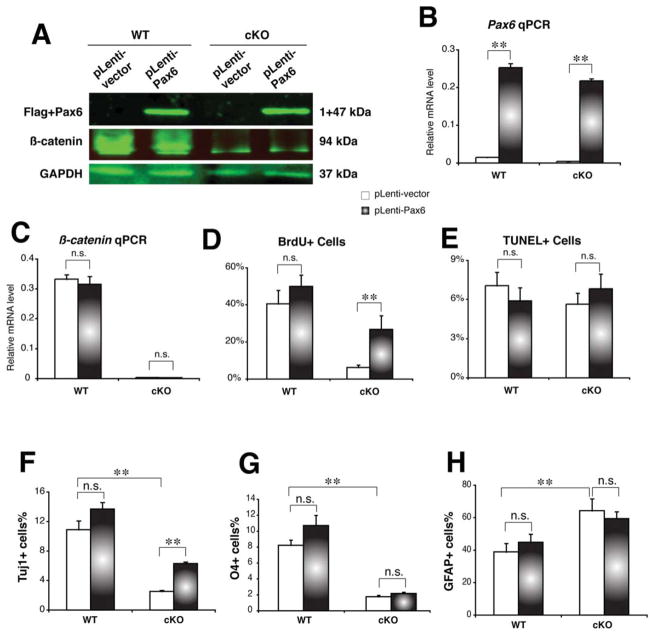
To determine whether Pax6 is a key downstream effector of β-catenin signaling in NSC proliferation and differentiation, we used a lentiviral transduction approach to ectopically express Pax6 cDNA in β-catenin-deficient NSCs. The EGFP expression experiments showed that nearly 100% of the wild-type or β-catenin-deficient neurospheres was efficiently transduced with the ectopic EGFP gene without significant impact on neural sphere formation (Supporting Information Fig. S5). Immunoblot and real-time PCR results revealed that the ectopic Pax6 (fused with the FLAG-tag) was highly expressed, but that did not affect normal or deficient β-catenin expression levels in the wild-type or in mutant neurospheres, respectively (Fig. 7A–7C). The percentage of BrdU(+) cells was significantly increased by 4.3-fold in β-catenin-deficient neurospheres after ectopic transduction of Pax6 (Fig. 7D and Supporting Information Fig. S6A). In contrast, the percentage of BrdU(+) cells was not significantly increased in the wild-type neurospheres after Pax6 transduction. There were no significant changes of the TUNEL(+) cell percentages in either wild-type or β-catenin-deficient neurospheres after Pax6 transduction (Fig. 7E and Supporting Information Fig. S6B). These results suggest that the forced expression of Pax6 partially rescued the defective proliferation of β-catenin-deficient NSCs.
Figure 7.
Lentiviral transduction of Pax6 rescues proliferation defects and repressed neurogenesis in the β-catenin-cKO neurospheres. (A): Immunoblots for the transduced Pax6 (detected by the fused Flag tag) or endogenous β-catenin in the wild-type or mutant neurosphere cells. The blank lentiviral vector was used as the negative transduction controls, and the GAPDH was detected for the protein loading controls. (B, C): Real-time PCR results for Pax6 or β-catenin mRNAs in the wild-type or mutant neurospheres transduced with either Pax6-expressing or blank lentiviruses. (D, E): Percentages of BrdU or TUNEL-positive cells in the wild-type or mutant neurospheres transduced with either Pax6-expressing or blank lentiviruses. (F-H): Percentages of Tuj1(+) neurons, O4(+) oligodendrocytes, and GFAP(+) astrocytes differentiated from the wild-type or mutant neurospheres transduced with either Pax6-expressing or blank lentiviruses. n.s., no statistical significance (p > .05); **, p < .01.
We then examined the effects of the lentiviral transduction of Pax6 on neural differentiation of the wild-type and the mutant neurosphere cells. Significantly, the percentage of the Tuj1(+) neurons increased by 2.5-fold, but neither the percentage of O4(+) oligodendrocytes nor that of GFAP(+) astrocytes were changed significantly (Fig. 7F–7H and Supporting Information Fig. S7). These results suggest that the forced expression of Pax6 can rescue impaired neurogenesis, but not oligodendrogenesis or astrogliogenesis in β-catenin-deficient NSCs. Taken together, our results suggest that the β-catenin/ Pax6 signaling exerts critical roles in self-renewal and neurogenesis of the cortical radial glia/NSCs.
Discussion
Wnt/β-catenin signaling pathway is known to regulate NSC functions in early gestation or in adult neurogenesis, but the underlying molecular mechanisms are not well understood. In this study, we demonstrate that Wnt/β-catenin signaling transcriptionally activates Pax6, and directs proliferation and neurogenesis of NSCs derived from cortical radial glia.
The structure of the cerebral cortex is precisely regulated by genetically encoded signaling molecules and transcription factors [56, 57]. We showed that mice exhibit defective radial glia and hypoplastic cortex from late gestation after conditional ablation of β-catenin with hGFAP-Cre. Previous studies have shown more dramatic cortical or forebrain defects and perinatal lethality in other β-catenin-deficient mutants. The entire forebrain or cerebral cortex are missing in the mutants when β-catenin is ablated with Wnt1-Cre or Foxg1-Cre in neuroepithelial cells and related neural tube derivatives [52, 58, 59]. Abnormal dorsoventral specification of the telencephalon is found in the mutants when β-catenin is ablated with Nes8-Cre or Nes11-Cre in neuroepithelial cells at E8.5 or E10.5, respectively [60]. Severely disrupted neuroepithelial organization is observed in the mutants when β-catenin is ablated with D6-Cre after E10.5 [30]. This study observed hypoplastic neocortex without severe disruption of cortical layer organization and patterning in the β-catenin;hGFAP-Cre mutants. However, cell adhesion defects of β-catenin-loss-of-function contribute to cortical cellular disorganizations during early gestation [58], which may also affect radial glia localization in the VZ/SVZ during late gestation as observed in this study. The underlying mechanisms will be addressed further in future studies. Together, these results indicate that β-catenin exerts age-specific functions in the neuroepithelial and radial glial stem cells for proper cortical patterning, organization, and/or neurogenesis during brain development. The hGFAP promoter is activated in the cortical radial glia around E13.5 [53, 61]. Thus, the conditional loss-of-function of β-catenin in radial glia at mid to late gestation has only a mild impact on early cortical patterning and layer organization, but a major impact on proliferation, differentiation, localization, and probably survival of radial glial stem cells during late gestation and postnatal development.
To address the cellular mechanisms of β-catenin signaling in cortical radial glia, this study used the neurosphere assay, which provides a quantitative in vitro model system to analyze neurogenesis and neural development and is a useful tool to study proliferation, self-renewal, and multipotency of NSCs [62, 63]. We detected dramatic deductions of sphere diameters and expansion of cell numbers from β-catenin-deficient neurospheres originated from the hGFAP-Cre-expressing cortical radial glia as demonstrated by the genetic fate mapping. Neurospheres consist of most-proliferating progenitors derived from the clonal stem cells. Our proliferation and apoptosis assays demonstrated that β-catenin ablation suppressed proliferation of sphere cells without affecting cell survival, further supporting the role of β-catenin in promoting proliferation of NSCs. This is contrary to a previous study which proposed that Wnt signaling inhibits the self-renewal capacity of mouse cortical neural precursors [64]. However, our results correlate with other findings, such as that β-catenin gain-of-function promotes the proliferation of neural progenitors in the developing cortex or adult SVZ [32, 65, 66], and that β-catenin loss-of-function caused early cell cycle exit and premature neurogenesis by the cortical progenitors [67, 68]. Our sphere-forming assays showed that the percentages of the sphere-forming cells are significantly lower in primary neurospheres and also in each successive passage of the mutant cultures than that in the control cultures. These results demonstrate an essential role of β-catenin in controlling self-renewal of NSCs.
Our in vivo and in vitro results showed that β-catenin-deficiency represses neuronal differentiation from the mutant radial glia or neurosphere cells. This is in concordance with previous studies demonstrating that ectopic Wnts or stabilized β-catenin promote neuronal differentiation of embryonic cortical precursors or intermediate progenitors [64, 69]. However, stabilized β-catenin in adult SVZ has been shown to inhibit neuronal differentiation [65], suggesting the age- and/or cell type-specific roles of Wnt/β-catenin signaling in neuronal differentiation.
At the molecular level, this study demonstrated that Pax6 is a transcriptional target and downstream effector of Wnt/β-catenin signaling in proliferation and neuronal differentiation of the cortical radial glia, a major NSC population in the developing cortex. Based on the sequential gene expression timing correlated with the region-specific cell fates during neurogenesis, a transcription factor cascade of Pax6-Ngn2-Tbr2-NeuroD-Tbr1, in the order of radial glia to intermediate progenitors to postmitotic neurons, has been defined in the embryonic neocortex [49, 70]. Pax6 is dominantly expressed in the proliferating radial glia in both mouse and human developing cortex [49, 71]. With in situ hybridization and real-time PCR, we showed a dramatic loss of Pax6 mRNAs in the cortical SVZ and the radial glia-derived neurosphere cells of the β-catenin-deficient mice. This is in agreement with a gain-of-function study that persistent activation of β-catenin in neuroepithelial cells leads to the expansion of Pax6 expression in the cortical SVZ with unknown mechanisms [66]. Pax6 plays important roles in radial glial function and neurogenesis. Pax6-deficiency by either genetic mutations in mice or gene-knockdown in human cells leads to defective proliferation and/or neuronal production of radial glia [47, 71, 72]. Thus, Pax6 is an earliest transcription factor in cortical neurogenesis and a potential downstream effector of β-catenin signaling in radial glia.
The WREs or Tcf/Lef1-binding sites are composed of a highly conserved consensus sequence 5′-(A/T)(A/T)CAA(A/T)G-3′ [73]. We found a Tcf/Lef1-binding site in the upstream promoter region of mouse Pax6. With ChIP and luciferase reporter assays, we demonstrate the specific DNA-binding and transcriptional activities of the Pax6 promoter targeted by the β-catenin signaling in both L cells and primary cortical cells, revealing a novel molecular mechanism by which Wnt/β-catenin signaling controls the expression of Pax6 in radial glia during cortical development. Interestingly, a recent study showed that Pax6 expression is also lost in the cortical radial glia of β-catenin gain-of-function mutants, after an initial expansion of Pax6 expression [74]. Although the underlying mechanism remains unclear, these results indicate that a balanced β-catenin signaling is required for proper Pax6 expression during cortical development.
Several previous studies report that Ngn1 and N-Myc are also potential downstream target genes of β-catenin signaling in cortical patterning and embryonic neurogenesis [64, 75]. The neurogenic transcription factors Ngn1/2 are expressed in the differentiated neuronal progenitors, and are potential downstream factors from Pax6. It is unclear whether β-catenin and Pax6 act sequentially or concurrently on the regulation of Ngn1/2 expression. In contrast, N-myc has been shown to promote the expansion of the neuronal progenitor cells and inhibit neuronal differentiation, particularly in the developing cerebellum [76]. Conditional ablation of N-myc in neuroepithelial cells leads to a mild repression of the cortical neurogenesis with no changes in Pax6 expression [75], suggesting that N-myc may cooperate or act after Pax6 on cortical neurogenesis. These results also suggest that β-catenin signaling plays age- and/or cell-type-specific roles through different sets of downstream effectors during cortical development.
With the lentiviral transduction approach, we further demonstrated that ectopic expression of Pax6 cDNA in β-catenin-deficient neurosphere cells rescued both defective proliferation and neurogenesis. Mammalian Pax6 gene encodes three isoforms: the canonical Pax6, Pax6(5a), and Pax6dPD [77]. The canonical Pax6 protein contains a paired domain (PD), a homeodomain, and a proline/serine/threonine-rich domain, while Pax6(5a) protein contains a 14-residue insertion in the PD domain, and the Pax6dPD protein lacks a PD domain [78–82]. These three isoforms have different expression levels in the brain, with the highest expression level of the canonical Pax6 during early neurogenesis [83, 84]. They also have different functions: the canonical Pax6 regulates both the cell proliferation and cell fate; Pax6(5a) affects the cell proliferation, but not cell fate; and the role of Pax6dPD in brain development remains unclear [85]. Based on their expression levels and functions in the brain, we cloned the canonical Pax6 cDNA for the lentiviral expression, which demonstrated partial rescue of the altered proliferation and neurogenesis in β-catenin-deficient neurosphere cells, suggesting that the other two isoforms of Pax6 may also play a role in the same processes.
Intriguingly, our results also demonstrated that lentiviral transduction of canonical Pax6 cDNA mildly promoted oligodendrocyte production in the wild-type, but could not rescue the defective oligodendrogenesis in β-catenin-deficient neurosphere cells. Moreover, elevated production of astrocytes was observed in the β-catenin-deficient mutants, which could not be rescued by ectopic expression of canonical Pax6 cDNA. The abnormal astrogliogenesis is partially supported by a recent study that excess astrogenesis is observed in the cortical NSC cultures from the Pax6 heterozygous mutant mice [72]. The underlying mechanisms of β-catenin signaling in oligodendrogenesis and the β-catenin/Pax6 cascade in astrogliogenesis need to be investigated in future studies.
Conclusion
Using conditional gene-targeting, immunohistochemistry, in situ hybridization, ChIP, promoter luciferase assays, neurosphere culture, lentiviral transduction rescue, and other related approaches, this study revealed that Pax6 is a transcriptional target and downstream effector of Wnt/β-catenin signaling in self-renewal and neuronal differentiation of radial glia, a major NSC population from midgestation to postnatal CNS development. Understanding the underlying molecular mechanisms of Wnt/β-catenin signaling in regulating self-renewal and differentiation of NSCs may provide the basis for the prevention and treatment of a wide range of neurodevelopmental disorders and related neurological disease.
Supplementary Material
Acknowledgments
We are grateful to Sylvia Evans (UC San Diego) and Sam Pleasure (UC San Francisco) for the active β-catenin or dnLef1 plasmids, and Jassimran Bainiwal and other members at the Zhou laboratory for assistance or discussion. Q.G. and T.Y. received postdoctoral stem cell training fellowships from the California Institute for Regenerative Medicine. This work was supported by grants from the Shriners Hospitals for Children (research Grants 86100 and 87500 to C.J.Z.) and NIH (1R01DE021696 to C.J.Z.).
Footnotes
Author Contributions
Q.G., A.L., and R.S.: conception and design, collection and/or assembly of data, and data analysis and interpretation; T.Y., A.S., and B.N.: collection of data; D.P., J.W., and H.W.C.: data analysis and interpretation; C.J.Z.: conception and design, collection and assembly of data, data analysis and interpretation, financial support, manuscript writing, and final approval of manuscript. Q.G., A.L., and R.S. contributed equally to this article.
Disclosure of Potential Conflicts of Interest
The authors have no potential conflicts of interest.
References
- 1.McKay R. Stem cells in the central nervous system. Science. 1997;276:66–71. doi: 10.1126/science.276.5309.66. [DOI] [PubMed] [Google Scholar]
- 2.Gage FH. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- 3.Temple S. The development of neural stem cells. Nature. 2001;414:112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- 4.Spassky N, Merkle FT, Flames N, et al. Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J Neurosci. 2005;25:10–18. doi: 10.1523/JNEUROSCI.1108-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panchision DM, McKay RD. The control of neural stem cells by morphogenic signals. Curr Opin Genet Dev. 2002;12:478–487. doi: 10.1016/s0959-437x(02)00329-5. [DOI] [PubMed] [Google Scholar]
- 6.Mu Y, Lee SW, Gage FH. Signaling in adult neurogenesis. Curr Opin Neurobiol. 2010;20:416–423. doi: 10.1016/j.conb.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang YZ, Plane JM, Jiang P, et al. Concise review: Quiescent and active states of endogenous adult neural stem cells: Identification and characterization. Stem Cells. 2011;29:907–912. doi: 10.1002/stem.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faigle R, Song H. Signaling mechanisms regulating adult neural stem cells and neurogenesis. Biochim Biophys Acta. 2013;1830:2435–2448. doi: 10.1016/j.bbagen.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 10.Nusse R. Wnt signaling and stem cell control. Cell Res. 2008;18:523–527. doi: 10.1038/cr.2008.47. [DOI] [PubMed] [Google Scholar]
- 11.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 12.Kuhl SJ, Kuhl M. On the role of Wnt/ beta-catenin signaling in stem cells. Biochim Biophys Acta. 2013;1830:2297–2306. doi: 10.1016/j.bbagen.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Sato N, Meijer L, Skaltsounis L, et al. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 14.Singla DK, Schneider DJ, LeWinter MM, et al. Wnt3a but not Wnt11 supports self-renewal of embryonic stem cells. Biochem Biophys Res Commun. 2006;345:789–795. doi: 10.1016/j.bbrc.2006.04.125. [DOI] [PubMed] [Google Scholar]
- 15.Ying QL, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buehr M, Meek S, Blair K, et al. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 17.ten Berge D, Kurek D, Blauwkamp T, et al. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat Cell Biol. 2011;13:1070–1075. doi: 10.1038/ncb2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reya T, Duncan AW, Ailles L, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 19.Otto A, Schmidt C, Luke G, et al. Canonical Wnt signalling induces satellite-cell proliferation during adult skeletal muscle regeneration. J Cell Sci. 2008;121:2939–2950. doi: 10.1242/jcs.026534. [DOI] [PubMed] [Google Scholar]
- 20.Marson A, Foreman R, Chevalier B, et al. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3:132–135. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lluis F, Pedone E, Pepe S, et al. Periodic activation of Wnt/beta-catenin signaling enhances somatic cell reprogramming mediated by cell fusion. Cell Stem Cell. 2008;3:493–507. doi: 10.1016/j.stem.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 22.Greco V, Chen T, Rendl M, et al. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4:155–169. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng YA, Nusse R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell. 2010;6:568–577. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaelidis TM, Lie DC. Wnt signaling and neural stem cells: Caught in the Wnt web. Cell Tissue Res. 2008;331:193–210. doi: 10.1007/s00441-007-0476-5. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L, Yang X, Yang S, et al. The Wnt/ beta-catenin signaling pathway in the adult neurogenesis. Eur J Neurosci. 2011;33:1–8. doi: 10.1111/j.1460-9568.2010.7483.x. [DOI] [PubMed] [Google Scholar]
- 27.Harrison-Uy SJ, Pleasure SJ. Wnt signaling and forebrain development. Cold Spring Harb Perspect Biol. 2012;4:a008094. doi: 10.1101/cshperspect.a008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SM, Tole S, Grove E, et al. A local Wnt-3a signal is required for development of the mammalian hippocampus. Development. 2000;127:457–467. doi: 10.1242/dev.127.3.457. [DOI] [PubMed] [Google Scholar]
- 29.Galceran J, Miyashita-Lin EM, Devaney E, et al. Hippocampus development and generation of dentate gyrus granule cells is regulated by LEF1. Development. 2000;127:469–482. doi: 10.1242/dev.127.3.469. [DOI] [PubMed] [Google Scholar]
- 30.Machon O, van den Bout CJ, Backman M, et al. Role of beta-catenin in the developing cortical and hippocampal neuroepithelium. Neuroscience. 2003;122:129–143. doi: 10.1016/s0306-4522(03)00519-0. [DOI] [PubMed] [Google Scholar]
- 31.Lie DC, Colamarino SA, Song HJ, et al. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437:1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- 32.Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- 33.Zhou CJ, Zhao C, Pleasure SJ. Wnt signaling mutants have decreased dentate granule cell production and radial glial scaffolding abnormalities. J Neurosci. 2004;24:121–126. doi: 10.1523/JNEUROSCI.4071-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou CJ, Borello U, Rubenstein JL, et al. Neuronal production and precursor proliferation defects in the neocortex of mice with loss of function in the canonical Wnt signaling pathway. Neuroscience. 2006;142:1119–1131. doi: 10.1016/j.neuroscience.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Kalani MY, Cheshier SH, Cord BJ, et al. Wnt-mediated self-renewal of neural stem/ progenitor cells. Proc Natl Acad Sci USA. 2008;105:16970–16975. doi: 10.1073/pnas.0808616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qu Q, Sun G, Li W, et al. Orphan nuclear receptor TLX activates Wnt/beta-catenin signalling to stimulate neural stem cell proliferation and self-renewal. Nat Cell Biol. 2010;12:31–40. doi: 10.1038/ncb2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang YZ, Yamagami T, Gan Q, et al. Canonical Wnt signaling promotes the proliferation and neurogenesis of peripheral olfactory stem cells during postnatal development and adult regeneration. J Cell Sci. 2011;124:1553–1563. doi: 10.1242/jcs.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alvarez-Buylla A, Garcia-Verdugo JM, Tramontin AD. A unified hypothesis on the lineage of neural stem cells. Nat Rev Neurosci. 2001;2:287–293. doi: 10.1038/35067582. [DOI] [PubMed] [Google Scholar]
- 39.Merkle FT, Tramontin AD, Garcia-Verdugo JM, et al. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc Natl Acad Sci USA. 2004;101:17528–17532. doi: 10.1073/pnas.0407893101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anthony TE, Klein C, Fishell G, et al. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- 41.Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 42.Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonaguidi MA, Wheeler MA, Shapiro JS, et al. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell. 2011;145:1142–1155. doi: 10.1016/j.cell.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noctor SC, Martinez-Cerdeno V, Ivic L, et al. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 45.Zhong W, Chia W. Neurogenesis and asymmetric cell division. Curr Opin Neurobiol. 2008;18:4–11. doi: 10.1016/j.conb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 46.Gotz M, Stoykova A, Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21:1031–1044. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 47.Heins N, Malatesta P, Cecconi F, et al. Glial cells generate neurons: The role of the transcription factor Pax6. Nat Neurosci. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- 48.Bibel M, Richter J, Schrenk K, et al. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat Neurosci. 2004;7:1003–1009. doi: 10.1038/nn1301. [DOI] [PubMed] [Google Scholar]
- 49.Englund C, Fink A, Lau C, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li VS, Ng SS, Boersema PJ, et al. Wnt signaling through inhibition of beta-catenin degradation in an intact Axin1 complex. Cell. 2012;149:1245–1256. doi: 10.1016/j.cell.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 51.Cadigan KM. TCFs and Wnt/beta-catenin signaling: More than one way to throw the switch. Curr Top Dev Biol. 2012;98:1–34. doi: 10.1016/B978-0-12-386499-4.00001-X. [DOI] [PubMed] [Google Scholar]
- 52.Brault V, Moore R, Kutsch S, et al. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- 53.Zhuo L, Theis M, Alvarez-Maya I, et al. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis. 2001;31:85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]
- 54.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 55.Malatesta P, Hack MA, Hartfuss E, et al. Neuronal or glial progeny: Regional differences in radial glia fate. Neuron. 2003;37:751–764. doi: 10.1016/s0896-6273(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 56.Sur M, Rubenstein JL. Patterning and plasticity of the cerebral cortex. Science. 2005;310:805–810. doi: 10.1126/science.1112070. [DOI] [PubMed] [Google Scholar]
- 57.Molyneaux BJ, Arlotta P, Menezes JR, et al. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 58.Junghans D, Hack I, Frotscher M, et al. Beta-catenin-mediated cell-adhesion is vital for embryonic forebrain development. Dev Dyn. 2005;233:528–539. doi: 10.1002/dvdy.20365. [DOI] [PubMed] [Google Scholar]
- 59.Wang Y, Song L, Zhou CJ. The canonical Wnt/beta-catenin signaling pathway regulates Fgf signaling for early facial development. Dev Biol. 2011;349:250–260. doi: 10.1016/j.ydbio.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 60.Backman M, Machon O, Mygland L, et al. Effects of canonical Wnt signaling on dorso-ventral specification of the mouse tel-encephalon. Dev Biol. 2005;279:155–168. doi: 10.1016/j.ydbio.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 61.Anthony TE, Heintz N. Genetic lineage tracing defines distinct neurogenic and gliogenic stages of ventral telencephalic radial glial development. Neural Dev. 2008;3:30. doi: 10.1186/1749-8104-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reynolds BA, Rietze RL. Neural stem cells and neurospheres—Re-evaluating the relationship. Nat Methods. 2005;2:333–336. doi: 10.1038/nmeth758. [DOI] [PubMed] [Google Scholar]
- 63.Jensen JB, Parmar M. Strengths and limitations of the neurosphere culture system. Mol Neurobiol. 2006;34:153–161. doi: 10.1385/MN:34:3:153. [DOI] [PubMed] [Google Scholar]
- 64.Hirabayashi Y, Itoh Y, Tabata H, et al. The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development. 2004;131:2791–2801. doi: 10.1242/dev.01165. [DOI] [PubMed] [Google Scholar]
- 65.Adachi K, Mirzadeh Z, Sakaguchi M, et al. Beta-catenin signaling promotes proliferation of progenitor cells in the adult mouse subventricular zone. Stem Cells. 2007;25:2827–2836. doi: 10.1634/stemcells.2007-0177. [DOI] [PubMed] [Google Scholar]
- 66.Wrobel CN, Mutch CA, Swaminathan S, et al. Persistent expression of stabilized beta-catenin delays maturation of radial glial cells into intermediate progenitors. Dev Biol. 2007;309:285–297. doi: 10.1016/j.ydbio.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woodhead GJ, Mutch CA, Olson EC, et al. Cell-autonomous beta-catenin signaling regulates cortical precursor proliferation. J Neurosci. 2006;26:12620–12630. doi: 10.1523/JNEUROSCI.3180-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Machon O, Backman M, Machonova O, et al. A dynamic gradient of Wnt signaling controls initiation of neurogenesis in the mammalian cortex and cellular specification in the hippocampus. Dev Biol. 2007;311:223–237. doi: 10.1016/j.ydbio.2007.08.038. [DOI] [PubMed] [Google Scholar]
- 69.Munji RN, Choe Y, Li G, et al. Wnt signaling regulates neuronal differentiation of cortical intermediate progenitors. J Neurosci. 2011;31:1676–1687. doi: 10.1523/JNEUROSCI.5404-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hevner RF. From radial glia to pyramidal-projection neuron: Transcription factor cascades in cerebral cortex development. Mol Neurobiol. 2006;33:33–50. doi: 10.1385/MN:33:1:033. [DOI] [PubMed] [Google Scholar]
- 71.Mo Z, Zecevic N. Is Pax6 critical for neurogenesis in the human fetal brain? Cereb Cortex. 2008;18:1455–1465. doi: 10.1093/cercor/bhm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sakayori N, Kikkawa T, Osumi N. Reduced proliferation and excess astrogenesis of Pax6 heterozygous neural stem/progenitor cells. Neurosci Res. 2012;74:116–121. doi: 10.1016/j.neures.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 73.Roose J, Clevers H. TCF transcription factors: Molecular switches in carcinogenesis. Biochim Biophys Acta. 1999;1424:M23–37. doi: 10.1016/s0304-419x(99)00026-8. [DOI] [PubMed] [Google Scholar]
- 74.Poschl J, Grammel D, Dorostkar MM, et al. Constitutive activation of beta-Catenin in neural progenitors results in disrupted proliferation and migration of neurons within the central nervous system. Dev Biol. 2013;374:319–332. doi: 10.1016/j.ydbio.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 75.Kuwahara A, Hirabayashi Y, Knoepfler PS, et al. Wnt signaling and its downstream target N-myc regulate basal progenitors in the developing neocortex. Development. 2010;137:1035–1044. doi: 10.1242/dev.046417. [DOI] [PubMed] [Google Scholar]
- 76.Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Osumi N, Shinohara H, Numayama-Tsuruta K, et al. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells. 2008;26:1663–1672. doi: 10.1634/stemcells.2007-0884. [DOI] [PubMed] [Google Scholar]
- 78.Epstein J, Cai J, Glaser T, et al. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J Biol Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- 79.Epstein JA, Glaser T, Cai J, et al. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- 80.Czerny T, Busslinger M. DNA-binding and transactivation properties of Pax-6: Three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5) Mol Cell Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jun S, Desplan C. Cooperative interactions between paired domain and homeodomain. Development. 1996;122:2639–2650. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]
- 82.Tang HK, Singh S, Saunders GF. Dissection of the transactivation function of the transcription factor encoded by the eye developmental gene PAX6. J Biol Chem. 1998;273:7210–7221. doi: 10.1074/jbc.273.13.7210. [DOI] [PubMed] [Google Scholar]
- 83.Anderson TR, Hedlund E, Carpenter EM. Differential Pax6 promoter activity and transcript expression during forebrain development. Mech Dev. 2002;114:171–175. doi: 10.1016/s0925-4773(02)00051-5. [DOI] [PubMed] [Google Scholar]
- 84.Pinson J, Mason JO, Simpson TI, et al. Regulation of the Pax6: Pax6(5a) mRNA ratio in the developing mammalian brain. BMC Dev Biol. 2005;5:13. doi: 10.1186/1471-213X-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haubst N, Berger J, Radjendirane V, et al. Molecular dissection of Pax6 function: The specific roles of the paired domain and homeodomain in brain development. Development. 2004;131:6131–6140. doi: 10.1242/dev.01524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.