Abstract
Adult mesenchymal stem cells, such as dental pulp stem cells, are of great interest for cell-based tissue engineering strategies because they can differentiate into a variety of tissue-specific cells, above all, into osteoblasts. In recent years, epigenetic studies on stem cells have indicated that specific histone alterations and modifying enzymes play essential roles in cell differentiation. However, although several studies have reported that valproic acid (VPA)—a selective inhibitor of histone deacetylases (HDAC)—enhances osteoblast differentiation, data on osteocalcin expression—a late-stage marker of differentiation—are limited. We therefore decided to study the effect of VPA on dental pulp stem cell differentiation. A low concentration of VPA did not reduce cell viability, proliferation, or cell cycle profile. However, it was sufficient to significantly enhance matrix mineralization by increasing osteopontin and bone sialoprotein expression. In contrast, osteocalcin levels were decreased, an effect induced at the transcriptional level, and were strongly correlated with inhibition of HDAC2. In fact, HDAC2 silencing with shRNA produced a similar effect to that of VPA treatment on the expression of osteoblast-related markers. We conclude that VPA does not induce terminal differentiation of osteoblasts, but stimulates the generation of less mature cells. Moreover, specific suppression of an individual HDAC by RNA interference could enhance only a single aspect of osteoblast differentiation, and thus produce selective effects.
Keywords: HDAC, Valproic acid, Osteogenesis, DPSC
Introduction
Bone regeneration is a major focus of current tissue engineering applications; the need for bone substitutes is particularly important. Dental pulp represents a promising source of mesenchymal stem cells (MSCs) for bone replacement therapies: the efficiency in obtaining stem cells from this niche is high, and it is relatively easy to access surgically. Dental pulp stem cells (DPSCs) have received extensive attention in the field of bone tissue engineering due to their distinct capability to differentiate into the osteogenic lineage [1–3] as well as into other cell types [4, 5]. The osteoblastic potential of DPSCs has been extensively demonstrated both in vitro and in vivo [6, 7]. Indeed, DPSCs were shown to differentiate into mature bone precursors and elements, and generate a three-dimensional (3D) fibrous bone tissue in vitro; when transplanted into murine models, these cells were capable of producing a lamellar bone tissue on account of codifferentiation of progenitors into osteoblasts and endothelial cells. Moreover, DPSCs were able to successfully repair mandible bone defects in humans when grafted upon a collagen sponge [8, 9], demonstrating that these cells could be an optimal choice for bone replacement therapies.
In recent years, investigations focused on stem cells have been supported by epigenetic studies. It is well known that the epigenetic regulation of gene expression is responsible for both the maintenance of a stem cell state and the determination of the cells, that is, their commitment. In fact, specific patterns of DNA methylation and histone modifications play an important role in the induction of MSC differentiation toward specific lineages. The specific gene expression patterns induced during stem cell differentiation are the result of elaborate control of activation/repression of large numbers of genes. Histone acetylation contributes to the formation of a more open, transcriptionally active chromatin structure. On the contrary, histone deacetylation is associated with a compact chromatin conformation and leads to transcriptional repression. The level of cellular histone acetylation is regulated by the opposing activities of histone acetyltransferases and histone deacetylases (HDACs) [10, 11].
The family of HDAC enzymes comprises at least 18 genes classified into four groups: class I (HDAC1, HDAC2, HDAC3, and HDAC8) and class II (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10) HDACs deacetylate histone tails and target other cellular proteins also [12]; class III HDACs (Sirtuins 1–7) were identified on the basis of their similarity with sir2, a yeast transcription repressor requiring NAD+ as a cofactor; HDAC11 is the only known class IV HDAC [13–15]. The inhibition of HDAC activity typically leads to derepression of transcription. Several classes of HDAC inhibitors (HDACi) have been identified, such as hydroxamic acids, cyclic peptides, short-chain fatty acids, and epoxides [16–18]. HDACi inhibit most class I and II HDACs, but have no effect on class III HDACs. In addition, they induce cell cycle arrest and differentiation and/or apoptosis of tumor cell lines in vitro and in vivo [19, 20]. Moreover, they have been demonstrated to enhance osteoblast differentiation in vitro [21] and new bone formation in vivo [22].
Valproic acid (VPA)—an FDA-approved short-chain fatty acid—has been widely used for more than 20 years for the treatment of different neurological disorders [23]. VPA acts as a potent HDAC inhibitor at the concentrations used clinically [24]. Therefore, since the safety of VPA in patients has been already tested, its clinical use for new molecular approaches is of great interest.
In this study, we aimed to observe the effects of VPA on osteogenic differentiation of DPSCs. We found that pretreatment of DPSCs and osteoblasts with VPA significantly improves mineralized matrix formation, enhancing expression of bone glycoproteins, such as osteopontin (OPN) and bone sialoprotein (BSP)—both involved in the formation of the mineralized matrix—but negatively affecting the expression of osteocalcin (OC), a late-stage marker of differentiation. Moreover, we ascertained that HDAC1 as well as HDAC2 are critical enzymes for osteoblast differentiation.
Materials and Methods
For reagents, cell culture and differentiation medium, RT-PCR, and quantitative RT-PCR see Supporting Information.
Growth Curves
Cell growth inhibition and cell viability were evaluated using trypan blue-dye exclusion. Cells were plated at 2.0 × 104 cells/well in a six-well plate and various concentrations of VPA (0.25, 0.5, 1, 2, or 5 mM) were added the next day. Cells without VPA treatment were considered as control. After 24, 48, 72, and 120 hours, cells were trypsinized and stained with 0.4% trypan blue (0.4% wt/vol). The total cell number and the proportion of viable cells were counted with the aid of a hemocytometer. The experiments were performed in triplicate.
Cell Cycle Analysis
Cells were detached from the plates by the standard trypsinization method and then fixed with ice-cold 80% ethanol. The cells were centrifuged and then stained with a solution of 50 µg/ml propidium iodide and 80 µg/ml RNase A for 60 minutes at 4°C in the dark. DNA content and cell cycle distribution were measured with a FACS ARIA II (BD Biosciences, San Jose, CA, http://www.bdbiosciences.com.), and the data were analyzed using Mod-Fit software (BD Biosciences, San Jose, CA).
Apoptosis Analysis
Apoptosis was assessed with a FITC-Annexin V Staining protocol (BD Biosciences, San Jose, CA). The percentage of apoptotic cells was determined by flow cytometry.
Histone Extraction and Western Blot
Cells (107 cells per milliliter) were lysed in Triton Extraction Buffer (TEB) containing 0.5% Triton X-100 (vol/vol), 2 mM phenylmethylsulfonyl fluoride, and 0.02% (wt/vol) NaN3 in phosphate buffered saline (PBS) for 10 minutes on ice with gentle stirring. After centrifugation at 6,500g for 10 minutes at 4°C, the supernatant was discarded and the pellet was washed in half the volume of TEB and centrifuged as before. The pellet was resuspended in 0.2 N HCl at a cell density of 4 × 107 cells per milliliter and acid extraction was carried out overnight at 4°C. Then, the samples were centrifuged at 6,500g for 10 minutes at 4°C, the supernatant removed, and protein content determined using Bradford assay. Equal amounts of histones (10 µg) were separated on a 15% SDS-polyacrylamide gel and then transferred to a polyvinylidene difluoride membrane. Membranes were incubated with primary antibodies to acetyl-histone H3 (#06-599: Millipore, Vimodrone [MI], Italy, http://www.millipore.com.) and acetyl-histone H4 (#06-598: Millipore, Vimodrone, Mi, Italy). Total histone H4 (ab31830: Abcam, Cambridge U.K., http://www.abcam.com) antibody was used for assessing loading. Proteins were detected according to manufacturer's protocols and visualized using chemiluminescence (GE Healthcare U.K. Limited, U.K., http://www.gehealthcare.com).
Flow Cytometry
Cells were detached using trypsin-EDTA (200 mg/l EDTA, 500 mg/l trypsin; Cambrex, East Rutherford, NJ, USA, http://www.cambrex.com). Intracellular staining for OC, BSP, and OPN was performed using FIX & PERM Cell Fixation & Cell Permeabilization Kit (Life Technologies Laboratories, Monza, Italy, http://www.lifetechnologies.com) according to the manufacturer's procedure. At least 300,000 cells were incubated with indirect or direct fluorescent-conjugated antibodies. The primary antibodies used were: Phycoerythrin (PE)-conjugated anti-h-OC and Carboxyfluorescein (CFS)-conjugated anti-h-OPN (R&D Systems, Minneapolis, MN), rabbit anti-HDAC1 and mouse anti-HDAC2 (SantaCruz, Santa Cruz, CA, http://www.scbt.com.), and rabbit anti-BSP (Abcam, Cambridge, UK). The secondary antibodies were anti-rabbit FITC and anti-mouse FITC, all purchased from Abcam. For negative controls, cells were stained with an isotype control antibody. Labeled cells were analyzed by flow cytometry using a FACS Aria II (BD Biosciences, San Jose, CA) and all data analyzed with FCS Express version 3 software.
Cell Seeding and Differentiation on a Collagen Scaffold in a Rotating System
In order to achieve 3D tissue formation, cells were seeded on a Gingistat (GABA VEBAS, Roma, Italy, http://www.gaba-info.it/) scaffold. This scaffold is a lyophilized collagen type I sponge. Collagen sponges were cut under sterile conditions into 5 × 5 × 5 mm3 cubes. Scaffold cubes were placed in six-well plates and a cell suspension of 1 × 106 cells contained in 200 µl medium was pipetted onto the top of each cube. Cells were allowed to adhere under a humidified atmosphere at 37°C and 5% CO2 for 4 hours. The seeded scaffolds were then placed in tubes containing osteogenic medium and cultured in a Wheaton roller culture apparatus (Wheaton Science Products, Millville, NJ, http://wheaton.com/.) for 30 days at 6 rpm in an incubator at 37°C and 5% CO2. Media were changed twice a week.
Cryostat Sectioning
After 3D culture, samples were fixed in 4% paraformaldehyde (PFA) and cryoprotected overnight at 4°C by immersion in a 30% (wt/vol) sucrose solution before being embedded in Tissue-Tek® O.C.T. Compound (Tissue-Tek; Sakura Finetek, Torrance, CA, http://www.sakuraus.com.) and frozen. Sections were cut 5-µm thick with a cryostat at −20°C and then processed for immunostaining.
Calcium Deposition and Alizarin Red Quantification
Calcium deposition was detected with Alizarin Red S staining. Briefly, cells cultured as above described and cryo-sectioned scaffolds were washed with cold PBS, fixed with 4% PFA for 15 minutes at room temperature, and stained with 2% Alizarin Red S solution for 20 minutes. Stained cells were extensively washed with deionized water to remove any nonspecific precipitation. Any positive red staining represented deposition of a calcified matrix. For the quantification of Alizarin Red S staining, 2 ml 10% (vol/vol) acetic acid was added to each well and the plate was incubated at room temperature for 30 minutes under gentle agitation [25]. The monolayer was scraped off the plate with a cell scraper and transferred to a 1.5 ml microcentrifuge tube after the addition of 1 ml 10% (vol/vol) acetic acid to the scraped cells. After vortexing for 30 seconds, the slurry was overlaid with 1.25 ml mineral oil (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com/), heated to 85°C for 10 minutes, and transferred to ice for 5 minutes. The slurry was then centrifuged at 20,000g for 15 minutes, and 500 µl of the supernatant then removed to a new 1.5 ml microcentrifuge tube. Two-hundred microliters of 10% (vol/vol) ammonium hydroxide was added to neutralize the acid. In some cases, the pH was measured at this point to ensure that it was between 4.1 and 4.5. Absorbance of aliquots (150 µl) of the supernatant was measured in triplicate at 405 nm in a 96-well format using opaque-walled, transparent-bottomed plates.
Immunohistochemistry/Immunofluorescence Analysis
Frozen sections or DPSCs cultured in six-well plates were fixed with 4% PFA in PBS for 20 minutes at room temperature. After washing in PBS, samples were permeabilized with 0.5% Triton X-100 for 5 minutes. Pretreatment of samples with 0.3% H2O2 followed by washing with PBS was necessary to inhibit endogenous peroxidase activity. Incubation with primary anti-OPN antibody (ab33046: Abcam, Cambridge, UK) was performed overnight at 4°C. Primary antibodies were detected using biotinylated goat anti-rabbit IgG (DAKO, Glostrip, Denmark, http://www.dako.com) and ExtrAvidin-peroxidase (Sigma-Aldrich, St. Louis, MO), applied subsequently for 30 minutes and followed by incubation with 3,30-diaminobenzidine tetra hydrochloride (DAB peroxidase substrate, Sigma-Aldrich, St. Louis, MO). For immunofluorescence analysis, OC (ab13418: Abcam, Cambridge, UK) was revealed using a PE-conjugated anti-mouse IgG secondary antibody.
Mallory's Trichrome Staining
Sections were first stained in 1% acid fuchsin (Sigma-Aldrich, St. Louis, MO) solution in distilled water then exposed to a 5% solution of phosphotungstic acid for 20 minutes, and finally stained in a mixture of 1% aniline blue (Sigma-Aldrich, St. Louis, MO), 2% orange G (Sigma-Aldrich, St. Louis, MO), and 2% oxalic acid (Sigma-Aldrich, St. Louis, MO) in distilled water for 30 minutes.
Transfection (shRNA Construct, Transfection, and Selection of Stable Selection)
shHDAC1, shHDAC2, and negative control shRNA (mock) neomycin-resistant SureSilencing shRNA plasmids were purchased from SABioscience (Qiagen, Milano, Italy, http://www.sabiosciences.com). Saos-2 cells were stably transfected with these constructs in an Amaxa Nucleofector device with the Amaxa Cell Line Nucleofector Kit V (Lonza GmbH, Cologne, Germany, http://www.lonza.com/), according to the manufacturer's instructions. Clones with downregulated expression of HDAC1 or HDAC2 were selected with 500 µg/ml G418. Clones were screened by quantitative RT-PCR and Western blot and then analyzed for BSP, OPN, and OC expression by flow cytometry or RT-qPCR analysis.
Chromatin Immunoprecipitation Assay
Protein–DNA cross-linking was performed by incubating cells (2.0 × 107 per condition) with 1% formaldehyde for 10 minutes at room temperature with gentle agitation. Glycine (0.125 M) was added to quench the reaction. Cells were rinsed with cold PBS and then harvested in 0.15 M NaCl–HEG buffer (1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES, pH 7.6). Cell pellets were then suspended in 2 ml of chromatin immunoprecipitation (ChIP) incubation buffer (0.15% SDS, 1% Triton X-100, 0.15 M NaCl–HEG) and sonicated to shear DNA to an average fragment size of 200–500 bp. Sonication was performed at maximum intensity for 30 seconds on and 30 seconds off for 15 cycles using a Bioruptor UCD-300 (Diagenode Liége, Belgium, http://www.diagenode.com) sonicator at 4°C.
Sonicated chromatin was centrifuged for 5 minutes and then incubated overnight with purified anti-AcH3 antibody (#06–599: Millipore, Vimodrone, Mi, Italy), or with rabbit IgG to provide controls, and Protein A/G PLUS-Agarose (Santa Cruz, Santa Cruz, CA). After several washings, DNA was eluted from beads using elution buffer (1% [wt/vol] SDS, 0.1 M NaHCO3, 2.5 µl/µg proteinase K) and crosslinking reversed by heating to 65°C for 3 hours. DNA was purified and subjected to qPCR with specific primers using a SYBR green kit (Applied Biosystems, Life Technologies Italia, Monza, Italy, http://www.lifetechnologies.com). Efficiency of ChIP was calculated as percentage of input (nonprecipitated control) corrected for background-(no antibody) precipitated DNA. PCR amplification was performed for OC promoter using Fw 5′-CCCAGCTCTGCTTGAACCTA-3′ and Rev 5′-TCAGCTCCAACTCACTCTTCC-3′ primers, and for BSP promoter using Fw 5′-ACATATTCGGAGCCCAAACTATTCA-3′ and Rev 5′-GAACGTGGATTCTCACCAGAAAAC-3′ primers.
Statistical Analysis
All experiments were carried out in triplicate and repeated at least three times. The results were presented as means ± SEM with p ≤ .05 considered as statistically significant.
Results
Effect of VPA on Cell Viability
Since HDACi cause growth arrest and apoptosis in tumor cells as well as in some nontumor cells [26, 27], we started by determining the noncytotoxic dose of VPA for DPSCs. To this end, DPSC cultures were exposed to different concentrations of VPA (0.25, 0.5, 1, 2, and 5 mM) for up to 72 hours. We found that untreated subconfluent DPSCs had a fibroblast-like, spindle-shaped appearance (Supporting Information Fig. S2A). When cultured with increasing concentrations of VPA, the morphology of the DPSCs changed progressively: in fact, they became increasingly lengthened and developed ever more extended cytoplasmic processes. This morphology resembled the typical phenotype of terminally differentiated cells. In addition, cultures became less confluent when exposed to the highest concentrations.
We then tested the effect of VPA on cell viability with the trypan blue dye exclusion method. For this, DPSCs were cultured with different concentrations of VPA for 24, 48, 72, and 120 hours (Supporting Information Fig. S2B). We did not detect appreciable differences in cell growth up to 48 hours of treatment and up to a concentration of 1 mM; however, 2 and 5 mM VPA for 48 hours reduced cell growth of DPSCs in a dose-and time-dependent manner. Similar results were observed in primary osteoblasts used as a reference control (Supporting Information Fig. S2C). Therefore, low concentrations of VPA (up to 1 mM) did not reduce cell viability of either DPSCs or primary osteoblasts for up to 48 hours of incubation.
Effect of VPA on Cell Cycle and Apoptosis
We then determined the effect of VPA on cell cycle by flow cytometry. No drastic changes in the cell cycle profiles were observed in DPSCs cultured with 1 mM VPA for 48 hours. However, a high concentration of VPA (5 mM) caused the appearance of a sub-G1 cell population, which corresponds to apoptotic cells (Supporting Information Fig. S3A).
The results obtained by flow cytometry were confirmed by RT-PCR for the expression of p21 (WAF1), an inhibitor of cyclin D/cdk4 and cyclin E/cdk2 complexes in the early G1 phase and the cyclin A/cdk2 complex in G2 [28, 29]. RT-PCR analysis revealed that p21 expression did not change within the range 0.25–5 mM VPA (Supporting Information Fig. S3B).
Furthermore, we analyzed apoptosis with fluorescence-activated cell sorting (FACS) after annexin V/propidium iodide labeling. There was significant apoptosis with 5 mM VPA (Supporting Information Fig. S3C). Apoptosis was low (approximately 3.4%) up to 2 mM VPA. Osteoblasts underwent only a minor induction of apoptosis, even at the highest concentration tested. Taken together, these results suggested that a low concentration of VPA (1 mM) did not negatively affect DPSC proliferation or cell cycle profile.
VPA and Histone Acetylation
Because VPA is an HDAC inhibitor, we evaluated modifications of histone acetylation on H3 and H4. DPSCs were cultured with different concentrations of VPA for 48 hours and Western blot analysis performed. A dose-dependent increase in acetylation of H3 and H4 was observed in DPSCs, confirming that VPA is able to inhibit HDACs in our cell model (Fig. 1).
Figure 1.
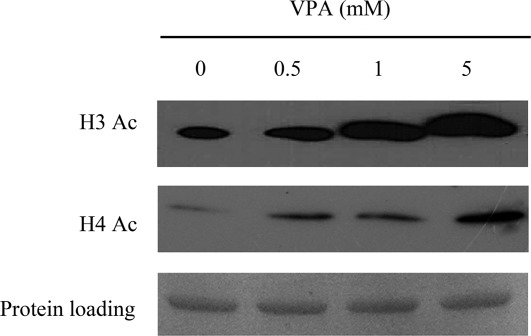
Effect of VPA on acetylation state of histones H3 and H4. Western blot analysis revealed a dose-dependent increase in acetylation of H3 and H4 in DPSCs stimulated in the presence of increasing concentrations of VPA. Abbreviation: VPA, valproic acid.
VPA Increases Expression of Bone Matrix Markers in DPSCs
We first determined whether VPA had effects on the expression of genes involved in stemness and osteogenic differentiation. Based on the results obtained above, DPSCs and primary osteoblasts were treated with 1 mM VPA for 48 hours. We noted drastic reductions in POU5F1 and NANOG expression in VPA-treated cells (Fig. 2A). In addition, VPA induced evident changes in osteogenic differentiation markers: in DPSCs, VPA increased bone-sialoprotein (BSP) and OPN expression, while decreasing OC expression. However, RUNX2 mRNA level was not altered. Similar results were observed in osteoblasts, in which mRNA levels of alkaline-phosphatase (bone ALP), BSP, and OPN were increased, while OC expression decreased; in addition, RUNX2 mRNA decreased in these cells (Fig. 2B).
Figure 2.
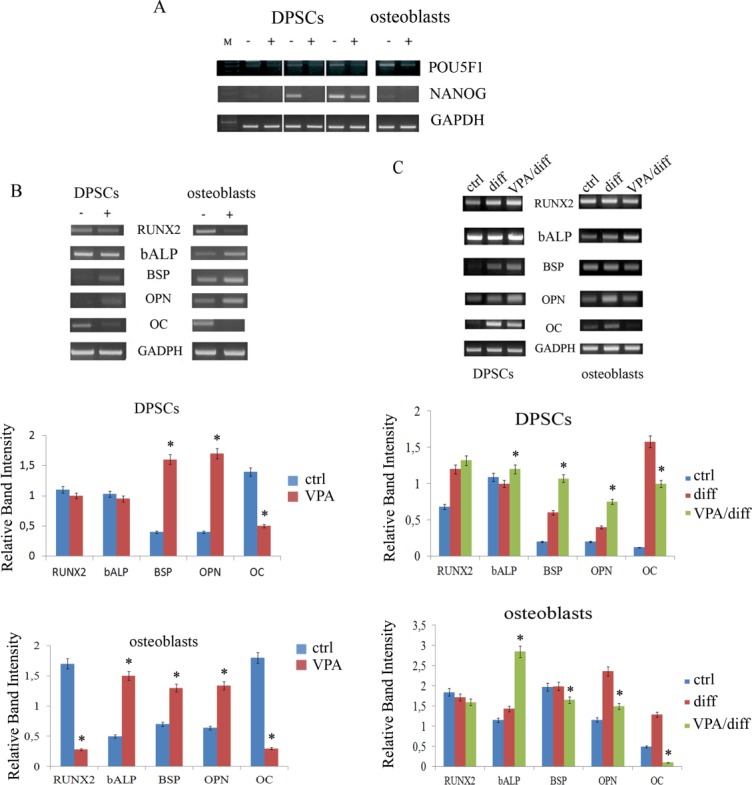
Modulation of stem cell marker expression and bone matrix markers in cells after VPA treatment. (A): RT-PCR in three different DPSC lines and osteoblasts revealed that expression of POU5F1 and NANOG was strongly reduced in VPA-treated cells (+) with respect to controls (−). (B): RT-PCR and relative densitometric analyses of bone-related markers after 48 hours of VPA treatment. VPA increased BSP and OPN expression, but decreased OC expression, in both DPSCs and osteoblasts. The level of RUNX2 mRNA was not altered in DPSCs, but it was lowered in osteoblasts. Expression of bone ALP was unchanged in DPSCs and increased in osteoblasts after VPA treatment. (C): RT-PCR and relative densitometric analyses of bone-related markers after osteogenic induction of VPA-treated cells. Osteogenic differentiation markers were more efficiently expressed in DPSCs that were pretreated with VPA (VPA/diff) when compared with DPSCs differentiated in only osteogenic medium (diff.). In primary osteoblasts, almost all the osteogenic markers in pretreated cells were decreased. Osteocalcin mRNA remained lower in both treated DPSCs and osteoblasts. All values are means ± SEM. *, p ≤ .05 versus relative control. Abbreviations: BSP, bone sialoprotein; DPSC, dental pulp stem cell; OC, osteocalcin; OPN, osteopontin; VPA, valproic acid.
We then evaluated the effect of VPA on DPSCs during osteogenic induction. DPSCs were treated with 1 mM VPA for 48 hours and then cultured in osteogenic medium for 21 days. RT-PCR analysis revealed that most osteogenic differentiation markers were more efficiently expressed in VPA-pretreated DPSCs than in DPSCs cultured in osteogenic medium alone (Fig. 2C). Indeed, we noted increased expression of those genes involved in the formation of extracellular matrix and in mineralization (bone ALP, BSP, and OPN) and a slightly increased expression of RUNX2; however, OC mRNA was less expressed. In contrast, all the osteogenic markers apart from bone ALP were less expressed in primary osteoblasts when these cells were pretreated with VPA. This could be due to the fact that osteoblasts are already committed and therefore express osteogenic markers in a temporal sequence different to that of DPSCs, which are considered to be more immature.
Protein expression analysis confirmed the RT-PCR studies: flow cytometry revealed significant upregulation of BSP and OPN, but not of OC, in VPA-treated DPSCs and osteoblasts (Fig. 3A) after 48 hours. Only after induction of osteogenic differentiation we noted downregulation of OC at the protein level (Fig. 3B). The molecular evidence for enhanced mineralized matrix formation during osteogenic differentiation in DPSCs treated with VPA was further sustained by Alizarin red staining of VPA-pretreated DPSCs and osteoblasts cultured in differentiation medium for 7, 14, and 21 days (Fig. 3C).
Figure 3.
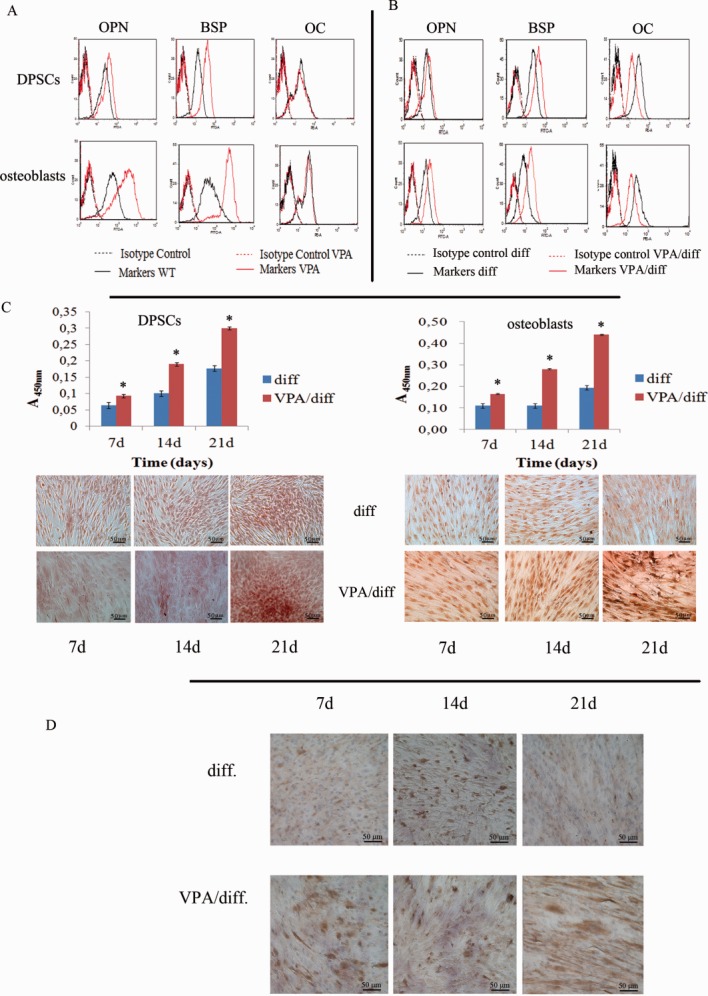
Effect of VPA on matrix mineralization. (A): Flow cytometry analysis revealed significant upregulation of BSP and OPN, but not of OC, in VPA-treated DPSCs and osteoblasts after 48 hours of VPA treatment. (B): Flow cytometry analysis after osteogenic induction. OC expression in pretreated cells was decreased. (C): Quantification of Alizarin red staining revealing enhanced mineralized matrix formation at different time points during osteogenic differentiation of DPSCs and osteoblasts treated with VPA. *, p ≤ .05 versus relative control. Scale bar = 50 µm. (D): Immunostaining for osteopontin in DPSCs at different time points during osteogenic differentiation. Immunoreactivity is more evident in VPA-pretreated DPSCs (VPA/diff) already at day 7, whereas in DPSCs cultured only in osteogenic medium (diff.), there is strong immunoreactivity only from day 14 of differentiation. Scale bar = 50 µm. Abbreviations: BSP, bone sialoprotein; DPSC, dental pulp stem cell; OC, osteocalcin; OPN, osteopontin; VPA, valproic acid.
In addition, we performed immunocytochemistry for OPN in DPSCs with or without VPA pretreatment and grown in osteogenic medium for 7, 14, and 21 days. Immunoreactivity was strong at 14 days in DPSCs cultured only in osteogenic medium; OPN immunoreactivity peaked earlier (day 7) and was more extensive when DPSCs were pretreated with VPA (Fig. 3D).
Effect of VPA on Osteogenic Differentiation of DPSCs in 3D Rotating Culture
We then investigated the response of DPSCs to VPA in an environment that more closely mimicked 3D bone tissue. To this end, DPSCs were seeded onto Gingistat collagen sponges and cultured in osteogenic medium for 30 days under rotating conditions. As can be seen from Figure 4, both untreated and VPA-pretreated DPSCs were not distributed homogeneously within the scaffold: cells were more present in superficial layers of the sponge and tended to become progressively less present toward the center of the structure. Differences in distribution and cellular morphology are more evident in Figure 4C, 4D: superficial cells were flattened and formed a compact layer, while cells that had migrated into the sponge were progressively more globular and/or polyhedric, a shape typical of more-differentiated osteoblasts.
Figure 4.
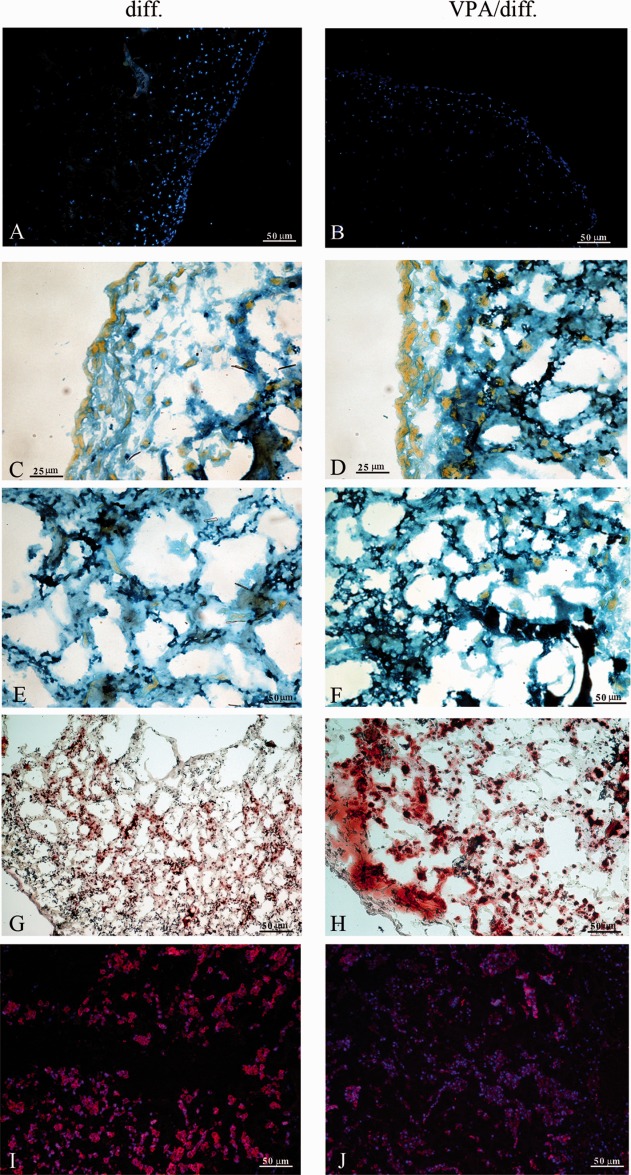
Three-dimensional rotating culture of dental pulp stem cells (DPSCs). (A, B): DAPI nuclear staining. The distribution density of untreated DPSCs and treated DPSCs was higher near the surface of the sponges and tended to decrease progressively toward the center of the scaffold. Scale bar = 50 µm. (C, D): More-superficial cells were flattened and formed a compact layer, while cells that had migrated into the sponge had a progressively more globular and/or polyhedric shape, typical of more-differentiated osteoblasts. Scale bar = 25 µm. (E, F): Mallory's trichrome stained newly synthesized collagen fibers in a deep blue color; these fibers were present on a different focal plane to that of the underlying scaffold material. Scale bar = 50 µm. In samples pretreated with VPA (F), the blue staining was much more intense and diffused compared with that in the controls (E). (G, H): Alizarin red staining revealing a higher concentration of calcium deposits in VPA-pretreated DPSCs than in control: the areas are more intensely colored and extended. Scale bar = 50 µm. (I, J): Osteocalcin immunofluorescence. Scale bar = 50 µm. VPA led to a reduction in osteocalcin expression in pretreated DPSCs (J). Abbreviation: VPA, valproic acid.
Although Mallory's trichrome staining evidenced also the collagen scaffold, newly synthesized collagen generated a more intense blue staining present on a different focal plane to that that of the scaffold (Fig. 4E, 4F). This observation was suggestive of apposition of new material. In the samples pretreated with VPA, the blue staining was much more intense and diffuse compared with that in control.
In addition, Alizarin red staining revealed that the concentration of calcium deposits was higher in VPA-pretreated DPSC than in untreated cells: areas were more intensely colored and extended (Fig. 4G, 4H). VPA also led to a reduction in OC expression in pretreated DPSCs (Fig. 4I, 4J).
HDAC Involvement During Osteogenic Differentiation of DPSCs
To determine whether the downregulation of OC expression by VPA was mediated by HDAC inhibition, DPSCs were cultured with 1 µM MS-275 for 48 hours. MS-275 preferentially inhibits HDAC1 (IC50: 300 nM for HDAC1, compared with 8 µM for HDAC3) and has no inhibitory effect against HDAC8 (IC50 [mt] 100 µM) [30, 31]. We found that similarly to VPA treatment, exposure to MS-275 decreased OC mRNA levels and enhanced expression of OPN and BSP in DPSCs (Fig. 5).
Figure 5.
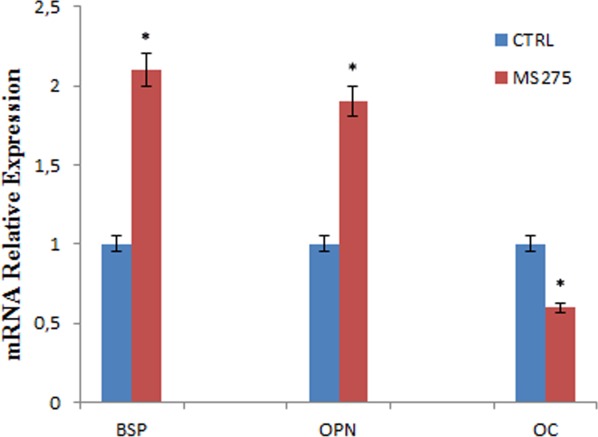
RT-qPCR for OPN, BSP, and OC in dental pulp stem cells treated with MS-275. MS-275 decreased osteocalcin mRNA levels and enhanced OPN and BSP expression. All values are means ± SEM. *, p ≤ .05 versus control. Abbreviations: BSP, bone sialoprotein; OC, osteocalcin; OPN, osteopontin.
Since VPA inhibits class I HDACs more efficiently than class II enzymes [24], and MS-275 has a similar effect on the pattern of osteoblast gene expression, we concentrated our attention to the study of HDAC1 and HDAC2 activities to identify which of these class I enzymes had a key role in osteogenic differentiation and on OC expression. We therefore compared the protein levels of the two HDACs in DPSCs (Fig. 6). We found that HDAC-1 and −2 were decreased in differentiated cells as well as in those pretreated with VPA.
Figure 6.
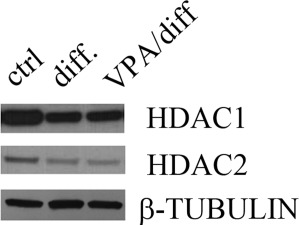
HDAC1 and HDAC2 expression during dental pulp stem cell (DPSC) differentiation. Western blot analysis of HDAC1 and HDAC2 in undifferentiated (ctrl) and differentiated DPSCs with or without VPA pretreatment (VPA/diff and diff., respectively). Expression of the two HDACs decreased during differentiation with or without pretreatment compared with control. Abbreviations: HDAC, histone deacetylase; VPA, valproic acid.
In order to clarify whether these changes in HDAC-1 and −2 expression were associated with the differentiation process, we evaluated the effect of knockdown of HDAC1 and HDAC2 on osteoblast markers expression. To this end, we used the osteoblast-like Saos-2 cell line—a model used to study events associated with osteoblast differentiation in humans [32, 33]—rather than DPSCs, which are empirically difficult to transfect due to their slow growth rate and vulnerability. Saos-2 cells responded to VPA similarly to DPSC and primary osteoblasts (Fig. 7A), and knockdown of HDAC1 with shRNA was effective (Supporting Information Fig. S4A). HDAC1 knockdown increased OPN and BSP expression compared with mock-treated cells; OC expression was slightly increased (Fig. 7B).
Figure 7.
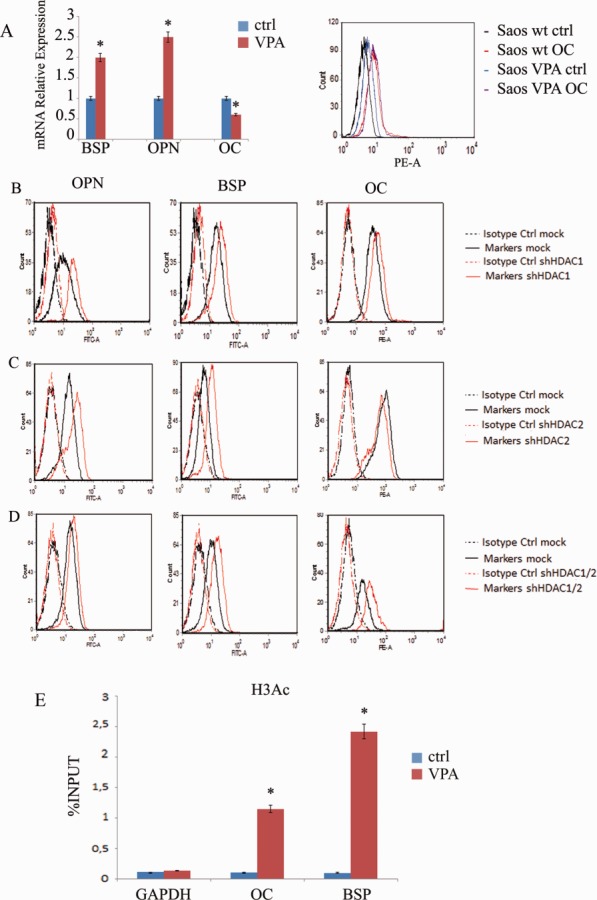
VPA treatment of Saos-2 cell culture after 48 hours of incubation. (A): Saos-2 cells had decreased osteocalcin mRNA and increased levels of BSP and OPN after treatment with VPA. All values are means ±SEM. *, p ≤ .05 versus control. No changes were found in osteocalcin expression by flow cytometry. (B): Effect of HDAC1 knockdown on osteoblast markers assessed by flow cytometry. HDAC1 knockdown increased expression of OPN and BSP compared with mock-treated cells; OC expression was slightly increased. (C): Effect of HDAC2 knockdown on osteoblast markers. Silenced cells had increased BSP and OPN expression but a decreased OC expression, as evidenced by flow cytometry. (D): OC was upregulated in cells silenced for both HDAC-1 and −2. (E): Chromatin immunoprecipitation (ChIP) assay on OC and BSP promoters. ChIP analysis revealed that there was a significant increase in histone H3 acetylation at both these promoters after VPA treatment. All values are means ± SEM. *, p ≤ .05 versus control. Abbreviations: BSP, bone sialoprotein; DPSC, dental pulp stem cell; OC, osteocalcin; OPN, osteopontin; VPA, valproic acid.
Subsequently, we performed silencing of HDAC2. Again, knockdown of HDAC2 was effective in decreasing the protein level without affecting HDAC1 expression (Supporting Information Fig. S4B). Surprisingly, OPN and BSP expression increased also with the silencing of HDAC2; however, OC expression decreased in this setting (Fig. 7C). Conversely, OC was upregulated in cells that had been silenced for both HDAC-1 and −2 (Fig. 7D).
ChIP Analysis
We then investigated whether the observed differential effects of VPA on OC and bone matrix markers expression were related to changes in histone acetylation in promoter regions. ChIP analysis was performed using antibodies to pan-acetylated histone H3 and primers for OC and BSP promoters. We found a significant increase in histone acetylation at both promoters after VPA treatment (Fig. 7E).
Discussion
This study focused on understanding the molecular basis of osteogenesis in DPSCs, endeavoring to apply this knowledge to tissue engineering. DPSCs have received extensive attention in the field of bone tissue engineering due to their distinct capability to differentiate into osteogenic lineages.
Numerous epigenetic modifications occur during osteogenic differentiation of MSCs. While much knowledge has been obtained on the epigenetic modifications responsible for MSC differentiation, further investigation to this end will improve our ability to use MSCs therapeutically. Indeed, it is conceivable that manipulation of epigenetic signatures associated with multipotency and pluripotency, as well as modifications associated with lineage-specific differentiation, could help the development of individualized therapies. The identification of the factors necessary to reprogram mesenchymal-derived somatic cells into more or less differentiated states can also provide insight into the regulation of MSC fate determination.
Stem cell differentiation is extremely sensitive to epigenetic changes. Therefore, application of epigenetic regulators, such as HDAC inhibitors, may be valuable for stem cell-based interventions [34]. Here, we investigated the effects of VPA on the differentiation of human DPSCs into osteoblasts. HDAC inhibitors have been reported to cause apoptotic cell death in a variety of cultured transformed cells [19, 26, 35, 36] and induce growth arrest in some normal cells [27]. Our results show that VPA does not induce toxic effects nor does it alter cell morphology or the cell cycle at the low (1 mM) concentration considered. However, at higher doses, its effect is related to the induction of apoptosis rather than to cell cycle alteration. We also investigated if the activity of VPA was indeed related to the ability of the drug to modify histone acetylation status. Western blot analysis of VPA-treated cells showed that total histone acetylation was increased, confirming that VPA is an HDAC inhibitor for DPSCs.
In our cell model, VPA increased expression of genes involved in the formation of mineralized matrix already at 48 hours of treatment. This effect was more marked when differentiation was induced in osteogenic medium, as evidenced by evaluating the expression of OPN and bone-sialoprotein as well as quantification of Alizarin red staining. In pretreated primary osteoblasts undergoing differentiation, decreases were observed for the mRNA but not the protein levels of BSP and OPN. These results, as mentioned above, were in line with the degree of maturity (commitment) of the cells.
It is well known that the progression of osteogenic differentiation occurs through temporal and spatial changes in the expression of bone-specific genes. During osteogenic differentiation, specific markers reach one or more peaks of expression, and this is strictly related to the cell maturation condition. A case in point is the expression of OPN, which not only appeared to be enhanced in VPA-treated cells, but also had two peaks in expression (at 7 and 21 days); the control peaked later, at 14 and 28 days (the latter data not shown). This effect could be due to the fact that VPA accelerates the maturation of osteoblasts in vitro [21]. Of note, VPA did not affect OC expression positively. Indeed, the mRNA level was lower in VPA-treated cells already at 48 hours of treatment, whereas protein expression was unchanged, as evidenced by flow cytometry. This could be due to the fact that the half-life of OC protein is more prolonged than that of its mRNA. Only after induction of osteogenic differentiation did we note a downregulation at the protein level.
On the basis of these findings, we tested the effect of VPA on DPSCs seeded onto a collagen sponge and in a 3D rotating culture. 3D cell culture is a superior system for performing stem cell differentiation because it improves bone differentiation. Dynamic flow positively affected cell distribution through the scaffold and further enhanced cell phenotypic expression and mineralized matrix synthesis.
Although there were no appreciable differences in the distribution of pretreated and untreated cells within the scaffolds, VPA-treated cells had a higher extracellular matrix (ECM) production, as evidenced by Mallory's trichrome and Alizarin red stainings. Moreover, untreated cells were uniformly and intensely immunoreactive for OC, while VPA-treated cells had a significantly lower or absent immunoreactivity for this marker. Thus, although we used a 3D system to improve bone differentiation, the expression of OC was once again affected negatively.
This study also aimed to understand the molecular mechanisms involved in osteogenic differentiation and in the downregulation of OC mRNA. To identify which HDAC had a key role in these processes, we treated DPSCs, primary osteoblasts, and Saos-2 cells with different HDACi (i.e., VPA and MS275). VPA preferentially inhibits class I HDACs [24], whereas MS-275 preferentially inhibits HDAC1 [29, 30]. We compared their effects on osteogenic marker expression and found that MS-275 increased OPN and BSP expression and decreased OC mRNA. Consequently, our attention focused on HDAC1 and HDAC2 activities. Previous studies identified a series of HDACs involved as regulators in the process of bone formation through the use of HDACi or RNA interference. HDAC3, HDAC6, and HDAC7 associate with Runx2, acting as transcriptional corepressors of Runx2 activity in osteoblasts [36–38]. HDAC4 and HDAC5 also negatively regulated Runx2 activity by deacetylating lysines in the Runx2 protein, leading to ubiquitin-mediated proteolysis [22]. Suppression of HDAC3 or HDAC1 expression by RNA interference accelerated the course of osteoblastic differentiation [36, 39].
In our study, we performed shRNA silencing of HDAC1 and, for the first time, HDAC2, in Saos-2 cells, a permanent cell line of human osteoblast-like cells. We found not only that HDAC1 is a key enzyme for osteogenic differentiation, as reported by Lee et al. [39], but that HDAC2 plays a role also. Indeed, HDAC2 silencing led to an increased expression of OPN and BSP, but downregulated OC mRNA, resembling the effect of VPA. However, this downregulation did not occur when we analyzed the cosilencing of HDAC1 and HDAC2 or the silencing of HDAC1 alone. Therefore, the inhibition of OC gene expression by VPA is most likely mediated through HDAC2 inhibition. However, further studies are required to determine whether HDAC2 inhibition directly suppresses OC gene expression given that histones were hyperacetylated in the promoter region, as evidenced by the ChIP assay. Histone acetylation, generally, is required for OC activation during osteoblast phenotype development [40]. VPA might affect a distinct subset of genes, some of which could act negatively on OC expression, and this effect could be due to the fact that in addition to selectively inhibiting the catalytic activity of class I HDACs, VPA induces proteasomal degradation of HDAC2 [41].
OC is the most abundant noncollagenous protein in bone. It is considered a late marker of bone differentiation [42], and it is of paramount significance in bone metabolism being also used as a clinical marker for bone turnover. Nevertheless, its exact function remains to be elucidated. Our data prompted us to hypothesize that VPA does not induce the terminal differentiation of osteoblasts, which usually express OC at high levels, but that it stimulates the generation of less mature osteoblasts expressing only OPN and bone-sialoprotein at the onset of the mineralization stage. Several clinical studies have already reported that VPA administered to patients as an anticonvulsant and mood-stabilizing drug is often associated with decreased bone mineral density, osteopenia, and osteoporosis [43]. Our hypothesis gives a possible explanation for these side effects since it has been described that osteocyte-ablated mice had fragile bones with intracortical porosity and microfractures [44].
Conclusions
We have performed silencing of HDAC2 for the first time, with the aim of evaluating the role of HDAC2 in osteogenic differentiation. We have demonstrated that HDAC2 silencing leads to an increased expression of OPN and BSP, and to a downregulated OC mRNA level, resembling the effect of VPA. Our study adds further information on the role of HDACs in osteogenic differentiation and indicates that the specific suppression of individual HDACs by RNA interference could enhance single aspects of osteoblast differentiation, and thus have selective effects.
Acknowledgments
This study was funded by EU PON n. 01_02834 and by MIUR PRIN (project of relevant interest) n.20102M7T8X_002.
Author Contributions
F.P.: conception and design, collection and/or assembly of data, data analysis and interpretation, and manuscript writing; M.L.N., V.T., V.D., P.N., and G.P.: collection and/or assembly of data; A.D.R. and L.L.: provision of study material or patients; L.A.: conception and design and collection and/or assembly of data; G.P.: conception and design, manuscript writing, and final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Gronthos S, Mankani M, Brahim J, et al. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci USA. 2000;97:13625–13630. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laino G, Graziano A, d'Aquino R, et al. An approachable human adult stem cell source for hard tissue engineering. J Cell Physiol. 2006;206:693–701. doi: 10.1002/jcp.20526. [DOI] [PubMed] [Google Scholar]
- 3.Laino G, d'Aquino R, Graziano A, et al. A new population of human adult dental pulp stem cells: A useful source of living autologous fibrous bone tissue (LAB) J Bone Mineral Res. 2005;20:1394–1402. doi: 10.1359/JBMR.050325. [DOI] [PubMed] [Google Scholar]
- 4.Paino F, Ricci G, Rosa De A, et al. Ecto-mesenchymal stem cells from dental pulp are committed to differentiate into active melanocytes. Eur Cell Mater. 2010;20:295–305. doi: 10.22203/ecm.v020a24. [DOI] [PubMed] [Google Scholar]
- 5.Zhang W, Walboomers XF, Shi S, et al. Multilineage differentiation potential of stem cells derived from human dental pulp after cryopreservation. Tissue Eng. 2006;12:2813–2823. doi: 10.1089/ten.2006.12.2813. [DOI] [PubMed] [Google Scholar]
- 6.Papaccio G, Graziano A, d'Aquino R, et al. Long-term cryopreservation of dental pulp stem cells (SBP-DPSCs) and their differentiated osteoblasts: A cell source for tissue repair. J Cell Physiol. 2006;208:319–325. doi: 10.1002/jcp.20667. [DOI] [PubMed] [Google Scholar]
- 7.d'Aquino R, Graziano A, Sampaolesi M, et al. Human postnatal dental pulp cells co-differentiate into osteoblasts and endotheliocytes: A pivotal synergy leading to adult bone tissue formation. Cell Death Differ. 2007;14:1162–1171. doi: 10.1038/sj.cdd.4402121. [DOI] [PubMed] [Google Scholar]
- 8.d'Aquino R, Rosa De A, Lanza V, et al. Human mandible bone defect repair by the grafting of dental pulp stem/progenitor cells and collagen sponge biocomplexes. Eur Cell Mater. 2009;18:75–83. doi: 10.22203/ecm.v018a07. [DOI] [PubMed] [Google Scholar]
- 9.Giuliani A, Manescu A, Langer M, et al. Three years after transplants in human mandibles, histological and in-line holotomography revealed that stem cells regenerated a compact rather than a spongy bone: Biological and clinical implications. Stem Cells Transl Med. 2013;2:316–324. doi: 10.5966/sctm.2012-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Legube G, Legube D. Regulating histone acetyltransferases and deacetylases. EMBO Rep. 2003;4:944–947. doi: 10.1038/sj.embor.embor941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 12.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.North BJ, Marshall BL, Borra MT, et al. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 14.Onyango P, Celic I, McCaffery JM, et al. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci USA. 2002;99:13653–13658. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Witt O, Deubzer HE, Milde T, et al. HDAC family: What are the cancer relevant targets? Cancer Lett. 2009;277:8–21. doi: 10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez-Menendez V, Tremolizzo L, Cavaletti G. Targeting cancer and neuropathy with histone deacetylase inhibitors: Two birds with one stone? Curr Cancer Drug Targets. 2008;8:266–274. doi: 10.2174/156800908784533508. [DOI] [PubMed] [Google Scholar]
- 17.Candido EP, Reeves R, Davie JR. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell. 1978;14:105–113. doi: 10.1016/0092-8674(78)90305-7. [DOI] [PubMed] [Google Scholar]
- 18.Saito A, Yamashita T, Mariko Y, et al. A synthetic inhibitor of histone deacetylase, MS-27–275, with marked in vivo antitumor activity against human tumors. Proc Natl Acad Sci USA. 1999;96:4592–4597. doi: 10.1073/pnas.96.8.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marks PA, Richon VM, Rifkind RA. Histone deacetylase inhibitors: Inducers of differentiation or apoptosis of transformed cells. J Natl Cancer Inst. 2000;92:1210–1216. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 20.Johnstone RW. Histone-deacetylase inhibitors: Novel drugs for the treatment of cancer. Nature Rev Drug Discov. 2002;1:287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- 21.Schroeder TM, Westendorf JJ. Histone deacetylase inhibitors promote osteoblast maturation. J Bone Miner Res. 2005;20:2254–2263. doi: 10.1359/JBMR.050813. [DOI] [PubMed] [Google Scholar]
- 22.Jeon EJ, Lee KY, Choi NS, et al. Bone morphogenetic protein-2 stimulates Runx2 acetylation. J Biol Chem. 2006;281:16502–16511. doi: 10.1074/jbc.M512494200. [DOI] [PubMed] [Google Scholar]
- 23.Loscher W. Basic pharmacology of valproate: A review after 35 years of clinical use for the treatment of epilepsy. CNS Drugs. 2002;16:669–694. doi: 10.2165/00023210-200216100-00003. [DOI] [PubMed] [Google Scholar]
- 24.Göttlicher M, Minucci S, Zhu P, et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gregory CA, Gunn WG, Peister A, et al. An Alizarin red-based assay of mineralization by adherent cells in culture: Comparison with cetylpyridinium chloride extraction. Anal Biochem. 2004;329:77–84. doi: 10.1016/j.ab.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Marks PA, Dokmanovic M. Histone deacetylase inhibitors: Discovery and development as anticancer agents. Expert Opin Investig Drugs. 2005;14:1497–1511. doi: 10.1517/13543784.14.12.1497. [DOI] [PubMed] [Google Scholar]
- 27.Qiu L, Burgess A, Fairlie DP, et al. Histone deacetylase inhibitors trigger a G2 checkpoint in normal cells that is defective in tumor cells. Mol Biol Cell. 2000;11:2069–2083. doi: 10.1091/mbc.11.6.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poon RYC, Jiang W, Toyoshima H, et al. Cyclin-dependent kinases are inactivated by a combination of p21 and Thr14/Tyr15 phosphorylation after UV-induced DNA damage. J Biol Chem. 1996;271:13283–13291. doi: 10.1074/jbc.271.22.13283. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Xiong Y, Beach D. Proliferating cell nuclear antigen and p21 are components of multiple cell cycle kinase complexes. Mol Biol Cell. 1993;4:897–906. doi: 10.1091/mbc.4.9.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu E, Dul E, Sung CM, et al. Identification of novel isoform-selective inhibitors within class I histone deacetylases. J Pharmacol Exp Ther. 2003;307:720–728. doi: 10.1124/jpet.103.055541. [DOI] [PubMed] [Google Scholar]
- 31.Khan N, Jeffers M, Kumar S, et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem J. 2008;409:581–589. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- 32.Rodan SB, Imai Y, Thiede MA, et al. Characterization of a human osteosarcoma cell line (Saos-2) with osteoblastic properties. Cancer Res. 1987;47:4961–4966. [PubMed] [Google Scholar]
- 33.Hausser HJ, Brenner RE. Phenotypic instability of Saos-2 cells in long-term culture. Biochem Biophys Res Commun. 2005;333:216–222. doi: 10.1016/j.bbrc.2005.05.097. [DOI] [PubMed] [Google Scholar]
- 34.Shafa M, Krawetz R, Rancourt DE. Returning to the stem state: Epigenetics of recapitulating pre-differentiation chromatin structure. Bioessays. 2010;32:791–799. doi: 10.1002/bies.201000033. [DOI] [PubMed] [Google Scholar]
- 35.Shao Y, Gao Z, Marks PA, et al. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2004;101:18030–18035. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroeder TM, Kahler RA, Li X, et al. Histone deacetylase 3 interacts with runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J Biol Chem. 2004;279:41998–42007. doi: 10.1074/jbc.M403702200. [DOI] [PubMed] [Google Scholar]
- 37.Westendorf JJ, Zaidi SK, Cascino JE, et al. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol Cell Biol. 2002;22:7982–7992. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jensen ED, Schroeder TM, Bailey J, et al. Histone deacetylase 7 associates with Runx2 and represses its activity during osteoblast maturation in a deacetylation-independent manner. J Bone Miner Res. 2008;23:361–372. doi: 10.1359/JBMR.071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee HW, Suh JH, Kim AY, et al. Histone deacetylase 1-mediated histone modification regulates osteoblast differentiation. Mol Endocrinol. 2006;20:2432–2443. doi: 10.1210/me.2006-0061. [DOI] [PubMed] [Google Scholar]
- 40.Shen J, Montecino M, Lian JB, et al. Histone acetylation in vivo at the osteocalcin locus is functionally linked to vitamin d-dependent, bone tissue-specific transcription. J Biol Chem. 2002;277:20284–20292. doi: 10.1074/jbc.M112440200. [DOI] [PubMed] [Google Scholar]
- 41.Krämer OH, Zhu P, Ostendorff HP, et al. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003;22:3411–3420. doi: 10.1093/emboj/cdg315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hauschka PV, Lian JB, Cole DE, et al. Osteocalcin and matrix Gla protein: Vitamin K-dependent proteins in bone. Physiol Rev. 1989;69:990–1047. doi: 10.1152/physrev.1989.69.3.990. [DOI] [PubMed] [Google Scholar]
- 43.Boluk A, Guzelipek M, Savli H, et al. The effect of valproate on bone mineral density in adult epileptic patients. Pharmacol Res. 2004;50:93–97. doi: 10.1016/j.phrs.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 44.Tatsumi S, Ishii K, Amizuka N, et al. Targeted ablation of osteocytes induces osteoporosis with defective mechanotransduction. Cell Metab. 2007;5:464–475. doi: 10.1016/j.cmet.2007.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


