Abstract
Context
The distribution of standard melanoma antibodies, S100, HMB45 and Melan-A, has been extensively studied. Yet, the overlap in their expression is less well-characterized.
Objective
To determine the joint distributions of the classic melanoma markers and to determine if classification according to joint antigen expression has prognostic relevance.
Design
S100, HMB45 and Melan-A were assayed by immunofluorescence-based immunohistochemistry on a large tissue microarray of 212 cutaneous melanoma primaries and 341 metastases. Positive expression for each antigen required ≥25% of melanoma cells displaying immunoreactivity. Marginal and joint distributions were determined across all markers. Bivariate associations with established clinicopathologic covariates and melanoma-specific survival analyses were conducted.
Results
Two hundred and ninety-five (91.6%), 203 (63.0%) and 236 (73.3%) of 322 assayable melanomas stained with S100, HMB45 and Melan-A, respectively. 27 melanomas, representing a diverse set of histopathologies, were S100-negative. Co-expression of all 3 antibodies was observed in 160 (49.7%) melanomas. Intensity of endogenous melanin pigment did not confound immunolabeling. Among primaries, associations with clinicopathologic parameters revealed a significant relationship only between HMB45 and microsatellitosis (P=.02). No significant differences among clinicopathologic criteria were observed across the HMB45/Melan-A joint distribution categories. Neither marginal HMB45 (P=.56) or Melan-A (P=.81) nor their joint distributions (P=.88), was associated with melanoma-specific survival.
Conclusion
Comprehensive characterization of the marginal and joint distributions for S100, HMB45 and Melan-A across a large series of cutaneous melanomas revealed diversity of expression across these antigens. However, these immunohistochemically-defined subclasses of melanomas do not significantly differ according to clinicopathologic correlates or outcome.
Introduction
Cutaneous malignant melanoma, with an estimated 70,230 new cases and 8,790 deaths expected in the United States during 2011 1, continues to be a major public health concern. In particular, localized melanomas >1.00 mm thick (Stage II), present a therapeutic challenge for clinicians as their prognosis is uncertain. While 10-year melanoma-specific mortality among Stage II patients following a curative resection approaches 50% 2, the adverse risk profile of available adjuvant chemotherapy supports its administration only among those with thick (>4.0 mm), ulcerated melanomas (Stage IIC) 3. The need for melanoma prognostic models capable of identifying those patients with the highest risk of recurrence at the time of diagnosis is well-established 4 and efforts to identify additional prognostic biomarkers are ongoing.
Melanoma-associated antigens (MAAs), due to their high sensitivity for cells of melanocytic origin, are routinely used in the clinic to discriminate melanomas from among other neoplastic lesions 5. Of the MAAs most commonly used in the clinic, S100 is most sensitive, staining approximately 95% of assayed lesions and, despite a specificity of 75%-87%, is considered the “gold standard” for immunohistochemical discrimination of melanocytic cells 6. Both S100 monoclonal and polyclonal antibodies stain primary and metastatic melanomas with equal efficiency and staining is noted in both the cytoplasm and nucleus of positive specimens 7. Furthermore, among positively-staining melanomas, S100 immunostain is more intense in melanomas with a higher Ki-67 fraction 8. The observed reduced specificity results from S100 expression on tumors of glial origin as well as on chondrocytes, adipocytes, dendritic cells and tumors derived from these tissues 6,9,10. Consequently, when morphology and S100 staining do not completely inform the diagnosis, such as occurs in unusual morphological variants of melanoma 11,12 or in the setting of melanoma mimics 6, staining with additional MAAs such as gp100, Melan-A/MART-1 or tyrosinase are conducted. While these antibodies’ sensitivity rarely exceeds 90%, their specificity for melanoma is typically >95% 6,9 and contributes to the differential diagnosis.
Despite that the immunohistochemical evaluation of melanocytic lesions during the diagnostic process typically involves simultaneous assay of multiple MAAs on serial tumor sections13 and that the marginal distributions for S100, HMB45 and Melan-A are well-established9, their joint distributions are only described for selected case series. The most robust of these data are reported by Jungbluth et al. where overlapping HMB45 and Melan-A immunoreactivity was noted among 53/65 melanoma metastases and 7/10 cutaneous primaries, exclusive Melan-A staining in 8/65 metastases and 1/10 primary and 11 metastases and 2 primaries non-reactive with both reagents 14. While the authors further commented that the dual-negative lesions included the assayed spindle and desmoplastic variants, S100 immunoreactivity was not reported for this series 14. Xu et al. evaluated a series of 30 S100+ melanomas, reporting on 8 HMB45+/Melan-A+, 9 HMB45−/Melan-A + and 13 HMB45−/Melan-A− lesions, the latter including 8 desmoplastic melanomas 15. Busam et al. reported Melan-A immunoreactivity on 26/26 S100+ epithelioid melanomas but only on 4/14 S100+/HMB45− spindled or desmoplastic melanomas 16. Kucher et al., reporting on a retrospective series of 40 sentinel lymph node biopsies positive for S100-immunoreactive melanoma noted 34/40 as HMB45+/Melan-A+, 1 as HMB45+/Melan-A− and 5 as HMB45−/Melan-A− 17. While other groups have published manuscripts where the methods include immunostaining of multiple MAAs on serial sections of melanoma lesions, the reported results are limited to either the assayed antibodies’ marginal distributions or the highlighting of certain expression categories (e.g., negative for all assayed antibodies) and do not offer the full joint distributions of the assayed MAAs 18-21. Taken together, while the majority of melanomas express multiple MAAs and, in particular, are positive for both HMB45 and Melan-A, there are still little data describing the prevalence and clinicopathologic correlates across joint MAA expression categories such as HMB45/Melan-A-discordant lesions.
To address this gap, we characterize the joint distributions of S100, HMB45 and Melan-A expression across a large series of 121 cutaneous melanoma primaries and 201 melanoma metastases. We define subcategories of melanomas according to their independent, individually-considered (i.e., “marginal”) and simultaneously combined (i.e., “joint”) distributions across the three assayed antigens and describe the associations between these subcategories with established melanoma clinicopathologic criteria and, among primary melanomas, with melanoma-specific survival. We not only consider those melanomas discordant for HMB45 and Melan-A expression but also describe the clinicopathologic characteristics and HMB45/Melan-A expression patterns for our sample of 27 S100-negative melanomas, one of the largest such series reported to date.
Materials and Methods
Patient tumor samples and tissue microarray construction
Three non-overlapping series of melanoma patients, 212 primary and 237 metastatic cutaneous melanomas surgically removed at Yale-New Haven Hospital during 1959-1994 and 104 additional metastatic melanomas surgically removed during 1995-2005 for which the formalin-fixed, paraffin-embedded (FFPE) tissue blocks were not exhausted during the diagnostic process and for which clinical information was available, were included in the analysis (Yale Human Investigations Committee protocol #8219). Demographic data, clinical course and follow-up through March 1st, 2011 were obtained through comprehensive review of the participant's medical record, the archives of the Connecticut Tumor Registry, the Social Security Death Index and the State of Connecticut Vital Records. Incomplete medical records resulted in missing clinical and demographic annotations.
A tissue microarray (TMA) representing single 0.6-mm diameter cores from each of the eligible specimens was constructed using the standard method 22. For internal quality control, 4 primary melanomas and 61 metastases were cored in duplicate. Duplicate cores of FFPE pellets constructed from each of 15 melanoma cell lines 23,24 were added as additional controls. Histopathologic annotation was conducted by pathologic review of included cases as previously described 25 with incomplete data fields arising from cases with missing tissue blocks/slides that prevented correct re-adjudication of the case. Assessment of melanin levels in each histospot was conducted by a single observer (XXX) on an H&E-stained cut of the TMA and graded on a semi-quantitative, 4-level scale with 0 representing no observable melanin and 3 indicating deep pigment in over 75% of the melanocytes.
Immunohistochemical staining and automated image capture
Two serial sections of the TMA were dewaxed in two exchanges of xylene and rehydrated using an ethanol gradient. Following antigen retrieval in supra-atmospheric pressured, boiling 6.5 mM sodium citrate (pH 6.0) for 10 minutes, endogenous peroxidase activity was blocked with 0.75% hydrogen peroxide and non-specific antigens were neutralized with 0.3% bovine serum albumin. Each of pre-diluted, neat HMB45 or anti-Melan-A A103 mouse monoclonal antibodies (Biogenex, Fremont, CA) were multiplexed with rabbit anti-S100 polyclonal (1:200; DAKO, Carpinteria, CA) were added to the TMA slide and incubated overnight at 4°C. Secondary antibodies, AlexaFluor-546-conjugated goat anti-rabbit (1:100, Life Technologies, Carlsbad, CA) diluted into anti-mouse Envision (neat, DAKO, Carpinteria, CA), were then added for 1 hour at room temperature followed by a 10-minute incubation with Cy5-tyramide (Perkin-Elmer Life Sciences, Waltham, MA) to label the anti-mouse Envision. Slides were coverslipped using Prolong Gold with 4’6-diamidino-2-phenylindole (DAPI, Life Technologies, Carlsbad, CA), the latter to visualize nuclei. Negative controls were obtained through omission of the primary antibody.
Automated image acquisition was done as described previously 23,26. Briefly, sets of monochromatic, high-resolution (1024 × 1024 pixel, 0.5 μm) images are captured for each histospot in each of the DAPI, AlexaFluor-546 and Cy5 fluorescent channels using a modified, computer-controlled epifluorescence microscope (Olympus BX-51 with xy stage and z controller, Center Valley, PA) illuminated by a high-pressure mercury bulb (Photonics Solutions, Edinburgh, United Kingdom) coupled with a high-resolution monochromatic camera (PCO-Tech, Romulus, MI).
Immunostaining evaluation and statistical analysis
Histospots containing <3% of tumor tissue were excluded from further analysis. Photomicrographs representing S100, HMB45 and Melan-A immunostaining were visually compared to a referent H&E for melanoma-specific immunostaining by 2 independent observers (XXX, XXX). A case was scored ‘positive’ for the selected antigen if ≥25% of the tumor cells, as defined by the corresponding area on the referent H&E, displayed immunofluorescence. For cases represented by more than 1 histospot, the case was designated as ‘positive’ if all histospots demonstrated ≥25% immunoreactivity. Cases with immunofluorescence covering <25% of the tumor region were classified as ‘negative’. Discrepancies in staining evaluation were resolved through consultation with a third investigator (XXX). Marginal as well as pair-wise and three-way joint distributions across the assayed melanoma antigens were determined using standard univariate statistics. Bivariate analyses comparing marginal and joint immunostaining patterns with individual clinicopathologic parameters were evaluated using chi-square analysis or analysis of variance, as appropriate. Associations with melanoma-specific survival were performed using Kaplan-Meier product-limit and Cox proportional hazards survival analyses. All statistical analyses were conducted using the Statview statistical package (SAS Institute, Cary, NC).
Results
Marginal and Joint Distributions of S100, HMB45 and Melan-A
To assess the marginal and joint distributions of S100, HMB45 and Melan-A/MART-1 in our large series of cutaneous melanomas, we stained two serial sections of our TMA, multiplexing either HMB45 or the anti-Melan-A/MART-1 A103 mouse monoclonal antibody with the DAKO anti-S100 rabbit polyclonal antibody on a single slide. The 322 surgical specimens (121 primaries and 201 metastases) with evaluable data across all three markers were included in this analysis. S100 staining that persisted over ≥25% of the tumor area was detected in 295/322 (91.6%) of the included histospots with immunofluorescent signal noted in both the nucleus and cytoplasm of positive specimens. By contrast, the remaining 27 melanomas were S100-negative with no evidence of S100 expression in >5% of spotted cells, consistent with the clinical definition of S100-negativity; focal S100 expression was not observed in our included histospots. Similarly, HMB45 stained ≥25% of the tumor area in 203/322 (63.0%) melanomas (73 primaries and 130 metastases) and Melan-A/MART-1 thusly stained 236 (73.3%) melanomas (84 primaries and 152 metastases). For HMB45 and Melan-A, focal expression in <25% of the assayed melanocytes was scored as a negative histospot.
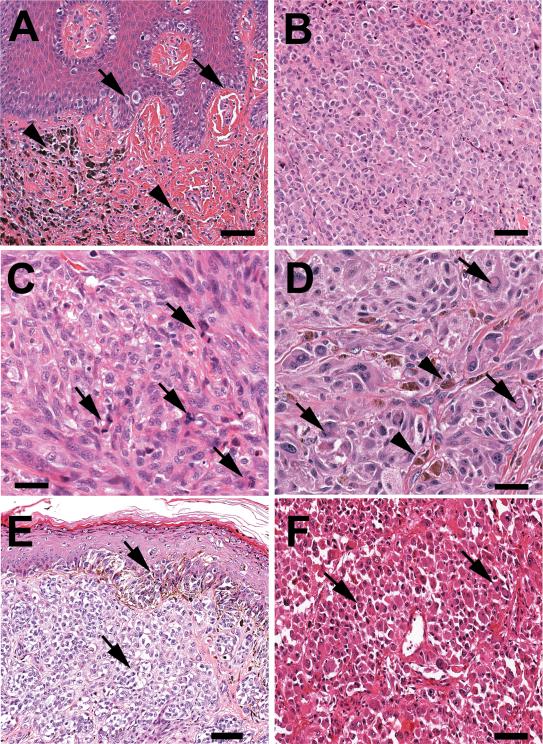
A total of 27 melanomas, including 21/201 metastases and 6/121 primaries (10.4% vs. 5.0%, P=.10) were S100-negative. The clinicopathologic characteristics of the S100-negative melanomas are displayed (Table 1). Seventeen (63.0%) of the S100-negative melanomas developed in male patients (P=.61). S100-negativity was observed across a broad spectrum of histopathologic subtypes among the primaries and among loco-regional as well as visceral disease among the metastases. Other than a higher but non-significant count of pan-MAA-negative melanomas noted among the metastases (5/201 (2.5%) vs. 1/121 (0.8%); Fisher's Exact P=.42), these melanomas were unremarkable with respect to clinicopathologic criteria including growth pattern, overall morphology and pigmentation. Review of their associated H&E sections revealed melanomas with unremarkable histology. S100-negative primary melanomas displayed solid-nested growth whereas S100-negative metastases were all solid with cell morphology that included epithelioid, spindled or hybrid morphology, typical of the disease (Figures 1A-1F).
Table 1.
Clinicopathologic characteristics of S100-negative melanomas
| Primary Melanomas (n=6; 5.0%) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Sample ID | Age at diagnosis | Gender | Resection year | Histopathologic subtype | Breslow (mm) | Growth pattern | Cell morphology | Pigmentation |
| 167 | 67 | Male | 1994 | Superficial spreading | 0.92 mm | Solid – nested | Epithelioid | 2: intense, focal |
| 173 | 77 | Male | 1978 | Nodular | 7.19 mm | Solid – sheets | Epithelioid | 0: absent |
| 229 | 77 | Female | 1992 | Acral lentiginous | 1.00 mm | Solid – nested | Epithelioid | 1: mild, focal |
| 2691 | 61 | Male | 1989 | Superficial spreading | 0.80 mm | Solid – nested | Epithelioid/Spindled | 1: moderate, focal |
| 283 | 34 | Male | 1978 | Superficial spreading | 3.49 mm | Solid – nested | Epithelioid/Spindled | 1: mild, focal |
| 490 | 71 | Male | 1976 | Nodular | 1.40 mm | Solid – sheets | Epithelioid | 2: moderate, focal |
| Metastatic Melanomas (n=21; 10.4%) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Sample ID | Age at diagnosis | Gender | Resection year | Metastatic site | Gross size | Growth pattern | Cell morphology | Pigmentation |
| 18 | 77 | Male | 1969 | Cutaneous | 1.7 cm | Solid | Epithelioid | 0: absent |
| 37 | 53 | Male | 1998 | Lymph node | 13.0 cm | Solid | Epithelioid | 2: intense, focal |
| 57 | 58 | Male | 1996 | Cutaneous | 5.2 cm | Solid | Epithelioid | 0: absent |
| 110 | 77 | Male | 1998 | Soft tissue | 1.0 cm | Solid | Epithelioid/Spindled | 2: intense, focal |
| 161 | 62 | Female | 1998 | Cutaneous | 1.0 cm | Solid | Epithelioid/Spindled | 1: moderate, focal |
| 217 | 58 | Female | 1978 | Cutaneous | Not available | Solid | Epithelioid/Spindled | 1: mild, focal |
| 224 | 43 | Female | 2001 | Lymph Node | 9.0 cm | Solid | Spindled | 3: intense, global |
| 252 | 81 | Female | 1991 | Cutaneous | 0.7 cm | Solid | Epithelioid | 0: absent |
| 255 | 75 | Male | 1991 | Visceral (colon) | 1.8 cm | Solid | Epithelioid | 0: absent |
| 2751 | 70 | Male | 1985 | Cutaneous | 2.8 cm | Solid | Spindled | 0: absent |
| 2871 | 53 | Female | 1999 | Cutaneous | 0.5 cm | Solid | Epithelioid | 0: absent |
| 299 | 26 | Male | 1982 | Lymph node | 10.0 cm | Solid | Spindled | 0: absent |
| 3111 | 68 | Male | 1971 | Cutaneous | 1.0 cm | Solid | Epithelioid | 2: intense, focal |
| 3691 | 36 | Male | 1982 | Cutaneous | 1.5 cm | Solid | Epithelioid | 0: absent |
| 3951 | 81 | Female | 1974 | Visceral (colon) | 10.8 cm | Solid | Epithelioid | 0: absent |
| 419 | 73 | Male | 1976 | Visceral (abdomen) | 3.0 cm | Solid | Epithelioid | 0: absent |
| 462 | 51 | Female | 1964 | Lymph node | 6.0 cm | Solid | Epithelioid | 3: intense, global |
| 521 | 64 | Male | 1990 | Cutaneous | 3.0 cm | Solid | Epithelioid | 1: mild, focal |
| 536 | 85 | Male | 1980 | Cutaneous | Not available | Solid | Epithelioid | 0: absent |
| 551 | 62 | Female | 1978 | Visceral (brain) | 2.5 cm | Solid | Epithelioid/Spindled | 0: absent |
| 563 | 21 | Male | 1973 | Lymph node | 1.0 cm | Solid | Epithelioid | 0: absent |
Melanoma showed no immunoreactivity to each of S100, HMB45 and Melan-A
Figure 1.
Histological characteristics of S100 negative and triple-antigen-negative melanomas. Overall, the lesions show a predominant solid growth pattern, are composed mainly of cells with epithelioid shape and present mild/focal pigmentation. (A) Case 490 (S100-negative): primary lesion showing atypical intraepidermal lentiginous proliferation of large melanoma cells with suprabasal pagetoid spread (arrows) and extensive dermal infiltration by epithelioid cells with nuclear atypia and focal pigmentation (arrowheads). (B) Case 369 (triple-antigen-negative): metastatic nodular lesion composed of a dense sheet of atypical epithelioid cells with hyperchromatic nuclei, ample eosinophilic cytoplasm and lack of pigmentation. (C) Case 275 (triple-antigen-negative): metastatic melanoma showing a solid proliferation of atypical predominant epithelioid cells with highly pleomorphic nuclei, prominent nucleoli, numerous mitotic figures (arrows) and absence of melanic-type pigment. (D) Case 161 (S100-negative): solid metastatic proliferation of large atypical cells with epithelioid morphology, prominent nucleoli, nuclear pseudoinclusions (arrows) and focal pigment deposition (arrowheads). (E) Case 269 (triple-antigen-negative): primary cutaneous lesion showing solid dermal sheets and intraepithelial proliferation of atypical cells with predominant clear cytoplasm and nuclear atypia (arrows). Note focal pigmentation. (F) Case 173 (S100-negative): Primary melanoma composed of highly atypical cells with some “rhabdoid” features displaying abundant eosinophilic cytoplasm, slight lateral nuclear displacement and focal nuclear vacuolation. Hematoxylin/eosin stain, 200x magnification (A, B, E and F) and 400x magnification (C and D).
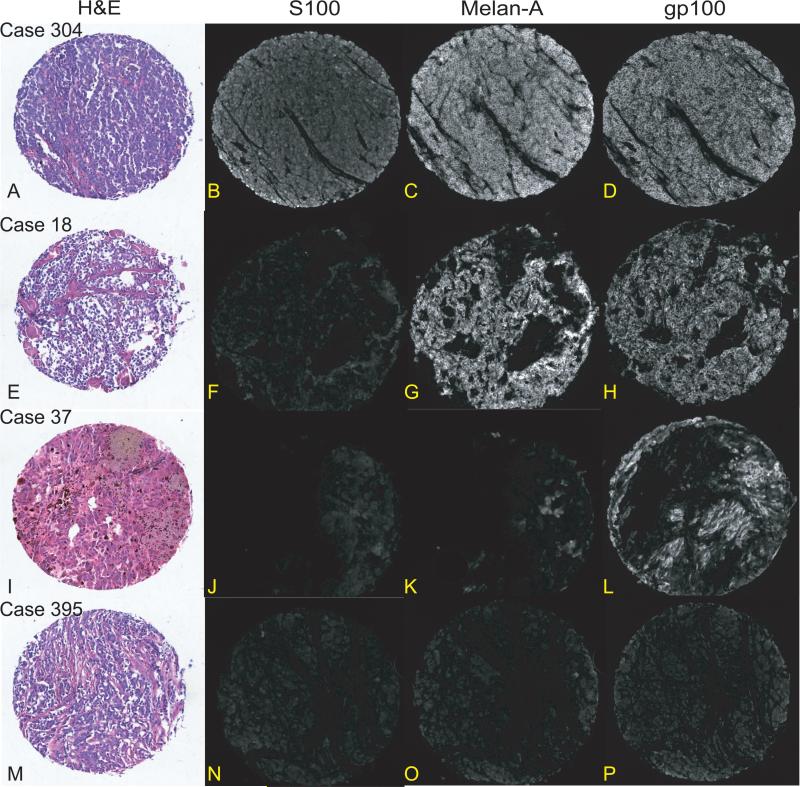
Next, we considered the joint distributions across all 3 antigens (Table 2, Figures 2A-2P). Twenty-five percent or more cellular immunoreactivity for all three assayed antigens was only observed in 160 melanomas (49.7%). By contrast, 156 melanomas expressed only 1 or 2 of the assayed antigens and all combinations of the three assayed antigens were observed among our dataset. Additional commonly occurring categories included S100+/HMB45-/Melan-A+ melanomas (n=61; 18.9%) and melanomas only expressing S100 (n=49; 15.2%). HMB45 or Melan-A immunoreactivity was also observed among the S100-negative melanomas, with each antigen present in a similar number of assayed lesions (18 vs. 15). Yet, 10 S100-negative melanomas demonstrated discordant staining patterns between HMB45 and Melan-A/MART-1. Finally, 1 primary melanoma and 5 metastatic melanomas did not react with any assayed antibody. The two-way cross-tabular joint analysis of HMB45 and Melan-A expression identified 172 (53.4%) melanomas positive for both antigens, 55 (17.1%) negative for both antigens and 95 (29.5%) discordant for the assayed MAAs with similar distributions noted among primaries and metastases (P=.65). Among melanomas discordant across HMB45 and Melan-A expression, a significantly larger number were Melan-A + (n=64) versus HMB45+ (n=31; P<.001).
Table 2.
Distribution of joint immunostaining across S100, HMB45 and Melan-A/MART-1
| n (%) | S100 | HMB45 | Melan-A |
|---|---|---|---|
| 160 (49.7%) | YES | YES | YES |
| 25 (7.8%) | YES | YES | NO |
| 61 (18.9%) | YES | NO | YES |
| 49 (15.2%) | YES | NO | NO |
| 12 (3.7%) | NO | YES | YES |
| 6 (1.9%) | NO | YES | NO |
| 3 (0.9%) | NO | NO | YES |
| 6 (1.9%) | NO | NO | NO |
Figure 2.
Index histospots showing brightfield hematoxylin and eosin and immunofluorescence photomicrographs depicting patterns of S100 (Rabbit polyclonal, DAKO, Carpinteria, CA), Melan-A (monoclonal A103, Biogenex, Fremont, CA) and HMB45 (Biogenex, Fremont, CA) from case examples demonstrating selected melanoma-associated antigen joint distribution patterns. Case 182 (images A-D) expresses all 3 melanoma-associated antigens. Case 18 (images E-H) is S100-negative but expresses both HMB45 and Melan-A. Case 37 (Images I-L) expresses only HMB45. Case 395 (images M-P) does not express any of the 3 assayed melanoma-associated antigens. 100x magnification (A-P).
We also assessed the association between antigen marginal and joint distributions and the semi-quantitative melanin pigment score. Whereas HMB45 staining was independent of pigmentation (P=.13), Melan-A staining was associated with pigmentation as a larger percentage of moderately or highly pigmented melanomas were Melan-A negative (melanin=2 or 3, n=17/42, 40.5%) compared with mildly or unpigmented lesions (melanin=1 or 0, n=62/260, 23.8%) (P=.01, Table 3). Of the 42 moderately or highly pigmented melanomas with S100 immunostaining data, only five were S100-negative where, by contrast, 22 S100-negative melanomas arose in unpigmented lesions. The association between joint HMB45/Melan-A distribution and degree of pigmentation yielded a highly significant result (P=.007; Table 3) with a smaller percentage of HMB45+/Melan-A+ lesions (35.7% vs. 51.7%-60.7%) and a larger percentage of doubly-negative lesions (28.6% vs. 9.8%-17.7%) among the highly pigmented lesions.
Table 3.
Independent and joint HMB45 and Melan-A immunoreactivity according to degree of pigmentation
| No pigment (n=209) | Mild pigment (n=51) | Moderate pigment (n=28) | High pigment (n=14) | P-value | |
|---|---|---|---|---|---|
| Marginal HMB45 expression | |||||
| HMB45 + | 123 (58.9%)1 | 35 (68.6%) | 22 (78.6%) | 10 (71.4%) | P=.13 |
| HMB45 − | 86 (41.1%) | 16 (31.4%) | 6 (21.4%) | 4 (28.6%) | |
| Marginal Melan-A expression | |||||
| Melan-A + | 157 (75.1%) | 41 (80.4%) | 20 (71.4%) | 5 (35.7%) | P=.012 |
| Melan-A − | 52 (24.9%) | 10 (19.6%) | 8 (28.6%) | 9 (64.3%) | |
| Joint HMB45/ Melan-A expression | |||||
| HMB45 + / Melan-A + | 108 (51.7%) | 30 (58.8%) | 17 (60.7%) | 5 (35.7%) | P=.0072 |
| HMB45 + / Melan-A − | 15 (7.2%) | 5 (9.8%) | 5 (17.9%) | 5 (35.7%) | |
| HMB45 − / Melan-A + | 49 (23.4%) | 11 (21.6%) | 3 (10.7%) | 0 (0.0%) | |
| HMB45 − / Melan-A − | 37 (17.7%) | 5 (9.8%) | 3 (10.7%) | 4 (28.6%) |
Percents may not total to 100% due to rounding
Significant at P<.05
For the subset of primary melanomas (n=121), we examined the associations between HMB45 and Melan-A marginal and joint expression with other clinicopathologic features as well as with melanoma-specific survival. Among single-marker bivariate comparisons, the only significant association revealed HMB45-negative melanomas as more likely to possess in-transit metastases at the time of diagnosis (P=.02); all of the remaining comparisons yielded null results (Table 4). We also noted no significant differences in the distribution of clinicopathologic criteria across the HMB45/Melan-A joint expression categories (Table 5). Lack of expression of either HMB45 (HR=1.20, 95% CI: 0.65-2.21; P=.56) or Melan-A (HR=1.09, 95% CI: 0.56-2.12; P=.81) was not associated with melanoma-specific mortality after adjusting for Breslow thickness (mm), stage at diagnosis and age at diagnosis. Survival analysis across the 4 joint distribution categories yielded overlapping product-limit survival curves on univariate analysis (P>.99; Figure 3) and no significant difference in a multi-variable proportional hazards model after adjusting for Breslow thickness and stage at diagnosis (P=.88; Table 6).
Table 4.
Individual HMB45 and Melan-A immunostaining associations with clinicopathologic parameters – primary melanomas
| Variable | HMB45 Positive (n=73) | HMB45 Negative (n=48) | P-value | Melan-A Positive (n=84) | Melan-A Negative (n=37) | P-value |
|---|---|---|---|---|---|---|
| Age at diagnosis (years) | 56.44 ± 16.54 | 60.33 ± 13.88 | P=.18 | 58.44 ± 16.24 | 57.00 ± 14.19 | P=.64 |
| Breslow thickness (mm) | 2.85 ± 2.24 | 2.48 ± 2.01 | P=.36 | 2.75 ± 2.06 | 2.61 ± 2.38 | P=.74 |
| Sex | ||||||
| Male | 37 (50.7%)1 | 22 (45.8%) | P=.60 | 39 (46.4%) | 20 (54.1%) | P=.44 |
| Female | 36 (49.3%) | 26 (54.2%) | 45 (53.6%) | 17 (45.9%) | ||
| Stage at diagnosis | ||||||
| Localized | 57 (85.1%) | 38 (88.4%) | P=.62 | 66 (85.7%) | 29 (87.8%) | P=.76 |
| Regional/Distant | 10 (14.9%) | 5 (11.6%) | 11 (14.3%) | 4 (12.1%) | ||
| Clark Level | ||||||
| II | 6 (8.2%) | 1 (2.0%) | P=.51 | 5 (6.0%) | 2 (5.4%) | P=.61 |
| III | 24 (32.9%) | 18 (37.5%) | 26 (31.0%) | 16 (43.2%) | ||
| IV | 32 (43.8%) | 23 (47.9%) | 41 (48.8%) | 14 (37.8%) | ||
| V | 11 (15.1%) | 6 (12.5%) | 12 (14.3%) | 5 (13.5%) | ||
| Ulceration | ||||||
| Yes | 24 (32.9%) | 17 (36.2%) | P=.71 | 32 (38.1%) | 9 (25.0%) | P=.17 |
| No | 49 (67.1%) | 30 (63.8%) | 52 (61.9%) | 27 (75.0%) | ||
| Microsatellitosis | ||||||
| Yes | 13 (17.8%) | 16 (34.0%) | P=.042 | 19 (22.6%) | 10 (27.8%) | P=.55 |
| No | 60 (82.2%) | 31 (66.0%) | 65 (77.4%) | 26 (72.2%) | ||
| Histopathologic subtype | ||||||
| Superficial spreading | 45 (61.6%) | 34 (72.3%) | P=.38 | 52 (61.9%) | 27 (75.0%) | P=.28 |
| Nodular | 16 (21.9%) | 6 (12.8%) | 15 (17.9%) | 7 (19.4%) | ||
| Lentigo maligna | 1 (1.4%) | 1 (2.1%) | 2 (2.4%) | 0 (0.0%) | ||
| Acral lentiginous | 6 (8.2%) | 1 (2.1%) | 7 (8.3%) | 0 (0.0%) | ||
| Other | 5 (6.8%) | 5 (10.6%) | 8 (9.5%) | 2 (5.4%) | ||
| Tumor-infiltrating lymphocytes | ||||||
| Absent/Sparse | 18 (24.7%) | 6 (12.5%) | P=.12 | 17 (20.2%) | 7 (18.9%) | P=.19 |
| Non-brisk | 44 (60.3%) | 29 (60.4%) | 54 (64.3%) | 19 (51.4%) | ||
| Brisk | 11 (15.1%) | 13 (27.1%) | 13 (15.5%) | 11 (29.7%) | ||
| Anatomic site of primary | ||||||
| Head/neck | 7 (9.6%) | 8 (18.2%) | P=.31 | 11 (13.3%) | 4 (12.1%) | P=.26 |
| Trunk | 21 (28.8%) | 16 (36.4%) | 22 (26.5%) | 15 (45.5%) | ||
| Upper Extremity | 11 (15.0%) | 6 (13.6%) | 12 (14.5%) | 5 (15.2%) | ||
| Lower Extremity | 34 (46.6%) | 14 (31.8%) | 38 (45.8%) | 10 (30.3%) |
Numbers may not sum to total due to missing values, percents may not sum to 100% due to rounding
Significant at P<.05
Table 5.
HMB45/Melan-A joint immunostaining associations with clinicopathologic parameters – primary melanomas
| Variable | HMB45 + / Melan-A + (n=60) | HMB45 + / Melan-A − (n=13) | HMB45 − / Melan-A + (n=24) | HMB45 − / Melan-A − (n=24) | P-value |
|---|---|---|---|---|---|
| Age at diagnosis (years) | 57.15 ± 15.89 | 53.23 ± 19.62 | 61.63 ± 16.98 | 59.04 ± 10.09 | P=.43 |
| Breslow thickness (mm) | 2.91 ± 2.24 | 2.57 ± 2.29 | 2.34 ± 1.45 | 2.63 ± 2.48 | P=.72 |
| Sex | |||||
| Male | 29 (48.3%)1 | 8 (61.5%) | 10 (41.7%) | 12 (50.0%) | P=.72 |
| Female | 31 (51.7%) | 5 (38.5%) | 14 (58.3%) | 12 (50.0%) | |
| Stage at diagnosis | |||||
| Localized | 46 (83.6%) | 11 (91.7%) | 20 (90.9%) | 18 (85.7%) | P=.79 |
| Regional/Distant | 9 (16.4%) | 1 (8.3%) | 2 (9.1%) | 3 (14.3%) | |
| Clark Level | |||||
| II | 4 (6.7%) | 2 (15.4%) | 1 (4.2%) | 0 (0.0%) | P=.72 |
| III | 19 (31.7%) | 5 (38.5%) | 7 (29.2%) | 11 (52.4%) | |
| IV | 28 (46.7%) | 4 (30.8%) | 13 (54.2%) | 10 (47.6%) | |
| V | 9 (15.0%) | 2 (15.4%) | 3 (12.5%) | 0 (0.0%) | |
| Ulceration | |||||
| Yes | 20 (33.3%) | 4 (30.8%) | 12 (50.0%) | 18 (78.3%) | P=.23 |
| No | 40 (66.7%) | 9 (69.2%) | 12 (50.0%) | 5 (21.7%) | |
| Microsatellitosis | |||||
| Yes | 11 (18.3%) | 2 (15.4%) | 8 (33.3%) | 8 (34.8%) | P=.24 |
| No | 49 (81.7%) | 11 (84.6%) | 16 (66.7%) | 15 (65.2%) | |
| Histopathologic subtype | |||||
| Superficial spreading | 36 (60.0%) | 9 (69.2%) | 16 (66.7%) | 18 (78.3%) | P=.62 |
| Nodular | 12 (20.0%) | 4 (30.8%) | 3 (12.5%) | 3 (13.0%) | |
| Lentigo maligna | 1 (1.7%) | 0 (0.0%) | 1 (4.2%) | 0 (0.0%) | |
| Acral lentiginous | 6 (10.0%) | 0 (0.0%) | 1 (4.2%) | 0 (0.0%) | |
| Other | 5 (8.3%) | 0 (0.0%) | 3 (12.5%) | 2 (8.7%) | |
| Tumor-infiltrating lymphocytes | |||||
| Absent/Sparse | 16 (26.7%) | 2 (15.4%) | 1 (4.2%) | 5 (20.8%) | P=.14 |
| Non-brisk | 37 (61.7%) | 7 (53.8%) | 17 (70.8%) | 12 (50.0%) | |
| Brisk | 7 (11.7%) | 4 (30.8%) | 6 (25.0%) | 7 (29.2%) | |
| Anatomic site of primary | |||||
| Head/neck | 6 (10.0%) | 1 (7.7%) | 5 (21.7%) | 3 (14.3%) | P=.61 |
| Trunk | 15 (25.0%) | 6 (46.2%) | 7 (30.4%) | 9 (42.9%) | |
| Upper Extremity | 9 (15.0%) | 2 (15.4%) | 3 (13.0%) | 3 (14.3%) | |
| Lower Extremity | 30 (50.0%) | 4 (13.8%) | 8 (34.8%) | 6 (28.6%) |
Numbers may not sum to total due to missing values, percents may not sum to 100% due to rounding
Figure 3.
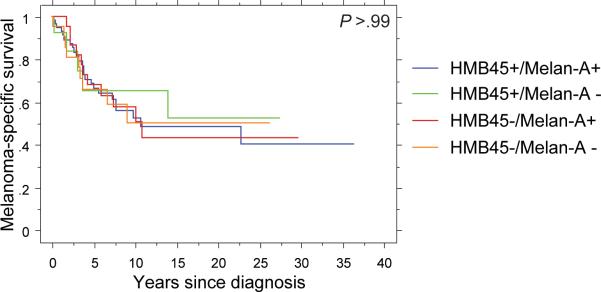
Differential melanoma-specific survival according to category of joint HMB45/Melan-A expression.
Table 6.
Joint HMB45/Melan-A immunostaining melanoma-specific survival hazard ratios
| Variable | Hazard Ratio (95% CI) | P-value |
|---|---|---|
| Breslow thickness (mm) | 1.18 (1.04 – 1.34) | P=.011 |
| Stage at diagnosis | ||
| Localized | 1.00 | P=.011 |
| Regional/Distant | 2.72 (1.24 – 5.95) | |
| HMB45/Melan-A joint distributions | ||
| HMB45 − / Melan-A − | 1.00 | P=.88 |
| HMB45 − / Melan-A + | 0.86 (0.34 – 2.16) | |
| HMB45 + / Melan-A − | 0.64 (0.20 – 2.09) | |
| HMB45+ / Melan-A + | 0.77 (0.35 – 1.73) |
Significant at P<.05
Discussion
The differential expression of S100, HMB45 and Melan-A/MART-1 has been extensively studied in cutaneous malignant melanoma. Yet, while trends for their marginal distributions are well-known, the overlap in their expression is less well-characterized with, to the best of our knowledge, no study evaluating the joint distributions of these antigens in samples of greater than 100 melanomas and no study considering the distribution of established melanoma clinicopathologic parameters in HMB45/Melan-A null or discordant lesions compared with the referent HMB45+/Melan-A+ tumor 14-17. To address this, we considered the marginal and joint expression of S100, HMB45 and Melan-A/MART-1 across a large TMA containing representative histospots from 213 primary melanomas and 342 melanoma metastases. Data from the 322 lesions (121 primaries and 201 metastases) with evaluable data for all three markers were included in our analysis.
Using immunofluorescence-based immunohistochemistry to assay our targeted antigens, 91.6% of our sample was S100+, 63.0% was HMB45+ and 73.3% were Melan-A/MART-1+, each registering slightly below the recognized sensitivities for each of these respective antigens 9 with similar rates of immunoreactivity occurring among primary and metastatic lesions. As we selected a cut-off of ≥25% immunolabeling across eligible melanocytes in each arrayed histospot, our decreased sensitivity might be due to more stringent criteria than the clinical standard of >5% positivity 27 such that lesions with patchy, focal staining would be alternatively classified as negative in our analysis. We also did not observe any association between HMB45 or Melan-A immunoreactivity with any assayed clinicopathologic factor except for an increased lack of HMB45 immunoreactivity among melanoma primaries with known microsatellitosis, consistent with the observation of less frequent HMB45 antigenicity among melanoma metastases 9,21. These data could suggest that lack of HMB45 immunoreactivity might be a property of primary melanomas more likely to metastasize as opposed to a phenotype acquired following metastasis. We also did not detect an association with melanoma-specific mortality with either HMB45 (P=.56) or Melan-A (P=.81) among our subset of primary melanomas after adjusting for Breslow thickness, stage at diagnosis or age at diagnosis. Although our data are consistent with previously published null results reported for Melan-A 28,29, the data for HMB45 are less straightforward with reports for significantly improved 29, significantly impaired30 and no association with survival 28 for HMB45 expression all reported using cut-offs of either 50% or 90% staining efficiency 31.
Interestingly, 27 (8.4%) of our assayed melanomas were S100-negative, representing, to the best of our knowledge, the largest single collection of S100-negative melanomas reported in the literature. To confirm this, we conducted a systematic search of the PubMed database through November 18th, 2011 using the keyword search (“S100-negative” AND “melanoma”). The search returned 4 manuscripts of which two 32,33 addressed melanoma clinical samples. Nine additional manuscripts were identified through review of cited references 7,8,34-40 including one manuscript 35 that contained a meta-analysis of 12 additional manuscripts all published prior to 1992. Although the meta-analysis published cumulative data describing S100 negativity in 13/230 primaries and 9/166 melanoma metastases, no single manuscript identified in our search reported on more than 20 S100-negative melanomas with all but two studies 32,36 describing fewer than five cases.
S100-negative lesions have been described more frequently among junctional nests on sun-damaged skin 33 and among metastatic lesions, even in the setting of a previously-documented S100-positive primary tumor 32. Lack of S100 expression may occur more frequently among ocular or acral melanomas 32,34 but has been observed across all histologic subtypes 35. Six of our S100-negative melanomas arose in primary lesions with the remaining 21 occurring in metastases. One study reporting on 11 S100-negative melanoma metastases with matched primary blocks suggested that loss of S100 antigenicity might be acquired along with metastatic competency as 9/11 had S100-positive primary lesions 32 with a second study that considered 12 S100-negative melanomas reporting equal rates between primaries and metastases 36. While a higher percentage of our assayed metastases (10.4%) were S100-negative compared to our primaries (5.0%), this difference trended towards but did not achieve statistical significance (p=0.10) most likely due to the small size of our S100-negative sample.
We also did not detect any significant associations between S100 immunoreactivity and other clinicopathologic criteria. Melanomas arising in males were as likely to be S100-negative as were melanomas arising in females. Among primary lesions, S100-negative melanomas occurred across a broad range of Breslow thicknesses and histopathologic subtypes. While one study noted a possibly increased rate of S100-negative lesions among acral lentiginous melanomas 34 and, in our study, two S100-negative lesions arose in acral lentiginous or amelanotic melanomas, the majority of our S100-negative lesions arose in superficial spreading or nodular melanomas. Among metastases, lack of S100 antigenicity was observed among cutaneous, soft tissue, lymph node and visceral lesions.
Six of our S100-negative melanomas, one primary and five metastases, also lacked expression of HMB45 and Melan-A. This “triple negative” pattern was also noted among 7/17 S100-negative metastases reported by Aisner et al. with 5/7 of these lesions arising from the back or shoulder 32. Interestingly, 5/6 of our metastatic “triple negatives” arose in cutaneous or subcutaneous soft tissue however our lesions derived from a more diverse set of primary tumor locations including the face, leg, and abdomen as well as the back. Application of next-generation sequencing to the exomes of S100-negative melanomas might be useful for identifying underlying molecular changes associated with both overall lack of S100 antigenicity and concordant lack of S100, HMB45 and Melan-A/MART-1 immunoreactivity.
While the marginal distributions for S100, HMB45 and Melan-A positivity and their associations with recognized melanoma clinicopathologic parameters are well-established 9, the literature describing their joint distributions is much more sparse. While MAA expression discordance can be expected in light of the well-documented differing sensitivities of the commonly used MAAs 9, there are very few published studies that describe the prevalence and clinical significance of MAA-concordant and discordant lesions. The largest of these studies evaluated overlapping HMB45/Melan-A immunoreactivity in 65 melanoma metastases and 10 cutaneous primaries 14. While the authors explored the distribution across all pair-wise antigen combinations, correlations with clinicopathologic criteria other than histologic subtype were not reported 14. Among their sample of 30 melanomas, Xu et al. observed some HMB45/Melan-A discordance with positive Melan-A expression occurring only in 9/14 S100+/HMB45- spindled or epithelioid melanomas 15. In their study of 17 S100-negative melanomas, Aisner et al. noted only concordance between HMB45 and Melan-A expression 32. To the best of our knowledge, our study of 322 assayable melanomas represents the largest series to date for which the joint distribution of HMB45 and Melan-A are considered and the only study to consider the relationship between joint HMB45/Melan-A expression and commonly reported clinicopathologic criteria among eligible primary lesions. The majority of our lesions were concordant for HMB45 and Melan-A expression with 53.4% expressing both antigens and 17.1% lacking expression in ≥25% of the arrayed melanoma. We also did observe melanomas discordant for MAA expression with 31 (9.6%) melanomas expressing HMB45 only and 64 (19.9%) melanomas expressing Melan-A only. Further bivariate analyses among the 121 assayed primary melanomas revealed no significant associations with any of the recognized clinicopathologic criteria and survival analyses revealed virtually overlapping survival curves across the 4 joint distribution categories to suggest that the prognostic impact MAA discordance may be small. While the significantly larger (P<.001) number of discordant lesions that express only Melan-A can be explained by Melan-A's recognized comparatively more diffuse and intense staining of melanomas that persists into the dermal layers of the assayed lesions 14,41 that possibly produced fewer false negatives among our sample of representative 0.6 mm histospots, we cannot exclude the possible role for genetic or epigenetic factors that underlie the development and distribution of the 4 classes of HMB45/Melan-A expression-defined melanomas and possible relationships with levels of MITF expression 42 may yield compelling insight into mechanisms relevant for melanocytic lesion development and progression.
While our study includes numerous strengths such as our large sample size, use of TMAs and automated image capture that eliminate the potential for laboratory drift by assaying all samples simultaneously in the same experimental batch and comprehensive specimen annotation to enable robust clinicopathologic associations, we also recognize several limitations with our experimental approach. First, although quantitative immunofluorescence is a well-established, unbiased measure of antigen expression across a broad spectrum of cancers 43,44, the presence of photo-reactive melanin might confound immunofluorescent readouts in melanoma 45. Not only does melanin exhibit broad spectral absorption that decreases monotonically with increasing wavelengths from 300-1100 nm 46,47, melanin has also recently been shown to display autofluorescence with separate excitations in the ranges of 370-470 nm and 785 nm and corresponding emissions at 540 nm and 890-900 nm, respectively 47,48 with the former in close proximity to the emission wavelength of our mask fluorophore (546 nm). While HMB45 expression was independent of melanin distribution with similar proportion of HMB45 negative lesions among all four categories of pigmentation, Melan-A immunostaining was associated with melanin levels where, despite similar proportions of Melan-A negative lesions observed in each of the unpigmented, mildly and moderately pigmented melanomas, among the highly pigmented melanomas an excess of Melan-A negative lesions were observed. This effect also translated into a similar significant association among the HMB45/Melan-A joint distribution categories. Because the association with melanin expression did not extend to all assayed MAAs, we cannot rule out an underlying biological mechanism that requires further elucidation.
A second limitation of our analysis is that our data were collected from a TMA that represented each melanoma with a single 0.6 mm histospot and not validated on whole-sections. Although current clinical diagnostic standards suggest whole-section analysis with MAA-positivity recognized with immunoreactivity in as few as 5% of cells 27, use of TMAs circumvent the risk for batch-to-batch variation that could be incurred during a whole-slide approach. However, recent TMA validation experiments have demonstrated that intra-tumoral heterogeneity can yield significant core-to-core variability of protein expression. The observed variance driven by both the lability of the target protein and the antibody selected for its measurement such that, in a marker-dependent fashion, as many as 11 independent histospots may be required to adequately capture potential heterogeneity 49. Specific TMA validation experiments conducted over a small series of antigens with prognostic potential suggest that, for melanoma, a minimum of three independent cores are needed to achieve 90% concordance for overall positive stain with whole sections 50-53. In the context of recognized focality of HMB45 immunostains and the association of decreased HMB45 and Melan-A stain in deeper dermal regions of primary melanomas, we cannot rule out some degree of measurement error that might increase our false-negative rate through our strategy of having sampled only 1 single 0.6mm histospot from each index lesion. Future work would include validating our findings across additional, redundant builds of the TMA.
Our experimental design is further limited by the need to sample HMB45 and Melan-A independently of each other on serial sections of our TMA and then to reconstruct joint distributions through the overlay of corresponding images across the serial sections. Although we forgo some precision by not being able to multiplex both HMB45 and Melan-A on the same section, our method for assessing joint distribution matches current clinical practice for evaluating occult micrometastases and malignancies of unknown primary in the surgical pathology suite 27.
In summary, we have comprehensively characterized the marginal and joint distributions, clinicopathologic correlates and prognostic potential for three clinically-relevant MAAs in a large series of primary and metastatic melanomas. Our study also describes the largest single series, to date of S100-negative melanomas. Future directions include transcriptome profiling and whole-exome sequencing of lesions representative of each joint distribution category as well as analysis of MITF and other MAA expression to further elucidate discriminating molecular characteristics that define the individual MAA-based melanoma subclasses.
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011 Jul-Aug;61(4):212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Balch CM, Gershenwald JE, Soong SJ, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009 Dec 20;27(36):6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garbe C, Peris K, Hauschild A, et al. Diagnosis and treatment of melanoma: European consensus-based interdisciplinary guideline. Eur J Cancer. 2010 Jan;46(2):270–283. doi: 10.1016/j.ejca.2009.10.032. [DOI] [PubMed] [Google Scholar]
- 4.Gimotty PA, Guerry D. Prognostication in thin cutaneous melanomas. Arch Pathol Lab Med. 2010 Dec;134(12):1758–1763. doi: 10.5858/2009-0653-RAR.1. [DOI] [PubMed] [Google Scholar]
- 5.Carlson JA, Ross JS, Slominski AJ. New techniques in dermatopathology that help to diagnose and prognosticate melanoma. Clin Dermatol. 2009 Jan-Feb;27(1):75–102. doi: 10.1016/j.clindermatol.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Busam KJ. The use and application of special techniques in assessing melanocytic tumours. Pathology. 2004 Oct;36(5):462–469. doi: 10.1080/00313020412331283824. [DOI] [PubMed] [Google Scholar]
- 7.Timar J, Udvarhelyi N, Banfalvi T, Gilde K, Orosz Z. Accuracy of the determination of S100B protein expression in malignant melanoma using polyclonal or monoclonal antibodies. Histopathology. 2004 Feb;44(2):180–184. doi: 10.1111/j.1365-2559.2004.01800.x. [DOI] [PubMed] [Google Scholar]
- 8.Sviatoha V, Tani E, Kleina R, Sperga M, Skoog L. Immunohistochemical analysis of the S100A1, S100B, CD44 and Bcl-2 antigens and the rate of cell proliferation assessed by Ki-67 antibody in benign and malignant melanocytic tumours. Melanoma Res. 2010 Apr;20(2):118–125. doi: 10.1097/CMR.0b013e3283350554. [DOI] [PubMed] [Google Scholar]
- 9.Ohsie SJ, Sarantopoulos GP, Cochran AJ, Binder SW. Immunohistochemical characteristics of melanoma. J Cutan Pathol. 2008 May;35(5):433–444. doi: 10.1111/j.1600-0560.2007.00891.x. [DOI] [PubMed] [Google Scholar]
- 10.Cochran AJ, Wen DR. S-100 protein as a marker for melanocytic and other tumours. Pathology. 1985 Apr;17(2):340–345. doi: 10.3109/00313028509063777. [DOI] [PubMed] [Google Scholar]
- 11.Banerjee SS, Harris M. Morphological and immunophenotypic variations in malignant melanoma. Histopathology. 2000 May;36(5):387–402. doi: 10.1046/j.1365-2559.2000.00894.x. [DOI] [PubMed] [Google Scholar]
- 12.Magro CM, Crowson AN, Mihm MC. Unusual variants of malignant melanoma. Mod Pathol. 2006 Feb;19(Suppl 2):S41–70. doi: 10.1038/modpathol.3800516. [DOI] [PubMed] [Google Scholar]
- 13.Prieto VG, Shea CR. Immunohistochemistry of melanocytic proliferations. Arch Pathol Lab Med. 2011 Jul;135(7):853–859. doi: 10.5858/2009-0717-RAR.1. [DOI] [PubMed] [Google Scholar]
- 14.Jungbluth AA, Busam KJ, Gerald WL, et al. A103: An anti-melan-a monoclonal antibody for the detection of malignant melanoma in paraffin-embedded tissues. Am J Surg Pathol. 1998 May;22(5):595–602. doi: 10.1097/00000478-199805000-00011. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Chu AY, Pasha TL, Elder DE, Zhang PJ. Immunoprofile of MITF, tyrosinase, melan-A, and MAGE-1 in HMB45-negative melanomas. Am J Surg Pathol. 2002 Jan;26(1):82–87. doi: 10.1097/00000478-200201000-00010. [DOI] [PubMed] [Google Scholar]
- 16.Busam KJ, Chen YT, Old LJ, et al. Expression of melan-A (MART1) in benign melanocytic nevi and primary cutaneous malignant melanoma. Am J Surg Pathol. 1998 Aug;22(8):976–982. doi: 10.1097/00000478-199808000-00007. [DOI] [PubMed] [Google Scholar]
- 17.Kucher C, Zhang PJ, Acs G, Roberts S, Xu X. Can Melan-A replace S-100 and HMB-45 in the evaluation of sentinel lymph nodes from patients with malignant melanoma? Appl Immunohistochem Mol Morphol. 2006 Sep;14(3):324–327. doi: 10.1097/00129039-200609000-00011. [DOI] [PubMed] [Google Scholar]
- 18.Blessing K, Sanders DS, Grant JJ. Comparison of immunohistochemical staining of the novel antibody melan-A with S100 protein and HMB-45 in malignant melanoma and melanoma variants. Histopathology. 1998 Feb;32(2):139–146. doi: 10.1046/j.1365-2559.1998.00312.x. [DOI] [PubMed] [Google Scholar]
- 19.Clarkson KS, Sturdgess IC, Molyneux AJ. The usefulness of tyrosinase in the immunohistochemical assessment of melanocytic lesions: a comparison of the novel T311 antibody (anti-tyrosinase) with S-100, HMB45, and A103 (anti-melan-A). J Clin Pathol. 2001 Mar;54(3):196–200. doi: 10.1136/jcp.54.3.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miettinen M, Fernandez M, Franssila K, Gatalica Z, Lasota J, Sarlomo-Rikala M. Microphthalmia transcription factor in the immunohistochemical diagnosis of metastatic melanoma: comparison with four other melanoma markers. Am J Surg Pathol. 2001 Feb;25(2):205–211. doi: 10.1097/00000478-200102000-00008. [DOI] [PubMed] [Google Scholar]
- 21.de Vries TJ, Fourkour A, Wobbes T, Verkroost G, Ruiter DJ, van Muijen GN. Heterogeneous expression of immunotherapy candidate proteins gp100, MART-1, and tyrosinase in human melanoma cell lines and in human melanocytic lesions. Cancer Res. 1997 Aug 1;57(15):3223–3229. [PubMed] [Google Scholar]
- 22.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998 Jul;4(7):844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 23.Moeder CB, Giltnane JM, Moulis SP, Rimm DL. Quantitative, fluorescence-based in-situ assessment of protein expression. Methods Mol Biol. 2009;520:163–175. doi: 10.1007/978-1-60327-811-9_12. [DOI] [PubMed] [Google Scholar]
- 24.Hoek K, Rimm DL, Williams KR, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004 Aug 1;64(15):5270–5282. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- 25.Gould Rothberg BE, Berger AJ, Molinaro AM, et al. Melanoma prognostic model using tissue microarrays and genetic algorithms. J Clin Oncol. 2009 Dec 1;27(34):5772–5780. doi: 10.1200/JCO.2009.22.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002 Nov;8(11):1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 27.Mathew R, Messina JL. Recent advances in pathologic evaluation and reporting of melanoma. Semin Oncol. 2012 Apr;39(2):184–191. doi: 10.1053/j.seminoncol.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Alonso SR, Ortiz P, Pollan M, et al. Progression in cutaneous malignant melanoma is associated with distinct expression profiles: a tissue microarray-based study. Am J Pathol. 2004 Jan;164(1):193–203. doi: 10.1016/s0002-9440(10)63110-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hofbauer GF, Burkhart A, Schuler G, Dummer R, Burg G, Nestle FO. High frequency of melanoma-associated antigen or HLA class I loss does not correlate with survival in primary melanoma. J Immunother. 2004 Jan-Feb;27(1):73–78. doi: 10.1097/00002371-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 30.Niezabitowski A, Czajecki K, Rys J, et al. Prognostic evaluation of cutaneous malignant melanoma: a clinicopathologic and immunohistochemical study. J Surg Oncol. 1999 Mar;70(3):150–160. doi: 10.1002/(sici)1096-9098(199903)70:3<150::aid-jso2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 31.Gould Rothberg BE, Bracken MB, Rimm DL. Tissue biomarkers for prognosis in cutaneous melanoma: a systematic review and meta-analysis. J Natl Cancer Inst. 2009 Apr 1;101(7):452–474. doi: 10.1093/jnci/djp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aisner DL, Maker A, Rosenberg SA, Berman DM. Loss of S100 antigenicity in metastatic melanoma. Hum Pathol. 2005 Sep;36(9):1016–1019. doi: 10.1016/j.humpath.2005.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson ML, Hossler EW, Ferringer TC, Elston DM. S100-negative junctional melanocytic proliferations. Am J Dermatopathol. 2011 May;33(3):327–329. doi: 10.1097/DAD.0b013e3181e031d1. [DOI] [PubMed] [Google Scholar]
- 34.Argenyi ZB, Cain C, Bromley C, et al. S-100 protein-negative malignant melanoma: fact or fiction? A light-microscopic and immunohistochemical study. Am J Dermatopathol. 1994 Jun;16(3):233–240. [PubMed] [Google Scholar]
- 35.Bishop PW, Menasce LP, Yates AJ, Win NA, Banerjee SS. An immunophenotypic survey of malignant melanomas. Histopathology. 1993 Aug;23(2):159–166. doi: 10.1111/j.1365-2559.1993.tb00474.x. [DOI] [PubMed] [Google Scholar]
- 36.Trefzer U, Rietz N, Chen Y, et al. SM5-1: a new monoclonal antibody which is highly sensitive and specific for melanocytic lesions. Arch Dermatol Res. 2000 Dec;292(12):583–589. doi: 10.1007/s004030000186. [DOI] [PubMed] [Google Scholar]
- 37.Ordonez NG, Ji XL, Hickey RC. Comparison of HMB-45 monoclonal antibody and S-100 protein in the immunohistochemical diagnosis of melanoma. Am J Clin Pathol. 1988 Oct;90(4):385–390. doi: 10.1093/ajcp/90.4.385. [DOI] [PubMed] [Google Scholar]
- 38.Orchard GE. Comparison of immunohistochemical labelling of melanocyte differentiation antibodies melan-A, tyrosinase and HMB 45 with NKIC3 and S100 protein in the evaluation of benign naevi and malignant melanoma. Histochem J. 2000 Aug;32(8):475–481. doi: 10.1023/a:1004192232357. [DOI] [PubMed] [Google Scholar]
- 39.Kaufmann O, Koch S, Burghardt J, Audring H, Dietel M. Tyrosinase, melan-A, and KBA62 as markers for the immunohistochemical identification of metastatic amelanotic melanomas on paraffin sections. Mod Pathol. 1998 Aug;11(8):740–746. [PubMed] [Google Scholar]
- 40.Zubovits J, Buzney E, Yu L, Duncan LM. HMB-45, S-100, NK1/C3, and MART-1 in metastatic melanoma. Hum Pathol. 2004 Feb;35(2):217–223. doi: 10.1016/j.humpath.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 41.Fetsch PA, Marincola FM, Abati A. The new melanoma markers: MART-1 and Melan-A (the NIH experience). Am J Surg Pathol. 1999 May;23(5):607–610. doi: 10.1097/00000478-199905000-00017. [DOI] [PubMed] [Google Scholar]
- 42.Mitra D, Fisher DE. Transcriptional regulation in melanoma. Hematol Oncol Clin North Am. 2009 Jun;23(3):447–465, viii. doi: 10.1016/j.hoc.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 43.Rimm DL. What brown cannot do for you. Nat Biotechnol. 2006 Aug;24(8):914–916. doi: 10.1038/nbt0806-914. [DOI] [PubMed] [Google Scholar]
- 44.Camp RL, Neumeister V, Rimm DL. A decade of tissue microarrays: progress in the discovery and validation of cancer biomarkers. J Clin Oncol. 2008 Dec 1;26(34):5630–5637. doi: 10.1200/JCO.2008.17.3567. [DOI] [PubMed] [Google Scholar]
- 45.Petty HR, Elner VM, Kawaji T, Clark A, Thompson D, Yang DL. A facile method for immunofluorescence microscopy of highly autofluorescent human retinal sections using nanoparticles with large Stokes shifts. J Neurosci Methods. 2010 Aug 30;191(2):222–226. doi: 10.1016/j.jneumeth.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kollias N, Baqer AH. Absorption mechanisms of human melanin in the visible, 400-720 nm. J Invest Dermatol. 1987 Oct;89(4):384–388. doi: 10.1111/1523-1747.ep12471764. [DOI] [PubMed] [Google Scholar]
- 47.Huang Z, Zeng H, Hamzavi I, et al. Cutaneous melanin exhibiting fluorescence emission under near-infrared light excitation. J Biomed Opt. 2006 May-Jun;11(3):34010. doi: 10.1117/1.2204007. [DOI] [PubMed] [Google Scholar]
- 48.Nighswander-Rempel SP, Riesz J, Gilmore J, Bothma JP, Meredith P. Quantitative fluorescence excitation spectra of synthetic eumelanin. J Phys Chem B. 2005 Nov 3;109(43):20629–20635. doi: 10.1021/jp053704+. [DOI] [PubMed] [Google Scholar]
- 49.Tolles J, Bai Y, Baquero M, Harris LN, Rimm DL, Molinaro AM. Optimal tumor sampling for immunostaining of biomarkers in breast carcinoma. Breast Cancer Res. 2011;13(3):R51. doi: 10.1186/bcr2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pacifico MD, Grover R, Richman PI, Buffa F, Daley FM, Wilson GD. nm23 as a prognostic marker in primary cutaneous melanoma: evaluation using tissue microarray in a patient group with long-term follow-up. Melanoma Res. 2005 Oct;15(5):435–440. doi: 10.1097/00008390-200510000-00012. [DOI] [PubMed] [Google Scholar]
- 51.Pacifico MD, Grover R, Richman PI, Daley FM, Buffa F, Wilson GD. Development of a tissue array for primary melanoma with long-term follow-up: discovering melanoma cell adhesion molecule as an important prognostic marker. Plast Reconstr Surg. 2005 Feb;115(2):367–375. doi: 10.1097/01.prs.0000148417.86768.c9. [DOI] [PubMed] [Google Scholar]
- 52.Pacifico MD, Grover R, Richman PI, Daley FM, Buffa F, Wilson GD. CD44v3 levels in primary cutaneous melanoma are predictive of prognosis: assessment by the use of tissue microarray. Int J Cancer. 2006 Mar 15;118(6):1460–1464. doi: 10.1002/ijc.21504. [DOI] [PubMed] [Google Scholar]
- 53.Pacifico MD, Grover R, Richman P, Daley F, Wilson GD. Validation of tissue microarray for the immunohistochemical profiling of melanoma. Melanoma Res. 2004 Feb;14(1):39–42. doi: 10.1097/00008390-200402000-00006. [DOI] [PubMed] [Google Scholar]