Abstract
Four independent regions within 8q24 near the MYC gene are associated with risk for prostate cancer. Here we investigated allelic imbalance at 8q24 risk variants and MYC gene DNA copy number (CN) in 27 primary prostate cancers. Heterozygotes were observed in 24 of 27 patients at one or more 8q24 markers and 27% of the loci exhibited AI in tumor DNA. 8q24 risk alleles were preferentially favored in the tumors. Increased MYC gene CN was observed in 33% of tumors, and the co-existence of increased MYC gene CN with AI at risk loci was observed in 86% (P<0.004 exact binomial test) of the informative tumors. No AI was observed in tumors which did not reveal increased MYC gene CN. Higher Gleason score was associated with tumors exhibiting AI (P=0.04), and also with increased MYC gene CN (P=0.02). Our results suggest that AI at 8q24 and increased MYC gene CN may both be related to high Gleason score in prostate cancer. Our findings also suggest that these two somatic alterations may be due to the same preferential chromosomal duplication event during prostate tumorigenesis.
Keywords: 8q24, allelic imbalance, MYC gene, DNA copy number, prostate cancer
Background
At least four independent susceptibility loci for prostate cancer risk have been identified on chromosome 8q24.21 among diverse ethnic populations [1–6]. Three of these regions are contiguous and independent, and are denoted as region 1 (Build 36, 128.54–128.62 Mb), region 2 (128.14–128.28 Mb), and region 3 (128.47–128.54 Mb). Common single nucleotide polymorphisms (SNPs) within these regions that have been consistently replicated in prostate cancer (Pca) risk are rs1447295 (region 1), rs16901979 (region 2) and rs6983267 (region 3), respectively [1–6]. The three risk-associated regions cluster within a 600-kb sequence that contains no known genes. In a previous case-control study of African Americans, we also confirmed these findings on 8q24.21 and further identified a novel independent locus (denoted as region 4) around SNP rs7008482 located on 8q24.13 [6], which lies within the NMSCE2 gene.
The association of 8q24 SNPs with prostate cancer risk has also been confirmed by many other follow-up studies [7–9]. Interestingly, SNPs within 8q24 are not only associated with prostate cancer risk, but also with the risk of colorectal, breast, bladder and kidney cancer [9–14]. The consistent finding of association between 8q24 SNPs and elevated risk of human cancers suggests that a common biological mechanism may contribute to development of various human cancers. However, our efforts to elucidate the molecular mechanisms that contribute to carcinogenesis have been slow since there are no known genes in the region. The closest and strongest candidate gene is the MYC oncogene which is approximately 200 kb from risk region 1. The established role of the MYC oncogene in various human cancers makes it the most promising candidate susceptibility gene in the 8q24 region. Recently, SNP rs9642880, 30 kb upstream of MYC was shown to be strongly associated with bladder cancer risk [12]. However, genetic variation in the MYC gene has been experimentally excluded as risk factors for prostate or colon cancer risk [1–4, 10–11].
It is unclear if the underlying 8q24 risk alleles interact or influence the MYC gene during prostatic carcinogenesis, yet it is known that MYC gene amplification and overexpression has been associated with human prostate cancer development [15–20]. Frequent MYC amplification has been reported in high-grade prostatic intraepithelial neoplasia (HGPIN), primary prostate cancer and metastases [15–20], and both MYC mRNA transcripts and nuclear MYC protein are elevated in some prostate cancers [20]. Furthermore, nuclear MYC protein is commonly found to be overexpressed in luminal cells of prostatic intraepithelial neoplasia, most primary tumors, and metastatic disease. However, MYC protein expression is not correlated with 8q24 amplification, suggesting alternative mechanisms for MYC overexpression [20].
Recently, allelic imbalance (AI) of the G allele of the 8q24 SNP, rs6983267 was reported in colorectal tumors, suggesting that germline genetic defect at this locus may play a role in the somatic evolution of human colorectal cancer [14]. However, similar findings have not been reported in prostate cancer. In this study, we investigated the relationship between MYC gene amplification and AI of 8q24 risk alleles and Pca clinical features. AI status was examined at SNPs in the four independent 8q24 regions associated with Pca risk in primary prostate tumor tissues and paired normal tissues collected from African American and European American patients. In addition, we also measured MYC gene DNA copy number (CN) and AI status within the MYC gene coding region.
Methods
Study subjects and DNA extraction
The study population originally consisted of 18 African American (AA) patients and 13 European American (EA) patients with prostate cancer. However, 3 AA samples and 1 EA sample were excluded in this study due to the lack of matched normal tissue samples. Surgical specimens collected at the Johns Hopkins Hospital between 1993 and 2007 were selected for processing due to relative homogeneity of nodules of both tumor tissue and benign tissue in the specimen. The tissue, frozen at the time of surgery, was sectioned on a cryostat and microscopically trimmed to isolate benign tissue from tumor tissue. Confirmatory Hematoxylin and Eosin (H&E) sections as well as slides for immunohistochemical staining were taken both prior to and after the sections that would be kept for processing. Genomic DNA was extracted from the frozen tissue sections using Trizol reagent. Processing of the samples was performed in a fashion blinded to ethnic origin and malignant composition.
Analysis of Sample Composition
Pathologic review was performed for each representative section of tumor and normal tissue before and after trimming of the surgical specimen was performed. Where H&E staining alone was insufficient to properly diagnose a set of sections, immunohistochemial analysis was undertaken. An estimate of the proportion of neoplastic cells was made for each benign and tumor sample. Each tumor sample was then assigned a Gleason grade. For the tumor tissue, 69.4% of the material processed contained over 70% prostate cancer cells with a Gleason score of 3+3 or greater. AI and MYC gene CN analysis were not conducted in tumor samples containing less than 50% neoplastic cells on final pathologic detection in this study. Ethnicity and clinical characteristics including age at diagnosis, Gleason score and tumor stages are listed in Table 1.
Table 1.
8q24 SNP genotypes, allelic imbalance and MYC gene copy number in 15 African American (AA) and 12 European American (EA) primary prostate tumor tissues.
| Case No. | Race | Clinical Features
|
8q24 SNP genotypes
|
MYC CN | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Age | Gleason Score | Stage | rs7008482 G/T | rs16901979 A/C | rs6983267 G/T | rs7014346 A/G | rs1447295 A/C | |||
| 21 | EA | 63 | 9 | T3BN0MX | ■ | C | T | G | ■ | 3.93 |
| 18 | EA | 55 | 7 | T2N0MX |

|
C | G | A | ■ | 3.47 |
| 12 | AA | 59 | 7 | T3BN0MX | G | A | G | ■ | ■ | 2.81 |
| 16 | EA | 70 | 7 | T3AN0MX | T | C | ■ | G | C | 2.48 |
| 13 | AA | 61 | 7 | T3AN0MX | G | C | □ | G | C | 2.46 |
| 5 | AA | 55 | 6 | T2N0MX | G | C | ■ | G | □ | 2.43 |
| 9 | AA | 65 | 9 | T3AN0MX | G | A | G | G | C | 2.24 |
| 1 | AA | 62 | 6 | T2N0MX | G |

|
G |

|
A | 2.2 |
| 6 | AA | 51 | 6 | T3BN0MX | G |

|
G | G |

|
2.2 |
| 3 | AA | 58 | 6 | T3AN0MX | G | □ | G | G | A | 2.18 |
| 24 | EA | 59 | 6 | T3AN0MX | □ | C | T | G | C | 2.11 |
| 26 | EA | 55 | 6 | T2N0MX | T | C | □ | □ | □ | 2.09 |
| 20 | EA | 61 | 6 | T2N0MX | □ | □ | □ | □ | C | 2.07 |
| 19 | EA | 55 | 6 | T2N0MX | □ | C | G | A | □ | 2.05 |
| 25 | EA | 65 | 7 | T3AN0MX | G | C | □ | □ | C | 2.04 |
| 15 | AA | 61 | 7 | T3AN0MX | □ | A | G | □ | □ | 1.93 |
| 17 | EA | 57 | 6 | T2N0MX | T | C | T | G | C | 1.92 |
| 14 | AA | 59 | 7 | T3AN0MX | G | C | G | A | C | 1.91 |
| 23 | EA | 43 | 6 | T2N0MX | □ | C | T | G | C | 1.91 |
| 27 | EA | 55 | 7 | T3BN0MX | G | C | G | □ | C | 1.89 |
| 22 | EA | 68 | 6 | T3AN0MX | □ | C | T | G | □ | 1.84 |
| 10 | AA | 59 | 7 | T3AN0MX | □ | C | □ | □ | C | 1.82 |
| 2 | AA | 56 | 6 | T2N0MX | G | □ | G | □ | □ | 1.77 |
| 11 | AA | 46 | 7 | T3AN0MX | □ | □ | G | □ | C | 1.76 |
| 8 | AA | 66 | 7 | T3AN0MX | G | A | G | □ | □ | 1.68 |
| 7 | AA | 48 | 7 | T3AN1MX | G | A | G | □ | □ | 1.63 |
| 4 | AA | 57 | 7 | TXN0MX | G |

|
G | G | □ | 1.34 |
□ Heterozygous without AI. ■ Heterozygous with high AI.
 Heterozygous with moderate AI.
Heterozygous with moderate AI.
SNP genotyping and AI analysis
Genotyping and AI assessment were carried out by PCR and direct DNA sequencing for 5 SNP markers at 4 independent risk-associated regions on 8q24 (Table 1). Primers used for PCR amplification and DNA sequencing were synthesized by IDT (Integrated DNA Technologies, Coralville, IA). All the sequences of primers are available upon request. The PCR mixture in a total volume of 20 μl consisted of 20 ng genomic DNA, 1 U AmpliTaq polymerase (Perkin Elmer, Foster City, CA), 10 × PCR buffer II (Perkin Elmer), 2.0 mM MgCl2, 0.6 mM each dNTP, and 12 pmol of each primer. Reaction conditions included an initial melting step at 95°C for 15 min, followed by 35 cycles of melting at 95°C for 30 s, annealing at 60°C for 30 s, and extending at 72°C for 30 s. A final extension was set at 72°C for 8 min. For DNA sequencing, PCR products were treated by using ExoSAP-IT (USB Co., Cleveland, OH) according to the user protocol, and then sequenced by using ABI 3730XL DNA sequencers (Applied Biosystems, Foster City, CA) at the Cancer Research Center DNA Sequencing Core Facility, the University of Chicago.
AI can only be identified in “informative” cases which carry a heterozygous genotype at a given SNP locus. AI of the 5 markers was scored by comparing the ratios of the allele peak heights in sequencing graphs between tumors and matched normal DNA samples as described previously [21]. The cutoff scores which denoted “high” AI were >1.67 or <0.60 [21], and the ratio scores that denote “moderate” AI were >1.25 or <0.80.
Quantitative PCR
Real-time PCR with the Mx3000P system (Stratagene, La Jolla, CA) and iQTMSYBR Green Supermix (Bio-Rad Laboratories) was used to quantify the number of copies of the MYC gene according to a similar protocol previously described [22]. Briefly, a 157-bp amplicon in the exon 3 of MYC gene was amplified with primers (forward: 5′-TGGATCACCTTCTGCTGGA-3′ and reverse: 5′-TCTGACACTGTCCAACTTGACC-3′). A 149bp amplicon in the LINE1 sequence was served as an internal control [23]. Real-time PCR for both MYC and LINE 1 were cycled 45 times at 95°C for 20s, 55°C for 1 min and 72°C for 30s after preheating at 95°C for 10 min. Measurement of both amplicons was repeated three times. MYC data was normalized to that of the LINE1 sequence (R1). The MYC/LINE1 ratio in the tumor DNA was then normalized to that of the normal tissue (R2), and the MYC gene copy number (CN) in each tumor sample was calculated by doubling the R2 value. In this study, the MYC gene was considered duplicated in tumor tissue when the copy number was ≥ 2.20.
MYC gene mutation screening
The coding region of the MYC gene was screened by using DNA sequencing in five selected prostate cancer tumor samples. Primers used in PCR reaction can be available under request. PCR conditions and DNA sequencing were the same as above.
Statistical Analysis
ANOVA analyses (F test) were used to detect associations between AI status and clinical features. The exact binomial test was performed to determine association between AI status and MYC CN. All analyses were two-sided. Analyses were performed for all subjects combined and separately for EA and AA ancestry.
Results
8q24 allelic imbalance analyses
Table 1 shows the genotypes and AI status of the five 8q24 SNP markers in our 27 AA and EA prostate tumor/normal DNA pairs. Forty-nine (49) heterozygous genotypes were identified and 13 of them (27%) exhibited allelic imbalance, including 7 high and 6 moderate (Table 1). Three subjects (cases 9, 14 and 17) were homozygous at all the 5 markers and could not be analyzed for AI.
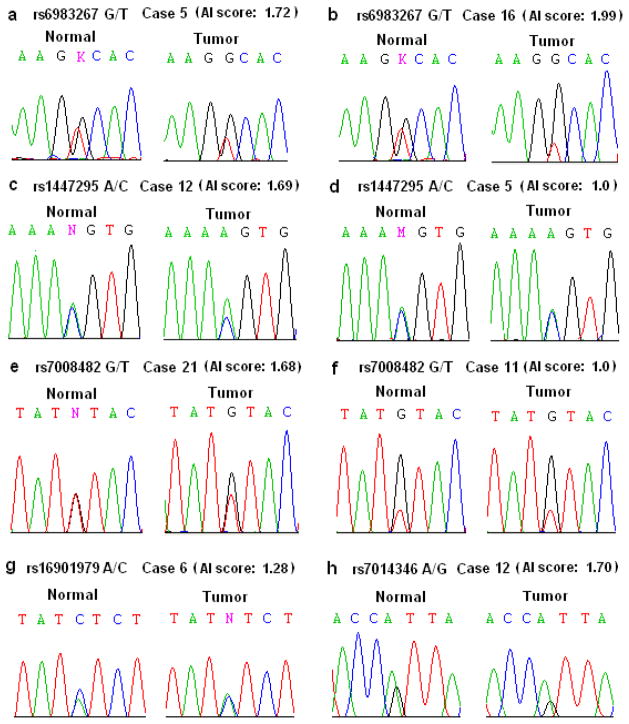
For the region 3 SNP rs6983267, a total of 7 heterozygous cases (3 AA and 4 EA patients) were observed, and two patients (the AA case 5 and the EA case 16) exhibited high AI at rs6983267 with AI scores of 1.72 and 1.99, respectively (Table 1, Figure 1a and 1b). It should be noted that in both of these two tumor tissues the rs6983267 risk allele ‘G’ was favored (Figure 1a and 1b). Because of the low heterozygosity of rs6983267 in the African descent population, we typed another region 3 SNP, rs7014346, which is about 11 kb telomic to rs6983267 and 60 kb away to rs1447295 (region 1). SNP rs7014346 which is in the strong linkage disequilibrium with rs6983267, exhibits higher heterozygosity among both the European and African populations and is associated with colorectal cancer risk (14). We observed 12 heterozygous patients for the rs7014346 A/G polymorphism (Table 1). One AA patient (case 12) showed high allelic imbalance at rs7014346 (AI score 1.70), and another AA patient (case 1) had moderate AI at rs7014346 with the risk allele A favored in tumor cells (Figure 1h).
Figure 1.
Allelic imbalance examples of heterozygous prostate cancer cases at the five typed 8q24 SNP markers. (a to h) Al scores obtained from sequence graphs of normal and tumor samples were shown at each 8q24 SNP locus for selected prostate cancer patients as indicated.
High allelic imbalance was also seen at the other three 8q24 regions. At rs1447295 in region 1, the closest risk-associated region to the MYC gene, 30.7% (4/13) of heterozygotes were observed with allelic imbalance (3 significant and 1 moderate) (Table 1). Notably, the risk allele A of rs1447295 was favored in all 4 (100%) of tumor samples with allelic imbalance. At rs16901979 in region 2, moderate allelic imbalance was observed in 3 of 6 African American heterozygous patients (case 1, 4 and 6) (Table 1 and Figure 1g). Interestingly, in case 6 the rs16901979 risk allele “A” was showed favored, but in the other two (case 1 and 4) the wild type allele “C” was favored.
The risk-associated SNP rs7008482, 2 Mb away from the MYC gene in region 4 is located in the intronic region of NSMCE2. The NSMCE2 gene (also named as hMMS21, NSE2) encodes a SUMO ligase which functions in DNA repair in human cells [24]. The risk allele G of rs7008482 is very common among European and African descendents. For SNP rs7008482, an EA patient (case 21) showed high allelic imbalance (AI score 1.68) (Table 1 and Figure 1e) while another EA patient (case 18) showed moderate AI. Of note, the risk allele rs7008482 G was found to be favored in both cases (100%).
In most informative patients allelic imbalance could be observed consistently at these informative SNP markers (Table 1). For example, in patient 21 high AI was observed at both rs7008482 and rs1447295, and for patient 12, AI was shown to be high for rs7014346 and rs1447295 as well. In AA case 5, however, significant AI was only shown at the heterozygous rs6983267 (Figure 1a) but not at SNP rs1447295 (Figure 1d).
MYC gene copy number analysis
Among these 27 tumor/normal DNAs, increased MYC gene CN (CN≥2.20) was found in 9 samples (6 AA patients and 3 EA patients) by using real-time PCR (Table 1). MYC gene CN alteration was observed in 40% (6/15) of AA tumors and in 25% (3/12) of EA tumors. Patient 21 showed the highest copy number of the MYC gene with a CN ratio at 3.93. Table 1 also reveals another AA patient (case 3) with slightly increased MYC gene CN with a CN score of 2.18. Of note, the MYC gene CN was decreased in the tumor sample from patient 4 (CN=1.34) when compared to the matched normal tissue, suggesting loss of the 8q24 region.
Correlation between 8q24 SNP AI, MYC gene copy number, and disease aggressiveness
The frequency of AI at the 8q24 markers was slightly higher among heterozygous loci in AAs (8/28 or 27%) than in EAs (5/21 or 24%). Among informative samples, tumor Gleason scores were higher in patients with high AI at two or more loci than in patients without AI (P=0.04). Additionally, increased MYC CN was associated with higher Gleason scores (P=0.02). No associations were observed for AI or MYC gene CN and tumor stage, race, or age at diagnosis.
The presence of both AI and increased MYC CN was observed in 88% of informative tumor/normal DNA pairs. MYC CN alteration was strongly associated with AI (P=0.0001). No allelic imbalance was observed in informative tumor/normal pairs without MYC gene copy number alteration.
Mutation detection in the MYC gene
The coding region of MYC gene was screened by DNA sequencing in 5 selected DNAs (case 5, 12, 16, 18, and 21) which showed significant AI and increased MYC gene copy number. No somatic mutations were identified in the MYC gene coding region in these 5 prostate tumors. However, two known SNPs, rs4645959 A/G and rs4645970 A/G, were observed in patient 16 and patient 12, respectively, and they both displayed high AI (Fig 2a and 2b). The SNP rs4645959, a missense variant (N26S) in MYC exon 2 with the S allele creating a PDK1 binding motif, has been shown correlation with familial breast cancer risk [25]. However, it was the wild type rs4645959 “A” allele that was overrepresented in the prostate tumor in this study (Figure 2b).
Figure 2.
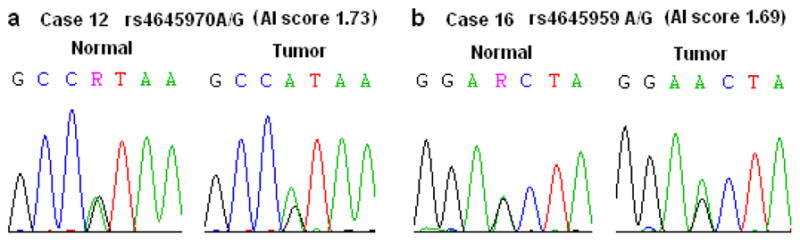
Allelic imbalance at two SNP markers within Myc gene. (a) AI ratio 1.73 at rs4645970 A/G within Myc exon-3 in patient 12 and (b) AI score 1.69 at rs4645959 A/G within MYC exon-2 in patients 16.
Discussion
In this study we observed that high allelic imbalance of 8q24 risk alleles was common (27%) in primary prostate cancers. Notably, the risk alleles of rs1447295 “A” (region 1), rs6983267 “G” (region 3) and rs70008482 “G” (region 4) are preferentially amplified in all prostate cancer samples when presenting with AI. This finding suggested that these selectively favored risk alleles in prostate tumor cells could play an oncogenic role during prostate carcinogenesis. We also found that increased MYC gene copy number is commonly accompanied with 8q24 allelic amplification in prostate tumor cells, indicating that the co-existence of 8q24 allelic imbalance and gain of MYC gene copy number might be due to the same event of 8q24 chromosomal amplification. Furthermore, we showed that both 8q24 allelic imbalance and MYC gene amplification may be associated with high Gleason score (>7) and possibly aggressive Pca. Similarly, previous studies using CGH assay and/or a dual-color labeling probe of MYC gene have shown that MYC gene copies tend to only slightly increase, mostly under 5 or less copies [17–19]. Our study further revealed that the increased MYC gene CN is likely due to preferential amplification of a large 8q24 chromosomal fragment in prostate cancer. Similar to our findings in prostate cancer, a recent 8q24 copy number analysis in colorectal cancers observed that a large segment of the rs6983267 G allele-bearing chromosome region is typically overrepresented 3–5 times in colorectal tumors [26].
Previous studies have shown that germline variation of the JAK2 oncogene predispose to increased risk of specific somatic mutations, suggesting that strong interactions may exist between somatic and germline changes within oncogenes [27]. Indeed we observed frequent MYC gene amplification and allelic imbalance in prostate cancers, suggesting that somatic MYC gene amplification in prostate cancer might not be equally contributed from the two parent chromosomes. To test whether MYC is involved in prostate cancer through the same mechanism as JAK2, we sequenced MYC coding region. However we could not detect any somatic mutations, indicating that MYC may be rarely mutated in prostate cancer, and the interaction of germline and somatic DNA changes might not exist in the MYC oncogenes involvement in prostate cancer. Given the small sample size we can not exclude the possibility that this mechanism may exist.
Our study is the first report on common allelic imbalance of 8q24 risk-associated markers in prostate cancers. However we acknowledge that this study has several important limitations. The sample size of prostate tumors we analyzed in this study is very small. Even though a differences in AI frequency and MYC DNA copy number alteration was observed between AA and EA patients, we may not have estimated the exact rates of AI and MYC gene CN alteration, nor the significance of our findings in the AA or EA populations when genetic background is concerned. Furthermore, due to the level of homozygosity for the five 8q24 markers, three patients were “uninformative” for AI assessment in this study, including one of our cases with the highest Gleason score. Therefore future studies should be conducted using more informative markers in a larger number of prostate tumor samples to access allelic imbalance at 8q24 risk-associated regions.
Conclusion
In this study we reported common and preferential allelic imbalance at 8q24 risk-associated regions and frequent MYC amplification in primary prostate tumors. Both 8q24 allelic imbalance and MYC amplification occur simultaneously and appear to be related to aggressive prostate cancers, however larger numbers of samples are needed to confirm our findings. These findings will help extend our understanding of the relevance of common genetic variation in the 8q24 region and MYC gene amplification in the etiology and pathology of prostate cancer.
Acknowledgments
The authors would like to thank all the men who volunteered to participate in this genetic study. This research was funded in part by the National Institutes of Health (S06GM08016) and the Department of Defense (DAMD W81XWH-07-1-0203 and DAMD W81XWH-06-1-0066).
Footnotes
Conflict of interest
The authors declare that they have no competing interests.
References
- 1.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 2.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 3.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 4.Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Witte JS. Multiple prostate cancer risk variants on 8q24. Nat Genet. 2007;39:579–580. doi: 10.1038/ng0507-579. [DOI] [PubMed] [Google Scholar]
- 6.Robbins C, Torres JB, Hooker S, Bonilla C, Hernandez W, et al. Confirmation study of prostate cancer risk variants at 8q24 in African Americans identifies a novel risk locus. Genome Res. 2007;17:1717–1722. doi: 10.1101/gr.6782707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, McDonnell SK, Slusser JP, Hebbring SJ, Cunningham JM, Jacobsen SJ, et al. Two common chromosome 8q24 variants are associated with increased risk for prostate cancer. Cancer Res. 2007;67:2944–2950. doi: 10.1158/0008-5472.CAN-06-3186. [DOI] [PubMed] [Google Scholar]
- 8.Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 9.Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24. 21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 11.Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 12.Kiemeney LA, Thorlacius S, Sulem P, Geller F, Aben KK, Stacey SN, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kupfer SS, Torres JB, Hooker S, Anderson JR, Skol AD, Ellis NA, et al. Novel single nucleotide polymorphism associations with colorectal cancer on chromosome 8q24 in African and European Americans. Carcinogenesis. 2009;30:1353–1357. doi: 10.1093/carcin/bgp123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuupanen S, Niittymaki I, Nousiainen K, Vanharanta S, Mecklin JP, Nuorva K, et al. Allelic imbalance at rs6983267 suggests selection of the risk allele in somatic colorectal tumor evolution. Cancer Res. 2008;68:14–17. doi: 10.1158/0008-5472.CAN-07-5766. [DOI] [PubMed] [Google Scholar]
- 15.Qian J, Jenkins RB, Bostwick DG. Detection of chromosomal anomalies and c-myc gene amplification in the cribriform pattern of prostatic intraepithelial neoplasia and carcinoma by fluorescence in situ hybridization. Mod Pathol. 1997;10:1113–1119. [PubMed] [Google Scholar]
- 16.Jenkins RB, Qian J, Lieber MM, Bostwick DG. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res. 1997;57:524–531. [PubMed] [Google Scholar]
- 17.Liu W, Xie CC, Zhu Y, Li T, Sun J, Cheng Y, et al. Homozygous deletions and recurrent amplifications implicate new genes involved in prostate cancer. Neoplasia. 2008;10:897–907. doi: 10.1593/neo.08428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lapointe J, Li C, Giacomini CP, Salari K, Huang S, Wang P, et al. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67:8504–8510. doi: 10.1158/0008-5472.CAN-07-0673. [DOI] [PubMed] [Google Scholar]
- 19.Ribeiro FR, Henrique R, Martins AT, Jeronimo C, Teixeira MR. Relative copy number gain of MYC in diagnostic needle biopsies is an independent prognostic factor for prostate cancer patients. Eur Urol. 2007;52:116–125. doi: 10.1016/j.eururo.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 20.Gurel B, Iwata T, Koh CM, Jenkins RB, Lan F, Van Dang C, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21:1156–1167. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canzian F, Salovaara R, Hemminki A, Kristo P, Chadwick RB, Aaltonen LA, et al. Semiautomated assessment of loss of heterozygosity and replication error in tumors. Cancer Res. 1996;56:3331–3337. [PubMed] [Google Scholar]
- 22.Liu W, Wu X, Zhang W, Montenegro RC, Fackenthal DL, Spitz JA, et al. Relationship of EGFR mutations, expression, amplification, and polymorphisms to epidermal growth factor receptor inhibitors in the NCI60 cell lines. Clin Cancer Res. 2007;13:6788–6795. doi: 10.1158/1078-0432.CCR-07-0547. [DOI] [PubMed] [Google Scholar]
- 23.Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 24.Potts PR, Yu H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol. 2005;25:7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wirtenberger M, Hemminki K, Forsti A, Klaes R, Schmutzler RK, Grzybowska E, et al. c-MYC Asn11Ser is associated with increased risk for familial breast cancer. Int J Cancer. 2005;117:638–642. doi: 10.1002/ijc.21225. [DOI] [PubMed] [Google Scholar]
- 26.Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 27.Kilpivaara O, Mukherjee S, Schram AM, Wadleigh M, Mullally A, Ebert BL, et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat Genet. 2009;41:455–459. doi: 10.1038/ng.342. [DOI] [PMC free article] [PubMed] [Google Scholar]