Summary
Proper microtubule polarity underlies overall neuronal polarity, but mechanisms for maintaining microtubule polarity are not well understood. Previous live imaging in Drosophila dendritic arborization (da) neurons showed that, while microtubules are uniformly plus-end out in axons, dendrites possess uniformly minus-end-out microtubules [1]. Thus, maintaining uniform microtubule polarity in dendrites requires that growing microtubule plus-ends entering branch points must be actively directed towards the cell body. A model was proposed in which EB1 tracks the plus-ends of microtubules growing into a branches and an associated kinesin-2 motor walks along a static microtubule to steer the plus-end toward the cell body. However, the fast plus-end binding dynamics of EB1 [2–5] appear at odds with this proposed mechanical function. To test this model in vitro, we reconstituted the system by artificially dimerizing EB1 to kinesin, growing microtubules from immobilized seeds, and imaging encounters between growing microtubule plus-ends and static microtubules. Consistent with in vivo observations, the EB1-kinesin complex actively steered growing microtubules. Thus EB1 kinetics and mechanics are sufficient to bend microtubules for several seconds. Other kinesins also demonstrated this activity, suggesting this is a general mechanism for organizing and maintaining proper microtubule polarity in cells.
Results and Discussion
Reconstructing +TIP-kinesin complex in vitro through chemically induced heterodimerization
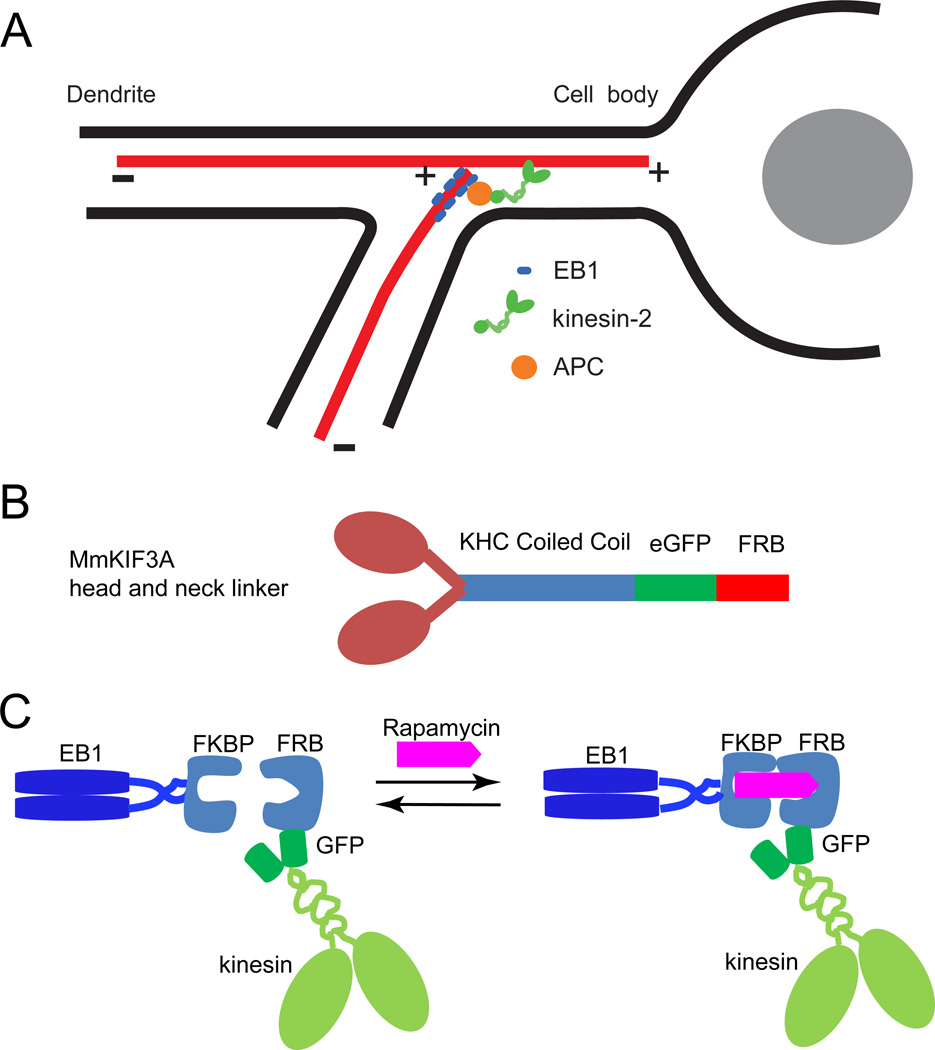
Based on previous work [1], it was hypothesized that the microtubule +-Tip Interacting Protein (+TIP) EB1 recruits the molecular motor kinesin-2 via the scaffolding protein Adenomatous polyposis coli (APC) to form a +TIP-kinesin complex at growing microtubule plus-ends. Microtubules growing into branch points are bent toward the plus-ends of stable microtubules at the junction by the motor activity of kinesin-2 (Figure 1A). To reconstruct the +TIP-kinesin complex in vitro, we linked kin2, a M. musculus kinesin-2 construct having similar motor properties to KIF3A/B heterodimer [6, 7], to human EB1 (Figure 1, B and C). EB1 and kin2 were fused at their C-termini to FKBP and FRB, respectively, which form a tight (KD ~ 12 nM) ternary complex in the presence of rapamycin [8, 9].
Figure 1.
In vitro reconstruction of EB1-kinesin complex. (A) Proposed model, based on live imaging, RNAi knockdowns and yeast two-hybrid screens, for maintaining uniform minus-end-out microtubule polarity in Drosophila dendrites. EB1 recruits kinesin-2 via APC to the plus-ends of microtubules growing into branch points, and kinesin-2 walks on existing microtubules to guide the growing microtubule towards the cell body. (B) Design of kin2 construct. The motor domain and neck linker of MmKIF3A were fused to the neck-coil and rod of Drosophila KHC [6, 7]. eGFP and the FRB tag were fused to the C-terminal, followed by a His6 tag. (C) Strategy for linking EB1, fused to FKBP at its C-terminal (EB1FKBP), to FRB-tagged kinesin (kinFRB) through rapamycin.
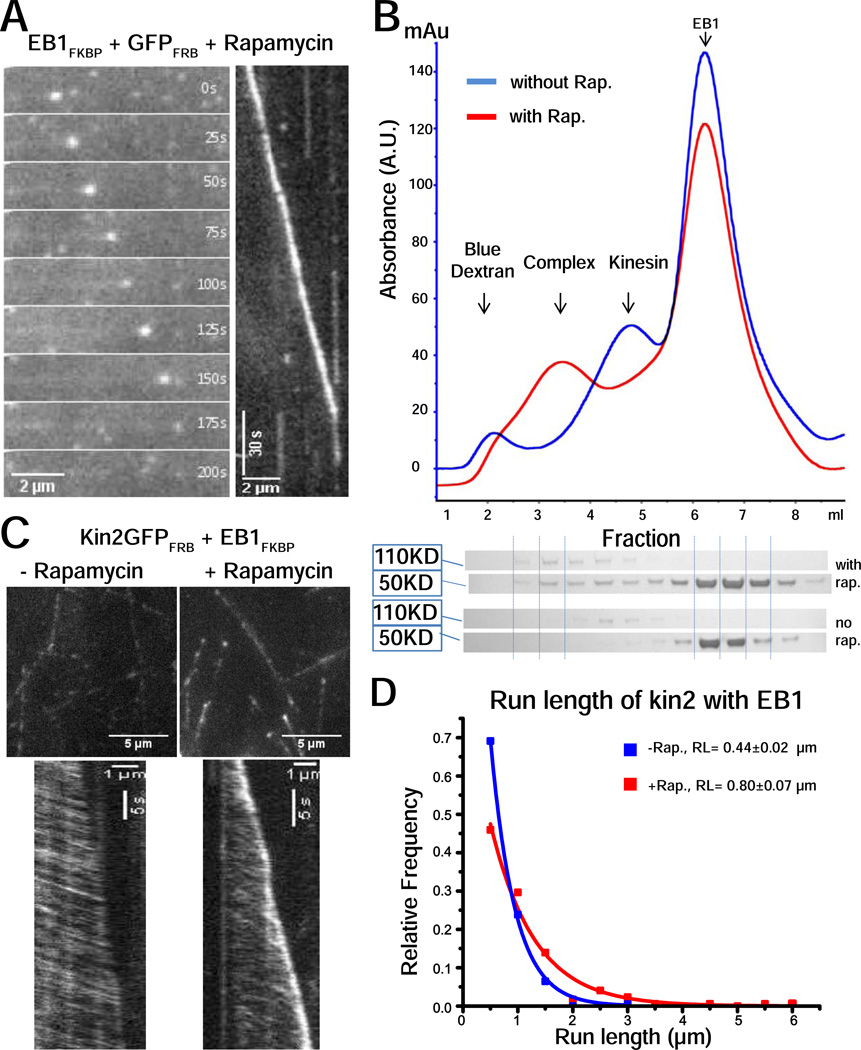
To confirm that the fusion tags did not alter EB1 or kin2 functions, we assessed their activities in TIRF-based functional assays. GFP-tagged kin2FRB moved processively along microtubules and EB1FKBP linked to GFPFRB through rapamycin clearly accumulated at growing microtubule plus-ends (Figure 2A and C). Because both kin2 and EB1 are dimers with each subunit containing a FKB or FRBP binding domain, addition of rapamycin could potentially generate a range of species beyond simple 1:1 complexes of dimers. It has been shown that linked kinesin dimers (such as kinesin-5 tetramers) can form a bridge between microtubules and slide one relative to the other [10], so we particularly wanted to avoid complexes containing multiple motors and large daisy-chained aggregates. To minimize the possibility that a single EB1 dimer could bind two kinesin dimers, we combined kin2, EB1 and rapamycin in a 1:5:5 ratio, and characterized the resulting complexes by gel filtration. In the absence of rapamycin, two clear peaks were observed, corresponding to the isolated species (Figure 2B, blue curve). Adding rapamycin reduced the peak corresponding to free EB1, completely eliminated the kin2 peak, and led to the emergence of a new single peak corresponding to the EB1-kinesin complex (Figure 2B, red curve). Gel densitometry analysis of the peak indicated a stoichiometry of 1.9 EB1 dimers per kinesin dimer, consistent with the expected 2:1 ratio. Hereafter, we refer to the kinesin-2-GFPFRB:EB1FKBP:rapamycin complex as the EB1-kinesin complex.
Figure 2.
(A) EB1FKBP-GFPFRB localizes to growing microtubule plus-ends. GMPCPP seeds were immobilized on silanized coverslips through biotin-neutravidin and free tubulin was added to generate dynamic microtubules. 150 nM GFPFRB was incubated with 750 nM EB1FKBP and 750 nM rapamycin, combined with 20 µM free tubulin, and introduced into the flow cell. +TIP tracking was observed by TIRF microscopy and is presented both as a montage (left) and a kymograph (right). (B) Hydrodynamic analysis of EB1-kinesin complex. 5 µM kin2GFPFRB and 25 µM EB1FKBP were incubated with (red) or without (blue) rapamycin on ice for 20 minutes before loading onto a gel filtration column. UV absorbance and Coomassie-stained SDS-PAGE gel of corresponding fractions are shown. (C) Localization of kin2GFPFRB on dynamic microtubules when incubated with EB1FKBP in the absence (left) and presence (right) of rapamycin. Upper panels show static views and lower panels show kymographs. (D) Run length of kin2GFPFRB on taxol-stabilized microtubules when incubated with EB1FKBP in the absence (blue, n = 201) or presence (red, n = 172) of rapamycin. Data were fit to single exponentials; mean run lengths with SE of fit are shown in legend.
EB1 recruits kinesin to growing microtubule plus-ends and increases its processivity
We next introduced the EB1-kinesin complex into a flow cell containing dynamic microtubules extending from surface-immobilized GMPCPP microtubule seeds. The EB1-kinesin complex consistently walked along microtubules indicating that formation of the complex did not affect kinesin motor activity. More importantly, EB1-kinesin complex also accumulated at growing microtubule plus-ends, which was not seen in the absence of rapamycin (Figure 2C). To ask whether EB1 interacts with the microtubule during kinesin stepping, we carried out single-molecule experiments on taxol-stabilized microtubules. Linking kin2 to EB1 increased its run length from 0.44 ± 0.02 µm to 0.80 ± 0.07 µm (mean ± SE from fit) (Figure 2D and S1), suggesting that EB1 acts as a tether to enhance kinesin-microtubule interactions.
EB1-kinesin complex is sufficient to bend growing microtubules
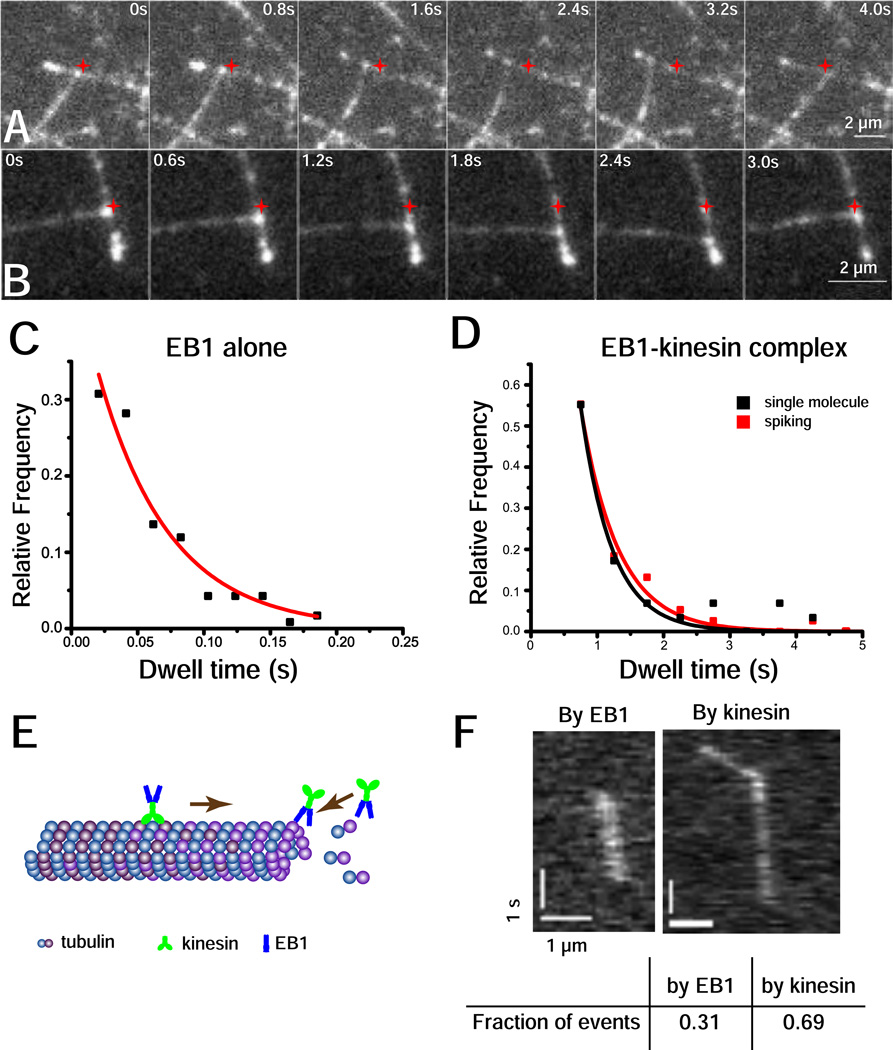
To test the ability of the EB1-kinesin complex to steer microtubules in vitro, we increased the surface density of GMPCPP seeds in our reconstitution assay to increase frequency of microtubule crossing events. If the EB1-kinesin complex is able to steer microtubule growth, then when one microtubule grows and encounters another microtubule laterally, the growing microtubule plus-end should be directed towards the plus-end of the static microtubule. Kin2GFPFRB and EB1FKBP were pre-incubated with rapamycin on ice for 20 minutes, added to the final extension solution containing 20 µM free tubulin, and the solution introduced into the flow cell. Kin2GFPFRB consistently walked along the microtubules, resulting in the entire length of the microtubules being highlighted. Microtubule plus-ends could be identified both by the direction of kinesin walking and by the accumulation of the EB1-kinesin complex at growing ends. Videos were recorded and analyzed for events in which the plus-end of a growing microtubule encounters the lattice of another microtubule. During these collision events we found that growing microtubule plus-ends, which were highlighted by the fluorescent EB1-kinesin complex, were bent and directed towards the plus-ends of the encountered microtubule (Figure 3, A and B; Movie S1 and S2). In the presence of rapamycin, 23 out of 60 encounters (38%) resulted in microtubule redirection, while in the absence of rapamycin, growing microtubule plus-ends all crossed over static microtubules without interacting (Movie S5).
Figure 3.
Microtubule steering by EB1-kin2 complex. (A) and (B): Two independent microtubule bending events are shown, imaging the GFP-labeled kinesin. The original encounter position is indicated by a red star. Kinesin, EB1 and rapamycin were incubated at ratio of 1:10:10 with 250 nM kin2GFPFRB. Montages are made from Movie S1 and S2, respectively. (C) and (D): EB1 dwell time at growing plus-ends. In (C), EB1FKBP was visualized by linking it to a streptavidin coated quantum dot (Qdot 565, Life Technologies) through biotinylated anti-his antibody (Qiagen) with 1:4:4 ratio of EB1:antibody:qdot and 3 nM of EB1 used; while in (D), EB1 was linked to kin2GFPFRB through rapamycin and visualized by GFP fluorescence at single-molecule concentrations alone (black) or spiked into 100-fold excess of unlabeled complex (red). (E): Diagram illustrating targeting of EB1-kinesin complexes to growing microtubule plus-ends either by direct EB1 binding or by kinesin walking. (F): Kymographs of EB1FKBP-kinesinGFPFRB targeting to growing microtubule plus-ends by the two mechanisms. Scale bars for both images are 1 second and 1 micron. Table shows fraction of events for each binding mode for data in D.
This result demonstrates that EB1-kinesin complexes at growing microtubule plus-ends are sufficient to direct the growth of microtubules along existing microtubules and lends strong support that this is a viable mechanism for maintaining uniform microtubule polarity in vivo. The entire bending process lasted up to several seconds and the microtubules eventually sprang back to their original relaxed position. In some cases, after the bent microtubule snapped back to its original position, the bright fluorescence at the plus-end continued to move along the static microtubule, suggesting that the point of failure was the link between EB1 and the growing microtubule plus-end and not the kinesin-microtubule link.
Linking to kinesin slows EB1 turnover at growing microtubule plus-ends
The relatively long microtubule deformations produced by the EB1-kinesin complex (bends lasting multiple seconds) appear at odds with the reported fast binding/unbinding kinetics of EB1 at growing microtubule plus ends (dwell times from 0.055 s to 0.81 s [2, 4, 11, 12]). To understand the dynamics of the system, it is important to characterize the residence time of EB1 and EB1-kinesin complexes at growing microtubule plus-ends. Using GFP fluorescence on dynamic microtubules in our assay buffer, dwell times of EB1 alone were too short for us to reliably measure. Therefore, we switched from dynamic microtubules to GTP-γ-S microtubules, which have been proposed to be faithful mimics of growing microtubule plus-ends [2], and labeled EB1 with quantum dots to increase our temporal resolution. The mean dwell time of individual EB1 dimers was 0.054 ± 0.007 s (mean ± SE of fit, n = 117, Figure 3C), corresponding to an off-rate of 18.5/s. To measure turnover rates of EB1-kinesin complexes at plus-ends, dynamic microtubules were extended from GMPCPP seeds as before, but very low concentrations (1 nM) of EB1-kinesin complex were introduced, enabling the visualization of individual complexes. An exponential fit to the data yielded a mean of 0.50 ± 0.079 s (n = 29, Figure 3D). The experiment was repeated using 2 nM labeled complex in the presence of 200 nM unlabeled complex and a similar duration of 0.57 ± 0.053 s (n = 38, Figure 3D) was found, indicating that crowding effects or cooperative interactions do not affect dwell time at the concentrations used in the microtubule bending assays. Hence, the EB1 residence time at growing microtubule ends is considerably shorter than the seconds-long observed bending durations.
The final question was: how are EB1-kinesin complexes targeted to growing microtubule plus-ends – by direct binding or by kinesin-driven transport (Figure 3E)? Targeting by kinesin-based transport was easily identified on kymographs as particles that moved rapidly along the microtubule until reaching the end and then continued at the slower microtubule growth rate (Figure 3F, right). However, events were also seen in which complexes bound directly to the growing plus end (Figure 3F, left). Interestingly, in both cases individual complexes tracked the growing plus-end, consistent with the kinesin domains generating plus-end movement and the EB1 domains maintaining plus-end association.
Microtubule steering ability is not restricted to kinesin-2
It is not known whether kinesin-2 motors have particular characteristics that make them uniquely suited for this microtubule steering function or whether this ability is common to all N-terminal kinesins. Even for kinesin-2 there is a coordination issue – microtubules polymerize at rates of several microns per minute, while kinesin-2 walks along microtubules at tens of microns per minute, suggesting that the growing microtubule would not be able to keep up with the rate of motor-induced bending.
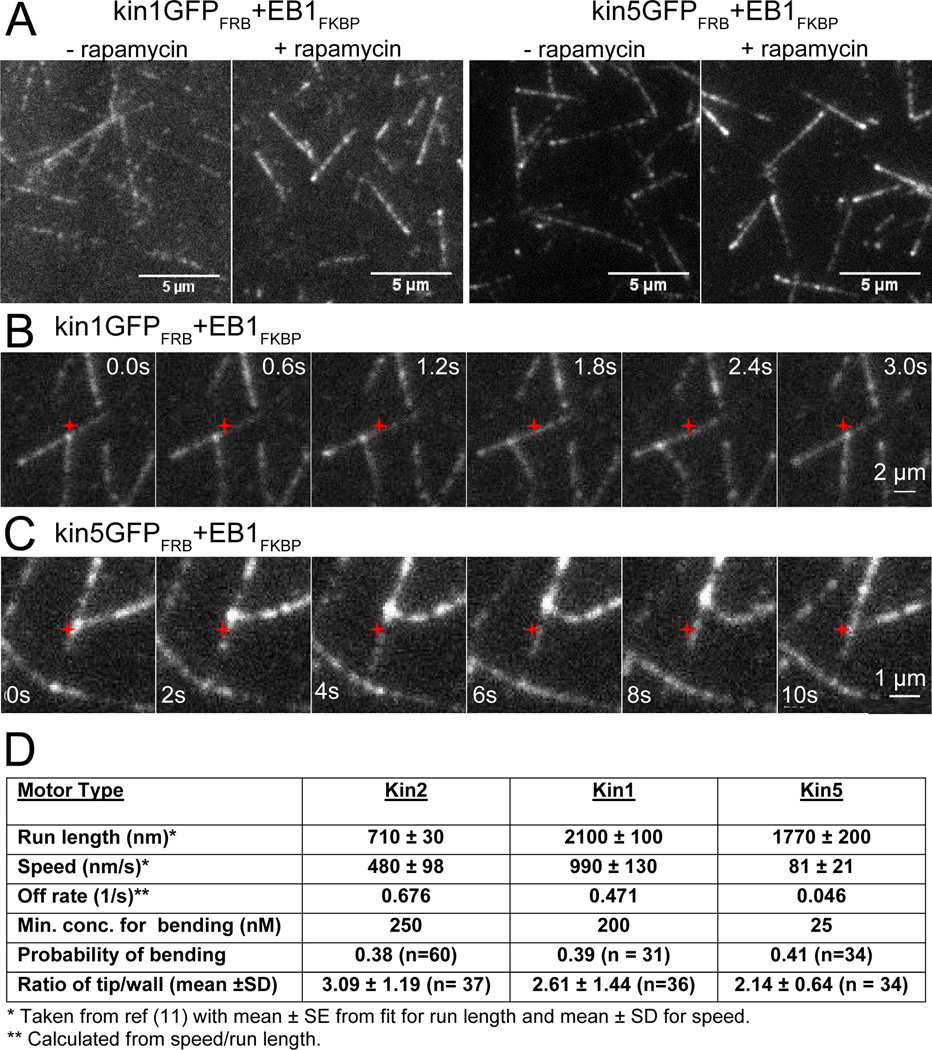
To address this question, we chose two recombinant kinesins that were characterized previously in single-molecule motility experiments – kin1, a tail-less Drosophila kinesin-1, that moves at twice the speed of kin2, and kin5, an engineered Xenopus kinesin-5 (KSP), that moves at one fifth the speed of kin2 [13]. The motors were engineered identically to kin2 (Figure 1B), and kin5 was additionally modified by shortening its neck linker such that it matched the processivity of kin1 [6, 7, 13]. Hence the two motors have nearly identical run lengths but roughly 10-fold different velocities.
Similar to kin2GFPFRB, both kin1GFPFRB and kin5GFPFRB accumulated at growing microtubule plus-ends when linked to EB1 (Figure 4A and S3). Strikingly, both EB1-kin1 and EB1-kin5 complex were able to direct microtubule growth in the same manner as kin2 (Figure 4, B and C; Movie S3 and S4). One difference between motors was the concentration of EB1-kinesin complex necessary for steering; the minimum concentration for reliable steering for kin2 was 250 nM, while kin5 required only 25 nM and kin1 was intermediate at 200 nM (Figure 4D). Interestingly, the concentration of motors required for bending scaled linearly with the microtubule off-rate (= speed ÷ run length), meaning that (assuming similar on-rates) the microtubule affinity and not the motor velocity is the principal determinant of microtubule bending. The fact that all three motors were able to steer growing microtubules indicates that this property is not unique to kinesin-2 and it could potentially be a general mechanism involving motors other than kinesin-2.
Figure 4.
Microtubule steering by kin1 and kin5-based complexes. (A) kin1GFPFRB and kin5GFPFRB accumulated at growing microtubule plus-ends only when incubated with EB1FKBP and rapamycin. Kymographs are shown in Figure S3. (B) and (C): Microtubule steering by EB1-kin1 and EB1-kin5 complex, respectively. The original encounter position is indicated by red star. Kinesin, EB1 and rapamycin were incubated at ratio of 1:10:10. 200 nM kin1GFPFRB and 25 nM kin5GFPFRB (with shortened neck linker to enhance processivity) were used. Montages were made from Movie S3 and S4, respectively. (D) Table of motor properties showing that minimum motor concentration for bending scales with motor off-rate and not velocity. Probability of bending is defined as the fraction of microtubule crossing events that resulted in the growing microtubule bending toward the plus-end of the static microtubule. Ratio of tip/wall is defined as the peak fluorescence intensity at the microtubule tip divided by the peak along the microtubule wall; see the Supplemental Data for details.
Microtubule organization in cells
By recruiting other binding partners to microtubule plus-ends, EB1 has been implicated in controlling microtubule dynamics [14, 15], bridging microtubule ends to cellular structures [16–18], and proper positioning of the mitotic spindle [19, 20]. The idea that EB1 has the ability to sustain mechanical forces at growing microtubule plus-ends has, until now, lacked direct experimental support. This question is of particular importance because a number of motor proteins capable of generating both pulling and pushing forces in microtubule networks can be targeted to growing microtubule plus-ends with the help of EB1 [15, 20–22].
Neurons are not the only polarized cells whose function requires uniformly oriented microtubule bundles or arrays. In fact, many if not most differentiated cells have cell-type-specific noncentrosomal microtubule networks [23, 24]. For instance, in epithelial cells, microtubules are aligned along the apico-basal axis with their minus-ends towards apical side and plus-ends towards the basal side [23, 24]. The molecular mechanisms that guide microtubule remodeling during epithelial differentiation and maintain proper microtubule polarity post-differentiation are still largely unknown. A recent study showed that septin binds both EB1 and microtubules, and that growing microtubule plus-ends track existing septin-coated microtubules in epithelial cells [25]. RNAi knockdown of septin leads to entangled microtubule plus-end trajectories, suggesting that septin and EB1 act together to co-align microtubules. In another study in epithelial cells, the homodimeric kinesin-2 motor KIF17 was reported to co-localize with EB1 and APC at growing microtubule plus-ends and play a role in proper epithelial polarization [21].
In addition to microtubule-microtubule interactions, there is also evidence that EB1 maintains proper microtubule organization in cells by linking growing microtubule plus-ends to actin filaments. Knockout of the microtubule-actin cross-linking factor ACF7 in keratinocytes led to a model in which EB1 and ACF7 coordinate their activities to guide growing microtubules to focal adhesions along existing actin filaments [26]. An even better analog to the EB1-APC-kinesin complex is found in yeast, where proper mitotic spindle orientation requires a myosin V motor (Myo2) bridged through the adaptor protein Kar9 to Bim1, the yeast EB1 homolog. The Bim1-Kar9-Myo2 complex localizes to microtubule plus-ends and guides microtubules along polarized actin filaments [20]. Together, these reports suggest that +TIP-motor complexes provide a general system for controlling microtubule organization in cells by directing the growth of microtubule plus-ends using existing cues. In this context, the present work demonstrating that a minimal system of just EB1 and kinesin is competent to steer microtubule growth provides vital biophysical support for these models.
Mechanical properties of EB1
The observed plus-end steering requires that EB1 proteins remain at the growing microtubule plus-end while kinesin walks along the lattice of an existing microtubule, meaning that EB1 must bear the mechanical forces generated by microtubule bending. While EB1-kinesin complexes had 500 msec plus-end dwell times, for a complex bridging two microtubules the upper limit for the duration of the interaction would more likely be defined by the 53 msec dwell time of isolated EB1 on GTP-γ-S microtubules. Depending on the motor type used, microtubules were bent for an average of between 3 and 11 seconds, or roughly 100-fold longer than the duration of a single EB1-microtubule interaction. Hence, this microtubule steering mechanism requires a pool of EB1-kinesin complexes (perhaps upwards of 100 based on the discrepancy in kinetics) that dynamically bind and unbind with kinetics much faster than the rate of microtubule bending.
While EB1 and kinesin were artificially dimerized in our in vitro assay, one question is whether EB1-APC and kinesin-APC interactions (neither of which has been characterized) are sufficiently strong or long-lived to sustain microtubule bending. As a first approximation, if their off-rates are slower than the 18 s−1 dissociation rate of EB1 from microtubules, then they should not be the weak link in the system. The fact that APC was replaceable in vitro supports the idea that APC acts as a scaffold, but because APC itself binds microtubules, it could play an important role in enhancing microtubule interactions in vivo. For instance, it may enhance the affinity of EB1 to the growing microtubule and may also act as one of perhaps many microtubule crosslinking proteins that stabilize the bent conformation. The membrane will also serve as an important mechanical barrier such that the small deflection of the growing plus-end is “locked in” by the barrier and further stabilized as the microtubule continues to grow.
In conclusion, we demonstrate that a complex of EB1 and kinesin is mechanically capable of force generation at microtubule plus-ends and these forces can be used to bend microtubules. This work expands the cellular functions of both kinesin motors and +TIPs.
Supplementary Material
Highlights.
In dendrites, microtubules growing into junctions must be steered toward cell body
EB1 and kinesin-2 have been shown to interact through APC
EB1-kinesin complex walks along microtubules and localizes to growing plus-ends
During encounters with static microtubule, EB1-kinesin actively bends filaments
Acknowledgments
The authors thank Shankar Shastry for assistance with microscope assays, David Arginteanu for protein expression and purification, Erkan Tuzel (Worcester Polytechnic Institute) for helpful discussions of molecular mechanics, and ARIAD Pharmaceuticals for the generous gift of FRB and FKBP plasmids. This work was supported by NIH R01GM076476 to W.O.H and R01GM100076 to W.O.H. and M.M.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Experimental Procedures can be found in Supplemental Data section
References
- 1.Mattie FJ, Stackpole MM, Stone MC, Clippard JR, Rudnick DA, Qiu Y, Tao J, Allender DL, Parmar M, Rolls MM. Directed microtubule growth, +TIPs, and kinesin-2 are required for uniform microtubule polarity in dendrites. Curr Biol. 2010;20:2169–2177. doi: 10.1016/j.cub.2010.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maurer SP, Bieling P, Cope J, Hoenger A, Surrey T. GTPgammaS microtubules mimic the growing microtubule end structure recognized by end-binding proteins (EBs) Proc Natl Acad Sci U S A. 2011;108:3988–3993. doi: 10.1073/pnas.1014758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieling P, Kandels-Lewis S, Telley IA, van Dijk J, Janke C, Surrey T. CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J Cell Biol. 2008;183:1223–1233. doi: 10.1083/jcb.200809190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buey RM, Mohan R, Leslie K, Walzthoeni T, Missimer JH, Menzel A, Bjelic S, Bargsten K, Grigoriev I, Smal I, et al. Insights into EB1 structure and the role of its C-terminal domain for discriminating microtubule tips from the lattice. Mol Biol Cell. 2011;22:2912–2923. doi: 10.1091/mbc.E11-01-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixit R, Barnett B, Lazarus JE, Tokito M, Goldman YE, Holzbaur EL. Microtubule plus-end tracking by CLIP-170 requires EB1. Proc Natl Acad Sci U S A. 2009;106:492–497. doi: 10.1073/pnas.0807614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shastry S, Hancock WO. Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Curr Biol. 2010;20:939–943. doi: 10.1016/j.cub.2010.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muthukrishnan G, Zhang Y, Shastry S, Hancock WO. The processivity of kinesin-2 motors suggests diminished front-head gating. Curr Biol. 2009;19:442–447. doi: 10.1016/j.cub.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 9.Banaszynski LA, Liu CW, Wandless TJ. Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc. 2005;127:4715–4721. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- 10.Kapitein LC, Peterman EJG, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- 11.Bieling P, Kandels-Lewis S, Telley IA, van Dijk J, Janke C, Surrey T. CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J Cell Biol. 2008;183:1223–1233. doi: 10.1083/jcb.200809190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixit R, Barnett B, Lazarus JE, Tokito M, Goldman YE, Holzbaur ELF. Microtubule plus-end tracking by CLIP-170 requires EB1. Proc Natl Acad Sci U S A. 2009;106:492–497. doi: 10.1073/pnas.0807614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shastry S, Hancock WO. Interhead tension determines processivity across diverse N-terminal kinesins. Proc Natl Acad Sci U S A. 2011;108:16253–16258. doi: 10.1073/pnas.1102628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tirnauer JS, Grego S, Salmon ED, Mitchison TJ. EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol Biol Cell. 2002;13:3614–3626. doi: 10.1091/mbc.E02-04-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manna T, Honnappa S, Steinmetz MO, Wilson L. Suppression of microtubule dynamic instability by the +TIP protein EB1 and its modulation by the CAP-Gly domain of p150glued. Biochemistry. 2008;47:779–786. doi: 10.1021/bi701912g. [DOI] [PubMed] [Google Scholar]
- 16.Lansbergen G, Grigoriev I, Mimori-Kiyosue Y, Ohtsuka T, Higa S, Kitajima I, Demmers J, Galjart N, Houtsmuller AB, Grosveld F, et al. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5 beta. Developmental Cell. 2006;11:21–32. doi: 10.1016/j.devcel.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 17.Kodama A, Karakesisoglou I, Wong E, Vaezi A, Fuchs E. ACF7: an essential integrator of microtubule dynamics. Cell. 2003;115:343–354. doi: 10.1016/s0092-8674(03)00813-4. [DOI] [PubMed] [Google Scholar]
- 18.Moseley JB, Bartolini F, Okada K, Wen Y, Gundersen GG, Goode BL. Regulated binding of adenomatous polyposis coli protein to actin. Journal of Biological Chemistry. 2007;282:12661–12668. doi: 10.1074/jbc.M610615200. [DOI] [PubMed] [Google Scholar]
- 19.Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- 20.Liakopoulos D, Kusch J, Grava S, Vogel J, Barral Y. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell. 2003;112:561–574. doi: 10.1016/s0092-8674(03)00119-3. [DOI] [PubMed] [Google Scholar]
- 21.Jaulin F, Kreitzer G. KIF17 stabilizes microtubules and contributes to epithelial morphogenesis by acting at MT plus ends with EB1 and APC. J Cell Biol. 2010;190:443–460. doi: 10.1083/jcb.201006044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai DW, McEwen DP, Martens JR, Meyhofer E, Verhey KJ. Single Molecule Imaging Reveals Differences in Microtubule Track Selection Between Kinesin Motors. Plos Biology. 2009;7 doi: 10.1371/journal.pbio.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luders J, Stearns T. Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol. 2007;8:161–167. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- 24.Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. Journal of Cell Science. 2006;119:4155–4163. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- 25.Bowen JR, Hwang D, Bai X, Roy D, Spiliotis ET. Septin GTPases spatially guide microtubule organization and plus end dynamics in polarizing epithelia. J Cell Biol. 2011;194:187–197. doi: 10.1083/jcb.201102076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X, Kodama A, Fuchs E. ACF7 regulates cytoskeletal-focal adhesion dynamics and migration and has ATPase activity. Cell. 2008;135:137–148. doi: 10.1016/j.cell.2008.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.