Abstract
The Orange Carotenoid Protein (OCP) plays a similar photoprotective role in cyanobacterial photosynthesis to that of non-photochemical quenching in higher plants. Under high-light conditions, OCP binds to the phycobilisome (PBS) and reduces energy transfer to the photosystems. The protective cycle starts from a light-induced activation of OCP. Detailed information on the molecular mechanism of this process as well as the subsequent recruitment of active OCP to the phycobilisome is not known. We report here our investigation on OCP photoactivation from the cyanobacterium Synechocystis sp. PCC 6803 by using a combination of native mass spectrometry (MS) and protein cross-linking. We demonstrate that Native MS is able to capture OCP with its intact pigment and further reveal that OCP undergoes a dimer-tomonomer transition upon light illumination. The reversion of activated form of OCP to inactive, dark form was also observed by using native MS. Furthermore, in vitro reconstitution of OCP and PBS allowed to perform protein chemical cross-linking experiments. LC-MS/MS analysis identified cross-linking species between OCP and the PBS core components. Our result indicates that the N-terminal domain of OCP is closely involved in the association with a site formed by two allophycocyanin trimers in the basal cylinders of the phycobilisome core. This report helps to understand the activation mechanism of OCP and the structural binding site of OCP during the cyanobacterial non-photochemical quenching process.
Cyanobacteria primarily collect light via the phycobilisome (PBS), a mega-dalton extramembrane antenna pigment-protein complex containing covalently bound bilin pigments 1. In addition to the pigmented phycobiliproteins, pigment-free linker proteins play important roles in assembly and in maintaining a functional core-and-rod structure 2. Energy collected by the PBS rapidly migrates from rods to the core and subsequently is transferred to a membrane-embedded reaction center of Photosystem II (PSII) or Photosystem I (PSI). Under saturating-light conditions, most cyanobacteria induce a photoprotective mechanism in which excess absorbed energy is dissipated as heat in a process called Non-Photochemical Quenching (NPQ), which decreases the energy arriving at the PSII and PSI reaction centers 3. Two proteins, the orange carotenoid protein (OCP) and the fluorescence recovery protein (FRP), are known to be involved in photoprotection and restoring full capacity of light harvesting function, although the mechanistic details of this process have not been elucidated 4, 5.
The OCP is a 35 kDa water-soluble protein. Krogmann’s group6, 7 first reported the isolation and characterization of OCP before its photoprotective function was established. Crystal structures of the OCP protein have been determined from Arthrospira maxima 8 and Synechocystis sp. PCC 6803 5. The OCP protein contains a single molecule of 3’-hydroxyechinenone (3’-hECN), a carotenoid having 11 conjugated carbon-carbon double bonds. It is the first photosensory protein discovered with carotenoid as the pigment4. Both N- and C-terminal domains bind the pigment (3’-hECN) such that it is almost buried in the crystal structure of the OCP 8. The OCP and photoprotective function in cyanobacteria were linked together by Kirilovsky’s group 9, 10. In darkness or non-saturated light conditions, the OCP appears orange (OCPo). The photoactivation of the OCP can be triggered by blue-green light, which converts the OCPo into a red form (OCPr). The active OCPr is metastable and quickly converts back to the inactive form (OCPo) in dark conditions. Previous spectroscopic studies demonstrated that both the OCP protein and pigment (3’-hECN) undergo dramatic conformational changes upon bluegreen light illumination10–13, indicative of the intimate interactions of the pigment and the apo-OCP. Genetic results demonstrated that in the absence of OCP, fluorescence of PBS cannot be quenched in cyanobacteria. In the presence of excess OCP, faster fluorescence quenching of PBS is observed14. In vitro reconstitution studies indicated that only the OCPr is competent to bind PBS and triggers the fluorescence quenching15. The FRP, a 13 kDa protein on an operon with OCP in most cyanobacterial species 16, can accelerate the release of OCP from PBS and thus recover the PBS fluorescence in dark conditions16.
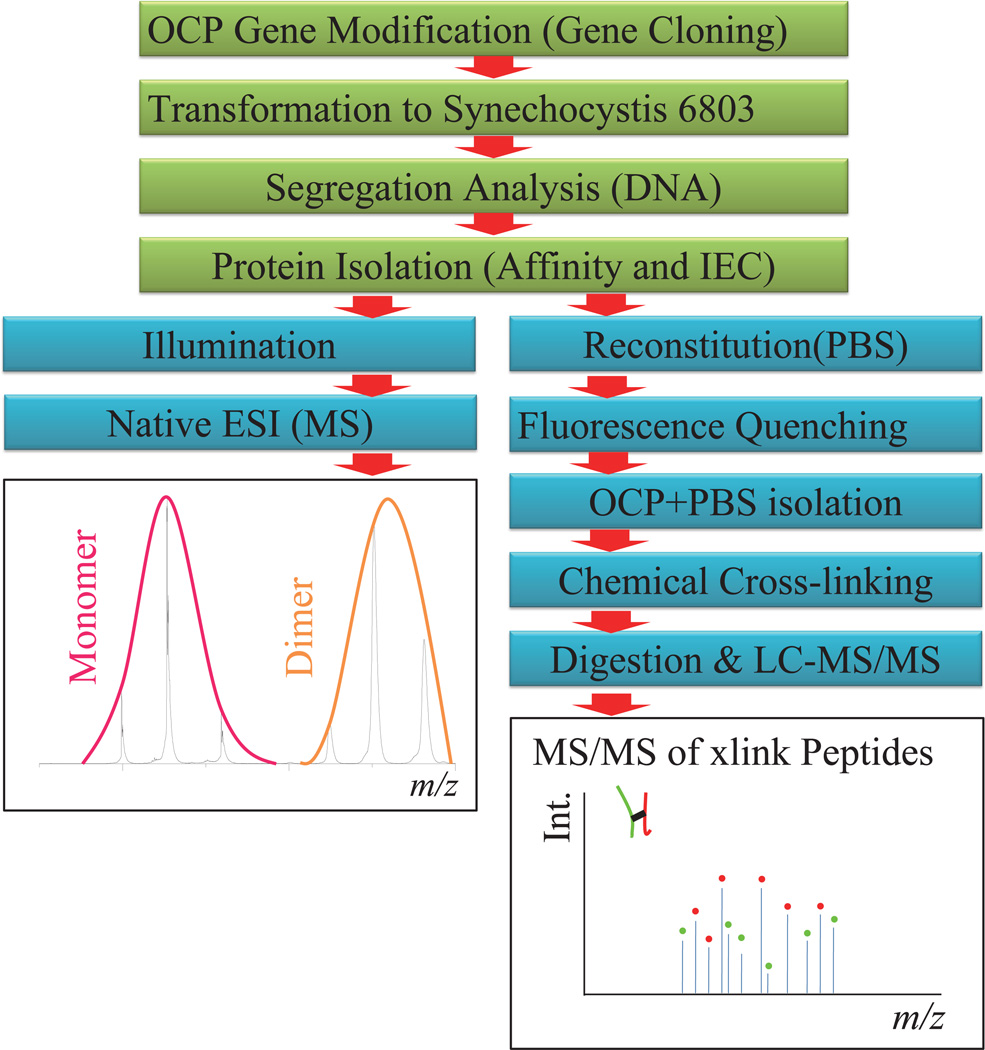
Even though structural models of OCP have been proposed 8, 17, there are still questions on how the transition between OCPo and OCPr takes place and whether there are any oligomeriztion state changes during the transition. Two MS based protein analysis approaches were employed in this research (Fig. 1). Here we report the real-time description of the OCPo-to-OCPr transition by using native MS, an emerging approach that allows for protein oligomerization state analysis under non-denaturing conditions 18, 19, as well as for non-protein cofactor stoichiometry analysis 20. When combined with ion-mobility MS (IM-MS), this approach can be used to probe structural transitions of protein complexes in their near-native states 21, 22.
Figure 1.
Schematic outline of the MS based approach in studies of photoactivation and interaction of the OCP.
We are also interested in the structural location of OCPr in PBS, which still remains an open question 23, 24, but is extremely important to understand the molecular quenching mechanism from the point of view of the structural orientation of 3’-hECN and the chromophores in the PBS core. Site directed mutagenesis suggested that R155 of the OCP in the N-terminal domain is involved in the stabilization of the OCP protein and in the interaction with PBS for fluorescence quenching 13. Consensus has been reached that OCP binds to the PBS core, however, it is still controversial to which APC (APC660 or APC680) OCPr is binding25, 26. Both APC trimers have been suggested as the OCP binding site by previous publications from different groups23, 25, 27, 28. Recently, MS based protein cross-linking became a robust tool in the investigation of protein-protein interaction among large protein complexes29, 30. In this study, a MS-based protein cross-linking experiment was also adopted to pinpoint the structural location of the OCP when it is bound to the PBS.
MATERIALS AND METHODS
Chemicals
Water, acetonitrile, formic acid and ammonium acetate were purchased from Sigma-Aldrich (St. Louis MO) with LC grade. Cross-linking reagents, bis-[sulfosuccinimidyl] suberate (BS3) and disuccinimidyl suberate (DSS), were purchased from Pierce (Thermo Scientific). HiTrap Q column was purchased from GE (Pittsburgh, PA, USA)
Growth of Synechocystis sp. PCC 6803 and Mutant Construction
Synechocystis 6803 strains, wild type and C-terminally His6-tagged OCP strain (OCP-His strain, see below) were grown in BG11 medium 31 at 30 °C under 30 Jmol photons m−2s−1 with supplemented antibiotics of gentamicin (5 µg/mL). Fusion PCR protocol 32 was used to construct a transformation cassette to replace the native OCP (Slr1963) in Synechocystis 6803 (Fig. S1). The OCP was purified as previously described 10 with minor modifications in a Bio-Rad FPLC system. Briefly, to construct a transformation cassette, a partial Slr1963 (OCP gene) sequence without the stop codon was amplified using the OCP1F (P1) and OCPGMR (P3) primers (Fig. S1 and Table S1). The PCR fragment contains the affinity tag (His6), which was achieved from the primer P3. The gentamicin cassette was amplified separately by PCR using primer P4 and P5. A fusion PCR product of partial Slr1963 and gentamicin was produced and fused to the Slr1963 downstream fragment. Segregation of the modified OCP was verified by PCR analysis using primer 2 and primer 5. The fully segregated strain was used for this study.
OCP and PBS Isolation
HiTrap Q HP column (Ion Exchange Chromatography, IEC) was used for the cleanup after Ni-NTA affinity chromatography isolation. The isolated OCP was identified by SDS-PAGE and Immunodetection (Fig.S2). The dark adapted OCP was illuminated by white light at intensity of ~ 2000 Jmol photons m−2s−1. To prevent buffer from warming up, the beam was first passed through ~10 cm water layer. Illumination resulted in rapid changes in the absorption spectra of the OCP as can be seen in Fig. S3.
Native MS of OCP
The OCP protein sample was buffer exchanged to 200 mM ammonium acetate by using a 10 kDa MWCO filter (Millipore Amicon Centrifugal Filters, Billerica, MA). Following buffer exchange, 10 µL (~50 µM) was loaded onto an offline electrospray capillary (GlassTip 2µm ID, New Objective, Woburn, MA). The protein sample solution was injected to a hybrid ion-mobility quadrupole time-of-flight mass spectrometer (Q-IM-TOF, SYNAPT G2 HDMS, Waters Inc., Milford, MA). The instrument was operated under gentle ESI conditions (capillary voltage 1.5–1.8 kV, sampling cone 20 V, extraction cone 2 V, source temperature 30°C). The collision energy at the trap and transfer region was adjusted from 8 to 58 V for dissociating the OCP- 3’-hECN complex. The pressure of the vacuum/backing region was 5.1–5.6 mbar. For the ion mobility measurements, the helium cell gas flow was 180 mL/min, the IMS gas flow was 90 mL/min, the IMS wave velocity was 650 m/s, and the IMS wave height was 40 V. Nitrogen was used as the mobility carrier gas. Each spectrum was acquired from m/z 1500–7500 every 1 s. The instrument was externally calibrated up to 8000 m/z with the clusters produced by ESI of a NaI solution. The peak picking and data processing was performed in Masslynx (v 4.1) and DriftScope software (Water Inc., Milford, MA).
PBS and OCP reconstitution and protein cross-linking experiment
The PBS-OCP complexes were prepared in 0.8 M phosphate buffer (pH 7.0) by illumination of isolated PBS with 2000 Jmol photons m−2s−1 of white light for 10 min at 23 °C in the presence of OCP (OCP:PBS ratio of 40:1) 15. To separate the resulting PBS-OCP complexes from the unbound OCP after illumination, the sample was loaded onto a 100 kDa filter (Millipore Amicon Centrifugal Filters, Billerica, MA) and centrifuged at 4,000×g at 23 °C for 20 min. Fresh 0.8 M phosphate buffer was applied three times to dilute the retentate. The composition of the blue retentate was checked by absorption and fluorescence emission spectroscopies (Fig. S4). The concentration of PBS was calculated based on the molar absorptivity formula 15. The cross-linking experiment and LC/MS/MS were used as previously described with minor changes 33. OCP-PBS sample was resuspended at 0.1 µM based on PB concentration calculation. BS3 and DSS cross-linking was performed according to the manufacturer’s protocol followed by desalting using Zeba columns (Thermo Scientific) with minor modifications. Modified samples were precipitated using acetone and the resuspended samples were directly subjected to trypsin digestion.
The trypsin digestion and LC-MS experiment
The digestion and LC-MS experiment protocol as well as the data processing were adapted from previous publication. The details are listed in the supplementary information.
RESULTS AND DISCUSSION
Native MS analysis of the transition between OCPo and OCPr
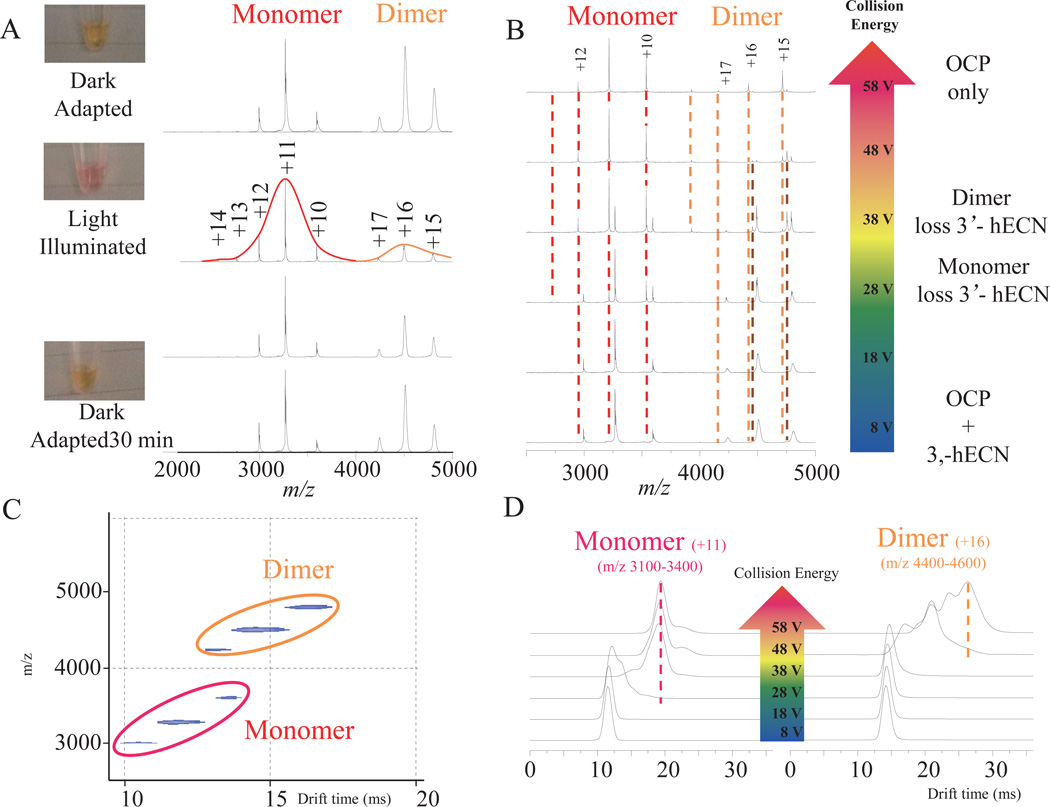
Under standard electrospray (ESI) condition, i.e. OCP in organic solvent, MS spectra show that OCP is denatured and the pigment (3’-hECN) is dissociated from the protein (Fig. S5). When OCPo was submitted to the native ESI, however, it carries nearly 30 fewer charges than in the denatured conformation (Fig. 2A and S5), suggesting retention of the near-native state. OCP samples with different concentrations (down to ~10 µM) have been analyzed. Two charge state distributions were observed. Two charge-state envelopes appear at relatively high m/z (from m/z 2,750 to 5,000). Molecular weight (MW) assignment indicates that the low m/z envelope (from 3,000 to 3,600) corresponds to an OCP monomer with one 3’-hECN (35.5 kDa, charge states from +10 to +12), whereas the high m/z envelope (from 4,200 to 5,000) corresponds to the OCP dimer with two 3’-hECNs (71 kDa, charge states from +15 to +17). The overall abundances of the two states are similar with a monomer: dimer ratio of ~1:1.
Figure 2.
Native MS spectra of the OCP. (A) Dark-adapted (sample is in orange color) OCP in 200 mM ammonium acetate was analyzed by native MS. Two charge envelopes were observed in the spectrum. The left envelope (labeled in red color) corresponds to the MW of monomeric OCP protein and one 3’-hECN. The right envelope (labeled in orange color) corresponds to the MW of dimeric OCP and two 3’-hECNs. The dark-adapted sample was illuminated with white light for 1.5 min. Two spectra were acquired when the OCP sample was restored to a dark environment for 5 and 30 min. (B) Native MS of the OCP with collision energy ramp. (C) Two dimensional IM-MS spectrum of OCP sample. The Y axis shows the mass spectrometry measurement (mass to charge ratio, m/z), while the X axis shows the ion mobility measurement (drift time, ms). Multiple charge states from two species were grouped into monomer (red) and dimer (orange). (D) Ion mobility measurement of OCP. Two charge states, one for OCP monomer (+11) and one for OCP dimer (+16), were monitored by ion mobility with the collision energy ramp.
This observation presents the opportunity to use native MS to examine the monomer-dimer equilibrium as a function of light. When the dark-adapted OCP sample was illuminated by white light, the expected orange-to-red transition from OCPo to active OCPr was observed (Fig. 2A and S3). The native mass spectrum, taken after illumination, shows a substantial loss of dimer; the monomer:dimer ratio became ~5:1 (Fig. 2A). Further, the charge-state distribution of the monomer OCP is extended higher (up to +14). We then stored the light-illuminated OCP sample in the dark to allow the reverse OCPr-to-OCPo transition to take place. After 5 min, the chargestate extension (up to +14) of the monomer disappeared, replaced by a concomitant increase of the dimer (monomer:dimer = 5:2). When we analyzed the same OCP sample after 30 min in the dark, the monomer:dimer had returned nearly to normal (~ 5:4), and the OCP sample had reassumed an orange color (Fig. 2A).
The oligomeric state of crystalline OCP is a dimer, as observed by X-ray crystallography 8. There is no consensus, however, on the oligomeric state of OCP in solution or in vivo. The relatively fast photochemical relaxation from OCPr to OCPo challenges traditional biochemistry approaches that monitor variations in stoichiometry such as biochemical preparation based technology, like FPLC or HPLC. However, in our native MS, only 30 seconds elapsed between the sample light illumination and real time MS data collection. The entire MS running time for one sample is only 1–2 minutes. Our native MS experiment can also be used to reveal real-time changes of the OCP oligomerization state that cannot be detected by denaturing, standard ESI (Fig. S5). Although the accurate quantitative information cannot be generated without isotope enriched internal standard, the conversation between monomer and dimer can be obtained by in native MS. The native MS results suggest that the monomeric form of OCP becomes the dominant species in the light-illuminated OCP sample, whereas the dark-adapted OCP sample is a mixture of monomer and dimer. Consistently, the dimeric oligomeric state of OCP can be restored by keeping the light-illuminated OCP sample in the dark (Fig. 2). The shift to higher charge state upon illumination of monomeric OCP is indicative of the presence of an “open” conformation of the OCP monomer. This “open” conformation may correspond to the OCPr that interacts with the PBS.
Collision-induced dissociation of OCP-pigment complex
We sought to characterize further the two oligomeric states found by native ESI of the dark-adapted OCP sample (OCPo) (Fig. 2B) by collisional activation of the gas-phase ions. When the collision energy of the mass spectrometer trap region was increased, the pigment dissociated from its protein binding site, allowing us to estimate the relative strengths of non-covalent carotenoid-OCP polypeptide interactions in both oligomeric states. The monomer shows only one mass-shift peak that corresponds to the loss of a single 3’-hECN, while the dimer shows two mass-shift peaks that correspond to the sequential loss of two 3’-hECNs. The 3’-hECNs are lost from the monomeric OCP at 28 V but at 38 V from the dimer. The majority of OCP, both monomer and dimer, have lost 3’-hECN at 58 V. These results indicate that the OCP dimer binds 3’-hECN more tightly or 3’-hECN is more protected than in OCP monomer and is more resistant to unfolding.
Ion mobility measurement of OCP monomer and dimer
We also monitored the dissociation of 3’-hECN from the OCP with ion mobility (IM) upon introduction by native MS (Fig. 2C and 2D). The OCP monomer and dimer, each containing three major charge states, are separable by ion-mobility. Comparing the drift-time spectra of each oligomeric state, +11 for monomer and +16 for dimer, against the collision energy ramp, we found that single drift-time distribution for each charge state at low collision energy (8 V) became broader, split, and shifted to longer times, caused by protein conformational changes 34. This result (Fig. 2D) indicates that the cross-section of OCP for both monomer and dimer increased upon activation under the increased collision energy and adopted a more “unfolded” (larger size) conformational state. The drift time shift first occurred for the monomer at 38 V, but at 48 V for the dimer.
Previous studies have indicated that conformational changes of the OCP take place during the transition between OCPo and OCPr as monitored by amide I and amide II vibrational changes 10. It was not known at that time whether transitions in the oligomerization state of OCP are involved in this process. This is now resolved by native MS, which captures the interconversion between monomeric and dimeric OCP. The CID and ion mobility experiments indicate that the pigment is more easily dissociated from monomer than the dimer. The monomer has a more solvent-exposed 3’-hECN. The results from native MS and IM indicate that there is a light-induced conversion between dimeric and monomeric OCP. Moreover, they suggest that this conversion is required in the transition of OCPo to OCPr and that the relatively “open” form of monomer (with extended charge state in native MS) is the active OCPr.
Protein cross-linking of the PBS-OCP complex
To locate the interface between OCP and PBS, we used chemical cross-linking of lysine residues coupled with LC-MS/MS detection. We took advantage of a recent breakthrough that the PBS-OCP interaction can be reconstituted in vitro 15. The OCP-mediated fluorescence quenching can also be observed from the in vitro reconstituted PBS-OCP sample (Fig. S4). In our reconstitution experiments, changes in the 400–550 nm range of the absorption spectrum of the light-treated PBS-OCP mixture, taken before and after ultrafiltration, indicate effective removal of unbound OCP (Fig. S4A), underlying the stoichiometric binding of the OCP and PBS. PBS fluorescence further demonstrated that upon fast removal of unbound OCP, the resulting PBS-OCP is still in a quenched state (Fig. S4B), indicating that the OCPr is bound to its functional site. This sample was immediately used for cross-linking reactions using bifunctional lysine specific reagents (See materials and methods). The cross-linked protein sample gave good sequence coverage of PBS components (ApcA, ApcB, ApcC, ApcE, ApcF, CpcA, CpcB, CpcC, CpcD and CpcG) as well as of OCP (Table S2).
Protein cross-linking in combination with mass spectrometry has established a detailed catalog of product ion species29, 35, 36. In our experiments, fifty-one mono-link peptides (attached to only one amino acid) were identified by LC-MS/MS (Fig. S6). There are 21 mono-link peptides from the rod of PBS, and 26 mono-link peptides from the core of the PBS. Four mono-link peptides were identified from the OCP. Loop-link peptides (where the cross linker is attached to two amino acids within one peptide) were identified for both PBS and OCP. The most informative cross-linking species are cross-link peptides (where the cross linker is attached to two different polypeptides), and 22 of these were identified. On the basis of those cross-link peptides (Fig. S7), we could map the interaction area between two protein fragments. The distance between two cross-linked sites can be elucidated with the consideration of the flexibility of protein conformations. For example, the distance could be 9 to 24 Å with 11.4 Å cross-linkers e.g. DSS BS335. We also identified several intra-link peptides (where the cross linker is attached to two amino acids in the same polypeptide) within subunits. Because there are multiple copies for most of the PBS subunits in one PBS complex, the intra-link peptides could be from one copy or two copies of the same subunit.
Given that the major goal of these experiments is to map the interaction between OCP and PBS, we focused on the cross-link peptides between OCP and PBS (Table S3). Among the crosslink peptides, the cross-linked lysines (K) are OPC:K167-ApcB:K58, OCP:K249-ApcB:K26, and OCP:K170-ApcE:K4 (Table S3). All these lysines are solvent-exposed, according to the crystal structures of APC (PDB ID, 4F0U) and OCP (PDB ID, 3MG1) from Synechococcus elongatus sp. PCC 7942 and Synechocystis sp. PCC 6803 respectively; the former protein sequence shares high identity with that of Synechocystis sp. PCC 6803 8, 17.
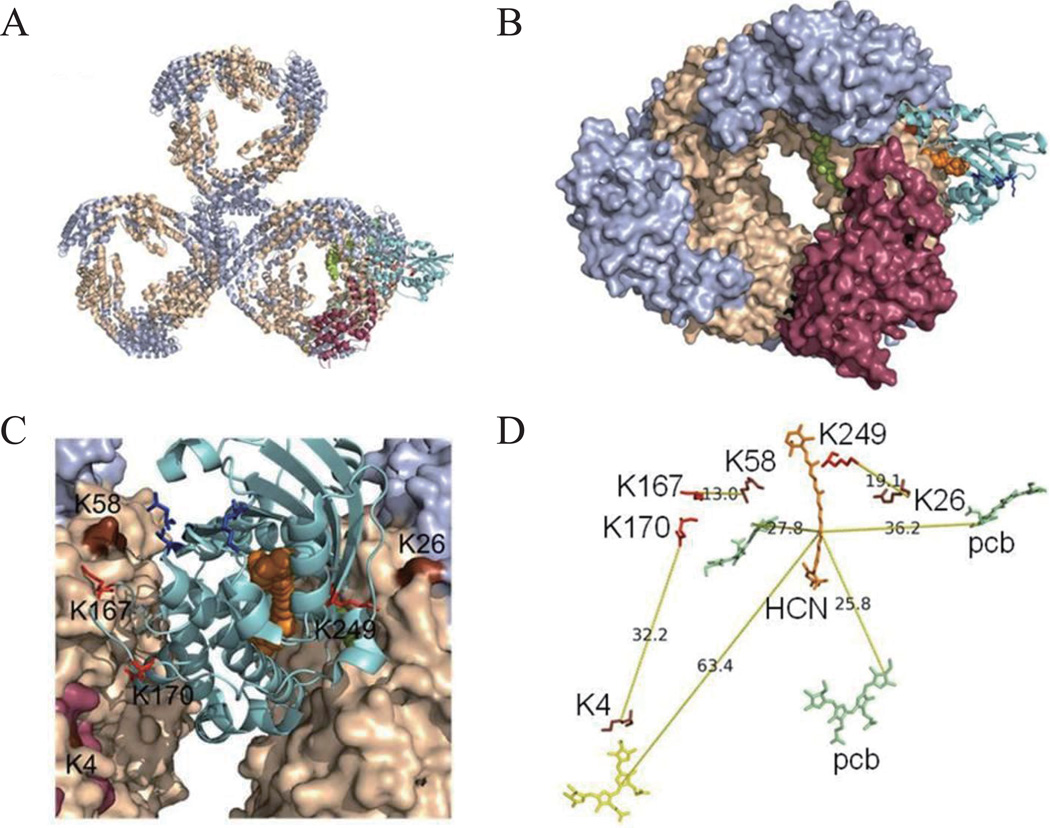
Although it is known that photoactivated OCPr protects the photosynthetic RCs by close interaction with the PBS and quenches the excess energy absorbed by PBS, the binding site remains elusive, and this greatly hampers the elucidation of the quenching mechanism. Based on our cross-linking experiments, we propose here a model of PBS-OCP interaction (Fig. 3) by using model program i-TASSER37. In this model, the OCP is located in the basal cylinder(s) of the PBS core instead of the upper one (Fig. 3A and B). The N-terminal domain (residue 15–165) 8, 17 is buried between two APC trimers, one APC660 and one APC680 28, leaving the C-terminal domain solvent-accessible (Fig. 3), with OPC:K167 and OCP:K249 located close to two adjacent ApcB proteins from APC680 and APC660 trimers, respectively. Our model suggests that there are only two OCP binding and, thus, quenching sites per PBS. These structural results based on LC MS/MS spectra are in contrast to the hypothesis that the OCP can bind to any APC660, which implies that there are more than two binding sites per PBS owing to the multiple copies of APC660 in PBS.
Fig. 3.
The structural location of the OCP in PBS. (A) The OCP (cyan) binds to the basal cylinder containing both APC660 trimers and APC680. ApcA (light blue), ApcB (wheat), ApcE (raserry). 3D structure of ApcE phycocyanobilin binding domain (raspberry) was predicted and was docked to one ApcA subunit 38, 39. (B) Close-up view of the OCP between two basal APC trimers. The N-terminal domain of the OCP is buried within two APC trimers, leaving the Cterminal domain solvent-exposed including D220, V232, F299 (stick, blue). (C) View of close association of the OCP to ApcB. Interlinks of ApcB:K58-OCP:K167 and ApcB:K26-OCP:K249 are labeled, with sticks (K167, K249 of the OCP) and surface (K58, K26). The interlink of ApcE:K4-OCP:K170 is rendered. 3’-hECN is presented by orange spheres. (D) Spatial relationships of 3’-hECN (HCN, stick, orange) and PCBs and lysine residues involved in the cross-linking. D220, V232, F299 are involved in the docking of Fluorescence Recovery Protein (FRP, stick, blue). K4 (ApcE, stick, brown), K26, K58 (ApcB, stick, brown).
The N- and C-terminal domains of the OCP are joined by a long (~25 amino acids) peptide chain. Even though this is the only region of the protein that is not strongly conserved, we found that the K167 and K170 in this linker are cross-linked to ApcB:K58 and ApcE:K4 respectively(Table S3). It seems that this flexible linker could pass conformational information from the C-terminal to the buried N-terminal domains of the protein or vice versa and thereby enable the energy coupling of 3’-hECN and phycocyanobilins (PCB). According to our model, PCB pigments from APC660 are located spatially closer to the pigment of OCP than is PCB from ApcE. Because spatial distance dictates the excitation energy transfer efficiency, we hypothesize that OCPr will preferentially quench the excitation energy from APC660. Similar conclusions about quenching sites were recently proposed based on spectroscopic methods and mutation studies 27 26. We think that our results constitute direct structural evidence to support this hypothesis. There are reports showing that ApcE (LCM) could also be the binding partner of the OCP, and that ApcE is directly involved in the OCP-mediated NPQ 23, 25. Our data do not exclude this hypothesis; however, if this were the case, the quenching efficiency could be very low in vivo, owing to the increased spatial distance between 3’-hECN and the terminal emitter on ApcE (Fig. 3). Additionally, non-physiological conditions (2 M urea, pH2.5 or 60 mM formic acid, pH3.0) used in the preparation of ApcE and in the reconstitution of ApcE-OCP in these two reports could lead to unexpected results 4.
It was proposed previously that the N-terminal domain of OCP that contains R155 is directly involved in the binding of OCPr to APC trimers based on site-directed mutagenesis and quenching analysis. Furthermore, breakage of the R155-E244 salt bridge could play an important role during the activation of OCP 13. There are multiple APC trimers, including multiple APC660 and at least two APC680, in the PBS core. To locate which APC trimer the OCPr is binding to will help to understand the detailed photoprotection mechanism in cyanobacteria. Our results narrow down the OCP binding site to the cleft formed between two APC trimers and thus will facilitate future experiments such as determining which amino acids on either OCP or ApcB are playing important roles in the stabilization or destabilization of the quenching complex.
The C-terminal domain of OCP (D220, V232, F299) was recently shown to be involved in the interactions between FRP 16, which in vivo is essential to recover the full capacity of the PBS light-harvesting function, presumably by playing a role in detaching OCPr. In our model, three amino acid residues (D220, V232, F299) are located on the C-terminal domain of the OCP, consistent with the docking and co-immunoprecipitation (Co-IP) analysis 16.
CONCLUSIONS
Evidence is presented here using native MS that the transient conversion of OCPo to OCPr involves the monomerization of the OCP, which is competent to bind to the PBS for fluorescence quenching and photoprotection. Protein cross-linking analysis further indicates that the Nterminal domain of the OCP is buried between the APC660 trimer and the APC680 trimer in the PBS core. This structural information of the OCP provides a basis for future studies on the detailed interactions between the 3’-hECN and chromophores in the PBS core and the regulation of light harvesting.
The potential of MS based approaches in studies of OCP related photoactivation has been demonstrated. Although MS based approaches cannot provide high-resolution structural information, the speed and sensitivity of MS will further benefit the investigation of OCP related photoactivation in cyanobacteria.
Supplementary Material
Acknowledgments
Funding Source Statement: The research was supported by the Photosynthetic Antenna Research Center, an Energy Frontier Research Center funded by the U.S. DOE, Office of Basic Energy Sciences (Grant No. DE-SC 0001035 to R.E.B) and National Institute of General Medical Science (Grant No. 8 P41 GM103422-35 to M.L.G). H.L., D.M.N, M.P. and J.J. were funded by the DOE grant, H.Z. was funded equally by the DOE and NIH grants, and instrumentation was made available from both the DOE and NIH-supported programs.
Footnotes
SUPPORTING INFORMATION
Supporting information contains experimental procedures and additional data. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Glazer AN. Phycobiliproteins. Methods Enzymol. 1988;167:291–303. doi: 10.1016/0076-6879(88)67034-0. [DOI] [PubMed] [Google Scholar]
- 2.Peschek G. Photosynthesis and Respiration of Cyanobacteria. In: Peschek G, Löffelhardt W, Schmetterer G, editors. The Phototrophic Prokaryotes. US: Springer; 1999. pp. 201–209. [Google Scholar]
- 3.Niyogi KK, Truong TB. Evolution of flexible non-photochemical quenching mechanisms that regulate light harvesting in oxygenic photosynthesis. Curr Opin Plant Biol. 2013;16:307–314. doi: 10.1016/j.pbi.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Kirilovsky D, Kerfeld CA. The Orange Carotenoid Protein: a blue-green light photoactive protein. Photochem Photobiol Sci. 2013;12:1135–1143. doi: 10.1039/c3pp25406b. [DOI] [PubMed] [Google Scholar]
- 5.Kirilovsky D, Kerfeld CA. The orange carotenoid protein in photoprotection of photosystem II in cyanobacteria. Bba-Bioenergetics. 2012;1817:158–166. doi: 10.1016/j.bbabio.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 6.Holt TK, Krogmann DW. A carotenoid-protein from cyanobacteria. Biochimica et biophysica acta. 1981;637:408–414. [Google Scholar]
- 7.Wu YP, Krogmann DW. The orange carotenoid protein of Synechocystis PCC 6803. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1997;1322:1–7. doi: 10.1016/s0005-2728(97)00067-4. [DOI] [PubMed] [Google Scholar]
- 8.Kerfeld CA, Sawaya MR, Brahmandam V, Cascio D, Ho KK, Trevithick-Sutton CC, Krogmann DW, Yeates TO. The crystal structure of a cyanobacterial water-soluble carotenoid binding protein. Structure. 2003;11:55–65. doi: 10.1016/s0969-2126(02)00936-x. [DOI] [PubMed] [Google Scholar]
- 9.Wilson A, Ajlani G, Verbavatz JM, Vass I, Kerfeld CA, Kirilovsky D. A soluble carotenoid protein involved in phycobilisome-related energy dissipation in cyanobacteria. The Plant cell. 2006;18:992–1007. doi: 10.1105/tpc.105.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson A, Punginelli C, Gall A, Bonetti C, Alexandre M, Routaboul JM, Kerfeld CA, van Grondelle R, Robert B, Kennis JT, Kirilovsky D. A photoactive carotenoid protein acting as light intensity sensor. Proc Natl Acad Sci U S A. 2008;105:12075–12080. doi: 10.1073/pnas.0804636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polivka T, Chabera P, Kerfeld CA. Carotenoid-protein interaction alters the S(1) energy of hydroxyechinenone in the Orange Carotenoid Protein. Biochimica et biophysica acta. 2013;1827:248–254. doi: 10.1016/j.bbabio.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Berera R, van Stokkum IH, Gwizdala M, Wilson A, Kirilovsky D, van Grondelle R. The photophysics of the orange carotenoid protein, a light-powered molecular switch. The journal of physical chemistry. B. 2012;116:2568–2574. doi: 10.1021/jp2108329. [DOI] [PubMed] [Google Scholar]
- 13.Wilson A, Gwizdala M, Mezzetti A, Alexandre M, Kerfeld CA, Kirilovsky D. The essential role of the N-terminal domain of the orange carotenoid protein in cyanobacterial photoprotection: importance of a positive charge for phycobilisome binding. The Plant cell. 2012;24:1972–1983. doi: 10.1105/tpc.112.096909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stadnichuk IN, Yanyushin MF, Zharmukhamedov SK, Maksimov EG, Muronets EM, Pashchenko VZ. Quenching of phycobilisome fluorescence by orange carotenoid protein. Doklady. Biochemistry and biophysics. 2011;439:167–170. doi: 10.1134/S1607672911040053. [DOI] [PubMed] [Google Scholar]
- 15.Gwizdala M, Wilson A, Kirilovsky D. In vitro reconstitution of the cyanobacterial photoprotective mechanism mediated by the Orange Carotenoid Protein in Synechocystis PCC 6803. The Plant cell. 2011;23:2631–2643. doi: 10.1105/tpc.111.086884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sutter M, Wilson A, Leverenz RL, Lopez-Igual R, Thurotte A, Salmeen AE, Kirilovsky D, Kerfeld CA. Crystal structure of the FRP and identification of the active site for modulation of OCP-mediated photoprotection in cyanobacteria. Proc Natl Acad Sci U S A. 2013;110:10022–10027. doi: 10.1073/pnas.1303673110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson A, Kinney JN, Zwart PH, Punginelli C, D'Haene S, Perreau F, Klein MG, Kirilovsky D, Kerfeld CA. Structural Determinants Underlying Photoprotection in the Photoactive Orange Carotenoid Protein of Cyanobacteria. J Biol Chem. 2010;285:18364–18375. doi: 10.1074/jbc.M110.115709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benesch JL, Ruotolo BT, Simmons DA, Robinson CV. Protein complexes in the gas phase: technology for structural genomics and proteomics. Chemical reviews. 2007;107:3544–3567. doi: 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]
- 19.Heck AJR, van den Heuvel RHH. Investigation of intact protein complexes by mass spectrometry. Mass spectrometry reviews. 2004;23:368–389. doi: 10.1002/mas.10081. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Cui W, Gross ML, Blankenship RE. Native mass spectrometry of photosynthetic pigment–protein complexes. FEBS Lett. 2013;587:1012–1020. doi: 10.1016/j.febslet.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong Y, Hyung SJ, Ruotolo BT. Ion mobility-mass spectrometry for structural proteomics. Expert review of proteomics. 2012;9:47–58. doi: 10.1586/epr.11.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uetrecht C, Rose RJ, van Duijn E, Lorenzen K, Heck AJR. Ion mobility mass spectrometry of proteins and protein assemblies. Chem Soc Rev. 2010;39:1633–1655. doi: 10.1039/b914002f. [DOI] [PubMed] [Google Scholar]
- 23.Stadnichuk IN, Yanyushin MF, Maksimov EG, Lukashev EP, Zharmukhamedov SK, Elanskaya IV, Paschenko VZ. Site of nonphotochemical quenching of the phycobilisome by orange carotenoid protein in the cyanobacterium Synechocystis sp. PCC 6803. Biochimica et biophysica acta. 2012;1817:1436–1445. doi: 10.1016/j.bbabio.2012.03.023. [DOI] [PubMed] [Google Scholar]
- 24.Kuzminov FI, Karapetyan NV, Rakhimberdieva MG, Elanskaya IV, Gorbunov MY, Fadeev VV. Investigation of OCP-triggered dissipation of excitation energy in PSI/PSII-less Synechocystis sp. PCC 6803 mutant using non-linear laser fluorimetry. Biochimica et Biophysica Acta - Bioenergetics. 2012;1817:1012–1021. doi: 10.1016/j.bbabio.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 25.Stadnichuk IN, Yanyushin MF, Bernat G, Zlenko DV, Krasilnikov PM, Lukashev EP, Maksimov EG, Paschenko VZ. Fluorescence quenching of the phycobilisome terminal emitter L from the cyanobacterium Synechocystis sp. PCC 6803 detected in vivo and in vitro. J Photochem Photobiol B. 2013;125 C:137–145. doi: 10.1016/j.jphotobiol.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 26.Jallet D, Gwizdala M, Kirilovsky D. ApcD, ApcF and ApcE are not required for the Orange Carotenoid Protein related phycobilisome fluorescence quenching in the cyanobacterium Synechocystis PCC 6803. Biochim Biophys Acta. 2012;1817:1418–1427. doi: 10.1016/j.bbabio.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 27.Tian LJ, Gwizdala M, van Stokkum IHM, Koehorst RBM, Kirilovsky D, van Amerongen H. Picosecond Kinetics of Light Harvesting and Photoprotective Quenching in Wild-Type and Mutant Phycobilisomes Isolated from the Cyanobacterium Synechocystis PCC 6803. Biophys J. 2012;102:1692–1700. doi: 10.1016/j.bpj.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jallet D, Gwizdala M, Kirilovsky D. ApcD, ApcF and ApcE are not required for the Orange Carotenoid Protein related phycobilisome fluorescence quenching in the cyanobacterium Synechocystis PCC 6803. Bba-Bioenergetics. 2012;1817:1418–1427. doi: 10.1016/j.bbabio.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 29.Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass spectrometry reviews. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 30.Petrotchenko EV, Borchers CH. Crosslinking combined with mass spectrometry for structural proteomics. Mass spectrometry reviews. 2010;29:862–876. doi: 10.1002/mas.20293. [DOI] [PubMed] [Google Scholar]
- 31.Allen MM, Stanier RY. Growth and division of some unicellular bluegreen algae. Journal of General Microbiology. 1968;51:199–202. doi: 10.1099/00221287-51-2-199. [DOI] [PubMed] [Google Scholar]
- 32.Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- 33.Herzog F, Kahraman A, Boehringer D, Mak R, Bracher A, Walzthoeni T, Leitner A, Beck M, Hartl FU, Ban N, Malmstrom L, Aebersold R. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 2012;337:1348–1352. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- 34.Politis A, Park AY, Hyung SJ, Barsky D, Ruotolo BT, Robinson CV. Integrating ion mobility mass spectrometry with molecular modelling to determine the architecture of multiprotein complexes. PloS one. 2010;5:e12080. doi: 10.1371/journal.pone.0012080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Molecular & cellular proteomics : MCP. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruce JE. In vivo protein complex topologies: sights through a cross-linking lens. Proteomics. 2012;12:1565–1575. doi: 10.1002/pmic.201100516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H, Zhang H, Niedzwiedzki DM, Prado M, He G, Gross ML, Blankenship RE. Phycobilisomes supply excitations to both photosystems in a megacomplex in cyanobacteria. Science. 2013 doi: 10.1126/science.1242321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.