Abstract
Hedgehog (Hh) signaling in cancer cells drives changes in the tumor microenvironment that are incompletely understood. Here we report that Hh- driven tumors exhibit an increase in myeloid-derived suppressor cells (MDSC) and a decrease in T cells, indicative of an immune suppressive tumor microenvironment. This change was associated with activated TGFβ signaling in several cell types in BCCs. We determined that TGFβ signaling in bone marrow (BM)-derived cells, not keratinocytes, regulates MDSC and promotes tumor development. Tgfbr2 deficiency in the BM-derived cells also reduced the size of previously developed tumors in mice. We identified CCL2 as the major chemokine attracting MDSC to tumor, whose expression was Tgfbr2-dependent, whereas its receptor CCR2 was highly expressed in MDSC population. CCL2 alone was sufficient to induce migration of MDSC. Moreover, the CCR2 inhibitors prevented MDSC migration towards skin cells in vitro, reduced MDSC accumulation and Hh signaling-driven tumor development in mice. Our results reveal a signaling network critical for Hh signaling in cancer cells to establish an effective immune suppressive microenvironment during tumor development.
Keywords: hedgehog, MDSC, immune suppressive tumor microenvironment, basal cell carcinomas
INTRODUCTION
Cancer cells do not develop in isolation, but rather require collaborative interactions with cells in the tumor microenvironment (TME) to regulate tumor angiogenesis, extracellular matrix remodeling, tumor invasion and metastasis, and to evade the immune surveillance system (1-4). Although all the signaling events leading to an immunosuppressive TME are not completely understood, there are several proposed mechanisms by which cancer cells help establish an immunosuppressive microenvironment (3, 5-7). For example, tumor cells can evade detection through loss of tumor specific surface antigens. Frequently, a subset of immune cells in the TME, including myeloid-derived suppressor cells (MDSC), regulatory T cells, some macrophages and dendritic cells, inhibit T cell proliferation, leading to an immunosuppressive microenvironment. MDSC are immunosuppressive immature myeloid cells with monocytic or granulocytic morphology (3). In mice, MDSC can be recognized by cell surface expression of CD11b and Gr1, and regulatory T cells display a CD4+CD25+Foxp3+ profile. Accumulation of these immunosuppressive cells at the tumor site is regulated by many inflammatory cytokines and chemokines. Mouse models of cancer with inducible gene expression provide a unique opportunity to investigate how the immunosuppressive tumor microenvironment is established during cancer development.
Mice are not susceptible to BCC development unless the hedgehog (Hh) pathway is activated, through tissue-specific deletion of Patched 1 (Ptch1) or inducible expression of activated Smoothened molecule SmoM2 (8-12), providing a robust system to investigate interactions between Hh signaling-activated keratinocytes and the surrounding cells in the TME. The Hh pathway is known to regulate embryonic development, cell proliferation, differentiation and carcinogenesis. At the present, it is not clear how activated Hh signaling alters TME and whether TME is critical for BCC development in mice (9, 12). Because activated Hh signaling is found in a variety of human cancers, the novel information gleaned from our present studies may help us to understand the role of Hh signaling in other cancer types.
In this study, we examined changes in cell populations during development of BCCs and rhabdomyosarcomas, and observed a significant increase of MDSC in SmoM2-dependent tumors. Our previous publication and the molecular analysis suggest that elevated TGFβ signaling may be responsible for this change. The functional significance of TGFβ signaling in the TME for BCC formation was further investigated by BM transplantation approaches to create TME chimeras. BM cells (with or without Tgfbr2) were transplanted into the mice with inducible expression of SmoM2 in keratinocytes, and the tumor development was monitored over time. To further delineate the molecular events, we examined expression of cytokines and chemokines, and determined their regulation by TGFβ signaling during tumor development. The significance of these factors for MDSC accumulation was further tested by cell migration analysis, and their effects on tumor formation were examined by specific inhibitors in tumor-bearing mice. We have determined that blocking TGFβ signaling in the BM-derived cells in the TME or reduced CCL2/CCR2 signaling inhibits Hh-mediated tumor development.
MATERIALS AND METHODS
Animal Studies
All animal studies have been approved by the Institutional Animal Care and Use Committee in Indiana University. K14-cre mice were obtained from the Emice Program in National Cancer Institute, K14-creER, Mx1-Cre, Tgfbr2flox/flox (also shown as Tgfbr2f/f), R26-SmoM2YFP and ROSAmT/mG mice were purchased from The Jackson Laboratory. Mice were maintained and mated under pathogen-free husbandry conditions. To obtain Tgfbr2 deletion, Tgfbr2flox/flox mice were mated with K14-creER and R26-SmoM2YFP mice separately. The resulting Tgfbr2flox+/−/K14-creER+ mice were mated with Tgfbr2flox+/−/R26-SmoM2YFP+ mice to obtain K14-creER+/R26-SmoM2YFP+/ Tgfbr2flox/flox mice. We also obtained Mx1-Cre+/R26-SmoM2YFP+/ Tgfbr2flox/flox mice using a similar procedure.
Genotyping of mice was performed by PCR with specific primers provided by the vendors using lysed tail from each mouse [0.3cm tail in 100 μl of PCRDirect (tail) solution (Viagen Inc.) with 1mg/ml proteinase K at 55°C overnight, then 85°C for 45min, and use 0.5 to 1 μl of the lysate for each 25μl PCR reaction].
Drug treatments in mice
Expression of SmoM2YFP in K14-creER+/R26-SmoM2YFP+ mice was induced by oral administration of tamoxifen (80mg/Kg body weight in 100 μl of vegetable oil in each feeding) for 5 consecutive days with a feeding needle. For Mx1 induction, we injected (via i.p.) 50μg of PolyI:C in 20 μl of PBS to new born mice 3 times (P1, 3 &5) or 200 μg of polyI:C in 100μl of PBS to adult mice 4 times (day 1, 3, 5 &12).
CCR2 antagonists RS-102895 (Sigma), RS504393 (R&D Systems) and CXCR4 antagonist AMD3100 (Sigma) were first dissolved in DMSO and diluted in 70% ethanol (1μM for RS102895 and AMD11070) for topical application and in PBS (2mg/kg for RS-504393) for oral gavage (13, 14). For topical application, chemicals in solution were applied onto the abdomen areas daily for 10 days with 10 μl of solution. Ethanol (70%) was used as a control. For oral administration, we applied RS-504393 at 2mg/Kg or PBS buffer twice a day for 7 days. Five mice per group on average were used in this study. At the end of the study, skin biopsies were collected for Hematoxylin & Eosin (H&E) staining.
Histology and microscopic BCC analyses
Histology was performed according to a previously published procedure (15). The proportion of tumor area to the total tissue area was quantified using Imagine-J. To avoid discrepancy from age and genetic backgrounds of the mice, we used littermates from the same mating cage for selection of treatment groups or genotypes. Because of the variation between back skin and abdomen skin in tumor development, we used the midline abdomen skin for histology studies.
Cell population analyses and cell sorting of skin tissues, peripheral blood and spleen
Mouse skins were submerged in Dispase® solution (Life technology at 5mg/ml in PBS) for 2 hours at 37°C to separate epidermis from dermis. One part of epidermis was used for total RNA extraction using TRI® reagent (Sigma). The rest of epidermis was digested with collagenase IV (1mg/ml in DMEM with 10% FBS) for 1 hour at 37°C. Well digested tissues were filtered through a cell strainer (pore size: 70μm) and then spun at 500g via bench top centrifugation to obtain single cells.
Specific cell populations were recognized by cell surface markers through specific antibody staining: CD11b+Gr1+ for MDSC population; T cell populations include CD3+CD4+, CD3+CD8+ and γδ (CD3+γδT+) T cells. To block non-specific binding, cells were first incubated cells with 10%FBS in PBS for 30 minutes on ice. Antibodies used in this study included PE conjugated anti-mouse CD11b (Biolegend, San Diego, CA, USA), APC conjugated anti-mouse GR1 (Biolegend), FITC conjugated anti-mouse CD3 (Biolegend), APC-Cy7 conjugated anti-mouse CD4 (eBioscience), PE conjugated anti-mouse CD8 (eBioscience), APC conjugated anti-mouse γδ T (eBioscience) and PE-Cy7 conjugated anti-mouse TCRβ (eBioscience), Alexa Fluor® 488 Conjugated anti-vimentin IgG (Cell Signaling Technology Inc., cat# 9853) and anti-phospho-SMAD2 (Cell Signaling Technology Inc., Cat# 8828). For cell labeling of peripheral blood and spleen cells, ammonium-chloride-potassium buffer (Gibco®) was used to lyse red blood cells before blocking the non-specific binding (10% FBS in PBS) and antibody labeling. DAPI staining was used to gate out dead cells for flow cytometry analyses. For intracellular staining, we used Cytofix/Cytoperm™ to permeabilize cells following the vendor's instruction (BD Biosciences). Stained cells were analyzed by BD FACSCalibur APC and Flow-jo. For cell sorting, stained cells were sorted on a BD FACSAria (Becton Dickinson, Franklin Lakes, NJ, USA) according to the fluorescence used.
T cell proliferation analysis
T cells from mouse spleen were isolated using Pan T cell isolation kit II (Miltenyi Biotec Inc.) in which no-target cells were retained on a MACS column while unlabeled T cells passed through and were collected for CFSE labeling using CellTrace™ CFSE cell proliferation kit (C34554) (Molecular Probes). Purified T cells were cultured in RPMI with 10% heat-inactivated FBS without antibiotics. To activate T cell and to stimulate T cell proliferation, T cells were cultured on CD3 antibody-coated plates (clone 145-2C11 from BioXcell at 8μg/ml for 2 hours at 37°C) with 1μg/ml CD28 antibodies (clone 37.51 from BD Pharmingen™) in the medium. The effects of CD11b+Gr1+ cells on T cell proliferation was assayed after addition of CD11b+Gr1+ cells for 4 days. The ratios of T cell: CD11b+Gr1+ cell were 10:1 or 20:1, depending on the availability of CD11b+Gr1+ cell number. In our studies, the two ratios gave similar results. CD11b+Gr1+ cells from mouse spleen and skin tumors were sorted after labeling with PE conjugated anti-mouse CD11b and APC conjugated anti-mouse GR1 (Biolegend). CFSE contents in T cells were analyzed by flow cytometric analysis. Low intensity of CFSE labeling indicated more proliferative whereas high intensity was suggestive of less proliferative. Each treatment group has triplets of samples and each experiment was repeated for three times with similar results.
Migration Assay
Cell migration was assessed as described (16) using CD11b+Gr1+ cells sorted form spleen in the upper chamber and CD3−Gr1−CD11b− cells, γδ T cell (CD3+γδT+) or chemokines CCL2/CCL7/CCl8 in the lower chamber. Chemokines CCL2, CCL7 and CLL8 were obtained from R&D Systems. CCR2 antagonist RS-102895 and CXCR4 antagonist AMD3100 were purchased from Sigma. To prevent chemokine receptor function, sorted CD11b+Gr1+ cells were incubated with RS-102895 (2μM), AMD3100 (1.25μM) or the solvent during migration assay based on previous studies (17-19).
RT-PCR and Real-time PCR
Total RNA was isolated from the tissues using TRI reagent (Sigma) according to the manufacturers’ instructions. One μg of total RNA was reverse transcribed into cDNAs using the first-strand synthesis kit (Roche). We performed real-time RT-PCR with a previously reported procedure (15).
Western Blotting, immunofluorescent staining and ELISA analysis
Epidermis was first lysed with a protein loading buffer in ultra sound bath for 5 min. Specific antibodies to Smad2, pSmad2, β-actin were purchased from Cell Signaling Technology Inc. Proteins were detected according to a procedure reported previously. Also, we used a previously published procedure (20) for immunofluorescent staining with specific antibodies to vimentin (Cell Signaling Inc., Cat# 9853), phospho-SMAD2 (Cell signaling Inc., Cat# 8828) and phospho-SMAD3 (Santa Cruz Biotechnology Inc, Cat# S130218).
ELISA was performed using a kit from Peprotech according to the manufacturer's protocol. The CCL2 protein concentration was calculated from the standard curve generated with CCL2 controls and the value from each samples. Triplicates of samples were used, and the assay was repeated 3 times with similar results. The data presented in Fig.6D was the average of these results with STDEV.
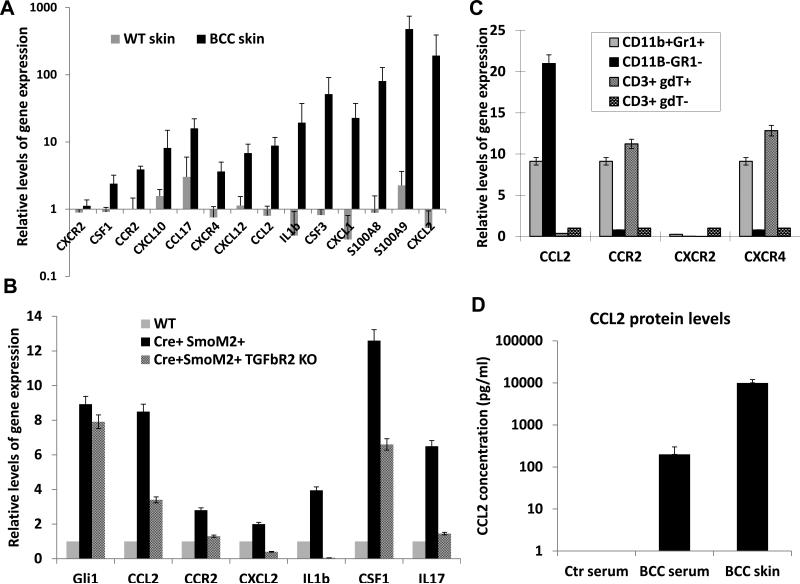
Figure 6. Expression of chemokines, cytokines and inflammatory factors in SmoM2-mediated skin tumors.
A shows altered expression of chemokines, cytokines and inflammatory factors in SmoM2-driven skin tumors. Comparison between K14-creER/R26-SmoM2 (as WT skin) mice and R26-SmoM2 mice (as BCC skin) showed significant elevation of gene expression (indicated by*) for Csf1, Csf3, Ccr2, Ccl2, cxcl10, Ccl17, Cxcr4, Cxcl12, Cxcl1, Cxcl2, Il1b, S100a8 and S100a9, but not Cxcr2. B shows examples of factors regulated by TGFβ signaling in skin tissues. Significant difference between normal and tumorous skin tissues was indicated as * whereas significant reduction between Mx1-cre/R26-SmoM2 and Mx1-cre/R26-SmoM2/Tgfbr2f/f mice was indicated as ** (p values< 0.05 by Student's t test). C shows expression of chemokines and their receptors in different cell populations of SmoM2-driven skin tumors. Significant elevation of gene expression was indicated by * (p values<0.05). D shows ELISA analysis of the CCL2 protein level in peripheral blood and skin tissues from tumor-bearing mice, with a comparison with the serum level of CCL2 from R26-SmoM2 mice (no tumor-bearing control). While control mice had nearly undetectable CCL2 protein in the serum, the serum level of CCL2 protein in tumor-bearing mice reached ~200pn/ml. The highest level was detected in the skin tumor (~1,000pg/ml). It thus appears that there is a CCL2 protein gradient from a low level in the serum to a high level in the tumor.
Bone Marrow Transplantation
One week before transplantation, mice were transported into the designated room suitable to house immune deficient mice and provided with acidified water containing neomycin (100mg/L). The animals received 1 × 7Gy total body irradiation plus 1 × 4Gy 4 hours later. Bone marrow was isolated from appropriate mice (polyI:C -stimulated Mx1cre/Tgfbr2f/f mice or control mice), and 5 × 106 cells/mouse in 100μl of serum-free RPMI were injected intravenously to rescue the hematopoietic system of the irradiated mice. Wet food was provided to mice for 4 weeks after transplantation.
Statistical Analyses
Data are presented as mean ± SD. Statistical analyses were performed using Mann-Whitney test or the Student's t test (two tailed) to compare the results, with P values of <0.05 indicating statistically significant difference.
RESULTS
Activation of Hh signaling is associated with MDSC accumulation
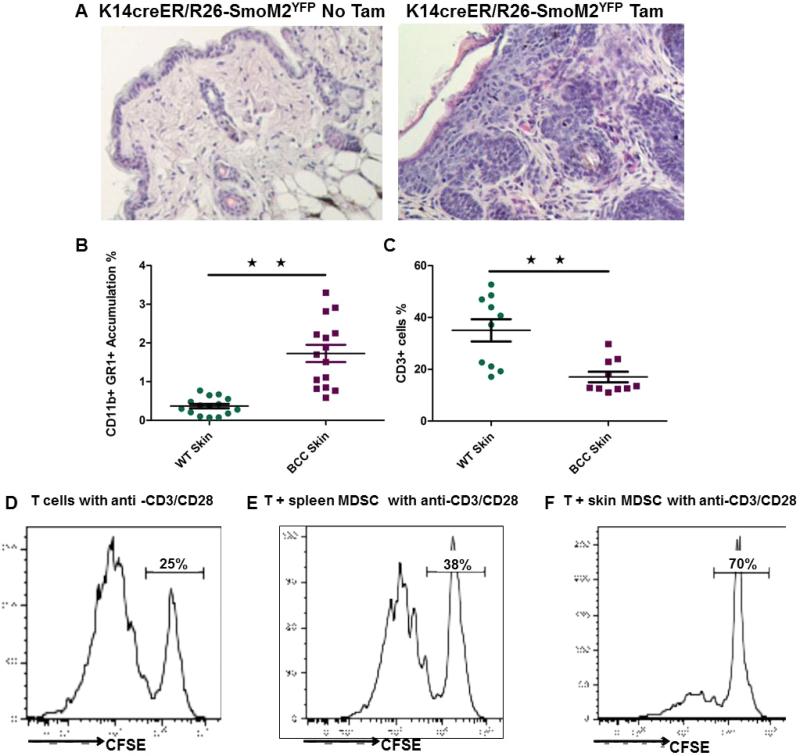
Aberrant activation of Hh signaling is the driver for development of BCCs but the molecular and cellular mechanisms by which Hh signaling mediates tumor development are not completely understood. Using K14creER and R26SmoM2YFP (21-23) mice, we established mouse models of BCCs (Fig. 1A) and examined changes in cell populations of the skin epidermis. As shown in Fig. 1B and 1C, we found accumulation of CD11b+Gr1+ cells and a decrease in CD3+ cells in the tumor, compared to normal skin. Similar changes of CD11b+Gr1+ were also observed in peripheral blood and spleen of tumor-bearing mice (Figure S1). We noticed that the severe skin phenotype (or with a high percentage of tumor area) is associated with a higher level of CD11b+Gr1+ cell population and a lower level of CD3+ cells. In addition to these populations, we observed an increase in the hair follicle stem cell population (20) as well as in tumor-associated fibroblasts (Figure S2), but a decrease in γδT cells (not shown). Regulatory T-cells (CD4+ CD25+ Foxp3+) are not detectable in the skin of these mice. CD11b and Gr1, which are markers for MDSC, are generally not expressed in normal skin tissues. MDSC are a group of cells with a suppressive activity on T cell proliferation (3). To confirm CD11b+Gr1+ cell population as MDSC, we purified CD11b+Gr1+ cells from spleen and tumors of K14creER+/R26SmoM2YFP mice and tested their effects on carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled T cell proliferation. Figure 1D-F showed suppression of T cell proliferation by CD11b+Gr1+ cells. Similar to several reports in the literature (24, 25), we found that CD11b+Gr1+ cells from tumors were more effective than those from spleen on T cell suppression. These results indicate that an immune suppressive tumor microenvironment was created during development of BCCs.
Figure 1. Accumulation of MDSC during SmoM2-mediated skin tumor development.
K14-creER/R26-SmoM2YFP mice can be induced to expression oncogenic SmoM2 in keratinocytes by oral administration of tamoxifen (see Methods). A shows H&E images from mice without (left) or with (right) tamoxifen treatment. B shows flow cytometric results of CD11b+Gr1+ cells, and C shows a summary of T cells (CD3+) in skin tissues. Each dot represented one mouse. D-F show suppressive activities of CD11b+Gr1+ cells isolated from spleen (E) and skin tumors (F) on proliferation of CFSE-labeled T cells. Purified T cells were first labeled with CFSE, and their proliferation was measured by the reduced intensity of CFSE contents after T cell activation with CD3 and CD28 antibodies in the presence or absence (D) of CD11b+Gr1+ cells (see Methods for details). Triplets of samples were used for each experiment and the experiment was repeated three times with similar results. D is the positive control for T cell activation by CD3 and CD28 antibodies.
The phenotype in BCCs was also observed in SmoM2-mediated rhabdomyosarcomas in CAGG-CreER/R26-SmoM2 mice (21) (Fig.S3), indicating that Hh signaling-mediated regulation on the immune suppressive tumor microenvironment is not BCC specific.
Abolishing TGFβ signaling reduces SmoM2-mediated tumor development
Previously, we have reported that TGFβ signaling is activated through up-regulation of TGFβ2 in SmoM2-mediated skin tumors and is required for tumor development (15). This regulation may be a direct one because addition of Smoothened agonist purmorphamine alone in Hh-responsive C3H10T1/2cells rapidly induced expression of TGFβ2 transcript, resulting in elevated SMAD2 protein phosphorylation, a marker for activated TGFβ signaling (15). Based upon review of the published literature (26-28), we hypothesized that TGFβ signaling is a good candidate pathway critical for MDSC recruitment in SmoM2-induced tumors.
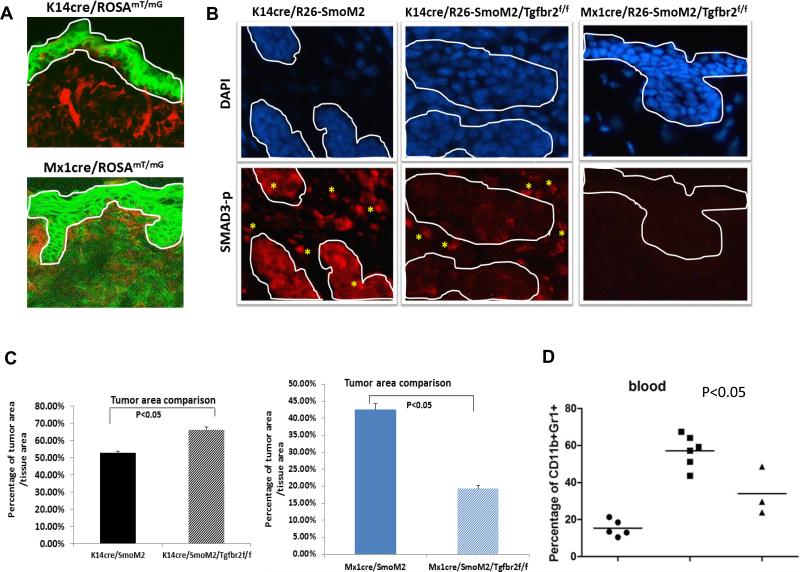
The role of TGFβ signaling in cancer is tissue context dependent (28). TGFβ signaling in the tumor cells is tumor suppressive in early tumor development but gains tumor promoting effects during metastasis. To examine the role of TGFβ signaling in the keratinocytes, we generated mice with tissue specific gene deletion of Tgfbr2 under control of the keratinocyte-specific K14 promoter (29). Confirming the specificity, the reporter mouse K14cre/ROSAmT/mG showed restricted expression of the GFP to epidermis (Fig.2A). Furthermore, SMAD3 phosphorylation was hardly detectable in the tumor but was not affected in the tumor stroma of K14cre/R26-SmoM2/Tgfbr2f/f mice (Fig.2B SMAD3-p staining in the middle panel, tumor was circulated by the white line), supporting the conclusion that TGFβ signaling is compromised in keratinocytes of these animals. We found that K14cre/R26-SmoM2/Tgfbr2f/f mice, that had keratinocyte-specific Tgfbr2 deletion, developed significantly larger tumors (measured by tumor areas) than Tgfbr2-expressing K14cre/R26-SmoM2 mice (Fig.2C left). These data demonstrate that TGFβ signaling in keratinocytes is tumor suppressive.
Figure 2. Genetic evidence for the role of TGFβ signaling during SmoM2-induced tumor development.
A shows expression of GFP and tomato red in K14cre/ROSAmT/mG and Mx1cre/ROSAmT/mG reporter mice. GFP expression indicates cre recombination. The epidermis was within the white line. As expected, GFP expression was only observed within the epidermis in K14cre/ROSAmT/mG mice whereas the epidermis from Mx1cre/ROSAmT/mG mice (after polyI:C stimulation) had GFP in both epidermis and dermis. B shows images with phosphorylated SMAD3 staining (indicated as Smad3-P) in skin tissues of different mice. As readout of TGFβ signaling, phosphorylated SMAD3 was detected both in epidermis/tumor and dermis in K14cre/R26-SmoM2 (left panel and positive staining in red, and tumors were circulated by white lines). In K14cre/R26-SmoM2/Tgfbr2f/f mice in which Tgfbr2 was knockout in keratinocytes, phosphorylated SMAD3 was only detected in the stroma (* indicates positive cells in red). In contrast, Mx1cre/R26-SmoM2/Tgfbr2f/f mice, which had Tgfbr2 knockout in both keratinocytes and stroma/dermis, had almost no detectable phosphorylated SMAD3. Epidermis and tumors were circled by white lines. C shows the percentage of tumor area/tissue area from multiple mice (n>3) in each group. P values below 0.05 were regarded as statistically significant between Tgfβr2-expressing and Tgfβr2 deficient mice. D shows the blood and spleen levels of CD11b+Gr1+ cells from Mx1-cre mice. The levels of CD11b+Gr1+ cells from blood and spleen are associated with the severity of skin tumor phenotypes. Mx1-cre/SmoM2/Tgfβr2f/f mice had a significant decrease of CD11b+Gr1+ cell population in blood and spleen in comparison with Tgfbr2 expressing Mx1-cre/SmoM2 mice (p<0.05).
The above results, which are consistent with the common view of the tumor suppressive effect of TGFβ signaling in tumor epithelial cells (28), are different from our previous results using the TGFβ signaling inhibitor SD208 that TGFβ signaling inhibition reduced tumor size (15). To address this discrepancy, we performed additional genetic studies using Mx1-cre, which is active both in keratinocytes and in bone marrow-derived cells (30, 31). The reporter mouse Mx1cre/ROSAmT/mG showed GFP expression both in the epidermis and the dermis (Fig.2A). Successful deletion of TGFβ signaling was achieved in Mx1-cre/SmoM2/Tgfbr2f/f mice as indicated by nearly undetectable SMAD3 phosphorylation in epidermis and dermis (Fig.2B, SMAD3-p staining in the right panel). In comparison with Tgfbr2-expressing Mx1cre/SmoM2 mice, we found a significant reduction of tumor area in Tgfbr2 deficient mice (Fig.2C right), indicating that TGFβ signaling is indeed required for SmoM2-mediated skin tumor development. Further analyses indicate that deletion of Tgfbr2 in Mx1-cre-expressing cells in this model decreased the level of CD11b+Gr1+ cell population (Fig.2D), suggesting a possible role of TGFβ signaling in regulation of MDSC accumulation in skin tumors. No phenotypes were observed in Mx1-cre/Tgfbr2f/f mice or K14-cre/Tgfbr2f/f mice, indicating that the effects of TGFβ signaling in skin tumor are SmoM2 dependent.
Comparing the data from K14-cre mice with those from Mx1-cre mice, we conclude that the tumor promoting effects of TGFβ signaling in the context of active Hh signaling come mainly from the tumor microenvironment, not the keratinocytes.
Activation of TGFβ signaling in multiple cell types in SmoM2-mediated cancer development
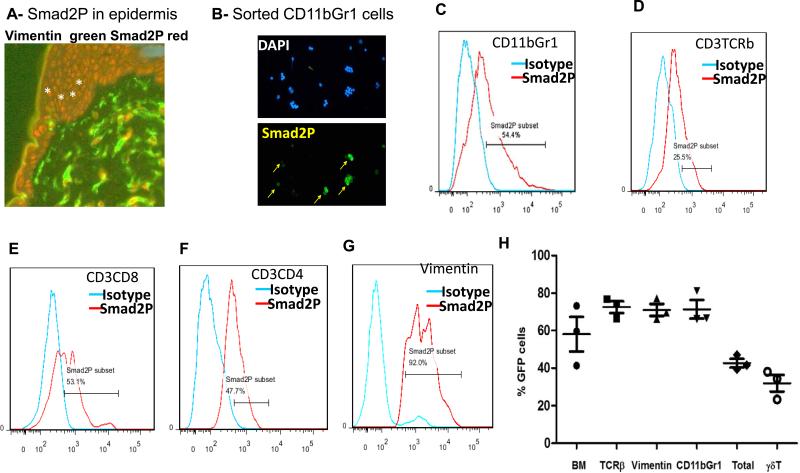
In order to show whether TGFβ signaling is responsible for MDSC accumulation in the TME, we first identified the specific stromal cell types with TGFβ signaling activation through detecting the level of SMAD2 phosphorylation, a marker for TGFβ signaling activation (27), by flow cytometry and immunofluorescent staining. As shown in Fig.3, we found that >60% of CD11b+Gr1+ cells have detectable SMAD2 phosphorylation (Fig.3B/C). SMAD2 phosphorylation was also detected in keratinocytes (Fig.3A, positive cells in red), T cells (Fig.3D-F) and fibroblasts (vimentin positive in Fig.3G). These results indicate that TGFβ signaling is activated in many cell types in the TME during BCC development and is associated with MDSC accumulation. However, with such widespread TGFβ signaling, it is impossible to genetically eliminate TGFβ-mediated signaling (via tissue-specific deletion of Tgfbr2) in all cell types.
Figure 3. TGFβ signaling activation in different cell populations in SmoM2-derived skin tumors.
The level of TGFβ signaling was indicated by the level of phospho-SMAD2 detected by either flow cytometry of immunofluorescent staining of sorted cell populations. A shows immunofluorescent (IF) staining of phosphorylated SMAD2 in skin of K14creER/R26-SmoM2 mice. Vimentin positive cells (green) indicate fibroblasts. Smad2P (red) was detectable in vimentin positive and vimentin negative cells. Keratinocytes (indicated by *) were mostly positive for Smad2P staining. B shows IF staining of phosphorylated SMAD2 (indicated as Smad2P in green) in sorted CD11b+Gr1+ cells. C shows flow cytometry comparing phosphorylated SMAD2 antibody staining with the isotype control antibody in CD11b+Gr1+ cells. D-G show flow data on Smad2P positive cells (%) in CD3+TCRβ+ (D), CD3+CD8+ (E), CD3+CD4+ (F) and vimentin positive (G) cell population respectively. H shows BM-derived cells in skin tumors as indicated by GFP positivity in different cell populations after transplantation of GFP-expressing BM cells into tamoxifen-treated K14-creER/R26-SmoM2YFP mice.
Our alternative approach was to perform bone marrow (BM) transplantation using WT or Tgfbr2 deficient bone marrow. Indeed, in our BM transplantation experiments using GFP-expressing BM cells (32) with the level of BM cell engraftment of 70% or more, we found 40% of GFP positive cells in the total epidermal cells (Fig.3H), indicating over half of the epidermal cells in the tumor were derived from BM cells in SmoM2-derived skin tumors. In addition, around 70% of CD3+CD4+, CD3+CD8+, MDSC, and vimentin+ cells were GFP positive (Fig.3H), suggesting that most of these cells are derived from BM. In contrast, less than 30% of γδT cells were GFP positive (Fig.3H), which is consistent with the literature that most γδT cells emerge from thymus in early postnatal stage mice (33).
Taken together, our BM transplantation studies indicated that numerous cell types in the tumor stromal compartment are derived from BM cells. Based on these data, we predicted that transplantation of BM cells with Tgfbr2 deficiency into K14creER/SmoM2 mice would allow us to assess the role of TGFβ signaling for the tumor microenvironment.
TGFβ signaling in BM-derived cells is required for Hh-induced skin cancer development
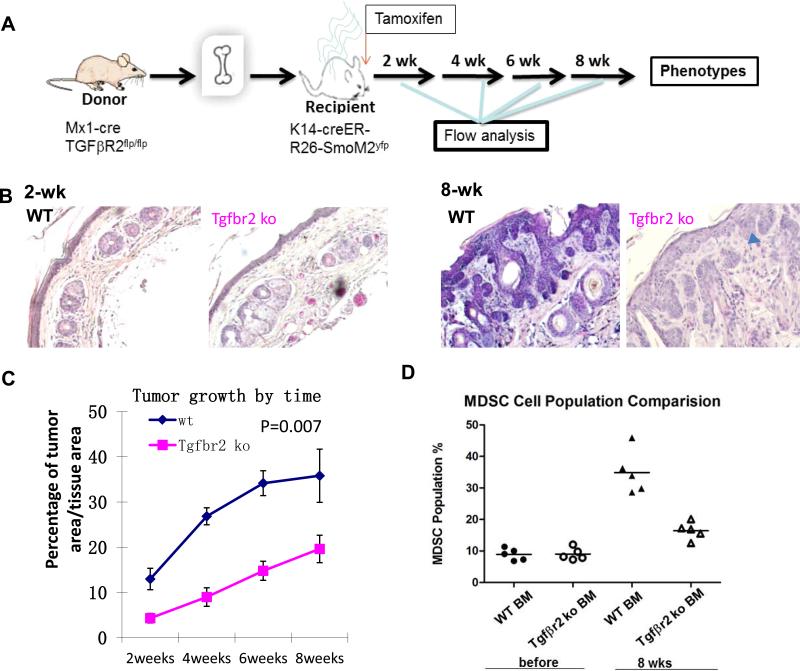
To examine the role of TGFβ signaling in TME for tumor initiation and development, we performed BM transplantation before induction of SmoM2 expression. The donor BM cells were either Tgfbr2 -deficient (from Poly I: C -treated Mx1-cre/Tgfbr2f/f mice) or wild type control (from Tgfbr2f/f mice) whereas the recipient mice were all K14-creER/R26-SmoM2. Expression of SmoM2 was induced by oral administration of tamoxifen (Fig.4A). Mice were monitored daily to examine the severity of skin tumor development. We found that while mice receiving wild type BM cells developed skin lesions and BCC-like tumors (Fig.4B), mice receiving Tgfbr2-deficient BM cells had only minimal cell proliferation in the skin (Fig.S4) and showed significantly slower tumor development (Fig.4C, p value=0.007). In fact, we observed only residual tumors in many skin sections (indicated by a blue arrowhead in Fig.4B at 8 weeks, also in Fig. S4A), indicating a slow tumor development. Consistent with these results, we found a high level of EDU incorporation in mice with control BM but a very low level in mice with Tgfbr2 deficient BM (Figure S4). Only a low level of phosphorylated SMAD2 staining was detected in mice with Tgfbr2-deficient BM (arrows in mouse #477, Figure S5). We also observed an association between reduced TGFβ signaling and decreased number of CD11b+Gr1+ cells in the peripheral blood (Fig.4D) and in the skin tumors (Fig.S6).
Figure 4. Effects of Tgfbr2 deficiency in BM-derived cells for SmoM2-induced tumor development.
A shows a diagram of the experimental design. Before BM transplantation, recipient mice were irradiated (1 ×7 Gy and 1 × 4 Gy with 4 hrs in between) before tail vein injection with 5 × 106 BM cells/mouse. Tamoxifen was used to induce cre expression in K14 positive cells (keratinocytes), peripheral blood and tail skin biopsies were collected at different time points for analyses. B shows H&E staining of skin sections from SmoM2-derived skin tumors with wild type (top panel) or Tgfbr2 deficient (bottom panel) BM-derived cells before SmoM2 induction. C shows tumor growth in mice with wild type (blue) or Tgfbr2 deficiency (pink) in BM-derived cells. The difference of tumor growth between the two groups was statistically significant [(p=0.007 by Student's t test (matched groups and two tailed)]. The time shown in this figure was the weeks after SmoM2 induction. D shows the level of CD11b+Gr1+ cells in peripheral blood of different mice.
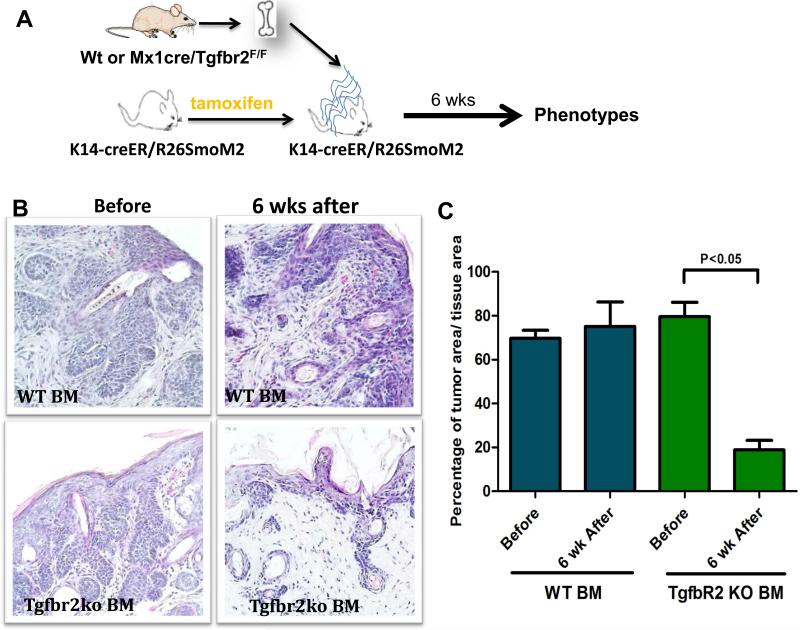
We then assessed whether deletion of Tgfbr2 in BM has any effects on tumors developed from SmoM2 expression. We first induced SmoM2 expression by tamoxifen administration and then performed BM transplantation (Fig.5A). We examined skin lesions receiving wild type or Tgfbr2 deficient BM cells, and found that mice receiving Tgfbr2 deficient BM cells had a significant reduction in tumor area (Fig. 5B/C). Irradiation during BM transplantation did not affect tumor development because no significant difference in tumor area was observed between irradiated mice and those without irradiation (not shown). Compared with mice receiving wild type control BM cells, we found that Tgfbr2 deletion in BM-derived cells decreased the tumor size (Fig.5C). These data indicate that stromal Tgfbr2 signaling plays a positive role in SmoM2-mediated tumor development as well as tumor maintenance.
Figure 5. Effects of Tgfbr2 knockout in BM-derived cells on existing SmoM2-derived tumors.
A shows a diagram of the experimental design. In brief, SmoM2 was first induced by tamoxifen before BM transplantation. Six weeks later, skin biopsies were collected for histology analyses. B shows H&E staining of skin sections with wild type (top panel) or Tgfbr2 deficiency (bottom panel) in the BM-derived cells after SmoM2 induction. C shows the percentage of tumor areas in different groups of mice.
To examine whether our data on TGFβ signaling are limited to BCCs or has a significant role in other type of tumors, we assessed the effects of Tgfbr2 deficiency in BM-derived cells on tumor growth of melanoma cell line B16-F10 (Fig.S7). First, we generated mice with wild type or Tgfbr2 deficient BM. Cell surface marker analyses indicated a nearly complete engraftment of transplanted BM. These mice were then implanted with B16-F10 cells via subcutaneous injection (2 × 105 cells/ mouse), and the tumor size was measured weekly. As shown in Fig. S7A, we found a significant reduction of tumor growth in mice receiving Tgfbr2-deficient BM cells. At the end of the study, we sacrificed the mice and obtained the tumor weight from all the mice. Tumors in mice receiving Tgfbr2 deficient BM cells had less tumor mass than those receiving wild type BM cells (Fig. S7B-C). These results demonstrate that the tumor promoting role of TGFβ signaling in the tumor environment appears not restricted to BCC.
The molecular basis for MDSC accumulation during skin carcinogenesis
To determine whether the increase of MDSC is a result of cell proliferation at the tumor site, we measured DNA synthesis of various cell populations in normal and tumorous skin tissues by EDU labeling. After EDU labeling for 14 hours in mice, we sorted various cell populations. As shown in Figure S8, we found that tumorous skin tissues have high levels of EDU labeling in keratinocytes, T cells, γδ T cells but very low labeling in CD11b+Gr1+ cells (Figure S8A). Further, we compared EDU labeling of CD11b+Gr1+ cells from different sites (tumors, peripheral blood and spleen). EDU labeling in CD11b+Gr1+ cells from spleen and peripheral blood was 50% and 15% respectively, but less than 5% in tumor-derived CD11b+Gr1+ cells (Fig.S8B). These results indicate that MDSC accumulation at the tumor site is not due to elevated cell proliferation, more likely a result of cell migration.
To elucidate the molecular mechanisms by which the MDSC population is recruited to the TME, we examined expression of cytokines and chemokines commonly expressed in normal or diseased skin. Of 30 relevant candidates, we found 12 of them with elevated expression in mouse BCCs. In particular, we found increased expression of Ccl2, Ccr2, Csf1, Cxcl2, Il17 and Il1b in skin tumors [Fig.6A, comparison between R26-SmoM2 (as WT skin) mice and K14-creER/R26-SmoM2 mice (as BCC skin)]. Similar results were also observed through comparison between Mx1-creER/R26-SmoM2 mice and R26SmoM2 mice (Fig.6B and Fig.S9). To determine the candidate chemokines and chemokine receptors, we tried to detect the chemokines and the matched receptors. We found elevated expression of Ccl2 and the gene transcript for its receptor CCR2 in the tumor (Fig.6B). Similarly, expression of Cxcl12 and the transcript for its receptor CXCR4 was also elevated in the tumor. In some instances, we did not detect expression of the receptor despite high levels of chemokines (e.g., Cxcl2, Cxcl4 and Cxcr2). We also examined whether their expression was regulated by TGFβ signaling through comparing Mx1-cre/R26-SmoM2/Tgfbr2+/+ with Mx1-cre/R26-SmoM2/Tgfbr2f/f mice (Fig.6B and Fig. S9). Those genes regulated by TGFβ signaling are likely the candidates for additional studies.
Next, we examined expression of these genes in distinct cell populations in mouse BCCs. For cell population analyses, we first obtained single cells from mice. Antibodies specific to T cells (CD3, CD4, CD8, TCRβ and TCRγδ), MDSCs (CD11b+Gr1+) and non-immune cells (CD11b−Gr1−CD3−) were used for cell sorting, and the sorted cells were analyzed for gene expression by real-time PCR. We found that Ccr2 was highly expressed in MDSC (Fig.6C) whereas Il17 was specifically expressed in γδT cells (Figure S10). Keratinocytes and fibroblasts do not have specific cell surface markers but were enriched in the CD3−CD11b−Gr1− subset. We found that Ccl2 was highly expressed in CD3−CD11b−Gr1− cells (Fig.6C).
High Ccr2 expression in CD11b+Gr1+ cells (Fig.6C) and high Ccl2 in CD3−CD11b−Gr1− skin cells (mainly keratinocytes and fibroblasts) suggested that the CCL2/CCR2 signaling axis may play an important role for MDSC recruitment into the tumor site. ELISA analysis showed that the serum level of CCL2 was barely detectable in mice without skin tumors, but was increased to ~200pg/ml in tumor bearing mice (Fig.6D). The level of CCL2 in the skin tissue of tumor-bearing mice reached 1,000pg/ml (Fig.6D). This CCL2 gradient (high in the skin and low in the peripheral blood) is consistent with our hypothesis that CCL2 is an important chemokine to recruit CCR2-expressing MDSCs.
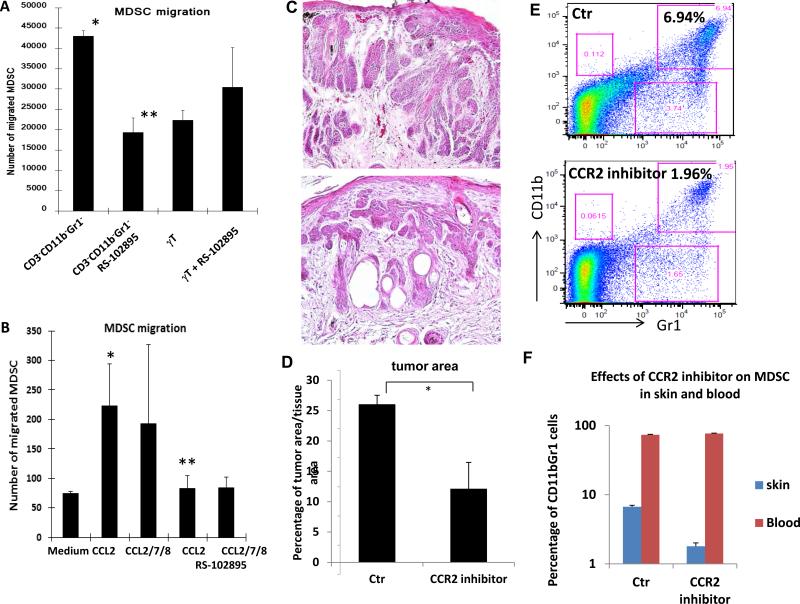
To test our hypothesis, we performed Boyden chamber chemotaxis assays with purified CD11b+Gr1+ cells in the top chamber with other purified cell population in the bottom chamber. We found that CD3−CD11b−Gr1− cells (high Ccl2 expression) were more effective than CD3+γδT+ cells (low Ccl2 expression) in promoting migration of CD11b+Gr1+ cells (high Ccr2 expression) (Fig.7A). When the CCR2 inhibitor RS102895 (34) was incubated with CD11b+Gr1+ cells during chemotaxis assay, CD3−CD11b−Gr1− cell-mediated cell migration was abolished (Fig.7A). This effect appears to be specific because the CXCR4 inhibitor AMD3100 (35) had little effects on CD3−CD11b−Gr1− cell-mediated cell migration of CD11b+Gr1+ cells (Fig.S11). Furthermore, We found that addition of CCL2 in the bottom chamber was sufficient to induce CD11b+Gr1+ migration, which was prevented in the presence of CCR2 antagonist RS102895 (Fig.7B). Addition of three chemokines CCL2, CCL7 and CCL8 had similar effects as CCL2, suggesting that CCL2 is the major player in MDSC recruitment. These results demonstrated that the CCL2/CCR2 signaling axis is capable of promoting MDSC migration.
Figure 7. Regulation of CD11b+Gr1+ migration, recruitment and tumor development by the CCL2/CCR2 signaling axis.
A shows effects of different cell populations (sorted and placed in the bottom chamber) on CD11b+Gr1+ cell migration. CD11b−Gr1− cells, which include keratinocytes and fibroblasts, are more effective in inducing migration of CD11b+Gr1+ cells towards the bottom chamber than γδ-T cells. When CCR2 antagonist RS102895 was used to incubate with CD11b+Gr1+ cells, migration of CD11b+Gr1+ cells was greatly reduced. B shows the effect of CCL2 for CD11b+Gr1+ cell migration. Migrated CD11b+Gr1+ cells (top chamber) into the bottom chamber in the presence of CCL2, CCL2/7/8 or the control media was examined using Boyden chambers. P value <0.05 was regarded as statistically significant (indicated as * in comparison with the medium control or as ** in comparison with the control treatment).
Tumor bearing K14-creER/R26-SmoM2YFP mice were treated with CCR2 antagonists RS102895 (topical application at 1μM once a day for 10 days), RS504393 (oral gavage at 2mg/kg body weight twice daily for 7 days) or the control solvent for each treatment. Tumor areas in each group were measured by ImageJ after H&E staining. C shows H&E pictures from RS-504393-treated mice (oral gavage), and D shows the average tumor areas in each group. RS-102895 has the similar results as RS-504393 (data not shown). E shows percentage of CD11b+Gr1+ cells at the tumor site of RS504393-treated mice. F shows the summary of CD11b+Gr1+ cell population in skin tissues (indicated as skin) and peripheral blood (indicated as blood) of tumor-bearing mice with (indicated as CCR2 inhibitor) or without RS504393 treatment (indicated as Ctr). The significant difference between the treatment group and the control was calculated by Student's t test (two samples), with p value< 0.05 as statistically significant (indicated as *). Significant reduction of MDSC population by CCR2 inhibitor in the skin tissues, not in the blood, was observed.
Inhibiting CCL2/CCR2 signaling reduced SmoM2-mediated carcinogenesis
To demonstrate the significance of the CCL2/CCR2 signaling axis for SmoM2-mediated tumor formation, we treated K14-cre/R26-SmoM2 mice with the specific CCR2 inhibitors RS102895 (1μM for topical application) or RS504393 (36) (2mg/kg for oral gavage twice daily) for 5 days (18, 19). After treatment, we compared tumor areas by ImageJ analysis of H&E slides from skin sections, and found a significant reduction of tumor area (>50% with p<0.05) (Fig.7C, 7D). This decrease in tumor was correlated with a significant reduction of CD11b+Gr1+ cells (from 5.8% to 2.2%, Fig.7E-F) in the TME (p value<0.05). In contrast, RS504393 had no significant effects on MDSC in the peripheral blood (Fig.7F), indicating that CCR2 is critical for recruitment of MDSC to the tumor site. We also noticed that the effect was more dramatic with oral gavage administration of the drug. These results indicated that specific inhibition of the CCL2/CCR2 signaling axis was sufficient to decrease the level of MDSC cell population at the tumor site and to reduce SmoM2-mediated skin tumor development.
DISCUSSION
Tumor-induced increase in MDSC population at the tumor site, in peripheral blood and in spleen of tumor-bearing mice is an important immune escape mechanism (37). It is believed that tumor secreted growth factors, chemokines and enzymes regulate MDSCs, but the exact signaling events responsible for MDSC expansion, recruitment and activation are not completely understood in many tumor models using genetically engineered mice. There are at least two models to explain how MDSCs are regulated during tumor development. The One Signal Model proposes that one signal is sufficient to regulate all aspects (generation, recruitment and activation) of MDSC regulation, while the Two Signal Model suggests at least two signals are needed to regulate these processes. More publications now support the Two Signal Model [reviewed in(13)]. Our data are in agreement with the Two Signal Model. In our study, we found a critical role of TGFβ signaling in bone marrow-derived cells for recruitment of MDSCs to the tumor site, which was accomplished by regulation of the CCL2/CCR2 signaling axis. In support of our hypothesis, we have shown that inhibition of CCR2 reduced the number of MDSCs in the skin tissue, but not MDSCs in peripheral blood (Fig.7F). These results demonstrate that the levels of MDSC in peripheral blood and at the tumor site were differentially regulated, with the CCL2/CCR2 signaling axis as the major regulator for MDSC population at the tumor site in our model.
In this study, we revealed an important role of TGFβ signaling in promoting tumor development. Our previous studies suggested direct regulation of TGFβ2 by hedgehog signaling (15). Using keratin 14 promoter-driven expression of oncogenic SmoM2, we detected activated TGFβ signaling in many cell types in addition to keratinocytes (see Fig.3). Many of these cells are derived from BM during SmoM2-mediated tumor formation. We showed that Tgfbr2 deficiency in keratinocytes accelerated tumor formation (Fig.2) whereas Tgfbr2 deletion in BM-derived cells (Fig.4 and Fig.5) decreased SmoM2-mediated tumor development, indicating a tumor-promoting effect of TGFβ signaling during BCC development. These studies are consistent with a recent report on myeloid cell-specific Tgfbr2 deletion in the mammary gland tumors (38). Our findings are BCC-specific because tumor growth of melanoma cell line B16F10 also required TGFβ signaling in BM-derived cells (Fig.S7). Because cell specific targeting for drug delivery in the clinical setting is very challenging if not impossible, our revelation of the overall tumor promoting function of TGFβ signaling for BCC has significant clinical implications. For example, topical application of TGFβ signaling inhibitors may be an effective way to treat skin cancer. We also want to point out that our conclusion may not be applicable for all other types of cancer because Tgfbr2 deletion appears to promote tumor development and progression in mammary gland malignancies (39, 40).
Our data indicate that the tumor promoting effect of TGFβ signaling in the mouse model of BCCs is in part through recruitment of MDSC population to the tumor site, allowing tumor cells to escape from the immune surveillance system of the host (3). While several studies have indicated a role of TGFβ signaling for recruitment of T-reg cells (41-43), T-reg cell accumulation at the tumor site in SmoM2-dependent tumors was hardly detectable in our mouse model, indicating that TGFβ signaling-mediated effect in BCCs may not be directly related to T-reg cells. Our data are consistent with previous results on direct regulation of MDSC by TGFβ signaling (3, 38, 44). We still don't know how tumor-secreted TGFβ molecules are transported to MDSC in the peripheral blood, and one possibility is that TGFβ molecules are transported through tumor exosomes, which can travel and fuse with a variety of cell types (34).
There are many factors known to be involved in MDSC regulation, including CXCL1, CXCL5, CCL2, CCL3 and GM-CSF (3, 14). In our studies, we detected a high level of Ccr2 in MDSC and elevated Ccl2 expression in the TME during tumor formation, and their expression is regulated by TGFβ signaling (Fig.6). Based on our data, we propose a model for the role of TGFβ and the CCL2/CCR2 signaling axis for Hh-mediated tumor development. In this model (see Fig.S12), activated Hh signaling induces TGFβ signaling activation, which functions as an immune suppressive cytokine (45) to induce more expression of CCL2 and CCR2. The high concentration of CCL2 at the tumor site helps recruitment of MDSC into the tumor site, and creates the immune suppressive tumor microenvironment.
In conclusion, our studies reveal a novel signaling network of Hh-TGFβ-CCL2/CCR2 in the recruitment of MDSC to the tumor site during Hh signaling-mediated tumor development (see Figure S12). Hh signaling in keratinocytes triggers elevated expression of TGFβ molecules, which turn on a cascade of signaling events to create an immune suppressive tumor microenvironment. This study may help understand the microenvironment in Hh signaling-associated tumors and provide new strategies for topical treatments of BCCs.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by National Cancer Institute R01CA155086, R01CA94160, The Wells Center for Pediatric Research and IU Simon Cancer Center.
Footnotes
The authors have no conflict of interest to disclose
REFERENCES
- 1.Shiao SL, Ganesan AP, Rugo HS, Coussens LM. Immune microenvironments in solid tumors: new targets for therapy. Genes Dev. 2011;25:2559–72. doi: 10.1101/gad.169029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–22. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 3.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nature reviews Immunology. 2009;9:162–74. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Kao J, Ko EC, Eisenstein S, Sikora AG, Fu S, Chen SH. Targeting immune suppressing myeloid-derived suppressor cells in oncology. Critical reviews in oncology/hematology. 2011;77:12–9. doi: 10.1016/j.critrevonc.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ostrand-Rosenberg S, Sinha P, Beury DW, Clements VK. Cross-talk between myeloid-derived suppressor cells (MDSC), macrophages, and dendritic cells enhances tumor-induced immune suppression. Seminars in cancer biology. 2012;22:275–81. doi: 10.1016/j.semcancer.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonda N, Chioda M, Zilio S, Simonato F, Bronte V. Transcription factors in myeloid-derived suppressor cell recruitment and function. Current opinion in immunology. 2011;23:279–85. doi: 10.1016/j.coi.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Epstein EH. Basal cell carcinomas: attack of the hedgehog. Nat Rev Cancer. 2008;8:743–54. doi: 10.1038/nrc2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng JM, Curran T. The Hedgehog's tale: developing strategies for targeting cancer. Nat Rev Cancer. 2011;11:493–501. doi: 10.1038/nrc3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ingham PW. Hedgehog signaling: a tale of two lipids. Science. 2001;294:1879–81. doi: 10.1126/science.1064115. [DOI] [PubMed] [Google Scholar]
- 11.Lum L, Beachy PA. The Hedgehog response network: sensors, switches, and routers. Science. 2004;304:1755–9. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- 12.Yang L, Xie G, Fan Q, Xie J. Activation of the hedgehog-signaling pathway in human cancer and the clinical implications. Oncogene. 2010;29:469–81. doi: 10.1038/onc.2009.392. [DOI] [PubMed] [Google Scholar]
- 13.Condamine T, Gabrilovich DI. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol. 2011;32:19–25. doi: 10.1016/j.it.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toh B, Wang X, Keeble J, Sim WJ, Khoo K, Wong WC, et al. Mesenchymal transition and dissemination of cancer cells is driven by myeloid-derived suppressor cells infiltrating the primary tumor. PLoS Biol. 2011;9:e1001162. doi: 10.1371/journal.pbio.1001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan Q, He M, Sheng T, Zhang X, Sinha M, Luxon B, et al. Requirement of TGF{beta} Signaling for SMO-mediated Carcinogenesis. J Biol Chem. 2010;285:36570–6. doi: 10.1074/jbc.C110.164442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen M, O'Connor KL. Integrin alpha6beta4 promotes expression of autotaxin/ENPP2 autocrine motility factor in breast carcinoma cells. Oncogene. 2005;24:5125–30. doi: 10.1038/sj.onc.1208729. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell LA, Henderson AJ, Dow SW. Suppression of vaccine immunity by inflammatory monocytes. J Immunol. 2012;189:5612–21. doi: 10.4049/jimmunol.1202151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nam BY, Paeng J, Kim SH, Lee SH, Kim do H, Kang HY, et al. The MCP-1/CCR2 axis in podocytes is involved in apoptosis induced by diabetic conditions. Apoptosis : an international journal on programmed cell death. 2012;17:1–13. doi: 10.1007/s10495-011-0661-6. [DOI] [PubMed] [Google Scholar]
- 19.Sun X, Charbonneau C, Wei L, Yang W, Chen Q, Terek RM. CXCR4-targeted therapy inhibits VEGF expression and chondrosarcoma angiogenesis and metastasis. Mol Cancer Ther. 2013;12:1163–70. doi: 10.1158/1535-7163.MCT-12-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu D, Fan Q, Zhang X, Xie J. A role for transcription factor STAT3 signaling in oncogene smoothened-driven carcinogenesis. J Biol Chem. 2012;287:38356–66. doi: 10.1074/jbc.M112.377382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao J, Ligon KL, Rakhlin EY, Thayer SP, Bronson RT, Rowitch D, et al. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 2006;66:10171–8. doi: 10.1158/0008-5472.CAN-06-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grachtchouk M, Pero J, Yang SH, Ermilov AN, Michael LE, Wang A, et al. Basal cell carcinomas in mice arise from hair follicle stem cells and multiple epithelial progenitor populations. J Clin Invest. 2011;121:1768–81. doi: 10.1172/JCI46307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang GY, Wang J, Mancianti ML, Epstein EH., Jr Basal cell carcinomas arise from hair follicle stem cells in Ptch1(+/−) mice. Cancer Cell. 2011;19:114–24. doi: 10.1016/j.ccr.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haverkamp JM, Crist SA, Elzey BD, Cimen C, Ratliff TL. In vivo suppressive function of myeloid-derived suppressor cells is limited to the inflammatory site. European journal of immunology. 2011;41:749–59. doi: 10.1002/eji.201041069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corzo CA, Condamine T, Lu L, Cotter MJ, Youn JI, Cheng P, et al. HIF-1alpha regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J Exp Med. 2010;207:2439–53. doi: 10.1084/jem.20100587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Connolly EC, Freimuth J, Akhurst RJ. Complexities of TGF-beta targeted cancer therapy. International journal of biological sciences. 2012;8:964–78. doi: 10.7150/ijbs.4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He W, Cao T, Smith DA, Myers TE, Wang XJ. Smads mediate signaling of the TGFbeta superfamily in normal keratinocytes but are lost during skin chemical carcinogenesis. Oncogene. 2001;20:471–83. doi: 10.1038/sj.onc.1204117. [DOI] [PubMed] [Google Scholar]
- 28.Massague J. TGFbeta signalling in context. Nature reviews Molecular cell biology. 2012;13:616–30. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biswas S, Chytil A, Washington K, Romero-Gallo J, Gorska AE, Wirth PS, et al. Transforming growth factor beta receptor type II inactivation promotes the establishment and progression of colon cancer. Cancer Res. 2004;64:4687–92. doi: 10.1158/0008-5472.CAN-03-3255. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, He XC, Tong WG, Johnson T, Wiedemann LM, Mishina Y, et al. Bone morphogenetic protein signaling inhibits hair follicle anagen induction by restricting epithelial stem/progenitor cell activation and expansion. Stem Cells. 2006;24:2826–39. doi: 10.1634/stemcells.2005-0544. [DOI] [PubMed] [Google Scholar]
- 31.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–9. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 32.Srour EF, Yoder MC. Flow cytometric analysis of hematopoietic development. Methods in molecular medicine. 2005;105:65–80. doi: 10.1385/1-59259-826-9:065. [DOI] [PubMed] [Google Scholar]
- 33.Macleod AS, Havran WL. Functions of skin-resident gammadelta T cells. Cell Mol Life Sci. 2011;68:2399–408. doi: 10.1007/s00018-011-0702-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mirzadegan T, Diehl F, Ebi B, Bhakta S, Polsky I, McCarley D, et al. Identification of the binding site for a novel class of CCR2b chemokine receptor antagonists: binding to a common chemokine receptor motif within the helical bundle. The Journal of biological chemistry. 2000;275:25562–71. doi: 10.1074/jbc.M000692200. [DOI] [PubMed] [Google Scholar]
- 35.Domanska UM, Timmer-Bosscha H, Nagengast WB, Oude Munnink TH, Kruizinga RC, Ananias HJ, et al. CXCR4 inhibition with AMD3100 sensitizes prostate cancer to docetaxel chemotherapy. Neoplasia. 2012;14:709–18. doi: 10.1593/neo.12324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yiakouvaki A, Dimitriou M, Karakasiliotis I, Eftychi C, Theocharis S, Kontoyiannis DL. Myeloid cell expression of the RNA-binding protein HuR protects mice from pathologic inflammation and colorectal carcinogenesis. The Journal of clinical investigation. 2012;122:48–61. doi: 10.1172/JCI45021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talmadge JE, Gabrilovich DI. History of myeloid-derived suppressor cells. Nat Rev Cancer. 2013;13:739–52. doi: 10.1038/nrc3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Novitskiy SV, Pickup MW, Chytil A, Polosukhina D, Owens P, Moses HL. Deletion of TGF-beta signaling in myeloid cells enhances their anti-tumorigenic properties. Journal of leukocyte biology. 2012;92:641–51. doi: 10.1189/jlb.1211639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forrester E, Chytil A, Bierie B, Aakre M, Gorska AE, Sharif-Afshar AR, et al. Effect of conditional knockout of the type II TGF-beta receptor gene in mammary epithelia on mammary gland development and polyomavirus middle T antigen induced tumor formation and metastasis. Cancer research. 2005;65:2296–302. doi: 10.1158/0008-5472.CAN-04-3272. [DOI] [PubMed] [Google Scholar]
- 40.Yang L, Huang J, Ren X, Gorska AE, Chytil A, Aakre M, et al. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer cell. 2008;13:23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, et al. TGF-beta-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer cell. 2012;22:291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strainic MG, Shevach EM, An F, Lin F, Medof ME. Absence of signaling into CD4(+) cells via C3aR and C5aR enables autoinductive TGF-beta1 signaling and induction of Foxp3(+) regulatory T cells. Nature immunology. 2013;14:162–71. doi: 10.1038/ni.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlenner SM, Weigmann B, Ruan Q, Chen Y, von Boehmer H. Smad3 binding to the foxp3 enhancer is dispensable for the development of regulatory T cells with the exception of the gut. The Journal of experimental medicine. 2012;209:1529–35. doi: 10.1084/jem.20112646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang X, Poliakov A, Liu C, Liu Y, Deng ZB, Wang J, et al. Induction of myeloid-derived suppressor cells by tumor exosomes. International journal of cancer Journal international du cancer. 2009;124:2621–33. doi: 10.1002/ijc.24249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flavell RA, Sanjabi S, Wrzesinski SH, Licona-Limon P. The polarization of immune cells in the tumour environment by TGFbeta. Nat Rev Immunol. 2010;10:554–67. doi: 10.1038/nri2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.