This work shows that translational regulation of gene expression in a recently formed allopolyploid is widespread, reduces transcriptional differences between the polyploid and its diploid progenitors, and correlates with the retention of genes from an older polyploidy event. These findings suggest that translational regulation is significant in both early and long-term responses to polyploidy.
Abstract
All flowering plants have experienced repeated rounds of polyploidy (whole-genome duplication), which has in turn driven the evolution of novel phenotypes and ecological tolerances and been a major driver of speciation. The effects of polyploidy on gene expression have been studied extensively at the level of transcription and, to a much lesser extent, at the level of the steady state proteome, but not at the level of translation. We used polysome profiling by RNA-Seq to quantify translational regulation of gene expression in a recently formed (∼100,000 years ago) allotetraploid (Glycine dolichocarpa) closely related to the cultivated soybean (Glycine max). We show that there is a high level of concordance between the allopolyploid transcriptome and translatome overall but that at least one-quarter of the transcriptome is translationally regulated. We further show that translational regulation preferentially targets genes involved in transcription, translation, and photosynthesis, causes regional and possibly whole-chromosome shifts in expression bias between duplicated genes (homoeologs), and reduces transcriptional differences between the polyploid and its diploid progenitors, possibly attenuating misregulation resulting from genome merger and/or doubling. Finally, translational regulation correlates positively with long-term retention of homoeologs from a paleopolyploidy event, suggesting that it plays a significant role in polyploid evolution.
INTRODUCTION
Polyploidy (whole-genome duplication [WGD]) is a pervasive feature of plant evolution. Duplication events occurred in the common ancestor of seed plants ∼319 million years ago (MYA), the common ancestor of flowering plants ∼192 MYA, and the common ancestor of core eudicots (the largest group of flowering plants) ∼117 MYA (Tang et al., 2008; Jiao et al., 2011, 2012). The majority of core eudicot lineages, and most major crops, have subsequently experienced one or more additional polyploidy events (Bowers et al., 2003; Blanc and Wolfe, 2004a; Schlueter et al., 2004; Jiao et al., 2011, 2012). Polyploidy has generated a wide range of novel and/or transgressive traits (Levin, 1983; Warner and Edwards, 1993; Pires et al., 2004; Ni et al., 2009; Jiao et al., 2011; Coate et al., 2012, 2013; Ilut et al., 2012), is a common mechanism of plant speciation (Wood et al., 2009), and may have played a central role in flowering plant diversification (Fawcett et al., 2009; Jiao et al., 2011).
Consequently, much effort has been invested in understanding how genome duplication, complicated by genome merger in polyploids formed through hybridization (allopolyploids), functions to give rise to new traits. A central focus of this effort has aimed at understanding the regulation of gene expression in massively duplicated genomes (Guo et al., 1996; Adams et al., 2003; Albertin et al., 2006, 2007; Wang et al., 2006; Flagel et al., 2008, 2009; Hegarty et al., 2008; Ni et al., 2009; Buggs et al., 2010; Coate and Doyle, 2010; Schnable et al., 2011; Ilut et al., 2012; Coate et al., 2013a). The vast majority of polyploid expression studies have measured transcript abundance, leaving open the question of how regulatory steps downstream of transcription impact these recurring patterns of gene expression. Variations in transcript abundance are not always biologically meaningful, and many functional patterns of gene expression are established posttranscriptionally (Keene, 2007; Joshi et al., 2011). Translation represents a major regulatory step in the gene expression pathway (Kawaguchi and Bailey-Serres, 2002; Schwanhäusser et al., 2011; Vogel and Marcotte, 2012). In plants, translational regulation plays a significant role in cell type–specific gene expression (Jiao and Meyerowitz, 2010; Mustroph et al., 2009; Mustroph and Bailey-Serres, 2010) and is pervasive in response to a wide range of stimuli (Kawaguchi et al., 2004; Branco-Price et al., 2005, 2008; Mustroph et al., 2009; Juntawong and Bailey-Serres, 2012; Park et al., 2012; Reynoso et al., 2012). Translation is often regulated in a sequence-specific manner (Sonenberg and Hinnebusch, 2009), and extensive posttranscriptional regulation of allele-specific expression has been demonstrated in yeast hybrids (Khan et al., 2012).
A handful of studies in polyploids have examined expression at the protein level (Albertin et al., 2006, 2007; Hu et al., 2011, 2013; Yao et al., 2011), but due to both technical and biological constraints, these studies have provided data on homoeolog-specific expression for only a small number of genes (Hu et al., 2011, 2013; Koh et al., 2012). Additionally, protein abundance is the net result of transcription, translation, and other regulatory steps (e.g., mRNA degradation) and does not therefore provide direct information about the contributions of these steps individually. Thus, although translational regulation likely represents a biologically and evolutionarily significant aspect of polyploid gene expression, it remains largely unexplored.
Polysome profiling takes advantage of the fact that transcripts bound by polyribosomes (polysomes) migrate further than unbound transcripts when centrifuged through a Suc density gradient. Consequently, polysomal RNA (which we define as the translatome) can be separated from nonpolysomal RNA. The concentration of transcripts from any given gene can then be compared between the two fractions, with differences indicating translational regulation of gene expression. Thus, polysome profiling is a relatively straightforward method for characterizing the translatome, providing a direct measure of translational regulation, and, when coupled with RNA-Seq, enabling high-throughput quantification of homoeolog usage. To determine the extent, nature, and significance of translational regulation in allopolyploidy, we performed polysome profiling with RNA-Seq on leaf tissue of Glycine dolichocarpa, an allotetraploid species related to soybean (Glycine max) formed via hybridization between the diploid species Glycine syndetika and Glycine tomentella ∼100,000 years ago (Doyle et al., 2004). We show that the translatome of G. dolichocarpa is highly correlated with its transcriptome but that nearly 25% of genes are significantly regulated at the translational level. We further show that translational regulation differs by transcript abundance and function of the underlying genes, causes regional and possibly whole-chromosome shifts in homoeolog expression bias, and reduces deviations between the transcriptomes of G. dolichocarpa and its diploid progenitors. Finally, we show that translational regulation correlates positively with long-term retention of homoeologous genes, suggesting that translational regulation plays a significant role in the evolution of polyploid genomes.
RESULTS
To estimate the extent to which gene expression is translationally regulated in an allotetraploid (G. dolichocarpa = T2), we isolated total mRNA, polysomal mRNA, and nonpolysomal mRNA from leaf tissue and performed RNA-Seq on each fraction. Total mRNA (designated “T”) is all of the mRNA in a cell (the transcriptome). Polysomal RNA (P) is the fraction of the transcriptome associated with polysomes (the translatome). Nonpolysomal RNA (NP) is the fraction of mRNA not associated with polysomes; thus, the fraction of standing mRNA not contributing to the translatome (Table 1 summarizes the abbreviations used frequently in this article). We also performed RNA-Seq on the leaf transcriptomes of each diploid progenitor species (G. tomentella [D3] and G. syndetika [D4]). We used these data to quantify and characterize translational regulation of the combined T2 transcriptome (combining the D3 and D4 homoeologous subtranscriptomes; “combined expression”), as well as translational regulation of each homoeologous subtranscriptome (“homoeolog expression”). In total, we analyzed two biological replicates of the T2 transcriptome, three biological replicates each of T2 polysomal RNA and T2 nonpolysomal RNA, and three biological replicates each of the D3 and D4 diploid progenitor transcriptomes.
Table 1. Abbreviations Used.
| Abbreviation | Definition |
|---|---|
| T2 | The allopolyploid, G. dolichocarpa |
| D3 | Diploid progenitor, G. tomentella |
| D4 | Diploid progenitor, G. syndetika |
| T | Total mRNA (the transcriptome) |
| P | Polysomal mRNA (the translatome) |
| NP | Nonpolysomal mRNA |
| PTR | Positive translational regulation |
| PTRD3 | PTR of the D3 homoeolog |
| PTRD4 | PTR of the D4 homoeolog |
| NTR | Negative translational regulation |
| NTRD3 | NTR of the D3 homoeolog |
| NTRD4 | NTR of the D4 homoeolog |
| DDP | Diploid distinguishing polymorphism |
| RPKM | Reads per kilobase per million reads |
| RPKMD3 | RPKMs derived from the D3 homoeolog |
| RPKMD4 | RPKMs derived from the D4 homoeolog |
Combined Expression
Reads were mapped to 40,553 genes in at least one of the three mRNA fractions (87% of the 46,430 high confidence gene models in the G. max reference genome, Glyma 1.0; Schmutz et al., 2010), with 34,197 genes (74%) expressed in all three fractions. Normalized expression estimates (reads per kilobase per million mapped reads [RPKM]) were highly correlated among replicates, though correlations were slightly higher in P (r2 = 0.93 to 0.98) than in NP (r2 = 0.93 to 0.95) and considerably higher than in T (r2 = 0.84) (Supplemental Figure 1). This is consistent with previous studies showing that transcription is more stochastic than translation and that translational regulation serves to remove biological noise inherent at the transcriptional level (Keene, 2007, Joshi et al., 2011).
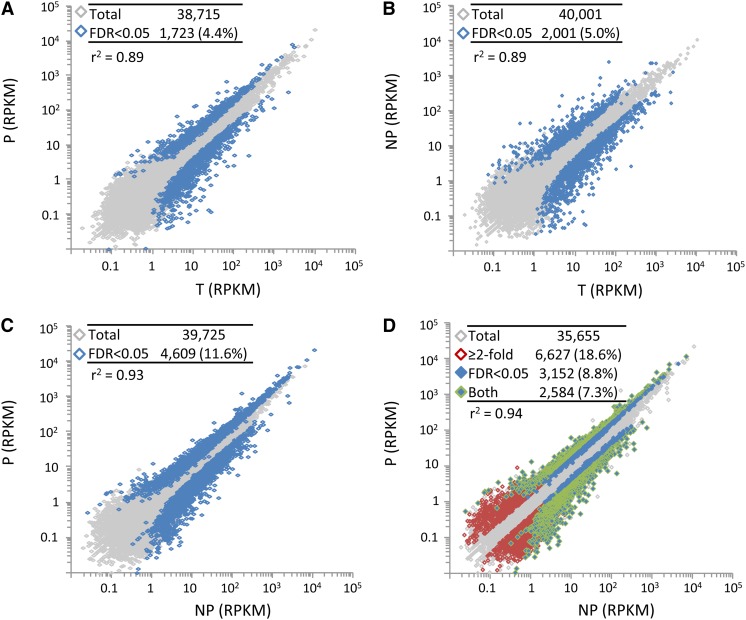
Overall, expression profiles were also highly correlated among the three RNA fractions (r2 ≥ 0.89; Figure 1). Globally, therefore, the translatome closely reflects the transcriptome. Nonetheless, 4 to 12% of genes differed in transcript concentration (false discovery rate [FDR] < 0.05) in each pairwise comparison (Figures 1A to 1C), indicating translational regulation for a subset of genes. We defined genes as positively translationally regulated (PTR) if transcript abundance was significantly higher in P than in NP with an intermediate value in T. Similarly, we defined genes as negatively translationally regulated (NTR) if transcript abundance was significantly higher in NP than in P with an intermediate value in T. By these criteria, 8.8% of genes were translationally regulated, with 1224 genes (3.4%) exhibiting positive translational regulation and 1928 genes (5.4%) exhibiting negative translational regulation (Figure 1D). Using more relaxed criteria of a 2-fold expression difference between P and NP (regardless of statistical significance), with an intermediate expression level in T, 18.6% of genes were translationally regulated (2152 genes [6.0%] PTR and 4475 [12.6%] NTR). Requiring that the difference in expression between P and NP be significant (FDR < 0.05) and ≥2-fold, 7.3% of genes were translationally regulated (881 [2.5%] PTR and 1703 [4.8%] NTR). Thus, roughly 7 to 19% of the T2 combined transcriptome was translationally regulated, of which approximately one-third were PTR and two-thirds were NTR.
Figure 1.
The T2 Transcriptome and Translatome Are Highly Correlated for Combined Expression.
Scatterplots of expression estimates (RPKM) for pairwise comparisons between the three mRNA pools. Tables indicate the total number of genes plotted (“Total”; shown in gray) and the number of genes that are differentially expressed at FDR < 0.05 (“FDR < 0.05”; shown in blue). In (D), genes with a ≥2-fold difference in expression (regardless of significance) are shown with red outline, and genes with FDR < 0.05 and with ≥2-fold difference in expression are shown with blue fill and green outline.
(A) Polysomal (P) versus total (T).
(B) Nonpolysomal (NP) versus total (T).
(C) Polysomal (P) versus nonpolysomal (NP).
(D) Polysomal (P) versus nonpolysomal (NP), excluding genes with no expression detected in T.
Homoeolog Expression
Based on RNA-Seq libraries from three D3 diploid accessions and three D4 diploid accessions, we identified sites that differed between the two diploid progenitor species of T2, which we call diploid distinguishing polymorphisms (DDPs; Ilut et al., 2012). Sites that were polymorphic within a diploid species were excluded from the DDP set. Wherever reads from T2 overlapped these sites at a read depth ≥5, we calculated the fraction of reads derived from each homoeolog. For each gene, we estimated homoeolog usage by calculating the weighted average of all DDPs in that gene.
In total, we identified 190,374 DDPs, of which 166,496 were distributed across 27,132 genes (an average of 6.1 DDPs/gene) and 23,878 were annotated as intergenic. To focus on genes with the most reliable estimates of homoeolog usage, we restricted our analysis to the subset for which estimates were obtained in all biological replicates of all three RNA fractions (T, P, and NP). This subset consisted of 3874 genes (hereafter referred to as the “homoeolog gene set”). Within this gene set, homoeolog usage estimates were derived from 31,154 DDPs (8.1 DDPs/gene on average). We observed both D3 and D4 sequences in 28,503 out of 31,154 DDPs (91.5%) and in 3862 out of 3874 homoeolog genes (99.7%) in at least one of the three T2 RNA fractions. This confirms that the vast majority of DDPs represent conserved differences between the D3 and D4 diploids that were inherited by T2. Estimates of homoeolog usage (fraction of reads) were multiplied by the total read count for each of the 3874 homoeolog genes to obtain homoeolog-specific read counts and RPKMs (RPKMD3 and RPKMD4). As with combined expression, biological replicates were highly correlated for homoeolog expression in P and NP (r2 ≥ 0.88) and to a lesser extent in T (r2 = 0.82).
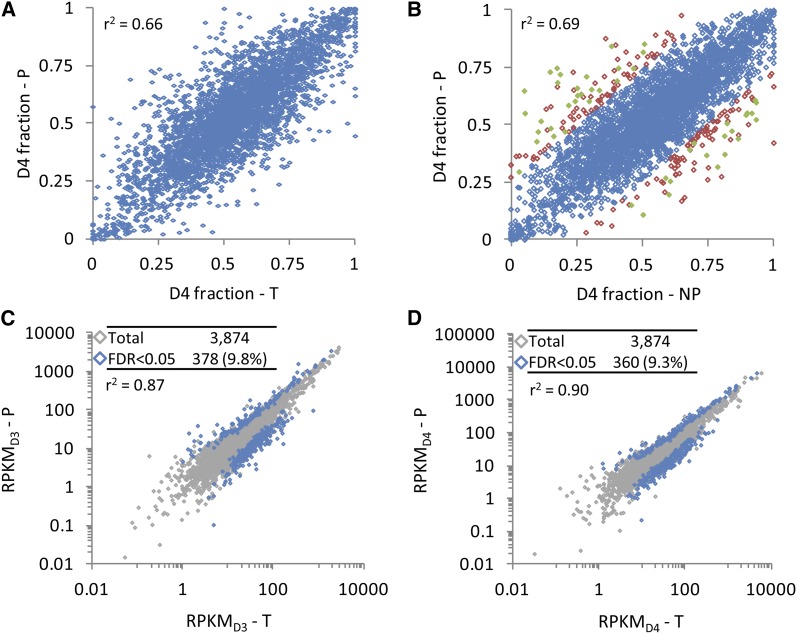
As with combined expression, the translatome was highly correlated with the transcriptome for relative expression of homoeologs (r2 ≥ 0.66; Figures 2A and 2B) and for homoeolog-specific RPKM (r2 ≥ 0.87; Figures 2C and 2D). Of 632 genes exhibiting a significant bias in homoeolog usage (RPKMD3 ≠ RPKMD4) in T (FDR < 0.05), 600 (94.9%) showed usage bias in the same direction in P and NP (Supplemental Figure 2). This bias was significant (FDR < 0.05) for 566 genes in P (89.6%), 534 genes in NP (84.5%), and 511 genes in both P and NP (80.9%). Only one gene exhibited significant homoeolog bias in T and significant opposite bias in P (Glyma11g13960; D4/D3 = 0.24 in T and 2.29 in P). Thus, extreme translational regulation of homoeolog usage was rare, and homoeolog usage at the translational level generally reflected homoeolog usage at the transcriptional level.
Figure 2.
The T2 Transcriptome and Translatome Are Highly Correlated for Relative Homoeolog Usage and for Expression within Each Subgenome.
(A) Scatterplot of relative homoeolog expression (proportion of combined expression derived from the D4 homoeolog) in the homoeolog gene set (n = 3874) for the polysomal (P) versus total (T) mRNA fractions.
(B) Scatterplot of relative homoeolog expression in the polysomal (P) versus nonpolysomal (NP) mRNA fractions. A total of 3669 of 3874 genes (94.7%) showed a shift in relative homoeolog usage of <25% (blue diamonds); of the remaining 205 genes (5.3%) that showed a ≥25% shift in relative homoeolog expression (red or green diamonds), 48 (1.2%; green diamonds) showed a statistically significant shift (translational regulation of relative homoeolog usage; TRRHU).
(C) Scatterplot of D3 homoeolog expression (RPKMD3) in P versus T.
(D) Scatterplot of D4 homoeolog expression (RPKMD4) in P versus T.
Tables in (C) and (D) indicate total number of genes plotted (“Total”; shown in gray) and the number of genes that are differentially expressed at FDR < 0.05 (“FDR < 0.05”; shown in blue).
Nonetheless, as with combined expression, we observed several cases of translational regulation of homoeolog usage. Such regulation can take two forms: (1) shifts in the proportion of combined expression derived from each homoeolog (D4/D3 homoeolog ratio = relative regulation; Figure 2B) and (2) absolute increases or decreases in subgenome-specific transcript abundance (P versus NP expression for D3 or D4 homoeolog = absolute regulation; Figures 2C and 2D).
We identified 48 genes (1.2%) exhibiting significant translational regulation of relative homoeolog usage (Figure 2B; Supplemental Data Set 1). For 28 genes, the proportion of D4 homoeolog transcripts increased in the polysomal fraction versus the transcriptome, and for 20 genes, the proportion of D4 homoeolog transcripts decreased. Using more relaxed criteria of a ≥25 percentage point shift regardless of statistical significance (e.g., polysomal D4 fraction = 40%; nonpolysomal D4 fraction = 65%), 205 genes (5.3%) were translationally regulated (Figure 2B). Thus, translational regulation of relative homoeolog usage (1 to 5%) appears to be less common than translational regulation of combined expression (7 to 19%).
We detected more cases of absolute regulation within each subgenome (significant differences in RPKMD3 and/or RPKMD4 between P and NP, with an intermediate value in T; Figures 2C and 2D). A total of 378 genes (9.8% of the homoeolog gene set) exhibited translational regulation of the D3 homoeolog. Of these, 148 (39.2%) were positively regulated (exhibited an enrichment of the D3 homoeolog in P versus NP; designated “PTRD3”), and 230 (60.8%) were negatively regulated (exhibited a depletion of the D3 homoeolog in P versus NP; “NTRD3”). Similarly, 360 genes (9.3%) exhibited translational regulation of the D4 homoeolog, of which 132 (36.7%) were positively regulated (PTRD4) and 228 (63.3%) were negatively regulated (NTRD4). Thus, the extent of translational regulation was roughly equal between the two homoeologous transcriptomes and similar to that of combined expression. Additionally, as with combined expression, negative TR was more prevalent than positive TR within subtranscriptomes.
Translational Regulation of the T2 Transcriptome Is Widespread
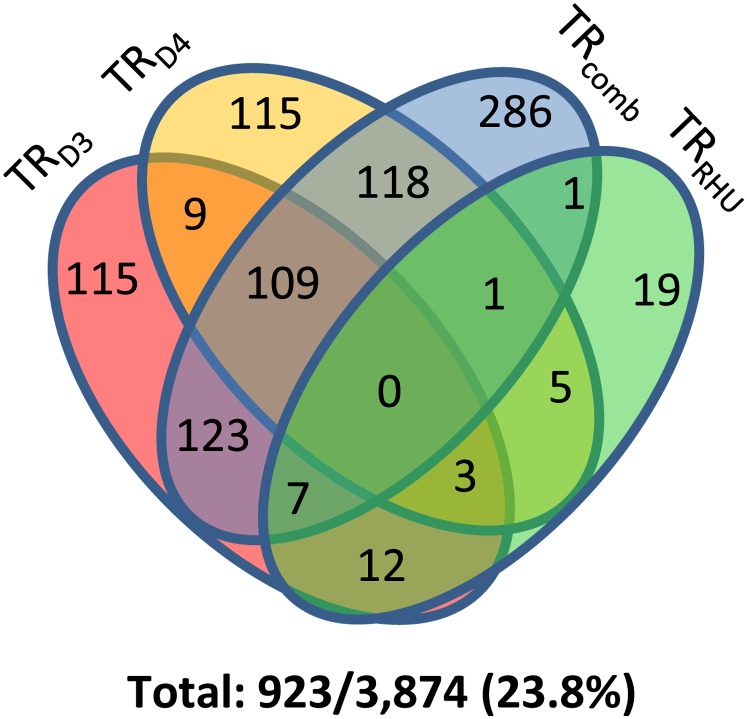
Within the homoeolog gene set, we detected one or more forms of translational regulation in 923 out of 3874 genes (23.8%; Figure 3). This most likely represents a minimum estimate of the true extent of translational regulation as it is likely that we would have detected additional cases with more biological replication. Though the homoeolog gene set includes only 8.3% of the high confidence genes in the soybean genome, the fraction of these exhibiting translational regulation of combined expression (645/3874; 16.6%) is similar to our estimate from the full gene set (7.3 to 18.6%), suggesting that the homoeolog genes are representative of the whole transcriptome. Thus, although there was a high level of concordance overall between the T2 transcriptome and translatome, we estimate that at least one-quarter of all genes are translationally regulated in one form or another. A total of 637 genes in the homoeolog gene set (16.4%) exhibited translational regulation at the homoeolog level (Figure 3). Extrapolating to the whole genome, we infer that ∼7600 genes (16.4% of the high confidence gene models) are translationally regulated at the homoeolog level in T2.
Figure 3.
Nearly One-Quarter of the Genes in the Homoeolog Gene Set Demonstrate Translational Regulation in T2.
Venn diagram of translationally regulated genes in the homoeolog gene set (n = 3874). TRD3, translational regulation of the D3 homoeolog; TRD4, translational regulation of the D4 homoeolog; TRcomb, translational regulation of combined (D3+D4) expression; TRRHU, translational regulation of relative homoeolog usage.
The fact that several hundred genes exhibited absolute translational regulation within subgenomes, but only 48 exhibited significant shifts in relative homoeolog usage, indicates that the two subgenomes are, in most cases, translationally regulated in concert. Indeed, 84.1% of genes exhibiting translational regulation of absolute homoeolog usage in one or both subgenomes responded in the same direction in both (Table 2). Additionally, only four genes exhibited significant opposite regulation between the two subgenomes in T2, each involving PTRD4 and NTRD3. Together, these data suggest that translational regulation tends to affect both homoeologous transcriptomes equally.
Table 2. Homoeologs Are Translationally Regulated in Concert.
| Homoeolog 2 |
|||
|---|---|---|---|
| Homoeolog 1 | Total | Samea | Oppositeb |
| PTR | 242 | 210 (86.8%) | 32 (13.2%) |
| NTR | 375 | 309 (82.4%) | 39 (10.4%) |
| Total | 617 | 519 (84.1%) | 71 (11.5%) |
For genes that are PTR or NTR for at least one homoeolog, the distribution of genes showing the same or opposite translational regulation of the other homoeolog. Values in parentheses indicate the number of genes exhibiting the specified form of translational regulation.
Both homoeologs are translationally regulated in the same direction (positive or negative).
D3 and D4 homoeologs are translationally regulated in opposite directions (one positive and the other negative).
Of the 48 genes exhibiting significant translational regulation of relative homoeolog usage (cis-translational regulation), 28 were also significantly translationally regulated in terms of absolute homoeolog usage, with 23 exhibiting NTR of one or the other homoeolog and only eight exhibiting PTR of one or the other homoeolog (three were PTRD4 and NTRD3). This is consistent with the observation that NTR is more prevalent than PTR, both for combined expression and for absolute regulation of homoeologs.
Translational Regulation Differs by Transcript Abundance
Mean expression in T for the PTR gene set was 111.7 RPKM, compared with 18.7 RPKM for the full set of genes expressed in T, and 11.7 RPKM for the NTR gene set. Median expression was 23.2 RPKM, 3.9 RPKM, and 5.4 RPKM for PTR, T, and NTR, respectively. Thus, the distribution of expression levels was shifted upwards for PTR genes relative to NTR (Supplemental Figure 3A). Comparable patterns were observed for each subtranscriptome (Supplemental Figures 3B and 3C). Therefore, it appears that translational regulation in T2 tends to promote protein synthesis from highly transcribed genes and to suppress protein synthesis from genes that are transcribed at lower levels.
Similar to the genes that were NTR for absolute expression, genes that exhibited regulation of relative homoeolog usage were generally transcribed at a lower level than those that did not (Supplemental Figure 3D). This is consistent with the observation that translational regulation of relative homoeolog usage is usually achieved via negative regulation of one of the two homoeologs. Expression levels were similar for genes in which translation favored D3 (mean = 34.4 RPKM, median = 23.0 RPKM, n = 20) or D4 (mean = 34.7 RPKM, median = 22.6 RPKM, n = 28).
Translational Regulation Differs by Gene Ontology and Subcellular Localization
Gene Ontology enrichment analysis indicated that the PTR gene sets (PTR, PTRD3, and PTRD4) were enriched for genes encoding ribosomal proteins or otherwise functioning in translation and depleted for genes functioning in transcription, whereas the NTR gene sets (NTR, NTRD3, and NTRD4) showed the opposite pattern (Supplemental Figure 4 and Supplemental Data Set 2). Consistent with these molecular process terms, the PTR sets were also enriched for genes encoding cytoplasmic proteins, whereas the NTR sets were enriched for genes encoding proteins localized to the nucleus. The PTR gene sets were also enriched for genes encoding plastid proteins, including genes involved in photosynthesis, whereas plastid- and photosynthesis-related Gene Ontology terms were underrepresented in the NTR gene sets (Supplemental Figure 4). Accordingly, targeting predictions from Predotar (http://urgi.versailles.inra.fr/predotar/predotar.html) indicated that the PTR gene set is enriched for genes targeted to plastids: 25.2% of the PTR genes are predicted to be chloroplast targeted compared with 7.8% of the high-confidence soybean genes (χ2 = 480, df = 1, P < 0.001) and 6.7% of the NTR genes (χ2 = 212.0, df = 1, P < 0.001).
Translational Regulation Has No Net Effect on D4 Genome Expression Dominance
Across the homoeolog gene set, mean homoeolog usage was biased slightly toward D4 in all three RNA fractions: 53.6% in T and P, and 53.7% in NP (Supplemental Figure 5A). By contrast, mean expression was 49.9% D4 at the diploid level (obtained by dividing expression [RPKM] in the D4 diploid by the sum of expression in D3 and D4). Thus, D4 homoeolog usage was significantly higher in all three tetraploid fractions (T, P, and NP) than is predicted by combining the diploid transcriptomes in silico (P < 0.001; t test). Similarly, all three fractions exhibited comparable numbers of genes with a D4 expression bias (χ2 ≤ 0.192, df = 1, P ≥ 0.662) and significantly more than expected based on the combined diploid transcriptomes (χ2 ≥ 49.9, df = 1, P < 0.001; Supplemental Figure 5B). Because the D4 homoeolog fractions in P and NP were comparable to that in T, there does not appear to be any global translational bias in homoeolog usage, consistent with our conclusion that translational regulation predominantly affects both homoeologs equally.
Translational Regulation Has Regional Effects on Homoeolog Dominance
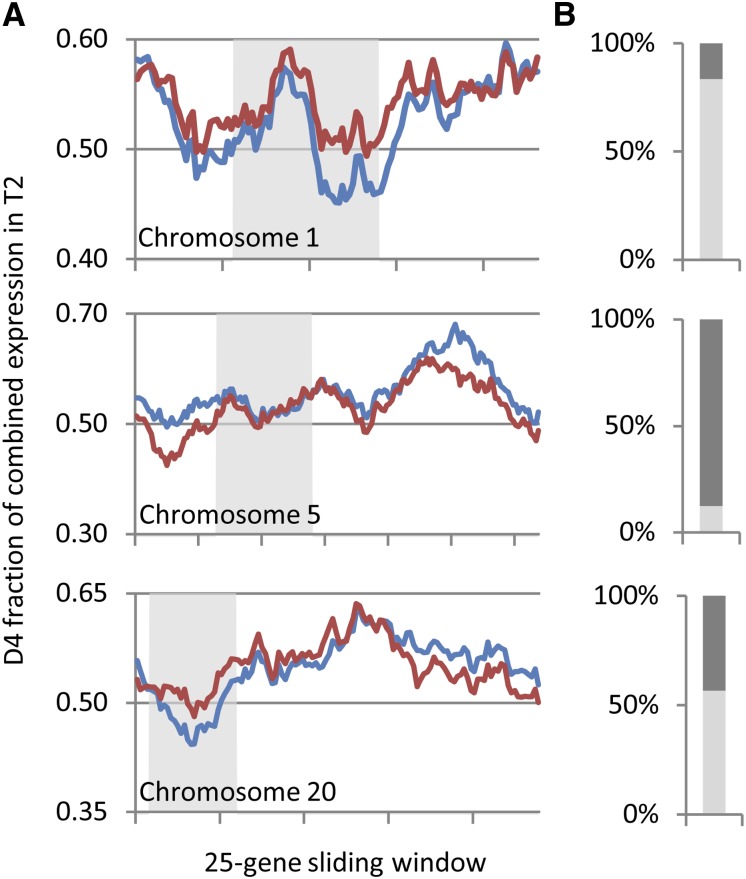
Unbalanced homoeolog expression bias at the transcriptional level, like that favoring D4 homoeologs in T2, has been observed in other allopolyploids (Flagel et al., 2008, 2009; Innes et al., 2008; Buggs et al., 2010; Lin et al., 2010; Schnable et al., 2011; Freeling et al., 2012; Grover et al., 2012) and has been proposed to facilitate biased fractionation (the disproportionate loss of genes from one homoeologous genome versus the other; Freeling and Thomas, 2006; Schnable et al., 2011; Freeling et al., 2012). Averaging D4 homoeolog usage over 25-gene windows along chromosomes of the soybean reference genome, we observed nearly uniform D4 homoeolog expression dominance at the transcriptional level (T) over most of the length of all 20 chromosomes (Figure 4; Supplemental Figure 6). The D4 homoeolog contributed >50% of combined expression in 2734 out of 3389 25-gene windows (80.7%).
Figure 4.
Translational Regulation Exerts Regional Effects on Homoeolog Usage in T2.
(A) Average D4 homoeolog fraction of combined expression in the T2 transcriptome (T; blue line) and translatome (P; red line) in 25-gene sliding windows across chromosomes 1, 5, and 20. Tick marks on the x axis indicate 25-gene increments (starting from the 25th gene). Gray shading delineates sliding windows in which the majority of genes are located in pericentromeric regions defined by Du et al. (2012).
(B) Percentage of 25-gene windows in which the D4 fraction is increased in P relative to T (light gray) or in which the D4 homoeolog fraction is decreased in P relative to T (dark gray).
Though the D4 bias was not amplified at the level of translation overall, we looked to see if physically linked genes show similar patterns of bias that might suggest translational regulation favors one subgenome over the other in localized genomic neighborhoods. Translational regulation acted to shift expression toward the D4 homoeolog across the majority of chromosomes 1 (Figure 4), 4, 16, and 18 (Supplemental Figure 6). By contrast, translation shifted expression toward the D3 homoeolog for most of chromosomes 5 (Figure 4) and 14 (Supplemental Figure 6). On chromosome 20, translation increased D4 homoeolog bias across 68 of the first 74 25-gene windows (covering ∼80% of the length of the chromosome) and decreased D4 bias across the remaining 46 25-gene windows (Figure 4). Similar mixed patterns were observed for the remaining chromosomes (2, 3, 6 to 13, 15, 17, and 19; Supplemental Figure 6). It is likely that genomic rearrangements have occurred between T2 (and/or its diploid progenitors) and the soybean reference genome, such that not all genes physically linked in soybean are linked in T2. Some of the mixed patterns observed here (e.g., chromosome 20) could therefore be an artifact of such rearrangements. Overall, however, synteny is likely to be largely conserved between T2 and soybean. Genetic linkage maps of nine chromosomes in Glycine latifolia, which is equally diverged from soybean as is T2, were nearly collinear with their homoeologous soybean chromosomes (Chang et al. 2013). Thus, translational regulation appears to have regional and/or chromosomal effects on expression bias, in some cases favoring the D3 subgenome, and in other cases favoring the D4 subgenome.
Because translational regulation generally promotes the expression of highly transcribed genes (Supplemental Figure 3), one possibility is that these regional effects are simply a byproduct of translation favoring the more highly transcribed homoeolog. This does not appear to be the case, however, because translation frequently shifted homoeolog usage in favor of the less transcribed homoeolog (Figure 4). For example, translation increased the D4 fraction of combined expression on chromosome 1 even where the D4 homoeolog was transcribed at a lower level than the D3 homoeolog. Additionally, despite modest effects on homoeolog usage overall, translational regulation reversed transcriptional bias (changed which homoeolog was expressed at >50% of combined expression) in 525 out of 3389 25-gene windows (15.5%) and in 773 out of 3874 genes (20.0%). Shifts toward the D3 homoeolog (310 25-gene windows) were more common than shifts toward the D4 homoeolog (215 25-gene windows) in absolute terms (P < 0.0001; two-tailed binomial test), though less frequent as a proportion of possible 25-gene windows (11.3% of 25-gene windows with D4 transcriptional dominance compared with 32.8% of 25-gene windows with D3 transcriptional dominance; χ2 = 186.0, df = 1, P < 0.001).
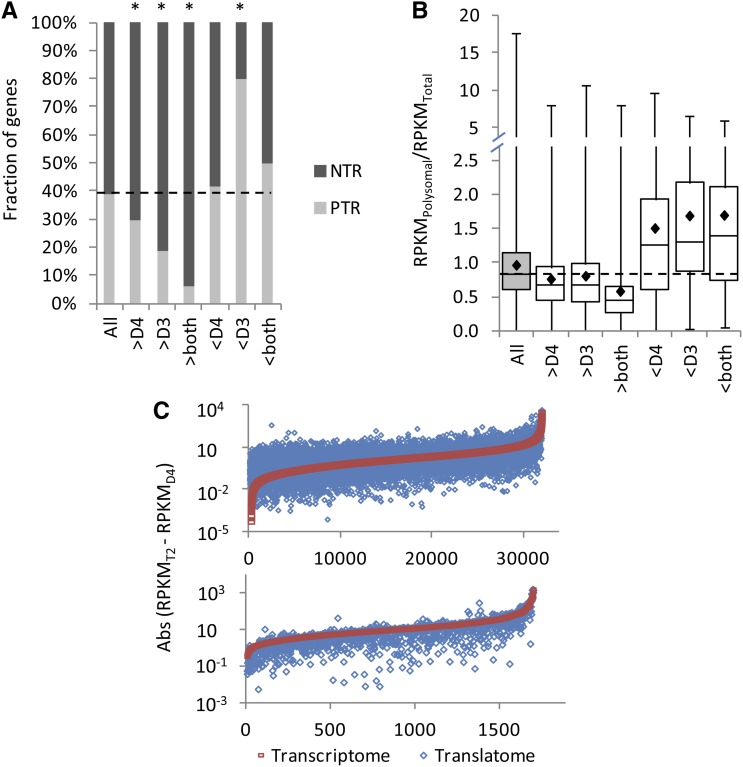
Translational Regulation Reduces Transcriptional Differences between T2 and Its Diploid Progenitors
We quantified the leaf transcriptomes of the diploid progenitor species of T2 (D3 and D4) by RNA-Seq. For genes exhibiting significant differences in combined expression at the transcriptional level between T2 and one or both diploids (FDR < 0.05), translation generally acted to reduce these differences. Genes that were transcribed at a significantly higher level in T2 than in the D3 and/or D4 diploids were more likely to be NTR than the genome-wide average (Figure 5A). Conversely, genes that were transcribed at a significantly lower level in T2 than in D3, and to a lesser extent D4, were more likely to be PTR than the genome-wide average (Figure 5A). A similar picture is observed when expression levels of individual genes are considered: genes that were overtranscribed in T2 versus either diploid were translationally repressed (lower RPKMP/RPKMT) relative to the whole genome, and genes that were undertranscribed in T2 versus either diploid exhibited enhanced translation (higher RPKMP/RPKMT) relative to the genome-wide average (Figure 5B). As a result, absolute differences in RPKM between T2 and either diploid were generally reduced in the T2 translatome relative to the T2 transcriptome (Figure 5C; Supplemental Table 1).
Figure 5.
Translational Regulation Reduces Differences in Combined Transcription between T2 and Its Diploid Progenitors.
(A) Fraction of translationally regulated genes that are PTR and NTR for the full genome (“All”) and for genes that are differentially transcribed in T2 versus one or both of its diploid progenitors (e.g., “>D4” indicates that transcript abundance [RPKM] in T2 is significantly higher than in the D4 diploid; “<D4” indicates that transcript abundance in T2 is significantly lower than in the D4 diploid). Asterisks indicate a ratio significantly different than the genome-wide ratio (χ2 P value < 0.05).
(B) Combined expression in polysomal mRNA (translatome) divided by combined expression in total mRNA (transcriptome) in T2 for the full genome and for the subsets of genes that are differentially transcribed between T2 and its progenitors. Black diamonds represent sample means. Error bars indicate minimum and maximum values.
(C) Scatterplot of differences in expression between T2 (transcriptome or translatome) and the D4 diploid transcriptome (absolute value [RPKMT2 − RPKMD4]) for the whole genome (top) and for genes that are differentially transcribed between T2 and D4 (bottom). The smaller the difference between T2 and D4, the closer the data point is to the x axis. Genes are ordered by differences between the T2 and D4 transcriptomes (red squares). Blue diamonds show the corresponding difference between the T2 translatome and D4 transcriptome. The differences between the T2 translatome and D4 are greater than or equal to the differences between the T2 transcriptome and D4 for the majority of genes overall (63.6%; blue diamonds at or above the red curve). By contrast, for genes that are differentially transcribed between T2 and D4, differences are smaller in the T2 translatome for the majority of genes (63.5%; blue diamonds below the red curve). Similar patterns were observed for T2 versus D3 and T2 versus D3 and D4 (data not shown).
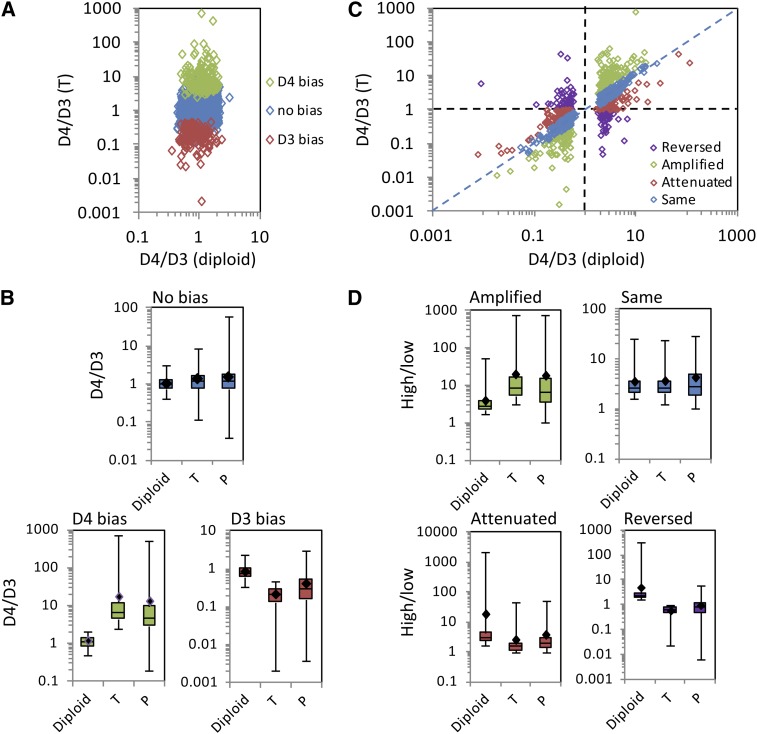
Translational regulation also tended to modulate homoeolog expression ratios to reflect diploid expression ratios more closely. Of the 3874 genes in the homoeolog gene set, 3044 were transcribed at equivalent levels in the D3 and D4 diploids. Of these, homoeolog usage within the T2 transcriptome was balanced for 2705 genes (88.9%) and deviated from equal expression for 339 genes (11.1%), with 163 showing a D3 homoeolog bias and 176 showing a D4 homoeolog bias (Figure 6A). For those genes with balanced homoeolog usage in T, consistent with equal expression at the diploid level, translational regulation had no directional effect on D4/D3 homoeolog ratios (Figure 6B). By contrast, for genes in which a homoeolog bias was introduced in the T2 transcriptome, translational regulation partially restores the balance in homoeolog expression (moved the D4/D3 ratio closer to 1; Figure 6B). The homoeolog ratio was more diploid-like in P than in T for 215 of 339 genes with a homoeolog bias in T (P < 0.0001; two-tailed binomial probability assuming an equal chance for P to be more or less diploid-like than T).
Figure 6.
Translational Regulation Reduces Differences in Homoeolog Usage between T2 and Its Diploid Progenitors.
(A) D4/D3 homoeolog ratios in the T2 transcriptome are plotted against D4/D3 diploid ratios for genes that are equally transcribed in the D3 and D4 diploids (n = 3044). Genes in green show significantly higher expression of the D4 homoeolog than of the D3 homoeolog (“D4 bias”) in the T2 transcriptome. Genes in red show significantly higher expression of the D3 homoeolog than of the D4 homoeolog (“D3 bias”) in the T2 transcriptome. Genes in blue show equal expression of D3 and D4 homoeologs in T2 (“No bias”), consistent with equal expression between D3 and D4 diploids.
(B) Box-and-whisker plots showing D4/D3 ratios in the diploids, the T2 transcriptome (T), and the T2 translatome (P) for each category (no bias, D4 bias, and D3 bias) shown in (A).
(C) D4/D3 homoeolog ratios in the T2 transcriptome are plotted against D4/D3 diploid ratios for genes that are differentially transcribed between D3 and D4 diploids (FDR < 0.05; n = 830). Genes are color coded based on whether the T2 transcriptome preserves (“Same”), amplifies, attenuates, or reverses the D4/D3 ratio relative to the diploids.
(D) Box-and-whisker plots showing ratios (highest expressed at the diploid level/lowest expressed at the diploid level) in the diploids, the T2 transcriptome (T), and the T2 translatome (P) for each category (amplified, same, attenuated, and reversed) shown in (C). In each case where homoeolog usage in the T2 transcriptome differs from relative expression in the diploids, translational regulation partially restores homoeolog usage to a more diploid-like level.
Conversely, 830 genes differed significantly in transcript abundance between diploid D3 and diploid D4 (FDR < 0.05), with 408 more highly expressed in D3 and 422 more highly expressed in D4. For these 830 genes, 128 homoeolog pairs (15.4%) exhibited significant bias (FDR < 0.05) in the T2 transcriptome that was comparable to the bias at the diploid level (<1.5-fold difference between the diploid D4/D3 ratio and the D4/D3 ratio in T; Figure 6C). Of the remaining genes, homoeolog bias was amplified in 213 (25.7%), attenuated in 176 (21.2%), and reversed in 125 (15.1%) relative to differences at the diploid level (Figure 6C). For the 128 genes showing a D4/D3 homoeolog ratio in T comparable to that of the diploids, the translatome had no directional effect on homoeolog usage (Figure 6D). By contrast, for genes in which the homoeolog ratio in T deviated from the diploid ratio, translational regulation tended to partially restore homoeolog bias to a more diploid-like level (Figure 6D). The homoeolog ratio was more diploid-like in P than in T for 321/514 genes (62.5%) among these genes (P < 0.0001; two-tailed binomial probability).
Overall, therefore, there was a propensity for translational regulation to partially restore homoeolog expression ratios in T to a more diploid-like level (Figures 6B and 6D). This was true whether or not genes were differentially expressed at the diploid level and regardless of the direction in which homoeolog usage deviated from relative expression in the diploids.
Positive Translational Regulation Correlates with Greater Retention of Homoeologs from the Most Recent Glycine Paleopolyploidy Event
The common ancestor of T2, D3, D4, and G. max experienced a WGD event 5 to 13 MYA (Shoemaker et al., 2006; Schmutz et al., 2010; Doyle and Egan, 2010; defined as the “A” WGD event in Coate et al., 2011). Based on conserved synteny within the G. max genome, Du et al. (2012) identified genes that have retained both homoeologs (“deletion-resistant” [delR], following Paterson et al., 2006) and genes that have fractionated (“duplication-resistant” [dupR]) following the “A” duplication.
Overall, translationally regulated genes were more likely to have retained duplicates from the “A” duplication event than the genome-wide average (Table 3), with PTR genes exhibiting slightly higher retention levels than NTR genes. With the exception of the genes that were NTR for combined expression, all sets of translationally regulated genes had significantly higher retention levels than the whole genome for “A” homoeolog pairs.
Table 3. Distribution of Retained Homoeologs (DelR) and Singletons (DupR) in the Soybean Genome from the Glycine “A” Polyploidy Event by Type of Translational Regulation.
| Gene Class | Gene Count | % of Total |
|||||
|---|---|---|---|---|---|---|---|
| DelRa | DupRb | Total | DelR | DupR | χ2 | P Value | |
| All | 28,066 | 12,965 | 41,031 | 68.4 | 31.6 | – | – |
| PTR | 876 | 296 | 1,172 | 74.7 | 25.3 | 21.3 | <0.001 |
| PTRD3 | 117 | 28 | 145 | 80.7 | 19.3 | 10.1 | 0.001 |
| PTRD4 | 98 | 29 | 127 | 77.2 | 22.8 | 4.5 | 0.034 |
| NTR | 1,242 | 530 | 1,772 | 70.1 | 29.9 | 2.2 | 0.134 |
| NTRD3 | 172 | 52 | 224 | 76.8 | 23.2 | 7.3 | 0.007 |
| NTRD4 | 168 | 50 | 218 | 77.1 | 22.9 | 7.5 | 0.006 |
| TRRHU | 40 | 8 | 48 | 83.3 | 16.7 | 5.0 | 0.026 |
TRRHU = translational regulation of relative homoeolog usage. Reported χ2 and P values are for comparisons of the corresponding gene set to the full genome (“All”).
Deletion-resistant (“A” homoeologs retained).
Duplication-resistant (“A” homoeologs fractionated).
High expression levels have been shown to correlate with homoeolog retention in soybean and other species (Jiang et al., 2013). Because PTR genes are also more highly expressed than the genome-wide average (Supplemental Figure 3), we looked to see if the higher retention rate for PTR genes was simply a function of higher average transcription level. Percentage of retention for PTR genes with transcript abundance at or below the genome-wide average (17.8 RPKM in T) was comparable to that of PTR genes with transcript abundance greater than the genome-wide average (74.4% versus 74.7%). Additionally, median transcript abundance was similar for PTR dupR genes and PTR delR genes (22.7 RPKM versus 24.0 RPKM). Thus, the correlation between PTR and higher retention following the “A” polyploidy event does not appear to be a function of higher transcript abundance. The fact that NTR genes, which were generally transcribed below the genome-wide average, also exhibited elevated retention of duplicates from the “A” polyploidy event further suggests that the correlation between translational regulation and retention is not simply a function of transcription level.
DISCUSSION
Though translatomic approaches have been used to study regulation of gene expression in plants (Kawaguchi et al., 2004; Jiao and Meyerowitz, 2010; Mustroph and Bailey-Serres, 2010; Juntawong and Bailey-Serres, 2012), few studies have examined gene expression downstream of transcription in recent polyploids (Albertin et al., 2006, 2007; Hu et al., 2011, 2013; Yao et al., 2011), and none have specifically examined translational regulation. Here, we have shown that the transcriptome and translatome of a recently formed allotetraploid (G. dolichocarpa T2) related to soybean were highly correlated overall but that ∼10% of genes in each duplicated subgenome (homoeologs) exhibited significant translational regulation, and 10% exhibited translational regulation of combined expression. Together, of the 3874 genes for which the contribution of the two homoeologous subgenomes could be distinguished with confidence (the homoeolog gene set), 923 (23.8%) exhibited at least one form of translational regulation (translational regulation of combined expression, absolute regulation of homoeolog usage, and/or relative regulation of homoeolog usage). It is likely that with additional biological replication (and, thereby, more statistical power), this number would have been higher. Thus, assuming that the homoeolog gene set is representative of the complete gene set in soybean, at least one-quarter of all genes in the leaf transcriptome are translationally regulated to some extent.
NTR was more prevalent than PTR, both for combined expression and homoeolog-specific expression, which appears to be a common pattern in plants. In unstressed Arabidopsis thaliana leaves, 118 genes exhibited polysomal over nonpolysomal (P/NP) expression ratios below the 95% confidence interval for the genome-wide distribution, compared with 91 exhibiting P/NP ratios above the 95% confidence interval (Kawaguchi et al., 2004). Translational repression was also dramatically more prevalent than PTR in various stress responses, such as dehydration (Kawaguchi et al., 2004) and hypoxia (Branco-Price et al., 2005, 2008).
Though ∼10% of the genes in each homoeologous genome were translationally regulated, the two subgenomes were mostly regulated in concert. Consequently, only ∼1 to 5% of genes exhibited translational regulation of relative homoeolog usage (a shift between the full transcriptome and the polysomal fraction in the proportion of expression derived from each homoeolog). Thus, homoeolog expression bias (or lack thereof) observed at the transcriptional level was largely preserved in the translatome. Consistent with these observations, Hu et al. (2013) found greater transcriptome/proteome concordance for homoeolog usage than for combined expression in allopolyploid Gossypium barbadense.
T2 exhibited expression bias favoring D4 at the transcriptional level with, on average, 53.6% of transcripts derived from the D4 homoeolog. Though subtle, this D4 homoeolog bias was quite consistent throughout the genome: 80.7% of 25-gene windows across the soybean reference genome exhibit an average D4 proportion of combined expression >50%. Similar levels of expression dominance have been observed in other allopolyploids, including cotton (Gossypium hirsutum; Flagel et al., 2008) and maize (Zea mays; Schnable et al., 2011).
We speculated that translational regulation might enhance D4 genome dominance but found that it had no net effect on homoeolog usage: The mean D4 homoeolog proportion of combined expression in the T2 translatome was identical to that in the transcriptome (53.6%), and 89% of 25-gene windows with a transcriptional bias favoring D4 homoeologs retained a D4 bias in the translatome. However, translational regulation did exert many regional effects on homoeolog usage. Strikingly, translation shifted expression toward the D3 subgenome over most of the length of two soybean reference chromosomes, as well as in several smaller genomic neighborhoods within chromosomes. It is not known to what extent synteny is conserved between T2 and soybean, so we cannot determine the precise geography of translational effects on homoeolog usage in T2. However, limited evidence suggests that gene order is largely conserved between the two genomes (Innes et al., 2008, Chang et al., 2013), and it appears clear that translational regulation imposes regional, and possibly chromosomal, shifts in homoeolog usage.
Flagel et al. (2009) presented evidence for coordinated transcriptional regulation of physically linked homoeologs, and translation has been shown to coordinately regulate functionally related genes via RNA regulons (Keene, 2007). Our data suggest that there is coordinated translational regulation of physically linked genes as well. The mechanism for such regulation is not readily apparent, given that transcripts are not bound by any sort of physical linkage, but linked genes could share similar cis-elements that function to coordinate translation (e.g., pyrimidine-rich translational elements common to the 5′ untranslated regions of most mTOR-regulated mammalian genes; Hsieh et al., 2012).
Overall, translational regulation reversed transcriptional bias in 15.5% of 25-gene windows and 20.0% of individual genes, with shifts toward the D3 homoeolog slightly more frequent than shifts toward the D4 homoeolog. Therefore, rather than amplifying and/or stabilizing D4 dominance genome-wide, translational regulation acted on a more local level, amplifying D4 bias in some genomic neighborhoods, but favoring D3 in others. Thus, though the effects of translation on homoeolog usage were generally modest, our data suggest that genome dominance may in fact be a chromosomal or segmental phenomenon, rather than being genome-wide as has been proposed (Schnable et al., 2011; Freeling et al., 2012). In maize and other paleopolyploids where unbalanced homoeolog expression bias has been observed, the diploid progenitors are extinct, making it impossible to definitively assign chromosomes to subgenomes. In such studies, the assumption was made that one subgenome comprises the less fractionated chromosomes and the other subgenome comprises the more fractionated chromosomes. If fractionation bias is dictated by homoeolog expression (Schnable et al., 2011; Freeling et al., 2012), our data suggest this assumption may be unwarranted and that fractionation bias occurs at finer scales than whole subgenomes.
Intriguingly, our data suggest that translational regulation plays a role in determining the long-term fates of homoeologous genes. Genes that were translationally regulated in our study exhibited significantly higher retention of duplicates from the Glycine-specific paleopolyploidy event 5 to 13 MYA (Doyle and Egan, 2010) than genes that were not, with PTR genes retained at a slightly higher rate than NTR genes.
PTR could facilitate retention by strengthening selection on absolute dosage (Conant and Wolfe, 2007). Recent studies have shown a significant association between the metabolic flux catalyzed by an enzyme and retention of the underlying genes following a paleopolyploidy event in Arabidopsis (Bekaert et al., 2011; Hudson et al., 2011). The authors speculated that increases in the protein products of some genes are beneficial (e.g., enzymes catalyzing rate-limiting steps in essential metabolic pathways). If protein abundance increases with gene copy number (a positive gene dosage response), selection would therefore act to preserve duplicates of genes encoding such proteins. Similarly, selection might favor positive translational regulation as another mechanism for increasing the cellular titer of such proteins.
PTR in G. dolichocarpa leaves preferentially targeted highly transcribed genes, which likely encode many of the proteins that are required at a high cellular titer. Additionally, genes with plastid functions and genes encoding translation-related proteins (ribosomal proteins and translation initiation factors) were preferentially PTR. Ribosomal proteins are among the most abundant proteins in the cell (Ishihama et al., 2008; Marguerat et al., 2012), and translation is rate limiting for protein synthesis of the most abundant proteins (Marguerat et al., 2012). Thus, it might be expected that an increase in ribosomal proteins would facilitate greater protein synthesis, particularly in a recently formed polyploid such as T2, which transcribes nearly twice as many genes as its diploid progenitors (Coate and Doyle, 2010). Similarly, photosynthetic proteins are among the most abundant proteins in green plant tissue and are often rate limiting to primary productivity (Coate et al., 2011; Coate and Doyle, 2013). The fact that ribosomal and photosynthetic proteins are more likely to be PTR than the genome-wide average in T2 and more likely to be retained in duplicate following polyploidy in a variety of species (Blanc and Wolfe, 2004b) supports the notion that they are under selection for increased absolute dosage.
Alternatively, it has been proposed that selection on relative dosage explains many recurring patterns of gene duplicate retention and loss (Papp et al., 2003; Birchler et al., 2007; Freeling, 2009; Birchler and Veitia, 2010; Coate et al., 2011). Specifically, many protein complexes and signaling cascades require a specific stoichiometry among the interacting proteins to function properly. To the extent that protein abundance is dictated by gene copy number, small-scale duplications (e.g., tandem duplications) that affect some but not all subunits of dosage sensitive complexes disrupt this stoichiometry and are therefore deleterious. By contrast, by duplicating all genes in a dosage sensitive complex, polyploidy is more likely to preserve proper stoichiometry. As a consequence, dosage-sensitive genes are preferentially retained following WGD and preferentially lost following small-scale duplications (Blanc and Wolfe, 2004b; Seoighe and Gehring, 2004; Maere et al., 2005; Paterson et al., 2006; Thomas et al., 2006; Coate et al., 2011). However, protein abundance is regulated at many other levels beyond gene copy number, and polyploidy has been shown to produce a wide range of gene dosage responses at the transcriptional level (Guo et al., 1996; Coate and Doyle, 2010) that potentially disrupt the stoichiometry of interacting proteins. Having additional layers of gene expression regulation on top of transcription could reduce unbalanced or ectopic expression resulting from duplication and provide an additional safeguard against dosage imbalance.
In many instances, translational regulation likely represents a mechanism for reducing transcriptional noise (Keene, 2007; Joshi et al., 2011), and we found that translational regulation tends to reduce differences in expression between the combined T2 transcriptome and the transcriptomes of its diploid progenitor species. Similarly, translation generally brings homoeolog usage in T2 more in line with expression levels in the two diploid progenitor species. These translational adjustments could represent corrections of transcriptional misregulation induced by genome merger or doubling. By correcting transcriptional misregulation resulting from gene duplication, we propose that translational regulation reduces selection against gene duplicates in dosage-sensitive complexes, thereby explaining higher long-term retention of homoeologs among translationally regulated genes than the genome as a whole.
Finally, the few cases of translational regulation of relative homoeolog usage could represent a form of expression sub- or neofunctionalization (Flagel et al., 2008). Following duplication, most genes experience relaxed selective constraint (Lynch and Conery, 2000, 2003). This, in turn, is thought to facilitate the evolution of new functions (neofunctionalization) or partitioning of ancestral functions (subfunctionalization). Genes that are sub- or neofunctionalized are then preserved via purifying selection. Relaxed selection could have enabled the acquisition of novel cis-regulatory elements or mutation of existing cis-regulatory elements that resulted in new or altered forms of translational regulation. Such sub- or neofunctionalization at the translational level could have then favored duplicate retention (Adams and Wendel, 2005).
In conclusion, though the leaf transcriptome and translatome of allopolyploid G. dolichocarpa T2 were highly correlated overall, translational regulation exerted considerable influence over gene expression, with nearly one-quarter of the transcriptome translationally regulated to some extent. This translational regulation appeared to tune the polyploid transcriptome to the exigencies of a doubled genome. T2 leaf mesophyll cells have larger transcriptomes (Coate and Doyle, 2010) and more chloroplasts than do D3 or D4 diploids (Coate et al., 2012). In response, translational regulation appeared to increase production of both translational machinery and plastid-targeted proteins. At the same time, translation reduced differences between the transcriptomes of T2 and its diploid progenitors, suggesting that it served to attenuate misregulation resulting from genome merger and/or doubling. We propose that these adjustments explain why translationally regulated genes are more likely to be retained in duplicate following older polyploidy events than genes that are not.
METHODS
Plant Material
Glycine dolichocarpa (T2; CSIRO accession number G1134) plants were grown from seed in a growth chamber under the following conditions: 12-h/12-h light/dark cycle, 22°C/18°C light/dark temperature, and a light intensity of 125 μmol m−2 s−1. Two weeks after germination, fully expanded leaflets were collected 1.5 to 2.0 h into the light period and immediately frozen in liquid nitrogen. Three separate pools of leaf tissue were collected for three biological replicates. For each biological replicate, leaflets were pooled from six individuals (one leaflet per individual).
Polysome Extraction
For each biological replicate, 0.2 g of leaf tissue was ground to a fine powder in liquid nitrogen and suspended in 1 mL of extraction buffer (0.2 M Tris-HCl, 0.2 M KCl, 35 mM MgCl2, 25 mM EGTA, 0.2 M Suc, 1% Triton X-100, 2% polyoxyethylene-10-tridecyl ether, 0.5 mg/mL heparin, 100 mM β-mercaptoethanol, 100 μg/mL chloramphenicol, and 25 μg/mL cyclohexamide). Debris was removed by centrifuging for 1 min at 4000 rpm and 4°C through a Pierce 2-mL centrifuge column (Fisher). The filtrate was incubated on ice for 10 min to solubilize membranes and centrifuged for 5 min at 14,000 rpm and 4°C. The supernatant was transferred to a new tube, combined with one-twentieth volume of sodium deoxycholate, incubated for 5 min on ice to complete microsomal membrane solubilization, and centrifuged again for 5 min at 14,000 rpm and 4°C.
For each of three biological replicates, the supernatant was divided into two equal fractions, and EDTA was added to one fraction to a final concentration of 20 mM to dissociate RNP complexes (EDTA control). The untreated fraction was layered onto a 15 to 62.5% Suc gradient (10 mL total volume) containing 40 mM Tris-HCl, 20 mM KCl, 10 mM MgCl2, 0.5 mg/mL heparin, 0.1 mg/mL chloramphenicol, and 0.025 mg/mL cyclohexamide. The EDTA control was layered onto an identical Suc gradient except that it contained 1 mM EDTA in place of 10 mM MgCl2. Gradients were centrifuged at 45,000 rpm for 65 min at 4°C and separated into 12 fractions by sequentially pipetting 410-μL aliquots from the top of the gradient. Each fraction was pipetted into a tube containing 50 μL of 0.2 M EDTA and 5% SDS, pH 8.0, and inverted to mix.
RNA Extraction
Phenol/chloroform/isoamyl (0.4 mL; 25:24:1) was added to each fraction, which was then vortexed and centrifuged at maximum speed for 4 min at room temperature. The upper phase was then transferred to a new tube and mixed with 1 mL 95% ethanol and then centrifuged at maximum speed for 15 min at room temperature. Pellets were dried and resuspended in 30 μL of ice-cold TE buffer.
To determine which fractions contained polysomal RNA, aliquots of each fraction were run on nondenaturing agarose gels. RNA/ribosome associations are disrupted in the EDTA-treated controls. Consequently, fractions from the EDTA control gradients for which rRNA bands could be visualized (fractions 1 to 6 in all cases) contained nonpolysomal RNA. rRNA bands were absent or nearly invisible in all heavier fractions of EDTA controls (Supplemental Figure 7). By contrast, rRNA bands were clearly visible in all fractions of untreated (EDTA-free) gradients (Supplemental Figure 7). Consequently, fractions 7 to 12 of the untreated gradients were assumed to contain polysomal RNA. This cutoff corresponds to those used in other studies (Kawaguchi et al., 2004) and was confirmed by quantifying RNA in each fraction by UV absorbance. For the EDTA-treated fractions, 84% of the total nucleic acids recovered was contained in fractions 1 to 6. The bulk of the remaining 16% was most likely genomic DNA, based on the presence of high molecular weight bands on the gels. RNA concentrations were more uniform across the EDTA-free fractions, and 41% of total nucleic acids recovered was in fractions 7 to 12.
For each biological replicate, fractions 1 to 6 from the EDTA-free gradients were pooled and designated “nonpolysomal” (NP). Fractions 7 to 12 were pooled and designated “polysomal” (P). Unfractionated RNA extracted using the RNeasy Kit (Qiagen) was designated “total” RNA (T). All RNA fractions were treated with DNase I (NEB).
Illumina Library Preparation and Sequencing
Single-end RNA-Seq libraries were constructed for three biological replicates each of polysomal and nonpolysomal RNA. Libraries were made following the Illumina mRNA sequencing protocol (Illumina), with the following modifications: (1) two rounds of poly(A) selection were performed using the Dynabeads mRNA DIRECT kit (Life Technologies); (2) RNA was fragmented for 2 min at 70°C using the RNA fragmentation reagents kit (Life Technologies); and (3) Illumina PE adapters were replaced with custom-made adapters containing 3-nucleotide barcodes in order to facilitate multiplexing of samples.
Sequencing was performed on the HiSequation 2000 platform (Illumina), generating100-nucleotide reads, respectively. Equimolar amounts of three barcoded libraries were combined and sequenced per channel. In addition to the polysomal and nonpolysomal libraries, the transcriptomes (total mRNA) of two biological replicates of T2 total and three biological replicates of each diploid progenitor, Glycine syndetika (D4; CSIRO accession numbers G1300, G2073, and G2321), and Glycine tomentella (D3; CSIRO accession numbers G1366, G1403, and G1820) were grown under identical conditions and prepared and sequenced as described by Coate et al. (2013).
Data Analysis
Multiplexed reads from FASTQ Sanger files were split into their respective libraries based on 5′ 3-nucleotide barcode sequences, then trimmed to remove barcodes and low quality 3′ bases. Reads were quality filtered using the FASTQ Quality Filter in the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/; default settings). Reads were mapped to the soybean (Glycine max) genome using Bowtie (Langmead et al., 2009) with the following parameters: -a,–best,–strata, -m 1, -n 3, -l 30, and -e 250 (78-nucleotide reads) or –e 288 (90-nucleotide reads). These settings ensure that only reads mapping unambiguously to a single locus in the soybean genome were used in this study. Read counts per gene were determined using HTSeq (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html) using the “–m intersection-noempty” setting. For each library, transcript abundance per transcriptome for a given gene was estimated as the number of reads unambiguously mapped to that gene per kilobase and per million unambiguously mapped reads generated by that library (RKPM). Genes differentially expressed (FDR < 0.05) in pairwise comparisons of T, P, and NP and in pairwise comparisons of T, D3 diploid, and D4 diploid were identified using DESeq (Anders and Huber, 2010). For D3 and D4, individual accessions (n = 3) were treated as biological replicates for the species. The contributions of D3 and D4 subgenomes to gene expression in T, P, and NP were determined as described by Ilut et al. (2012), except that three accessions per diploid species were sequenced instead of one. Sites that were polymorphic among accessions within either diploid species were excluded from the list of DDPs.
Gene ontology enrichment analysis was performed using the AgriGO Web tool (http://bioinfo.cau.edu.cn/agriGO/; Du et al., 2010). Significance was determined using Fisher’s exact test with the Yekutieli adjustment for multiple comparisons. For combined expression, enrichment was determined relative to all genes expressed in T and in P and/or NP (n = 35,655). For homoeolog-specific expression, enrichment was determined relative to the homoeolog gene set (n = 3874).
Accession Numbers
Sequence data (FASTQ files from RNA-Seq experiments) from this article can be found in the National Center for Biotechnology Information Sequence Read Archive under accession numbers SRX131445-46, SRX134815-16, SRX134818, SRX134820, SRX134822, SRX134824, SRX134827, SRX316786, and SRX316881 to SRX316885.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Correlations of Combined Expression Estimates among Biological Replicates.
Supplemental Figure 2. Strong Transcriptional Biases in Homoeolog Usage Are Preserved in the Translatome.
Supplemental Figure 3. Distribution of Transcript Levels for Genes That Are Translationally Regulated and Those That Are Not.
Supplemental Figure 4. Relative Enrichment or Depletion of Plant GOSlim Terms among Genes That Are Translationally Regulated.
Supplemental Figure 5. D4 Homoeolog Expression Dominance in T2.
Supplemental Figure 6. Average D4 Homoeolog Fraction of Combined Expression in the T2 Transcriptome and Translatome in 25-Gene Sliding Windows across Each Soybean Reference Chromosome.
Supplemental Figure 7. Nondenaturing Gels of RNA Samples Extracted from Suc Gradient Fractions.
Supplemental Table 1. Translation Reduces Transcript-Level Differences in Expression between T2 and Its Diploid Progenitors.
Supplemental Data Set 1. Genes Exhibiting Significant Translational Regulation of Relative Homoeolog Usage.
Supplemental Data Set 2. Gene Ontology Terms Overrepresented among Translationally Regulated Gene Sets.
Supplementary Material
Acknowledgments
We thank Amber Hotto for help with the polysome profiling protocol. This work was supported by grants from the U.S. National Science Foundation (0939423, 0744306, and 0822258).
AUTHOR CONTRIBUTIONS
J.J.D. and J.E.C. designed the research. J.E.C. performed the research. J.E.C., H.B., and J.J.D. analyzed the data. J.E.C. and J.J.D. wrote the article.
Glossary
- MYA
million years ago
- RPKM
reads per kilobase per million mapped reads
- FDR
false discovery rate
- PTR
positively translationally regulated
- NTR
negatively translationally regulated
- DDP
diploid distinguishing polymorphism
- WGD
whole-genome duplication
Footnotes
Online version contains Web-only data.
References
- Adams K.L., Cronn R., Percifield R., Wendel J.F. (2003). Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100: 4649–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams K.L., Wendel J.F. (2005). Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8: 135–141 [DOI] [PubMed] [Google Scholar]
- Albertin W., Alix K., Balliau T., Brabant P., Davanture M., Malosse C., Valot B., Thiellement H. (2007). Differential regulation of gene products in newly synthesized Brassica napus allotetraploids is not related to protein function nor subcellular localization. BMC Genomics 8: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertin W., Balliau T., Brabant P., Chèvre A.M., Eber F., Malosse C., Thiellement H. (2006). Numerous and rapid nonstochastic modifications of gene products in newly synthesized Brassica napus allotetraploids. Genetics 173: 1101–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekaert M., Edger P.P., Pires J.C., Conant G.C. (2011). Two-phase resolution of polyploidy in the Arabidopsis metabolic network gives rise to relative and absolute dosage constraints. Plant Cell 23: 1719–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Veitia R.A. (2010). The gene balance hypothesis: Implications for gene regulation, quantitative traits and evolution. New Phytol. 186: 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Yao H., Chudalayandi S. (2007). Biological consequences of dosage dependent gene regulatory systems. Biochim. Biophys. Acta 1769: 422–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Wolfe K.H. (2004a). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Wolfe K.H. (2004b). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J.E., Chapman B.A., Rong J.K., Paterson A.H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Branco-Price C., Kaiser K.A., Jang C.J.H., Larive C.K., Bailey-Serres J. (2008). Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J. 56: 743–755 [DOI] [PubMed] [Google Scholar]
- Branco-Price C., Kawaguchi R., Ferreira R.B., Bailey-Serres J. (2005). Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann. Bot. (Lond.) 96: 647–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggs R.J.A., Chamala S., Wu W., Gao L., May G.D., Schnable P.S., Soltis D.E., Soltis P.S., Barbazuk W.B. (2010). Characterization of duplicate gene evolution in the recent natural allopolyploid Tragopogon miscellus by next-generation sequencing and Sequenom iPLEX MassARRAY genotyping. Mol. Ecol. 19 (suppl. 1): 132–146 [DOI] [PubMed] [Google Scholar]
- Chang S., Hartman G.L., Singh R.J., Lambert K.N., Hobbs H.A., Domier L.L. (2013). Identification of high-quality single-nucleotide polymorphisms in Glycine latifolia using a heterologous reference genome sequence. Theor. Appl. Genet. 126: 1627–1638 [DOI] [PubMed] [Google Scholar]
- Coate, J.E., and Doyle, J.J. (2013). Genomics and transcriptomics of photosynthesis in polyploids. In Polyploid and Hybrid Genomics, Z.J. Chen and J.A. Birchler, eds (Hoboken, NJ: Wiley-Blackwell), pp. 153–169. [Google Scholar]
- Coate J.E., Doyle J.J. (2010). Quantifying whole transcriptome size, a prerequisite for understanding transcriptome evolution across species: An example from a plant allopolyploid. Genome Biol. Evol. 2: 534–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coate J.E., Luciano A.K., Seralathan V., Minchew K.J., Owens T.G., Doyle J.J. (2012). Anatomical, biochemical, and photosynthetic responses to recent allopolyploidy in Glycine dolichocarpa (Fabaceae). Am. J. Bot. 99: 55–67 [DOI] [PubMed] [Google Scholar]
- Coate J.E., Powell A.F., Owens T.G., Doyle J.J. (2013). Transgressive physiological and transcriptomic responses to light stress in allopolyploid Glycine dolichocarpa (Leguminosae). Heredity (Edinb) 110: 160–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coate J.E., Schlueter J.A., Whaley A.M., Doyle J.J. (2011). Comparative evolution of photosynthetic genes in response to polyploid and nonpolyploid duplication. Plant Physiol. 155: 2081–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant G.C., Wolfe K.H. (2007). Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol. Syst. Biol. 3: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J.J., Doyle J.L., Rauscher J.T., Brown A.H.D. (2004). Evolution of the perennial soybean polyploid complex (Glycine subgenus Glycine): A study of contrasts. Biol. J. Linn. Soc. Lond. 82: 583–597 [Google Scholar]
- Doyle J.J., Egan A.N. (2010). Dating the origins of polyploidy events. New Phytol. 186: 73–85 [DOI] [PubMed] [Google Scholar]
- Du J., Tian Z., Sui Y., Zhao M., Song Q., Cannon S.B., Cregan P., Ma J. (2012). Pericentromeric effects shape the patterns of divergence, retention, and expression of duplicated genes in the paleopolyploid soybean. Plant Cell 24: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett J.A., Maere S., Van de Peer Y. (2009). Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc. Natl. Acad. Sci. USA 106: 5737–5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L., Udall J., Nettleton D., Wendel J. (2008). Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol. 6: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L.E., Chen L., Chaudhary B., Wendel J.F. (2009). Coordinated and fine-scale control of homoeologous gene expression in allotetraploid cotton. J. Hered. 100: 487–490 [DOI] [PubMed] [Google Scholar]
- Freeling M. (2009). Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 60: 433–453 [DOI] [PubMed] [Google Scholar]
- Freeling M., Thomas B.C. (2006). Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 16: 805–814 [DOI] [PubMed] [Google Scholar]
- Freeling M., Woodhouse M.R., Subramaniam S., Turco G., Lisch D., Schnable J.C. (2012). Fractionation mutagenesis and similar consequences of mechanisms removing dispensable or less-expressed DNA in plants. Curr. Opin. Plant Biol. 15: 131–139 [DOI] [PubMed] [Google Scholar]
- Grover C.E., Gallagher J.P., Szadkowski E.P., Yoo M.J., Flagel L.E., Wendel J.F. (2012). Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol. 196: 966–971 [DOI] [PubMed] [Google Scholar]
- Guo M., Davis D., Birchler J.A. (1996). Dosage effects on gene expression in a maize ploidy series. Genetics 142: 1349–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty M.J., Barker G.L., Brennan A.C., Edwards K.J., Abbott R.J., Hiscock S.J. (2008). Changes to gene expression associated with hybrid speciation in plants: Further insights from transcriptomic studies in Senecio. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363: 3055–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh A.C., et al. (2012). The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485: 55–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Houston N.L., Pathak D., Schmidt L., Thelen J.J., Wendel J.F. (2011). Genomically biased accumulation of seed storage proteins in allopolyploid cotton. Genetics 189: 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Koh J., Yoo M.J., Grupp K., Chen S., Wendel J.F. (2013). Proteomic profiling of developing cotton fibers from wild and domesticated Gossypium barbadense. New Phytol. 200: 570–582 [DOI] [PubMed] [Google Scholar]
- Hudson C.M., Puckett E.E., Bekaert M., Pires J.C., Conant G.C. (2011). Selection for higher gene copy number after different types of plant gene duplications. Genome Biol. Evol. 3: 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilut D.C., Coate J.E., Luciano A.K., Owens T.G., May G.D., Farmer A.D., Doyle J.J. (2012). A comparative transcriptomic study of an allotetraploid and its diploid progenitors illustrates the unique advantages and challenges of RNA-seq in plant species. Am. J. Bot. 99: 383–396 [DOI] [PubMed] [Google Scholar]
- Innes R.W., et al. (2008). Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean. Plant Physiol. 148: 1740–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y., Schmidt T., Rappsilber J., Mann M., Hartl F.U., Kerner M.J., Frishman D. (2008). Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Liu Y., Xia E., Gao L. (2013). Prevalent role of gene features in determining evolutionary fates of whole-genome duplication duplicated genes in flowering plants. Plant Physiol. 161: 1844–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100 [DOI] [PubMed] [Google Scholar]
- Jiao Y., et al. (2012). A genome triplication associated with early diversification of the core eudicots. Genome Biol. 13: R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Meyerowitz E.M. (2010). Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol. Syst. Biol. 6: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A., Van de Peer Y., Michoel T. (2011). Structural and functional organization of RNA regulons in the post-transcriptional regulatory network of yeast. Nucleic Acids Res. 39: 9108–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P., Bailey-Serres J. (2012). Dynamic light regulation of translation status in Arabidopsis thaliana. Front. Plant Sci. 3: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R., Bailey-Serres J. (2002). Regulation of translational initiation in plants. Curr. Opin. Plant Biol. 5: 460–465 [DOI] [PubMed] [Google Scholar]
- Kawaguchi R., Girke T., Bray E.A., Bailey-Serres J. (2004). Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J. 38: 823–839 [DOI] [PubMed] [Google Scholar]
- Keene J.D. (2007). RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 8: 533–543 [DOI] [PubMed] [Google Scholar]
- Khan Z., Bloom J.S., Amini S., Singh M., Perlman D.H., Caudy A.A., Kruglyak L. (2012). Quantitative measurement of allele-specific protein expression in a diploid yeast hybrid by LC-MS. Mol. Syst. Biol. 8: 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh J., Chen S., Zhu N., Yu F., Soltis P.S., Soltis D.E. (2012). Comparative proteomics of the recently and recurrently formed natural allopolyploid Tragapogon mirus (Asteraceae) and its parents. New Phytol. 196: 292–305 [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D.A. (1983). Poly ploidy and novelty in flowering plants. Am. Nat. 122: 1–25 [Google Scholar]
- Lin J.Y., Stupar R.M., Hans C., Hyten D.L., Jackson S.A. (2010). Structural and functional divergence of a 1-Mb duplicated region in the soybean (Glycine max) genome and comparison to an orthologous region from Phaseolus vulgaris. Plant Cell 22: 2545–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J.S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Lynch M., Conery J.S. (2003). The evolutionary demography of duplicate genes. J. Struct. Funct. Genomics 3: 35–44 [PubMed] [Google Scholar]
- Maere S., De Bodt S., Raes J., Casneuf T., Van Montagu M., Kuiper M., Van de Peer Y. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 102: 5454–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S., Schmidt A., Codlin S., Chen W., Aebersold R., Bähler J. (2012). Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151: 671–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A., Bailey-Serres J. (2010). The Arabidopsis translatome cell-specific mRNA atlas: Mining suberin and cutin lipid monomer biosynthesis genes as an example for data application. Plant Signal. Behav. 5: 320–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A., Zanetti M.E., Jang C.J.H., Holtan H.E., Repetti P.P., Galbraith D.W., Girke T., Bailey-Serres J. (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z., Kim E.D., Ha M., Lackey E., Liu J., Zhang Y., Sun Q., Chen Z.J. (2009). Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B., Pál C., Hurst L.D. (2003). Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197 [DOI] [PubMed] [Google Scholar]
- Park S.H., Chung P.J., Juntawong P., Bailey-Serres J., Kim Y.S., Jung H., Bang S.W., Kim Y.K., Do Choi Y., Kim J.K. (2012). Posttranscriptional control of photosynthetic mRNA decay under stress conditions requires 3′ and 5′ untranslated regions and correlates with differential polysome association in rice. Plant Physiol. 159: 1111–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A.H., Chapman B.A., Kissinger J.C., Bowers J.E., Feltus F.A., Estill J.C. (2006). Many gene and domain families have convergent fates following independent whole-genome duplication events in Arabidopsis, Oryza, Saccharomyces and Tetraodon. Trends Genet. 22: 597–602 [DOI] [PubMed] [Google Scholar]
- Pires J.C., Zhao J., Schranz M.E., Leon E.J., Quijada P.A., Lukens L.N., Osborn T.C. (2004). Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae). Biol. J. Linn. Soc. Lond. 82: 675–688 [Google Scholar]
- Reynoso M.A., Blanco F.A., Bailey-Serres J., Crespi M., Zanetti M.E. (2012). Selective recruitment of mRNAs and miRNAs to polyribosomes in response to rhizobia infection in Medicago truncatula. Plant J. 73: 289–301 [DOI] [PubMed] [Google Scholar]
- Schlueter J.A., Dixon P., Granger C., Grant D., Clark L., Doyle J.J., Shoemaker R.C. (2004). Mining EST databases to resolve evolutionary events in major crop species. Genome 47: 868–876 [DOI] [PubMed] [Google Scholar]
- Schmutz J., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schnable J.C., Springer N.M., Freeling M. (2011). Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 108: 4069–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011). Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Seoighe C., Gehring C. (2004). Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet. 20: 461–464 [DOI] [PubMed] [Google Scholar]
- Shoemaker R.C., Schlueter J., Doyle J.J. (2006). Paleopolyploidy and gene duplication in soybean and other legumes. Curr. Opin. Plant Biol. 9: 104–109 [DOI] [PubMed] [Google Scholar]
- Sonenberg N., Hinnebusch A.G. (2009). Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Wang X., Bowers J.E., Ming R., Alam M., Paterson A.H. (2008). Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 18: 1944–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B.C., Pedersen B., Freeling M. (2006). Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 16: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C., Marcotte E.M. (2012). Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13: 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tian L., Lee H.S., Wei N.E., Jiang H., Watson B., Madlung A., Osborn T.C., Doerge R.W., Comai L., Chen Z.J. (2006). Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner D.A., Edwards G.E. (1993). Effects of polyploidy on photosynthesis. Photosynth. Res. 35: 135–147 [DOI] [PubMed] [Google Scholar]
- Wood T.E., Takebayashi N., Barker M.S., Mayrose I., Greenspoon P.B., Rieseberg L.H. (2009). The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. USA 106: 13875–13879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Kato A., Mooney B., Birchler J.A. (2011). Phenotypic and gene expression analyses of a ploidy series of maize inbred Oh43. Plant Mol. Biol. 75: 237–251 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.