This work reports that sro1 (for allelic variation of a wheat SRO gene SRO1) showed higher poly(ADP ribose) polymerase activity than SRO1 due to two residue substitutions and that this protein is involved in both seedling vigor and tolerance to abiotic stress by modulating redox homoeostasis and maintaining genome stability.
Abstract
Plant growth inhibition is a common response to salinity. Under saline conditions, Shanrong No. 3 (SR3), a bread wheat (Triticum aestivum) introgression line, performs better than its parent wheat variety Jinan 177 (JN177) with respect to both seedling growth and abiotic stress tolerance. Furthermore, the endogenous reactive oxygen species (ROS) was also elevated in SR3 relative to JN177. The SR3 allele of sro1, a gene encoding a poly(ADP ribose) polymerase (PARP) domain protein, was identified to be crucial for both aspects of its superior performance. Unlike RADICAL-INDUCED CELL DEATH1 and other Arabidopsis thaliana SIMILAR TO RCD-ONE (SRO) proteins, sro1 has PARP activity. Both the overexpression of Ta-sro1 in wheat and its heterologous expression in Arabidopsis promote the accumulation of ROS, mainly by enhancing the activity of NADPH oxidase and the expression of NAD(P)H dehydrogenase, in conjunction with the suppression of alternative oxidase expression. Moreover, it promotes the activity of ascorbate-GSH cycle enzymes and GSH peroxidase cycle enzymes, which regulate ROS content and cellular redox homeostasis. sro1 is also found to be involved in the maintenance of genomic integrity. We show here that the wheat SRO has PARP activity; such activity could be manipulated to improve the growth of seedlings exposed to salinity stress by modulating redox homeostasis and maintaining genomic stability.
INTRODUCTION
Genetic analysis of the stress response of model plants has identified a number of key genes involved (Shinozaki and Yamaguchi-Shinozaki, 2000; Zhu, 2002; Munns and Tester, 2008), but few of these genes have as yet been genetically manipulated for crop improvement. Most attempts to enhance stress tolerance using a transgenic approach have failed because the transgene has typically exerted a negative effect on plant growth (Skirycz et al., 2011). Thus, to identify genes involved in both stress tolerance and growth improvement is important for crop breeding.
Salinity, in common with several other agents of abiotic stress, induces the production of toxic reactive oxygen species (ROS), which if not controlled, can ultimately trigger cell death (Dat et al., 2000). However, relatively low levels of ROS are implicated in stress tolerance. There is a substantial degree of support for the suggestion that maintaining a low level of endogenous H2O2 enhances tolerance to a number of abiotic stresses (Mittler, 2002; Neill et al., 2002; Apel and Hirt, 2004). On the other hand, some reports support the idea that constitutive elevation of H2O2 in plants can improve multistress tolerance in plants (Van Breusegem et al., 2008; Huang et al., 2009). However, whether elevated ROS content can simultaneously increase both plant growth and abiotic stress tolerance is unknown.
The enzyme poly(ADP ribose) polymerase (PARP) is part of the early response to DNA breakage caused by radiative and oxidative stress (Briggs and Bent, 2011). PARP-like genes are present in many eukaryotes, and the PARP catalytic domain is characteristically associated with a range of other domains (Amé et al., 2004; Jaspers et al., 2010). A prominent example is represented by the Arabidopsis thaliana gene RADICAL-INDUCED CELL DEATH1 (RCD1), which contains a WWE domain and an RST (for RCD-SRO-TAF4) domain, both of which are responsible for ensuring the protein–protein interactions, and its expression in Saccharomyces cerevisiae strain WYT could overcome the oxidative stress sensitive phenotype of these yeast cells (Belles-Boix et al., 2000; Aravind, 2001). The combination of a PARP and an RST domain is specific to plants, and proteins carrying both have been termed SIMILAR TO RCD-ONE (SRO) (Jaspers et al., 2010). A loss-of-function mutation in At-RCD1 results in an enhanced sensitivity to salt, Glc, and apoplastic ROS but increased resistance to freezing and chloroplastic superoxide formation by methyl viologen (MV) (Overmyer et al., 2000; Ahlfors et al., 2004; Fujibe et al., 2004; Katiyar-Agarwal et al., 2006; Teotia and Lamb, 2009). Double mutants in both RCD1 and SRO1 have severe developmental defects and behave differently in several developmental events and abiotic stress responses (Jaspers et al., 2009; Teotia and Lamb, 2009), which are similar to those seen in the stress-induced morphological response (Teotia et al., 2010), which is known to be associated with changes in redox balance (Potters et al., 2009). The Arabidopsis genome encodes at least three putative PARPs and six SROs. The former have high levels of sequence similarity with mammalian PARPs at the peptide level, but not all PARPs have poly(ADP ribosyl)ation activity; some show mono(ADP ribose) transferase (mART) activity and others appear to be enzymatically inactive (Citarelli et al., 2010). Although the ability to bind NAD+ is necessary for the activity of PARP, At-RCD1 does not readily bind NAD+ and has no detectable poly(ADP ribosyl)ation activity and mART activity (Jaspers et al., 2010). Other Arabidopsis SROs and rice (Oryza sativa) SRO1c were predicted to lack poly(ADP ribosyl)ation activity based on bioinformatics (Jaspers et al., 2010; You et al., 2013), and the Arabidopsis SROs were also reported to lack mART activity (Wang et al., 2011). Whether SROs in other plants are enzymatically active remains to be elucidated.
The salinity-tolerant bread wheat (Triticum aestivum) cultivar Shanrong No. 3 (SR3) is a derivative of an asymmetric somatic hybrid between bread wheat and tall wheatgrass (Thinopyrum ponticum) (Xia et al., 2003). This novel cultivar has been shown to be higher yielding than its wheat parent Jinan 177 (JN177) when grown in saline soil and has been planted commercially in a number of areas where the soil suffers from alkaline salinity (Xia, 2009). The genetic basis of much of its salinity tolerance has been shown to rely on ROS homeostasis based on the transcriptomic and proteomic outcomes in response to salinity stress (Peng et al., 2009; Liu et al., 2012). Here, we report the superior traits of SR3 and demonstrate that a candidate gene, which is the allele of the parent JN177 via point mutations, encodes an SRO protein containing a N-terminal WWE domain, a PARP-like domain, and a C-terminal RST domain. We found that the wheat SRO has PARP activity and DNA binding ability and is involved in redox homeostasis regulation and genomic stability maintenance. Our findings demonstrated that sro1 could be effectively used to improve plant growth and stress tolerance.
RESULTS
The Higher Salinity Tolerance of Cultivar SR3
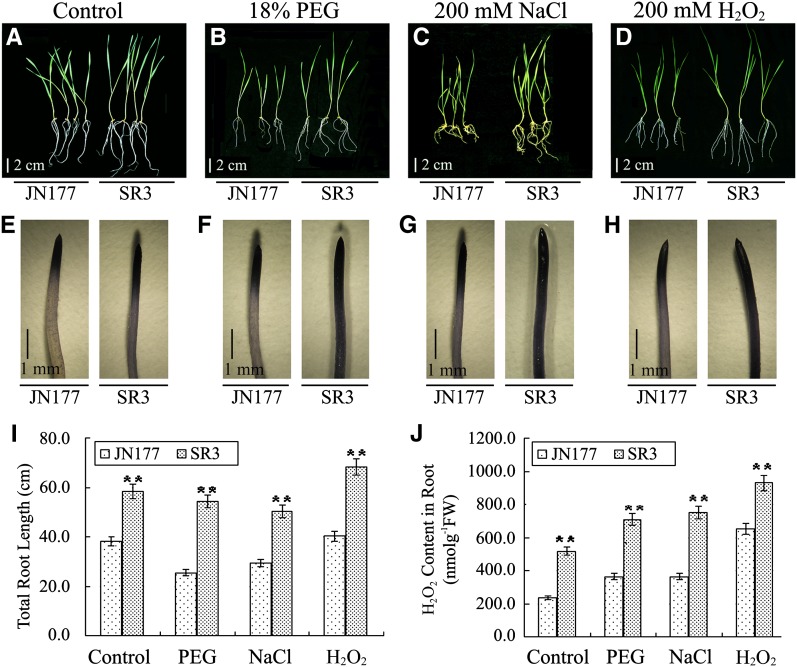
Compared with JN177, SR3 seedlings were larger in the normal field and kept their growth advantage under saline field conditions. More seedlings became yellow, wilted, and died in JN177 than in SR3 in the 0.5% salinity fields. To investigate SR3’s abiotic stress responses more efficiently, we further investigated its phenotypes of growth and stress resistance at the seedling stage grown in culture solution. SR3 seedlings grew more vigorously than those of its bread wheat parent cultivar JN177, under both nonstressed and stressed (polyethylene glycol [PEG], NaCl, or H2O2) conditions (Figures 1A to 1D and 1I; Supplemental Figure 1A). Alterations in cellular ROS levels are known to affect the growth of plants, especially the root system (Finkemeier et al., 2005; Olmos et al., 2006; Miller et al., 2007). Therefore, we examined the ROS content in the roots of SR3 and JN177 under both nonstressed and stressed (PEG, NaCl, or H2O2) conditions. Consistently, the ROS content of SR3 roots was higher than that of JN177, irrespective of the treatment (Figures 1E to 1H and 1J; Supplemental Figure 1B) and was positively correlated with root length (Figure 1I), implying that the gene(s) responsible for ROS accumulation in SR3 is also responsible for the vigorous root growth under stressed conditions.
Figure 1.
The Abiotic Stress Tolerance of Cultivar SR3.
(A) to (D) Phenotype of 1-week-old JN177 and SR3 under control (A), 18% PEG (B), 200 mM NaCl (C), or 200 mM H2O2 (D). Nonstressed or stressed seedlings grown for 2 weeks. JN177 is a bread wheat cultivar. The salinity tolerant bread wheat cultivar SR3 is a derivative of an asymmetric somatic hybrid between JN177 and tall wheatgrass.
(E) to (H) Root tips of 1-week-old JN177 and SR3 seedlings under control (E), 18% PEG (F), 200 mM NaCl (G), or 200 mM H2O2 (H) stained with nitroblue tetrazolium. The strength of the color showed the concentration of H2O2 in the root tips.
(I) Total root length of JN177 and SR3 under stress conditions.
(J) The root H2O2 content in the above seedlings. All data are given as mean ± sd. The double asterisks represent significant differences as determined by Student’s t test at P < 0.01 (n = 60). FW, fresh weight.
Bars = 2 cm in (A) to (D) and 1 mm in (E) to (H).
[See online article for color version of this figure.]
Isolation of a Stress-Responsive Gene Belonging to the SRO Family
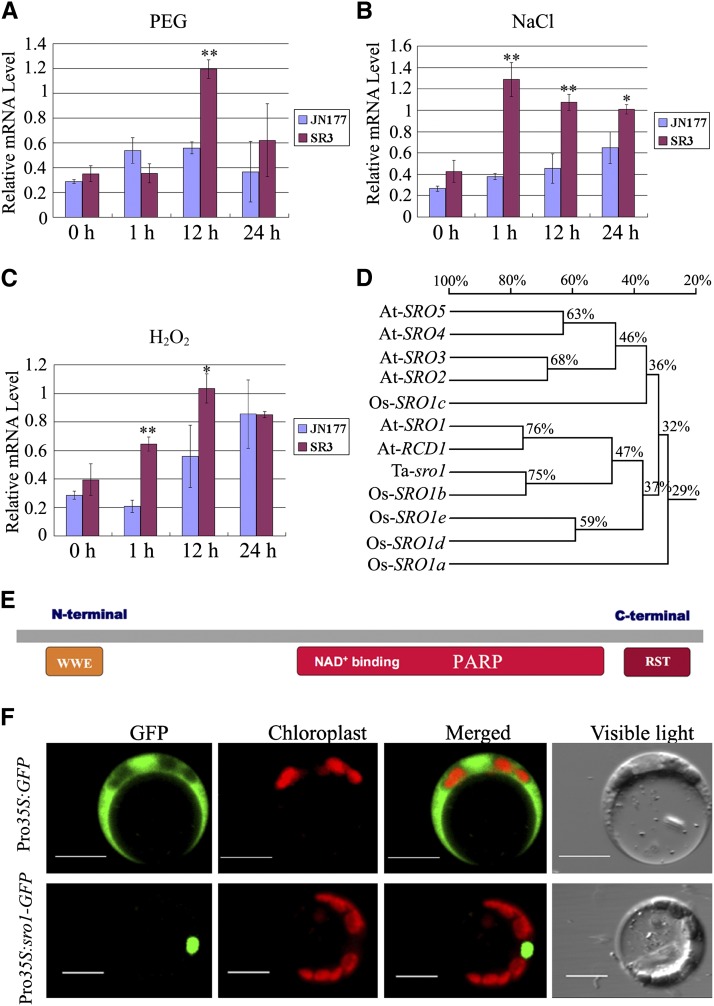
A set of 226 genes that were differentially expressed (by at least 2-fold) in SR3 seedlings under stress (200 mM NaCl or 18% PEG) was identified by microarray analysis (Liu et al., 2012). Out of these genes, those related to ROS homeostasis are suggested to play an important role in the generation of the above salinity-tolerant phenotype of cultivar SR3. Among the 64 genes related to oxidative stress (Supplemental Table 1), one shared a substantial degree of homology with At-RCD1. Copies of this gene have been located on the homoeologous group 5 chromosomes (http://wheat.pw.usda.gov/GG2/blast.shtml), which is also the site of the major stress tolerance locus in SR3 (our unpublished data). Expression analysis of this gene, based on quantitative RT-PCR, showed that both salinity and H2O2 stress induced its expression within 1 h, earlier than the dehydration stress (PEG), and its expression remained upregulated for at least 24 h under all three treatments. Transcript abundance was consistently higher in the roots of SR3 than in those of JN177 (Figures 2A to 2C).
Figure 2.
Expression, Gene Structure, and Subcellular Localization of Ta-sro1.
(A) to (C) Analysis of transcript differences between SR3 and JN177 exposed to 18% PEG (A), 200 mM NaCl (B), or 200 mM H2O2 (C) by quantitative RT-PCR. All data are given as mean ± sd of three independent biological replicates. The asterisks and double asterisks represent significant differences as determined by Student’s t test at P < 0.05 and P < 0.01, respectively.
(D) Phylogenetic analysis of Ta-sro1 and its homologous genes. This tree is based on a multiple alignment that includes the full-length cDNA sequences. The percentage given on the phylogeny shows the similarity between sequences. Alignments used to generate the phylogeny are shown in Supplemental Data Set 1.
(E) Predicted structure of Ta-sro1. Ta-sro1 contains the PARP signature and two protein–protein interaction domains: WWE and RST.
(F) Subcellular localization of Ta-sro1 protein. Images were captured using the following wavelengths: GFP (excitation, 488 nm; emission, 509 nm) and chlorophyll autofluorescence (excitation, 448 nm; emission, 647 nm). Bars = 10 μm.
The full-length cDNA sequences of both the SR3 and JN177 alleles comprised a 1734-bp open reading frame, encoding a 578-residue 63.8-kD protein. The nucleotide sequences differed from one another at three positions (Gly-585, Gly-749, and Gly-1027 in JN177 to Ala-585, Thr-749, and Ala-1027 in SR3), resulting in two residues being converted from Gly-250 and Ala-343 in JN177 to Val-250 and Thr-343 in SR3 (Supplemental Figure 2). The nucleotide sequence of its closest rice putative ortholog Os-SRO1b was 75% similar, while the Arabidopsis genes At-RCD1 and At-SRO1 (involved in the response to oxidative and osmotic stress; Teotia and Lamb, 2009) were 47% similar (Figure 2D). The gene product included both an RST and a PARP-like domain at its C terminus and a WWE domain at its N terminus (Figure 2E) and thus belongs to the SRO protein family. By comparing all other SRO genes from SR3, JN177, and tall wheatgrass, the two genes show the closest relationship, which indicates that the SR3 allele is derived from the JN177 SRO1 via point mutation; therefore, we designated JN177 SRO1 Ta-SRO1 and the SR3 allele Ta-sro1.
To determine where Ta-sro1 functions in cells, we transiently expressed Ta-sro1 with a green fluorescent protein (GFP) tag under the control of a constitutive cauliflower mosaic virus 35S promoter in Arabidopsis mesophyll protoplasts. In contrast with the GFP control that diffused in both the nucleus and cytoplasm, the Ta-sro1-GFP fusion protein exclusively localized to the nucleus (Figure 2F).
Constitutive Expression and Suppression of Ta-sro1 in Wheat
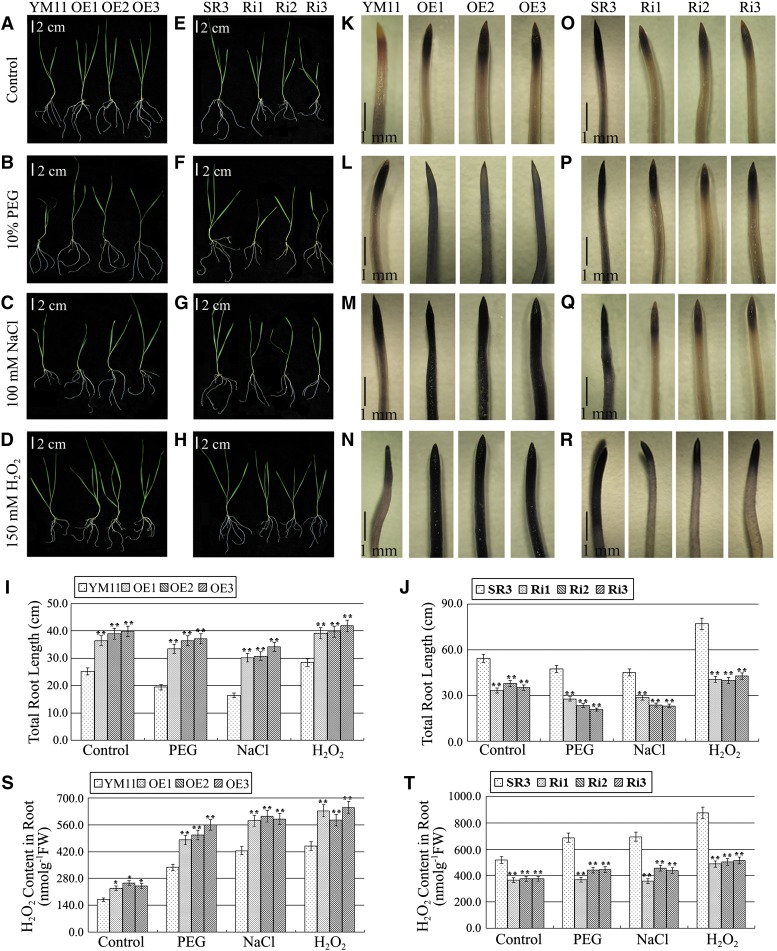
To determine if Ta-sro1 is the crucial gene that enhances both seedling growth and abiotic stress resistance in SR3, the function of Ta-sro1 was extensively explored both by constitutively expressing it in the salinity sensitive wheat cultivar Yangmai 11 and by abolishing its expression in SR3 (Supplemental Figure 3). As both wild-type Yangmai 11 and the SR3 Ta-sro1 RNA interference (RNAi) lines wilted rapidly when challenged with moderate levels of stress (18% PEG, 200 mM NaCl, and 200 mM H2O2 applied for SR3 and JN177), a lower level (10% PEG, 100 mM NaCl, and 150 mM H2O2) was applied to test these lines. The constitutive expression of Ta-sro1 increased the plant height as well as the root length under both control and stress conditions (Figures 3A to 3D and 3I; Supplemental Figure 4A), while its abolition had the opposite effect (Figures 3E to 3H and 3J; Supplemental Figure 4B). Accordingly, the ROS content of the former set of plants was higher (Figures 3K to 3N and 3S; Supplemental Figure 4C) and that of the latter was lower than the nonstressed controls (Figures 3O to 3R and 3T; Supplemental Figure 4D). Our investigations indicate that Ta-sro1 was largely responsible for the seedling’s vigorous growth and the stress tolerance of SR3.
Figure 3.
The Phenotype of Transgenic Wheat Lines under Abiotic Stress Treatment at the Seedling Stage.
(A) to (D) Phenotype of 1-week-old transgenic plants compared with YM11 under control (A), 10% PEG (B), 100 mM NaCl (C), or 150 mM H2O2 (D). OE1, OE2, and OE3 are transgenic wheat lines constitutively expressing Ta-sro1 cDNA from SR3 in wheat cultivar YM11.
(E) to (H) Phenotype of 1-week-old transgenic plants compared with SR3 under control (E), 10% PEG (F), 100 mM NaCl (G), or 150 mM H2O2 (H). Nonstressed or stressed seedlings grown for 2 weeks. Ri1, Ri2, and Ri3 are independent RNAi transgenic wheat lines knocked down for the expression of Ta-sro1 cDNA from SR3 in wheat cultivar SR3. YM11 and SR3 are nontransgenic controls.
(I) and (J) Total root length of the above seedlings.
(K) to (N) Comparison of root tips of 1-week-old seedlings under control (K), 10% PEG (L), 100 mM NaCl (M), or 150 mM H2O2 (N) stained with nitroblue tetrazolium between Ta-sro1 OE transgenic plants and YM11.
(O) to (R) Comparison of root tips of 1-week-old seedlings under control (O), 10% PEG (P), 100 mM NaCl (Q), or 150 mM H2O2 (R) stained with nitroblue tetrazolium between Ta-sro1 RNAi transgenic plants and SR3. The strength of the color showed the concentration of H2O2 in the root tips.
(S) and (T) The root H2O2 content in the above seedlings. All data are given as mean ± sd. The double asterisks represent significant differenced determined by Student’s t test at P < 0.01 (n = 15). FW, fresh weight.
Bars = 2 cm in (A) to (H) and 1 mm in (K) to (R).
[See online article for color version of this figure.]
The Ta-sro1 Protein Has PARP Activity
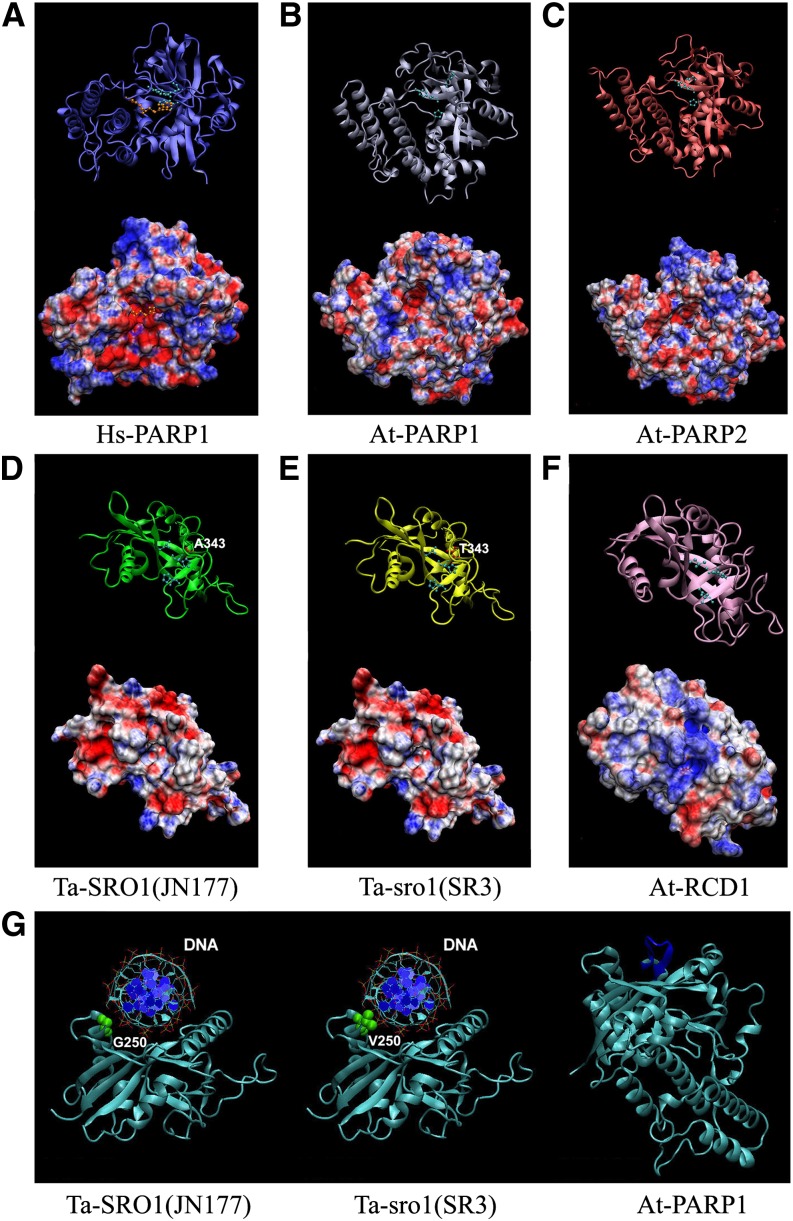
Unlike well-studied PARP enzymes, such as Homo sapiens Hs-PARP1, Hs-PARP2, At-PARP1, and At-PARP2, which showed conserved PARP activities, some PARP catalytic domain–containing proteins such as At-RCD1 did not possess the PARP activity (Jaspers et al., 2010). To further characterize the function mechanisms of the PARP catalytic domains of the SR3 Ta-sro1 and JN177 Ta-SRO1 proteins, we constructed three-dimensional models of SR3 Ta-sro1, JN177 Ta-SRO1, At-RCD1, At-PARP1, and At-PARP2 and compared them together with the reported crystal structure of Hs-PARP1 (PDB code: 1UK0) (Kinoshita et al., 2004). The resulting models were evaluated by two methods to make an integrative assessment of the structure quality, considering the geometries, stereochemistries, and energy distributions. The templates used for modeling these catalytic domains are shown in Supplemental Table 2. The evaluation results are indicative of a good quality of all five models (Supplemental Table 2). A structural analysis of Hs-PARP1 has shown that the active site pocket of its catalytic domain is negatively charged (Figure 4A) and includes the specific catalytic triad His862-Tyr896-Glu988, which is conserved in all PARP enzymes (Citarelli et al., 2010). These homology models revealed that the catalytic domains of Ta-SRO1, Ta-sro1, At-PARP1, and At-PARP2 possess an analogous negatively charged pocket (Figures 4B to 4E) to hold the positive NAD+ substrate, carrying the catalytic triads Leu312-His344-His407, Leu312-His344-His407, His833-Tyr867-Glu960, and His486-Tyr520-Glu614, respectively. The corresponding pocket of At-RCD1 is positively charged and contains the unusual triad variant Leu345-His377-Asn440 (Figure 4F), which may well explain its lack of catalytic ability. In addition, the predicted structure of Ta-sro1 includes an arc-shaped face on the opposite side of the catalytic domain, allowing for the localized accumulation of positive charge. The shape, size, and charge distribution over this surface of the predicted structure of Ta-sro1 imply that it could function as a DNA binding site (Figure 4G).
Figure 4.
Structural Models of the Catalytic Domains of PARP-Containing Proteins.
(A) to (F) Top diagrams: Representations of the protein backbone of Hs-PARP1 (A), At-PARP1 (B), At-PARP2 (C), Ta-SRO1 (JN177) (D), Ta-sro1 (SR3) (E), and At-RCD1 (F) with the catalytic triad residues labeled with light-blue spheres and sticks. The inhibitor molecule of human Hs-PARP1 (A) is shown by orange spheres and sticks. The Ala-343 residue of Ta-SRO1 (D) and Thr-343 residue of Ta-sro1 (E) are marked by red spheres and sticks. Bottom diagrams: molecular surface electrostatic potential representations. Positively charged areas shown in blue and negatively charged ones in red.
(G) Hypothetical models of interactions between the catalytic domain and DNA. The Gly-250 of Ta-SRO1 and Val-250 of Ta-sro1 on the lateral protrusion of the arc-shaped face are marked by green spheres.
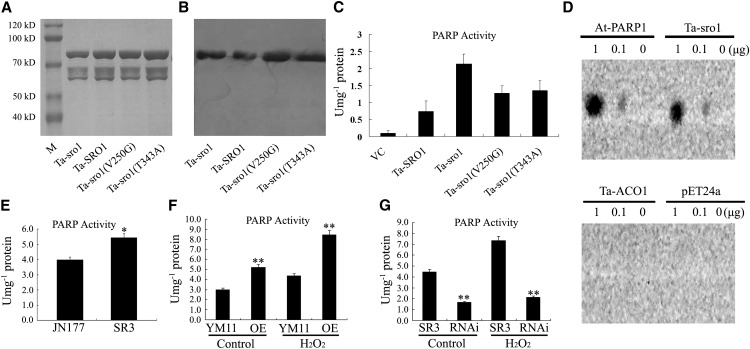
To determine whether Ta-SRO1 and Ta-sro1 have PARP enzyme activity in vitro, we expressed and purified the Ta-SRO1-His6, Ta-sro1-His6, Ta-sro1 (V250G)-His6, and Ta-sro1 (T343A)-His6 recombinant proteins from Escherichia coli cells (Figures 5A and 5B). The affinity-purified Ta-sro1 protein showed a higher level of PARP activity than did Ta-SRO1 (Figure 5C). However, reduced PARP activity could be detected in Ta-sro1 (V250G) and Ta-sro1 (T343A) (Figure 5C), in which the Val residue (V) at position 250 and Thr residue (T) at the position 343 of SR3 Ta-sro1 were replaced with the JN177 Ta-SRO1 residue Gly (G) and Ala (A), respectively. To demonstrate the ability of recombinant Ta-sro1-His6 and Ta-SRO1-His6 protein to bind to the damaged DNA in vitro, a dot blotting assay using a biotin-streptavidin system was performed. As shown in Figure 5D, Ta-sro1 exhibited significant binding toward the calf thymus DNA fragments. By contrast, Ta-SRO1 only showed weak binding to DNA fragments (Supplemental Figure 5). Taken together, these data indicate that SR3 Ta-sro1 is indeed a functional PARP enzyme and it shows higher activity than JN177 Ta-SRO1 due to the two point mutations.
Figure 5.
The in Vitro and in Vivo Poly(ADP Ribosyl)ation Activity of Ta-sro1.
(A) Photograph of purified Ta-SRO1-His6, Ta-sro1-His6, Ta-sro1 (V250G)-His6, and Ta-sro1 (T434A)-His6 separated by SDS-PAGE and stained with Coomassie Brilliant Blue R 250. M, Blue Plus II protein marker (TransGen Biotech).
(B) Immunoblot analysis of the above purified proteins. Different His fusion proteins were detected using an anti-His antibody.
(C) In vitro PARP activity assay. Total proteins from E. coli cells containing pET24a VC induced for 3 h by 1 mM isopropyl β-d-1-thiogalactopyranoside were analyzed for comparison.
(D) In vitro DNA binding activity of Ta-sro1-His6. Purified At-PARP1 protein was used as the positive control, while purified Ta-ACO1-His6 (for wheat aminocyclopropane-1-carboxylate oxidase 1) protein and pET24a total protein were used as negative controls.
(E) PARP activity in JN177 and SR3.
(F) PARP activity in wheat plants constitutively expressing Ta-sro1. The PARP activity of Ta-sro1 OE lines was higher than the nontransgenic control YM11 both under stress (YM11-H2O2 and OE-H2O2) and nonstress (YM11-C and OE-C) conditions.
(G) PARP activity in wheat RNAi knockdown plants of Ta-sro1. The PARP activity of Ta-sro1 RNAi lines was lower than that of SR3 both under stress (SR3-H2O2 and OE-H2O2) and nonstress (SR3-C and RNAi-C) conditions. All data are given as mean ± sd. The asterisks and double asterisks represent significant differences determined by Student’s t test at P < 0.05 and P < 0.01, respectively.
To investigate whether the PARP activity of Ta-sro1 is required for its biological functions in vivo, we systematically compared the PARP activity between different plant combinations. PARP activity in SR3 was considerably higher than that in JN177 (Figure 5E), a result mirrored by the behavior of the transgenic Yangmai 11 constitutively expressing Ta-sro1 and the SR3 Ta-sro1 RNAi knockdown lines (Figures 5F and 5G). These results suggest that both overall growth vigor and stress tolerance may result from the PARP activity of SR3 Ta-sro1.
Constitutive Expression of Ta-sro1 Increased ROS Production and Enhanced Stress Tolerance in Arabidopsis
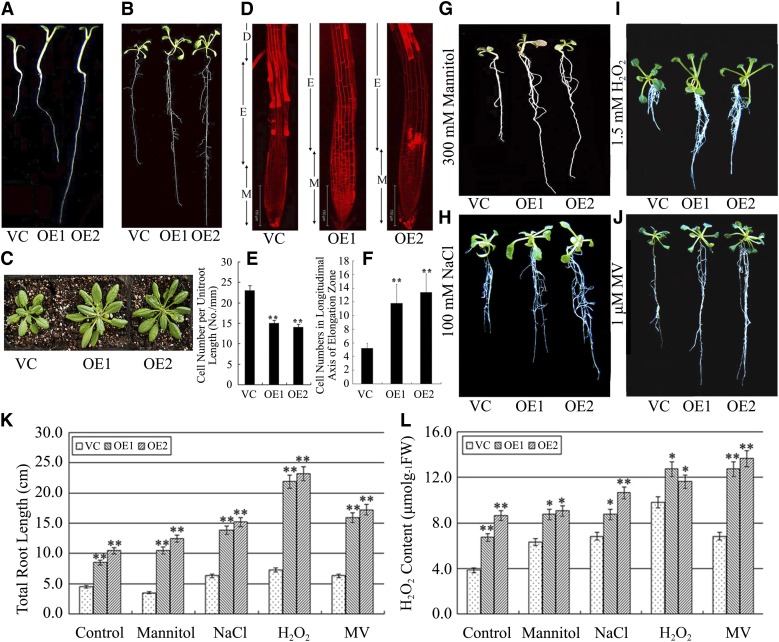
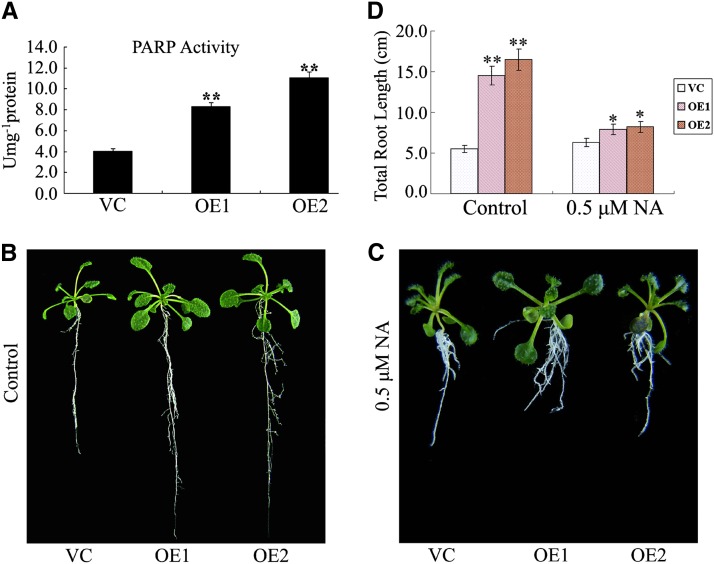
To gain further insight into the function of Ta-sro1 and the possible signaling pathways in which it participates, constitutive lines of Ta-sro1 were generated in Arabidopsis (Ta-sro1 overexpression [OE] lines). Two lines that accumulated detectable amounts of Ta-sro1 message (Supplemental Figure 6) were selected for further analysis. Root lengths of 1-week-old, 3-week-old and 8-week-old Ta-sro1 OE seedlings were substantially enhanced compared with those in the transgenic lines carrying an empty vector control (VC) lines (Figures 6A to 6C), which is caused by an enhanced cell division rate and larger cell size, as determined by comparing the cell numbers in the elongation zone and the root cell length (Figures 6D to 6F). The Ta-sro1 OE lines also displayed a notable improvement in their stress tolerance. This was particularly demonstrated by their superior root growth in the presence of 300 mM mannitol, 100 mM NaCl, 1.5 mM H2O2, and 1 μM MV, respectively (Figures 6G to 6K). A colorimetric assay showed that the Ta-sro1 OE lines accumulated more ROS than the VC lines under both nonstressed and stressed conditions (Figure 6L). Additionally, the Ta-sro1 OE lines exhibited more PARP activity than did the VC plants (Figure 7A), and root elongation was more severely compromised by the presence of the PARP inhibitor nicotinamide (NA) (Figures 7B to 7D), which further proved that these phenotypes conferred by the Ta-sro1 variant are not simply due to its higher level of expression but due to its higher PARP activity. Collectively, these data indicated that the heterologous expression of Ta-sro1 in Arabidopsis showed consistent phenotypes with wheat Ta-sro1 OE plants and SR3 and thus that Arabidopsis Ta-sro1 OE lines can be used for further research into the mechanism by which Ta-sro1 enhances seedling growth and abiotic stress resistance.
Figure 6.
Phenotype of Arabidopsis Plants Constitutively Expressing Ta-sro1.
(A) to (C) Phenotypes of 1-week-old (A), 3-week-old (B), and 8-week-old (C) transgenic plants compared with VC plants. The VC plants are a transgenic line carrying an empty vector. OE1 and OE2 are transgenic Arabidopsis lines constitutively expressing Ta-sro1 cDNA from SR3.
(D) Root tips of 1-week-old seedlings as visualized by confocal laser scanning microscopy after staining with propidium iodide. The elongation zone of OE lines was significantly longer than that of the VC. M, meristematic zone; E, elongation zone; D, differentiation zone.
(E) Cell number statistics in roots of 1-week-old seedlings, where the larger size of OE line root elongation zone cells resulted in fewer cells per unit root length.
(F) Cell number statistics in the longitudinal axis of the root elongation zone of 1-week-old seedlings. The elongation zone of OE lines contains more cells than that of the VC.
(G) to (J) Phenotype of 1-week-old transgenic seedlings stressed for 2 weeks with either 300 mM mannitol (G), 100 mM NaCl (H), 1.5 mM H2O2 (I), or 1 μΜ MV (J) compared with the VC.
(K) and (L) Total root length (K) and root H2O2 content (L) in stressed seedlings. All data are given as mean ± sd. The asterisks and double asterisks represent significant differences, as determined by Student’s t test at P < 0.05 and P < 0.01, respectively (n = 30). FW, fresh weight.
Figure 7.
PARP Activity and the Effect of the PARP Inhibitor NA on Root Growth in Arabidopsis Ta-sro1 OE Lines.
(A) Comparison of PARP activity between VC and Ta-sro1 OE lines.
(B) and (C) Seedling phenotype of transgenic Arabidopsis lines compared with control plants under normal conditions (B) and 0.5 μM NA treatment (C).
(D) Effect of NA treatment on root growth. All data are given as mean ± sd. The asterisks and double asterisks represent significant differences, as determined by Student’s t test at P < 0.05 and P < 0.01, respectively (n = 30).
[See online article for color version of this figure.]
To confirm that Ta-sro1 enhances stress tolerance by increasing ROS level, we introduced the Pro35S:sro1 transgene into the At-rbohF mutant background, in which the primary root growth is repressed (Kwak et al., 2003). We found that Ta-sro1 could rescue the shorter root of At-rbohF (Supplemental Figure 7) under nonstressful conditions. When exposed to 0.6 mM H2O2, the root elongation of the At-rbohF mutant was more severely inhibited, whereas Ta-sro1 overexpression in the At-rbohF/35S:sro1 transgenic line rescued the root length of the At-rbohF mutant, resulting in plants with roots that were even a little longer than those of the wild type (Supplemental Figure 7). Consistently, the Ta-sro1 OE line showed the longest root (Supplemental Figure 7). These results indicated that the higher ROS levels of Ta-sro1 OE lines was responsible for its stress tolerance.
ROS Accumulation in the Arabidopsis Ta-sro1 OE Lines
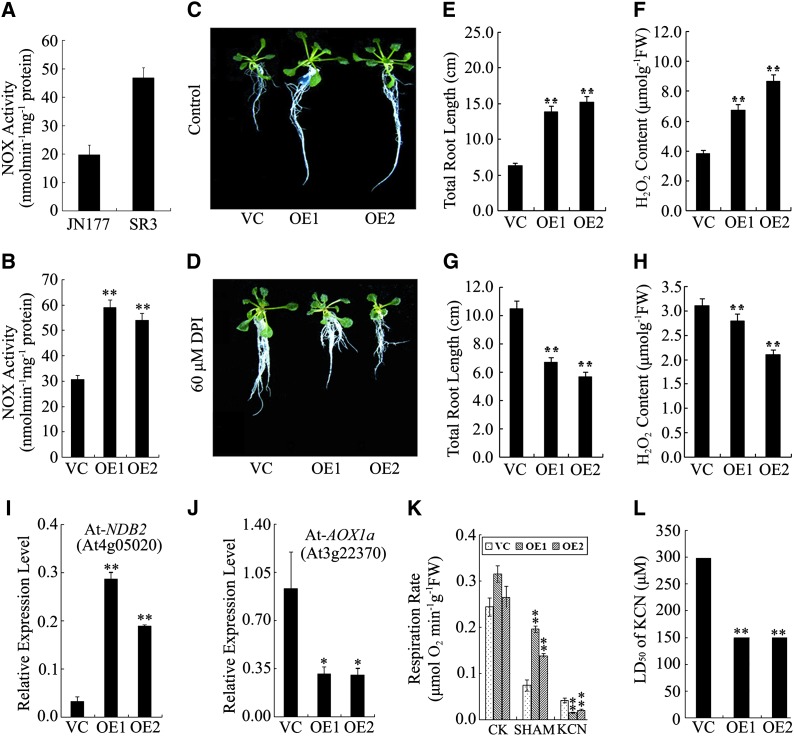
As NADPH oxidase (NOX) activity has been implicated in ROS formation (Foreman et al., 2003; Mittler et al., 2004) and Ta-sro1 can complement the At-rbohF (an important gene of NOX) mutant phenotype, NOX activity was compared between SR3 and JN177. Consistent with their contrasting accumulation of ROS, NOX activity was higher in the former (Figure 8A). Consistent with our expectation, the Ta-sro1 OE lines also showed higher NOX activity than VC lines (Figure 8B). When the plants were treated with the NOX inhibitor diphenylene iodonium (DPI), a measurable decrease in ROS accumulation and root elongation was observed for the Ta-sro1 OE lines (Figures 8C to 8H). A further source of ROS (particularly H2O2) is the mitochondrial electron transport chain, which in plants includes alternative oxidase (AOX) and various NAD(P)H dehydrogenases. The most ubiquitously expressed AOX gene in Arabidopsis is At-AOX1a, while At-NDB2 encodes an NAD(P)H dehydrogenase frequently coexpressed with At-AOX1a as part of the stress response (Clifton et al., 2006). Expression analysis indicated that At-AOX1a was more markedly downregulated in the Ta-sro1 OE line than in the VC plants, while At-NDB2 was upregulated (Figures 8I and 8J). The AOX-dependent respiration rate was measured either in the presence of potassium cyanide (KCN; a cytochrome inhibitor) or salicyl hydroxamic acid (SHAM; an AOX inhibitor). No significant difference between respiration rate of Ta-sro1 OE and VC was detectable prior to inhibitor treatment, but respiration was significantly increased in the former in the presence of SHAM and reduced by KCN (Figure 8K), suggesting that the Ta-sro1 transgene downregulates AOX. Furthermore, the LD50 (half lethal dose of KCN) for KCN was 150 μM for the Ta-sro1 OE lines but 300 μM for VC lines (Figure 8L). Taken together, the enhanced NOX activity and altered expression of At-AOX1a and At-NDB2 genes in the Ta-sro1 OE lines may be responsible for higher ROS accumulation in roots.
Figure 8.
Characterization of ROS Production Elements in Wheat and Arabidopsis Plants Constitutively Expressing Ta-sro1.
(A) Comparison of NOX activity between JN177 and SR3.
(B) Comparison of NOX activity between Ta-sro1 OE lines and VC plants.
(C) and (D) Phenotype of VC, Ta-sro1 OE1, and Ta-sro1 OE2 transgenic plants grown in normal conditions (C) or in the presence of 60 μM NOX inhibitor DPI (D).
(E) to (H) Root growth and H2O2 content in seedlings grown in the absence ([E] and [F]) or presence of DPI ([G] and [H]). FW, fresh weight.
(I) and (J) Expression levels of At-NDB2 [NAD(P)H dehydrogenase] (I) and At-AOX1a (J) between Ta-sro1 OE lines and VC plants.
(K) and (L) respiration rate (K) and LD50 (half lethal dose of KCN) for KCN (L) between Ta-sro1 OE lines and VC plants. All data are given as mean ± sd. The double asterisks represent significant differences, as determined by Student’s t test at P < 0.01 (n = 30).
[See online article for color version of this figure.]
ROS Scavenging in the Arabidopsis Ta-sro1 OE Lines
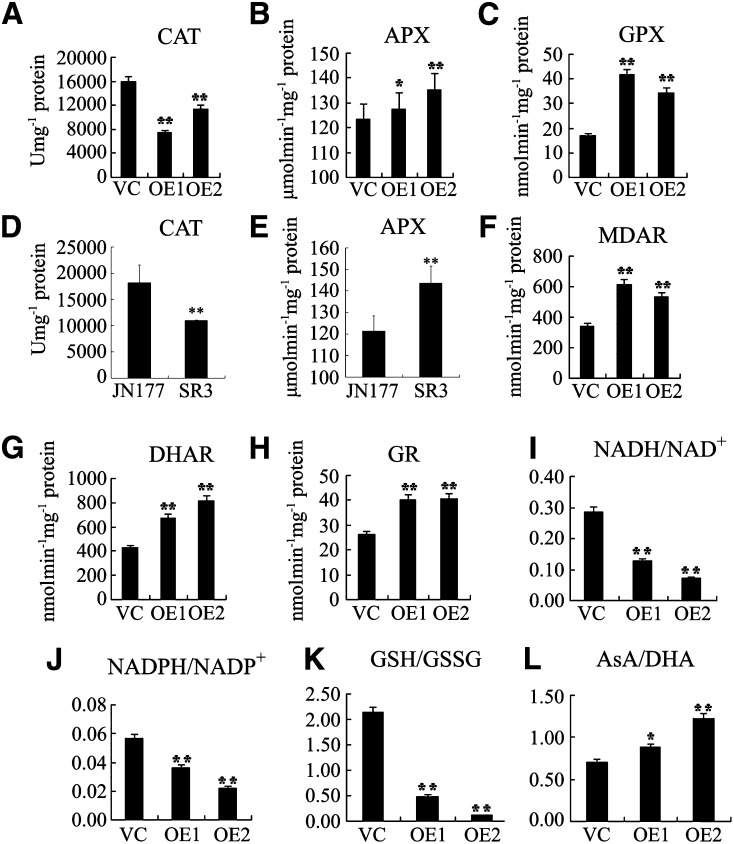
To address the possibility that ROS were detoxified more rapidly in the Ta-sro1 OE lines or that metabolism of these lines is more tolerant of elevated ROS content, the activities of various key enzymes (superoxide dismutase [SOD], peroxidase [POD], catalase [CAT], ascorbate peroxidase [APX], and glutathione peroxidase [GPX]) were determined. No significant difference was noted between the Ta-sro1 OE and VC lines with respect to either SOD or POD activity, but the level of both APX and GPX (involved in the removal of hydrogen peroxide) activity was higher in the Ta-sro1 OE lines, and CAT activity was lower (Figures 9A to 9C). To characterize ROS scavenging enzymes in SR3 and JN177, we also determined the activity of some key enzymes in wheat. Consistent with the enzyme activities in the Ta-sro1 OE and VC lines, SR3 also showed higher APX and lower CAT activity than JN177 (Figures 9D and 9E). No significant difference in SOD and POD activity was found between JN177 and SR3. These results suggest that the Ta-sro1 OE lines were more effective than the VC lines in terms of ROS detoxification.
Figure 9.
Cellular Redox Balance Is Affected in Wheat and Arabidopsis Plants Constitutively Expressing Ta-sro1.
(A) to (C) Comparison of enzyme activities of CAT (A), APX (B), and GPX (C) between Ta-sro1 OE lines and VC plants.
(D) and (E) Comparison of enzyme activities of CAT (D) and APX (E) between JN177 and SR3.
(F) to (H) Comparison of enzyme activities of MDAR (F), DHAR (G), and GR (H) between Ta-sro1 OE lines and VC plants.
(I) to (L) Comparison of the ratios of NADH/NAD+ (I), NADPH/NADP+ (J), GSH/GSSG (K), and AsA/DHA (L) between Ta-sro1 OE lines and VC plants. All data are given as mean ± sd. The asterisks and double asterisks represent significant differences, as determined by Student’s t test at P < 0.05 and P < 0.01, respectively.
The same approach was taken regarding redox balance maintenance, where the activities of enzymes in the ascorbate (AsA)-GSH cycle (monodehydroascorbate reductase [MDAR], dehydroascorbate reductase [DHAR], and glutathione reductase [GR]) and the presence of the four antioxidant couples AsA/dehydroascorbate (DHA), GSH/GSSG, NADH/NAD+, and NADPH/NADP+ were monitored. MDAR, DHAR, and GR activity was greater in the Ta-sro1 OE lines than in the VC lines (Figures 9F to 9H). While the AsA/DHA ratio was maintained more effectively in the Ta-sro1 OE lines, those for NADH/NAD+, NADPH/NADP+, and GSH/GSSG ratios were all reduced (Figures 9I to 9L). Thus, we can conclude that Ta-sro1 promotes the activity of AsA-GSH cycle enzymes and GPX cycle enzymes, which control ROS content and cellular redox homeostasis.
Increased Genomic Integrity in the Arabidopsis Ta-sro1 OE Lines
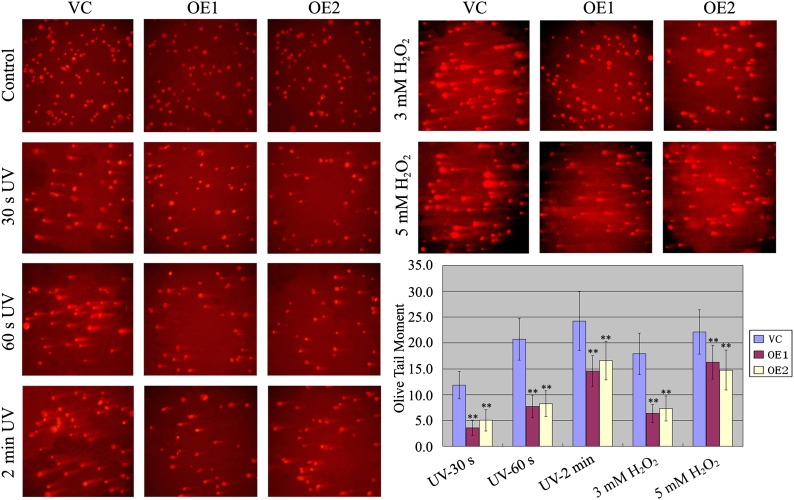
PARPs have been implicated in DNA damage repair and genomic integrity (Kim et al., 2005; Schreiber et al., 2006; Hassa and Hottiger, 2008). The involvement of Ta-sro1 in ROS homeostasis, PARP activity, and the nuclear localization of Ta-sro1 prompted us to test whether Ta-sro1 also functions in this process. We therefore detected the damage to genomic DNA of Ta-sro1 OE and VC lines under UV irradiation and H2O2 treatments by single-cell gel electrophoresis (SCGE). As shown in Figure 10, Ta-sro1 OE protoplasts were more resilient than those of the VC. Similarly, when exposed to UV irradiation, the protoplasts of SR3 showed less DNA damage than JN177 (Supplemental Figure 8), which further proved that Ta-sro1 may be largely responsible for the stress tolerance of SR3. Taken together, these results imply that Ta-sro1 plays an important role in DNA damage repair and genomic integrity through its PARP activity, by which it copes with the abiotic stresses and maintains plant growth.
Figure 10.
SCGE Analysis of the Protoplasts of Wild-Type and Arabidopsis Plants Constitutively Expressing Ta-sro1 Exposed to UV or H2O2.
DNA damage in protoplasts of Arabidopsis plants constitutively expressing Ta-sro1 and VC plants exposed to UV for 30 s, 60 s, and 2 min or 3 mM H2O2 or 5 mM H2O2 as indicated. Olive tail moment (OTM) represents the product of the percentage of total DNA in the tail (TD) and the tail length (TL): OTM = TL × TD. Abbreviations are as in Figure 5. All data are given as mean ± sd of four independent experiments. The double asterisks represent significant differences, as determined by Student’s t test at P < 0.01.
DISCUSSION
The PARP proteins are widely distributed among the eukaryotes (although are absent from yeast) and appear to be involved in processes ranging from DNA repair and the regulation of chromatin and telomere structure to transcriptional regulation, the response to abiotic and biotic stress, and the activation of cell death (Beneke and Bürkle, 2007; Citarelli et al., 2010). Surprisingly, this important protein family has received little attention from plant scientists to date (Skirycz et al., 2011). Here, we described the functional characterization of a wheat PARP and showed that it underpins both seedling vigor and tolerance to abiotic stress, through its catalytic activity and modulation of redox homoeostasis.
The PARP Activity of Ta-sro1 in Relation to the DNA Integrity of Ta-sro1 OE Lines
Eukaryotic organisms express multiple PARP proteins, which although recognized by the presence of a conserved PARP catalytic domain, do not all display PARP activity (Citarelli et al., 2010). The presence of a negatively charged pocket able to retain the NAD+ substrate, together with its LHH catalytic triads (although not a classical one) (Figure 4E), suggested that SR3 Ta-sro1 would have PARP activity, which was borne out experimentally (Figure 5C). The greater PARP activity of Ta-sro1 may well flow from substitution from Ala-343 to Thr-343 of Ta-sro1 because the hydrophobic Ala residue does not contribute to the interaction with the substrate, whereas the hydrophilic Thr residue does so by presenting both a methyl and a hydroxyl group to the catalytic surface, which encourages interaction with the substrate (Figures 4D and 4E). Unlike the well-characterized animal and plant PARPs, which feature at least one DNA binding domain, SRO proteins have no such domains (Citarelli et al., 2010). The predicted structure of Ta-sro1 includes an arc-shaped face on the far side of the catalytic domain (opposite to the catalytic pocket), allowing for the localized accumulation of positive charge. The shape, size, and charge distribution over this surface of the predicted structure of Ta-sro1 suggested that this surface may function as a DNA binding site (Figure 4G); therefore, it is reasonable to find that the Ta-sro1 protein has DNA binding ability (Figure 5D). Intriguingly, a second allelic difference between the SR3 Ta-sro1 and JN177 Ta-SRO1 proteins (Val-250/Gly-250) lies on a lateral protrusion from this surface (Figure 4G), which implies that the more hydrophobic Val in the SR3 version of the protein increases its DNA binding capacity over that achieved by the JN177 version (Supplemental Figure 5). This may contribute to the fact that Ta-sro1 (V250G) protein has decreased PARP enzyme activity compared with Ta-sro1 (Figure 5C). By contrast, Hs-PARP and At-PARP both comprise a two-stranded β-sheet above the arc-shaped face, giving rise to a structure that mitigates against DNA binding at this site (Figure 4G).
Reduced PARP activity has been reported to improve tolerance to a broad range of abiotic stresses, through its reduction in NAD+ consumption (De Block et al., 2005; Vanderauwera et al., 2007). The strong activation of PARP and poly(ADP ribosyl)ation can alter the cellular redox state, leading to a depletion in the availability of ATP, which in severe cases induces cell death (Schraufstatter et al., 1986). On the contrary, the constitutive expression of the SR3 allele of Ta-sro1 in Arabidopsis, despite the increased PARP activity of its gene product (Figure 7A) and decreased ATP content (Supplemental Figure 9), enhanced both plant growth and stress tolerance. This is explainable. Under normal conditions, heterologous expression of SR3 Ta-sro1 in Arabidopsis resulted in higher PARP activity and, thus, more ATP depletion. Upon exposure to abiotic stress (NaCl or H2O2), the ATP content of Ta-sro1 OE lines increased compared with the decrease or less of an increase ATP level in the wild type (Supplemental Figure 9). It is therefore possible that Ta-sro1 represses the alternative pathway by repressing AOX1a expression, thereby promoting the efficiency of ATP synthesis. This also implies that energy homeostasis is the basis of the stress tolerance.
PARP is also one of the earliest response factors to DNA breakage and plays an important role in maintaining DNA integrity (Kim et al., 2005). In mouse, both Parp1 and Parp2 knockouts are hypersensitive to alkylating agents as a result of their defective DNA excision repair machinery (Trucco et al., 1998; Schreiber et al., 2002; Kim et al., 2005; Schreiber et al., 2006; Hassa and Hottiger, 2008). The constitutive expression of Arabidopsis Parp in soybean (Glycine max) cells subjected to a mild level of oxidative stress reduced the frequency of ROS-induced DNA damage, while abolishing it produced the opposite effect (Amor et al., 1998). Here, SCGE analysis suggested that the higher PARP activity (Figure 7A) in Ta-sro1 OE transgenic plants helped maintain DNA integrity by enhancing the activity of the DNA repair machinery (Figure 10). In mammalian cells, PARP-1 could interact directly with ATM (for Ataxia-Telangiectasia mutated), an important sensor of ROS (Guo et al., 2010), and form a large protein complex to function in DNA repair and maintenance of genomic integrity through its PARP activity (Aguilar-Quesada et al., 2007; Gagné et al., 2008). In our study, At-ATM had a higher expression level in Arabidopsis Ta-sro1 OE lines than in the VC line under both NaCl and H2O2 treatments (Supplemental Figure 10). In addition, the PARP domain of Ta-sro1 is accompanied by two different putative protein–protein interaction domains, WWE and RST. Thus, Ta-sro1 could possibly act as a scaffold protein that assembles DNA damage repair proteins for their posttranslational modification, relocalization, or degradation.
Ta-sro1 Promotes Growth and Abiotic Stress Tolerance by Regulating Redox Homoeostasis
We demonstrated that the constitutive expression of Ta-sro1 in wheat and Arabidopsis enhanced the accumulation of ROS and promoted plant growth and stress tolerance (Figures 3 and 6). The production of ROS by the NOX homolog (Rboh) was first discovered in the defense reactions against pathogens, and the discovery of new functions for plant Rbohs underlined diverse roles for NOX-generated ROS, including hormone synthesis, the regulation of stomatal closure, and root hair growth (Hammond-Kosack and Jones, 1996; Sagi and Fluhr, 2001; Foreman et al., 2003; Kwak et al., 2003). The discovery that NOX activity was substantially increased in SR3 and Ta-sro1 OE lines (Figures 8A and 8B) and that Ta-sro1 could complement the absence of At-rbohF (Supplemental Figure 7) suggested that NOX-generated ROS also acts in plant growth and abiotic stress. NAD(P)H dehydrogenase is a non-proton-pumping respiratory enzyme that functions in parallel with complex I to transfer electrons from NAD(P)H to ubiquinone. Together, the ubiquinone radical and NAD(P)H dehydrogenase are the main generators of ROS in the mitochondrion (Dat et al., 2000), so it is reasonable to suggest that in the Ta-sro1 OE lines, the upregulation of the NAD(P)H dehydrogenase NDB2 (Figure 8H) would increase the production of ROS. In addition, the upregulation of both NOX and NAD(P)H dehydrogenase would likely decrease the NAD(P)H/NAD(P)+ ratio via the oxidation of NAD(P)H (Figures 9I and 9J). AOX activity helps to regulate oxygen homoeostasis within the mitochondria, and its suppression results in the elevation of ROS content (Maxwell et al., 1999). Both the gene expression and respiration inhibition experiments indicated a repression of AOX in the Ta-sro1 OE lines (Figure 8), which can explain, at least in part, the extra accumulation of ROS.
The dual function of ROS implies that its cellular concentration has to be tightly controlled (Apel and Hirt, 2004). It has been proposed that AsA and GSH act as ROS scavengers either interdependently (AsA-GSH cycle) or independently (GPX cycle), and the enzymes include APX, GPX, MDAR, DHAR, and GR (Foyer and Noctor, 2011). It was possible to show that most of the extra ROS induced by the constitutive expression of Ta-sro1 was scavenged through enzymes of the GPX cycle and AsA-GSH cycle and not by POD, SOD, or CAT (Figure 9). Note that both of these ROS processing pathways need GSH and NAD(P)H as reducing agents, thereby reducing the GSH/GSSG and NAD(P)H/NAD(P)+ ratios (Figures 9I to 9K). The efficient antioxidant system ensures redox homeostasis, which can be indicated by the lower malondialdehyde (MDA) content in Ta-sro1 overexpression lines of Arabidopsis under salinity (Supplemental Figure 11), in agreement with that in SR3 (Liu et al., 2012).
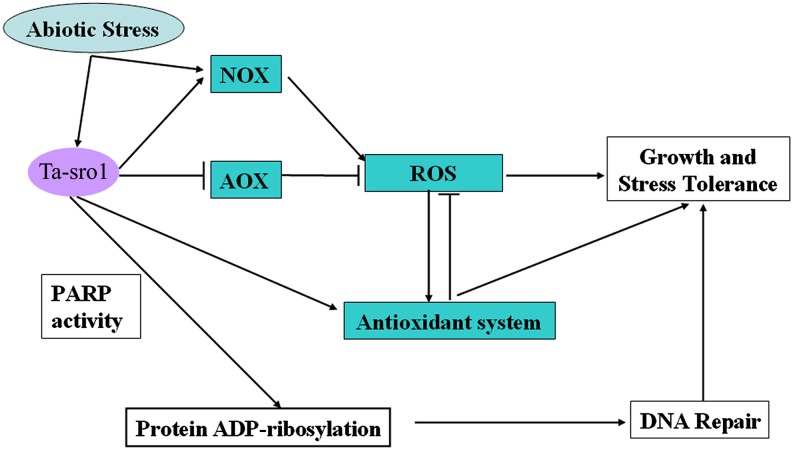
In conclusion, we demonstrated that Ta-sro1 is involved in seedling vigor and tolerance to abiotic stress, acting via the modulation of redox homoeostasis and the maintenance of genome stability (Figure 11). A possible outline mechanism is that its constitutive expression enhances NOX activity and NAD(P)H dehydrogenase expression, while at the same time repressing AOX expression. As a result, the plant accumulates a higher level of H2O2 than does the wild type. It also enhances the regulation exerted by the GPX cycle and AsA-GSH cycle over H2O2 concentration and redox homoeostasis. Meanwhile, the PARP enzyme activity of Ta-sro1 helps to maintain DNA integrity. Notably, based on the presented data we cannot exclude the possibility that Ta-sor1 acts by other mechanisms; however, all of the above properties make Ta-sro1 an excellent candidate for plant genetic improvement.
Figure 11.
A Proposed Model for the Role of Ta-sro1 in the Regulation of Abiotic Stress Tolerance and Plant Growth.
Abiotic stress induced the expression of Ta-sro1, which promoted the accumulation of ROS, mainly by enhancing NOX activity and NAD(P)H dehydrogenase expression in conjunction with the suppression of AOX expression. At the same time, Ta-sro1 promoted the activities of enzymes in the ROS scavenging system to regulate cellular redox homeostasis and maintain the ROS content to a higher level, through which stress tolerance and plant growth are controlled. Meanwhile, the PARP activity of Ta-sro1 could enhance the genome stability of plants by modulating DNA damage repair, which also improves stress tolerance and plant growth.
[See online article for color version of this figure.]
METHODS
Plant Materials and Growing Conditions
The bread wheat (Triticum aestivum) cultivar SR3 was derived from an asymmetric somatic hybrid between the cultivar JN177 and Thinopyrum ponticum. SR3, the salinity sensitive wheat cultivar Yangmai 11 and wild-type Arabidopsis thaliana ecotype Columbia-0 are all maintained in our laboratory. For germination, wheat grains were first surface sterilized in 2% (w/v) sodium hypochlorite (NaClO) and then laid on moistened filter paper for 2 d at 20°C. Seedlings of uniform size were transferred to a 12-h photoperiod regime (light/dark temperature 22/18°C) under 300 μmol m−2 s−1 illumination and a relative humidity of 50% and were grown hydroponically in half-strength Hoagland solution. Abiotic stress was applied to SR3 and JN177 by the addition of 18% (w/v) PEG, 200 mM NaCl, or 200 mM H2O2 to the hydroponic solution after 1 week. A lower level of abiotic stress was applied to the Ta-sro1 OE lines and RNAi lines together with control lines by the addition to the hydroponic solution of 10% (w/v) PEG, 100 mM NaCl, or 150 mM H2O2 after 1 week. Arabidopsis seeds were surface sterilized by immersion in 0.1% (w/v) mercuric chloride and plated on a 1% agar medium supplemented with Murashige and Skoog salts and 3% (w/v) Suc. After keeping seeds in the dark at 4°C for 3 d, the plates were removed to a 16-h photoperiod, 22°C, and 70% relative humidity regime (light intensity 170 to 200 μmol m−2 s−1). One-week-old seedlings were exposed to either 300 mM mannitol, 100 mM NaCl, 1.5 mM H2O2, 1 μM MV, or 50 to 600 μM KCN for 2 weeks.
Transgenic Constructs of Ta-sro1 and Genetic Transformation
To generate the RNAi construct, we searched the wheat genome sequence database (http://www.wheatgenome.org) for SRO and PARP homologous genes and identified 10 wheat SRO and eight PARP homologous sequences. Among them, only three sequences (chr5bl_10790093, chr4al_7142517, and chr5dl_4543936 referring to the three Ta-SRO1 alleles) located on 5B, 4A, and 5D genomes of wheat showed high similarity with Ta-sro1. The RNAi construct was designed in the 3′ end of Ta-sro1, which contains the RST domain and 3′-untranslated region of the above 3 SRO1 sequences. A 410-bp fragment was amplified using primers Ta-sro1Ri-F and Ta-sro1Ri-R and inserted conversely into the vector pTCK303 (under the control of the Zm-Ubiquitin promoter) to knock down Ta-sro1 and the other two alleles of SR3.
To generate the Pro35S:sro1 construct for overexpression in Arabidopsis and the At-rbohF mutant, Ta-sro1 coding DNA sequence (CDS) was amplified with primers Ta-sro1AOE-F and Ta-sro1AOE-R, and the XbaI-BamHI–digested PCR product was then ligated into the binary vector pBI121. To generate the ProUbi:sro1 plasmid for overexpression in wheat, the Ta-sro1 CDS was amplified using primers Ta-sro1WOE-F and Ta-sro1WOE-R and digested with BamHI and SacI for ligation into the binary vector pTCK303. The binary plasmids were transferred into the Agrobacterium tumefaciens strain AGL1 using the freeze-thaw method. Wheat plants were transformed using the shoot apical meristem method (Zhao et al., 2006), while Arabidopsis and At-rbohF mutant plants were transformed by the floral dip method (Clough and Bent, 1998). The primer sequences used for the above studies are listed in Supplemental Table 3.
RNA Extraction, cDNA Isolation, RT-PCR, and Real-Time Quantitative RT-PCR
Total RNA was extracted using the TRIzol reagent according to the user manual (Invitrogen). The first strand cDNA was synthesized using an oligo(dT) primer and SuperScript II reverse transcriptase (Invitrogen). The expression of Ta-sro1 transgene in wheat and Arabidopsis was confirmed by RT-PCR with the gene-specific primers. Real-time quantitative RT-PCR was performed in an iCycler thermal cycler and detected using the iCycler iQ real-time PCR detection system (Bio-Rad). The reaction mixture comprised 1 μL each of template, forward and reverse primer (0.3 μM each), and FastStart Universal SYBR Green Master (Rox; Roche). The relevant primers are given in Supplemental Table 3. For wheat, Actin was used as the internal control and for Arabidopsis Tubulin. All reactions were run in triplicate.
Subcellular Localization of Ta-sro1
To generate Pro35S:sro1-GFP, a BamHI-HindIII fragment containing the coding region of Ta-sro1 was amplified by the primers Ta-sro1LF and Ta-sro1LR (Supplemental Table 3) and subcloned into the BamHI and HindIII sites of Pro35S:GFP (Lin et al., 2009). The plasmids Pro35S:GFP and Pro35S:sro1-GFP were introduced into Arabidopsis mesophyll protoplasts as described (Sheen, 2001). After overnight incubation in the dark, the GFP signal and chlorophyll autofluorescence were examined under a confocal microscope at excitation wavelengths of 488 and 647 nm, respectively (FluoView 1000; Olympus).
Quantification of Root Length and H2O2 Content
The root length of treated and untreated control seedlings was measured using an image analysis system (WinRHIZO; Régent Instruments). Prior to H2O2 measurement, the wheat roots were excised and immersed for 1 h in PBS solution containing 0.5 mg mL–1 nitroblue tetrazolium. The staining reaction was stopped by the addition of an excess of 70% (v/v) methanol. The roots were rinsed in fresh 70% (v/v) methanol (Jones et al., 2007) and then observed and photographed under a stereomicroscope (SMZ-800; Nikon). H2O2 content was determined following the protocol of the H2O2 Colorimetric Assay Kit (Beyotime). A 150- to 300-mg sample of fresh tissue was snap-frozen in liquid nitrogen and powdered in a mortar together with 1.5 mL of frozen 5% (w/v) trichloroacetic acid (TCA) and 45 mg of activated charcoal. The homogenate was centrifuged at 18,000g for 10 min at 0°C, and the resulting supernatant filtered through a 45-µm nylon filter. The pH of the filtrate was adjusted to 8.4 with NH4OH. After refiltration, a 100-μL aliquot was mixed with 100 μL of the colorimetric reagent, and the reaction mixture was held for 30 min at 30°C. The intensity of the color that developed was spectrophotometrically assessed at 560 nm.
SCGE
For UV treatment, the Arabidopsis and wheat protoplasts were irradiated with UV-C (380 µW cm−2) for 30 s, 1 min, and 2 min and used directly for SCGE analysis. For H2O2 treatment, 3 or 5 mM H2O2 was added to the suspension and cultured for 12 h before protoplast isolation. These cells were mixed with 0.65% (w/v) low melting point agarose in 1× TBE (Tris/Borate/EDTA), and 70 μL was pipetted onto a microscope slide that had previously been coated with 1.5% (w/v) standard agarose and dried in an oven. The preparation was covered with a 22 × 22-mm cover slip and placed on ice for 5 min. After removing the cover slip, the slide was immersed in 2 M NaCl, 100 mM EDTA, 10 mM Tris, pH 10, 1% (w/v) N-lauroyl sarcosine sodium, 6% (v/v) DMSO, and 1% (v/v) Triton X-100 at 4°C for 3 h, washed twice with deionized water, and then transferred to 300 mM NaOH and 1 mM Na2EDTA, pH 13, for 40 min. The contents of the slide were subjected to electrophoresis in 1× TBE for 20 min at 2 V/cm and stained in 5 μg mL−1 ethidium bromide for 10 min. The distribution of fluorescence in the head and tail of the resulting comet of 30 to 50 cells (a measurement that correlates positively with the extent of DNA breakage) (Olive et al., 1990) was assessed. Olive tail moment (OTM) was used to estimate the DNA damage, which represents the product of the percentage of total DNA in the tail (TD) and the tail length (TL), measured from the center of the nucleus toward the end of the tail (OTM = TL × TD). Each assay was performed in triplicate.
LD50 for KCN and Respiration Rate in Arabidopsis OE Lines
One-week-old seedlings grown on Murashige and Skoog solid medium were transferred to the same medium supplemented with 50 to 600 μM KCN and scored for survival after 14 d. The respiration rate of the roots of Ta-sro1 OE and VC plants was obtained from 0.1 to 0.2 g (fresh weight) detached root tissue sealed in an airtight cuvette containing phosphate buffer, pH 6.8, and a Clark-type oxygen electrode (Oxy-lab; Hansatech). The capacity of the alternative and cytochrome C pathways was deduced from the difference between respiration in the presence of 0.5 mM KCN or 3 mM SHAM and residual respiration. The latter was measured in the presence of inhibitors of both 3 mM SHAM and 0.5 mM KCN (Bingham and Farrar, 1989).
Determination of PARP Activity
To express and purify Ta-SRO1, Ta-sro1, Ta-sro1 (V250G), and Ta-sro1 (T343A) proteins for PARP enzyme activity assay in vitro, the Ta-SRO1 and Ta-sro1 CDS were amplified using the primer set listed in Supplemental Table 3. Ta-sro1 (V250G) was generated by site-directed mutagenesis PCR (Ho et al., 1989) using primer pairs Ta-sro1PF/V250GR (5′-TGAATCACCAGGTTGGCC-3′) and V250GF (5′-GGCCAACCTGGTGATTCA-3′)/Ta-sro1PR. Ta-sro1 (T343A) was amplified using primer pairs Ta-sro1PF/T343AR (5′-TGCACCAATGCCATACACTG-3′) and T343AF (5′-CAGTGTATGGCATTGGTGCA-3′)/Ta-sro1PR. After sequencing, the DNA fragment containing the entire CDS of Ta-sro1, Ta-SRO1, Ta-sro1 (V250G), or Ta-sro1 (T343A) was cloned into the BamHI and NotI sites of the prokaryotic expression vector pET24a. The expression cassette for the fusion protein His6-tagged Ta-sro1 was transferred into Escherichia coli strain BL21 (DE3), and the subsequently expressed fusion protein was purified from homogenized bacterial cells using a His-tagged protein purification kit (R&D Systems) according to the manufacturer's protocol. The Quick Start Bradford protein assay (Bio-Rad) was used to determine the concentration of purified proteins. Affinity-purified His-fused proteins were used for PARP activity determination using the PARP Universal Colorimetric Assay Kit (Trevigen) following the manufacturer’s protocol.
To explore the PARP activity in vivo, roots of the wheat cultivars SR3 and JN177 and whole Arabidopsis seedlings were snap-frozen in liquid nitrogen and macerated in frozen 1× PARP buffer (Trevigen) containing 0.4 mM phenylmethylsulfonyl fluoride, 0.4 M NaCl, and 1% (w/v) Triton X-100. The homogenate was centrifuged at 18,000g for 10 min at 4°C, and the supernatant was used to determine the total PARP activity using the PARP Universal Colorimetric Assay Kit.
In Vitro Protein-DNA Binding Assay (Dot Blotting)
Dot blotting was performed as described previously with slight modifications (Tao et al., 2008). Ta-sro1-His6, Ta-SRO1-His6, At-PARP1-His6, and Ta-ACO1-His6 proteins were first blotted onto a polyvinylidene difluoride membrane. The membrane was blocked with 5% (w/v) nonfat milk in 15 mL of PBST (137 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 8 mM Na2HPO4, pH 7.4, and 0.1% [v/v] Tween 20) at room temperature for 2 h. The blocked membrane was then washed with 15 mL of PBST buffer and preincubated with 15 mL of DNA probe dilution buffer (0.3% BSA and 1% goat serum in PBST) for 1 h. For DNA probe preparation, the calf thymus DNA (Sigma-Aldrich) was covalently labeled using EZ-Link TFPA-PEG3-Biotin (Pierce), following the manufacturer’s protocol. The biotinylated calf thymus DNA was then added to the probe dilution buffer to a final concentration of 10 μg/mL, followed by incubation with the membrane at room temperature for 1 h. After washing three times with PBST buffer, the membrane was incubated with streptavidin–horseradish peroxidase conjugate (Pierce) in PBST buffer for 30 min. After three washes, the specific protein-DNA binding was detected with ECL Plus Western Blotting Detection Reagents (Pierce), and images were captured by Image Quant 400 (GE Healthcare).
Determination of Enzyme Activity
To determine CAT, APX, GPX, MDAR, and DHAR activity, ∼0.15 g of plant material was homogenized in 1 mL 50 mM KH2PO4, 0.1 mM EDTA, and 0.3% (w/v) Triton X-100. For GR and NOX, the Triton X-100 was omitted (Grace and Logan, 1996). Total protein was determined using a BCA protein assay kit (Beyotime) following the kit protocols. CAT activity was determined using the procedure of Aebi (1984), monitoring the decrease in absorbance at 240 nm of H2O2 for 1 min at 25°C. APX activity was measured spectrophotometrically at 290 nm according to the method of Nakano and Asada (1981), applying an AsA extinction coefficient of 2.8 mM−1 cm−1. GPX activity was measured according to Benabdellah et al. (2009), in which the decrease in absorbance at 340 nm of a reaction containing 100 mM Tris-HCl, pH 7.4, 0.5 mM EDTA, 1.0 mM NaN3, 2 mM H2O2, 0.25 mM NADPH, 2.25 mM GSH, and 1.0 unit GR was monitored. A molar extinction coefficient for NADPH of 6.22 × 103 M−1 cm−1 was assumed (Sebastiani et al., 2007). MDAR activity was assayed according to Miyake and Asada (1992) in which the decrease in absorbance at 340 nm of a reaction containing 50 mM HEPES-KOH, pH 7.6, 0.1 mM NADH, 2.5 mM AsA, and 0.3 unit AsA oxidase was monitored. The same molar extinction coefficient for NADPH as above was assumed. DHAR activity was determined according to Dalton et al. (1986) in which the decrease in absorbance at 265 nm of a reaction containing 100 mM HEPES-KOH, pH 7.0, 0.1 mM EDTA, 2.5 mM GSH, and 0.2 mM AsA was monitored. A molar extinction coefficient for AsA of 14 × 103 M−1 cm−1 was assumed. GR activity was measured based on the method of Grace and Logan (1996) in which the decrease in absorbance at 340 nm of a reaction containing 100 mM Tris-HCl, pH 8.0, 1 mM EDTA, 1 mM GSSG, and 0.2 mM NADPH was monitored. The same molar extinction coefficient for NADPH as above was assumed. NOX activity was measured as for GR activity, omitting GSSG in the reaction (Grace and Logan, 1996).
Determination of AsA/DHA, GSH/GSSG, and NAD(P)+/NAD(P)H Ratios
A modification of the method of Okamura (1980) was used to determine AsA and DHA content. Snap-frozen tissue (0.1 g) was ground in 1 mL of 5% (w/v) TCA and centrifuged at 15,000g for 15 min. Each extract was divided into two, with one part used for the AsA assay, and the other to assess the total AsA content. The DHA concentration was calculated from the difference between the latter and the former. AsA was quantified by first adding 100 μL of 5% (w/v) TCA and 100 μL ethanol to 100 μL of the TCA extract. Then the following were added sequentially: 50 μL of 0.4% (v/v) H3PO4-ethanol, 100 μL of 0.5% bathophenanthroline-ethanol, and 50 μL of 0.03% (w/v) FeCl3-ethanol. The solution was then left to stand for 90 min at 30°C, and the absorbance was read at 534 nm. To assess the total AsA content, a 25-μL aliquot of 1.8 M triethanolamine was added to 100 μL of the TCA extract to neutralize the pH. After incubation at room temperature for 10 min, 50 μL of 0.5% (v/v) N-ethylmaleimide was added to block any excess reducing reagent, followed by 50 μL of 20% (w/v) TCA to obtain a pH of 1 to 2. Thereafter, the protocol followed the one used for AsA determination.
Total GSH was measured by the 5,5′-dithiobis(2-nitrobenzoic acid)-GR recycling assay (Rahman et al., 2006). A 0.1-g plant sample was homogenized in 1 mL of ice-cold 5% (w/v) sulfosalicylic acid and centrifuged at 15,000g for 15 min. The supernatant was neutralized by the addition of 7.5 M triethanolamine. A 200-μL aliquot was then used to determine total GSH (GSH+GSSG) content, while 4 μL of 2-vinylpyridine was added to a second 200-μL aliquot, which was then held at 20°C for 60 min. A volume of 700 μL of 0.3 mM NADPH, 100 μL of 10 mM 5,5′-dithiobis (2-nitrobenzoic acid), and 150 μL of 125 mM NaH2PO4 in 6.3 mM EDTA, pH 6.5, was mixed with 50 μL of each of the two aliquots above, and 10 μL of 50 units/mL of GR was then added. Both reactions were monitored spectrophotometrically at 412 nm.
NAD(P)+ and NAD(P)H were quantified by methods adapted from Gibon and Larher (1997). A 0.1-g sample was homogenized in 1 mL of ice-cold 0.1 M HCl (for NAD+ and NADP+) or in 1 mL of 0.1 M NaOH (NADH and NADPH). The homogenates were boiled for 5 min and then cooled on ice and centrifuged (10,000g, 4°C, 10 min). The resulting supernatants were neutralized with either 0.1 M NaOH or 0.1 M HCl (as appropriate) and then recentrifuged (10,000g, 4°C, 10 min). These supernatants were kept on ice during a reduction reaction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide catalyzed by phenazine methosulfate in the presence of ethanol and either alcohol dehydrogenase (for NAD+ and NADH) or Glc-6-phosphate and Glc-6-phosphate dehydrogenase (for NADP+ and NADPH).
Determination of ATP and MDA Content
ATP was extracted at 2°C from 250 mg of frozen plant tissues by homogenization with 6 mL of 6% perchloric acid for 30 s in a Potter glass homogenizer. After centrifugation for 15 min at 20,000g at 2°C, the supernatant was neutralized with 5 M ice-cold K2CO3 (∼0.36 mL) to pH 7.75 and then precipitated for 1 h in an ice bath. KClO4 was removed by centrifugation for 15 min at 20,000g at 2°C, and the supernatant was used to determine ATP content immediately with the ENLITEN ATP assay system (Promega) according to the manufacturer's protocol. MDA content was determined according to Liu et al. (2012).
Three-Dimensional Model Construction and Analysis
Structural templates for the catalytic domains of the Ta-SRO1, Ta-sro1, At-RCD1, At-PARP1, and At-PARP2 proteins were determined using pGenThreader (Lobley et al., 2009). All recognized templates were assessed as “high” or “certain” by pGenThreader. The initial three-dimensional coordinates of the models were generated with MODELER 9v3 (Fiser et al., 2000). Thereafter, the models were iteratively evaluated with ProQ (Wallner and Elofsson, 2003) and PROCHECK (Laskowski et al., 1993) and locally optimized with ModLoop (Fiser and Sali, 2003), until satisfactory evaluation scores were obtained. The molecular surface electrostatic potential of the resulting models was calculated from the Adaptive Poisson-Boltzmann Solver software plug-in of the Visual Molecular Dynamics program (Humphrey et al., 1996).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative, GenBank/EMBL databases, or the Rice Genome Annotation Project database (http://rice.plantbiology.msu.edu) under the following accession numbers: Ta-sro1 (JN202574), Ta-ACO1 (KF014124), Ta-Actin (AB181991), At-RCD1 (At1g32230), At-SRO1 (At2g35510), At-SRO2 (At1g23550), At-SRO3 (At1g70440), At-SRO4 (At3g47720), At-SRO5 (At5g62520), At-AOX1a (At3g22370), At-NDB2 (At4g05020), At-RbohF (At1g64060), At-Tublin (At1g04820), At-ATM (At3g48190), Os-SRO1a (Os10g42710), Os-SRO1b (Os3g63770), Os-SRO1c (Os3g12820), Os-SRO1d (Os6g13860), and Os-SRO1e (Os4g57640).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Variation of Total Root Length and root H2O2 Content in SR3 under Stress Conditions.
Supplemental Figure 2. Sequence Alignment of Coding DNA Sequences and Protein Sequences of SRO1 and sro1.
Supplemental Figure 3. PCR and qRT-PCR Test of Transgenic Wheat Plants.
Supplemental Figure 4. Variation of Total Root Length and Root H2O2 Content in Transgenic Wheat Lines under Stress Conditions.
Supplemental Figure 5. In Vitro DNA Binding Activity of Ta-sro1-His6 and Ta-SRO1-His6 Proteins.
Supplemental Figure 6. Transgenic Test of the Constitutive Expression of Ta-sro1 in Wild-Type Arabidopsis.
Supplemental Figure 7. Overexpression of Ta-sro1 in the At-rbohF Mutant Background Promoted the Oxidative Stress Tolerance of the Mutant.
Supplemental Figure 8. Single-Cell Gel Electrophoresis Analysis of the Protoplasts of JN177 and SR3 Exposed to UV Treatment.
Supplemental Figure 9. The ATP Content between Vector Control and Ta-sro1 Overexpression Arabidopsis Lines under Normal and Stress Conditions.
Supplemental Figure 10. Expression of At-ATM in Vector Control and Ta-sro1 Overexpression Arabidopsis Lines.
Supplemental Figure 11. The Malondialdehyde Content of Vector Control and Ta-sro1 Overexpression Arabidopsis Lines under Salinity Stress.
Supplemental Table 1. Oxidative Stress–Related Genes Showing at Least a Twofold Induction or Repression upon Exposure to Abiotic Stress.
Supplemental Table 2. Model Evaluations and Templates (PDB Codes) Used for Modeling.
Supplemental Table 3. Primers Used for PCR, RT-PCR, and Real-Time RT-PCR Analysis.
Supplemental Data Set 1. Alignments Used to Generate the Phylogeny Presented in Figure 2D.
Supplementary Material
Acknowledgments
This work was supported by funds of the Major Program of the Natural Science Foundation of China (No. 31030053), the National Basic Research 973 Program of China (2012CB114204), and the National Transgenic Project (Grants 2013ZX08002002 and 2013ZX08002003).
AUTHOR CONTRIBUTIONS
G.X. designed the experiment. S.T.L. performed most of the experiments. S.W.L. and Mei W. performed some key experiments. T.W. performed three-dimensional model construction and analysis. C.M. characterized transgenic plants. Meng W. performed the SCGE experiment. G.X., S.T.L., S.W.L., and Mei W. wrote the article.
Glossary
- ROS
reactive oxygen species
- PARP
poly(ADP ribose) polymerase
- RST
RCD-SRO-TAF4
- MV
methyl viologen
- mART
mono(ADP ribose) transferase
- SR3
Shanrong No. 3
- JN177
Jinan 177
- PEG
polyethylene glycol
- RNAi
RNA interference
- OE
overexpression
- NA
nicotinamide
- NOX
NADPH oxidase
- DPI
diphenylene iodonium
- AOX
alternative oxidase
- KCN
potassium cyanide
- SHAM
salicyl hydroxamic acid
- POD
peroxidase
- CAT
catalase
- APX
ascorbate peroxidase
- GPX
glutathione peroxidase
- MDAR
monodehydroascorbate reductase
- DHAR
dehydroascorbate reductase
- GR
glutathione reductase
- SCGE
single-cell gel electrophoresis
- CDS
coding DNA sequence
- TCA
trichloroacetic acid
- VC
vector control
- AsA
ascorbate
- DHA
dehydroascorbate
- MDA
malondialdehyde
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Aebi, H. (1984). Catalase. In Methods in Enzymology, L. Packer, ed (Orlando, FL: Academic Press), pp. 121–126. [Google Scholar]
- Aguilar-Quesada R., Muñoz-Gámez J.A., Martín-Oliva D., Peralta A., Valenzuela M.T., Matínez-Romero R., Quiles-Pérez R., Menissier-de Murcia J., de Murcia G., Ruiz de Almodóvar M., Oliver F.J. (2007). Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol. Biol. 8: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlfors R., et al. (2004). Arabidopsis RADICAL-INDUCED CELL DEATH1 belongs to the WWE protein-protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell 16: 1925–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amé J.C., Spenlehauer C., de Murcia G. (2004). The PARP superfamily. Bioessays 26: 882–893 [DOI] [PubMed] [Google Scholar]
- Amor Y., Babiychuk E., Inzé D., Levine A. (1998). The involvement of poly(ADP-ribose) polymerase in the oxidative stress responses in plants. FEBS Lett. 440: 1–7 [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Aravind L. (2001). The WWE domain: A common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 26: 273–275 [DOI] [PubMed] [Google Scholar]
- Belles-Boix E., Babiychuk E., Van Montagu M., Inzé D., Kushnir S. (2000). CEO1, a new protein from Arabidopsis thaliana, protects yeast against oxidative damage. FEBS Lett. 482: 19–24 [DOI] [PubMed] [Google Scholar]
- Benabdellah K., Merlos M.A., Azcón-Aguilar C., Ferrol N. (2009). GintGRX1, the first characterized glomeromycotan glutaredoxin, is a multifunctional enzyme that responds to oxidative stress. Fungal Genet. Biol. 46: 94–103 [DOI] [PubMed] [Google Scholar]
- Beneke S., Bürkle A. (2007). Poly(ADP-ribosyl)ation in mammalian ageing. Nucleic Acids Res. 35: 7456–7465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham I.J., Farrar J.F. (1989). Activity and capacity of respiratory pathways in barley roots deprived of inorganic nutrients. Plant Physiol. Biochem. 27: 847–854 [Google Scholar]
- Briggs A.G., Bent A.F. (2011). Poly(ADP-ribosyl)ation in plants. Trends Plant Sci. 16: 372–380 [DOI] [PubMed] [Google Scholar]
- Citarelli M., Teotia S., Lamb R.S. (2010). Evolutionary history of the poly(ADP-ribose) polymerase gene family in eukaryotes. BMC Evol. Biol. 10: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton R., Millar A.H., Whelan J. (2006). Alternative oxidases in Arabidopsis: A comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim. Biophys. Acta 1757: 730–741 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dalton D.A., Russell S.A., Hanus F.J., Pascoe G.A., Evans H.J. (1986). Enzymatic reactions of ascorbate and glutathione that prevent peroxide damage in soybean root nodules. Proc. Natl. Acad. Sci. USA 83: 3811–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dat J., Vandenabeele S., Vranová E., Van Montagu M., Inzé D., Van Breusegem F. (2000). Dual action of the active oxygen species during plant stress responses. Cell. Mol. Life Sci. 57: 779–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Block M., Verduyn C., De Brouwer D., Cornelissen M. (2005). Poly(ADP-ribose) polymerase in plants affects energy homeostasis, cell death and stress tolerance. Plant J. 41: 95–106 [DOI] [PubMed] [Google Scholar]
- Finkemeier I., Goodman M., Lamkemeyer P., Kandlbinder A., Sweetlove L.J., Dietz K.J. (2005). The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J. Biol. Chem. 280: 12168–12180 [DOI] [PubMed] [Google Scholar]
- Fiser A., Do R.K., Sali A. (2000). Modeling of loops in protein structures. Protein Sci. 9: 1753–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser A., Sali A. (2003). ModLoop: Automated modeling of loops in protein structures. Bioinformatics 19: 2500–2501 [DOI] [PubMed] [Google Scholar]
- Foreman J., Demidchik V., Bothwell J.H., Mylona P., Miedema H., Torres M.A., Linstead P., Costa S., Brownlee C., Jones J.D., Davies J.M., Dolan L. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G. (2011). Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 155: 2–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujibe T., Saji H., Arakawa K., Yabe N., Takeuchi Y., Yamamoto K.T. (2004). A methyl viologen-resistant mutant of Arabidopsis, which is allelic to ozone-sensitive rcd1, is tolerant to supplemental ultraviolet-B irradiation. Plant Physiol. 134: 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagné J.P., Isabelle M., Lo K.S., Bourassa S., Hendzel M.J., Dawson V.L., Dawson T.M., Poirier G.G. (2008). Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 36: 6959–6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y., Larher F. (1997). Cycling assay for nicotinamide adenine dinucleotides: NaCl precipitation and ethanol solubilization of the reduced tetrazolium. Anal. Biochem. 251: 153–157 [DOI] [PubMed] [Google Scholar]
- Grace S.C., Logan B.A. (1996). Acclimation of foliar antioxidant systems to growth irradiance in three broad-leaved evergreen species. Plant Physiol. 112: 1631–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Kozlov S., Lavin M.F., Person M.D., Paull T.T. (2010). ATM activation by oxidative stress. Science 330: 517–521 [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack K.E., Jones J.D. (1996). Resistance gene-dependent plant defense responses. Plant Cell 8: 1773–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa P.O., Hottiger M.O. (2008). The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 13: 3046–3082 [DOI] [PubMed] [Google Scholar]
- Huang X.Y., Chao D.Y., Gao J.P., Zhu M.Z., Shi M., Lin H.X. (2009). A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 23: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. (1996). VMD: Visual molecular dynamics. J. Mol. Graph. 14: 33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59 [DOI] [PubMed] [Google Scholar]
- Jaspers P., Blomster T., Brosché M., Salojärvi J., Ahlfors R., Vainonen J.P., Reddy R.A., Immink R., Angenent G., Turck F., Overmyer K., Kangasjärvi J. (2009). Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J. 60: 268–279 [DOI] [PubMed] [Google Scholar]
- Jaspers P., Overmyer K., Wrzaczek M., Vainonen J.P., Blomster T., Salojärvi J., Reddy R.A., Kangasjärvi J. (2010). The RST and PARP-like domain containing SRO protein family: Analysis of protein structure, function and conservation in land plants. BMC Genomics 11: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.A., Raymond M.J., Yang Z., Smirnoff N. (2007). NADPH oxidase-dependent reactive oxygen species formation required for root hair growth depends on ROP GTPase. J. Exp. Bot. 58: 1261–1270 [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S., Zhu J., Kim K., Agarwal M., Fu X., Huang A., Zhu J.K. (2006). The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 18816–18821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.Y., Zhang T., Kraus W.L. (2005). Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 19: 1951–1967 [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Nakanishi I., Warizaya M., Iwashita A., Kido Y., Hattori K., Fujii T. (2004). Inhibitor-induced structural change of the active site of human poly(ADP-ribose) polymerase. FEBS Lett. 556: 43–46 [DOI] [PubMed] [Google Scholar]
- Kwak J.M., Mori I.C., Pei Z.M., Leonhardt N., Torres M.A., Dangl J.L., Bloom R.E., Bodde S., Jones J.D., Schroeder J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26: 283–291 [Google Scholar]
- Lin H., Wang R., Qian Q., Yan M., Meng X., Fu Z., Yan C., Jiang B., Su Z., Li J., Wang Y. (2009). DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21: 1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Li S., Wang M., Xia G. (2012). A transcriptomic analysis reveals the nature of salinity tolerance of a wheat introgression line. Plant Mol. Biol. 78: 159–169 [DOI] [PubMed] [Google Scholar]
- Lobley A., Sadowski M.I., Jones D.T. (2009). pGenTHREADER and pDomTHREADER: New methods for improved protein fold recognition and superfamily discrimination. Bioinformatics 25: 1761–1767 [DOI] [PubMed] [Google Scholar]
- Maxwell D.P., Wang Y., McIntosh L. (1999). The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. USA 96: 8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Suzuki N., Rizhsky L., Hegie A., Koussevitzky S., Mittler R. (2007). Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses. Plant Physiol. 144: 1777–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7: 405–410 [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Miyake, C., and Asada, K. (1992). Thylakoid-bound ascorbate peroxidase scavenges hydrogen peroxide photoproduced: Photoreduction of monodehydroascorbate radical. In Research in Photosynthesis, N. Murata, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), 563–566. [Google Scholar]
- Munns R., Tester M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Nakano Y., Asada K. (1981). Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 22: 867–880 [Google Scholar]
- Neill S., Desikan R., Hancock J. (2002). Hydrogen peroxide signalling. Curr. Opin. Plant Biol. 5: 388–395 [DOI] [PubMed] [Google Scholar]
- Okamura M. (1980). An improved method for determination of L-ascorbic acid and L-dehydroascorbic acid in blood plasma. Clin. Chim. Acta 103: 259–268 [DOI] [PubMed] [Google Scholar]
- Olive P.L., Banáth J.P., Durand R.E. (1990). Detection of etoposide resistance by measuring DNA damage in individual Chinese hamster cells. J. Natl. Cancer Inst. 82: 779–783 [DOI] [PubMed] [Google Scholar]
- Olmos E., Kiddle G., Pellny T., Kumar S., Foyer Ch. (2006). Modulation of plant morphology, root architecture, and cell structure by low vitamin C in Arabidopsis thaliana. J. Exp. Bot. 57: 1645–1655 [DOI] [PubMed] [Google Scholar]
- Overmyer K., Tuominen H., Kettunen R., Betz C., Langebartels C., Sandermann H., Jr, Kangasjärvi J. (2000). Ozone-sensitive arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 12: 1849–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z., Wang M., Li F., Lv H., Li C., Xia G. (2009). A proteomic study of the response to salinity and drought stress in an introgression strain of bread wheat. Mol. Cell. Proteomics 8: 2676–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potters G., Pasternak T.P., Guisez Y., Jansen M.A. (2009). Different stresses, similar morphogenic responses: Integrating a plethora of pathways. Plant Cell Environ. 32: 158–169 [DOI] [PubMed] [Google Scholar]
- Rahman I., Kode A., Biswas S.K. (2006). Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 1: 3159–3165 [DOI] [PubMed] [Google Scholar]
- Sagi M., Fluhr R. (2001). Superoxide production by plant homologues of the gp91(phox) NADPH oxidase. Modulation of activity by calcium and by tobacco mosaic virus infection. Plant Physiol. 126: 1281–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraufstatter I.U., Hyslop P.A., Hinshaw D.B., Spragg R.G., Sklar L.A., Cochrane C.G. (1986). Hydrogen peroxide-induced injury of cells and its prevention by inhibitors of poly(ADP-ribose) polymerase. Proc. Natl. Acad. Sci. USA 83: 4908–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V., Amé J.C., Dollé P., Schultz I., Rinaldi B., Fraulob V., Ménissier-de Murcia J., de Murcia G. (2002). Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 277: 23028–23036 [DOI] [PubMed] [Google Scholar]
- Schreiber V., Dantzer F., Ame J.C., de Murcia G. (2006). Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 7: 517–528 [DOI] [PubMed] [Google Scholar]
- Sebastiani M., et al. (2007). Induction of mitochondrial biogenesis is a maladaptive mechanism in mitochondrial cardiomyopathies. J. Am. Coll. Cardiol. 50: 1362–1369 [DOI] [PubMed] [Google Scholar]
- Sheen J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127: 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2000). Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3: 217–223 [PubMed] [Google Scholar]
- Skirycz A., et al. (2011). Survival and growth of Arabidopsis plants given limited water are not equal. Nat. Biotechnol. 29: 212–214 [DOI] [PubMed] [Google Scholar]
- Tao Z., Gao P., Hoffman D.W., Liu H.W. (2008). Domain C of human poly(ADP-ribose) polymerase-1 is important for enzyme activity and contains a novel zinc-ribbon motif. Biochemistry 47: 5804–5813 [DOI] [PubMed] [Google Scholar]
- Teotia S., Lamb R.S. (2009). The paralogous genes RADICAL-INDUCED CELL DEATH1 and SIMILAR TO RCD ONE1 have partially redundant functions during Arabidopsis development. Plant Physiol. 151: 180–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teotia S., Muthuswamy S., Lamb R.S. (2010). Radical-induced cell death1 and similar to RCD one1 and the stress-induced morphogenetic response. Plant Signal. Behav. 5: 143–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trucco C., Oliver F.J., de Murcia G., Ménissier-de Murcia J. (1998). DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 26: 2644–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Breusegem F., Bailey-Serres J., Mittler R. (2008). Unraveling the tapestry of networks involving reactive oxygen species in plants. Plant Physiol. 147: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera S., De Block M., Van de Steene N., van de Cotte B., Metzlaff M., Van Breusegem F. (2007). Silencing of poly(ADP-ribose) polymerase in plants alters abiotic stress signal transduction. Proc. Natl. Acad. Sci. USA 104: 15150–15155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner B., Elofsson A. (2003). Can correct protein models be identified? Protein Sci. 12: 1073–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Liang Q., Cao K., Ge X. (2011). Endogenous protein mono-ADP-ribosylation in Arabidopsis thaliana. Planta 233: 1287–1292 [DOI] [PubMed] [Google Scholar]
- Xia G. (2009). Progress of chromosome engineering mediated by asymmetric somatic hybridization. J. Genet. Genomics 36: 547–556 [DOI] [PubMed] [Google Scholar]
- Xia G., Xiang F., Zhou A., Wang H., Chen H. (2003). Asymmetric somatic hybridization between wheat (Triticum aestivum L.) and Agropyron elongatum (Host) Nevishi. Theor. Appl. Genet. 107: 299–305 [DOI] [PubMed] [Google Scholar]
- You J., Zong W., Li X., Ning J., Hu H., Li X., Xiao J., Xiong L. (2013). The SNAC1-targeted gene OsSRO1c modulates stomatal closure and oxidative stress tolerance by regulating hydrogen peroxide in rice. J. Exp. Bot. 64: 569–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T.J., Zhao S.Y., Chen H.M., Zhao Q.Z., Hu Z.M., Hou B.K., Xia G.M. (2006). Transgenic wheat progeny resistant to powdery mildew generated by Agrobacterium inoculum to the basal portion of wheat seedling. Plant Cell Rep. 25: 1199–1204 [DOI] [PubMed] [Google Scholar]
- Zhu J.K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.