Abstract
Low doses of UV-B light (280 to 315 nm) elicit photomorphogenic responses in plants that modify biochemical composition, photosynthetic competence, morphogenesis, and defense. UV RESISTANCE LOCUS8 (UVR8) mediates photomorphogenic responses to UV-B by regulating transcription of a set of target genes. UVR8 differs from other known photoreceptors in that it uses specific Trp amino acids instead of a prosthetic chromophore for light absorption during UV-B photoreception. Absorption of UV-B dissociates the UVR8 dimer into monomers, initiating signal transduction through interaction with CONSTITUTIVELY PHOTOMORPHOGENIC1. However, much remains to be learned about the physiological role of UVR8 and its interaction with other signaling pathways, the molecular mechanism of UVR8 photoreception, how the UVR8 protein initiates signaling, how it is regulated, and how UVR8 regulates transcription of its target genes.
INTRODUCTION
Light is of paramount importance in promoting the growth and development of plants. In addition to its role as the energy source for photosynthesis, light regulates numerous aspects of plant form and function throughout the life cycle, from seedling establishment to flowering time. These developmental responses to light, termed photomorphogenesis, are complex in that plants respond to several different facets of their radiation environment, in particular its spectral quality, the amount of light (defined as the photon fluence rate), and the duration and direction of illumination. To detect and respond to light, plants employ a suite of photoreceptors coupled to a network of signaling components and transcriptional effectors (Jiao et al., 2007; Kami et al., 2010). Whereas a wealth of information has been obtained about the photoreceptors and signaling mechanisms that plants use to detect light in the UV-A to far-red regions of the spectrum, much less is known about the perception of UV-B light (280 to 315 nm), which mediates numerous regulatory responses in plants (Jordan, 1996; Jansen, 2002; Frohnmeyer and Staiger, 2003; Ulm and Nagy, 2005; Jenkins, 2009).
UV-B radiation has a major impact on virtually all organisms, despite being a very small fraction of the daylight spectrum. Most UV-B is absorbed by the stratospheric ozone layer, and only wavelengths above ∼295 nm reach the surface of our planet. Because of its relatively high energy, UV-B radiation has the potential to damage molecules, such as DNA, and consequently to impair cellular processes, which ultimately may cause death. Thus, organisms have evolved strategies both to avoid and repair damage by UV-B. In plants, protection includes the production of reflective surface waxes and hairs and the synthesis of UV-absorbing phenolic “sun-screen” compounds that accumulate in the epidermal layer and reduce transmittance to cells below (Caldwell et al., 1983; Jordan 1996; Rozema et al., 1997; Frohnmeyer and Staiger, 2003; Jenkins, 2009). Damage caused by UV-B exposure is ameliorated in several ways, in particular by antioxidant systems and enzymes that repair DNA damage. An important feature of these protective responses is that they are stimulated when plants are exposed to UV-B.
Studies in the last century established that UV-B has regulatory effects on plant growth, morphology, and biochemical content (Klein, 1978). It was discovered that low doses of UV-B initiate photomorphogenic responses that could not be explained by the action of known photoreceptors and were not a consequence of DNA damage (Wellmann, 1976, 1983). The action spectrum for DNA damage peaks at ∼260 nm, whereas action spectra for photomorphogenic UV-B responses peak at ∼295 to 300 nm (Wellmann, 1976; Ensminger, 1993; Jenkins, 2009; Jiang et al., 2012a). Photomorphogenic responses to UV-B include the suppression of both hypocotyl extension and root growth in the low fluence range and the promotion of cotyledon opening (Wellmann, 1976; Ballaré et al., 1995; Kim et al., 1998; Boccalandro et al., 2001; Suesslin and Frohnmeyer, 2003; Shinkle et al., 2004; Tong et al., 2008; Conte et al., 2010). However, perhaps the most extensively characterized photomorphogenic response to UV-B is the biosynthesis of flavonoid compounds, which are constituents of the UV-absorbing sun screen (Caldwell et al., 1983; Hahlbrock and Scheel, 1989; Rozema et al., 1997; Ryan et al., 2001; Winkel-Shirley, 2002). Low doses of UV-B strongly stimulate the transcription of flavonoid biosynthesis genes, including that encoding the key enzyme chalcone synthase (CHS) (Hahlbrock and Scheel, 1989; Frohnmeyer et al., 1999; Jenkins et al., 2001; Hartmann et al., 2005). In addition, low doses of UV-B regulate the expression of numerous other genes associated with various plant processes (Ulm et al., 2004; Brown et al., 2005; Favory et al., 2009).
The discovery that plants exhibit photomorphogenic responses to UV-B suggested the existence of a UV-B photoreceptor comparable to other known photoreceptors. However, research over several decades failed to shed much light on the nature of the UV-B photoreceptor or the mechanisms of photomorphogenic UV-B signaling. Biochemical and genetic studies showed that photomorphogenic UV-B responses are distinct from phytochrome- and cryptochrome-mediated responses (Ballaré et al., 1995; Christie and Jenkins, 1996; Frohnmeyer et al., 1998; Boccalandro et al., 2001; Suesslin and Frohnmeyer, 2003) and that DNA damage signaling, reactive oxygen species signaling, and wound and defense signaling pathways, which are important in mediating some responses to UV-B (Jenkins, 2009), are not the mechanism of photomorphogenic UV-B perception (Boccalandro et al., 2001; Jenkins et al., 2001; Ulm et al., 2004; Gadjev et al., 2006; Jenkins, 2009; González Besteiro et al., 2011). Progress in the identification of a UV-B photoreceptor came from the application of a genetic approach. Kliebenstein et al. (2002) isolated an Arabidopsis thaliana mutant of UV RESISTANCE LOCUS8 (UVR8), which was subsequently shown to act as a UV-B photoreceptor (Rizzini et al., 2011). More recently, structural studies have revealed the molecular basis of photoreception by UVR8 (Christie et al., 2012; Wu et al., 2012). This article will focus on the physiological role of UVR8, its structure, and the mechanisms of photoreception, signal transduction, and regulation. Further discussion of UVR8 can be found in other recent reviews (Jenkins, 2009; Heijde and Ulm, 2012; Jiang et al., 2012a; Li et al., 2013; Tilbrook et al., 2013).
PHYSIOLOGICAL ROLE OF UVR8
The exploration of UVR8 function in vivo is still at an early stage. Initial studies focused on gene regulation by UVR8 and demonstrated its importance in UV protection. Subsequently, additional aspects of UVR8 function have emerged (Table 1). Present research is starting to reveal how UVR8 is integrated with other photoreceptor pathways in mediating responses in vivo. However, it is important to remember that virtually everything we know about UVR8 is derived from studies with Arabidopsis, so much remains to be learnt about its physiological role in other, diverse species.
Table 1. Responses to UV-B Mediated by UVR8.
| Response | Referencesa |
|---|---|
| Gene regulation | 1–10 |
| UV-B tolerance | 1–4 |
| Flavonoid biosynthesis | 1, 4, 6, 10, 13 |
| Hypocotyl growth suppression | 4, 12, 14 |
| Leaf/epidermal cell expansion | 4, 10, 11 |
| Endoreduplication in epidermal cells | 11 |
| Stomata per epidermal cell | 11 |
| Entrainment of circadian clock | 7 |
| Increased photosynthetic efficiency | 8 |
| Tolerance of B. cinerea infection | 13 |
1, Kliebenstein et al. (2002); 2, Brown et al. (2005); 3, Brown and Jenkins (2008); 4, Favory et al. (2009); 5, Brown et al. (2009); 6, Grüber et al. (2010); 7, Fehér et al. (2011); 8, Davey et al. (2012); 9, Lang-Mladek et al. (2012); 10, Morales et al. (2013); 11, Wargent et al. (2009); 12, Cloix et al. (2012); 13, Demkura and Ballaré (2012); 14, Huang et al. (2013).
Gene Regulation Underpins UVR8 Function
The Arabidopsis uvr8-1 mutant was isolated in a screen for plants hypersensitive to UV-B (Kliebenstein et al., 2002). This type of screen had previously identified mutants defective in either phenolic sunscreen biosynthesis (Lois and Buchanan, 1994) or the repair of DNA damage (Harlow et al., 1994; Jiang et al., 1997; Landry et al., 1997; Jenkins and Brown, 2007). However, uvr8-1 differs from these other mutants in that it exhibits altered gene regulation following UV-B exposure. The mutant has greatly reduced expression of the gene encoding the flavonoid biosynthesis enzyme CHS and reduced levels of protective flavonoids. In addition, uvr8-1 shows increased expression of PATHOGENESIS-RELATED1 (PR1) and PR5 proteins, suggesting a stress response. Based on the phenotype of the mutant, Kliebenstein et al. (2002) suggested, perceptively, that UVR8 might have a role in UV-B signaling. As discussed in detail below, the UVR8 gene does not encode a protein known to be involved in DNA repair or production of phenolic pigments. Rather, UVR8 encodes a seven-bladed β-propeller protein (Figure 1B) with moderate similarity to human REGULATOR OF CHROMATIN CONDENSATION1 (RCC1), a guanine nucleotide exchange factor involved in nucleocytoplasmic transport and the regulation of cell cycle progression and mitosis (Renault et al., 1998). However, this similarity is purely structural, and there is no evidence of functional homology between UVR8 and RCC1 (Brown et al., 2005).
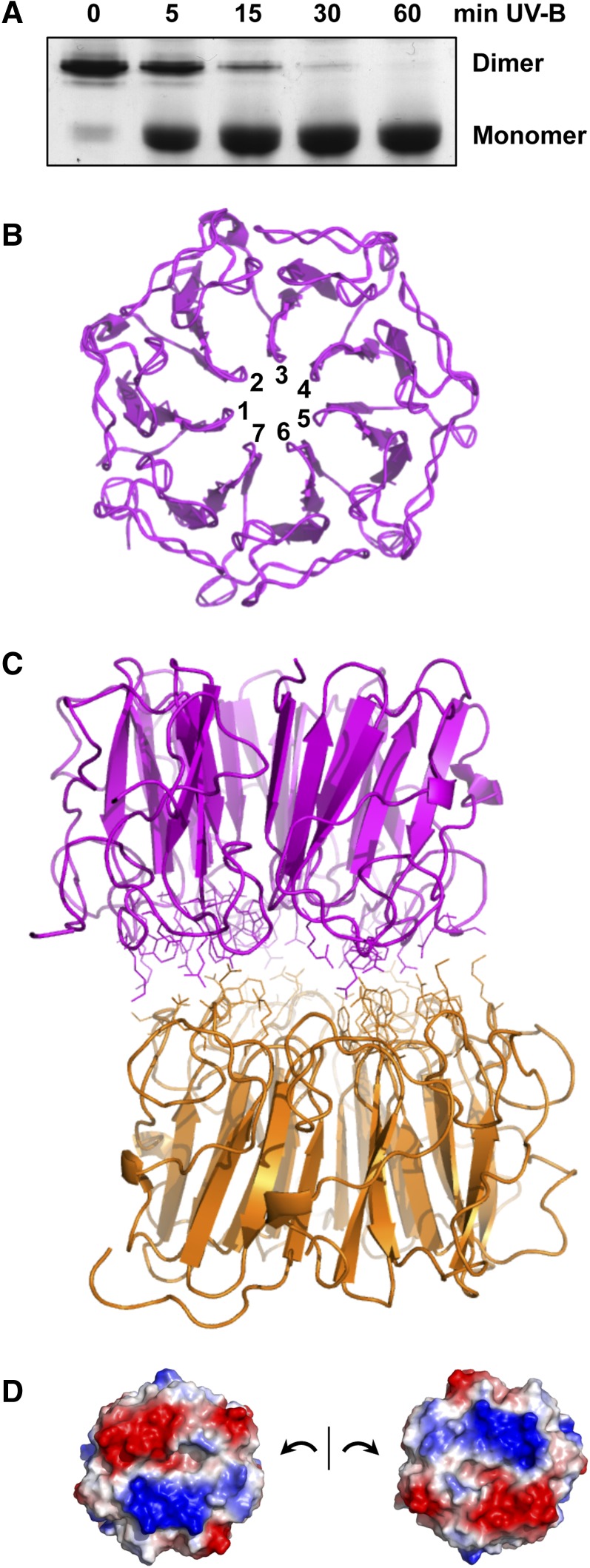
Figure 1.
UVR8 Dimer Structure and Monomerization.
(A) UV-B induces monomerization of UVR8. Coomassie blue–stained SDS-PAGE gel of purified UVR8 exposed for the times shown to 1.5 μmol m−2 s−1 narrowband UV-B (λmax 311 nm). Samples were prepared for electrophoresis without boiling. The UVR8 dimer and monomer are indicated.
(B) Seven-bladed β-propeller structure of the UVR8 monomer. The structure is shown for amino acids 14 to 380.
(C) Structure of the UVR8 dimer showing residues at the dimer interaction surface.
(D) The dimer interaction surfaces of two UVR8 monomers displayed to show patches of complementary electrostatic potential. Basic (blue) and acidic (red) amino acids contribute positive and negative charges, respectively.
Images in (B) to (D) were produced using PyMOL. (All panels produced from data presented by Christie et al. [2012].)
The initial characterization of uvr8-1 did not establish whether the mutant was altered specifically in responses to UV-B and did not explain the basis of its hypersensitivity to UV-B. Subsequently, Brown et al. (2005) isolated additional uvr8 alleles and showed that CHS expression in the mutant was only affected by UV-B and not by several other stimuli, indicating that UVR8 acts in a UV-B–specific manner. Given that UVR8 functions as a UV-B photoreceptor, it is unlikely to be involved directly in mediating responses to other stimuli. Nevertheless, as discussed below, UVR8 could influence responses to other stimuli through crosstalk with light and non-light signaling pathways, and it is also conceivable that other environmental factors could modulate the action of UVR8.
UVR8 acts by regulating transcription of a set of genes. Using microarray analysis, Brown et al. (2005) showed that UVR8 regulates expression of over 70 genes in response to UV-B in mature Arabidopsis leaf tissue, including those involved in flavonoid biosynthesis, DNA repair, and the amelioration of oxidative damage. These genes are associated with the prevention and repair of UV damage, and their impaired expression in uvr8 mutant plants accounts for the UV-B hypersensitivity phenotype. UVR8 regulates additional genes, including those involved with other aspects of metabolism, signaling, and transcriptional regulation. Furthermore, several UVR8-regulated genes encode chloroplast proteins, which likely explains why uvr8 exhibits impaired capacity for photosynthesis compared with wild-type plants under elevated UV-B (Davey et al., 2012). A subsequent microarray analysis (Favory et al., 2009) indicated that several hundred genes are regulated by UVR8 in Arabidopsis seedlings, many in common with those regulated by UVR8 in leaf tissue (Brown et al., 2005). In these experiments, seedlings were exposed to a narrowband UV-B source lacking the shorter UV-B wavelengths present in the broadband source used by Brown et al. (2005), facilitating identification of UVR8-specific transcripts.
Although there are differences in the numbers and identities of genes regulated by UVR8 between seedlings and leaf tissue and between different UV-B treatments, it is evident that UVR8 mediates the regulation of a substantial set of genes and therefore has a broad impact on plant processes. The full extent of the physiological role of UVR8 in Arabidopsis is not yet clear because knowledge of target gene function is incomplete. Moreover, almost all studies have been confined to plants growing in growth chambers; therefore, it is interesting that Morales et al. (2013) found that UVR8 has an important role in regulating gene expression and biochemical composition in Arabidopsis in natural sunlight. The gene expression responses mediated by UVR8 evidently have an important function in acclimating plants to UV-B so that they can subsequently tolerate relatively high levels of exposure. Experiments with plants growing in a sun simulator show that the uvr8 mutant is susceptible to damage by UV-B, with curled and chlorotic leaves, whereas plants overexpressing UVR8 are tolerant (Favory et al., 2009). Furthermore, experiments in natural sunlight support the idea that early acclimation mediated by UVR8 promotes growth under ambient solar UV-B (Morales et al., 2013).
The regulation of biochemical composition by UVR8 affects several plant processes apart from UV protection. For instance, various phenolic compounds that accumulate in leaf tissue exposed to UV-B are involved in deterring attack by insect pests and pathogens (Ballaré et al., 2012). UVR8 is involved in stimulating the expression of genes concerned with combating herbivore attack in natural light conditions (Morales et al., 2013). Demkura and Ballaré (2012) reported that UV-B treatment of Arabidopsis plants enhanced resistance to infection by the necrotrophic fungus Botrytis cinerea and this effect was reduced in uvr8 mutant plants. The protective effect of UV-B was absent in mutant plants defective in the sinapate biosynthesis enzyme ferrulic acid 5-hydroxylase. These observations suggest that UV-B, detected by UVR8, reduces B. cinerea infection by stimulating sinapate biosynthesis. Whether biochemical changes regulated by UVR8 confer tolerance to other pathogens or pests remains to be established.
Brown and Jenkins (2008) showed that UVR8 mediates gene expression responses over a wide range of UV-B fluence rates, extending to the very low fluence rates characteristic of photomorphogenic responses. Several other genes, not regulated by UVR8, required a 10-fold higher fluence rate of UV-B for induction. Thus, genes are regulated by UVR8-dependent and UVR8-independent UV-B signaling pathways with different characteristics. It is not known how many UV-B signaling pathways operate in plants. A number of pathways are not specific to UV-B, including those involved in DNA damage signaling and wound and defense signaling, and these pathways are likely to operate mainly at higher fluence rates of UV-B (Jenkins, 2009). Furthermore, a number of responses to low fluence rates of UV-B may be independent of UVR8, so the possibility of additional UV-B photoreceptor(s) cannot be excluded.
UVR8 Regulates Morphology
Low fluence rates of UV-B modulate several photomorphogenic responses in seedlings, including cotyledon opening, hypocotyl extension, and phototropism (Ballaré et al., 1995; Kim et al., 1998; Boccalandro et al., 2001; Eisinger et al., 2003; Suesslin and Frohnmeyer, 2003; Shinkle et al., 2004; Conte et al., 2010) and additionally regulate several aspects of morphogenesis in older plants, including leaf expansion, stem extension, and branching (Hectors et al., 2007; Wargent et al., 2009). It is not known how many of these UV-B responses are regulated, at least in part, via UVR8 signaling. Favory et al. (2009) showed that uvr8 mutant plants are impaired in the suppression of hypocotyl extension by UV-B. Consistent with this finding, overexpression of UVR8 in Arabidopsis produces dwarf plants in the presence of UV-B. UVR8 also influences leaf area. UV-B suppresses leaf expansion through a UVR8-independent reduction in epidermal cell number, but Arabidopsis uvr8 plants have smaller leaves than the wild type because they lack a compensatory stimulation of epidermal cell area by UV-B that is mediated by UVR8 (Wargent et al., 2009). In addition, UV-B promotes an increase in stomatal index (stomatal number per epidermal cell) in wild-type plants, and this stimulatory effect is impaired in uvr8. Thus, UVR8 is required for the regulation of epidermal cell expansion and differentiation under UV-B illumination.
The mechanisms underlying the regulation of cell extension and expansion by UVR8 are unknown. Various molecules are important in controlling extension growth, including auxin, brassinosteroids, gibberellins, and DELLA proteins, but no direct link between UVR8 and these regulators has yet been reported. Auxin has been implicated in regulating morphogenic responses to UV-B (Jansen, 2002), but the molecular basis of its involvement is not known. One potential mechanism by which UVR8 could regulate cell expansion is the control of endoreduplication, which results in endopolyploidy of cells and promotes cell growth under certain conditions (De Veylder et al., 2011). Epidermal cells of the uvr8 mutant are impaired in progression to higher ploidy levels compared with the wild type under UV-B (Wargent et al., 2009), and this may contribute to their reduced expansion. Further research is needed to investigate these aspects.
Integration of UVR8 with Other Pathways
Very little research has been undertaken to investigate how UVR8 integrates with other signaling pathways, including light signaling pathways. Light signaling is initiated by several different photoreceptors, the cryptochromes (cry1 and cry2), phototropins (phot1 and phot2) and Zeitlupe family proteins, which mediate responses to UV-A/blue light, and the phytochromes (phyA, B, C, D, and E in Arabidopsis), which mediate responses principally to red and far-red light. There is evidence that UV-B induction of CHS expression, mediated by UVR8, is negatively regulated by phyB and synergistically enhanced by UV-A and blue light (Wade et al., 2001), but the molecular basis of these interactions is unknown. Further evidence of interactions between UVR8 and other photoreceptor pathways was obtained recently by Morales et al. (2013), who studied transcriptome and metabolite profiles in wild-type and uvr8 mutant plants in natural sunlight, where the levels of UV-A light and total radiation are much higher than in growth cabinet experiments. In sunlight, UVR8 mediates a number of responses predicted from previous studies, such as the expression of genes concerned with UV-B protection, but unexpectedly, UVR8 has both positive and negative effects on gene expression and metabolite accumulation in response to UV-A light. Since cryptochromes mediate gene expression responses to UV-A, it is possible that UVR8 interacts with cryptochrome signaling under natural environmental conditions.
The phytochrome and cryptochrome photoreceptors mediate the input of light signals that entrain the circadian clock (Jiao et al., 2007). Fehér et al. (2011) found that exposure to low-fluence-rate UV-B entrains the circadian clock in Arabidopsis and UVR8 mediates this response. Entrainment entails the regulation of genes encoding components of the clock, and UVR8 mediates an increase in transcripts of several such genes in response to UV-B. Moreover, similar to other photoreceptor responses, the extent of responsiveness to UV-B is restricted (gated) by the clock, so that the increase in transcripts is maximal at particular times during the circadian cycle. Several genes in output pathways from the clock, including flavonoid biosynthesis and protection from oxidative stress, are subject to circadian regulation in response to UV-B. Thus, UVR8 acts with other light signaling pathways to ensure the appropriate timing of physiological processes.
UVR8 IS A UV-B PHOTORECEPTOR
Initial studies showed that UVR8 has properties expected of the elusive UV-B photoreceptor (Jenkins, 2009). In particular, UVR8 is UV-B specific and acts at low fluence rates to initiate classical UV-B photomorphogenic responses, such as the induction of flavonoid biosynthesis and hypocotyl growth suppression. Moreover, extensive genetic screens designed to identify UV-B photoreception or signaling components isolated only uvr8 and constitutively photomorphogenic1 (cop1) alleles (Brown et al., 2005; Oravecz et al., 2006; Favory et al., 2009). COP1, which is known to function as a component of an E3 ubiquitin ligase complex, acts as a positive regulator of UV-B responses and is the primary signaling partner of UVR8 (Oravecz et al., 2006; Favory et al., 2009; see section on UVR8 signaling below). In order to obtain convincing evidence that a protein functions in photoreception, it is necessary to demonstrate that light of a particular spectral quality directly affects the putative photoreceptor protein and that the effect is coupled to photoreceptor action. The ultimate proof is to show that a mutation of the protein that alters photoreception in vitro has an equivalent effect on responsiveness in vivo.
Rizzini et al. (2011) discovered that UV-B exposure of plants induces the conversion of UVR8 from a homodimer to a monomeric form (Figures 1A and 3C). Monomerization is accompanied by a physical change to the protein that exposes an epitope at the C terminus that is recognized by an anti-UVR8 antibody. Furthermore, bimolecular fluorescence complementation and coimmunoprecipitation experiments show that UVR8 physically interacts with COP1 in a UV-B–dependent manner in plants (Favory et al., 2009; Rizzini et al., 2011; Cloix et al., 2012; Figure 3B). Rizzini et al. (2011) showed that this interaction occurs when plant extracts are illuminated with UV-B and its UV-B dependence requires the presence of UVR8. These experiments support the hypothesis that UVR8 is a UV-B photoreceptor but do not prove it conclusively; the possibility that some other plant protein acts in photoreception, provided UVR8 is present, could not be completely excluded. It is therefore crucial that UVR8 was shown to mediate a UV-B response in heterologous systems. UVR8 interacts with COP1 in a UV-B–dependent manner when both proteins are expressed in yeast (Rizzini et al., 2011; Cloix et al., 2012) and mammalian cells (Rizzini et al., 2011; Crefcoeur et al., 2013). Yeast does not possess functional homologs of UVR8 or COP1 and does not mediate the UV-B–dependent interaction through DNA damage signaling (Cloix et al., 2012). The UV-B dependence of the interaction in yeast requires intact UVR8 but not intact COP1, indicating that UVR8 is responsible for photoreception (Rizzini et al., 2011; Cloix et al., 2012). Rizzini et al. (2011) further showed that UV-B stimulates monomerization of UVR8 in yeast.
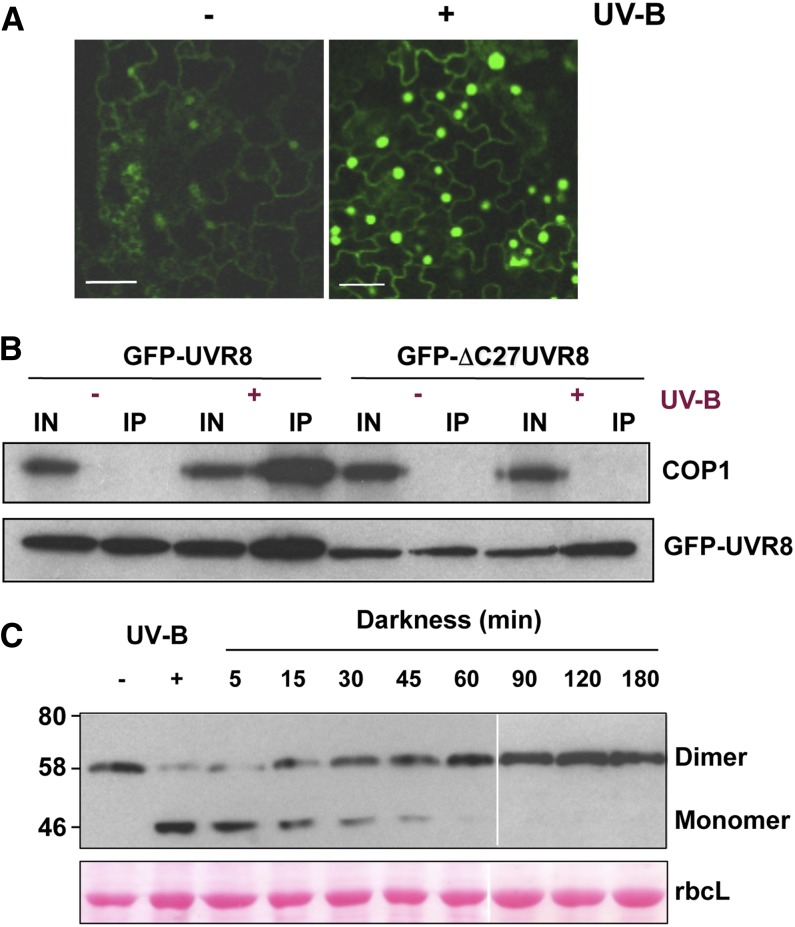
Figure 3.
UVR8 Signaling and Regulation.
(A) UV-B–induced nuclear accumulation of UVR8. Confocal image of epidermal cells of Arabidopsis uvr8-1 plants expressing UVR8pro:GFP-UVR8. Plants were grown in 20 μmol m−2 s−1 white light lacking UV-B (−) and exposed to 3 μmol m−2 s−1 broadband UV-B for 4 h (+). Bar = 20 μm. (Modified from Kaiserli and Jenkins [2007], Figure 3A.)
(B) Interaction of UVR8 and COP1 in plants. Whole-cell protein extracts were obtained from uvr8-1/UVR8pro:GFP-UVR8 and uvr8-1/UVR8pro:GFP-ΔC27UVR8 plants treated (+) or not (−) with 3 μmol m−2 s−1 narrowband UV-B. Coimmunoprecipitation assays were performed under the same illumination conditions. Input samples (15 μg; IN) and eluates (IP) were fractionated by SDS-PAGE, and protein gel blots were probed with anti-COP1 and anti-GFP antibodies. (Reprinted from Cloix et al. [2012], Figure 3A.)
(C) Regeneration of the UVR8 dimer. Immunoblot of whole-cell protein extracts from wild-type Landsberg erecta plants probed with anti-UVR8 antibody. Plants were exposed to 2.5 μmol m−2 s−1 narrowband UV-B for 3 h (+ UV-B) and then transferred to darkness for the indicated time periods before extracts were made. Extract samples were prepared for electrophoresis without boiling prior to SDS-PAGE and immunoblotting. The UVR8 dimer and monomer are indicated. Ponceau staining of Rubisco large subunit (rbcL) is shown as a loading control. (Reprinted from Heilmann and Jenkins [2013], Figure 1A.)
The experiments in yeast and plants together provide compelling evidence that UVR8 senses UV-B, causing monomerization and interaction with COP1. Moreover, studies with purified recombinant UVR8 unequivocally demonstrate its photoreceptor action. Purified UVR8 exists as a dimer that rapidly monomerizes when exposed to doses of UV-B that initiate responses in plants (Christie et al., 2012; Wu et al., 2012; Figure 1A). Furthermore, as discussed below, mutations that affect UV-B photoreception by UVR8 in vitro have equivalent effects on responses in vivo.
UVR8 STRUCTURE AND MOLECULAR FUNCTION
Arabidopsis UVR8 is a 440–amino acid protein with a molecular mass of ∼47 kD. Recently, biochemical and biophysical studies were undertaken with the purified recombinant protein expressed in Escherichia coli (Christie et al., 2012; Wu et al., 2012). The intact protein did not form crystals that could be used for x-ray crystallography, likely due to the flexibility of its N- and C-terminal regions. A high-resolution crystal structure was obtained for the UVR8 protein core lacking 11 amino acids at the N terminus and 59 amino acids at the C terminus (Christie et al., 2012; Wu et al., 2012). Nevertheless, the truncated protein is functional in UV-B photoreception and converts from dimer to monomer like the full-length protein. The crystal structure was obtained only for the dimeric protein because the monomer reassociates to form a dimer under conditions used for crystallization. Consistent with the structure predicted from the amino acid sequence (Kliebenstein et al., 2002), UVR8 was found to be a seven-bladed β-propeller protein (Figures 1B and 1C).
Salt Bridge Interactions Maintain the UVR8 Dimer
A key region of the UVR8 dimer structure is the interface where the two monomers are in contact. This interface is rich in aromatic and charged amino acids that are crucial for both the structure and photoreceptor function of UVR8. The structure reveals that charged amino acids at the dimer interface are grouped such that patches of complementary electrostatic potential are present on the opposing monomers (Figure 1D). The amino acids in these regions, mainly basic Arg and acidic Asp and Glu residues, form a network of salt bridges across the dimer interface that hold the monomers together. Numerous amino acids potentially contribute to maintaining the dimer, but some are likely to be more important than others because they can form double hydrogen–bonded rather than single hydrogen–bonded salt bridges. Notably, Arg-286 (R286) forms double and single hydrogen–bonded salt bridges with Asp residues 107 and 96 (D107 and D96), respectively, on the opposing monomer. In addition, a double hydrogen–bonded salt bridge is formed between R146 and Glu-182 (E182) and single hydrogen bonds are formed between a number of residues, for example, R338 and D44 and R354 and E53. The network of salt bridges is evidently strong because the dimer does not dissociate into monomers even in the presence of relatively high concentrations of SDS, provided the sample is not boiled (Rizzini et al., 2011; Christie et al., 2012; Wu et al., 2012). However, if the salt bridges are neutralized, the dimer dissociates; thus, purified UVR8 appears constitutively monomeric when the pH of the medium is decreased (Christie et al., 2012) or its ionic strength is increased (Wu et al., 2012).
The importance of particular salt bridges in maintaining the dimer was investigated by site-directed mutagenesis and examination of the purified mutant proteins (Christie et al., 2012; Wu et al., 2012). Wild-type UVR8 that has not been exposed to UV-B appears as a dimer when examined by SDS-PAGE with nonboiled samples, whereas the UV-B–treated protein is monomeric (Figure 1A). However, even slight weakening of the salt bridges makes the dimer susceptible to disruption by SDS, so many salt bridge mutants appear as monomers in the above assay. Size exclusion chromatography is a more rigorous and sensitive method to assess dimer/monomer status. Using this assay, it is clear that some mutants, such as R146 mutated to the small neutral amino acid Ala (UVR8R146A), form a dimer in vitro that monomerizes following UV-B exposure. Nevertheless, the dimer is weakened because the mutant protein appears monomeric in the nonboiled-SDS-PAGE assay (Christie et al., 2012). A key finding is that mutations that disrupt the R286-D107/D96 salt bridges make UVR8 constitutively monomeric, indicating that these interactions are crucial to maintaining the dimer. If these salt bridges cannot form, the remaining salt bridges are not sufficiently strong to form a dimer under the in vitro conditions used. Similarly, Wu et al. (2012) reported that UVR8R338A is constitutively monomeric, suggesting that R338 is also important in maintaining the dimer.
The molecular environment of UVR8 in vivo clearly differs from that used in the in vitro experiments, for example, with respect to ionic conditions and the presence of other proteins. It is therefore essential to test the importance of salt bridges in maintaining the UVR8 dimer in vivo.
Specific Trps Act as UV-B Chromophores for UVR8
In the decades preceding the discovery of the UVR8 photoreceptor, various authors speculated on the possible mechanism of UV-B photoreception. Some suggested that photoreception might be achieved by aromatic amino acids in a protein absorbing UV-B (Ensminger, 1993; Ballaré et al., 1995; Gerhardt et al., 2005), and this has turned out to be correct. Photoreceptor proteins normally employ bound cofactors as chromophores to absorb light of particular wavelengths; for example, phytochrome binds phytochromobilin and phototropin flavin mononucleotide as chromophores. By contrast, purified UVR8 does not have a prosthetic cofactor to act as a chromophore for UV-B, but instead employs specific amino acid residues for UV-B photoreception. The amino acid Trp (W) strongly absorbs UV-B wavelengths, and Arabidopsis UVR8 has 14 Trp residues, one in the C-terminal region (W400), six in the β-propeller core of the protein, and seven in the dimer interface (Figure 2A). The six Trp residues in the core (W39, W92, W144, W196, W300, and W352) are each in different blades of the propeller structure and, together with a Tyr (Y248) in the remaining blade, form a ring of aromatic residues that help to maintain the core structure by forming hydrogen bonds and hydrophobic interactions between adjacent propeller blades (Figure 2B). Mutation of three of these core Trp amino acids (W39, W144, and W352) to Ala results in unstable or nonfunctional UVR8 proteins in vivo, whereas mutation to the aromatic Phe (F) or Tyr (Y) does not, presumably because these residues form hydrophobic interactions that help to maintain structure (O’Hara and Jenkins, 2012).
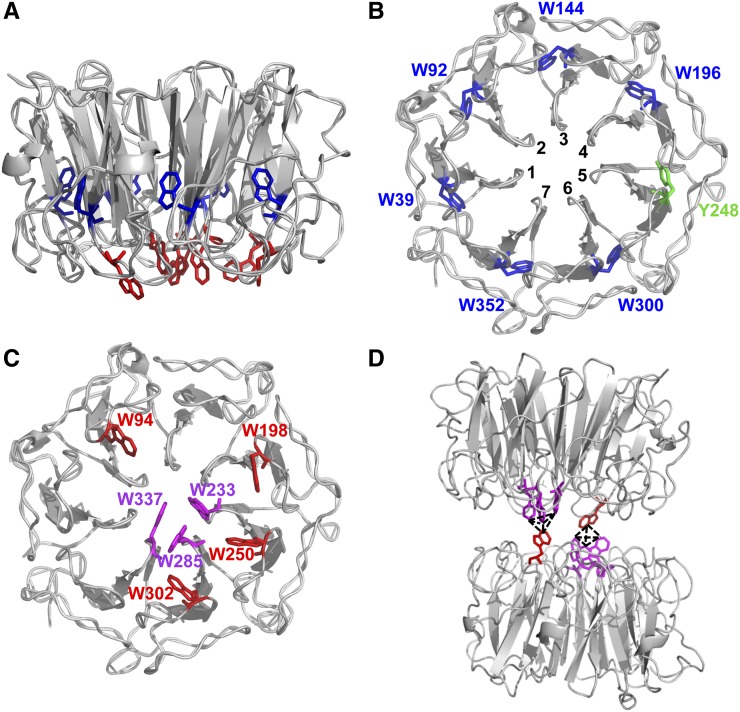
Figure 2.
UVR8 Has Distinct Groups of Trps.
(A) The arrangement of all UVR8 Trps (except W400) in the monomer viewed from the side. Trps in the protein core and at the dimer interaction surface are shown in blue and red, respectively.
(B) The Trps in the core viewed from the dimer interaction surface. Each Trp is associated with a different propeller blade (numbered). Y248 from blade 5 completes the ring of aromatic residues.
(C) The Trps at the dimer interaction surface. The triad Trps are shown in magenta.
(D) The UVR8 dimer has two pyramid clusters of excitonically coupled Trps, each consisting of the triad Trps together with W94 on the opposing monomer.
The images were produced using PyMOL. ([A] to [C] are reprinted from O’Hara and Jenkins [2012], Figures 1A, 1B, and 1D, respectively; [D] is produced from data presented in Christie et al. [2012].)
Of the seven Trp residues located in the dimer interface, W233, W285, and W337 are in a highly conserved GWRHT motif and are only ∼4 Å apart, enabling their electronic orbitals to overlap (Figure 2C). These Trp amino acids are referred to as the “triad.” W94 is close to the triad on the opposing monomer, forming a cross-dimer pyramid arrangement; there are two such pyramids in each dimer (Christie et al., 2012; Figure 2D). The remaining three Trp amino acids (W198, W250, and W302) are positioned at the periphery of the dimer interface (Figure 2C) and, together with Tyr residues Y201 and Y253 and F305, form an aromatic shield around W233 and W285 (Christie et al., 2012). The functional significance of this arrangement is not yet clear.
The functions of Trp residues in the dimeric interface have been investigated using purified mutant proteins in vitro. Size exclusion chromatography shows that UVR8W285F and UVR8W285A form dimers that are unaffected by UV-B exposure. However, UVR8W285A evidently forms a weak dimer as it appears constitutively monomeric when examined by SDS-PAGE with nonboiled samples (Christie et al., 2012; Wu et al., 2012). Similarly, UVR8W233F and UVR8W233A are dimers that do not respond to UV-B, whereas the corresponding mutants of W94 and W337 are dimers that monomerize in response to UV-B (Christie et al., 2012; Wu et al., 2012).
When Trp amino acids are sufficiently close together that they show excitonic coupling, they give a characteristic signal in the far-UV circular dichroism (CD) spectrum (Grishina and Woody, 1994). This signal is observed for purified UVR8, with a peak at 234 nm and trough at 221 nm. When purified wild-type UVR8 is exposed to UV-B, the CD signal becomes much reduced, indicating that excitonic coupling is lost. One factor contributing to this loss of signal will be separation of the monomers, which breaks the pyramidal arrangement. Indeed, some of the constitutively monomeric salt bridge mutants, such as UVR8D96N,D107N, have a much reduced CD signal (Christie et al., 2012). In addition, other changes associated with photoreception may contribute to the loss of the CD signal, such as spatial reorientation of coupled Trps or a redistribution of excited electrons.
Mutation of each of the pyramid Trps, either to Ala or Phe, reduces the magnitude of the CD signal prior to UV-B exposure to varying extents, with the greatest effect observed for UVR8W233F. These observations support the conclusion drawn from the crystal structure that the pyramid Trps are involved in excitonic coupling. Furthermore, mutation of W233 and W285 to either Ala or Phe abolishes the effect of UV-B exposure on the CD signal, implicating these Trp residues in photoreception of UV-B. Mutation of W337 has a lesser impact on UV-B perception and mutation of W94 essentially no impact.
Trp gives rise to a characteristic fluorescence following UV-B absorption, and it is likely that UV-B photoreception will lead to changes in fluorescence emission by chromophore Trp residues. Wu et al. (2012) monitored Trp fluorescence following UV-B excitation of wild-type UVR8. UV-B illumination initiates a rise in fluorescence at 335 nm followed by a gradual decline. The UVR8W285F and UVR8W285A mutants do not show this rise in fluorescence, indicating that they are impaired in UV-B photoreception. Similar results are obtained for UVR8W233F and UVR8W233A. However, several other Trp mutants, including UVR8W337F and UVR8W94F, behave like wild-type UVR8. These observations are consistent with the CD spectroscopy data (Christie et al., 2012) and indicate that W285 and W233 are the principal UV-B chromophores for UVR8.
Christie et al. (2012) found that mutation of W285 to Phe alters the spectral sensitivity of the photoreceptor. Phe absorbs UV-C wavelengths, maximally at 257 nm. Although UVR8W285F fails to absorb UV-B, it weakly absorbs UV-C, initiating monomerization and reducing the CD signal. This observation further demonstrates the key role of W285 as a chromophore for UVR8. Thus, UVR8 is different from all other known photoreceptors in using intrinsic amino acids as chromophores for spectral sensitivity.
The Mechanism of UVR8 Photoreception Is Not Clear
Although the key amino acids involved in maintenance of the UVR8 dimer structure and in UV-B photoreception have been identified, details of the mechanism by which photoreception leads to disruption of salt bridge interactions and, hence, monomerization are not yet known. The chromophore Trps are intimately associated with salt-bridging Arg residues; for instance, W285 is adjacent to R286. Furthermore, cation–Pi interactions between the aromatic rings of the Trps, in particular W233 and W285, and the side chains of adjacent Arg residues strengthen the interactions between the triad and salt-bridging residues. Wu et al. (2012) suggested that photoreception could disrupt these interactions and hence weaken intermolecular salt bridges, causing monomerization. An additional possibility is that the excitation of electrons in the Trp triad leads to the effective transfer of an electron to a salt-bridging Arg, such as R286, neutralizing the salt bridges (Christie et al., 2012). By applying biophysical methods to monitor the rapid molecular events associated with photoreception, it should be possible to test these alternative mechanisms and delineate the processes that lead to monomerization.
In Vivo Studies of UVR8 Photoreception
Prior to the availability of the crystal structure, modeling of UVR8 based on sequence similarity with RCC1 and other proteins (Rizzini et al., 2011; Wu et al., 2011) suggested that the clustering of the triad Trps and their presence in the conserved GWRHT motifs might have functional significance. Hence, functional analysis of Trp and other residues in the GWRHT motif was initiated by examination of UVR8 mutants in yeast and transgenic plants (Rizzini et al., 2011; O’Hara and Jenkins, 2012). Subsequently, resolution of the structure of UVR8 informed further mutational analyses.
Experiments in yeast examined the effect of Trp mutations on the dimer/monomer status of UVR8 and its interaction with COP1. When analyzed by SDS-PAGE with nonboiled samples (Rizzini et al., 2011), UVR8W285F is a dimer that does not respond to UV-B, whereas UVR8W337F is a dimer that monomerizes following UV-B exposure, consistent with the size exclusion chromatography results obtained with purified proteins. UVR8W285A, UVR8W337A, and UVR8W233F expressed in yeast appear as constitutive monomers in the SDS-PAGE assay, indicating a weakening of the dimer structures. Phe and Tyr mutants of the triad Trps do not interact with COP1 in yeast, but each of the Ala mutants interacts with COP1 both in the presence and absence of UV-B (Rizzini et al., 2011; O’Hara and Jenkins, 2012). The Ala mutation most likely causes exposure of regions of the protein that bind to COP1, as discussed further below (see UVR8 signaling section).
O’Hara and Jenkins (2012) extended the analysis of Trp mutants in yeast to experiments with transgenic Arabidopsis. Function was determined by complementation of the impaired gene expression and hypocotyl extension phenotypes of uvr8-1. Mutation of several Trp residues located in either the core of the protein or at the periphery of the dimer interface has little or no effect on UVR8 function in these assays, in agreement with findings in yeast. By contrast, function is affected by mutation of the triad Trp residues. UVR8W285F and UVR8W285A do not complement the impaired response to UV-B of uvr8-1. However, UVR8W233A shows a low level of responsiveness to UV-B, which is not evident from in vitro studies. UVR8W337A shows little loss of function compared with wild-type UVR8 and UVR8W94A no apparent loss of function. An important caveat is that the function of these mutant proteins in Arabidopsis has only been studied using saturating UV-B exposures in controlled environments. It is possible that different conclusions might be drawn under limiting doses of UV-B and under natural conditions.
In transgenic Arabidopsis, UVR8W285F forms a dimeric protein that does not respond to UV-B, whereas UVR8W285A appears constitutively monomeric, consistent with the findings in yeast (O’Hara and Jenkins, 2012; Huang et al., 2013; Heijde et al., 2013). UVR8W233A and UVR8W337A form weak dimers in the absence of UV-B and both respond to UV-B to produce monomers (O’Hara and Jenkins, 2012). UVR8W285F does not interact with COP1, but the Ala mutants UVR8W285A, UVR8W233A, and UVR8W337A all interact constitutively with COP1, as found in yeast. Since the Ala mutants of UVR8 triad Trps bind COP1 constitutively, it is possible that plants expressing these proteins would display a cop mutant phenotype, with short hypocotyls and expanded cotyledons in darkness. This was not observed by O’Hara and Jenkins (2012) but is reported by Huang et al. (2013) and Heijde et al. (2013). The extent of the cop phenotype is dependent on the level of transgene expression. The transgenic lines selected by O’Hara and Jenkins (2012) had relatively low levels of transgene expression, similar to that of a GFP-UVR8 fusion that functionally complements uvr8. Heijde et al. (2013) observed a mild cop phenotype in plants showing moderate (∼4-fold) overexpression of UVR8W285A but an obvious cop phenotype in plants with 30- to 40-fold overexpression. These plants do not lack COP1 but accumulate relatively high levels of the protein, which is stabilized by interaction with UVR8W285A. Interestingly, the high UVR8W285A overexpressors show activation of photomorphogenic UV-B responses in the absence of UV-B, with strong accumulation of the ELONGATED HYPOCOTYL5 (HY5) transcription factor, which is an important effector of UVR8-mediated responses (see section on UVR8 signaling). The mechanism by which overexpression of UVR8W285A leads to HY5 accumulation is not clear but likely involves posttranslational stabilization rather than transcriptional control.
In summary, the in vivo studies of UVR8 Trp function largely support and extend the in vitro observations and lead to the conclusion that W285, in particular, and W233 function as the principal UV-B absorbing chromophores of UVR8. W337 and W94 appear to have minor roles in that UVR8 can function in photoreception when they are mutated. Thus, the pyramid Trp residues are not equally important in photoreception. Although W285 and W233 appear to be the main UV-B chromophores of UVR8, it is interesting that they do not function in a redundant manner; the other Trps cannot compensate for the loss of W285, and W285 cannot completely compensate for mutation of W233. Since the chromophore Trps are not functionally redundant, it appears that they each have distinct roles in maintaining the integrity of the triad and in photoreception. The exact nature of these roles remains to be determined.
UVR8 Photoreception in Natural Daylight
The work undertaken to date has laid the foundations for further in vivo studies. In particular, it is important to understand how UVR8 functions in plants growing in natural conditions. The action spectrum for UVR8 activity in stimulating HY5 transcript accumulation (Brown et al., 2009) shows a strong peak at 280 nm, as expected from the absorption maximum of Trp, with action extending to at least 315 nm. Since the shortest wavelength UV-B that reaches the Earth’s surface is ∼295 nm, only the longer wavelength action of UVR8 is physiologically relevant. Many laboratory experiments now employ narrowband UV-B sources, which emit maximally at 311 nm, but studies are needed in natural daylight. It is important to know how the dimer/monomer status of UVR8 is controlled in natural conditions, how it is influenced by other environmental factors, and how monomerization correlates with gene regulation. Moreover, it will be interesting to discover how UVR8 acts in different species and in genotypes adapted to different light environments.
Evolutionary Conservation
UVR8 is highly conserved in plant evolution. Amino acid sequences with strong identity to Arabidopsis UVR8 are found not only in angiosperms, but in the moss Physcomitrella patens, the lycopod Selaginella moellendorffii, and the green algae Chlamydomonas reinhardtii and Volvox carteri (Rizzini et al., 2011; Wu et al., 2011). An important observation is that the number and position of key residues, including Trps and salt-bridging Args, are conserved, indicating that these proteins are likely to function with an equivalent molecular mechanism to higher plant UVR8. The conservation of UVR8 sequences suggests that the protein appeared early in plant evolution, before the appearance of vascular land plants. At that time, levels of UV-B would have exceeded those presently on earth because the ozone layer was not fully developed (Rozema et al., 1997). UV-B penetrates into water and would have been potentially damaging to photosynthetic algae; photosystem II is particularly susceptible to damage by UV-B (Jansen et al., 1996; Takahashi et al., 2010). Thus, UVR8 may have evolved to stimulate UV-protective mechanisms in early photosynthetic plants. Further insights into the evolution of UVR8 are likely to emerge from the analysis of algal genome sequences.
UVR8 SIGNALING
UVR8 Signaling Is Focused in the Nucleus
The cellular localization of UVR8 was studied initially using transgenic plants expressing the protein fused to green fluorescent protein (GFP) (Brown et al., 2005; Kaiserli and Jenkins, 2007). Confocal microscopy shows that GFP-UVR8 is present in both the nucleus and cytosol, and calculations of cellular distribution suggest that most is in the cytosol in plants that have not been exposed to UV-B. Similar results were obtained by immunodetection of native UVR8 in cellular fractions (Kaiserli and Jenkins, 2007). Nonetheless, UVR8 function is focused in the nucleus. Illumination of plants with a low fluence rate of UV-B stimulates the accumulation of UVR8 in the nucleus; ∼90% of nuclei show GFP fluorescence and, moreover, the brightness of the fluorescence increases strongly in UV-B–illuminated plants (Figure 3A). Nuclear accumulation of GFP-UVR8 is specific to UV-B light and is rapid, being detectable within 5 min of exposure. COP1 also accumulates in the nucleus in plants exposed to UV-B, and transient expression experiments in mustard hypocotyl cells suggest that CFP-UVR8 colocalizes with yellow fluorescent protein–COP1 in nuclear bodies following UV-B illumination (Favory et al., 2009). However, GFP-UVR8 is not observed in nuclear bodies in transgenic plants (Brown et al., 2005; Kaiserli and Jenkins, 2007).
It is not clear whether nuclear accumulation of UVR8 involves active translocation into the nucleus. An alternative possibility is that the protein is able to move between the nucleus and cytosol but is retained in the nucleus when it becomes active in the regulation of transcription in response to UV-B. Deletion of 23 amino acids from the N terminus of UVR8 prevents nuclear accumulation. This region does not contain an obvious nuclear localization signal, although it is conceivable that it interacts with a protein that mediates translocation into the nucleus, analogous to the action of FHY and FHL with phyA (Genoud et al., 2008). However, the 23–amino acid deletion may impact on the integrity of the first propeller blade of UVR8 and hence impair protein function, which in turn might prevent accumulation. Studies of the effects of additional deletions within the N-terminal region and of the mechanism of UVR8 movement into and out of the nucleus would therefore be informative.
An important point is that once UVR8 has accumulated in the nucleus following UV-B exposure, it appears to be retained; in plants placed in darkness for 24 h following UV-B treatment, ∼70% of nuclei still contain GFP-UVR8 (Kaiserli and Jenkins, 2007). Hence, in plants growing in natural photoperiodic cycles, UVR8 is likely to accumulate in the nucleus.
Given the importance of nuclear localization to UVR8 function, it is not known why so much of the protein is present in the cytosol. It is possible that the high level of cytosolic UVR8 is an aberration of using plants grown in the absence of UV-B, since UV-B exposure in nature will probably drive most UVR8 to accumulate in the nucleus. Alternatively, perhaps UVR8 moves very slowly into the nucleus following synthesis and therefore accumulates in the cytosol. A further possibility is that cytosolic UVR8 is involved in other cellular processes, although there is no evidence for such a role at present.
Although UVR8 acts in the nucleus, its presence in the nucleus in not sufficient for function; UV-B exposure is still required, consistent with its photoreceptor action. Attachment of a nuclear localization signal to UVR8 causes GFP-UVR8 to be constitutively nuclear. However, the nuclear localized protein is only able to stimulate gene expression if it is exposed to UV-B (Kaiserli and Jenkins, 2007).
COP1 Acts with UVR8 in Photomorphogenic UV-B Responses
As stated previously, COP1 is the primary signaling partner of UVR8 and it interacts with UVR8 in a UV-B–dependent manner (Favory et al., 2009; Rizzini et al., 2011; Cloix et al., 2012; Figure 3B). Genetic screens in plants and in yeast have so far failed to identify additional positive signaling components. Oravecz et al. (2006) showed that mutants in cop1 are impaired in the regulation of gene expression in response to UV-B. The genes regulated by COP1 are essentially the same as those regulated by UVR8 in seedlings (Favory et al., 2009), indicating that COP1 and UVR8 act together to mediate the photomorphogenic UV-B response. This positive action of COP1 contrasts sharply with its well-established function as a repressor of photomorphogenesis (Lau and Deng, 2012). In dark-grown seedlings, COP1 acts in an E3 ubiquitin ligase complex, targeting positive regulators of photomorphogenesis, such as HY5, for destruction (Osterlund et al., 2000; Lau and Deng, 2012), whereas in response to UV-B, COP1 promotes HY5 expression, along with numerous other genes (Oravecz et al., 2006). Although in principle COP1 could act positively in UV-B responses by initiating degradation of a negative regulator of UVR8 action, there is no direct evidence for such a scenario. COP1 has several domains that can mediate protein–protein interactions, so in principle it could facilitate the interaction of UVR8 with other proteins to promote transcription. However, at present the mechanism by which COP1 acts in the UV-B response is poorly understood.
The four Arabidopsis SUPPRESSOR OF PHYA-105 (SPA) proteins are WD40-domain proteins that associate with COP1 to form E3 ubiquitin ligase complexes (Laubinger et al., 2004; Zhu et al., 2008; Lau and Deng, 2012). Huang et al. (2013) report that SPA proteins act redundantly as positive regulators of UVR8 signaling; an impaired response to UV-B is seen in plants with multiple spa mutations. This phenotype was not observed in a previous study (Oravecz et al., 2006), although different experimental conditions were used. Huang et al. (2013) found that UV-B exposure reduces the association of COP1 and SPA proteins with the CULLIN4 (CUL4)–DAMAGED DNA BINDING PROTEIN1 (DDB1) E3 ubiquitin ligase complex. Following UV-B exposure, SPA proteins are found in a complex with COP1 and UVR8, although they do not appear to interact directly with UVR8. Similar findings were reported by Heijde et al. (2013), who identified proteins associated with UVR8W285A using a tandem affinity purification strategy. Thus, the UVR8-COP1-SPA protein complex acts to positively regulate photomorphogenic UV-B responses, although the exact role of the SPA proteins is not clear.
The physical interaction of COP1 with UVR8 may inactivate the E3 ubiquitin ligase activity of COP1. The COP1-SPA1 complex is known to interact with the cry1 and cry2 photoreceptors, leading to inactivation of its E3 ubiquitin ligase activity in blue light (Yang et al., 2001; Wang et al., 2001; Liu et al., 2011; Lian et al., 2011; Zuo et al., 2011). The UV-B–stimulated interaction of COP1 with UVR8 could provide an additional mechanism for promoting photomorphogenesis through inactivation of COP1. There is evidence that UV-B exposure protects the HY5 transcription factor from degradation (Favory et al., 2009; Huang et al., 2013). In particular, the presence of the UVR8-COP1-SPA complex under UV-B conditions stabilizes HY5 protein (Huang et al. 2013). In the absence of UV-B, COP1-SPA is recruited into the CUL4-DDB1 complex, which may promote HY5 degradation (Huang et al., 2013). However, the situation appears more complex because HY5 shows more degradation after UV-B exposure in cop1 plants than in the wild type and this degradation is inhibited by the proteasome inhibitor MG132, suggesting that an E3 ubiquitin ligase other than COP1 is involved in HY5 degradation. Thus, the UVR8-COP1-SPA protein complex promotes HY5 accumulation following UV-B exposure through posttranslational stabilization in addition to the stimulation of transcription.
Interaction of UVR8 with COP1 requires a 27–amino acid region of UVR8 located toward the C terminus of the protein (amino acids 397 to 423). Deletion of this region, referred to as C27, prevents interaction with COP1 both in yeast two-hybrid assays and in plants (Cloix et al., 2012; Figure 3B). In consequence, GFP-ΔC27UVR8 fails to stimulate gene expression and hypocotyl growth suppression in response to UV-B when expressed in uvr8-1. COP1 interacts with UVR8 via its WD40 domain (Rizzini et al., 2011; Cloix et al., 2012) and may recognize a short motif within C27 that is similar to COP1 binding sites in other proteins (Cloix et al., 2012; Wu et al., 2013). Yeast two-hybrid assays indicate that C27 is sufficient for interaction with the WD40 region of COP1. In this case, the interaction is not dependent on UV-B because the photoreceptive core of UVR8 is absent. Although C27 is both necessary and sufficient for interaction with COP1, it is entirely possible that one or more other regions of UVR8 are also involved in the interaction.
Further understanding of the interaction of UVR8 with COP1 requires information about the location of the C-terminal region of UVR8 and any changes in its conformation and, hence, availability for interaction with COP1 following photoreception. Since the UVR8 crystal structure does not provide information on the location of the C terminus, other methods will be needed to determine its structure and any UV-B–induced conformational changes.
A further dimension to the involvement of COP1 in photomorphogenic UV-B responses is that its own expression and accumulation is stimulated by UV-B. The association of COP1 with UVR8 leads to stabilization and accumulation of COP1 (Favory et al., 2009; Heijde et al., 2013). In addition, the UV-B stimulation of COP1 transcription is mediated by the HY5 and FAR-RED ELONGATED HYPOCOTYL3 (FHY3) transcription factors, which are able to bind to elements within the COP1 promoter (Huang et al., 2012). COP1 and HY5 produce a positive feedback loop, in that HY5 expression is stimulated by COP1 acting with UVR8; in turn, HY5 promotes COP1 expression. FHY3 expression is stimulated by UV-B, and a fhy3 mutant is impaired in UV-B photomorphogenic gene expression responses, hypocotyl growth suppression, and tolerance of damaging levels of UV-B (Huang et al., 2012). Thus, FHY3 is an important positive regulator of UV-B photomorphogenic responses through its stimulation of COP1 transcription.
HY5 Transcription Factor Is a Key Effector of UVR8-Mediated Responses
The HY5 transcription factor is one of the most important effectors of photomorphogenic responses mediated by different photoreceptors (Ang et al., 1998; Jiao et al., 2007; Lee et al., 2007). HY5 and the closely related protein HY5 HOMOLOG (HYH) act downstream of UVR8 and COP1 in UV-B responses. UV-B–dependent expression of HY5 and HYH is controlled by UVR8 (Brown et al., 2005; Favory et al., 2009). The UV-B induction of HY5 and HYH transcripts is very rapid and occurs in response to low doses of UV-B exposure (Ulm et al., 2004; Brown et al., 2005, 2009; Brown and Jenkins 2008). Transcriptome analysis of the hy5 mutant in comparison to the wild type (Brown et al., 2005; Oravecz et al., 2006) shows that HY5 regulates numerous gene targets of the UV-B photomorphogenic pathway. Expression analysis in hy5 and hyh mutants in comparison to hy5 hyh double mutant plants (Brown and Jenkins, 2008) reveals a degree of functional redundancy between the transcription factors in the regulation of some genes, but HY5 is the major effector of UVR8-mediated gene expression, controlling many of the downstream target genes. Consistent with this, hy5 is hypersensitive to elevated levels of UV-B, similar to uvr8, whereas hyh is more tolerant (Brown et al., 2005; Brown and Jenkins 2008).
Some of the photomorphogenic UV-B response genes regulated by HY5 are concerned with growth responses, since the hy5 mutant is impaired in hypocotyl growth suppression mediated by UVR8 (Oravecz et al., 2006; Cloix et al., 2012). These genes have not been identified but are likely to include those involved, for instance, in gibberellin or auxin signaling. Some of the UV-B response genes regulated by HY5 include other transcription factors, such as MYB12 involved in flavonol biosynthesis (Stracke et al., 2010), indicating a hierarchical cascade of transcriptional regulation. However, not all UVR8-regulated genes are controlled by HY5/HYH. For instance, the UV-B induction of the CCA1 and PRR9 genes, associated with the circadian clock, is unaffected in hy5 hyh mutant plants and the circadian gating of responsiveness to UV-B is also not dependent on HY5/HYH (Fehér et al., 2011).
HY5 action in UV-B responses is negatively regulated by the B-BOX ZINC FINGER PROTEIN24/SALT TOLERANCE (BBX24/STO) protein. This protein was originally reported to confer salt tolerance when expressed in yeast (Lippuner et al., 1996) but subsequently was found to be a repressor of photomorphogenic responses mediated by different photoreceptors (Indorf et al., 2007). Jiang et al. (2012b) reported that BBX24/STO represses hypocotyl growth, primary root growth, and gene expression responses to UV-B. Hence, the bbx24/sto mutant is hypersensitive to UV-B and adopts a dwarf phenotype. The expression of BBX24/STO is stimulated by UV-B and the protein accumulates under UV-B conditions, whereas in darkness, it is degraded, most likely by COP1. Jiang et al. (2012b) found that COP1 interacts with BBX24/STO following UV-B exposure and that COP1 promotes BBX24/STO accumulation in UV-B. In addition, BBX24/STO interacts with HY5 in both yeast and plants in both the presence and absence of UV-B. BBX24/STO suppresses HY5 accumulation specifically under UV-B illumination (Jiang et al., 2012b), although it is not clear whether this occurs as a result of transcriptional and/or posttranslational regulation. Interestingly, BBX24/STO appears to inhibit the ability of HY5 to stimulate transcription of a reporter construct in response to UV-B in a transient expression assay in Arabidopsis. This finding suggests that BBX24/STO can affect HY5 transcriptional activity directly, but the mechanism involved is not known.
BBX24/STO interacts with the RADICAL-INDUCED CELL DEATH1 (RCD1) protein (Belles-Boix et al., 2000), which is an additional negative regulator of UV-B signaling. The rcd1 mutant is tolerant of elevated levels of UV-B and has enhanced expression of HY5 and a number of UVR8-regulated genes under UV-B conditions as well as increased hypocotyl growth suppression (Jiang et al., 2009). RCD1 reduces the expression of BBX24/STO under UV-B exposure (Jiang et al., 2009). It therefore appears that RCD1 and BBX24/STO work together to negatively regulate HY5 in UVR8 signaling.
How Does UVR8 Regulate Transcription?
At present, there is very little information about events that occur following the interaction of UVR8 with COP1 that lead to the regulation of transcription.
There is evidence that UVR8 associates with chromatin through its ability to bind histones. UVR8 binds strongly to histone-agarose beads in vitro, requiring quite high salt concentrations for elution (Brown et al., 2005). In competition experiments, H2B is more effective than other histones in reducing binding of UVR8 to histone-agarose, suggesting that UVR8 interacts preferentially with histone H2B (Cloix and Jenkins, 2008). In agreement with these in vitro observations, UVR8 can readily be detected in plant chromatin preparations. When GFP-UVR8 is immunoprecipitated from chromatin preparations using an anti-GFP antibody, histones are present in the immunoprecipitated material (Cloix and Jenkins, 2008). Together, these findings indicate that UVR8 associates with chromatin in vivo via histones. However, the dynamics of UVR8 interaction with chromatin are unknown. Chromatin immunoprecipitation (ChIP) experiments suggest that UVR8 can associate with chromatin in both the absence and presence of UV-B (Cloix and Jenkins, 2008), but since quantitative data are lacking, it is not clear whether UV-B stimulates chromatin association.
Although it is likely that UVR8 interacts with other proteins in a chromatin context, no such proteins have yet been identified. There is no evidence that COP1 binds to chromatin, although there are technical difficulties in establishing indirect associations with chromatin mediated by other proteins (Zeng et al., 2006). It is therefore unknown whether COP1 interacts with UVR8 bound to chromatin. COP1 may not be required for UVR8 binding to chromatin, since the association is still observed in cop1-4 (Favory et al., 2009) and in plants expressing GFP-ΔC27UVR8, which does not bind COP1 (Cloix et al., 2012). Hence, the exact role that COP1 plays in regulating transcription with UVR8 remains a mystery.
ChIP experiments provide evidence that UVR8 interacts with chromatin at some genes it regulates. In experiments with plants expressing GFP-UVR8, an anti-GFP antibody immunoprecipitates chromatin fragments containing the promoter regions of the UVR8-regulated HY5, MYB12, and CRYD genes, but not the control ACTIN2 gene (Brown et al., 2005; Cloix and Jenkins, 2008). Similar results are obtained using anti-UVR8 antibodies with wild-type plants. With respect to the HY5 gene, association is seen in the promoter, coding and 3′ noncoding regions, but not 6 kb either side of the gene. The ChIP data raise the possibility that UVR8 might interact directly with chromatin at target gene loci to promote transcription (Brown et al., 2005), although the mechanism by which it would associate with specific genes is unknown. However, the situation is more complex because the promoter regions of some genes regulated by UVR8 (e.g., HYH and CHS) are not found in chromatin associated with UVR8 in ChIP experiments (Cloix and Jenkins, 2008). There is no obvious reason why some target genes and not others are associated with UVR8 in the ChIP experiments. However, the ChIP assays are subject to technical limitations because the signal is not very strong and obtaining quantitative data is therefore difficult. Thus, at present, the specificity of the ChIP interactions is not clear. Further studies, such as ChIP sequencing, may help to resolve this issue.
Although the mechanism by which UVR8 regulates transcription is obscure, it is likely that it promotes the activation or recruitment of transcription factors that regulate target genes and/or the remodeling of chromatin at selected gene loci. There is good evidence in maize (Zea mays) that UV-B regulates chromatin remodeling associated with transcriptional regulation. Genotypes from high altitude sites that are naturally exposed to elevated levels of UV-B have increased expression of transcripts encoding putative chromatin remodeling proteins (Casati et al., 2006). Moreover, when expression of genes encoding selected chromatin remodeling proteins is knocked down using RNA interference, plants become hypersensitive to UV-B and show altered expression of several UV-B–regulated genes. It is well established that histone modifications are important in recruiting regulatory proteins to particular sites in chromatin, and there is evidence that UV-B exposure causes histone modifications that correlate with altered transcription. ChIP experiments with maize show that acetylation of histones H3 and H4 is associated with increased transcription of several genes in response to UV-B (Casati et al., 2008). Similarly, in Arabidopsis, promoter regions of the UVR8-regulated HY5 and ELIP1 genes become enriched in chromatin containing diacetyl-histone H3(K9/K14) following UV-B exposure, indicating that this histone mark is associated with increased transcription in response to UV-B mediated by UVR8 (Cloix and Jenkins, 2008). Since there are many potential histone modifications, including several associated with responses to light (Fisher and Franklin, 2011), it will be valuable to identify all the modifications involved in responses to UV-B and whether their appearance is regulated by UVR8.
REGULATION OF UVR8
Negative Regulation of UVR8 Signaling
In many signaling pathways, the action of positive regulators is kept in check by one or more repressor proteins. UVR8 signaling is no exception. Two closely related proteins, REPRESSOR OF UV-B PHOTOMORPHOGENESIS1 (RUP1) and RUP2 act as redundant negative regulators of UVR8 action, effectively to ensure that the response to UV-B is balanced and not excessive. Grüber et al. (2010) found that a rup1 rup2 double mutant is hyperresponsive to UV-B, with enhanced hypocotyl growth suppression, increased UVR8-mediated HY5 and CHS gene expression, and elevated levels of flavonoids compared with wild-type plants under UV-B. By contrast, overexpression of RUP2 in transgenic plants suppresses expression of HY5 and CHS in response to UV-B. The RUP1 and RUP2 genes are stimulated by UV-B exposure under the control of UVR8, COP1, and HY5 (Grüber et al., 2010). Hence, the RUP proteins provide negative feedback regulation of UV-B signaling, preventing the exaggerated response to UV-B characteristic of plants overexpressing UVR8.
RUP1 and RUP2 are small WD40-repeat proteins with sequence similarity to the WD40 domains of COP1 and the SPA proteins. Bimolecular fluorescence complementation assays with proteins transiently expressed in mustard (Sinapis alba) seedling hypocotyls show that both RUPs interact with UVR8 (Grüber et al., 2010). Moreover, this interaction is seen both in the presence and absence of UV-B. Similar results are seen in yeast two-hybrid assays, although the interaction appears stronger under UV-B (Cloix et al., 2012). These findings show that negative regulation by the RUP proteins is achieved through physical interaction with UVR8. But how does interaction with RUPs impair UVR8 signaling? It is interesting that both RUP proteins interact with the C27 region of UVR8, which is also required for binding COP1. This finding suggests that RUP proteins may inhibit UVR8 signaling by impairing the binding of COP1. One possibility is that the RUP proteins have higher affinity for interaction with the C-terminal region of UVR8 than COP1. Alternatively, their affinity for binding to UVR8 may be similar to that of COP1, but their abundance may exceed that of COP1 as a result of expression following UV-B exposure, resulting in an increased probability of the RUPs binding to UVR8. Accurate measurements of the binding affinities and relative abundance of the proteins is required to resolve this point.
Although the RUP proteins evidently have an important role in regulating UVR8, they have additional functions. RUP1 and RUP2 are also known as EARLY FLOWERING BY OVEREXPRESSION1 (EFO1) and EFO2, respectively, because transgenic lines overexpressing these genes have early flowering phenotypes (Wang et al., 2011). In addition, the overexpressors have elongated hypocotyls. Mutants in efo2 (rup2) and efo1 efo2 (rup1 rup2) have reduced hypocotyl length and are also altered in leaf formation and expansion. The light sources used in these experiments likely contain either no or extremely little UV-B, so the EFO phenotype appears to have no relationship to UV-B perception by UVR8. Expression of RUP/EFO genes appears to be stimulated by light generally and not specifically by UV-B and is subject to circadian regulation. Further study showed that EFO2/RUP2 is a repressor of flowering, and it reduces expression of the floral inducer FLOWERING LOCUS T (FT) (Wang et al., 2011). FT is regulated by CONSTANS, a transcription factor whose proteolytic destruction is controlled by COP1 (Liu et al., 2008). Although EFO2/RUP2 does not regulate CO transcript levels, CO is nevertheless required for the effect of EFO2/RUP2 on flowering. Wang et al. (2011) speculated that EFO2/RUP2 may act in some way with COP1 to degrade CO. Whether the RUP proteins have a role in targeted proteolysis similar to the way the related SPA proteins act with COP1 remains to be seen. Like COP1 and the SPA proteins, the RUP proteins contain a conserved DWD (DDB1 binding WD40) motif (Lee et al., 2008) and may therefore be able to interact with CUL4-DDB1.
A recent study by Huang et al. (2013) suggests that CUL4 acts as a negative regulator of UVR8 signaling because the UV-B–induced expression of some UVR8-regulated transcripts (but not HY5) is enhanced in transgenic plants expressing reduced levels of CUL4. These plants have increased levels of HY5 protein, particularly under UV-B conditions, suggesting that CUL4 is involved in repressing HY5 accumulation, presumably by mediating proteolysis.
Regulation of Dimer/Monomer Status
An important question for any photoreceptor is how the photoreceptive form is regenerated after photoreception has occurred. In the case of UVR8, how is the dimer regenerated after formation of the monomer? Two possible mechanisms can be considered. First, monomers could simply reassociate to form the dimer, but this might not happen spontaneously. Second, monomers could be degraded after they initiate signaling, with the dimer replaced by de novo synthesis; this mechanism would require rapid turnover of the protein. With the latter alternative, inhibition of degradation is predicted to lead to accumulation of the protein, while inhibition of synthesis should prevent its reappearance. Experiments with Arabidopsis plants grown in the presence of either the protein synthesis inhibitor cycloheximide (Heijde and Ulm, 2013; Heilmann and Jenkins, 2013) or MG132 (Heilmann and Jenkins, 2013), which inhibits degradation by the proteasome, do not support the rapid turnover hypothesis. In both cases, the level of UVR8 protein stays relatively constant, indicating a low rate of turnover. Experiments with purified UVR8 show that monomers can reassociate to form dimers that can respond to UV-B (Christie et al., 2012; Wu et al., 2012). However, this process is very slow, taking over 24 h for most of the dimer to reform. By contrast, the reversion of monomers to dimers is very rapid in vivo, being complete within ∼1 h (Heijde and Ulm, 2013; Heilmann and Jenkins, 2013; Figure 3C).
That reversion to the dimer is much more rapid in vivo indicates that the process is facilitated. Dimer regeneration requires intact cells because the rate of dimer formation in UV-B–illuminated plant cell extracts is just as slow as for the purified protein (Heilmann and Jenkins, 2013). Protein synthesis is required to maximize the rate of reversion to the dimer, as reversion slowed in the presence of cycloheximide. In addition, reversion is slower in the cop1 mutant and in plants expressing UVR8 lacking the C27 region (Heilmann and Jenkins, 2013). Together, these observations indicate a requirement for expression of UV-B–induced proteins and the interaction of one or more proteins with the C-terminal domain of UVR8 to maximize the kinetics of reversion. In principle, COP1 could be involved in either or both processes. It is important to note, however, that additional factors are required because the rate of reversion in vivo in the presence of cycloheximide and in cop1 and GFP-ΔC27UVR8 plants is not as slow as in vitro. A likely explanation is that residual levels of the protein(s) that mediates dimer reversion are sufficient to promote regeneration in the absence of new protein synthesis.
Heijde and Ulm (2013) obtained evidence that RUP proteins regulate UVR8 dimer regeneration. The involvement of the RUP proteins is consistent with the findings of Heilmann and Jenkins (2013), in that the RUPs are UV-B induced and interact with the C27 region of UVR8. In rup1 rup2 double mutant plants, the rate of reversion to the dimer following UV-B–induced monomerization is much slower than in the wild type, indicating that the RUP proteins facilitate dimer formation. Conversely, plants overexpressing RUP2 have reduced levels of UVR8 monomer, which can be explained by enhanced dimer reversion limiting monomer accumulation. During the period of dimer reversion, COP1 dissociates from UVR8, but RUP-mediated redimerization does not depend on this process. Indeed, RUP action appears to be independent of COP1 in that dimer formation occurs much more slowly in a triple cop1 rup1 rup2 mutant than in cop1 alone (Heijde and Ulm, 2013). Additional experiments are now required to further define the role of the RUP proteins in dimer reversion, COP1 dissociation, and UVR8 negative regulation.
MODEL OF PHOTOMORPHOGENIC UV-B RESPONSES
The model of UVR8 photoreception and signaling that has emerged in recent years is that the dimer absorbs UV-B, monomerizes, and then binds COP1 to initiate signaling. Fundamentally, this is an accurate basic description of events, but some elaboration is needed to explain all observations (Figure 4). In wild-type UVR8, COP1 interaction is dependent on photoreception and binding requires the C27 region. Therefore, photoreception initiates some change that enables COP1 to bind. This could be a conformational change that exposes the C terminus. It is also possible that some additional region of the protein interacts with COP1 and this becomes accessible following photoreception. Several mutants with weak dimers, such as the Ala mutants of triad Trps, bind COP1 constitutively so possibly in these proteins the C27 region is exposed as a result of the mutation. An understanding of the events following UVR8 photoreception is hampered by the lack of information on the location of the C terminus in the crystal structure. Studies of the above UVR8 mutant proteins in vivo indicate that neither monomer formation nor binding to COP1 is sufficient to initiate a response when the mutant protein is expressed at levels similar to wild-type UVR8 (O’Hara and Jenkins, 2012). This suggests that photoreception initiates an additional process that converts the photoreceptor into an active state, although at present we do not understand what this is. The requirement for COP1 binding and activation is illustrated in Figure 4. Based on recent evidence, COP1 is shown bound to SPA proteins (Huang et al., 2013; Heijde et al., 2013). Once the active monomer bound to COP1 is produced, it initiates gene expression, through both transcription and stabilization of effectors, in particular HY5. The presumed increase in RUP protein accumulation, which has yet to be assayed in vivo, is proposed to negatively regulate UVR8 through displacing COP1 and stimulating redimerization. It is not clear when, if at all, RUP proteins dissociate from the UVR8 dimer. No doubt this model will be modified as new information is obtained.
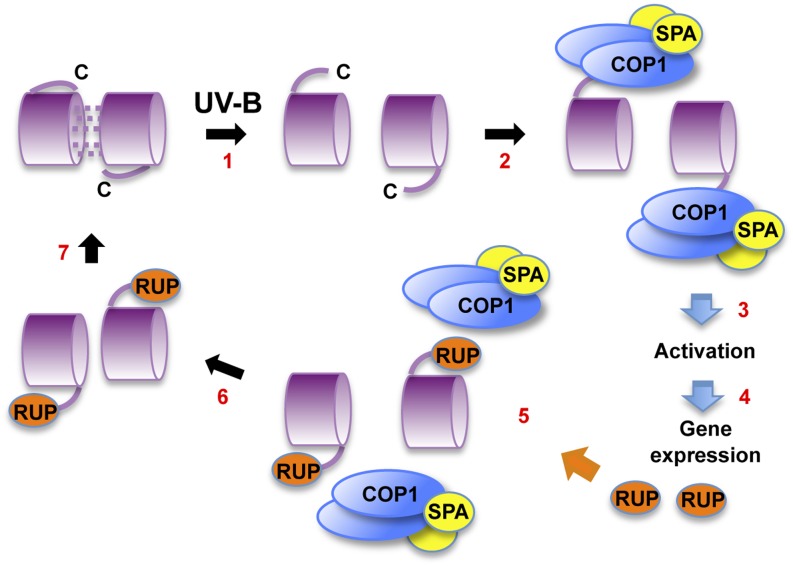
Figure 4.
Model of UVR8 Action.
(1) UV-B photoreception by dimeric UVR8 forms monomers. A proposed conformational change makes the C terminus of the protein available for interaction with COP1. However, the location of the C terminus is not known. (2) COP1, bound to SPA proteins, binds to UVR8 via the C27 region; it is not known whether other regions of UVR8 are involved in the interaction. (3) UVR8 bound to COP1/SPA adopts an active conformation ready to initiate gene expression. (4) UVR8 together with COP1/SPA regulates transcription of target genes, leading to photomorphogenic UV-B responses. (5) Among the genes induced are those encoding the RUP proteins, which negatively regulate UVR8. RUP1 and RUP2 bind to the C27 region of UVR8 and displace COP1. (6) The RUP proteins facilitate reassociation of UVR8 monomers to form the dimer. (7) The dimer is regenerated for photoreception.
SUMMARY AND PERSPECTIVES
Much has been learned about the physiological role and molecular function of UVR8 since its discovery in 2002. However, we are still at the beginning of the UVR8 story. Much remains to be discovered about the physiological significance of UVR8, particularly in species other than Arabidopsis, its integration with other signaling pathways, and the molecular mechanism of action of the photoreceptor.
Elucidation of the crystal structure of UVR8 has provided the basis for a more detailed understanding of the protein. The novel Trp-based mechanism of photoreception is remarkable, but not fully understood. Although the chromophore Trps have been identified, it is not clear how photoreception leads to disruption of the salt bridges that maintain the dimer. The application of appropriate biophysical methods is required to identify how excited states of the Trps neutralize the key salt bridges.
The structure of UVR8 is only partly characterized. The locations of the C and N termini and how conformation of the protein changes in going from dimer to monomer are unknown. This is crucial to understanding interactions with other proteins. Monomer formation exposes the dimer interaction surface, which is an important site for protein–protein interactions in other β-propeller proteins (Stirnimann et al., 2010). To date, only COP1 and the RUP proteins are known to interact directly with UVR8 in plants; further research may identify additional interactors. Both COP1 and the RUP proteins interact with the C terminus of UVR8, but the interaction of COP1 is UV-B dependent, whereas that of the RUP proteins is not. It is important to understand the basis of this difference. It is likely that photoreception initiates conformational changes to UVR8, particularly with respect to the C terminus, to promote COP1 binding. An additional possibility is that COP1 interacts with another region of the protein that is exposed following photoreception.
Signal transduction leads to transcriptional regulation, but this key aspect of UVR8 function is the least understood. The significance of UVR8 interaction with chromatin is not entirely clear. Does UVR8 form part of a complex on chromatin that regulates target genes? If so, how does it selectively associate with these genes? And is COP1 also part of the complex or does it dissociate from UVR8 prior to chromatin interaction? If the interaction of UVR8 with chromatin is nonspecific, then we need to understand how it can recruit transcription factors to selected target genes. The regulation of HY5 accumulation is crucial. UVR8 photoreception stimulates HY5 transcription but, in addition, binding of COP1 to UVR8 leads to stabilization of HY5, promoting its accumulation. However, the mechanisms involved in HY5 transcriptional regulation and stabilization of HY5 protein are poorly understood. Clearly, much work is required to address these questions.
In addition to understanding the fundamental basis of UVR8 function, it is important to consider potential applications. Plant responses to UV-B are important in agriculture (Wargent and Jordan, 2013), and UVR8 regulates plant biochemical composition, which can affect nutritional quality and defense against herbivores and pathogens. Moreover, UVR8 regulates morphogenesis. Hence, manipulation of the amount and biochemical properties of UVR8 may have applications in crop improvement. UVR8 may have further applications in synthetic biology and medicine through its potential as an optogenetic light switch. Optogenetics employs photoreceptors to manipulate processes in various organisms; UVR8 has several properties that make it attractive for such applications, and its potential in this respect has already been demonstrated (Crefcoeur et al., 2013; Müller et al., 2013).
Acknowledgments
I thank John Christie, Eirini Kaiserli, Brian Smith, and members of the Jenkins Lab for discussion of UVR8 and comments on the article. Research in my laboratory is supported by the UK Biotechnology and Biological Sciences Research Council, The Leverhulme Trust, and the University of Glasgow.
AUTHOR CONTRIBUTIONS
G.I.J. wrote the article.
References
- Ang L.H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., Deng X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1: 213–222 [DOI] [PubMed] [Google Scholar]
- Ballaré C.L., Barnes P.W., Flint S.D. (1995). Inhibition of hypocotyl elongation by ultraviolet-B radiation in de-etiolating tomato seedlings. 1. The photoreceptor. Physiol. Plant. 93: 584–592 [Google Scholar]
- Ballaré C.L., Mazza C.A., Austin A.T., Pierik R. (2012). Canopy light and plant health. Plant Physiol. 160: 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belles-Boix E., Babiychuk E., Van Montagu M., Inzé D., Kushnir S. (2000). CEO1, a new protein from Arabidopsis thaliana, protects yeast against oxidative damage. FEBS Lett. 482: 19–24 [DOI] [PubMed] [Google Scholar]
- Boccalandro H.E., Mazza C.A., Mazzella M.A., Casal J.J., Ballaré C.L. (2001). Ultraviolet B radiation enhances a phytochrome-B-mediated photomorphogenic response in Arabidopsis. Plant Physiol. 126: 780–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B.A., Cloix C., Jiang G.H., Kaiserli E., Herzyk P., Kliebenstein D.J., Jenkins G.I. (2005). A UV-B-specific signaling component orchestrates plant UV protection. Proc. Natl. Acad. Sci. USA 102: 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B.A., Headland L.R., Jenkins G.I. (2009). UV-B action spectrum for UVR8-mediated HY5 transcript accumulation in Arabidopsis. Photochem. Photobiol. 85: 1147–1155 [DOI] [PubMed] [Google Scholar]
- Brown B.A., Jenkins G.I. (2008). UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol. 146: 576–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell M.M., Robberecht R., Flint S.D. (1983). Internal filters: prospects for UV-acclimation in higher plants. Physiol. Plant. 58: 445–450 [Google Scholar]
- Casati P., Campi M., Chu F., Suzuki N., Maltby D., Guan S., Burlingame A.L., Walbot V. (2008). Histone acetylation and chromatin remodeling are required for UV-B-dependent transcriptional activation of regulated genes in maize. Plant Cell 20: 827–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P., Stapleton A.E., Blum J.E., Walbot V. (2006). Genome-wide analysis of high-altitude maize and gene knockdown stocks implicates chromatin remodeling proteins in response to UV-B. Plant J. 46: 613–627 [DOI] [PubMed] [Google Scholar]
- Christie J.M., Arvai A.S., Baxter K.J., Heilmann M., Pratt A.J., O’Hara A., Kelly S.M., Hothorn M., Smith B.O., Hitomi K., Jenkins G.I., Getzoff E.D. (2012). Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335: 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie J.M., Jenkins G.I. (1996). Distinct UV-B and UV-A/blue light signal transduction pathways induce chalcone synthase gene expression in Arabidopsis cells. Plant Cell 8: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloix C., Jenkins G.I. (2008). Interaction of the Arabidopsis UV-B-specific signaling component UVR8 with chromatin. Mol. Plant 1: 118–128 [DOI] [PubMed] [Google Scholar]
- Cloix C., Kaiserli E., Heilmann M., Baxter K.J., Brown B.A., O’Hara A., Smith B.O., Christie J.M., Jenkins G.I. (2012). C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. Proc. Natl. Acad. Sci. USA 109: 16366–16370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte M., de Simone S., Simmons S.J., Ballaré C.L., Stapleton A.E. (2010). Chromosomal loci important for cotyledon opening under UV-B in Arabidopsis thaliana. BMC Plant Biol. 10: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crefcoeur R.P., Yin R., Ulm R., Halazonetis T.D. (2013). Ultraviolet-B-mediated induction of protein-protein interactions in mammalian cells. Nat. Commun. 4: 1779. [DOI] [PubMed] [Google Scholar]
- Davey M.P., Susanti N.I., Wargent J.J., Findlay J.E., Paul Quick W., Paul N.D., Jenkins G.I. (2012). The UV-B photoreceptor UVR8 promotes photosynthetic efficiency in Arabidopsis thaliana exposed to elevated levels of UV-B. Photosynth. Res. 114: 121–131 [DOI] [PubMed] [Google Scholar]
- Demkura P.V., Ballaré C.L. (2012). UVR8 mediates UV-B-induced Arabidopsis defense responses against Botrytis cinerea by controlling sinapate accumulation. Mol. Plant 5: 642–652 [DOI] [PubMed] [Google Scholar]
- De Veylder L., Larkin J.C., Schnittger A. (2011). Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 16: 624–634 [DOI] [PubMed] [Google Scholar]
- Eisinger W.R., Bogomolni R.A., Taiz L. (2003). Interactions between a blue-green reversible photoreceptor and a separate UV-B receptor in stomatal guard cells. Am. J. Bot. 90: 1560–1566 [DOI] [PubMed] [Google Scholar]
- Ensminger P.A. (1993). Control of development in plants and fungi by far-UV radiation. Physiol. Plant. 88: 501–508 [Google Scholar]
- Favory J.J., et al. (2009). Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 28: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehér B., Kozma-Bognár L., Kevei E., Hajdu A., Binkert M., Davis S.J., Schäfer E., Ulm R., Nagy F. (2011). Functional interaction of the circadian clock and UV RESISTANCE LOCUS 8-controlled UV-B signaling pathways in Arabidopsis thaliana. Plant J. 67: 37–48 [DOI] [PubMed] [Google Scholar]
- Fisher A.J., Franklin K.A. (2011). Chromatin remodelling in plant light signalling. Physiol. Plant. 142: 305–313 [DOI] [PubMed] [Google Scholar]
- Frohnmeyer H., Bowler C., Zhu J.K., Yamagata H., Schäfer E., Chua N.H. (1998). Different roles for calcium and calmodulin in phytochrome- and UV-regulated expression of chalcone synthase. Plant J. 13: 763–772 [Google Scholar]
- Frohnmeyer H., Loyall L., Blatt M.R., Grabov A. (1999). Millisecond UV-B irradiation evokes prolonged elevation of cytosolic-free Ca2+ and stimulates gene expression in transgenic parsley cell cultures. Plant J. 20: 109–117 [DOI] [PubMed] [Google Scholar]
- Frohnmeyer H., Staiger D. (2003). Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiol. 133: 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev I., Vanderauwera S., Gechev T.S., Laloi C., Minkov I.N., Shulaev V., Apel K., Inzé D., Mittler R., Van Breusegem F. (2006). Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud T., Schweizer F., Tscheuschler A., Debrieux D., Casal J.J., Schäfer E., Hiltbrunner A., Fankhauser C. (2008). FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet. 4: e1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt K.E., Wilson M.I., Greenberg B.M. (2005). Ultraviolet wavelength dependence of photomorphological and photosynthetic responses in Brassica napus and Arabidopsis thaliana. Photochem. Photobiol. 81: 1061–1068 [DOI] [PubMed] [Google Scholar]
- González Besteiro M.A., Bartels S., Albert A., Ulm R. (2011). Arabidopsis MAP kinase phosphatase 1 and its target MAP kinases 3 and 6 antagonistically determine UV-B stress tolerance, independent of the UVR8 photoreceptor pathway. Plant J. 68: 727–737 [DOI] [PubMed] [Google Scholar]
- Grishina I.B., Woody R.W. (1994). Contributions of tryptophan side chains to the circular dichroism of globular proteins: Exciton couplets and coupled oscillators. Faraday Discuss. 99: 245–262 [DOI] [PubMed] [Google Scholar]
- Grüber H., Heijde M., Heller W., Albert A., Seidlitz H.K., Ulm R. (2010). Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 20132–20137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahlbrock K., Scheel D. (1989). Physiology and molecular biology of phenylpropanoid metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40: 347–369 [Google Scholar]
- Harlow G.R., Jenkins M.E., Pittalwala T.S., Mount D.W. (1994). Isolation of uvh1, an Arabidopsis mutant hypersensitive to ultraviolet light and ionizing radiation. Plant Cell 6: 227–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U., Sagasser M., Mehrtens F., Stracke R., Weisshaar B. (2005). Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 57: 155–171 [DOI] [PubMed] [Google Scholar]
- Hectors K., Prinsen E., De Coen W., Jansen M.A.K., Guisez Y. (2007). Arabidopsis thaliana plants acclimated to low dose rates of ultraviolet B radiation show specific changes in morphology and gene expression in the absence of stress symptoms. New Phytol. 175: 255–270 [DOI] [PubMed] [Google Scholar]
- Heijde M., Ulm R. (2012). UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 17: 230–237 [DOI] [PubMed] [Google Scholar]
- Heijde M., Ulm R. (2013). Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc. Natl. Acad. Sci. USA 110: 1113–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde M., Binkert M., Yin R., Ares-Orpel F., Rizzini L., Van De Slijke E., Persiau G., Nolf J., Gevaert K., De Jaeger G., Ulm R. (2013). Constitutively active UVR8 photoreceptor variant in Arabidopsis. Proc. Natl. Acad. Sci. USA 110: 20326–20331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann M., Jenkins G.I. (2013). Rapid reversion from monomer to dimer regenerates the ultraviolet-B photoreceptor UV RESISTANCE LOCUS8 in intact Arabidopsis plants. Plant Physiol. 161: 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Ouyang X., Yang P., Lau O.S., Chen L., Wei N., Deng X.W. (2013). Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc. Natl. Acad. Sci. USA 110: 16669–16674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Ouyang X., Yang P., Lau O.S., Li G., Li J., Chen H., Deng X.W. (2012). Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell 24: 4590–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indorf M., Cordero J., Neuhaus G., Rodríguez-Franco M. (2007). Salt tolerance (STO), a stress-related protein, has a major role in light signalling. Plant J. 51: 563–574 [DOI] [PubMed] [Google Scholar]
- Jansen M.A.K. (2002). Ultraviolet-B radiation effects on plants: Induction of morphogenic responses. Physiol. Plant. 116: 423–429 [Google Scholar]
- Jansen M.A.K., Gaba V., Greenberg B.M., Mattoo A.K., Edelman M. (1996). Low threshold levels of UV-B in a background of photosynthetically acive radiation trigger rapid degradation of the D2 protein of photosystem-II. Plant J. 9: 693–699 [Google Scholar]
- Jenkins G.I. (2009). Signal transduction in responses to UV-B radiation. Annu. Rev. Plant Biol. 60: 407–431 [DOI] [PubMed] [Google Scholar]
- Jenkins, G.I., and Brown, B.A. (2007). UV-B perception and signal transduction. In Light and Plant Development, G.C. Whitelam and K.J. Halliday, eds (Oxford, UK: Blackwell Publishing), pp. 155–182. [Google Scholar]
- Jenkins G.I., Long J.C., Wade H.K., Shenton M.R., Bibikova T.N. (2001). UV and blue light signaling: Pathways regulating chalcone synthase gene expression in Arabidopsis. New Phytol. 151: 121–131 [DOI] [PubMed] [Google Scholar]
- Jiang C.Z., Yee J., Mitchell D.L., Britt A.B. (1997). Photorepair mutants of Arabidopsis. Proc. Natl. Acad. Sci. USA 94: 7441–7445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Wang Y., Björn L.O., Li S. (2009). Arabidopsis radical-induced cell death1 is involved in UV-B signaling. Photochem. Photobiol. Sci. 8: 838–846 [DOI] [PubMed] [Google Scholar]
- Jiang L., Wang Y., Li Q.F., Björn L.O., He J.X., Li S.S. (2012b). Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res. 22: 1046–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Wang Y., Olof Björn L., He J.-X., Li S.-S. (2012a). Sensing of UV-B radiation by plants. Plant Signal. Behav. 7: 999–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Lau O.S., Deng X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Jordan B.R. (1996). The effects of ultraviolet-B radiation on plants: A molecular perspective. Adv. Bot. Res. 22: 97–162 [Google Scholar]
- Kaiserli E., Jenkins G.I. (2007). UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell 19: 2662–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami C., Lorrain S., Hornitschek P., Fankhauser C. (2010). Light-regulated plant growth and development. Curr. Top. Dev. Biol. 91: 29–66 [DOI] [PubMed] [Google Scholar]
- Kim B.C., Tennessen D.J., Last R.L. (1998). UV-B-induced photomorphogenesis in Arabidopsis thaliana. Plant J. 15: 667–674 [DOI] [PubMed] [Google Scholar]
- Klein R.M. (1978). Plants and near-ultraviolet radiation. Bot. Rev. 44: 1–127 [Google Scholar]
- Kliebenstein D.J., Lim J.E., Landry L.G., Last R.L. (2002). Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 130: 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry L.G., Stapleton A.E., Lim J., Hoffman P., Hays J.B., Walbot V., Last R.L. (1997). An Arabidopsis photolyase mutant is hypersensitive to ultraviolet-B radiation. Proc. Natl. Acad. Sci. USA 94: 328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang-Mladek C., Xie L., Nigam N., Chumak N., Binkert M., Neubert S., Hauser M.-T. (2012). UV-B signaling pathways and fluence rate dependent transcriptional regulation of ARIADNE12. Physiol. Plant. 145: 527–539 [DOI] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Laubinger S., Fittinghoff K., Hoecker U. (2004). The SPA quartet: A family of WD-repeat proteins with a central role in suppression of photomorphogenesis in arabidopsis. Plant Cell 16: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H.Y., Lee I., Deng X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Terzaghi W., Gusmaroli G., Charron J.B., Yoon H.-J., Chen H., He Y.J., Xiong Y., Deng X.W. (2008). Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20: 152–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yang L., Jin D., Nezames C.D., Terzaghi W., Deng X.-W. (2013). UV-B-induced photomorphogenesis in Arabidopsis. Protein Cell 4: 485–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H.L., He S.B., Zhang Y.C., Zhu D.M., Zhang J.Y., Jia K.P., Sun S.X., Li L., Yang H.Q. (2011). Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippuner V., Cyert M.S., Gasser C.S. (1996). Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J. Biol. Chem. 271: 12859–12866 [DOI] [PubMed] [Google Scholar]
- Liu B., Zuo Z., Liu H., Liu X., Lin C. (2011). Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.J., Zhang Y.C., Li Q.H., Sang Y., Mao J., Lian H.L., Wang L., Yang H.Q. (2008). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois R., Buchanan B.B. (1994). Severe sensitivity to ultraviolet-radiation in an Arabidopsis mutant deficient in flavonoid accumulation. 2. Mechanisms of UV-resistance in Arabidopsis. Planta 194: 504–509 [Google Scholar]
- Morales L.O., Brosché M., Vainonen J., Jenkins G.I., Wargent J.J., Sipari N., Strid A., Lindfors A.V., Tegelberg R., Aphalo P.J. (2013). Multiple roles for UV RESISTANCE LOCUS8 in regulating gene expression and metabolite accumulation in Arabidopsis under solar ultraviolet radiation. Plant Physiol. 161: 744–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K., Engesser R., Schulz S., Steinberg T., Tomakidi P., Weber C.C., Ulm R., Timmer J., Zurbriggen M.D., Weber W. (2013). Multi-chromatic control of mammalian gene expression and signaling. Nucleic Acids Res. 41: e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara A., Jenkins G.I. (2012). In vivo function of tryptophans in the Arabidopsis UV-B photoreceptor UVR8. Plant Cell 24: 3755–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravecz A., Baumann A., Máté Z., Brzezinska A., Molinier J., Oakeley E.J., Adám É., Schäfer E., Nagy F., Ulm R. (2006). CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Renault L., Nassar N., Vetter I., Becker J., Klebe C., Roth M., Wittinghofer A. (1998). The 1.7 A crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature 392: 97–101 [DOI] [PubMed] [Google Scholar]
- Rizzini L., Favory J.-J., Cloix C., Faggionato D., O’Hara A., Kaiserli E., Baumeister R., Schäfer E., Nagy F., Jenkins G.I., Ulm R. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Rozema J., van de Staaij J., Björn L.O., Caldwell M. (1997). UV-B as an environmental factor in plant life: Stress and regulation. Trends Ecol. Evol. (Amst.) 12: 22–28 [DOI] [PubMed] [Google Scholar]
- Ryan K.G., Swinny E.E., Winefield C., Markham K.R. (2001). Flavonoids and UV photoprotection in Arabidopsis mutants. Z. Naturforsch., C, J. Biosci. 56: 745–754 [DOI] [PubMed] [Google Scholar]
- Shinkle J.R., Atkins A.K., Humphrey E.E., Rodgers C.W., Wheeler S.L., Barnes P.W. (2004). Growth and morphological responses to different UV wavebands in cucumber (Cucumis sativum) and other dicotyledonous seedlings. Physiol. Plant. 120: 240–248 [DOI] [PubMed] [Google Scholar]
- Stracke R., Favory J.-J., Gruber H., Bartelniewoehner L., Bartels S., Binkert M., Funk M., Weisshaar B., Ulm R. (2010). The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 33: 88–103 [DOI] [PubMed] [Google Scholar]
- Stirnimann C.U., Petsalaki E., Russell R.B., Müller C.W. (2010). WD40 proteins propel cellular networks. Trends Biochem. Sci. 35: 565–574 [DOI] [PubMed] [Google Scholar]
- Suesslin C., Frohnmeyer H. (2003). An Arabidopsis mutant defective in UV-B light-mediated responses. Plant J. 33: 591–601 [DOI] [PubMed] [Google Scholar]
- Takahashi S., Milward S.E., Yamori W., Evans J.R., Hillier W., Badger M.R. (2010). The solar action spectrum of photosystem II damage. Plant Physiol. 153: 988–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilbrook K., Arongaus A.B., Binkert M., Heijde M., Yin R., Ulm R. (2013). The UVR8 UV-B photoreceptor: perception, signaling and response. The Arabidopsis Book 11: e0164, /. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H., Leasure C.D., Hou X., Yuen G., Briggs W., He Z.H. (2008). Role of root UV-B sensing in Arabidopsis early seedling development. Proc. Natl. Acad. Sci. USA 105: 21039–21044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R., Baumann A., Oravecz A., Máté Z., Adám E., Oakeley E.J., Schäfer E., Nagy F. (2004). Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R., Nagy F. (2005). Signalling and gene regulation in response to ultraviolet light. Curr. Opin. Plant Biol. 8: 477–482 [DOI] [PubMed] [Google Scholar]
- Wade H.K., Bibikova T.N., Valentine W.J., Jenkins G.I. (2001). Interactions within a network of phytochrome, cryptochrome and UV-B phototransduction pathways regulate chalcone synthase gene expression in Arabidopsis leaf tissue. Plant J. 25: 675–685 [DOI] [PubMed] [Google Scholar]
- Wang H., Ma L.G., Li J.M., Zhao H.Y., Deng X.W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Wang W., Yang D., Feldmann K.A. (2011). EFO1 and EFO2, encoding putative WD-domain proteins, have overlapping and distinct roles in the regulation of vegetative development and flowering of Arabidopsis. J. Exp. Bot. 62: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Wargent J.J., Gegas V.C., Jenkins G.I., Doonan J.H., Paul N.D. (2009). UVR8 in Arabidopsis thaliana regulates multiple aspects of cellular differentiation during leaf development in response to ultraviolet B radiation. New Phytol. 183: 315–326 [DOI] [PubMed] [Google Scholar]
- Wargent J.J., Jordan B.R. (2013). From ozone depletion to agriculture: Understanding the role of UV radiation in sustainable crop production. New Phytol. 197: 1058–1076 [DOI] [PubMed] [Google Scholar]
- Wellmann E. (1976). Specific ultraviolet effects in plant morphogenesis. Photochem. Photobiol. 24: 659–660 [DOI] [PubMed] [Google Scholar]
- Wellmann, E. (1983). UV radiation in photomorphogenesis. In Encyclopedia of Plant Physiology New Series, Vol. 16B, W. Shropshire Jr. and H. Mohr, eds (Berlin: Springer), pp. 745–756. [Google Scholar]
- Winkel-Shirley B. (2002). Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5: 218–223 [DOI] [PubMed] [Google Scholar]
- Wu D., Hu Q., Yan Z., Chen W., Yan C., Huang X., Zhang J., Yang P., Deng H., Wang J., Deng X., Shi Y. (2012). Structural basis of ultraviolet-B perception by UVR8. Nature 484: 214–219 [DOI] [PubMed] [Google Scholar]
- Wu M., Eriksson L.A., Strid A. (2013). Theoretical prediction of the protein–protein interaction between Arabidopsis thaliana COP1 and UVR8. Theor. Chem. Acc. 132: 1371 [Google Scholar]
- Wu M., Grahn E., Eriksson L.A., Strid A. (2011). Computational evidence for the role of Arabidopsis thaliana UVR8 as UV-B photoreceptor and identification of its chromophore amino acids. J. Chem. Inf. Model. 51: 1287–1295 [DOI] [PubMed] [Google Scholar]
- Yang H.Q., Tang R.H., Cashmore A.R. (2001). The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng P.-Y., Vakoc C.R., Chen Z.-C., Blobel G.A., Berger S.L. (2006). In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. Biotechniques 41: 694–698, 696, 698 [DOI] [PubMed] [Google Scholar]
- Zhu D., Maier A., Lee J.-H., Laubinger S., Saijo Y., Wang H., Qu L.-J., Hoecker U., Deng X.W. (2008). Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20: 2307–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z., Liu H., Liu B., Liu X., Lin C. (2011). Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 21: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]