The transcriptional coactivator ANGUSTIFOLIA3 (AN3) stimulates cell division during Arabidopsis leaf development. It is shown that AN3 associates with SWI/SNF chromatin remodeling complexes to regulate the expression of important downstream transcription factors and that the module SWI/SNF-AN3 is a major player in the transition from cell division to cell expansion in developing leaves.
Abstract
The transcriptional coactivator ANGUSTIFOLIA3 (AN3) stimulates cell proliferation during Arabidopsis thaliana leaf development, but the molecular mechanism is largely unknown. Here, we show that inducible nuclear localization of AN3 during initial leaf growth results in differential expression of important transcriptional regulators, including GROWTH REGULATING FACTORs (GRFs). Chromatin purification further revealed the presence of AN3 at the loci of GRF5, GRF6, CYTOKININ RESPONSE FACTOR2, CONSTANS-LIKE5 (COL5), HECATE1 (HEC1), and ARABIDOPSIS RESPONSE REGULATOR4 (ARR4). Tandem affinity purification of protein complexes using AN3 as bait identified plant SWITCH/SUCROSE NONFERMENTING (SWI/SNF) chromatin remodeling complexes formed around the ATPases BRAHMA (BRM) or SPLAYED. Moreover, SWI/SNF ASSOCIATED PROTEIN 73B (SWP73B) is recruited by AN3 to the promoters of GRF5, GRF3, COL5, and ARR4, and both SWP73B and BRM occupy the HEC1 promoter. Furthermore, we show that AN3 and BRM genetically interact. The data indicate that AN3 associates with chromatin remodelers to regulate transcription. In addition, modification of SWI3C expression levels increases leaf size, underlining the importance of chromatin dynamics for growth regulation. Our results place the SWI/SNF-AN3 module as a major player at the transition from cell proliferation to cell differentiation in a developing leaf.
INTRODUCTION
After Arabidopsis thaliana seeds have germinated, new leaves arise from the shoot apical meristem as rod-like primordia that develop into mature leaves. Initially, leaf primordia contain only dividing cells, and this proliferation phase is followed by the transition phase that forms a bridge to the cell expansion phase, where cells exit the mitotic cell cycle and start differentiation (Donnelly et al., 1999; Beemster et al., 2005). During the transition phase, dividing and expanding cells coexist in the leaf and comprise the basal and apical leaf parts, respectively. The boundary between them, termed the cell cycle arrest front, establishes rapidly and disappears abruptly at the end of the transition phase (Kazama et al., 2010; Andriankaja et al., 2012).
ANGUSTIFOLIA3 (AN3)/GRF-INTERACTING FACTOR1 (GIF1), a member of the GIF family of transcriptional coactivators along with GIF2 and GIF3, plays a key role in Arabidopsis shoot development (Kim and Kende, 2004). AN3 and GIF2 are important for cotyledon identity establishment during embryogenesis (Kanei et al., 2012), and ectopic expression of AN3, GIF2, and GIF3 increases leaf size due to an increase in cell number (Horiguchi et al., 2005; Lee et al., 2009). On the other hand, loss of AN3 function results in smaller and narrower leaves with fewer cells (Kim and Kende, 2004; Horiguchi et al., 2005). Whereas gif2 and gif3 leaves are almost identical to wild-type leaves, double and triple gif mutations synergistically reduce cell number, revealing the overlapping and redundant functions of these genes (Lee et al., 2009). AN3 is also involved in the determination of adaxial/abaxial leaf polarity (Horiguchi et al., 2011). Furthermore, moving from the leaf mesophyll, where it is synthesized, to the epidermis, the AN3 protein itself is proposed to coordinate epidermal cell proliferation with proliferation in the leaf mesophyll (Kawade et al., 2013).
GIFs, as their name reveals, were first identified by the interaction with GROWTH REGULATING FACTOR1 (GRF1) (Kim and Kende, 2004), a transcription factor that is part of a family comprising nine members (GRF1 to GRF9) (Kim et al., 2003). Like GIFs, GRFs likely stimulate leaf cell proliferation since overexpression enhances leaf growth and cell division, as shown for GRF1, GRF2, and GRF5 (Horiguchi et al., 2005; Kim and Lee, 2006). A reduction in leaf cell number has only been shown for grf4 and grf5 single mutants, whereas functional redundancy becomes apparent from the different double, triple, or quadruple combinations of grf1, grf2, grf3, grf4, or grf5 mutations that synergistically diminish leaf growth (Kim et al., 2003; Horiguchi et al., 2005; Kim and Lee, 2006).
Because of their similar functions, physical interaction, and synergistic defects in leaf size (Kim and Kende, 2004), GIFs and GRFs are thought to form a functional transcriptional coactivator/transcription factor complex that affects gene expression for correct lateral organ development. Molecular data on components of the GIF/GRF signaling pathway are only beginning to emerge, as seven GRFs are predicted targets of microRNA396 (miR396), which restricts GRF expression to the basal part of the leaf (Liu et al., 2009; Rodriguez et al., 2010). However, the transcriptional network directly downstream of the AN3/GRF module has yet to be uncovered.
DNA binding transcription factors often cooperate with transcriptional coactivators, and they both promote transcription in similar ways, such as by stimulating general complex formation around RNA polymerase II or by recruiting chromatin remodelers. The N-terminal domain of GIF proteins is homologous to the SNH domain of human SYNOVIAL TRANSLOCATION (SYT) (Kim and Kende, 2004; Horiguchi et al., 2005), which was shown to interact with human BRAHMA (BRM) and BRAHMA RELATED GENE1 (BRG1), the two human SWITCH/SUCROSE NONFERMENTING (SWI/SNF) chromatin remodeling ATPases (Nagai et al., 2001; Perani et al., 2003). Given this sequence homology, GIF transcriptional coactivators are likely to promote transcription by association with SWI/SNF chromatin remodelers.
SWI/SNF are high molecular weight complexes that use the energy derived from ATP hydrolysis to change interactions between histone octamers and the DNA (Clapier and Cairns, 2009). Evolutionary conservation allowed for the description of various SWI/SNF complex subunits in Arabidopsis based on sequence similarity with metazoan subunits and include four SWI2/SNF2 ATPases (BRM, SPLAYED [SYD], MINU1/CHR12, and MINU2/CHR23), four SWI3 proteins (SWI3A to SWI3D), two SWI/SNF ASSOCIATED PROTEINS 73 (SWP73A/CHC2 and SWP73B/CHC1), two ACTIN RELATED PROTEINS predicted to belong to SWI/SNF complexes (ARP4 and ARP7), and a single protein termed BUSHY (BSH) (The Chromatin Database, www.chromdb.org; Meagher et al., 2005; Jerzmanowski, 2007; Kwon and Wagner, 2007; Sang et al., 2012). The complexes are assembled around one central ATPase, and differences in complex composition further result from the incorporation of distinct paralogous subunit family members and the more transient, often tissue-specific, interactions with other proteins like transcriptional coactivators and transcription factors (Clapier and Cairns, 2009; Hargreaves and Crabtree, 2011).
Genetic analysis has proven the importance of the subunits of SWI/SNF complexes in the transcriptional regulation of key developmental processes. Mutation of BRM, SYD, SWI3C, and SWI3D and silencing of SWP73B, BSH, and ARP4 results in severely dwarfed plants that have reduced leaf and stem size, and perturbed flowering time and flower development, often leading to sterility (Brzeski et al., 1999; Wagner and Meyerowitz, 2002; Kandasamy et al., 2005b; Sarnowski et al., 2005; Hurtado et al., 2006; Crane and Gelvin, 2007). Moreover, loss of function of SWI3A, SWI3B, and ARP7 causes embryonic lethality, just as in the minu1 minu2 double mutation (Kandasamy et al., 2005a; Sarnowski et al., 2005; Sang et al., 2012). The molecular mechanisms by which the SWI/SNF complexes execute these roles are starting to be understood. For example, BRM, SYD, and SWI3 proteins were demonstrated to be involved in the regulation of transcription factors that determine cotyledon boundary establishment (Kwon et al., 2006) and shoot apical meristem maintenance (Kwon et al., 2005), in the inhibition of cytokinin responses to promote leaf maturation (Efroni et al., 2013), and in the stimulation of gibberellin responses in the plant (Archacki et al., 2013; Sarnowska et al., 2013). During reproductive development, both the transition to flowering and the expression of flower homeotic genes depend on BRM, SYD, and SWI3 activity (Wagner and Meyerowitz, 2002; Sarnowski et al., 2005; Hurtado et al., 2006; Su et al., 2006; Farrona et al., 2011; Wu et al., 2012). In addition, seed storage protein–encoding genes and genes involved in stress signaling mediated by abscisic acid, jasmonate, and ethylene were shown to be regulated by BRM or SYD (Tang et al., 2008; Walley et al., 2008; Han et al., 2012). As has been reported for mammals, plant SWI/SNF complexes play a role in pluripotency and cell fate determination, and different complexes composed of paralogous subunits can have overlapping, but also unique, roles (Bezhani et al., 2007; Hargreaves and Crabtree, 2011).
Here, we identify important transcription factors that are regulated by AN3, extending the network downstream of the GIF/GRF module. We also report the identification of Arabidopsis SWI/SNF complexes that are associated with AN3 and provide evidence that chromatin remodeling activity is involved in the transcriptional regulation of downstream AN3 targets, suggesting that AN3 functions to recruit SWI/SNF complexes to promote cell division during leaf development.
RESULTS
Induction of AN3 Activity Enhances Leaf Growth and CYCB1;1 Expression
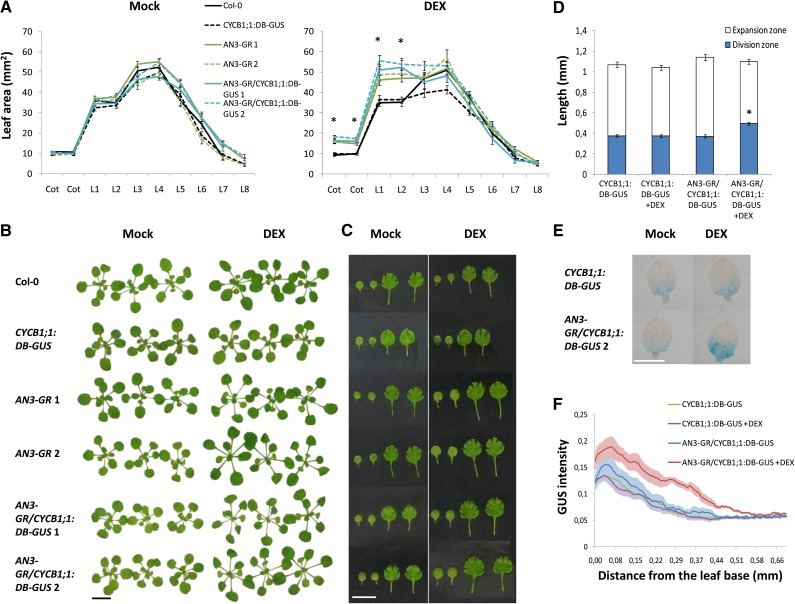
To gain insight into the molecular pathways downstream of AN3, plants containing an inducible gain-of-function construct, 35S:AN3-GR, hereafter designated AN3-GR, were generated. Fusion to the rat glucocorticoid receptor (GR) domain allows translocation of transcriptional regulators to the nucleus only after application of a glucocorticoid hormone, such as dexamethasone (DEX), thereby activating the downstream transcriptional responses.
Wild-type Columbia-0 (Col-0) plants and plants expressing CYCB1;1:D-Box-GUS-GFP (CYCB1;1:DB-GUS) (Eloy et al., 2011), a cell division marker, were transformed with the AN3-GR construct and independent homozygous lines were obtained. Without DEX application, the size of individual AN3-GR leaves at 21 d after stratification (DAS) was indistinguishable from wild-type and CYCB1;1:DB-GUS leaves (Figures 1A to 1C). Growth of AN3-GR plants on 25 µM DEX from germination onwards led to the development of larger cotyledons and leaves 1 and 2, compared with mock-treated transformants and DEX-treated control plants (Figures 1A to 1C). This phenotype is reminiscent of plants overexpressing AN3 (Horiguchi et al., 2005), confirming the functionality of the construct.
Figure 1.
Induction of AN3 Activity Enhances Leaf Growth and CYCB1;1 Expression.
(A) to (C) Twenty-one-day-old plants, grown on control medium (Mock) or medium supplemented with DEX.
(A) Leaf area of cotyledons (Cot) and leaves 1 to 8 (L1 to L8), measured from leaf series. Error bars are se (n ≥ 12). Asterisks indicate significant difference from the wild type (Col-0) (P < 0.01, Student’s t test).
(B) Rosettes.
(C) Cotyledons and leaves 1 and 2. Bars = 10 mm in (B) and (C).
(D) to (F) GUS staining of CYCB1;1:DB-GUS and AN3-GR/CYCB1;1:DB-GUS leaves 1 and 2. Plants were transferred at 9 DAS to mock medium or medium supplemented with DEX for 24 h.
(D) GUS-stained and nonstained regions, indicating the division and expansion zones, respectively, measured along the length of the leaf. Error bars are se (n ≥ 22). Asterisks indicate significant difference from DEX-treated control plants (P < 0.01, Student’s t test).
(E) Leaves 1 and 2 were mounted on slides for picture taking. Bar = 1 mm.
(F) GUS staining was measured with Image J in a defined area along the leaf length. Error bars are se (n ≥ 18).
The CYCB1;1:DB-GUS construct allows for quantitative analysis of mitotic activity in developing plants. AN3-GR/CYCB1;1:DB-GUS plants were grown for 9 DAS and subsequently transferred to medium supplemented with 10 µM DEX for 24 h, after which the first leaves were analyzed for GUS staining. At this stage, mitotic activity is only present in the basal region in CYCB1;1:DB-GUS leaves (Figures 1D and 1E). Induction of AN3 activity extended the region of GUS staining measured along the length of the leaf while total leaf length was unaffected by 24-h DEX treatment (Figures 1D and 1E). In addition, the GUS intensity in the stained region was increased in AN3-GR/CYCB1;1:DB-GUS leaves compared with untreated and CYCB1;1:DB-GUS leaves (Figures 1E and 1F), indicating a function for AN3 in both the duration and the rate of cell proliferation.
Analysis of Downstream AN3 Responses
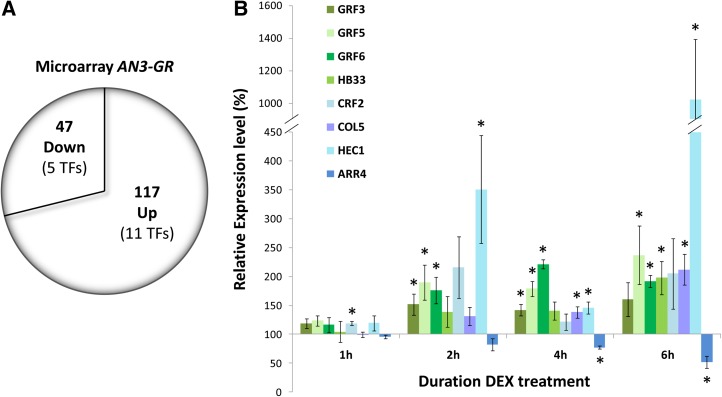
How does AN3, described to be a transcriptional coactivator, positively regulate leaf cell division? To answer this question, developing first leaves of AN3-GR and wild-type plants were subjected to transcript profiling using Affymetrix ATH1 microarrays after transfer at 8 DAS to DEX-containing medium for 8 h. At this time point, a substantial amount of cells in leaves 1 and 2 is proliferating, while some cells start transitioning from cell proliferation to cell expansion. In addition, wild-type AN3 expression is associated with proliferating cells and decreases at 8 DAS (Horiguchi et al., 2005, 2011). Steroid activation of AN3 function in AN3-GR leaves resulted in the induction of 117 genes and the repression of 47 genes, including 11 and 5 transcription factors, respectively, compared with DEX-treated wild-type leaves, with a false discovery rate < 0.05 (Figure 2A, Table 1; Supplemental Data Set 1). Strikingly, four members of the GRF family were upregulated: GRF3, GRF5, GRF6, and GRF8.
Figure 2.
Identification of Transcription Factors Rapidly Regulated by AN3, by Time-Course Analysis of Expression Levels.
(A) Wild-type and AN3-GR plants were grown for 8 d and transferred to medium supplemented with 5 µM DEX for 8 h. The number of upregulated and downregulated genes with P value < 0.05 is shown, and differentially expressed transcription factors (TFs) are indicated in parentheses.
(B) Transcription factors differentially expressed in AN3-GR leaves 1 and 2 compared with wild-type leaves 1, 2, 4, or 6 h after DEX treatment. qRT-PCR expression levels were normalized to DEX-treated wild-type expression levels, which are set at 100% for each time point. Error bars are se of three biological replicates. Asterisks indicate significant difference from DEX-treated wild-type plants (P < 0.1, Student’s t test).
Table 1. Transcription Factors Differentially Expressed after Induction of AN3 Activity.
| AGI Codea | Annotation | FC | P Value | Wild Type 9 to 10 DAS | an3 | 35S:GRF5 | TChAP-Seq AN3-HBH |
|---|---|---|---|---|---|---|---|
| AT5G28640 | AN3 | 8.26 | 0.00031 | Down | |||
| AT5G67060 | HEC1 | 2.48 | 0.04199 | Up | X | ||
| AT3G13960 | GRF5 | 2.32 | 0.01151 | Down | Up | ||
| AT1G75240 | HB33 | 1.94 | 0.01975 | Down | Up | ||
| AT2G06200 | GRF6 | 1.92 | 0.03288 | ||||
| AT4G39780 | AP2 domain–containing transcription factor, putative | 1.55 | 0.04592 | ||||
| AT2G36400 | GRF3 | 1.54 | 0.04199 | Down | Down | ||
| AT2G42870 | PAR1 (PHY RAPIDLY REGULATED1) | 1.54 | 0.04161 | ||||
| AT4G24150 | GRF8 | 1.51 | 0.04865 | ||||
| AT4G23750 | CRF2 | 1.46 | 0.04161 | X | |||
| AT1G51700 | ADOF1 (Dof zinc-finger protein) | −1.65 | 0.04161 | ||||
| AT1G71030 | MYBL2 (MYB-LIKE2) | −1.64 | 0.03820 | Up | |||
| AT5G47640 | NF-YB2 (NUCLEAR FACTOR Y, SUBUNIT B2) | −1.52 | 0.03288 | X | |||
| AT1G10470 | ARR4 | −1.49 | 0.05260 | Upb | |||
| AT1G28370 | ERF11 (ERF DOMAIN PROTEIN11) | −1.48 | 0.04085 | X | |||
| AT5G66070 | Zinc-finger (C3HC4-type RING finger) family protein | −1.43 | 0.04536 |
Affymetrix ATH1 transcript profiles of AN3-GR leaves 1 and 2 compared with wild-type leaves 8 h after DEX treatment. Differentially expressed transcription factors with P value < 0.05 are shown, ordered according to fold change (FC). In addition, the intersection with publicly available microarray data sets of wild-type leaf 3 between days 9 and 10 (Andriankaja et al., 2012), an3 leaves 1 and 2 (Horiguchi et al., 2011), and 35S:GRF5 shoots (Gonzalez et al., 2010) is indicated, and the last column shows the presence in the AN3-HBH TChAP-seq data set.
Arabidopsis Genome Initiative.
The P value of ARR4 is not < 0.05, but the gene is upregulated in an3 and independently confirmed, as shown in Supplemental Figure 7.
Functional enrichment analysis for MapMan categories with PageMan (Usadel et al., 2006) revealed an overrepresentation among the upregulated genes of categories, including RNA processing and RNA regulation of transcription, DNA synthesis and chromatin structure, amino acid activation pseudouridylate synthesis, ribosomal protein synthesis, and proteins not assigned to a functional category, including ABC1 family proteins and pentatricopeptide repeat–containing proteins (Supplemental Figure 1A). In addition, Gene Ontology (GO) overrepresentation analysis of subcellular localization with PLAZA (Van Bel et al., 2012) uncovered the presence of proteins predominantly in the nucleus, the nucleolus, and the intracellular organelle lumen (Supplemental Figure 1C). The downregulated genes were primarily enriched in categories of sulfur-containing secondary metabolism and transport (Supplemental Figure 1B). Taken together, these microarray data suggest a role for AN3 in the regulation of the general processes that sustain the high metabolic rate of dividing cells.
Comparison of the differentially expressed genes with recently published transcriptome sets of leaf 3 from day 8 to day 13, covering the subsequent phases of cell proliferation, transition, and expansion (Andriankaja et al., 2012), revealed a significant overlap between the AN3-upregulated genes and the genes whose expression went down between days 9 and 10, concomitant with a sharp transition from cell proliferation to expansion (Supplemental Figure 2A and Supplemental Data Set 1; Table 1). Both data sets are enriched for similar functional categories, namely, RNA processing and RNA regulation of transcription, DNA synthesis and chromatin remodeling, and ribosomal protein synthesis. An albeit smaller, but significant, overlap was also found between the genes downregulated after AN3-GR induction and the genes upregulated during development of leaf 3 between days 9 and 10 (Supplemental Figure 2B and Supplemental Data Set 1; Table 1). Moreover, expression of most genes in both overlaps gradually decreases or increases, respectively, during leaf 3 development between days 8 and 13 (Supplemental Figures 3A and 3B), confirming that AN3 functions in activating gene transcription that favors cell proliferation, while to a lesser extent, also inhibiting expression of genes that promote differentiation.
In addition, the intersection was analyzed with microarray data from an3 mutant leaves 1 and 2 (Horiguchi et al., 2011) and 35S:GRF5 rosettes at stage 1.03 (Gonzalez et al., 2010). Although the number of genes in the intersections was small, the AN3-GR downregulated genes significantly overlapped with genes regulated in the same direction in 35S:GRF5 plants and in the opposite direction in an3 mutant plants. Also, upregulated genes from DEX-treated AN3-GR leaves significantly overlapped with the upregulated genes in 35S:GRF5 seedlings (Supplemental Figures 2A and 2B). Transcription factors with higher expression in both AN3-GR and 35S:GRF5 leaves encoded the basic helix-loop-helix transcriptional regulator HECATE1 (HEC1) and HOMEOBOX33 (HB33) (Table 1).
Identification of Transcription Factors Rapidly Regulated by AN3
During 8-h treatment with DEX, AN3-GR activation triggered the expression of numerous genes. To analyze this in more detail, a time-course experiment was conducted, in which RNA levels were quantified with quantitative RT-PCR (qRT-PCR) at 1, 2, 4, and 6 h after transfer of AN3-GR and wild-type plants to DEX-containing medium. The transcripts of all differentially expressed transcription factors were analyzed, as well as the remaining GRFs. Expression levels of the latter were analyzed to investigate the preference of AN3 to regulate certain GRFs and because GRF4 and GRF9 are not represented on the ATH1 array.
GRF3, GRF5, and GRF6 were induced significantly from 2 h onwards (Figure 2B). The transcript levels of GRF7 and GRF8 were in general low and variable in the first leaves and not significantly affected by DEX treatment. Also, the expression of the other GRFs was not markedly changed (Supplemental Figure 4A), in accordance with the microarray data, suggesting that AN3 directly and specifically activates the transcription of GRF3, GRF5, and GRF6. AN3 was previously shown to interact with GRF5 in yeast two-hybrid (Y2H) assays (Kim and Kende, 2004; Horiguchi et al., 2005), and by coimmunoprecipitation (Co-IP) from cell suspension cultures, we confirm the physical interaction of AN3 with GRF5 (Supplemental Figure 5). Furthermore, GRF5 and GRF6 expression levels were downregulated in 12-d-old an3 rosettes, although GRF3 expression was unchanged compared with the wild type (Supplemental Figure 6). Taken together, the above suggests that AN3/GRF complexes could activate their own transcription.
Analysis of the expression kinetics of the remaining upregulated transcription factors revealed that four were induced earlier than 8 h after DEX treatment (Figure 2B): CYTOKININ RESPONSE FACTOR2 (CRF2) was transiently significantly upregulated 1 h after transfer to DEX, HEC1 after 2, 4, and 6 h, CONSTANS-LIKE5 (COL5) after 4 and 6 h, and HB33 after 6 h (Figure 2B).
None of the significantly downregulated transcription factors appeared to be repressed earlier than 8 h (Supplemental Figure 4B), which suggests that they are likely not early targets of the AN3 transcriptional response. We also analyzed the expression of A-type ARABIDOPSIS RESPONSE REGULATOR4 (ARR4), despite its false discovery rate of 0.0526 (Table 1). ARR4 is of interest because it was found to be upregulated in the an3 mutant and downregulated in 35S:GRF5 plants (Table 1; Supplemental Figure 7). ARR4 transcript levels were significantly lower in AN3-GR leaves 4 and 6 h after induction compared with the wild type (Figure 2B), confirming its rapid repression by AN3 in developing leaves.
In conclusion, the time-course experiment identified GRF3, GRF5, GRF6, CRF2, HEC1, COL5, HB33, and ARR4 as genes whose expression is rapidly changed upon AN3 activation.
Genome-Wide Determination of AN3 Binding Sites
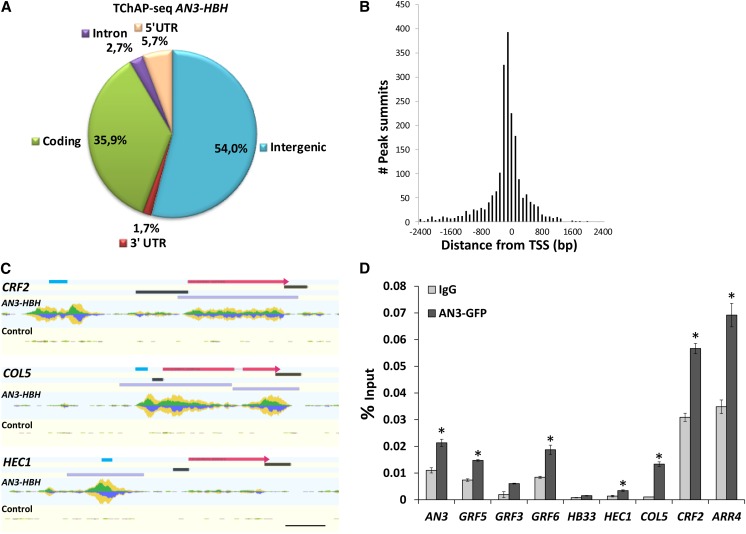
Next, we aimed to identify direct targets of AN3 among the rapidly up- and downregulated transcription factors by analyzing the presence of AN3 at their genomic regions. Thereto, tandem chromatin affinity purification (TChAP), a variant of chromatin immunoprecipitation (ChIP; see Methods), was performed and followed by sequencing (TChAP-seq). Because the TChAP protocol requires relatively large amounts of input material, Arabidopsis cell suspension cultures were used as starting material. A TChAP-purified cell culture overexpressing HBH-tagged AN3 was compared with a TChAP-purified wild-type PSB-D cell culture. A total of 23.47 million reads were obtained for the AN3-HBH TChAP sample after Illumina sequencing of the purified DNA, and 27.30 million reads for the wild-type TChAP control sample. After discarding redundant reads and reads that did not map uniquely to the genome, 2836 peaks were called using model-based analysis of ChIP-Seq (MACS; Zhang et al., 2008), corresponding to 2702 genes in the Arabidopsis genome (Supplemental Data Set 2).
A relatively even distribution of peaks was observed across the five chromosomes, with the exception of the gene-poor centromeric regions, which can be expected for a transcriptional regulator (Supplemental Figure 8A). Analysis of the peak locations revealed the presence of 54% in the intergenic regions, including the promoters (Figure 3A). While 35.9% of peaks was assigned to coding regions, only 2.7% was located in the introns and 7.4% in the untranslated regions (UTRs) (Figure 3A). When compared with the fractions of these genomic regions in the full Arabidopsis genome, the percentage of peaks in coding regions is similar to what can be expected (36%). However, the number of peaks in intergenic regions and UTRs are increased given the genome-wide fractions of 41 and 5%, respectively, while the introns are depleted of peaks (expected fraction 16%). Next, the location of the peak summits in relation to the position of the start codon (translation start site) was analyzed for the peaks that mapped closest to the 5′ end of the neighboring gene. Of these 2040 peaks, more than half had a summit located between −200 and +200 bp from the translation start site with a maximum between −100 and 0 bp (Figure 3B), illustrating the molecular function of AN3 as a transcriptional coactivator in the regulation of gene expression. Additionally, a search for motifs using RSAT peak motifs (Thomas-Chollier et al., 2012) led to the identification of two significantly enriched motifs in the peak sequences: the tgaCACGTGgca motif containing the core G-box sequence (CACGTG) and the GAGA motif (GAGAGAGA) (Supplemental Figures 8B and 8C), a putative element of Arabidopsis core promoters (Yamamoto et al., 2009). Within the peak sequences, which have a median peak length around 1200 bp, the distribution of both motifs was enriched near the peak summits (Supplemental Figures 8B to 8D).
Figure 3.
Genome-Wide Determination of AN3 Binding Sites and Identification of Direct AN3 Target Transcription Factors by TChAP-Seq and ChIP-qPCR.
(A) Genome-wide distribution of the location of the peaks called by MACS (Zhang et al., 2008), after TChAP on AN3-HBH–transformed cell cultures followed by sequencing.
(B) Distance in base pairs of the peak summits relative to the translation start site (TSS) of the nearest gene. Peak summits (2040) were included which are located closest to 5′ gene ends.
(C) GenomeView representation (Abeel et al., 2012) of the TChAP-seq results for CRF2, COL5, and HEC1 loci, showing read coverage in the TChAP-purified AN3-HBH versus the wild-type control samples. The reads are piled up with forward reads above the axis displayed in green and reverse reads below the axis in blue. Total coverage is indicated in yellow. Scaling was done relative to the maximum number of reads. The coding regions are indicated as pink boxes, the UTRs as black boxes, and the peaks as purple boxes above the reads. The regions amplified with ChIP-qPCR are indicated in light blue. Bar = 0.4 kb.
(D) ChIP with anti-IgG and anti-GFP antibody on 14-d-old plants expressing GFP-tagged AN3. Enrichment was determined with qPCR and for each locus normalized against the input. For diagrams of the loci including the amplified regions, see Supplemental Figure 9. Error bars are sd of two biological replicates. Asterisks indicate significant difference from wild-type plants (P < 0.05, Student’s t test).
AN3 Is Present at the Genomic Loci of Downstream Transcriptional Regulators
Subsequently, the AN3-HBH TChAP-seq data set, obtained from cell cultures, was searched for the presence of peaks mapping to the loci encoding transcription factors that were differentially expressed upon AN3-GR induction in proliferating leaves. Five out of 20 genes found in the overlap between the AN3-HBH TChAP-seq and the AN3-GR microarray data sets are transcription factors (Table 1), revealing a significant enrichment (P = 2.35E-3, χ2 test). Also, the complete AN3-HBH TChAP-seq data set was found to be enriched for the presence of transcription factors (P = 2.07E-22, χ2 test), pointing toward a function for AN3 as a key regulator of an extended downstream transcriptional network.
Among the rapidly differentially expressed transcription factors (Figure 2B), the CRF2, COL5, and HEC1 loci were found to be associated with AN3. Peaks could be detected along the coding regions of the CRF2 and COL5 AN3-HBH sample, while the number of reads was low in the corresponding genomic regions of the control sample (Figure 3C), resulting in a 35-fold enrichment of reads at both loci. Furthermore, AN3 occupancy was detected in the promoter and 5′ UTRs of the COL5 locus with a 29-fold increase in reads, and a steep peak corresponding to a fold change of around 19 was observed in the promoter region of HEC1 (Figure 3C). The regulatory DNA regions of COL5 contain the G-box–derived motif and the GAGA motif, and the GAGA motif was found in the HEC1 promoter (Supplemental Data Set 2). Although an 11-fold increase in reads was also observed in the CRF2 promoter region, no peak was called (Figure 3C). In addition, MACS did not identify peaks in the GRF3, GRF5, GRF6, HB33, nor ARR4 loci, possibly due to the different plant material used for microarray analysis and TChAP-seq. However, reads piled up at the end of the ARR4 coding region and the 3′ UTR resulting in a 15-fold enrichment compared with the control sample (Supplemental Figure 9). Likewise, an almost 19-fold increase in the number of reads was observed in the AN3 coding region, although no peak was called (Supplemental Figure 9).
Since TChAP-seq was performed on cell cultures, AN3 association with the transcription factor loci was further investigated in planta by ChIP. Chromatin was isolated with anti-GFP antibody from 14-d-old plants constitutively expressing a C-terminal fusion of AN3 to GFP (35S:AN3-GFP) and compared with anti-IgG ChIP-purified samples. With quantitative PCR (qPCR), a 2- to 10-fold enrichment of the promoter regions of CRF2, COL5, and HEC1 was detected, confirming AN3 occupancy at these loci. Moreover, the presence of AN3 was also shown at its own promoter and the promoters of GRF5, GRF6, and ARR4 (Figure 3D).
Taken together, AN3 rapidly activates or represses the expression and is able to bind the genomic loci of GRF5, GRF6, CRF2, COL5, HEC1, and ARR4, likely rendering them direct targets of AN3 transcriptional regulation. Furthermore, AN3 was shown to associate with its own promoter, arguing in favor for the AN3/GRF complexes to regulate their own expression.
AN3 Associates with SWI/SNF Chromatin Remodeling Complexes
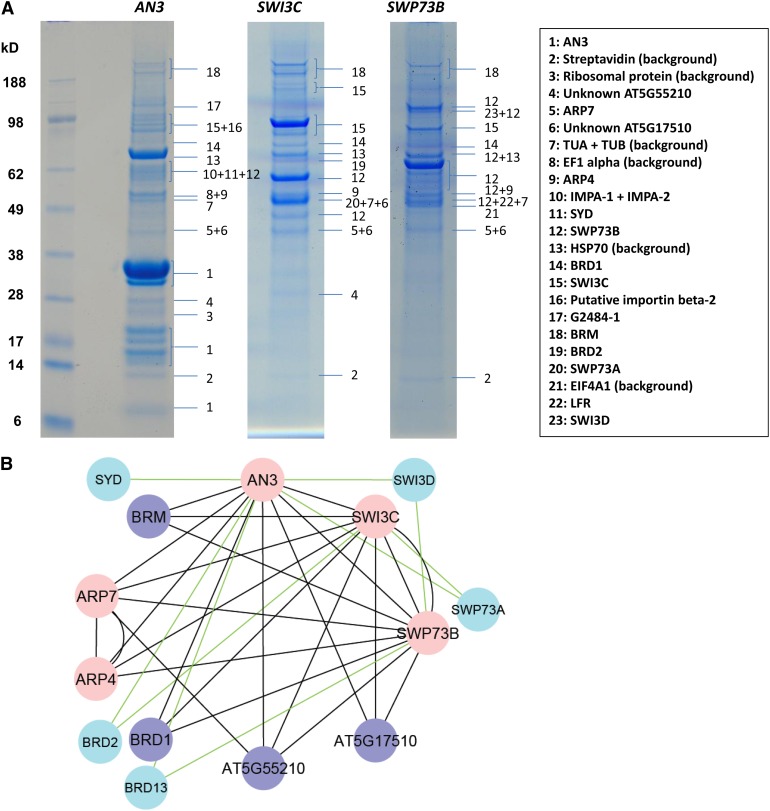
To identify additional interacting partners of AN3 besides the GRFs, AN3 was used as a bait for tandem affinity purification (TAP) followed by mass spectrometry analysis (TAP/MS), a powerful method to isolate and identify protein complexes (Van Leene et al., 2007; see Methods for experimental details). Both C- and N-terminal fusions of AN3 to the GS TAP tag were expressed under the control of the 35S promoter in Arabidopsis cell cultures. Eight independent TAP experiments resulted in the identification of 14 proteins, including AN3 (Table 2). Furthermore, TAP from 6-d-old Arabidopsis seedlings expressing C-terminal GS-tagged AN3 from the CDKA;1 promoter confirmed 11 out of the 14 proteins previously isolated from cell cultures. In addition, five other preys were identified (Table 2).
Table 2. Tandem Affinity Purification with AN3, SWI3C, SWP73B, ARP7, and ARP4 as Baits.
| AN3 |
AN3 Planta |
SWI3C |
SWP73B |
ARP7 |
ARP4 |
|||
|---|---|---|---|---|---|---|---|---|
| AGI Codea | Annotation | ChromDB ID | Eight Exps. | One Exp. | Five Exps. | Five Exps. | Four Exps. | Two Exps. |
| AT5G28640 | AN3 | – | 4 (bait) | 1 (bait) | ||||
| AT2G28290 | SYD | CHR3 | 8 | 1 | ||||
| AT2G46020 | BRM | CHR2 | 6 | 1 | 5 | 3 | ||
| AT1G21700 | SWI3C | CHB4 | 4 | 1 | 5 (bait) | 4 | ||
| AT4G34430 | SWI3D | CHB3 | 1 | 4 | ||||
| AT3G01890 | SWP73A | CHC2 | 1 | 5 | ||||
| AT5G14170 | SWP73B | CHC1 | 4 | 1 | 5 | 5 (bait) | ||
| AT1G18450 | ARP4 | ARP4 | 4 | 1 | 5 | 3 | 1 | 2 (bait) |
| AT3G60830 | ARP7 | ARP7 | 7 | 1 | 5 | 4 | 4 (bait) | 2 |
| AT1G20670 | BRD1 | BRD1 | 1 | 1 | 5 | 5 | ||
| AT1G76380 | BRD2 | BRD2 | 1 | 4 | ||||
| AT5G55040 | BRD13 | BRD13 | 1 | 2 | ||||
| AT5G55210 | Unknown protein | – | 4 | 1 | 4 | 4 | 2 | |
| AT5G17510 | Unknown protein | – | 3 | 1 | 5 | 4 | ||
| AT3G22990 | LFR | – | 1 | 4 | ||||
| AT4G17330 | G2484-1 | – | 6 | 1 | ||||
| AT4G16143 | IMPA-2 | – | 5 | |||||
| AT3G06720 | IMPA-1 | – | 4 | |||||
| AT5G53480 | Putative importin β-2 | – | 4 | |||||
| AT3G17590 | BSH | CHE1 | 4 | |||||
| AT2G47620 | SWI3A | CHB1 | 5 | |||||
| AT2G33610 | SWI3B | CHB2 | 4 | |||||
| AT5G45600 | TAF14B | YDG1 | 2 | |||||
| AT5G14240 | Thioredoxin superfamily protein | – | 2 | 2 | ||||
| AT4G22320 | Unknown protein | – | 1 | 1 | 4 | |||
| AT1G47128 | RD21 | – | 4 | |||||
| AT1G32730 | Unknown protein | – | 4 | |||||
| AT1G06500 | Unknown protein | – | 3 | |||||
| AT3G18380 | Homeobox transcription factor | – | 2 |
TAP was performed on Arabidopsis cell cultures and for AN3 on Arabidopsis seedlings (AN3 planta). The presence in The Chromatin Database (www.chromdb.org) is shown (ChromDB ID). The numbers indicate the number of experiments (Exps) in which the protein was identified.
Arabidopsis Genome Initiative.
Strikingly, several plant homologs of SWI/SNF complex subunits were repeatedly purified from cell culture and seedlings, including SYD, BRM, SWI3C, SWP73B, ARP4, and ARP7 (Figure 4A). SWI3D and SWP73A were isolated from seedlings as well. Hence, AN3 associates with plant SWI/SNF complexes.
Figure 4.
Tandem Affinity Purification Reveals Interaction of AN3 with SWI/SNF Complexes.
(A) Images of denaturing gels of TAP experiments with C-terminal GS-tagged AN3 and N-terminal GS-tagged SWI3C and SWP73B in cell cultures. Only interactors that could be distinguished as a band are indicated.
(B) Cytoscape (Shannon et al., 2003) protein interaction networks are based on the TAP experiments shown in Table 2. Pink nodes indicate proteins used as bait and purple nodes those that were pulled down with at least three out of five baits. Paralogous proteins identified by AN3 TAP are represented by blue nodes. Black edges are used when the proteins were identified by at least three baits and green edges for proteins identified by one or two bait proteins.
Reverse TAP experiments with SWI3C, SWP73B, ARP7, and ARP4 (Table 2, Figure 4A) allowed the reconstruction of putative SWI/SNF complexes around BRM and AN3 (Figure 4B). The network edges do not necessarily represent direct protein–protein interactions because TAP does not allow the distinction between direct and indirect interactions. Nevertheless, the reciprocal TAPs provide useful information on the architecture of plant SWI/SNF complexes (Table 2, Figure 4B). First, the SWI3C TAP fusion pulled down BRM as a single ATPase, and other SWI3 proteins were absent, confirming the preferred coexistence of SWI3C with BRM (Hurtado et al., 2006; Archacki et al., 2009). Second, TAP with SWP73B yielded BRM, BSH, and all SWI3 proteins, but lacked SWP73A, while both SWP73 proteins were detected by AN3 and SWI3C TAP experiments. This indicates that SWP73A and SWP73B are mutually exclusive but show a rather low specificity for a certain subunit composition. Third, ARP4 and ARP7 were detected in all experiments, proving their coexistence; finally, three unknown proteins encoded by At5g55210, At5g17510, and At4g22320 represent high-confidence plant SWI/SNF-interacting proteins given their purification by at least three baits (Table 2, Figure 4).
Furthermore, the reciprocal isolation of bromodomain proteins BRD1, BRD2, and/or BRD13 with AN3, SWI3C, and SWP73B as baits stood out (Table 2, Figure 4B) because of homology with animal polybromo proteins. These are signature proteins that define the pBAF (for POLYBROMO-ASSOCIATED BAF) SWI/SNF class and distinguish it from the BAF SWI/SNF class that is characterized by the presence of one or more AT-RICH INTERACTION DOMAIN (ARID)–like proteins (Hargreaves and Crabtree, 2011). In addition, AN3 and SWP73B TAP isolated LEAF AND FLOWER RELATED (LFR) (Table 2), a protein that shows 28% homology to human ARID2/BAF200 (Wang et al., 2009), a signature protein of pBAF complexes, indicating that complexes around AN3 show more resemblance to animal pBAF complexes.
Taken together, we isolated SWI/SNF chromatin remodeling complexes from Arabidopsis, revealing that AN3 may recruit pBAF-like complexes, including those containing BRM or SYD; SWI3C and/or SWI3D; SWP73A or SWP73B; and ARP4 and ARP7.
Binding of SWI/SNF Proteins SWP73B and BRM to AN3 Target Promoters and a Function for AN3 in SWP73B Recruitment
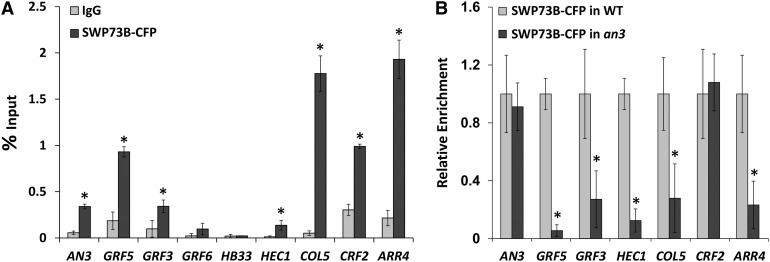
AN3 binds SWI/SNF chromatin remodeling complexes most likely to recruit the complexes to genomic regions of the downstream target genes, where they move the nucleosomes and thereby modulate the accessibility of cis-regulatory elements. One way to investigate whether SWI/SNF complexes are involved in the regulation of transcription of the AN3 target genes is to demonstrate the presence of the chromatin remodeling complexes at the target loci. Thereto, ChIP was used to analyze if two SWI/SNF proteins purified by AN3 TAP, SWP73B and BRM, occupy the promoters of the downstream transcription factors and the AN3 promoter itself.
Plants were transformed with CFP-tagged SWP73B constructs (35S:SWP73B-CFP) and grown until 14 DAS. Chromatin was precipitated with an anti-GFP antibody from Arabidopsis rosettes, and enrichment of selected DNA sequences was determined by qPCR. Primers annealing to the promoter regions revealed significant enrichments for the AN3, GRF3, GRF5, CRF2, COL5, HEC1, and ARR4 loci compared with ChIP-qPCR from 35S:SWP73B-CFP plants with anti-IgG antibody (Figure 5A). Remarkably, a strong overlap with AN3-GFP ChIP-qPCR could be observed, apart from the differences in GRF3 and GRF6 promoter occupancy (Figure 3D).
Figure 5.
AN3 Is Essential for Optimal Binding of SWP73B to a Subset of their Target Promoters.
ChIP with anti-IgG and anti-GFP antibody on Col-0 plants expressing CFP-tagged SWP73B (A) and with anti-GFP antibody on Col-0 (WT) and an3 mutants expressing CFP-tagged SWP73B (B). Enrichment was determined with qPCR and for each locus normalized against the input. In addition, for (B), the enrichment in the wild type was set arbitrarily to 1. For diagrams of the loci including the amplified regions, see Figure 3D and Supplemental Figure 9. Error bars are sd (n = 2). Asterisks indicate significant difference from wild-type plants (P < 0.05, Student’s t test).
BRM ChIP was performed with an anti-HA antibody on 9-d-old transgenic plants expressing biologically active HA-tagged BRM (Han et al., 2012). A strong enrichment was observed for the promoter of HEC1 in BRM-HA shoots, while no significant differences were found for AN3, GRF3, GRF5, GRF6, or HB33 promoter regions compared with anti-HA ChIP from wild-type plants (Supplemental Figure 10).
To provide more solid proof of the involvement of AN3 in recruiting SWI/SNF complexes to its target loci, association of SWP73B to the target promoters was analyzed in the an3 mutant background. Anti-GFP ChIP was performed on Col-0 and an3 plants expressing CFP-tagged SWP73B. Relative to 35:SWP73B-CFP control plants, the binding of SWP73B to the GRF5, GRF3, HEC1, COL5, and ARR4 promoter regions was significantly reduced in 35:SWP73B-CFP/an3 plants (Figure 5B), indicating that AN3 is essential for the optimal binding of SWP73B to these loci. No differences in enrichment could be observed for the AN3 and CRF2 loci in the absence of AN3 (Figure 5B).
Thus, both SWP73B and BRM physically associate with the HEC1 promoter and SWP73B is present at the promoter of AN3, GRF3, GRF5, CRF2, COL5, and ARR4 in young Arabidopsis seedlings. Furthermore, a role for AN3 in the recruitment of SWP73B to a subset of common target loci is demonstrated.
BRM Is Essential for Expression of the AN3 Target Genes
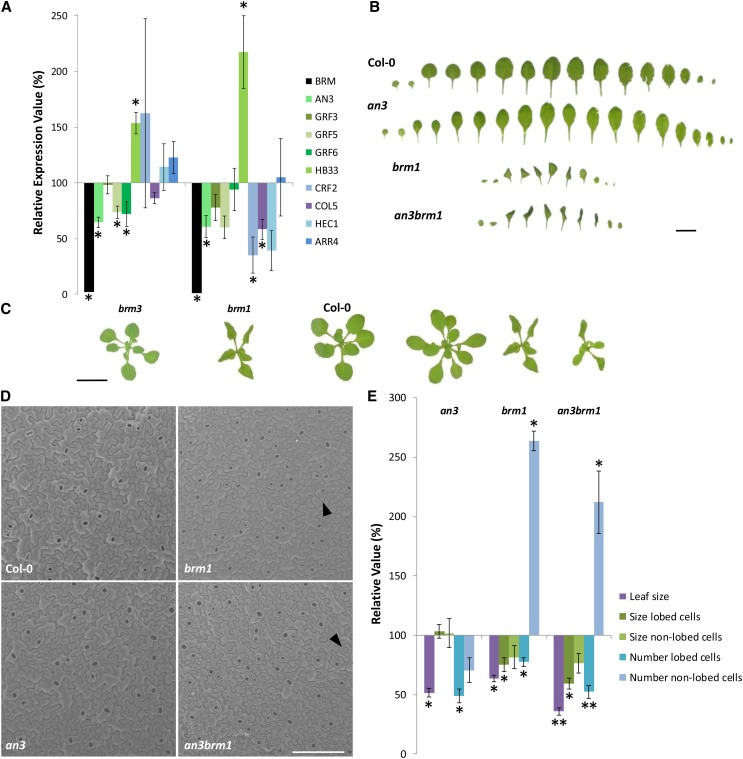
To investigate whether SWI/SNF complexes formed around the ATPase BRM are necessary for proper activation or repression of AN3 and the genes regulated by AN3, their expression was analyzed in brm mutants. The brm1 mutant has a T-DNA insert in the first exon, resulting in severe developmental defects, such as small spiral-shaped leaves with downward curling edges (Figure 6C; Hurtado et al., 2006). The brm3 mutant shows only a mild reduction in leaf growth (Figure 6C), since here T-DNA insertion gives rise to a truncated protein missing the C-terminal bromo and DNA binding domains, which does not seem to interfere with complex assembly (Farrona et al., 2007). Because the switch to reproductive development is affected in brm mutants (Hurtado et al., 2006; Farrona et al., 2011), shoots were harvested at early time points in long-day or noninductive short-day conditions to determine the role of BRM in gene expression specifically related to leaf development. As such, qRT-PCR expression levels of AN3, GRF5, and GRF6 were found to be significantly reduced in 8-d-old brm3 shoots grown in long-day conditions compared with wild-type seedlings (Figure 6A). Transcription of AN3 was also significantly reduced in brm1 rosettes grown for 22 d in short-day conditions. In addition, CRF2 and COL5 were downregulated in brm1 shoots. GRF3, HEC1, and ARR4 were not differentially expressed when brm was mutated, whereas HB33, in contrast with our expectations, was upregulated (Figure 6A). Thus, the correct transcription of AN3 and several genes regulated by AN3 seems to depend on BRM, suggesting that BRM is recruited by AN3 to remodel the chromatin at the respective regulatory DNA regions.
Figure 6.
BRM Is Involved in Regulation of Transcription of AN3 Target Genes and Genetically Interacts with AN3.
(A) Expression levels determined by qRT-PCR in brm3 rosettes of 8-d-old plants grown in long-day (16 h light/8 h dark) conditions and in brm1 rosettes of 22-d-old plants grown in short-day (8 h light/16 h dark) conditions. Normalization of expression levels was done relative to those of the wild type (Col-0), which are set at 100% for each gene. Error bars are se (n = 3). Asterisks indicate significant difference from wild-type plants (P < 0.1, Student’s t test).
(B) Leaf series of 24-d-old wild-type, brm1, an3, and an3 brm1 plants grown in long-day conditions. Bar = 10 mm.
(C) Rosettes of brm3, brm1, the wild type, an3, and an3 brm1 plants at 22 DAS. Bar = 10 mm.
(D) Scanning electron microscopy pictures of the abaxial epidermis of 22-d-old leaves 1 and 2 of wild-type, an3, brm1, and an3 brm1 plants. Examples of small nonlobed cells in brm1 and an3 brm1 leaves are indicated by arrowheads. Bar = 150 µm.
(E) Leaf size, pavement cell sizes, and pavement cell numbers of 22-d-old leaves 1 and 2. Nonlobed and lobed cells are defined as follows: nonlobed cells < 25 µm2 < lobed cells. Normalization was done relative to the wild type (Col-0), which is set at 100% for each measurement. Error bars are se (n = 5). Single asterisks indicate significant difference from wild-type plants, and double asterisks indicate significant difference from brm1 plants (P < 0.01, Student’s t test).
BRM and AN3 Genetically Interact
In comparison with the brm1 null mutant, the an3 deletion mutant displays a milder leaf growth defect characterized by smaller and narrower leaves and a shorter plastochron (Figures 6B and 6C; Horiguchi et al., 2005). To examine the genetic interaction between AN3 and BRM during leaf development, double an3 brm1 mutants were generated. Since brm1 plants are sterile, heterozygous brm1 were crossed with homozygous an3 plants. After selection and self-pollination of AN3BRM1/an3brm1 heterozygotes, the F2 progeny was searched for an3 brm1 double mutants. The three different wild-type, an3, and brm1 shoot phenotypes could be distinguished, but no additional rosette phenotypes were observed, suggesting that an3 brm1 plants have either the an3 or the brm1 phenotype. Subsequent genotyping revealed that an3 brm1 phenotypes strongly resembled the more severe brm1 shoot phenotypes. To confirm these results, the F3 progeny of an3BRM1/an3brm1 F2 plants was analyzed. The presence of plants among the F3 offspring with the brm1 phenotype that lacked both an3 and brm1 transcripts indeed demonstrated that the visible phenotype of an3 brm1 double homozygous shoots is indistinguishable from the brm1 single mutant shoot phenotype (Figures 6B and 6C). Although 25% of the an3BRM1/an3brm1 offspring is expected to be double homozygous, only around 16% had the brm1 phenotype (Supplemental Table 1). The segregation with a reduced brm1 homozygous progeny was shown in previous reports, which demonstrated a reduced male and female gametophytic transmission of the brm1 mutant allele (Hurtado et al., 2006; Archacki et al., 2009).
The shoot phenotype of the brm1 mutant visually does not appear to be enhanced by simultaneous an3 loss of function (Figures 6B and 6C). However, the curled morphology of the brm1 and an3 brm1 leaves prevents the detection of potential subtle phenotypic differences. Therefore, leaves 1 and 2 were analyzed in more detail. Measurements of the flattened leaf areas at 22 DAS revealed a more or less equal reduction in an3 and brm1 single mutants compared with wild-type plants, while the first leaves of an3 brm1 plants were further reduced in size (Figure 6E).
Examination of the pavement cells using scanning electron microscopy revealed a clear distinction between Col-0 and an3 cells on the one hand and brm1 and an3 brm1 cells on the other hand. Whereas most pavement cells of Col-0 and an3 leaves obtained the characteristic puzzle shape, brm1 and an3 brm1 leaves contained an increased percentage of small cells, which appear to have divided recently (Figure 6D). This is an indication that mutation of brm1 delays the development of individual leaves, which is consistent with the severely reduced number of leaves in 3-week-old plants (Figures 6B and 6C). To make a relevant comparison at the cellular level between leaves at different developmental stages, the pavement cells were subdivided in two categories. A cutoff was determined based on the size distribution of the pavement cells resulting in a lobed category and a nonlobed category, where lobed > 25 µm2 > nonlobed, corresponding to their visual appearance. As such, it was confirmed that mutation of an3 significantly reduced the number of lobed cells (Figure 6E). brm1 leaves have a decreased size and number of lobed cells, while the number of nonlobed cells was strongly increased. Comparable increases in nonlobed cell number and comparable decreases in lobed cell size were observed in the an3 brm1 double mutant, while the number of lobed cells was further decreased relative to the brm1 mutant (Figure 6E). Hence, despite the absence of brm1 and the corresponding severe phenotype, mutation of an3 results in a subtle reduction in the number of lobed cells. Similar results were observed when pavement cells of leaves 1 and 2 were analyzed at 14 DAS (Supplemental Figure 11).
Taken together, the strong pleiotropic brm1 phenotype is fully penetrant in an3 brm1 leaves, although the effect of an3 mutation is not entirely absent. Given their molecular functions and protein complex formation, this suggests that AN3 works together with BRM to perform at least part of its function during leaf development.
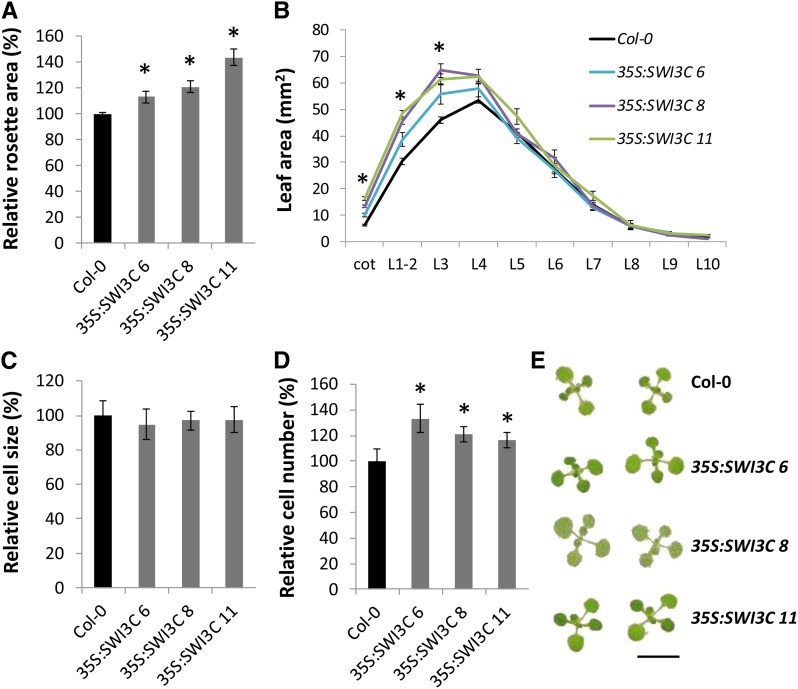
Overexpression of SWI3C Enhances Leaf Growth
Since knockout and knockdown of SWI/SNF subunits results in severely dwarfed plants, we wondered if increased expression could lead to the development of larger organs. To answer this question, SWI3C was ectopically expressed using the 35S promoter in wild-type background. Independent transformants were obtained, expression levels were quantified, and individual leaf and rosette areas were measured. This revealed that increased expression of SWI3C frequently led to an increase in rosette area (Figures 7A and 7E; Supplemental Figure 12A). Four out of 12 35S:SWI3C lines had significantly larger rosette sizes. Although rosette area was similar to that of the wild type in five other 35S:SWI3C lines, closer inspection of their individual leaves revealed that four lines showed a strong increase in the size of the first leaves, whereas younger leaves were similar to or smaller than those of the wild type (Supplemental Figures 12A and 12B). In contrast, the remaining three 35S:SWI3C lines appeared to be smaller compared with wild-type plants (Supplemental Figure 12A), but analyses of transgene expression levels revealed a correlation with the phenotype. Only when mRNA levels were increased more than 3-fold did overexpression of SWI3C enhance leaf size (Supplemental Figure 12C).
Figure 7.
Overexpression of SWI3C Enhances Leaf Growth.
(A) Total rosette area calculated from individual leaf sizes from 21-d-old 35S:SWI3C plants. Error bars are se (n ≥ 10). Asterisks indicate significant difference from the wild type (Col-0) (P < 0.05, Student’s t test).
(B) Individual cotyledon (Cot) and leaf areas (L1 to L10) measured from leaf series made at 21 DAS from plants with increased leaf growth indicated in (A). Error bars are se (n = 8). Asterisks indicate significant difference from the wild type (Col-0) (P < 0.05, Student’s t test).
(C) and (D) Pavement cell area (C) and pavement cell number (D) of 21-d-old leaves 1 and 2. Error bars are se (n = 6). Asterisks indicate significant difference from the wild type (Col-0) (P < 0.01, Student’s t test).
(E) Rosettes of 15-d-old Col-0 and 35S:SWI3C lines showing enhanced leaf growth. Bar = 10 mm.
At 21 DAS, a significant increase in the size of the cotyledons and leaves 1, 2, and 3 was measured in three selected independent 35S:SWI3C lines (Figure 7B). To elucidate the cellular nature of the increase in leaf growth, pavement cell size and number of the first leaves were determined in the three 35S:SWI3C lines. Significant increases in cell number but not cell size were observed, indicating that enhanced cell proliferation, and not cell expansion, was responsible for the increased leaf area when SWI3C is overexpressed (Figures 7C and 7D). This indicates that the SWI/SNF component SWI3C is important for the stimulation of cell division during leaf development.
DISCUSSION
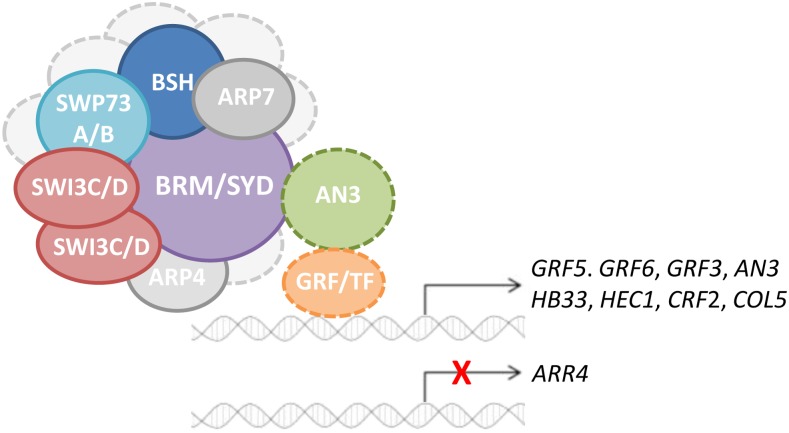
By the identification of the transcriptional coactivator AN3 as an interacting protein of SWI/SNF complexes in Arabidopsis, this work provides an explanation for the necessity of SWI/SNF chromatin remodeling during leaf development. We hypothesize that AN3 forms a bridge between SWI/SNF complexes and GRFs or possibly other transcription factors to direct BRM or SYD ATPase activity for efficient target gene transcription (Figure 8). This acts to delay the exit from the mitotic cell cycle, thereby simultaneously delaying the start of differentiation. Microarray analysis of developing first leaves in which AN3 is activated revealed induction of genes that are downregulated and repression of genes that are upregulated during the transition from cell proliferation to expansion, supporting the proposed role of the SWI/SNF-AN3 complex in the regulation of leaf growth.
Figure 8.
Model for AN3 Mode of Action.
AN3 associates with SWI/SNF chromatin remodeling complexes formed around a central ATPase, BRM or SYD, including SWP73A or SWP73B, SWI3C and/or SWI3D, and ARP4 and ARP7. The presence of BSH remains to be elucidated, and other putative subunits are depicted as light-gray circles. AN3 binds GRFs or possibly other, yet to be identified, transcription factors to recruit chromatin remodeling activity to induce or repress expression of downstream target genes.
Ribosome-Related Processes Downstream of AN3
Our findings show that enhanced AN3 activity increases the cell division rate and the duration of the cell division phase, which is consistent with the lower maximum proliferation rate and the early loss of mitotic activity in the an3 mutant (Ferjani et al., 2007; Lee et al., 2009; Horiguchi et al., 2011). In order to maintain high cell proliferation rates, massive protein synthesis by ribosomes is required, and a large portion of carbon and energy is recruited for protein synthesis and ribosome biogenesis during plant growth (Piques et al., 2009). The upregulated genes following AN3 activation were enriched for genes involved in the synthesis of ribosomes and located in the nucleolus where ribosome biogenesis and assembly starts, indicating that AN3 might contribute to the stimulation of ribosome biogenesis. In concert, ribosomal and ribosome-related proteins have been described to regulate leaf development in conjunction with AN3. The combination of mutations in an3 and oli2, the latter most likely responsible for rRNA processing, synergistically reduces leaf cell number (Fujikura et al., 2009). Expression of OLI2 and another closely related OLI2-like gene (At4g26600) was induced in AN3-GR leaves after DEX treatment, and the OLI2 locus was identified by AN3-HBH TChAP-seq, providing evidence for the molecular basis of the synergism between AN3 and OLI2. Previous studies also showed in an3 mutants a downregulation of genes encoding the histone deacetylases HDT1 and HDT2 (Horiguchi et al., 2011), and, consistently, expression of HDT1 and HDT2 was induced in AN3-GR leaves. Moreover, HDT1 was shown to be involved in histone deacetylation of rRNA genes (Lawrence et al., 2004), underlining the importance of chromatin modifications and ribosome function for AN3-stimulated leaf growth.
A Model for AN3/GRF Action
By qRT-PCR, we showed that GRF3, GRF5, and GRF6 were significantly induced 2 h after AN3 activation, which makes them putative immediate downstream targets of AN3. AN3 lacks DNA binding capacity but has been demonstrated by Y2H to interact with GRF1, GRF2, GRF4, GRF5, and GRF9 (Kim and Kende, 2004; Horiguchi et al., 2005). In addition, we confirmed the interaction of AN3 with GRF5 by Co-IP. Furthermore, GRF5 and GRF6 were downregulated in an3 mutant rosettes and AN3 was shown to be present at the GRF5, GRF6, and AN3 promoters. This leads to the hypothesis that AN3/GRF complexes themselves activate GRF, as well as AN3 transcription, as is reported for many other transcription factor/coactivator complexes. The downregulation of AN3, GRF5, and GRF6 by overexpression of miR396 strengthens this hypothesis, since GRF5 and GRF6 do not contain a miR396-target site, in contrast with the other GRFs (Liu et al., 2009; Rodriguez et al., 2010; Wang et al., 2011). More specifically, AN3/GRF complexes most likely activate transcription of only GRF3, GRF5, and GRF6 in proliferating leaf cells, since no other GRFs were differentially expressed. This supports the likelihood that besides overlapping functions, GRFs have unique specialized functions that are needed for normal leaf development, corroborated by the decrease in leaf size of single grf mutants (Horiguchi et al., 2005; Kim and Lee, 2006).
Our results show that AN3 also binds the genomic regions of CRF2, HEC1, COL5, and ARR4, and whereas CRF2 is transiently induced at 1 h and HEC1 is induced after 2 h of DEX treatment, differences in COL5 and ARR4 expression levels only become apparent from 4 h onwards. Likely, CRF2 and HEC1 are primary targets of AN3. Because HEC1, ARR4, and HB33 were also differentially expressed in 35S:GRF5 shoots, we propose a model where a complex of AN3/GRF5 regulates transcription of a subset of target genes (HEC1, ARR4, and possibly HB33), while AN3 binds to other co-occurring GRFs, likely, GRF3 and GRF6, to modulate the expression of additional targets like CRF2 and COL5. Alternatively, AN3 can also regulate the expression of the target genes independently of the GRFs by associating with other transcription factors (Figure 8).
SWI/SNF Chromatin Remodeling Complexes Associated with AN3
TAP/MS with AN3, SWI3C, SWP73B, ARP7, and ARP4 as baits resulted in the identification of SWI/SNF complexes from Arabidopsis cell cultures and seedlings. It revealed the co-occurrence of multiple complexes composed of different subunits, of which homologs define mammalian SWI/SNF complexes, underlining their evolutionary conservation (Jerzmanowski, 2007; Hargreaves and Crabtree, 2011). Mutually exclusive ATPases (BRM and SYD) and SWP73 proteins (SWP73A and B) were copurified, indicating that, as in mammals, combinatorial assembly might contribute to increase gene regulation, generating greater functional diversity (Wilson and Roberts, 2011).
Predicted from sequence similarity before (Kim and Kende, 2004; Horiguchi et al., 2005), association of AN3 with SWI/SNF chromatin remodelers is now experimentally confirmed, both in cell cultures and seedlings. Either SYD or BRM constitute the central ATPase subunit, and based on the described interaction of the human homolog SYT with BRM and BRG1 (Nagai et al., 2001; Perani et al., 2003), AN3 most likely binds BRM or SYD directly. Moreover, if stoichiometry is conserved among plants and mammals, the AN3-containing complexes harbor SWP73A or SWP73B, ARP4 and ARP7, two SWI3 proteins, SWI3C and/or SWI3D, and a BSH protein (Figure 8). Previous studies based on Y2H and in vitro pull-down experiments identified pairwise interactions between SYD, BRM, BSH, and SWI3 proteins. It was hypothesized that, to recruit BSH, SWI/SNF complexes around BRM or SYD have to include SWI3A or SWI3B, while SWI3D can only be recruited by SWI3B (Sarnowski et al., 2002, 2005; Farrona et al., 2004; Hurtado et al., 2006; Bezhani et al., 2007). However, TAP-tagged SWI3C only pulled down SWI3C, and AN3 TAP copurified SWI3D as well, but never SWI3A or SWI3B, nor BSH. Judging from the frequent identification of all SWI3 proteins and BSH with SWP73B TAP, our results suggest that SWI3C may be the sole SWI3-type protein in a subset of SWI/SNF complexes or can co-occur with SWI3D, while BSH can be absent. However, as BSH is the only Arabidopsis homolog of the SNF5 core subunit, it likely makes integral part of the complexes. Its absence in most TAP experiments could be due to, for instance, interference of the TAP tag with stable BSH binding to the complex and concomitant loss of BSH during purification. Conversely, the previously used experimental systems in vitro or in yeast may not have allowed for identification of all interactions, demonstrating the need for complementary techniques in planta.
The copurification of multiple BRD proteins confirms the previously postulated hypothesis that plant proteins with one bromodomain associate and act as functional homologs of animal polybromo proteins (Jerzmanowski, 2007). Furthermore, the co-occurrence of BRD proteins and LFR, which are putative pBAF-like signature proteins, and the absence of BAF-defining proteins with an ARID domain, while several such proteins are present in the Arabidopsis genome (Jerzmanowski, 2007), suggest a subdivision in the class of plant SWI/SNF complexes similar to animals. The complexes associating with AN3 resemble human pBAF. Interestingly, BRD2 was found to be misregulated in brm and syd mutants (Bezhani et al., 2007), strengthening the notion that the expression of genes encoding associated proteins, like AN3, could be regulated by the SWI/SNF complex itself.
SWI/SNF Complexes Regulate Expression of AN3 and Its Downstream Target Genes
Several lines of evidence support that, as a transcriptional coactivator, AN3 modulates transcription by means of interaction with SWI/SNF complexes. First, the promoters of AN3 and direct target transcription factors are also shown to be physically bound by SWP73B and/or BRM, two SWI/SNF complex members purified by AN3. As such, the presence of both BRM and SWP73B is shown at the HEC1 promoter, while SWP73B occupies AN3, GRF3, GRF5, CRF2, COL5, and ARR4 loci. Second, AN3 is essential for the recruitment of SWP73B to the promoter regions of GRF3, GRF5, COL5, and ARR4. Third, functional BRM is shown to be necessary for the correct expression of CRF2, COL5, and HB33. Moreover, AN3 and GRF5 expression are also dependent on BRM, corroborating that AN3/GRFs likely regulate their own transcription by recruiting SWI/SNF complexes. Fourth, the effect of the an3 mutation is reduced in the absence of brm1, suggesting that AN3 associates with SWI/SNF complexes around BRM to perform part of its functions during vegetative leaf formation. AN3-TAP also identified SYD, which was shown to function only partially redundant with BRM in Arabidopsis seedlings (Bezhani et al., 2007). Therefore, the additional decrease in cell number in the an3 brm1 leaves compared with brm1 leaves likely results from the association of AN3 with SYD-containing chromatin remodeling complexes that substitute for BRM activity. Fourth, similar to overexpression of AN3 and GRF5 (Horiguchi et al., 2005; Gonzalez et al., 2010), overexpression of SWI3C results in increased leaf growth due to enhanced cell division, although additional experiments are needed to prove the involvement of AN3 and its target genes.
In addition, at the genome-wide level, AN3 binding sites are distributed with a higher than random frequency in intergenic regions and UTRs, with a maximum number of sites located between −100 and 0 bp from the start codon. Compared with the binding profiles of Arabidopsis transcription factors determined by ChIP-seq and ChIP-ChIP (Oh et al., 2009; Ouyang et al., 2011), a much higher percentage of AN3 binding sites is located in the coding regions and the UTRs, at the cost of intergenic and intron localizations. However, this is consistent with the peak distributions obtained with ChIP-seq of metazoan homologs of BRM, SWI3, and BSH proteins. ChIP-seq of mouse BRG1 and BAF155, for example, results in the presence of peaks in the gene body, besides the peak enrichment over the transcription start site (Ho et al., 2009). Similarly, the regions bound by human BRG1, BAF155, BAF177, and Ini1 are enriched for 5′ gene ends and RNA Polymerase II and III binding sites, which also target the coding sequence (Euskirchen et al., 2011). Moreover, the preference for nucleosome positioning in the exons over the introns (Andersson et al., 2009; Tilgner et al., 2009) might result in a reduced need for SWI/SNF remodeling activity in the introns, possibly explaining the depletion of AN3 binding sites in the introns relative to the exons.
A Function for CRF2, ARR4, COL5, HEC1, and HB33 Downstream of AN3
The identification of CRF2, which is induced by cytokinins (Rashotte et al., 2006), as a putative direct AN3 target gene hints at a role for the cytokinin response pathway downstream of SWI/SNF-AN3. At the same time, repression of ARR4 by AN3 could be a general mechanism to reduce the negative feedback inhibition on B-type ARRs, thereby reinforcing cytokinin signaling, known to stimulate leaf cell proliferation (Werner and Schmülling, 2009; Holst et al., 2011). Likewise, BRM was recently demonstrated to directly affect transcription of ARR16 together with the TCP4 transcription factor. In this case, however, ARR16 was induced to repress cytokinin responses promoting cell differentiation during leaf development (Efroni et al., 2013). Of note, ARR4 expression was not reduced in brm mutants, suggesting different roles for AN3 and BRM in regulation of this cytokinin response regulator. Together, our data imply that SWI/SNF complexes balance proliferation with differentiation, as previously reported in metazoans (Hargreaves and Crabtree, 2011).
A function in the regulation of leaf growth for two other direct AN3 targets, HEC1 and the putative transcription factor COL5, has not been described thus far. However, HEC1 and COL5 were shown to affect female reproductive tract development and the transition to flowering, respectively (Gremski et al., 2007; Hassidim et al., 2009; Crawford and Yanofsky, 2011). In addition, COL5 contains the G-box–derived motif, tgaCACGTGgca, which was found to be significantly enriched near the summits of the AN3 TChAP-seq peaks. The core G-box sequence CACGTG was shown to be involved in gene regulation in response to light and daylength (Oh et al., 2009; Spensley et al., 2009), conditions also influencing flowering. In fact, flowering time and flower development are affected in an3 and double/triple gif mutants (Lee et al., 2009), and BRM also has been demonstrated to regulate flowering (Farrona et al., 2004, 2007, 2011; Wu et al., 2012), together suggesting a putative function for AN3 during flowering and flower development by the regulation of HEC1 and COL5 transcription through SWI/SNF activity. Interestingly, COL5 and HEC1 were shown by Y2H to interact with BRM and SWI3C (Efroni et al., 2013). Extended research on HEC1 and COL5 loss- and gain-of-function mutants is required to shed light on their exact role during AN3-stimulated vegetative leaf development.
Overexpression of HB33 on the other hand was shown to enhance leaf growth (Hong et al., 2011), which is consistent with a positive role for HB33 downstream in the AN3/GRF5 signaling cascade. Although enhanced HB33 expression was associated with AN3 activation, the gene was not repressed by brm mutation as expected, but induced. The reason for this is currently unknown, but might involve feedback mechanisms and the activity of other transcriptional regulators.
In conclusion, AN3 associates with pBAF-type SWI/SNF complexes around BRM or SYD on the one hand, while interacting with DNA binding transcription factors on the other hand, thereby likely recruiting the ATPase activity to specific genomic regions necessary for efficient target gene regulation. As such, AN3 activates a broad spectrum of downstream responses to regulate the transition from leaf cell proliferation to cell expansion.
METHODS
Cloning, Construction of Transgenic Plants, and Plant Materials
DNA of Arabidopsis thaliana ecotype Col-0 was used to amplify all coding regions. The 35S:AN3-GR construct was made based on the pBI-ΔGR vector (Lloyd et al., 1994), from which the GR domain was amplified. Through Multisite Gateway cloning (Invitrogen), the 35S:AN3-GR construct was introduced into pK7m34GW and pH7m34GW (Karimi et al., 2007a, 2007b) and transformed by floral dip with Agrobacterium tumefaciens strain C58C1 (pMP90).
For overexpression, SWI3C cDNA was introduced into pK7WG2 containing the 35S promoter, and for ChIP-qPCR, AN3 was introduced into pK7FWG2 generating 35S:AN3-GFP (Karimi et al., 2007b). For Co-IP and TChAP, 35S:AN3-HBH, 35S:GRF5-HA, and 35S:GFP-HA constructs were obtained by Multisite Gateway cloning in the destination vector pK7GW43D (Karimi et al., 2007a).
GRF5-overexpressing plants and an3-4 mutants were kindly provided by Hirokazu Tsukaya and Gorou Horiguchi (Horiguchi et al., 2005). brm1 and brm3 mutants were obtained from the Salk collection (http://signal.salk.edu/) and described before: brm1 (SALK_030046) (Hurtado et al., 2006; Kwon et al., 2006) and brm3 (SALK_088462) (Farrona et al., 2007).
Growth Conditions and Growth Measurements
Plants were grown in vitro in sterile plates containing half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with 1% Suc at 21°C under a 16-h-day/8-h-night regime. Leaf areas were measured with ImageJ (http://rsb.info.nih.gov/ij/) after dissection of individual leaves. Rosette areas were calculated as the sum of the individual leaf areas.
For scanning electron microscopy, dental imprints were made from the abaxial epidermis, covered with nail polish that was carefully peeled off, and imaged by the Tabletop TM-1000 scanning electron microscope (Hitachi). Cell drawings were made with GIMP2 software (http://www.gimp.org/). Abaxial epidermis cells of SWI3C lines were drawn with a microscope (Leica) equipped with differential interference contrast optics and a drawing tube. Image analysis to obtain the cellular parameters was done as previously described (Andriankaja et al., 2012).
For transcript profiling and GUS staining, the plates containing medium were overlaid with nylon meshes (Prosep) of 20-µm pore size, after which seeds were sown. Seedlings were transferred to plates containing mock medium or medium supplemented with DEX (D4902-1G; Sigma-Aldrich) by lifting the nylon mesh with forceps.
GUS Staining and Analysis
Seedlings were GUS-stained for 8 h according to a protocol described by Andriankaja et al. (2012). Leaf length and GUS staining were measured with the ImageJ software (NIH). Leaves 1 and 2 were dissected and imaged in a horizontal position, the background was subtracted with a rolling ball radius adjustment of 50, and a defined area along the length of the leaf was selected with the rectangle tool. Next, the color intensity in the rectangle was measured with a one-pixel resolution along the horizontal axis using the plot profile function. The data points were then calibrated by adjusting the distance from pixels to millimeters, and the color intensities were normalized to an arbitrary scale of 0 to 1 with one indicating highest GUS expression.
RNA Extraction
Rosettes were harvested in liquid nitrogen. For microdissections of leaves 1 and 2, seedlings were harvested in RNAlater solution (AM7021; Ambion), incubated at 4°C for at least one night, microdissected on a cold plate under a stereomicroscope, and frozen in liquid nitrogen. RNA was extracted according to a combined protocol of TRI reagent RT (Molecular Research Center) and the RNeasy kit (Qiagen) with on-column DNase (Qiagen) digestion.
ATH1 Expression Profiling and Data Analysis
RNA of three biological replicates of AN3-GR and wild-type leaves 1 and 2 was hybridized to single Affymetrix ATH1 Genome arrays at the VIB Nucleomics Core (Leuven, Belgium). Data analysis was done as previously described (Gonzalez et al., 2010), and Ath1121501attairgcdf_14.0.0 was used as the chip definition file (http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/CDF_download.asp). Three differentially expressed genes, At1g35670, At4g14680, and At5g24240, were removed from Supplemental Data Set 1 because they were identified as false positives. Differentially expressed genes were investigated with PageMan (Usadel et al., 2006) and PLAZA (Van Bel et al., 2012) to calculate the functional overrepresentation of MapMan and Gene Ontology categories, respectively. Overlap with public microarray data was calculated with Fisher exact tests (fisher.test function in R) followed by Bonferroni P-value correction (Hochberg, 1988).
Expression Analysis
qRT-PCR was performed as previously described (Vercruyssen et al., 2011). In short, LightCycler 480 SYBR Green I Master (Roche) was used, and relative expression levels were determined by the method of Livak and Schmittgen (2001). Three technical replicates were performed for each reaction, and two or three biological replicates were done, as stated in the corresponding figure legends. Primer sequences are listed in Supplemental Table 2. ARR4 expression levels were measured using an nCounter Analysis System (NanoString Technologies) by the VIB Nucleomics Core as described (Geiss et al., 2008). The nCounter code set contained probe pairs for 108 Arabidopsis genes, including 10 housekeeping genes. The data were normalized by a two-step procedure with internal spike-in controls and the three most stable reference genes included in the probe set (CDKA;1, UBC, and CBP20).
Co-IP
Proteins were extracted from 2-d-old PSB-D cell suspension cultures and cotransformed with 35S:AN3-HBH and 35S:GRF5-HA, or 35S:AN3-HBH and 35S:GFP-HA. Thereto, cell cultures were ground in liquid nitrogen in homogenization buffer (25 mM Tris-Cl, pH 7.6, 75 mM NaCl, 15 mM MgCl2, 15 mM EGTA, 15 mM p-nitrophenylphosphate, 60 mM β-glycerophosphate, 1 mM DTT, 0.1% Nonidet P-40, 0.1 mM Na3VO4, 1 mM NaF, and protease inhibitor cocktail P9599 [Sigma-Aldrich]).
For immunoprecipitations, 500 μg of total protein in homogenization buffer was incubated at 4°C for 2 h with 50 μL of 50% (v/v) anti-HA affinity matrix (Roche). Beads were washed three times with 500 μL homogenization buffer and used for protein gel blot analysis.
Proteins were separated by 12% SDS-PAGE and blotted onto Immobilon-P membranes (Millipore). Filters were blocked in 3% (v/v) milk powder in 25 mM Tris-Cl, pH 8, 150 mM NaCl, and 0.05% Tween 20 for at least 1 h at room temperature and incubated overnight at 4°C with HA (1/1000) (Roche) or His (1/2000) (Qiagen) antibody in blocking buffer. Antigen-antibody complexes were detected with horseradish peroxidase–conjugated IgG diluted 1/10,000 (Amersham Biosciences) with a chemiluminescence system (Perkin-Elmer).
TChAP-Seq
TChAP was performed on 2-d-old exponentially growing 35S:AN3-HBH and wild-type PSB-D cell cultures. Maintenance and stable transformation of Arabidopsis cell suspension cultures was done according to Van Leene et al. (2007). In short, chromatin, isolated from formaldehyde-treated cell cultures was tandem affinity purified on Ni-NTA Superflow (Qiagen) and Streptavidin Sepharose (GE Healthcare), respectively, followed by ChIP protocol reverse cross-linking, deproteinization, and DNA purification. Full details on the TChAP protocol are provided in Supplemental Methods 1.
The 35S:AN3-HBH and wild-type PSB-D TChAP DNA libraries were prepared according to the protocol of Illumina and sequenced on a Genome II Analyzer (Illumina). The quality control of the sequencing data was performed by means of FastQC (v0.10.0; http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/). Overrepresented sequences were removed using fastx-clipper from the fastx toolkit (v0.0.13; http://hannonlab.cshl.edu/fastx_toolkit/). The reads were mapped to the unmasked TAIR10 reference genome of Arabidopsis (TAIR10_chr_all.fas; ftp.arabidopsis.org) using default settings (v0.5.9) (Li and Durbin, 2009). Reads that could not be assigned to a unique position in the genome were removed using SAMtools (v0.1.18) (Li et al., 2009) by setting the mapping quality threshold (-q) at 1. Redundant reads were removed, retaining only one read per start position, using Picard tools (v1.56; http://picard.sourceforge.net).
Peak calling was performed using MACS 1.4.2 (Zhang et al., 2008). The genome size (-g) was set at 1.0e8, and the MFOLD parameter (-m) was set at 5.40 to accommodate enough peaks for the peak model. Other parameters were set at their default values. Peak regions were annotated based on the location of their summits with respect to genes close by, as annotated in the TAIR10 release present in the PLAZA2.5 database (Van Bel et al., 2012). A peak was assigned to the closest gene, taking into account both up- and downstream regions of the peak. When a peak is located within the boundaries of a gene, it was assigned to this gene.
De novo motif finding was performed using peak motifs (Thomas-Chollier et al., 2012). The complete peak regions were submitted to the algorithm (default settings). The P value for motif enrichment in the peak set compared with the genomic background was calculated empirically. All motifs from peak motifs were mapped in 100 random sets of peaks of the same size and length distribution with matrix scan (Thomas-Chollier et al., 2011), using the same parameters as used in peak motifs. For a set of redundant motifs, one representative was chosen based on the lowest P value.
TAP–Liquid Chromatography–Tandem MS Analysis
Cloning of tag-fused transgenes and transformation of Arabidopsis cell suspension cultures were performed as previously described (Van Leene et al., 2007). TAP of protein complexes was done using the GS tag (Bürckstümmer et al., 2006), followed by protein precipitation and separation according to Van Leene et al. (2008). For the proteolysis and peptide isolation, acquisition of mass spectra by a 4800 MALDI TOF/TOF proteomics analyzer (AB SCIEX), and MS-based protein homology identification, we refer to Van Leene et al. (2010). The in planta TAP was analyzed on an LTQ Orbitrap Velos. Experimental background proteins were subtracted based on ∼40 TAP experiments on wild-type cultures and cultures expressing TAP-tagged mock proteins GUS, RFP, and GFP (Van Leene et al., 2010). Full details on the liquid chromatography–tandem MS analysis are provided in Supplemental Methods 2, and protein identification details are provided in Supplemental Data Sets 3 and 4.
ChIP
AN3 and SWP73B ChIP assays were performed on in vitro–grown seedlings using anti-GFP (Clontech) and anti-IgG (Millipore) antibodies, modified from Gendrel et al. (2005). Briefly, after plant material fixation in 1% (v/v) formaldehyde, tissues were homogenized, nuclei isolated, and lysed. Cross-linked chromatin was sonicated using a water bath Bioruptor UCD-200 (Diagenode) (15 s on/15 s off pulses; 15 times). The complexes were immunoprecipitated with 1 µg antibody, overnight at 4°C with gentle shaking, and incubated for 1 h at 4°C with 50 μL of Dynabeads Protein A (Invitrogen). Immunoprecipitated DNA was then recovered using the IPure kit (Diagenode) and analyzed by qPCR. An aliquot of untreated sonicated chromatin was processed in parallel to use as the total input DNA control. Seedlings transformed with HA-tagged BRM (BRM:BRM-HA) (Han et al., 2012) were cross-linked according to a method described previously (Winter et al., 2011). Chromatin was immunoprecipitated with 20 μL of anti-HA antibody (Roche) and quantified by comparing the threshold cycle values between ChIP DNA and a dilution series of input DNA with qPCR. The percentage of input values of the ChIP DNA was further normalized over the value obtained for the retrotransposon TA3 (Johnson et al., 2002). Primer sequences are listed in Supplemental Table 2.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the accession numbers listed in Tables 1 and 2. The microarray data have been submitted to the Gene Expression Omnibus database (accession number GSE42875), and the TChAP-seq data sets have been submitted to the National Center for Biotechnology Information Short Read Archive sequence database (Project ID PRJNA183696).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Overrepresentation Analysis of Differentially Expressed Genes after AN3 Induction.
Supplemental Figure 2. Comparison of the Differentially Expressed Genes after AN3 Induction with Publicly Available Microarray Data Sets.
Supplemental Figure 3. Expression Profiles of Selected Genes during Leaf 3 Development from Proliferation to Expansion.
Supplemental Figure 4. Transcription Factors Not Differentially Expressed 1, 2, 4, or 6 h after AN3 Induction.
Supplemental Figure 5. Coimmunoprecipitation of AN3 and GRF5.
Supplemental Figure 6. AN3 and GRF Expression in an3 Plants.
Supplemental Figure 7. ARR4 Expression in an3 and 35S:GRF5 Leaves.
Supplemental Figure 8. Distribution, Lengths, and Motif Analysis of AN3 Binding Sites Determined by TChAP-Sequencing.
Supplemental Figure 9. TChAP-Sequencing Results for AN3, GRF, HB33, and ARR4 Loci.
Supplemental Figure 10. BRM Binds the HEC1 Promoter.
Supplemental Figure 11. Cellular Analysis of an3 brm1 Leaves at 14 DAS.
Supplemental Figure 12. 35S:SWI3C Lines.
Supplemental Table 1. Phenotype of the F3 Progeny of Different Self-Pollinated Parent Plants Carrying the an3 and/or brm1 Allele(s).
Supplemental Table 2. qPCR and ChIP Primer Sequences.
Supplemental Methods 1. Tandem Chromatin Affinity Purification.
Supplemental Methods 2. LC-MS/MS Analysis.
Supplemental Data Set 1. Differentially Expressed Genes after Induction of AN3 Activity.
Supplemental Data Set 2. AN3-HBH TChAP-Sequencing results.
Supplemental Data Set 3. Protein Identification Details Obtained with the 4800 MALDI TOF/TOF Proteomics Analyzer after TAP from Arabidopsis Cell Cultures.
Supplemental Data Set 4. Protein Identification Details Obtained with the LTQ Orbitrap Velos after TAP from Arabidopsis Seedlings.
Supplementary Material
Acknowledgments
We thank all colleagues of the Systems Biology of Yield group for fruitful discussions and Annick Bleys for help in preparing the article. This work was supported by Ghent University (“Bijzonder Onderzoeksfonds Methusalem” Project BOF08/01M00408 and Multidisciplinary Research Partnership “Biotechnology for a Sustainable Economy” Project 01MRB510W), the Belgian Science Policy Office (IUAP VI/33), and the European Union FP6 (“AGRON-OMICS” Grant LSHG-CT-2006-037704). K.V. is supported by Ghent University (Multidisciplinary Research Partnership “Bioinformatics: from nucleotides to networks”). D.W. and S.-K.H. are supported by a National Institutes of Health grant (R01 GM64650-01). R.A. was funded by a National Science Centre grant (2011/01/D/NZ1/01614). A.V. and K.S.H. are supported by the Agency for Innovation by Science and Technology in Flanders. T.A., S.D.B., and J.V.L. are postdoctoral fellows of the Research Foundation-Flanders. S.D. is supported by the “Special Research Fund (BOF)” of Ghent University.
AUTHOR CONTRIBUTIONS
L.V., A.V., N.G., K.V., G.D.J., and D.I. designed the research. L.V., A.V., N.G., D.E., M.A., L.D.M, M.V., and K.M. performed the research. L.V., A.V., N.G., K.S.H., D.E., R.A., J.V.L., S.D.B., T.A., F.C., S.D., K.G., A.J., and K.V. analyzed data. T.J., M.B., S.-K.H., and D.W. provided the ChIP-qPCR data. L.V., N.G., and D.I. wrote the article.
Glossary
- DEX
dexamethasone
- Col-0
Columbia-0
- DAS
days after stratification
- qRT-PCR
quantitative RT-PCR
- Y2H
yeast two-hybrid
- Co-IP
coimmunoprecipitation
- TChAP
tandem chromatin affinity purification
- ChIP
chromatin immunoprecipitation
- TChAP-seq
TChAP followed by sequencing
- MACS
model-based analysis of ChIP-Seq
- UTR
untranslated region
- qPCR
quantitative PCR
- TAP
tandem affinity purification
- MS
mass spectrometry
Footnotes
Online version contains Web-only data.
References
- Abeel T., Van Parys T., Saeys Y., Galagan J., Van de Peer Y. (2012). GenomeView: A next-generation genome browser. Nucleic Acids Res. 40: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R., Enroth S., Rada-Iglesias A., Wadelius C., Komorowski J. (2009). Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 19: 1732–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriankaja M., Dhondt S., De Bodt S., Vanhaeren H., Coppens F., De Milde L., Mühlenbock P., Skirycz A., Gonzalez N., Beemster G.T.S., Inzé D. (2012). Exit from proliferation during leaf development in Arabidopsis thaliana: A not-so-gradual process. Dev. Cell 22: 64–78. [DOI] [PubMed] [Google Scholar]
- Archacki R., et al. (2013). BRAHMA ATPase of the SWI/SNF chromatin remodeling complex acts as a positive regulator of gibberellin-mediated responses in Arabidopsis. PLoS ONE 8: e58588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archacki R., Sarnowski T.J., Halibart-Puzio J., Brzeska K., Buszewicz D., Prymakowska-Bosak M., Koncz C., Jerzmanowski A. (2009). Genetic analysis of functional redundancy of BRM ATPase and ATSWI3C subunits of Arabidopsis SWI/SNF chromatin remodelling complexes. Planta 229: 1281–1292. [DOI] [PubMed] [Google Scholar]
- Beemster G.T.S., De Veylder L., Vercruysse S., West G., Rombaut D., Van Hummelen P., Galichet A., Gruissem W., Inzé D., Vuylsteke M. (2005). Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol. 138: 734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezhani S., Winter C., Hershman S., Wagner J.D., Kennedy J.F., Kwon C.S., Pfluger J., Su Y.H., Wagner D. (2007). Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell 19: 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeski J., Podstolski W., Olczak K., Jerzmanowski A. (1999). Identification and analysis of the Arabidopsis thaliana BSH gene, a member of the SNF5 gene family. Nucleic Acids Res. 27: 2393–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürckstümmer T., Bennett K.L., Preradovic A., Schütze G., Hantschel O., Superti-Furga G., Bauch A. (2006). An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods 3: 1013–1019. [DOI] [PubMed] [Google Scholar]
- Clapier C.R., Cairns B.R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78: 273–304. [DOI] [PubMed] [Google Scholar]
- Crane Y.M., Gelvin S.B. (2007). RNAi-mediated gene silencing reveals involvement of Arabidopsis chromatin-related genes in Agrobacterium-mediated root transformation. Proc. Natl. Acad. Sci. USA 104: 15156–15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford B.C.W., Yanofsky M.F. (2011). HALF FILLED promotes reproductive tract development and fertilization efficiency in Arabidopsis thaliana. Development 138: 2999–3009. [DOI] [PubMed] [Google Scholar]
- Donnelly P.M., Bonetta D., Tsukaya H., Dengler R.E., Dengler N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215: 407–419. [DOI] [PubMed] [Google Scholar]
- Efroni I., Han S.K., Kim H.J., Wu M.F., Steiner E., Birnbaum K.D., Hong J.C., Eshed Y., Wagner D. (2013). Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 24: 438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloy N.B., de Freitas Lima M., Van Damme D., Vanhaeren H., Gonzalez N., De Milde L., Hemerly A.S., Beemster G.T.S., Inzé D., Ferreira P.C.G. (2011). The APC/C subunit 10 plays an essential role in cell proliferation during leaf development. Plant J. 68: 351–363. [DOI] [PubMed] [Google Scholar]
- Euskirchen G.M., Auerbach R.K., Davidov E., Gianoulis T.A., Zhong G., Rozowsky J., Bhardwaj N., Gerstein M.B., Snyder M. (2011). Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 7: e1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona S., Hurtado L., Reyes J.C. (2007). A nucleosome interaction module is required for normal function of Arabidopsis thaliana BRAHMA. J. Mol. Biol. 373: 240–250. [DOI] [PubMed] [Google Scholar]
- Farrona S., Hurtado L., Bowman J.L., Reyes J.C. (2004). The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131: 4965–4975. [DOI] [PubMed] [Google Scholar]
- Farrona S., Hurtado L., March-Díaz R., Schmitz R.J., Florencio F.J., Turck F., Amasino R.M., Reyes J.C. (2011). Brahma is required for proper expression of the floral repressor FLC in Arabidopsis. PLoS ONE 6: e17997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferjani A., Horiguchi G., Yano S., Tsukaya H. (2007). Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol. 144: 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura U., Horiguchi G., Ponce M.R., Micol J.L., Tsukaya H. (2009). Coordination of cell proliferation and cell expansion mediated by ribosome-related processes in the leaves of Arabidopsis thaliana. Plant J. 59: 499–508. [DOI] [PubMed] [Google Scholar]
- Geiss G.K., et al. (2008). Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 26: 317–325. [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Martienssen R., Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218. [DOI] [PubMed] [Google Scholar]
- Gonzalez N., et al. (2010). Increased leaf size: Different means to an end. Plant Physiol. 153: 1261–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremski K., Ditta G., Yanofsky M.F. (2007). The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 134: 3593–3601. [DOI] [PubMed] [Google Scholar]
- Han S.K., Sang Y., Rodrigues A., Wu M.F., Rodriguez P.L., Wagner D. (2012). The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 24: 4892–4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves D.C., Crabtree G.R. (2011). ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res. 21: 396–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassidim M., Harir Y., Yakir E., Kron I., Green R.M. (2009). Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta 230: 481–491. [DOI] [PubMed] [Google Scholar]
- Ho L.N., Jothi R., Ronan J.L., Cui K.R., Zhao K.J., Crabtree G.R. (2009). An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. USA 106: 5187–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y. (1988). A sharper Bonferroni procedure for multiple tests of significance. Biometrika 75: 800–802. [Google Scholar]
- Holst K., Schmülling T., Werner T. (2011). Enhanced cytokinin degradation in leaf primordia of transgenic Arabidopsis plants reduces leaf size and shoot organ primordia formation. J. Plant Physiol. 168: 1328–1334. [DOI] [PubMed] [Google Scholar]
- Hong S.-Y., Kim O.-K., Kim S.-G., Yang M.-S., Park C.-M. (2011). Nuclear import and DNA binding of the ZHD5 transcription factor is modulated by a competitive peptide inhibitor in Arabidopsis. J. Biol. Chem. 286: 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G., Kim G.-T., Tsukaya H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43: 68–78. [DOI] [PubMed] [Google Scholar]
- Horiguchi G., Nakayama H., Ishikawa N., Kubo M., Demura T., Fukuda H., Tsukaya H. (2011). ANGUSTIFOLIA3 plays roles in adaxial/abaxial patterning and growth in leaf morphogenesis. Plant Cell Physiol. 52: 112–124. [DOI] [PubMed] [Google Scholar]
- Hurtado L., Farrona S., Reyes J.C. (2006). The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol. Biol. 62: 291–304. [DOI] [PubMed] [Google Scholar]
- Jerzmanowski A. (2007). SWI/SNF chromatin remodeling and linker histones in plants. Biochim. Biophys. Acta 1769: 330–345. [DOI] [PubMed] [Google Scholar]
- Johnson L.M., Cao X.F., Jacobsen S.E. (2002). Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12: 1360–1367. [DOI] [PubMed] [Google Scholar]
- Kandasamy M.K., Deal R.B., McKinney E.C., Meagher R.B. (2005b). Silencing the nuclear actin-related protein AtARP4 in Arabidopsis has multiple effects on plant development, including early flowering and delayed floral senescence. Plant J. 41: 845–858. [DOI] [PubMed] [Google Scholar]
- Kandasamy M.K., McKinney E.C., Deal R.B., Meagher R.B. (2005a). Arabidopsis ARP7 is an essential actin-related protein required for normal embryogenesis, plant architecture, and floral organ abscission. Plant Physiol. 138: 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanei M., Horiguchi G., Tsukaya H. (2012). Stable establishment of cotyledon identity during embryogenesis in Arabidopsis by ANGUSTIFOLIA3 and HANABA TARANU. Development 139: 2436–2446. [DOI] [PubMed] [Google Scholar]
- Karimi M., Bleys A., Vanderhaeghen R., Hilson P. (2007b). Building blocks for plant gene assembly. Plant Physiol. 145: 1183–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Depicker A., Hilson P. (2007a). Recombinational cloning with plant gateway vectors. Plant Physiol. 145: 1144–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawade K., Horiguchi G., Usami T., Hirai M.Y., Tsukaya H. (2013). ANGUSTIFOLIA3 signaling coordinates proliferation between clonally distinct cells in leaves. Curr. Biol. 23: 788–792. [DOI] [PubMed] [Google Scholar]
- Kazama T., Ichihashi Y., Murata S., Tsukaya H. (2010). The mechanism of cell cycle arrest front progression explained by a KLUH/CYP78A5-dependent mobile growth factor in developing leaves of Arabidopsis thaliana. Plant Cell Physiol. 51: 1046–1054. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Choi D., Kende H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36: 94–104. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Kende H. (2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 13374–13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Lee B.H. (2006). GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J. Plant Biol. 49: 463–468. [Google Scholar]
- Kwon C.S., Chen C., Wagner D. (2005). WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 19: 992–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C.S., Hibara K., Pfluger J., Bezhani S., Metha H., Aida M., Tasaka M., Wagner D. (2006). A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development 133: 3223–3230. [DOI] [PubMed] [Google Scholar]
- Kwon C.S., Wagner D. (2007). Unwinding chromatin for development and growth: A few genes at a time. Trends Genet. 23: 403–412. [DOI] [PubMed] [Google Scholar]
- Lawrence R.J., Earley K., Pontes O., Silva M., Chen Z.J., Neves N., Viegas W., Pikaard C.S. (2004). A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 13: 599–609. [DOI] [PubMed] [Google Scholar]
- Lee B.H., Ko J.-H., Lee S., Lee Y., Pak J.-H., Kim J.H. (2009). The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 151: 655–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., , and 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Song Y., Chen Z., Yu D. (2009). Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plant. 136: 223–236. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lloyd A.M., Schena M., Walbot V., Davis R.W. (1994). Epidermal cell fate determination in Arabidopsis: Patterns defined by a steroid-inducible regulator. Science 266: 436–439. [DOI] [PubMed] [Google Scholar]
- Meagher R.B., Deal R.B., Kandasamy M.K., McKinney E.C. (2005). Nuclear actin-related proteins as epigenetic regulators of development. Plant Physiol. 139: 1576–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Nagai M., Tanaka S., Tsuda M., Endo S., Kato H., Sonobe H., Minami A., Hiraga H., Nishihara H., Sawa H., Nagashima K. (2001). Analysis of transforming activity of human synovial sarcoma-associated chimeric protein SYT-SSX1 bound to chromatin remodeling factor hBRM/hSNF2 alpha. Proc. Natl. Acad. Sci. USA 98: 3843–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., Choi G. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X.H., et al. (2011). Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23: 2514–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perani M., Ingram C.J.E., Cooper C.S., Garrett M.D., Goodwin G.H. (2003). Conserved SNH domain of the proto-oncoprotein SYT interacts with components of the human chromatin remodelling complexes, while the QPGY repeat domain forms homo-oligomers. Oncogene 22: 8156–8167. [DOI] [PubMed] [Google Scholar]
- Piques M., Schulze W.X., Höhne M., Usadel B., Gibon Y., Rohwer J., Stitt M. (2009). Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol. Syst. Biol. 5: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte A.M., Mason M.G., Hutchison C.E., Ferreira F.J., Schaller G.E., Kieber J.J. (2006). A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. USA 103: 11081–11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R.E., Mecchia M.A., Debernardi J.M., Schommer C., Weigel D., Palatnik J.F. (2010). Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137: 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y., Silva-Ortega C.O., Wu S., Yamaguchi N., Wu M.F., Pfluger J., Gillmor C.S., Gallagher K.L., Wagner D. (2012). Mutations in two non-canonical Arabidopsis SWI2/SNF2 chromatin remodeling ATPases cause embryogenesis and stem cell maintenance defects. Plant J. 72: 1000–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowska E.A., et al. (2013). DELLA-interacting SWI3C core subunit of switch/sucrose nonfermenting chromatin remodeling complex modulates gibberellin responses and hormonal cross talk in Arabidopsis. Plant Physiol. 163: 305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowski T.J., Ríos G., Jásik J., Swiezewski S., Kaczanowski S., Li Y., Kwiatkowska A., Pawlikowska K., Koźbiał M., Koźbiał P., Koncz C., Jerzmanowski A. (2005). SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during Arabidopsis development. Plant Cell 17: 2454–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowski T.J., Swiezewski S., Pawlikowska K., Kaczanowski S., Jerzmanowski A. (2002). AtSWI3B, an Arabidopsis homolog of SWI3, a core subunit of yeast Swi/Snf chromatin remodeling complex, interacts with FCA, a regulator of flowering time. Nucleic Acids Res. 30: 3412–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spensley M., Kim J.Y., Picot E., Reid J., Ott S., Helliwell C., Carré I.A. (2009). Evolutionarily conserved regulatory motifs in the promoter of the Arabidopsis clock gene LATE ELONGATED HYPOCOTYL. Plant Cell 21: 2606–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Kwon C.S., Bezhani S., Huvermann B., Chen C., Peragine A., Kennedy J.F., Wagner D. (2006). The N-terminal ATPase AT-hook-containing region of the Arabidopsis chromatin-remodeling protein SPLAYED is sufficient for biological activity. Plant J. 46: 685–699. [DOI] [PubMed] [Google Scholar]
- Tang X.R., Hou A.F., Babu M., Nguyen V., Hurtado L., Lu Q., Reyes J.C., Wang A.M., Keller W.A., Harada J.J., Tsang E.W.T., Cui Y.H. (2008). The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol. 147: 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M., Defrance M., Medina-Rivera A., Sand O., Herrmann C., Thieffry D., van Helden J. (2011). RSAT 2011: Regulatory sequence analysis tools. Nucleic Acids Res. 39: W86–W91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M., Herrmann C., Defrance M., Sand O., Thieffry D., van Helden J. (2012). RSAT peak-motifs: motif analysis in full-size ChIP-seq datasets. Nucleic Acids Res. 40: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H., Nikolaou C., Althammer S., Sammeth M., Beato M., Valcárcel J., Guigó R. (2009). Nucleosome positioning as a determinant of exon recognition. Nat. Struct. Mol. Biol. 16: 996–1001. [DOI] [PubMed] [Google Scholar]
- Usadel B., Nagel A., Steinhauser D., Gibon Y., Bläsing O.E., Redestig H., Sreenivasulu N., Krall L., Hannah M.A., Poree F., Fernie A.R., Stitt M. (2006). PageMan: An interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bel M., Proost S., Wischnitzki E., Movahedi S., Scheerlinck C., Van de Peer Y., Vandepoele K. (2012). Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 158: 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., et al. (2010). Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 6: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., et al. (2007). A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol. Cell. Proteomics 6: 1226–1238. [DOI] [PubMed] [Google Scholar]
- Van Leene J., Witters E., Inzé D., De Jaeger G. (2008). Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci. 13: 517–520. [DOI] [PubMed] [Google Scholar]
- Vercruyssen L., Gonzalez N., Werner T., Schmülling T., Inzé D. (2011). Combining enhanced root and shoot growth reveals cross talk between pathways that control plant organ size in Arabidopsis. Plant Physiol. 155: 1339–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D., Meyerowitz E.M. (2002). SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr. Biol. 12: 85–94. [DOI] [PubMed] [Google Scholar]
- Walley J.W., Rowe H.C., Xiao Y.M., Chehab E.W., Kliebenstein D.J., Wagner D., Dehesh K. (2008). The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog. 4: e1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Gu X., Xu D., Wang W., Wang H., Zeng M., Chang Z., Huang H., Cui X. (2011). miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J. Exp. Bot. 62: 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.J., Yuan T.T., Yuan C., Niu Y.Q., Sun D.Y., Cui S.J. (2009). LFR, which encodes a novel nuclear-localized Armadillo-repeat protein, affects multiple developmental processes in the aerial organs in Arabidopsis. Plant Mol. Biol. 69: 121–131. [DOI] [PubMed] [Google Scholar]
- Werner T., Schmülling T. (2009). Cytokinin action in plant development. Curr. Opin. Plant Biol. 12: 527–538. [DOI] [PubMed] [Google Scholar]
- Wilson B.G., Roberts C.W.M. (2011). SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 11: 481–492. [DOI] [PubMed] [Google Scholar]
- Winter C.M., et al. (2011). LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev. Cell 20: 430–443. [DOI] [PubMed] [Google Scholar]
- Wu M.F., Sang Y., Bezhani S., Yamaguchi N., Han S.K., Li Z.T., Su Y.H., Slewinski T.L., Wagner D. (2012). SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc. Natl. Acad. Sci. USA 109: 3576–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y.Y., Yoshitsugu T., Sakurai T., Seki M., Shinozaki K., Obokata J. (2009). Heterogeneity of Arabidopsis core promoters revealed by high-density TSS analysis. Plant J. 60: 350–362. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.