This work examined the function of the GARP family transcription factor KANADI1 (KAN1), finding that KAN1 binds a GNATA(A/T) cis-motif to repress transcription of target genes involved in auxin biosynthesis, auxin transport, and the auxin response. KAN1 affects auxin responses in opposition to the HD-ZIPIII transcription factor REVOLUTA.
Abstract
The formation of leaves and other lateral organs in plants depends on the proper specification of adaxial-abaxial (upper-lower) polarity. KANADI1 (KAN1), a member of the GARP family of transcription factors, is a key regulator of abaxial identity, leaf growth, and meristem formation in Arabidopsis thaliana. Here, we demonstrate that the Myb-like domain in KAN1 binds the 6-bp motif GNATA(A/T) and that this motif alone is sufficient to squelch transcription of a linked reporter in vivo. In addition, we report that KAN1 acts as a transcriptional repressor. Among its targets are genes involved in auxin biosynthesis, auxin transport, and auxin response. Furthermore, we find that the adaxializing HD-ZIPIII transcription factor REVOLUTA has opposing effects on multiple components of the auxin pathway. We hypothesize that HD-ZIPIII and KANADI transcription factors pattern auxin accumulation and responsiveness in the embryo. Specifically, we propose the opposing actions of KANADI and HD-ZIPIII factors on cotyledon formation (KANADI represses and HD-ZIPIII promotes cotyledon formation) occur through their opposing actions on genes acting at multiple steps in the auxin pathway.
INTRODUCTION
Leaves begin as bumps, or leaf primordia, that grow out from the shoot apical meristem (SAM). The region of the primordium closest to the center of the meristem, the adaxial domain, develops into the upper half of the leaf. The region of the primordium furthest from the center of the meristem, the abaxial domain, develops into the lower half of the leaf. With regard to the geometry of the leaf, then, the terms adaxial and upper are synonymous and the terms abaxial and lower are synonymous.
Subdivision of the primordium into ad- and abaxial domains is important not only because the upper and lower halves of leaves have specialized roles in photosynthesis but also because the establishment of ad- and abaxial domains generate the ad/abaxial boundary. This boundary is both necessary and sufficient to define the site of outgrowth of the leaf blade (Waites and Hudson, 1995; Evans, 2007).
Genetic studies in Arabidopsis thaliana indicate that the KANADI genes (KAN1 to KAN4) have overlapping functions in the promotion of abaxial fate in lateral organs (Eshed et al., 1999, 2001, 2004; Kerstetter et al., 2001; Emery et al., 2003; Pekker et al., 2005; McAbee et al., 2006; Izhaki and Bowman, 2007). Mutations in any single KAN gene cause relatively mild defects in leaf development (Kerstetter et al., 2001). However, plants lacking several of these genes exhibit conspicuous defects in embryos, lateral organs, and vascular patterning that can be attributed to the loss of abaxial, or peripheral, identity. For example, kan1 kan2 double mutants have reduced blade expansion and form ectopic leaf-like outgrowths on the abaxial blade surface (Eshed et al., 2001), whereas kan1 kan2 kan3 triple mutants have almost no blade expansion and produce nearly cylindrical, adaxialized leaves with radialized stem vasculature (Eshed et al., 2004). Mutations in KAN4/ABERRANT TESTA SHAPE cause defects in the polarity and growth of ovule integuments but in combination with kan1 and kan2 mutations cause significant changes in auxin distribution and major defects in embryo patterning (Leon-Kloosterziel et al., 1994; McAbee et al., 2006; Izhaki and Bowman, 2007).
Ectopic expression of individual KAN genes causes profound abaxialization of lateral organs and disrupted vascular patterning (Kerstetter et al., 2001; Eshed et al., 2001, 2004; Emery et al., 2003). The complementary loss- and gain-of-function KAN phenotypes indicate that abaxial fate depends on the level and pattern of KAN gene expression during organogenesis.
KAN genes encode members of the GARP family of MYB-like transcription factors expressed in the abaxial domains of lateral organs and in the abaxial/peripheral domains of the embryo (Eshed et al., 2001, 2004; Kerstetter et al., 2001; Hosoda et al., 2002; Izhaki and Bowman, 2007). In the single case where a target has been identified, KAN acts to repress the transcription of the target ASYMMETRIC2 locus (Wu et al., 2008). Consistent with the action of KAN as a repressor of transcription, Causier et al. (2012) found that KAN1 protein physically interacts in yeast with the TOPLESS corepressor protein.
The KAN genes act in opposition to the HD-ZIPIII loci: The former act to promote abaxial (lower) fates in organs, while the latter act to promote adaxial (upper) fates (McConnell and Barton, 1998; McConnell et al., 2001; Emery et al., 2003). HD-ZIPIII genes encode homeodomain-leucine zipper containing transcription factors expressed primarily in the adaxial domains of organs, throughout the SAM and in the developing vasculature (Baima et al., 1995, 2001; McConnell et al., 2001; Kang et al., 2002; Prigge et al., 2005).
In addition to having opposing roles in polarization of the leaf along the ad/abaxial dimension, HD-ZIPIII genes have opposing roles to KAN in the promotion of growth of new SAMs: HD-ZIPIII proteins promote the formation of new SAMs, while KAN activity represses their formation (Talbert et al., 1995; McConnell and Barton, 1998; Kerstetter et al., 2001).
The one case in which HD-ZIPIII and KAN proteins act in the same direction, to promote growth, is in the establishment of the leaf blade. Specification of the adaxial leaf domain and abaxial leaf domains generates an ad/abaxial boundary at which the leaf blade is formed. Once leaves are formed, HD-ZIPIII and KAN proteins are required to coordinate growth of the upper and lower sides of the leaf: In the absence of HD-ZIPIII function, leaves curl down, while in the absence of KAN function, leaves curl up.
Several observations have linked the REVOLUTA (REV)/KAN ad/abaxial regulators to the control of patterned signaling by the plant hormone auxin. Izhaki and Bowman (2007) observed ectopic auxin accumulation at the site of ectopic outgrowths from the hypocotyl of kan1 kan2 kan4 triple mutant embryos. Since regions of high auxin accumulation are hypothesized to be responsible for, and the site of, the formation of new organs (Reinhardt et al., 2000, 2003; Heisler et al., 2005), Izhaki and Bowman proposed that KAN proteins control the correct spatial accumulation and sensing of auxin. However, the mechanism through which this occurs was not explored.
In keeping with a role for KAN in auxin sensing, Kelley et al. (2012) observed that ARF3 and KAN proteins physically interact. ARF3, also called ETTIN, is a member of the AUXIN RESPONSE FACTOR family of auxin-stimulated transcription factors and is required for ectopic KAN to fully abaxialize lateral organs (Pekker et al., 2005). Ectopic KAN causes the formation of radialized leaves with abaxial characteristics around their circumference. When ARF3 is removed by mutation, these KAN overexpressing organs are able to produce blade. Further evidence of a role for KAN in the regulation of auxin action was found by Brandt et al. (2012), who identified the auxin biosynthetic genes TAA1 and YUC5 as targets of HD-ZIPIII (activation) and KAN regulation (repression). Finally, the PIN1 and PIN2 auxin transporters are expressed at lower levels, and tip-to-base transport of auxin is reduced in loss-of-function (abaxialized) mutants of the HD-ZIPIII REV gene (Zhong and Ye, 2001). REV is referred to in this work as IFL, showing that HD-ZIPIII function is required for polar auxin transport in the shoot.
To better understand the mechanism through which KAN regulates polarity and growth in the plant, we define an optimal KAN binding site and identify a set of genes targeted by KAN in planta that act to control auxin biosynthesis, transport, and signaling. These results show that KAN and HD-ZIPIII factors act in opposition at several steps in the auxin pathway.
RESULTS
Identification of an in Vitro Binding Site for KAN
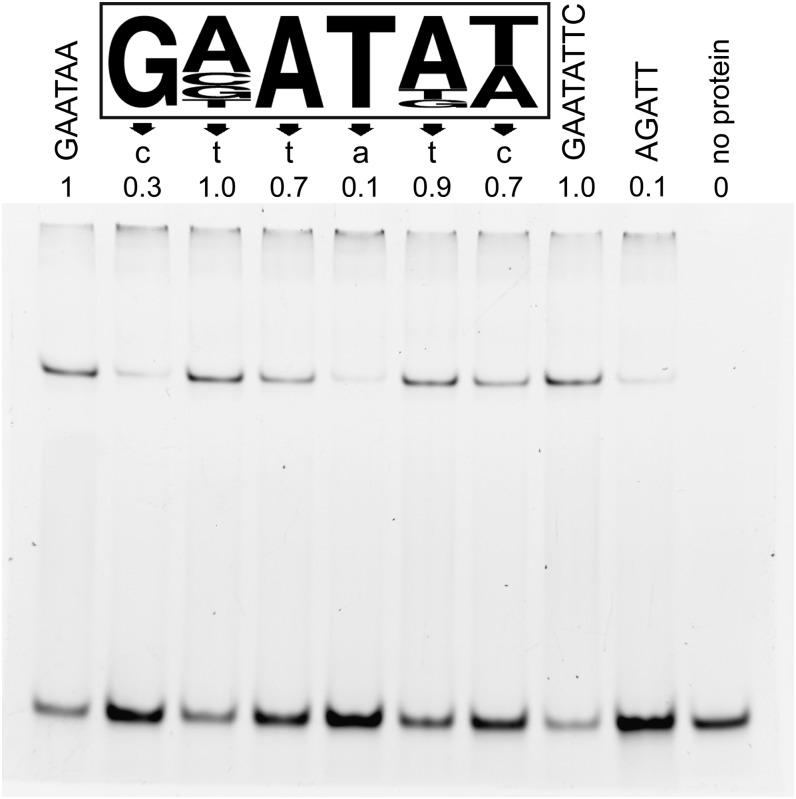
A binding site for KAN1 was identified upstream of ASYMMETRIC LEAVES2 (AS2) based on a mutation that causes ectopic expression of AS2 (Wu et al., 2008). To define more generally the binding site for KAN, we performed oligonucleotide selection experiments using purified KAN1 protein. The full-length KAN1 protein proved toxic when expressed in Escherichia coli (data not shown), so we instead generated a recombinant protein consisting of the predicted KAN1 DNA binding domain (KAN1bd) fused to glutathione S-transferase (GST). KAN1bd-GST was affinity purified and used for electrophoretic mobility shift assay (EMSA)–based PCR-assisted oligonucleotide selection. This experiment produced 50 nonredundant oligonucleotide sequences that contained one or more instances of the partly degenerate 6-bp motif GNATA(T/A), which we termed the KANADI box (KBX) (Figure 1; Supplemental Figure 1). To clarify the contributions of individual bases of KBX to KAN1 binding, we performed EMSA with double-stranded oligonucleotides bearing point mutations throughout this sequence. Nucleotides at the first, third, fourth, and sixth positions were critical for high affinity binding in vitro (Figure 1). KAN1bd-GST bound equally well to the 6-bp consensus sequence GAATAA and to an 8-bp palindrome, GAATATTC, that appeared in 6 of the 50 selected sequences (Figure 1; Supplemental Figure 1). By contrast, the protein showed little affinity for the consensus binding site (AGATT) of the GARP protein ARR10 (Hosoda et al., 2002) (Figure 1). These results demonstrate that the KAN1 GARP domain selectively binds DNA and define a novel binding site for this member of the GARP family of transcription factors.
Figure 1.
EMSA Reveals KAN1 DNA Binding Characteristics in Vitro.
The in vitro KAN1 DNA binding site (KBX; boxed) was identified using affinity-purified KAN1db-GST protein in EMSA-based oligonucleotide selection (Supplemental Figure 1). The height of each nucleotide letter is proportional to its representation. Effects of mutating individual sites within the consensus DNA binding site are shown immediately below each position in the consensus KBX site. The mean fraction of bound DNA in three independent replicates was calculated relative to the consensus (GAATAA, lane 1), which was arbitrarily set to 1.0. EMSA of KAN1db-GST bound to a perfect palindrome of KBX (GAATATT, lane 8) was similar to that of that of a single site. The KAN1db-GST showed little affinity for the consensus binding site for the GARP protein ARR10 (AGATT, lane 9) (Hosoda et al., 2002).
In order to determine if the KBX sequence is sufficient to mediate KAN1-regulated expression in planta, synthetic promoter constructs were generated by fusing repeats of KBX (or mKBX in which the fourth T was converted into A, a critical base pair change that disrupts KAN1 binding in vitro) upstream of a minimal transcription start site and the reporter gene β-glucuronidase (GUS). Furthermore, the KBX (or mKBX) repeats were inserted downstream of 6× UAS (upstream activation sequence) repeats that are able to activate GUS expression only in the presence of the yeast-derived GAL4-VP16 transcriptional activator. This system allowed the effects of KBX to be examined both in the absence and presence of GAL4-VP16 to reveal whether KBX confers any tissue-specific gene activation or repression respectively.
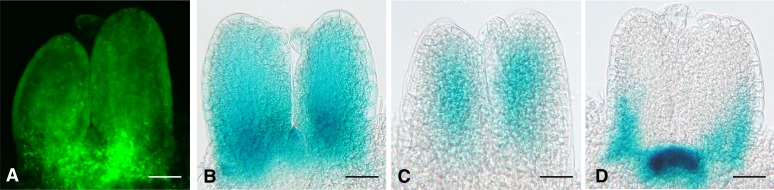
In transgenic Columbia (Col) plants, UAS:GUS, UAS-KBX:GUS, and UAS-mKBX:GUS constructs did not produce detectable GUS activity (data not shown). However, when these transgenic lines were crossed to E100, a GAL4-GFP (for green fluorescent protein) enhancer trap line that displays strong GAL4-GFP expression in rapidly dividing tissues, such as SAM and leaf primordia (Figure 2), GUS expression was observed in distinct patterns. E100>>UAS-mKBX:GUS lines showed GUS activity throughout the SAM and young leaf primordia in a pattern that mirrored GFP in E100 (Figure 2). By contrast, E100>>UAS-KBX:GUS plants lacked GUS expression in the SAM and the abaxial side of young leaf primordia (Figure 2), a pattern complementary to where pKAN1:GUS is expressed in plants (Figure 2). This result shows that KBX repeats are sufficient to direct tissue-specific repression in the context of an otherwise constitutive promoter, which strongly supports the biological significance of this motif in vivo.
Figure 2.
KBX Confers Tissue-Specific Repression of the Downstream Gene in Planta.
(A) GAL4 GFP enhancer trap line E100 exhibits GFP expression in the SAM and young leaf primordia.
(B) The GUS expression pattern in E100>>UAS-mKBX:GUS mirrors the GFP pattern in E100.
(C) In E100>>UAS-KBX:GUS, the GUS stain was only detected on the adaxial side of the leaf primordia and absent in the SAM and on the abaxial side of leaves.
(D) KAN1:GUS is expressed on the abaxial side of leaf primordia and in the SAM, a pattern complementary to the E100>>UAS-KBX:GUS expression.
Bars = 20 μm.
Generation of an Inducible KAN Protein
Transgenic plants that constitutively express KAN1 usually fail to produce a SAM and arrest as seedlings (Eshed et al., 2001; Kerstetter et al., 2001). In order to characterize the effects of ectopic KAN1 expression later in shoot development, we produced an inducible form of this protein by fusing the regulatory domain of the rat glucocorticoid receptor (GR) to the C-terminal end of KAN1 (Wagner et al., 1999). Transgenic plants expressing KAN1-GR under the regulation of the cauliflower mosaic virus 35S promoter grew slightly slower than normal but were otherwise morphologically normal (Figure 3; Supplemental Figure 2). By contrast, 35S:KAN1-GR (KAN1-GR) seedlings grown on media containing 10 μM dexamethasone (DEX) were strikingly similar to 35S:KAN1 plants (Eshed et al., 2001; Kerstetter et al., 2001); in addition to having narrow cotyledons, the first true leaves of these plants emerged as small, radialized, peg-like structures, and no subsequent leaves were formed indicating arrest of SAM activity (Figure 3; Supplemental Figure 2). By contrast, soil-grown KAN1-GR plants treated with DEX every other day had relatively mild developmental defects: Petioles were shortened and leaves curled downward. Similar morphology is seen in as1 and as2 mutants (Serrano-Cartagena et al., 1999) (Figure 3; Supplemental Figure 2) consistent with the ability of KAN to repress as2 transcription (Wu et al., 2008).
Figure 3.
Posttranslational Activation of KAN1-GR Produces Defects in Leaf Polarity and Meristem Function.
Continuous exposure of KAN1-GR seedlings to 10 μM DEX (B) on media for 9 d led to loss of cotyledon blade expansion, formation of partially radialized leaf primordia, and inhibition of further shoot meristem activity consistent with strong KAN1 overexpression. Mock-treated KAN1-GR seedlings (A) resembled mock- or DEX- treated Col seedlings (Supplemental Figure 2). By contrast, soil-grown KAN1-GR seedlings exposed every other day to 10 μM DEX ([D] and [F]) displayed reduced petiole and blade expansion with strong epinasty leading to leaves with an asymmetric appearance ([G]; bottom) that was not evident in mock-treated plants ([C], [E], and [G]; top). Plants were photographed at 14 ([C] and [D]) and 29 d old ([E] to [G]).
Identification of KAN1 Target Genes
To identify KAN1 target (KANT) genes that regulate leaf polarity, we performed a microarray analysis of gene expression in mock- and DEX-treated KAN1-GR seedlings using the Affymetrix ATH1 GeneChip and RNA isolated from 9-d-old seedlings following 4-h mock or DEX treatments. After removing loci whose expression was affected by DEX in wild-type controls, we identified 222 loci that displayed at least a 1.8-fold difference (P < 0.005; Supplemental Data Set 1) in mock- and DEX-treated KAN1-GR plants. Of these, 133 loci were downregulated and 89 were upregulated. The number of KBX sites in the promoters of these genes was compared with their frequency in all promoters. The sequence GNATA(A/T) or its complement occurs on average six times in the upstream 1000 bp of the 31,407 annotated Arabidopsis genes (TAIR 6.0) but appeared an average of seven times in the promoters of the 222 responsive loci (P < 0.005; Table 1). When up- and downregulated promoters were examined independently, it became apparent that the downregulated genes possessed, on average, eight KBX sequences (P < 0.005), whereas upregulated genes were not significantly different from the genome average (Table 1). This correlation indicates that KAN1-GR may function primarily as a transcriptional repressor.
Table 1. Enrichment of KBX Sites in KAN-Responsive Promoters.
| Loci (Promoters)a | GNATA(A/T) Sites | Mean (±sd) | P Valueb | |
|---|---|---|---|---|
| Nuclear genes | (31,128) | 180,827 | 5.8 ± 2.9 | |
| DEX-responsive genes | 222 (231) | 1,644 | 7.1 ± 3.1 | 9 × 10−12 |
| Repressed | 133 (137) | 1,059 | 7.7 ± 3.0 | 1 × 10−14 |
| Direct targets | 82 (84) | 646 | 7.7 ± 2.9 | 3 × 10−9 |
| Indirect targets | 38 (39) | 287 | 7.4 ± 3.0 | 5 × 10−4 |
| Induced | 89 (94) | 585 | 6.2 ± 3.0 | 0.087 |
| Direct targets | 22 (23) | 122 | 5.3 ± 2.4 | 0.206 |
| Indirect targets | 61 (64) | 421 | 6.6 ± 3.1 | 0.019 |
Promoters (upstream 1000 bp) of differentially expressed genes were analyzed for the occurrence of KAN1 binding sites using Promomer (http://bbc.botany.utoronto.ca/). Some array element loci recognize more than one expressed sequence. The number of promoters represented is indicated in parentheses.
Student’s t test with two-tailed distribution
To identify potential direct targets of KAN1-GR, microarray analyses were performed in the presence of cycloheximide (CHX), a potent inhibitor of protein synthesis. Genes directly regulated by KAN1-GR are expected to be insensitive to CHX because protein synthesis is not required for the effect of DEX on KAN1-GR activity (Pratt et al., 2004). CHX treatment had a dramatic effect on global gene expression; nearly one-third of the transcripts in wild-type seedlings were affected by a 4-h exposure to CHX (data not shown). Genes that showed a significant expression difference (P < 0.005) in DEX+CHX versus mock+CHX seedlings, which was in the same direction as in the DEX- versus mock-treated seedlings, were considered to be direct targets of KAN1-GR. Genes that were differentially expressed in DEX but not DEX+CHX-treated seedlings were considered to be indirect targets of KAN-GR. Using these parameters, a majority (61.7%, 82 of 133) of downregulated loci appeared to be direct targets of KAN-GR, whereas only a minority (24.7%, 22 of 89) of upregulated genes was in this category (Supplemental Data Set 1). This result suggests that activated KAN1-GR primarily functions as a repressor. The existence of genes upregulated by KAN1-GR in response to DEX in the presence of CHX may indicate that, in a minority of cases, KAN1-GR acts as a transcriptional activator, but it is also possible that KAN1-GR represses microRNA generating loci that target the apparently upregulated genes.
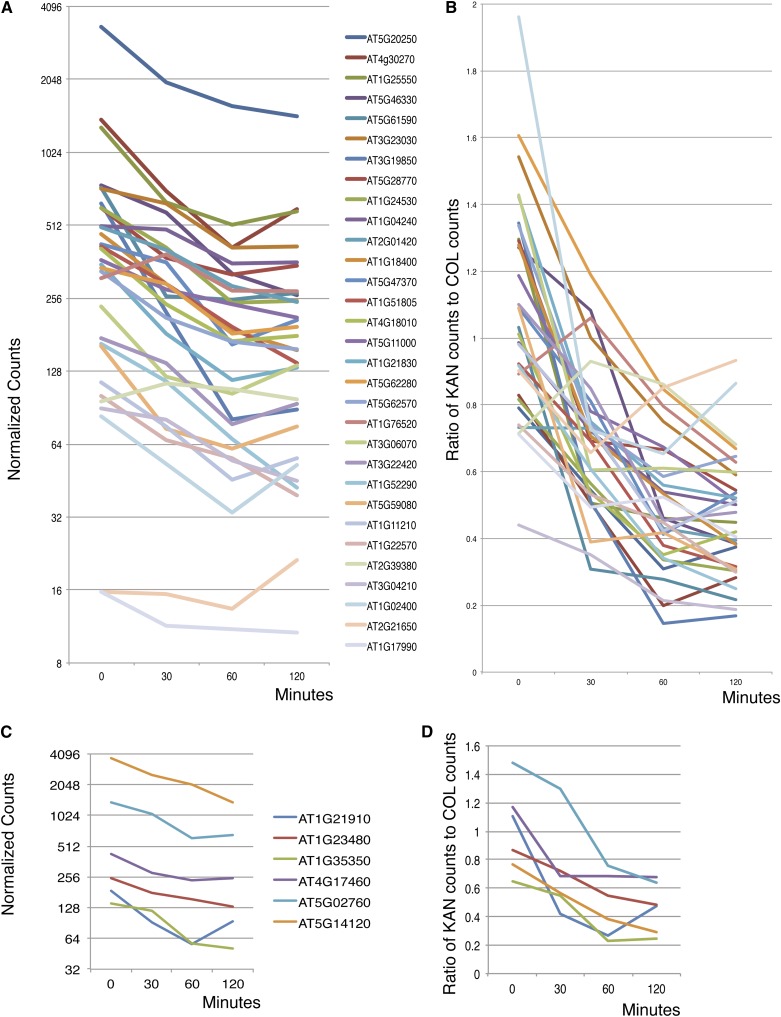
We performed RT-PCR with limiting number of amplification cycles to validate targets identified in the microarray experiment (Supplemental Figure 3). Of 38 genes tested, 32 showed evidence of downregulation by KAN1-GR in the RT-PCR assays (Supplemental Figure 3). We also mined data for these 38 genes from a parallel study done using RNA-seq as a tool to measure transcript levels. Using this technique, all but one (AT2G39380) of the 32 genes testing positive by RT-PCR showed downregulation by KAN1-GR (Figures 4A and 4B). Interestingly, examination of RNA-seq data on the six genes that failed the RT-PCR test reveals that expression of all six decreases following KAN1-GR activation by DEX (Figures 4C and 4D). Thus, RT-PCR, especially when limited to one time point, may not be as sensitive a technique in the detection of downregulated targets as RNA-seq on a full time course.
Figure 4.
Validation of Microarray/RT-PCR Identified KAN1 Target Genes by RNA-Seq.
(A) Transcripts that tested positive by RT-PCR test. Data plotted as number of normalized counts after DEX treatment.
(B) Transcripts that tested positive by RT-PCR test. Data plotted as ratio of number of counts in DEX treated KAN1-GR samples versus DEX-treated Col samples.
(C) Transcripts that tested negative by RT-PCR test. Data plotted as number of normalized counts after DEX treatment.
(D) Transcripts that tested negative by RT-PCR test. Data plotted as ratio of number of counts in DEX treated KAN1-GR samples versus DEX-treated Col samples. Minutes = minutes of DEX treatment.
The RNA-seq data in Figure 4 show that, as a group, KAN1-GR downregulated genes display similar expression profiles. Active KAN1-GR reduces transcript levels by 2- to 4-fold over the first 60 min, indicating a typical transcript half life of 30 to 45 min. Most transcripts have plateaued or begun to increase by the 120-min time point. This could be because early genes activated by KAN1-GR inhibit its activity. Alternatively, it could be because the activated KAN1-GR protein is destroyed or otherwise inactivated by this time.
Identification of Direct Target Genes
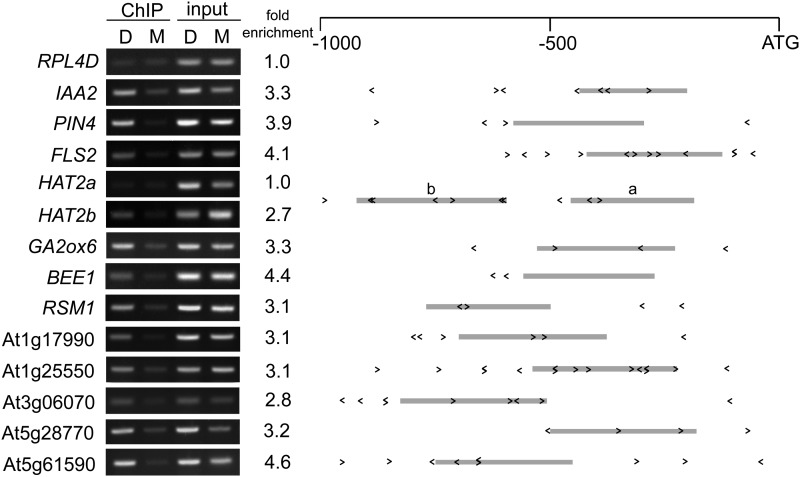
To test whether KAN1 binds directly to the promoters of these putative KAN target genes, we performed chromatin immunoprecipitation (ChIP) assays on KAN1-GR seedlings using an anti-GR antibody. Because DEX treatment promotes the translocation of GR fusion proteins to the nucleus (Pratt et al., 2004), chromatin fragments bound by KAN1-GR are expected to be enriched in DEX-treated relative to mock-treated samples. We were particularly intrigued by the fact that many putative KAN1 target genes are transcription factors or have been implicated in phytohormone signaling or biosynthesis; therefore, we chose to focus on these genes. Promoter fragments of 12 KAN1 target genes that we examined were enriched in DEX-treated KAN1-GR samples (Figure 5; Supplemental Table 1). Direct interaction between KAN1 and these promoter elements also revealed the in vivo function of KBX because the PCR-amplified portions representing these promoters always include or flank one or more KBXs. Our result also shows that the enrichment in KAN1-ChIP was dependent on the specific KBX fragment tested; for example, only one of two KBX-containing regions of the HAT2 promoter (HAT2b) appeared to be associated with KAN1-GR (Figure 5), which suggests that KBX alone is not sufficient for KAN1 binding. ChIP experiments performed with wild-type plants did not reveal detectable differences between mock- and DEX-treated samples, confirming that the DEX-dependent enrichment of these fragments in KAN1-GR plants depends on KAN1-GR (Supplemental Figure 4). We conclude that most of the genes identified as repressed by DEX both in the presence and absence of CHX in the microarray analysis are direct targets of KAN1-GR.
Figure 5.
ChIP Confirms DEX-Dependent Association of KAN-GR with the Promoters of KANT Genes.
ChIP was performed on 9-d-old transgenic Arabidopsis seedlings using antibodies specific for GR. Immunoprecipitated genomic DNA from mock (M) and DEX (D) treated KAN-GR and wild-type control seedlings (Supplemental Figure 4) was amplified with primers specific for the indicated promoters. Fold enrichment was calculated by normalizing PCR product intensities to a negative control, the RIBOSOMAL PROTEIN L4D (RPL4D) coding region, followed by calculating the ratio DEX IP/input to mock IP/input. The mean of at least two independent IP experiments (Supplemental Table 2) with technical replicates is reported as fold enrichment. Schematics of the gene promoters are shown with the positions and orientations of KBX sites indicated by < or > and the amplified region represented by a gray bar.
KAN and REV Oppositely Regulate Genes Involved in Auxin Biosynthesis, Transport, and Signaling
Among the genes identified above as directly repressed by KAN are FLS2 and PIN-FORMED4 (PIN4). PIN4 is an auxin efflux carrier (Friml et al., 2002), and FLS2 mediates flagellin-induced expression of miR393a, a microRNA that in turn targets the auxin receptor gene TIR1 (Navarro et al., 2006). This suggests that the mechanism through which KAN suppresses cotyledon formation is through transcriptional repression of genes involved in auxin transport and signaling.
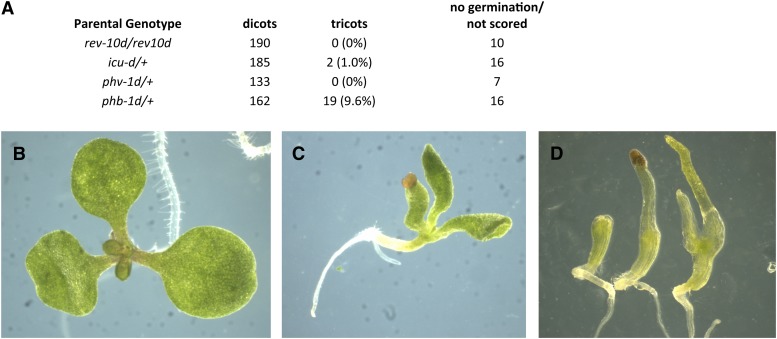
HD-ZIPIII genes have opposite roles to KAN genes. Instead of repressing cotyledon outgrowth, they promote cotyledon formation: Embryos triply mutant for the HD-ZIPIII genes PHABULOSA/ATHB-14, PHAVOLUTA/ATHB-9, and REV fail to form either one or both cotyledons (Figure 6; Emery et al., 2003; Prigge et al., 2005). Furthermore, we observed that overexpression of the PHABULOSA and INCURVATA4/CORONA/ATHB-15 HD-ZIPIII genes (due to mutations in the microRNA complementary sites) leads to extra cotyledon formation (Figure 6). Thus, increased HD-ZIPIII activity leads to extra cotyledon formation, while decreased activity leads to loss of cotyledon formation.
Figure 6.
Cotyledon Numbers Are Altered in HD-ZIPIII Gain- and Loss-of-Function Mutants.
(A) Frequency of tricots in self progeny of gain of function mutants of REV, PHABULOSA, PHAVOLUTA, and INCURVATA4.
(B) incurvata4-d mutant tricot with normal cotyledon blades.
(C) phabulosa-1d tricot with tube formed adaxialized cotyledons.
(D) ph phb rev triple mutants with no (left), one (middle), or two (right) radialized, abaxialized cotyledons.
To determine if additional auxin-related genes are regulated by these opposing factors, we surveyed genes involved in the auxin pathway for their response to induced REV (HD-ZIPIII) or KAN1 action. We first assayed genes for their regulation by GR-REV and KAN1-GR in a parallel microarray study (Reinhart et al., 2013). Supplemental Table 2 shows genes involved in auxin biosynthesis, transport, and signaling with associated P values for comparisons between GR-REV and wild-type Col, KAN1-GR and wild type Col, and GR-REV and KAN1-GR. Genes showing evidence for regulation in the microarray experiment were then surveyed for their regulation in an independent experiment we performed in which RNA-SEQ was used to measure transcript abundance instead of microarray (Table 2; Supplemental Figures 5 to 10).
Table 2. Regulation of Auxin Pathway Genes by REV and KAN.
| Gene Title | Transcript ID | p(TxG)GR-REV versus KAN1-GRa |
p(TxG) GR-REV versus Cola |
p(TxG)KAN1-GR versus Cola |
Regulation | |||
|---|---|---|---|---|---|---|---|---|
| MAb | SEQ | MAb | SEQ | MAb | SEQ | |||
| Auxin Biosynthetic Enzyme Genes | ||||||||
| YUCCA5 | AT5G43890 | 1.5E-02 | <0.0001 | 1.1E-02 | <0.0001 | 8.4E-01 | 1.8E-01 | REV - U |
| TAA1 | AT1G70560 | 5.5E-05 | <0.0001 | 6.3E-03 | 6.0E-04 | 7.5E-02 | 5.4E-01 | REV - U |
| AUX1 Family of Influx Transporters | ||||||||
| LAX1 | AT5G01240 | 2.9E-02 | 1.2E-02 | 5.4E-01 | 3.4E-01 | 1.4E-01 | 8.5E-01 | REV/KAN - U |
| LAX2 | AT2G21050 | 3.5E-02 | 2.0E-01 | 8.9E-01 | 4.7E-02 | 7.6E-01 | 7.0E-01 | REV - U |
| LAX3 | AT1G77690 | 2.0E-02 | 5.9E-01 | 3.1E-01 | 3.7E-02 | 9.9E-01 | 4.3E-02 | REV - U; KAN - U |
| PIN Family of Auxin Transport Facilitators | ||||||||
| PIN3 | AT1G70940 | 4.7E-02 | 6.3E-02 | 8.7E-01 | 8.4E-02 | 1.3E-01 | 2.3E-01 | – |
| PIN4 | AT2G01420 | 5.1E-04 | 4.4E-03 | 9.6E-01 | 8.4E-01 | 8.4E-04 | 2.1E-01 | KAN - D |
| PGP Family of Auxin Transport Facilitators | ||||||||
| PGP6 | AT2G39480 | 3.6E-02 | 9.1E-01 | 9.7E-01 | 2.7E-01 | 1.5E-01 | 5.2E-01 | – |
| PGP19 | AT3G28860 | 1.6E-02 | 3.5E-01 | 2.8E-03 | 6.9E-02 | 4.5E-01 | 4.7E-01 | – |
| PGP21 | AT3G62150 | 2.6E-01 | 2.8E-01 | 2.7E-02 | 1.1E-01 | 2.5E-02 | 2.7E-01 | – |
| NPH3-Like BTB-POZ Domain Proteins | ||||||||
| AT1G52770 | AT1G52770 | 8.2E-05 | 1.0E-04 | 1.3E-07 | <.0001 | 9.7E-01 | 8.6E-01 | REV - U |
| AT1G50280 | AT1G50280 | 7.8E-03 | 3.1E-02 | 5.7E-03 | 1.5E-01 | 4.6E-01 | 9.6E-01 | REV - U |
| RPT2 | AT2G30510 | 1.8E-03 | NA | 6.4E-01 | NA | 3.1E-02 | NA | – |
| AT3G08570 | AT3G08570 | 1.6E-01 | 3.4E-02 | 5.9E-01 | 4.8E-01 | 3.4E-02 | 2.8E-02 | KAN - U |
| AT3G19850 | AT3G19850 | 4.2E-05 | 1.0E-04 | 1.1E-01 | 8.4E-01 | 2.3E-07 | 1.2E-03 | KAN - D |
| AT3G15570 | AT3G15570 | 3.0E-05 | 4.2E-02 | 7.4E-01 | 5.4E-01 | 6.0E-06 | 5.2E-01 | KAN - D |
| ENP1/NPY1 | AT4G31820 | 2.5E-02 | 3.7E-01 | 4.2E-01 | 1.3E-02 | 5.2E-02 | 7.6E-01 | REV - U |
| NPY2 | AT2G14820 | NA | 6.8E-01 | NA | 3.2E-01 | NA | 6.5E-02 | – |
| NPY3 | AT5G67440 | 4.7E-02 | 1.1E-02 | 9.2E-01 | 8.0E-01 | 4.0E-03 | 3.7E-02 | KAN - D |
| NPY4 | At2G23050 | NA | 5.5E-01 | NA | 5.6E-01 | NA | 1.7E-01 | – |
| NPY5 | AT4G37590 | 1.1E-02 | 1.4E-03 | 8.8E-01 | 2.2E-01 | 5.5E-03 | 1.7E-01 | KAN - D |
| AT5G47800 | AT5G47800 | 1.7E-02 | 5.0E-04 | 9.2E-01 | 4.2E-03 | 2.4E-01 | 8.2E-02 | REV - U; KAN - D |
| AGC KINASE Encoding Genes | ||||||||
| WAG1 | AT1G53700 | 1.5E-03 | 9.7E-01 | 2.1E-05 | 6.0E-01 | 4.3E-01 | 6.8E-01 | REV - U |
| WAG2 | AT3G14370 | 8.8E-03 | 3.1E-01 | 1.6E-01 | 4.4E-01 | 1.9E-01 | 9.7E-01 | – |
| PHOT1 | AT3G45780 | 4.0E-03 | 8.5E-01 | 3.6E-02 | 6.5E-02 | 2.2E-01 | 7.8E-01 | – |
| PINOID | AT2G34650 | NA | 7.2E-01 | NA | 7.7E-01 | NA | 4.0E-01 | – |
| PINOID2 | AT2G26700 | NA | 2.0E-01 | NA | 8.6E-01 | NA | 9.8E-01 | – |
| IAA Protein Coding Genes | ||||||||
| SHY2/IAA3 | AT1G04240 | 3.6E-02 | 1.8E-01 | 8.8E-01 | 4.0E-01 | 2.1E-02 | 1.2E-01 | – |
| IAA18 | AT1G51950 | 5.2E-02 | 2.9E-03 | 8.0E-01 | 4.8E-01 | 9.9E-02 | 7.9E-03 | KAN - D |
| IAA13 | AT2G33310 | 1.1E-02 | 7.0E-02 | 5.1E-02 | 9.9E-02 | 2.9E-01 | 3.6E-01 | – |
| IAA2 | AT3G23030 | 1.9E-01 | 1.0E-04 | 8.4E-01 | 2.0E-01 | 3.7E-01 | 2.7E-03 | KAN - D |
| IAA11 | AT4G28640 | 2.2E-02 | 3.6E-02 | 1.0E-01 | 2.0E-01 | 3.4E-02 | 7.3E-01 | KAN - U |
| ARF Genes | ||||||||
| ARF10 | AT2G28350 | 6.1E-02 | 1.4E-01 | 7.5E-02 | 9.2E-01 | 1.1E-04 | 3.4E-01 | – |
| ETT/ARF3 | AT2G33860 | 3.1E-02 | 2.5E-01 | 3.5E-02 | 1.3E-02 | 4.5E-01 | 6.5E-01 | REV - U |
| Auxin Signaling | ||||||||
| FLS2 | AT5G46330 | 3.1E-04 | 4.3E-02 | 6.7E-01 | 1.4E-01 | 2.5E-06 | 2.0E-04 | KAN - D |
NA, not assayed; MA, measured by ATH1 microarray; Seq, measured by RNA-seq; D, downregulated; U, upregulated.
Values at P < 0.05 are in bold.
Data from Reinhart et al. (2013).
Comparing these analyses, and using the criterion that a gene had to show statistical significance in at least one comparison from each type of experiment (microarray and RNA-seq), we found evidence for regulation by KAN1-GR and/or GR-REV of two genes encoding auxin biosynthetic enzymes (YUCCA5 and TAA1), three genes encoding auxin influx transporters (LAX1, LAX2, and LAX3), one gene encoding a PIN family auxin efflux transporter (PIN4), nine genes encoding NPH-like BTP POZ domain proteins (At1g52770, At1g50280, At3g08570, At3g19850, At3g15570, ENP1/NPY1, NPY3, NPY5, and At5g47800), two genes encoding indole-3-acetic acid (IAA) family auxin signal transducers (IAA11 and IAA18), and one gene encoding an ARF family transcriptional regulator (ARF3) (Table 2, Figure 7; Supplemental Figures 5 to 8).
Figure 7.
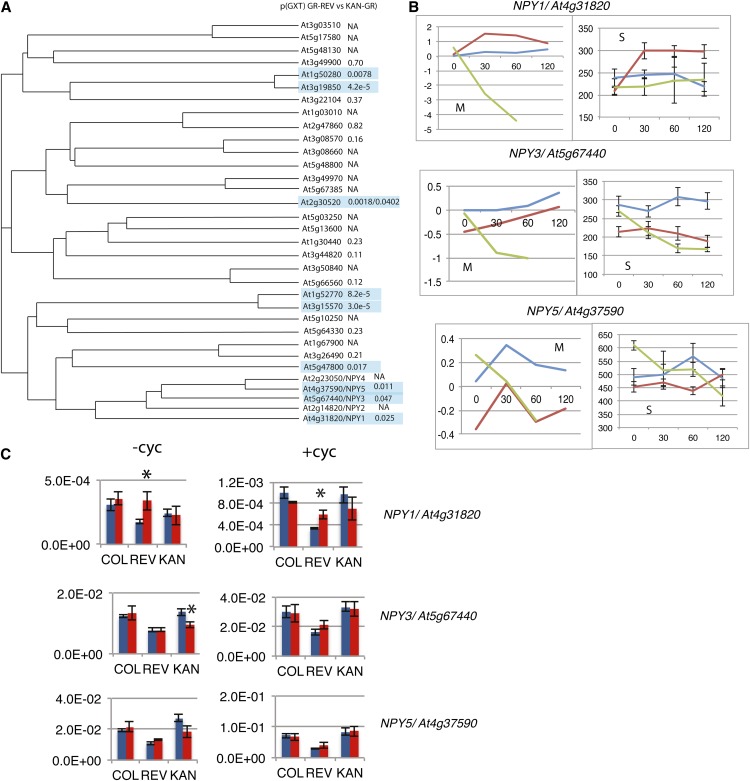
Differential Regulation of Members of the NPY/MEL Gene Family by REV and KAN.
(A) Phylogenetic tree of members of the NPH3-like family of genes. Values are probabilities for genotype by time of treatment interaction in a two-way ANOVA (microarray experiment) comparing GR- REV lines treated with DEX to KAN1-GR lines treated with DEX. NA, not assayed.
(B) Graphs of transcript levels for wild-type (blue), GR-REV (red), and KAN1-GR (green) lines treated with DEX. M, data from microarray experiment. y axis is normalized expression in log2 units. S, data from RNA-seq experiment. y axis is normalized counts. Error bars are se.
(C) qRT-PCR experiments on cDNAs made from DEX-treated seedlings for 1 h in the presence and absence of CHX. Three biological replicates were tested for each bar. (Three technical replicates were tested for each biological replicate.) Expression is relative to actin. Asterisk indicates significant difference relative to Col.
Most striking was the extensive regulation of members of the NPH3-like family of genes (Table 2; Supplemental Figure 6). Nine of 18 NPH3-like genes assayed showed evidence of differential regulation by REV relative to KAN in the microarray and RNA-seq experiments. The regulated NPH3 genes are distributed in small clusters throughout branches of the phylogenetic tree (Figure 7, Table 2). They are either upregulated by GR-REV or downregulated by KAN1-GR, with the exception of At3g08570, which is upregulated by KAN1-GR (Supplemental Figure 6).
Of particular interest are the NPY1/MAB4/ENP1, NPY2/MEL4, NPY3/MEL2, NPY4/MEL3, and NPY 5/MEL1 genes. Mutations in genes within this clade disrupt cotyledon formation (Furutani et al., 2007, 2011; Cheng et al., 2008). Moreover, this subclade of NPH3-like proteins, together with type 3 AGC protein kinases, has been implicated in the control of polar localization of auxin efflux carriers within the cell (Dhonukshe et al., 2010; Furutani et al., 2011), suggesting that these genes promote cotyledon formation by determining which face of the cell PIN proteins are directed and, therefore, the direction of auxin transport.
Three of the five NPY genes, NPY1, NPY3, and NPY5, showed statistically significant responses to GR-REV versus KAN1-GR (Table 2, Figure 7). Probes for NPY2 and NPY4 were not present on the microarray. However, NPY2 and NPY4 expression could be assayed by RNA-seq (Supplemental Figure 6) and by quantitative RT-PCR (qRT-PCR; data not shown), and in neither case were transcript levels significantly changed in response to GR-REV or KAN1-GR. It is notable that the three genes that show regulation, NPY1, NPY3, and NPY5, all show higher levels of expression (normalized counts are in the mid hundreds), while those that do not are expressed at roughly 10-fold lower levels (normalized counts in the mid tens; Supplemental Figure 6).
A third technique, qRT-PCR, on independent samples confirmed statistically significant upregulation of NPY1 by GR-REV in the presence and absence of CHX, indicating that NPY1 is likely a direct target of REV activation. NPY1 showed downregulation by KAN1-GR in the microarray experiment, but this was not repeated in either the RNA-SEQ or qRT-PCR experiments. NPY3 transcripts showed statistically significant downregulation in response to KAN1-GR in all three experiments: microarray, qRT-PCR, and RNA-seq (Table 2, Figure 7; Supplemental Figure 6), but NPY3 levels did not respond to KAN1-GR in the presence of CHX, indicating that KAN1-GR downregulation of NPY3 is likely an indirect effect. NPY5 transcript levels were decreased by KAN1-GR in both microarray and RNA-seq experiments but were unchanged in the qRT-PCR experiments (Table 2, Figure 7; Supplemental Figure 6). It is unclear whether this is due to variation between experiments or to the limited number of time points assayed in the qRT-PCR experiment. In summary, REV increases transcription, most likely by direct activation, of NPY1, while KAN decreases transcript levels, probably indirectly, of NPY3.
Among the genes encoding transcriptional regulators, IAA11 and IAA18 showed reproducible downregulation by KAN1-GR and ARF3/ETTIN showed reproducible upregulation by GR-REV. We also reexamined the expression of the Aux/IAA transcriptional regulator IAA2 by RT-PCR since this gene was identified as a potential KAN target in the original microarray experiment based on a 4-h DEX treatment (Supplemental Data Set 1) and since this gene showed high statistical significance for KAN1-GR downregulation in the RNA-seq experiment. We found that IAA2 was dramatically repressed by DEX in the presence of CHX, suggesting that it is a direct target of KAN-GR (Supplemental Figure 3). IAA2 was also positive in ChIP experiments (Figure 4). In order to determine if IAA2 is misregulated in kan1 mutants, we examined IAA2 expression by qRT-PCR and found that IAA2 was upregulated in kan1 and further upregulated in kan1 kan2 mutant seedlings (Supplemental Figure 8).
Similarly, we followed up on the WAG1 and WAG2 genes with qRT-PCR since the microarray and RNA-seq experiments yielded different results (Table 2; Supplemental Figures 9 and 10). qRT-PCR on an independent set of samples showed upregulation of WAG1 by GR-REV both in the presence and absence of CHX and downregulation of WAG2 by KAN1-GR but only in the absence of CHX. These experiments are consistent with REV acting as a direct regulator of WAG1 and KAN1 acting as an indirect regulator of WAG2. However, the very different patterns of WAG1 and WAG2 expression in the microarray and RNA-seq experiments (Supplemental Table 2 and Supplemental Figure 10) urge caution in drawing conclusions from these results.
DISCUSSION
KAN as a Repressor of Gene Function
Like other members of the GARP family of transcription factors, KAN proteins contain a single MYB-like DNA binding domain. KAN proteins have been placed into GARP subgroup 1 together with the cytokinin response ARR proteins and the mesophyll cell differentiation factor G2 based on their lack of a coiled coil domain (Hosoda et al., 2002). The lack of a coiled coil domain and the identification of the nonpalindromic binding site AGATT led Hosoda et al. (2002) to hypothesize that ARR10 binds to DNA as a monomer. This is in contrast with GARP subgroup 2 proteins, which contain a coiled coil domain and, in the case of PHR1, bind to a palindromic sequence (Rubio et al., 2001). Consistent with this, the KAN1 binding site we identified is short (6 bp) and nonpalindromic.
While both KAN1 and ARR proteins bind DNA as a monomer, in our experiments, KAN1 did not show affinity for the ARR binding site (AGATT) in vitro and instead bound to the sequence GNATA. Of the eight amino acids in the ARR recognition helix, only three are conserved in KAN1. Interestingly, the KAN1 binding site is identical to the half site of the palindromic sequence bound by the PHR protein involved in phosphate starvation (GNATATNC; Rubio et al., 2001). Comparison of the PHR and KAN1 binding sites reveals that both PHR and KAN1 have Lys-228 in common where ARR10 has Ala-228. In ARR10, Ala-228 contacts the first AT base pair in the AGATT binding site, making this residue a good candidate for altering specificity to a GC base pair at that position (GNATA).
The sequence we identified through in vitro studies is identical to the KAN1 binding site upstream of the AS2 locus (Wu et al., 2008). This binding site was identified via a dominant mutation, as2-5d, which causes ectopic expression of AS2 due to failure of KAN binding. The higher frequency of this binding site upstream of KAN1-regulated transcripts than the genome average provides additional support for the importance of this sequence.
The studies on the KAN1 binding site upstream of AS2 showed it was required for KAN1 repression. In this study, we found that the KAN1 binding site, when present in the context of a GAL4 driven reporter expressed in the SAM, is sufficient to confer reporter downregulation in the abaxial tissues expressing KAN1. However, because KAN1 does not bind to all KAN1 binding sites as determined by ChIP and the functional KAN1 binding controlling AS2 expression is just upstream of and adjacent to a second KAN1 binding site that appears to lack function (Wu et al., 2008), the context of KAN1 binding sites is important in determining whether they are functional.
All evidence to date points to KAN1 proteins as negative regulators of transcription; so far, all genes that behave like direct targets are downregulated in response to KAN1. KAN1 protein has recently been shown to interact with the TOPLESS corepressor (Causier et al., 2012). Thus, KAN1 may cause repression by interacting with TOPLESS, thereby recruiting chromatin repressive enzymes. The nature of these repressive enzymes is not yet known but may include histone deacetylases (Wang et al., 2013).
Patterning of Auxin Response by KAN and the Oppositely Acting HD-ZIPIII Transcription Factors
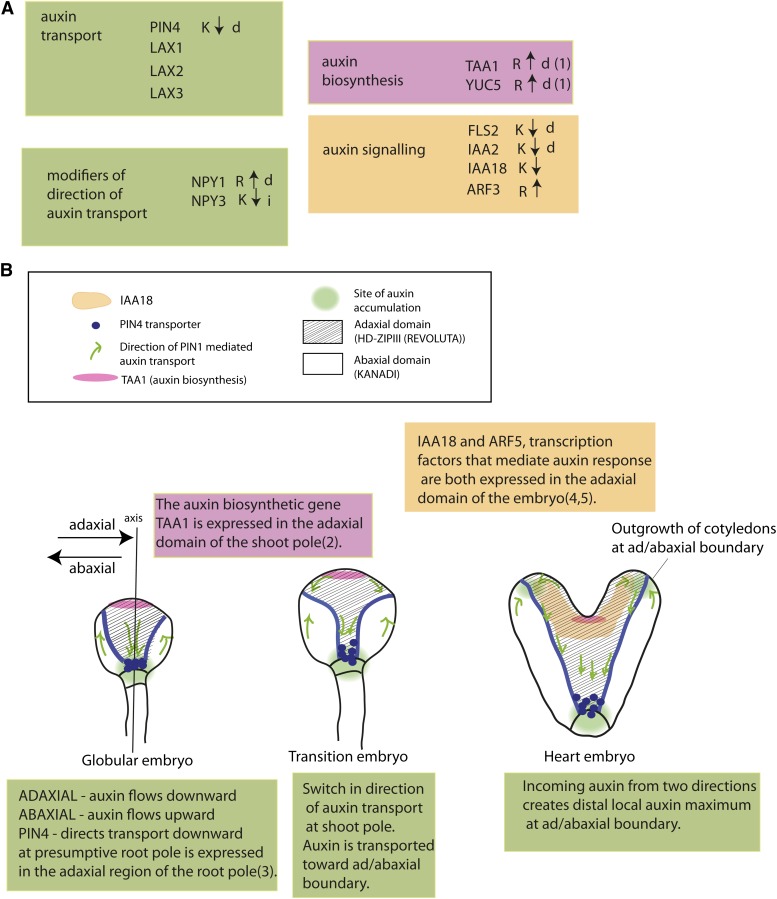
One of our findings is that REV and KAN1 regulate genes that control auxin action at several steps: biosynthesis, transport, regulation of transport, and signal transduction. These experiments suggest the mechanism of REV and KAN1 action on auxin-mediated developmental events is through additive effects on several genes (Figure 8B) rather than on a single downstream target gene. Consistent with this, analysis of combinations of mutants in the auxin biosynthetic genes, NPY, and type 3 AGC kinase mutants show that decreased levels of each gene type can act additively to affect cotyledon formation (Cheng et al., 2007b, 2008; Furutani et al., 2007).
Figure 8.
Model for Embryonic Patterning of Auxin Biosynthesis, Transport, and Reception by the Ad/Abaxial Regulators REV and KAN.
(A) Summary of REV- and KAN-regulated auxin pathway genes. Arrow indicates direction of regulation. i = indirectly regulated. Data for direct regulation of TAA1 and YUC5 by REV is from Brandt et al. (2012) (1), Stepanova et al. (2008) (2), Friml et al. (2002) (3), Ploense et al. (2009) (4), and Hamman et al. (1999) (5).
(B) Location of KAN- and REV-regulated components of the auxin pathway during embryogenesis.
Mutations in the HD-ZIPIII genes and in the KAN genes cause either failure to form organs (loss of HD-ZIPIII function; Emery et al., 2003; Prigge et al., 2005), the formation of extra or ectopic organs (gain of function HD-ZIPIII; this work), or loss of KAN function (Izhaki and Bowman, 2007). Similarly, mutations in the auxin pathway cause loss of cotyledon formation (Cheng et al., 2007a, 2007b, 2008; Furutani et al., 2007) or development of extra cotyledons (Christensen et al., 2000). To date, it has been unclear how these two regulatory pathways interact with one another in the control of organ positioning and outgrowth. Our results suggest that the ad/abaxial pathway plays a major role in patterning auxin transport and response in the Arabidopsis embryo.
The ad/abaxial patterning network and the auxin transport and signaling network both play a role in organ (leaf) formation in vegetatively growing plants as well as in embryos. Because the embryo is simpler in its structure and mutations in either system cause defects in cotyledon formation, we focus here on the role of HD-ZIPIII and KAN patterning auxin transport and responsiveness in the embryo. We assume that similar connections between the ad/abaxial signaling system and the auxin system hold for vegetative SAMs as well.
The pattern of auxin transport becomes increasingly complex as the embryo adds new cells and cell types (reviewed in Möller and Weijers, 2009). In two-cell-stage embryos, auxin is pumped upward into the apical cell from, or through, the basal cell. The direction of transport reverses in the globular embryo where it is transported downward from the embryo proper into the suspensor. By late globular stage, transport forms a circuit in the embryo, traveling down through the center of the embryo and back up along the outer cells (Figure 8). This difference in transport direction between cells in the inner and outer regions of the embryo is an early characteristic that differentiates adaxial (inner) and abaxial (outer) cells in the globular embryo.
As the apical portion of the globular embryo becomes partitioned into SAM and cotyledon domains, transport patterns shift again. New pathways transport auxin away from the incipient SAM (apically and centrally located) toward the sites of new cotyledon formation in epidermal cells (Figure 8). These new streams of outwardly directed auxin collide with the streams flowing upward through the basal, abaxial regions of the plant. At the collision point, a high local concentration of auxin, called an auxin maximum, forms. The auxin maxima occur at the positions where the cotyledons will form and are thought to drive their formation. Auxin is transported down and away from the auxin maxima at the developing cotyledon tips along what will become the primary vascular strand of the developing cotyledon (Figure 8). Note that the auxin maximum and the developing vascular strand are located at, or very close to, the ad/abaxial boundary.
Auxin alters cellular transcription by binding to the TIR1 auxin receptor, causing it to target IAA proteins for degradation (Dharmasiri et al., 2005). Destruction of IAA proteins leads to release of inhibition of ARF-type transcription factors. IAA family members differ in their affinity for auxin. Therefore, tissue-specific expression of particular IAA proteins may determine sensitivity to auxin (Calderón Villalobos et al., 2012). IAA18 and BODENLOS/IAA12 are good candidates for mediating the auxin signal required for cotyledon development since mutations that render these proteins auxin resistant cause inhibition of cotyledon formation (Hamann et al., 1999; Ploense et al., 2009). IAA18 is expressed on the adaxial side of cotyledons. Repression of IAA18 by KAN1-GR may be responsible for this pattern of expression.
Auxin transport in the embryo occurs through PIN1, PIN3, PIN4, and PIN7 auxin efflux carriers (Friml et al., 2003). Loss-of-function mutations in the corresponding genes, when combined, cause defects in cotyledon growth. Promoters for these genes direct expression in different subdomains of the plant (Benková et al., 2003). The factors that control PIN expression are largely unknown. The PLETHORA transcription factors were suggested to directly activate PIN1 transcription in the incipient leaf primordium and thereby control leaf primordium formation in the SAM (Prasad et al., 2011). More recent work by Pinon et al. (2013) has cast doubt on these experiments and instead suggested that PLETHORA controls leaf placement through activation of auxin biosynthetic genes in the SAM.
In this study, we found that PIN4 is repressed by KAN1. PIN4 is expressed in the basal end of the embryo where it acts to direct auxin in a basal/rootward direction. Its expression is limited to cells in the basal, adaxial region of the embryo, consistent with a model in which KAN inhibits rootward auxin flow by repressing PIN4 in the outer cells of the embryo (Figure 8). Consistent with this, globular embryos (32 cells) are the first stage at which an auxin-related phenotype is apparent in kan1 kan2 kan4 triple mutants. In wild-type embryos, PIN1 protein levels are at increased levels in the 16 cells that make up the apical epidermis (Figures 5A and 5B in Izhaki and Bowman, 2007). In kan1 kan2 kan4 triple mutants, PIN1 levels remain low throughout the embryo. Since PIN1 protein levels are an indicator of auxin levels (Heisler et al., 2005), low PIN1 levels may be an indicator that auxin does not accumulate in the apical epidermis in kan1 kan2 kan4 triple mutants rather than a direct consequence of lack of KAN action. Decreased auxin and PIN1 levels can be explained if PIN4 is ectopically expressed in kan triple mutants and auxin is therefore inappropriately directed downward in these mutants.
The mechanism that determines which pole of the cell PIN transporters localize to, and therefore in which direction auxin is transported, involves members of the family of type 3 AGC3 kinases and their partners, the NPY (NPH3-like BTB-POZ domain) proteins. PIN proteins localize to the rootward end of the cell by default but, in the presence of NPY-AGC, switch to the shootward end of the cell (Friml et al., 2004). This occurs in part through the phosphorylation of the PIN proteins by the PID kinases (Dhonukshe et al., 2010). NPY proteins localize to the same pole as PIN proteins, where they may act as stable markers or perhaps determinants of cell polarity (Furutani et al., 2011).
REV and KAN1 have opposing effects on NPY family genes. REV acts as an apparent direct activator of NPY1, while KAN1 acts as an indirect repressor of NPY3. The opposing effects of REV and KAN1 on NPY transcript levels are predicted to result in the targeting of PIN proteins to opposite sides of the cell in REV- and KAN-expressing domains. This is indeed what is observed in the epidermis of the embryo where PIN proteins are localized away from the apex in the adaxial domain and toward the apex in the abaxial domain resulting in opposing streams of auxin colliding at the epidermal ad/abaxial boundary.
REV and KAN1 have opposing effects on TAA1 transcription (Brandt et al., 2012; this article). TAA1 encodes an auxin biosynthetic enzyme, the expression of which is limited in its expression to an apical central domain of the embryo (Stepanova et al., 2008), as expected if it is activated by REV and repressed by KAN. It is also limited to a central region of the SAM (supplemental data in Yadav et al., 2009). Experiments using auxin transport inhibitors in embryos show that auxin accumulates in the adaxial domain when auxin transport is blocked (Benková et al., 2003), consistent with auxin biosynthesis occurring within the adaxial domain and excluded from the abaxial domain.
In summary, the ad/abaxial regulatory HD-ZIPIII and KAN factors regulate genes at all steps that lead to the presence of auxin maxima at the site of cotyledon outgrowth. As such, and given the known phenotypic consequences for cotyledon formation of HD-ZIPIII and KAN mutations, these results suggest that the ad/abaxial pathway plays an important role in patterning auxin synthesis, transport, accumulation, and sensing in the embryo. However, additional experiments beyond the scope of this work will be needed to determine the relative importance of these steps in establishing new organ primordia in the embryo and at the SAM.
While this study was under revision, a related study of global KAN1 targets was published (Merelo et al., 2013). Merelo et al. also found that KAN1 regulates a number of genes in the auxin pathway. Many genes are in common between the two studies, but there are also important differences. For instance, we have not observed regulation of either PIN1 (At1g73590) or PINOID (At2g34650), two genes known to play major roles in auxin transport, by KAN1-GR. Differences in results between the two studies may be due to differences in experimental design (e.g., number of replicates, choice of controls, and time points), normalization, and/or statistical analysis.
METHODS
Plant Materials and Growth Conditions
All plants used in this study were in the Col background and were grown at 22°C under long-day (16 h) illumination. The kan2-5 and kan2-6 alleles were identified in an ethyl methanesulfonate–mutagenized population homozygous for kan1-11. DNA sequencing of PCR products from genomic DNA of the mutant plants revealed that kan2-5 carries the same nucleotide lesion as kan2-1 (Eshed et al., 2001), resulting in a stop codon in the first exon, and the kan2-6 allele carries a G-to-A change in the 5′ splice site of the second intron.
Transgenic Plants and Enhancer Trap Lines
In order to make the synthetic UAS constructs, the UAS promoter (6×UAS) from pUAS:mGFP5-er (a gift from Jim Haseloff) was amplified using primers UASf and UASr, digested with AvrII, and cloned into binary vector pCAMBIA1381Z (Cambia) to generate pUAS1381Z as the vector for inserting KBX and mKBX repeats. KBX and mKBX repeats were synthesized by first annealing the complementary single-stranded oligos for KBX and mKBX (Supplemental Table 3), then treating the synthesized double-stranded DNA with restriction enzymes SpeI and AvrII, followed by ligation of the digested fragments. The resulting concatemers were separated on an agarose gel. The DNA bands with the size of 7 to 14 repeats were individually collected. DNA from these bands was purified using the gel purification protocol (Qiagen) and cloned into pBluescript II SK+/−. Concatemers containing the tandem repeats were selected by digestion with SpeI and AvrII because only repeats in the same orientation are resistant to both SpeI and AvrII. The candidates picked up from this screening were further confirmed by sequencing. The two concatemers finally chosen contain seven repeats of KBX and eight repeats of mKBX. These two concatemers were released from pBluescript by SpeI and XbaI and cloned into pUAS1381Z described above. Both KBX and mKBX repeats were inserted between the UAS elements and the minimal transcriptional start site of pUAS1381Z. The resulting constructs UAS-KBX:GUS and UAS-mKBX:GUS, as well as the empty vector (termed UAS:GUS), were transformed into Col plants using the floral dip method. T1 transformants were selected using hygromycin B and crossed directly to E100, a GAL4-GFP enhancer trap line obtained from the Haseloff and Poethig collections (http://www.arabidopsis.org). The GAL4-GFP enhancer trap lines contain a construct comprising a GAL4-VP16 transcriptional activator and a modified GFP gene (mGFP5ER) under the control of GAL4 upstream activation sequences (UAS). The construct is randomly located in the genome and reports the activity of endogenous enhancer elements in the vicinity of reporter gene insertion (Haseloff, 1999; Laplaze et al., 2005; Gardner et al., 2009). The GAL4-GFP enhancer trap in E100 was reportedly inserted upstream of and in the same orientation as At3g09630, the 60S ribosomal protein L4/L1 (RPL4A). Analysis of the flanking sequence revealed that the T-DNA left border is inserted in the 5′ untranslated region of RPL4A, three bases upstream of the translational start site (ATG). E100 was reported to have strong constitutive GFP expression, particularly in embryos, meristems, and young leaf primordia. The F1 seedlings from the crosses between E100 and UAS-KBX:GUS (or UAS-mKBX:GUS) were analyzed for GUS expression. As a control line, pKAN1:GUS transgenic plants were generated as described by Wu et al. (2008)
In the microarray, RT-PCR, and ChIP experiments, the KAN-GR construct (Wu et al., 2008) was transformed into Col plants with the same floral dip method as described above, and transformants were also selected on hygromycin B media. A line homozygous for the KAN-GR T-DNA that was phenotypically normal in the absence of DEX and showed a strong and consistent response on DEX-containing media was selected for all subsequent experiments. For continuous DEX treatments, seeds homozygous for KAN-GR were germinated on media containing half-strength Murashige and Skoog (MS) salts and 0.8% agar supplemented either with 10 μM DEX or 0.05% ethanol for mock treatment. For intermittent treatment, soil-grown seedlings were painted every second day either with 10 μM DEX or mock (0.05% ethanol) plus 0.015% Silwet L-77. For RNA and chromatin isolation on KAN1-GR lines, 9-d-old seedlings were grown on half-strength MS basal medium without Suc before being submerged for 4 h with gentle agitation in liquid half-strength MS plus 1% Suc containing one or more of the following: 0.05% ethanol (mock), 10 μM DEX, and 10 μM CHX. The exception to this were the CHX and DEX treatments of GR-REV and KAN1 lines in which NPY and AGC3 kinase transcripts were measured. These experiments were done on seedlings grown in liquid culture as described by Reinhart et al. (2013).
Plasmid Construction for KAN1bd Purification
The putative DNA binding domain of KAN1 was amplified from the cDNA clone (Kerstetter et al., 2001) using the following primers and cycling conditions: 5′-ATTCggatCcAAGATGCCGACAAAGCGAAGC-3′ and 5′-AAGCgaattcCTTGTTAGTGGTCTTAACAGTTCG-3′ (lowercase letters represent mismatched bases), 94°C for 20 s, 54°C for 20 s, and 72°C for 15 s for 34 cycles. Amplified DNA and the vector pGEX-2TK (Amersham Biosciences) were digested with BamHI and EcoRI and recombined such that the amino acids 210 to 280 of KAN1 were fused in frame with GST to form pGEX-KAN1bd.
Purification of KAN1bd Protein
Escherichia coli BL21 (DE3) cells carrying pGEX-KAN1bd were grown at 37°C in Luria-Bertani medium to OD600 between 0.6 and 0.8, harvested 2.5 h after protein expression, and induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside. Cell pellets were resuspended and lysed by sonication in 1× PBS at 4°C then centrifuged at 12,000g for 30 min to pellet debris. Soluble recombinant protein was purified using MicroSpin GST Purification Modules (Amersham Biosciences) according to the manufacturer’s instructions. Purified KAN1bd protein was subsequently dialyzed against 20 mM Tris-HCl, pH 8.0, and 80 mM KCl to remove reduced glutathione. Proteins were prepared for EMSA by treating with 0.07 units/μL DNase I (Fermentas) on ice for 1 h to remove contaminating E. coli DNA, and DNase I was inactivated with 2 mM EDTA.
EMSA
Complementary 54-bp oligonucleotides (Supplemental Table 3) containing a KBX or its variants were synthesized (IDT) and annealed, and 2 pmol of DNA was incubated with 10 pmol of purified protein for 1 h at 4°C. DNA-protein complexes were resolved on a 9% native polyacrylamide gels in Tris-borate-EDTA buffer at 4°C and stained with SYBR Safe (Invitrogen). The fluorescence intensity of each DNA fragment was measured using Kodak Molecular Imaging Software 4.0 (Eastman Kodak). Bands were normalized using Gaussian curve with background subtraction. Mean fluorescence intensities and standard errors were calculated from at least three independent EMSA experiments.
Histology
GUS staining was performed according to Donnelly et al. (1999) with modifications. Five-day-old plants were fixed in 80% acetone at −20°C for 20 min and then stained with 2 mM X-Gluc in 1× GUS buffer (9 mM potassium ferrocyanide and potassium ferricyanide) overnight in 30°C. After removing the chlorophyll with ethanol series, the first two leaves were dissected from the shoot and observed under a compound microscope.
Microarray Analysis
After DEX or mock treatments, seedlings were frozen in liquid nitrogen. Total RNA was extracted from tissue ground with mortar and pestle using TRIzol reagent (Invitrogen) followed by subsequent purification using the RNeasy plant mini kit (Qiagen). For each genotype and treatment, two independently obtained RNA extracts from plants grown and treated at different times (biological replicates) were labeled and hybridized to ATH1 microarrays at the Transcriptional Profiling Shared Resource of the Cancer Institute of New Jersey (New Brunswick, NJ) with Affymetrix reagents and protocols. Analysis was performed using the Affymetrix Microarray Suite v5 with a median target intensity of 150. Only significant (P < 0.005) n-fold changes of at least 1.8 in DEX- versus mock-treated KAN-GR samples are reported in Supplemental Data Set 1, although smaller changes were sometimes significant.
RNA-Seq Analysis
Fifty seeds per flask were grown in 250 mL of sterile MS medium plus Suc on a platform shaker in 24 h of cool white fluorescent light. At 7 d after germination, DEX was added to each flask to a final concentration of 50 μM. Plants were harvested at 0, 30, 60, and 120 min after DEX treatment. Six biological replicates were done for each wild-type time point, and three biological replicates were done for each transgenic time point except for GR-REV at 30 min where two biological replicates were done. Libraries were made and samples run by Otogenetics using Illumina chemistry. Sequenced reads were mapped back to the Arabidopsis thaliana genome using DNA Nexus Classic software. The resulting counts were normalized using DESEQ (Bioconductor). Two-way ANOVA tests were done on selected genes using VassarStats online calculator (http://vassarstats.net/anova2u.html).
ChIP Analysis
Nine-day-old DEX- or mock-treated KAN-GR and wild-type seedlings were harvested, washed with deionized water, and cross-linked with 1% formaldehyde. Cross-linking was quenched with 0.125 M Gly. Procedures for nuclear extracts and immunoprecipitation were adapted (Gendrel et al., 2002) with following modifications: Conditions for sonication of nuclear extracts were empirically determined to obtain an average DNA fragment size of 600 bp. Sonication was performed on ice with four pulses of 12 s with 1-min pauses at power setting 6 (40% duty cycle and 20% input; Heat System-Ultrasonics). After chromatin shearing, 10 μL of anti-GR P-20 (Santa Cruz Biotechnology) was added to each sample to immunoprecipitate KAN-GR proteins. After reversing cross-links, DNA was purified by phenol:chloroform extraction, ethanol precipitated, and resuspended in 50 μL of 10 mM Tris and 1 mM EDTA, pH 8. One microliter of immunoprecipitated DNA was used in ChIP PCR. Input DNA was diluted 120 times to achieve PCR product band intensities comparable to ChIP samples. Primers recognizing different regions in the promoters and the control gene RPL4D can be found in Supplemental Table 3. PCR conditions were as follows: 33 cycles, 94°C for 30 s, 56°C for 30 s, and 72°C for 30 s. DNA band intensity was measured using the Gaussian Curve method with background subtraction with Molecular Imaging Software 4.0 (Eastman Kodak). The abundance ratio of promoter fragments in DEX- versus mock-treated ChIP and input samples were normalized by dividing by the ratio of the negative-control gene RPL4D in DEX- or mock-treated samples. The enrichment of each gene in DEX- versus mock-treated samples results from dividing normalized DEX- to mock-treated ChIP ratios by the DEX- to mock-treated input ratios.
PCR-Assisted in Vitro DNA Binding Site Selection
KAN1bd DNA binding site selection was performed essentially as described (Hosoda et al., 2002). A mixture of 54-base oligonucleotides was synthesized (IDT) with the central 16 bases consisting of random sequences. Oligonucleotides were converted into double-stranded DNA using Klenow fragment (Fermentas) and primer BSSr (Supplemental Table 3). Double-stranded DNA (2 nmol) was incubated with 20 pmol of purified protein in dialysis buffer for 1 h at 4°C. Sample buffer (2% SDS, 10% glycerol, 60 mM Tris, 5% β-mercaptoethanol, and 0.01% bromophenol blue, pH 6.8) was added, and the protein/DNA mixture was separated on two identical 9% native polyacrylamide gels in 0.5× Tris-borate-EDTA buffer at 4°C for 120 V hours. One gel was stained with SYBR Safe (Invitrogen) to visualize DNA in the DNA-protein complex, and the other gel was stained with E-Zinc reversible stain kit (Pierce) to visualize protein in the complex. A single band that was retarded relative to free DNA or free protein was excised from the gel, and DNA was purified by phenol:chloroform extraction and then amplified using primers BSSf and BSSr (Supplemental Table 3). PCR products were precipitated and used in subsequent rounds of oligonucleotide selection. SYBR Safe and E-Zinc visualized EMSA selections were performed independently. After six cycles of selection, the resulting DNAs were cloned into pGEM-T Easy (Promega) and sequenced. Sequence comparison and motif identification were performed using Web implementations of MEME (Bailey et al., 2006) (http://meme.nbcr.net/meme/intro.html) and the Gibbs Motif Sampler (Thompson et al., 2003) (http://bayesweb.wadsworth.org/gibbs/gibbs.html).
RT-PCR
Two micrograms of total RNA was treated with DNaseI (Fermentas) and then reverse transcribed into first-strand cDNA using iScript (Bio-Rad) in 20-μL reactions. One microliter of a 10-fold dilution was used as template for PCR. The number of cycles for each primer set was determined empirically. PCRs were repeated at least thrice with independent cDNA synthesis reactions. Quantitative PCR was performed on a Rotor-Gene 3000 (Corbett Life Science) with IQ SYBR Green Supermix (Bio-Rad).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: REV, At5g60690; KAN1, At5g16560; INCURVATA4, At1g52150; PHABULOSA, At2g34710; PHAVOLUTA, At1g30490; AS2, At1g65620; and ACTIN2, At3g18780.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of 50 Sequences Obtained from PCR-Assisted Binding Site Selection.
Supplemental Figure 2. Table of Phenotypes in Loss-of-Function Mutants and Posttranslationally Activated KAN1-GR Plants.
Supplemental Figure 3. Expression of Candidate KAN1 Target Genes Was Confirmed Using RT-PCR.
Supplemental Figure 4. Chromatin Immunoprecipitation Experiments on Mock- and DEX-Treated Col Seedlings Reveal No Enrichment.
Supplemental Figure 5. Transcript Levels for Arabidopsis Genes Encoding Auxin Biosynthetic Enzymes (YUC5 and TAA1) and Auxin Transporters in Response to DEX Treatment of GR-REV and KAN1-GR Transgenic Plants.
Supplemental Figure 6. Transcript Levels for Arabidopsis Genes Encoding Members of the NPH3 Family of BTB-POZ Domain Proteins in Response to DEX Treatment of GR-REV and KAN1-GR Transgenic Plants.
Supplemental Figure 7. Transcript Levels for Arabidopsis Genes Encoding Auxin-Responsive Transcriptional Regulators.
Supplemental Figure 8. IAA2 Expression Is Upregulated in Loss-of-Function kan Mutants and Downregulated in Posttranslationally Activated KAN-GR.
Supplemental Figure 9. Differential Regulation of Members of the AGC3 Gene Family by REVOLUTA and KAN.
Supplemental Figure 10. Transcript Levels for Arabidopsis Genes Encoding AGC Kinases in Response to DEX Treatment of GR-REV and KAN1-GR Transgenic Plants.
Supplemental Table 1. Fold Enrichment in the KAN1-GR CHIP Experiment.
Supplemental Table 2. Auxin-Regulated Genes by REV and KAN.
Supplemental Table 3. Sequences of Primers Used in This Work.
Supplemental Data Set 1. KAN-Regulated Genes.
Supplemental Data Set 2. Alignment Corresponding to the Phylogenetic Analysis in Figure 7.
Supplemental Data Set 3. Alignment Corresponding to the Phylogenetic Analysis in Supplemental Figure 9.
Supplementary Material
Acknowledgments
We thank Doris Wagner for pBS-GR, G.O. Romero for quantitative PCR analysis on IAA2, and Curtis Krier and Hao Liu at the Transcriptional Profiling Shared Resource of the Cancer Institute of New Jersey for microarray services. Adam Longhurst helped with statistical analysis. This work was supported by grants from the National Science Foundation (IBN-0343179 to R.A.K and IOS-0929413 to M.K.B.) and the Charles and Johanna Busch Foundation to R.A.K., Y.H., and T.H.
AUTHOR CONTRIBUTIONS
R.A.K. conceived and designed the experiments, made the constructs, and generated transgenic lines. T.H. and Y.H. characterized the transgenic plants. Y.H. performed the microarray experiments. C.L. performed RT-PCR to confirm expression results. T.H. performed the oligonucleotide selection and ChIP experiments. B.R. designed experiments and helped generate and analyze data. N.R.N. performed RT-PCR experiments and phenotypic characterization. S.A.H. analyzed RNA-seq data. F.T.-R. generated the RNAs for the RNA-seq experiment. M.K.B. designed experiments, analyzed data, and wrote the article. R.A.K. analyzed data and wrote the article.
Glossary
- GST
glutathione S-transferase
- EMSA
electrophoretic mobility shift assay
- KBX
KANADI box
- GUS
β-glucuronidase
- Col
Columbia
- DEX
dexamethasone
- ChIP
chromatin immunoprecipitation
- CHX
cycloheximide
- IAA
indole-3-acetic acid
- qRT-PCR
quantitative RT-PCR
- MS
Murashige and Skoog
Footnotes
Online version contains Web-only data.
References
- Bailey T.L., Williams N., Misleh C., Li W.W. (2006). MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34: W369–W373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baima S., Nobili F., Sessa G., Lucchetti S., Ruberti I., Morelli G. (1995). The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121: 4171–4182 [DOI] [PubMed] [Google Scholar]
- Baima S., Possenti M., Matteucci A., Wisman E., Altamura M.M., Ruberti I., Morelli G. (2001). The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126: 643–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Brandt R., et al. (2012). Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J. 72: 31–42 [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos L.I.A., et al. (2012). A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8: 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B., Ashworth M., Guo W., Davies B. (2012). The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 158: 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2007a). Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Qin G., Dai X., Zhao Y. (2007b). NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 18825–18829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Qin G., Dai X., Zhao Y. (2008). NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 21017–21022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S.K., Dagenais N., Chory J., Weigel D. (2000). Regulation of auxin response by the protein kinase PINOID. Cell 100: 469–478 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Huang F., Galvan-Ampudia C.S., Mähönen A.P., Kleine-Vehn J., Xu J., Quint A., Prasad K., Friml J., Scheres B., Offringa R. (2010). Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137: 3245–3255 [DOI] [PubMed] [Google Scholar]
- Donnelly P.M., Bonetta D., Tsukaya H., Dengler R.E., Dengler N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Eshed Y., Baum S.F., Bowman J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99: 199–209 [DOI] [PubMed] [Google Scholar]
- Eshed Y., Baum S.F., Perea J.V., Bowman J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Eshed Y., Izhaki A., Baum S.F., Floyd S.K., Bowman J.L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131: 2997–3006 [DOI] [PubMed] [Google Scholar]
- Evans M.M.S. (2007). The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell 19: 46–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J., Benková E., Blilou I., Wisniewska J., Hamann T., Ljung K., Woody S., Sandberg G., Scheres B., Jürgens G., Palme K. (2002). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673 [DOI] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Friml J., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865 [DOI] [PubMed] [Google Scholar]
- Furutani M., Kajiwara T., Kato T., Treml B.S., Stockum C., Torres-Ruiz R.A., Tasaka M. (2007). The gene MACCHI-BOU 4/ENHANCER OF PINOID encodes a NPH3-like protein and reveals similarities between organogenesis and phototropism at the molecular level. Development 134: 3849–3859 [DOI] [PubMed] [Google Scholar]
- Furutani M., Sakamoto N., Yoshida S., Kajiwara T., Robert H.S., Friml J., Tasaka M. (2011). Polar-localized NPH3-like proteins regulate polarity and endocytosis of PIN-FORMED auxin efflux carriers. Development 138: 2069–2078 [DOI] [PubMed] [Google Scholar]
- Gardner M.J., Baker A.J., Assie J.M., Poethig R.S., Haseloff J.P., Webb A.A. (2009). GAL4 GFP enhancer trap lines for analysis of stomatal guard cell development and gene expression. J. Exp. Bot. 60: 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Yordan C., Colot V., Martienssen R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297: 1871–1873 [DOI] [PubMed] [Google Scholar]
- Hamann T., Mayer U., Jürgens G. (1999). The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126: 1387–1395 [DOI] [PubMed] [Google Scholar]
- Haseloff J. (1999). GFP variants for multispectral imaging of living cells. Methods Cell Biol. 58: 139–151 [DOI] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hosoda K., Imamura A., Katoh E., Hatta T., Tachiki M., Yamada H., Mizuno T., Yamazaki T. (2002). Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14: 2015–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izhaki A., Bowman J.L. (2007). KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19: 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Tang J., Donnelly P., Dengler N. (2002). Primary vascular pattern and expression of ATHB8 in shoots of Arabidopsis. New Phytol. 158: 443–454 [DOI] [PubMed] [Google Scholar]
- Kelley D.R., Arreola A., Gallagher T.L., Gasser C.S. (2012). ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 139: 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter R.A., Bollman K., Taylor R.A., Bomblies K., Poethig R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411: 706–709 [DOI] [PubMed] [Google Scholar]
- Laplaze L., Parizot B., Baker A., Ricaud L., Martinière A., Auguy F., Franche C., Nussaume L., Bogusz D., Haseloff J. (2005). GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J. Exp. Bot. 56: 2433–2442 [DOI] [PubMed] [Google Scholar]
- Leon-Kloosterziel K.M., Keijzer C.J., Koornneef M. (1994). A seed shape mutant of Arabidopsis that is affected in integument development. Plant Cell 6: 385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAbee J.M., Hill T.A., Skinner D.J., Izhaki A., Hauser B.A., Meister R.J., Venugopala Reddy G., Meyerowitz E.M., Bowman J.L., Gasser C.S. (2006). ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. Plant J. 46: 522–531 [DOI] [PubMed] [Google Scholar]
- McConnell J.R., Barton M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942 [DOI] [PubMed] [Google Scholar]
- McConnell J.R., Emery J., Eshed Y., Bao N., Bowman J., Barton M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- Merelo P., Xie Y., Brand L., Ott F., Weigel D., Bowman J.L., Heisler M.G., Wenkel S. (2013). Genome-wide identification of KANADI1 target genes. PLoS ONE 8: e77341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller B., Weijers D. (2009). Auxin control of embryo patterning. Cold Spring Harb. Perspect. Biol. 1: a001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L., Dunoyer P., Jay F., Arnold B., Dharmasiri N., Estelle M., Voinnet O., Jones J.D. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439 [DOI] [PubMed] [Google Scholar]
- Pekker I., Alvarez J.P., Eshed Y. (2005). Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17: 2899–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon V., Prasad K., Grigg S.P., Sanchez-Perez G.F., Scheres B. (2013). Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proc. Natl. Acad. Sci. USA 110: 1107–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploense S.E., Wu M.F., Nagpal P., Reed J.W. (2009). A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development 136: 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K., et al. (2011). Arabidopsis PLETHORA transcription factors control phyllotaxis. Curr. Biol. 21: 1123–1128 [DOI] [PubMed] [Google Scholar]
- Pratt W.B., Galigniana M.D., Morishima Y., Murphy P.J. (2004). Role of molecular chaperones in steroid receptor action. Essays Biochem. 40: 41–58 [DOI] [PubMed] [Google Scholar]
- Prigge M.J., Otsuga D., Alonso J.M., Ecker J.R., Drews G.N., Clark S.E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17: 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Mandel T., Kuhlemeier C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Liu T., Newell N.R., Magnani E., Huang T., Kerstetter R., Michaels S., Barton M.K. (2013). Establishing a framework for the ad/abaxial regulatory network of Arabidopsis: Ascertaining targets of class III homeodomain leucine zipper and KANADI regulation. Plant Cell 25: 3228–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V., Linhares F., Solano R., Martín A.C., Iglesias J., Leyva A., Paz-Ares J. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Cartagena J., Robles P., Ponce M.R., Micol J.L. (1999). Genetic analysis of leaf form mutants from the Arabidopsis Information Service collection. Mol. Gen. Genet. 261: 725–739 [DOI] [PubMed] [Google Scholar]
- Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.Y., Dolezal K., Schlereth A., Jürgens G., Alonso J.M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Talbert P.B., Adler H.T., Parks D.W., Comai L. (1995). The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121: 2723–2735 [DOI] [PubMed] [Google Scholar]
- Thompson W., Rouchka E.C., Lawrence C.E. (2003). Gibbs Recursive Sampler: Finding transcription factor binding sites. Nucleic Acids Res. 31: 3580–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D., Sablowski R.W., Meyerowitz E.M. (1999). Transcriptional activation of APETALA1 by LEAFY. Science 285: 582–584 [DOI] [PubMed] [Google Scholar]
- Waites R., Hudson A. (1995). phantastica: A gene required for dorsiventrality of leaves in Antirrhinum majus. Development 121: 2143–2154 [Google Scholar]
- Wang L., Kim J., Somers D.E. (2013). Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. USA 110: 761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Lin W.C., Huang T., Poethig R.S., Springer P.S., Kerstetter R.A. (2008). KANADI1 regulates adaxial-abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2. Proc. Natl. Acad. Sci. USA 105: 16392–16397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R.K., Girke T., Pasala S., Xie M., Reddy G.V. (2009). Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc. Natl. Acad. Sci. USA 106: 4941–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Ye Z.H. (2001). Alteration of auxin polar transport in the Arabidopsis ifl1 mutants. Plant Physiol. 126: 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.