Disruption of the host-cell actin cytoskeleton promotes the susceptibility of plants to diverse microbes. However, the signaling cascades and regulatory proteins linking the perception of microbes to cytoskeletal remodeling remain largely uncharacterized. This study implicates a key actin binding protein, ACTIN DEPOLYMERIZING FACTOR4, in plant innate immune signaling.
Abstract
Conserved microbe-associated molecular patterns (MAMPs) are sensed by pattern recognition receptors (PRRs) on cells of plants and animals. MAMP perception typically triggers rearrangements to actin cytoskeletal arrays during innate immune signaling. However, the signaling cascades linking PRR activation by MAMPs to cytoskeleton remodeling are not well characterized. Here, we developed a system to dissect, at high spatial and temporal resolution, the regulation of actin dynamics during innate immune signaling in plant cells. Within minutes of MAMP perception, we detected changes to single actin filament turnover in epidermal cells treated with bacterial and fungal MAMPs. These MAMP-induced alterations phenocopied an ACTIN DEPOLYMERIZING FACTOR4 (ADF4) knockout mutant. Moreover, actin arrays in the adf4 mutant were unresponsive to a bacterial MAMP, elf26, but responded normally to the fungal MAMP, chitin. Together, our data provide strong genetic and cytological evidence for the inhibition of ADF activity regulating actin remodeling during innate immune signaling. This work is the first to directly link an ADF/cofilin to the cytoskeletal rearrangements elicited directly after pathogen perception in plant or mammalian cells.
INTRODUCTION
The innate immune system is a critical line of defense to protect the host from infection by potential pathogens; this includes both nonspecific basal and inducible responses. Much of our understanding of innate immune responses comes from studies of toll-like receptor (TLR) signaling pathways in mammalian cells. TLRs trigger cellular responses by serving as specific receptors for extracellular molecules containing pathogen- or microbe-associated molecular patterns (MAMPs; Janeway and Medzhitov, 2002; Matzinger, 2002; Stuart et al., 2013). The resulting signal transduction cascades stimulate de novo gene transcription and regulate changes in lipid content, membrane ruffling, and endocytosis; the latter events have been correlated with changes in actin cytoskeletal dynamics during basal immune signaling (Granucci et al., 2001; Huang et al., 2001). In dendritic cells (DCs), TLR signaling results in the destabilization of actin-rich podosomes or actin bundles, presumably providing a pool of actin monomers necessary to build the new actin arrays involved in the uptake of activated MAMP receptors (West et al., 2004; Irving et al., 2012). Actin cytoskeletal remodeling in mammalian cells is a critical facet of innate immunity that functions in phagocytosis, the arrangement and endocytosis of cell surface receptors, vesicular transport of antimicrobial compounds, membrane ruffling, and motility (Gordón-Alonso et al., 2010). Hundreds of actin binding proteins regulate the behavior of actin arrays in eukaryotic cells, and many of these are capable of transducing signals from intracellular second messengers (Pollard and Cooper, 2009). A recent study of DCs responding to MAMPs demonstrates a requirement for gelsolin, which was shown to regulate actin dynamics through an antiviral kinase (Irving et al., 2012). Whether other actin binding proteins serve as targets for innate immune signaling and what molecular mechanisms underlie actin cytoskeletal rearrangements are still largely unknown.
Unlike mammals, plants lack DCs, macrophages, or an acquired immune system, and therefore rely on innate immunity to perceive and respond to both harmful and beneficial microbes (Chisholm et al., 2006; Jones and Dangl, 2006; Tsuda and Katagiri, 2010). Since plants are sessile, innate immunity and the associated defense responses need to occur in virtually all cells to thwart pathogen proliferation. In plants, the innate immune response is known as pattern-triggered immunity (PTI; Chisholm et al., 2006; Jones and Dangl, 2006; Tsuda and Katagiri, 2010). Hallmark events associated with PTI include changes in cellular reactive oxygen species (ROS) and pH, increased defense gene transcription, and the deposition of callose to fortify the cell wall (Tsuda and Katagiri, 2010). Recently, we demonstrated that both pathogenic bacteria and MAMP peptides trigger a transient increase in actin filament abundance during the innate immune response of plant epidermal cells (Henty-Ridilla et al., 2013a). This actin-based response does not require contributions from a type-three secretion system or bacterial effector proteins (Henty-Ridilla et al., 2013a). Additionally, when actin polymerization is blocked with the inhibitor latrunculin B (LatB), plants are more susceptible to pathogenic and nonpathogenic bacteria (Henty-Ridilla et al., 2013a). Even during interactions between plant cells and mutualistic bacteria, such as root-infecting Rhizobium spp, there are prominent increases in actin filament abundance (Crdenas et al., 1998). This remodeling requires the Arp2/3 complex and SCAR/WAVE actin nucleation machinery (Yokota et al., 2009; Miyahara et al., 2010). However, the entire complement of molecular machinery that senses and transduces immune signaling to actin cytoskeleton remodeling is still largely unknown in both plant and animal systems.
The cortical actin array of plant epidermal cells represents an excellent model system for studies of cytoskeletal dynamics and responses to biotic stress (Day et al., 2011; Henty-Ridilla et al., 2013b). Variable-angle epifluorescence microscopy (VAEM) affords high spatial and temporal resolution imaging of cytoskeletal turnover and allows quantitative analyses of bundles and single actin filaments (Staiger et al., 2009; Blanchoin et al., 2010; Staiger et al., 2010; Henty-Ridilla et al., 2013b). Single actin filaments are faint and ephemeral structures that are also highly dynamic; they exist in the cytoplasm on average for ∼20 s (Staiger et al., 2009; Smertenko et al., 2010). Single actin filament elongation is fast (∼2 µm/s), and growing filaments originate de novo in the cytoplasm, as branches from the side of preexisting filaments, or from recently severed filament ends (Staiger et al., 2009). Filament disassembly occurs through prolific severing activity, rather than filament depolymerization from pointed ends (Staiger et al., 2009). Moreover, this model system for dissecting actin turnover is amenable to both genetic and pharmacological approaches (Henty et al., 2011; Li et al., 2012; Tóth et al., 2012; Henty-Ridilla et al., 2013b). For example, an ACTIN DEPOLYMERIZING FACTOR (ADF) knockout mutant in Arabidopsis thaliana displays a significant decrease in filament severing activity, as well as increases in maximum filament lengths and lifetimes (Henty et al., 2011).
Based on knowledge of actin turnover mechanisms, we predict that changes in actin filament abundance during innate immunity occur by increasing the extent of actin polymerization, by suppressing filament disassembly, or both. Similar to studies with animal TLRs, much is known about the pattern recognition receptors (PRRs) that initiate signaling pathways during plant innate immunity (Day et al., 2011); however, the molecular machinery that links perception of MAMPs to actin cytoskeleton remodeling are poorly understood. Here, we develop a model system using epidermal cells of the dark-grown hypocotyl to dissect innate immune signaling in response to MAMP perception. We provide genetic and cytological evidence that inhibition of ADF4 during plant innate immune signaling regulates actin dynamics in order to execute key events associated with PTI, such as cell wall fortification and transcriptional activation of defense gene markers.
RESULTS
Cortical Actin Arrays Respond Rapidly to MAMP Perception
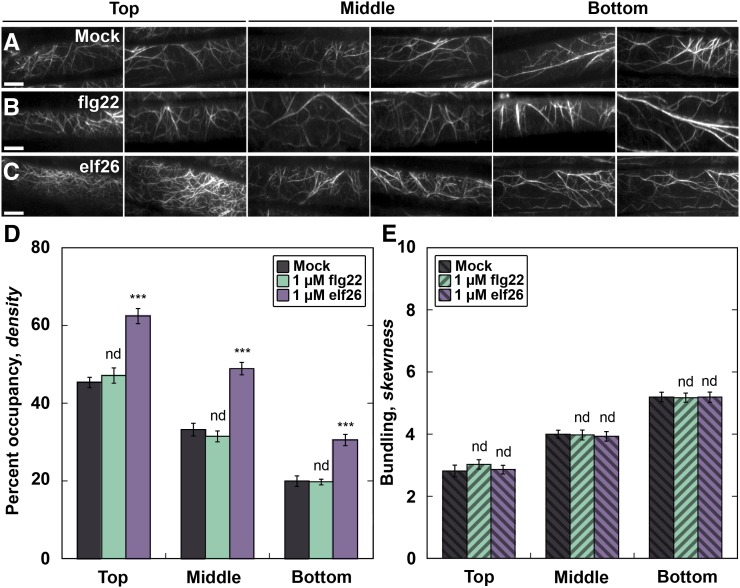
Upon perception of pathogenic and nonpathogenic microbes, plant epidermal cells display an increase in actin filament abundance, which can be mimicked by MAMP peptide treatments (Henty-Ridilla et al., 2013a). Increased actin abundance is an important feature of plant innate immunity, as plants infiltrated with the actin polymerization inhibitor LatB are more susceptible to the growth of the bacterial phytopathogen Pseudomonas syringae pv tomato DC3000 (Henty-Ridilla et al., 2013a). To investigate the signaling cascades and response regulators that elicit increased actin filament abundance during innate immunity, we treated Arabidopsis hypocotyls expressing the actin reporter green fluorescent protein (GFP)-fABD2 with fungal and bacterial MAMP mimics and measured changes to the architecture of actin arrays in the cortical cytoskeleton of epidermal cells. We detected a dose- and time-dependent increase in actin filament abundance within minutes of treatment with a conserved 26–amino acid peptide from bacterial elongation factor (EF-Tu), elf26, in epidermal cells throughout the hypocotyl (Figures 1C and 1D; Supplemental Figure 1). This result is similar, but much faster, than the response of epidermal cells from light-grown cotyledons following treatment with microbes or a synthetic peptide mimic of P. syringae flagellin, flg22 (Henty-Ridilla et al., 2013a). However, the receptor that perceives flg22, FLAGELLIN SENSING2 (FLS2), is not expressed in dark-grown hypocotyls (Supplemental Figure 2; Ma et al., 2005). Thus, we did not observe a measureable change in actin architecture following flg22 treatment of hypocotyls (Figures 1B, 1D, and 1E; Supplemental Figure 1). Subsequent analyses were performed only at the base of hypocotyls with 1 µM MAMP peptide, as this region showed the greatest increase in actin filament abundance (Figure 1D).
Figure 1.
Actin Filament Density Increases following elf26 Treatment.
(A) to (C) VAEM images of hypocotyl epidermal cells expressing GFP-fABD2. Hypocotyls were treated with mock (A), 1 μM flg22 (B), or 1 μM elf26 (C) peptides for 5 min prior to imaging. Images are from individual representative hypocotyls following different treatments, with cells located near the cotyledon (top) at the left and cells located near the root (bottom) at the right. Bars = 10 μm.
(D) Actin filament abundance, or percentage of occupancy, was measured and binned into three equal regions along the length of the hypocotyl. All epidermal cells treated with elf26 had significantly increased filament density throughout the hypocotyl when compared with mock-treated seedlings. However, cells treated with flg22 were not significantly different from mock.
(E) The extent of actin filament bundling, or skewness, was measured from the same images used for (D). There was no significant difference between treatments with flg22, elf26, or mock.
Values given are means ± se (n = 450 cells per region, from at least 30 hypocotyls). Asterisks represent significant differences by ANOVA, with Tukey HSD posthoc analysis (nd = not significantly different from mock; ***P < 0.001).
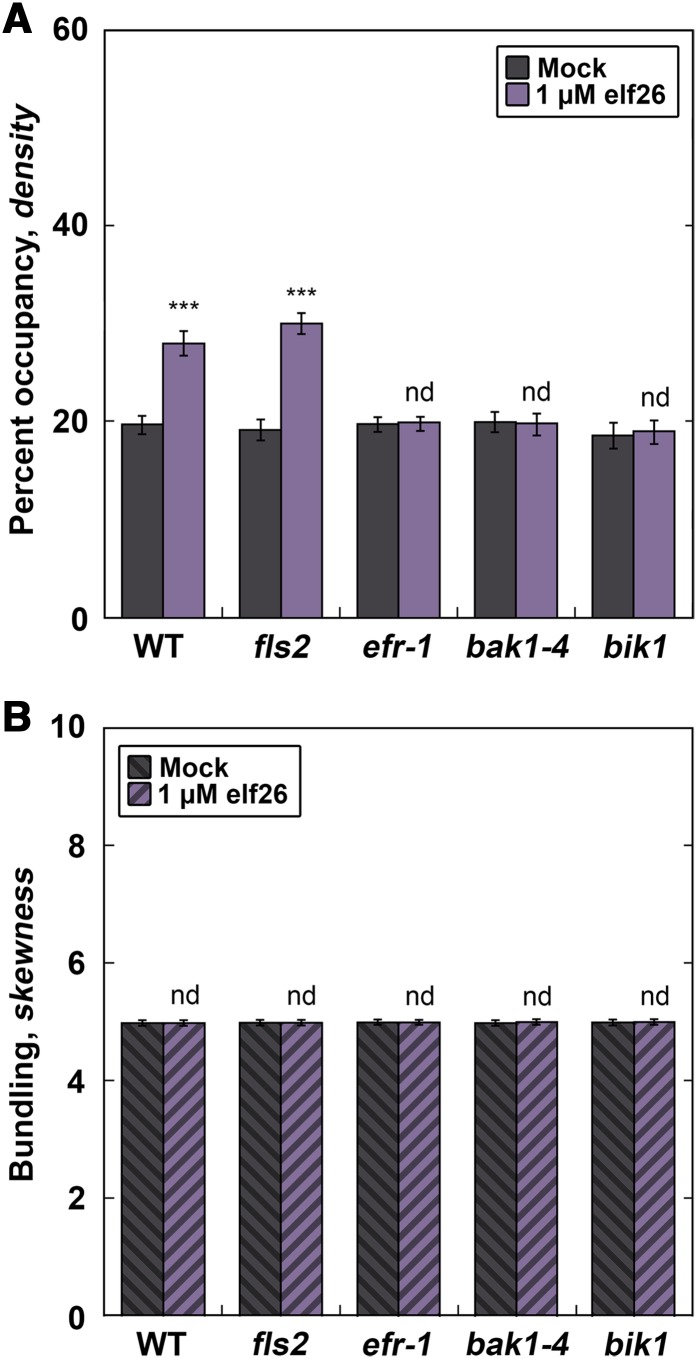
The rapid increase in actin filament abundance following elf26 treatment suggests that perception of MAMPs at the plasma membrane is important for cytoskeletal rearrangements. To test whether components of a MAMP receptor complex regulate actin filament abundance, we performed actin architecture analysis on several Arabidopsis knockout mutants that have disruptions in the EF-Tu signaling pathway (Figure 2). The bacterial MAMP elf26 is recognized by a plasma membrane–associated PRR complex, comprising the leucine-rich repeat–receptor kinases EF-Tu RECEPTOR (EFR) and BRI1-ASSOCIATED KINASE (BAK1) as well as the cytoplasmic kinase BOTRYTIS-INDUCED KINASE (BIK1) (Backues et al., 2010; Boudsocq et al., 2010; Mao et al., 2011; Zipfel et al., 2006). As expected, epidermal cells from wild-type plants and the fls2 mutant (lacking the PRR for flg22, FLS2) treated with elf26 for 5 min showed significantly enhanced filament abundance (Figure 2A). However, the homozygous mutant seedlings efr-1, bak1-4, and bik1 all lacked a significant increase in filament abundance following elf26 treatment (Figure 2A). Additionally, no significant changes to the extent of filament bundling were observed in the wild type or any of the mutants (Figure 2B). In summary, treatment with elf26 peptide is sufficient to induce rapid increases in actin filament abundance in epidermal cells and this requires the EFR-PRR complex.
Figure 2.
Recognition of elf26 by the EFR Receptor Complex Is Required for the Increase in Actin Abundance.
(A) Arabidopsis knockout mutants define the early signaling steps required for increased actin abundance in hypocotyl epidermal cells. Filament abundance was measured in epidermal cells from homozygous signaling mutant seedlings expressing GFP-fABD2 following 5 min of treatment with 1 µM elf26 or mock. Columbia-0 expressing GFP-fABD2 was used as the wild-type control. Knockout lines for EFR and components of the PRR complex, BAK1 and BIK1, were used to define the host-signaling components required for eliciting the actin response. Wild-type and fls2 homozygous mutant seedlings exhibit enhanced filament abundance following 1 µM elf26 treatment, whereas knockout mutants for the EFR-PRR complex (i.e., efr-1, bak1-4, and bik1) did not have a measurable change compared with the mock control.
(B) The extent of actin filament bundling was not significantly different from mock-treated controls. Images used for analysis in (A) were measured for filament bundling.
Values given are means ±se (n = 300 cells per genotype from at least 30 hypocotyls). Asterisks represent significant differences by ANOVA, with Tukey HSD posthoc analysis (nd = not significantly different from mock; ***P < 0.001).
Single Actin Filament Turnover Is Altered following elf26 Treatment
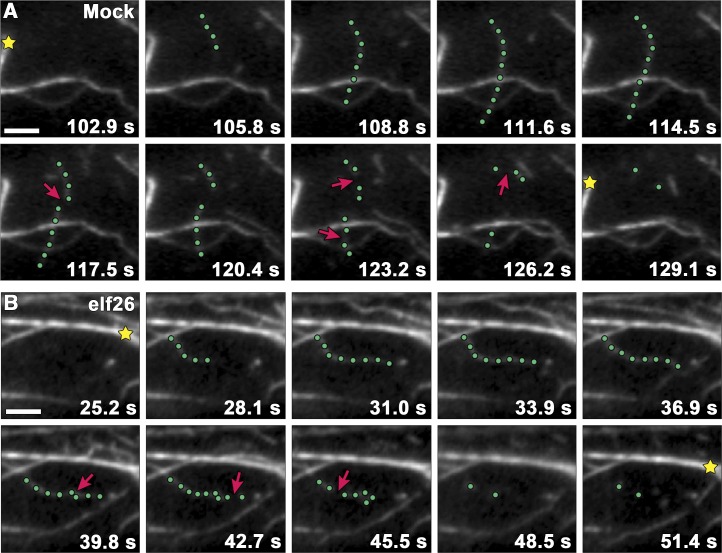
Examination of single actin filament dynamics can provide detailed insights about the molecular mechanism of actin filament assembly and turnover in living plant cells (Staiger et al., 2009; Smertenko et al., 2010). Furthermore, genetic and pharmacological perturbations of single actin filament behavior often correlate with changes in the architecture of actin arrays (Henty et al., 2011; Li et al., 2012; Tóth et al., 2012; Henty-Ridilla et al., 2013b). To understand the molecular mechanism of actin remodeling associated with MAMP-induced increases in actin filament abundance, we used time-lapse microscopy to track dynamic filaments in live epidermal cells following treatment with elf26 (Figure 3, Table 1). Growing actin filaments in mock-treated epidermal cells originated mostly from the side (46%) of preexisting filaments, or de novo in the cytoplasm (34%), and elongated at a mean rate of 1.7 µm s−1 to an average maximum filament length of ∼13 µm (Table 1). Filaments were present for ∼20 s before disassembling and disappearing completely as a result of severing activity (Figure 3A, Table 1). Most severed ends did not regrow (∼3%) or anneal to other filaments (∼2%).
Figure 3.
Filament Severing Is Significantly Reduced following elf26 Treatment.
(A) Time-lapse series of VAEM images shows actin filament turnover in the cortical cytoplasm of a mock-treated epidermal cell. The highlighted filament (green dots) elongated at 1.72 µm s−1 before suffering several breaks (arrows). Actin filament bundles (stars) remained relatively stationary throughout the time series.
(B) A representative growing filament (green dots) from a wild-type cell treated with 1 μM elf26 peptide for 5 min displayed fewer severing events (arrows) compared with the mock-treated cell.
Micrographs in (A) and (B) were collected at 1.5-s intervals, and every other image is presented in each montage. Bars = 5 μm.
Table 1. Actin Dynamics Parameters from Mock- and elf26-Treated Epidermal Cells.
| Stochastic Dynamics Parameters | Mock | 1 µM elf26 | 1 µM flg22 |
|---|---|---|---|
| Elongation rate (µm s−1) | 1.7 ± 0.1 | 1.9 ± 0.2nd | 1.7 ± 0.1nd |
| Severing frequency (breaks µm−1 s−1) | 0.015 ± 0.002 | 0.009 ± 0.001*** | 0.016 ± 0.001nd |
| Max. filament length (µm) | 12.8 ± 0.7 | 18.3 ± 0.8*** | 12.8 ± 0.5nd |
| Max. filament lifetime (s) | 20 ± 1 | 25 ± 2*** | 20 ± 1nd |
| Regrowth of severed ends (%) | 2.7 ± 0.1 | 2.5 ± 0.1nd | 3.0 ± 0.1nd |
| Annealing of severed ends (%) | 2.3 ± 0.2 | 9.0 ± 0.4*** | 2.3 ± 0.2nd |
| Filament origin (% per cell) | |||
| De novo | 33.6 ± 1.4 | 28.3 ± 1.1** | 35.0 ± 1.3nd |
| Ends | 20.7 ± 1.3 | 20.6 ± 1.1nd | 20.8 ± 1.4nd |
| Side | 46.0 ± 2.1 | 51.1 ± 1.3* | 44.2 ± 1.9nd |
nd, not significantly different from mock control value by Student's t test (P value > 0.05). Significantly different from mock control value by Student's t test: *P value ≤ 0.05, **P value ≤ 0.01, and ***P value ≤ 0.001. Values given are means ±se, with n > 50 filaments from n > 30 epidermal cells and at least 10 hypocotyls per treatment.
A specific subset of these actin filament dynamics parameters changed significantly after 5 min of treatment with elf26 (Table 1). Notably, there was a significant decrease in the proportion of filaments originating de novo and a corresponding increase in filaments originating from the side of nascent filaments (Figure 3B, Table 1). Treatment with elf26 also elicited an increase in the average maximum length and lifetime of growing filaments, a 4-fold increase in filament–filament annealing, and a 2-fold reduction in severing activity (Figure 3B, Table 1). However, filament elongation rate and regrowth frequency were not significantly altered following elf26 treatment. Treatment with 1 µM flg22, a negative control, was not significantly different from mock for any parameter measured (Table 1). Therefore, treatment with the MAMP elf26 elicits specific changes to actin filament turnover that lead to a rapid increase in the overall abundance of actin filaments during plant innate immune signaling.
Actin Architecture and Dynamics in an adf4 Mutant Fail to Respond to elf26 Treatment
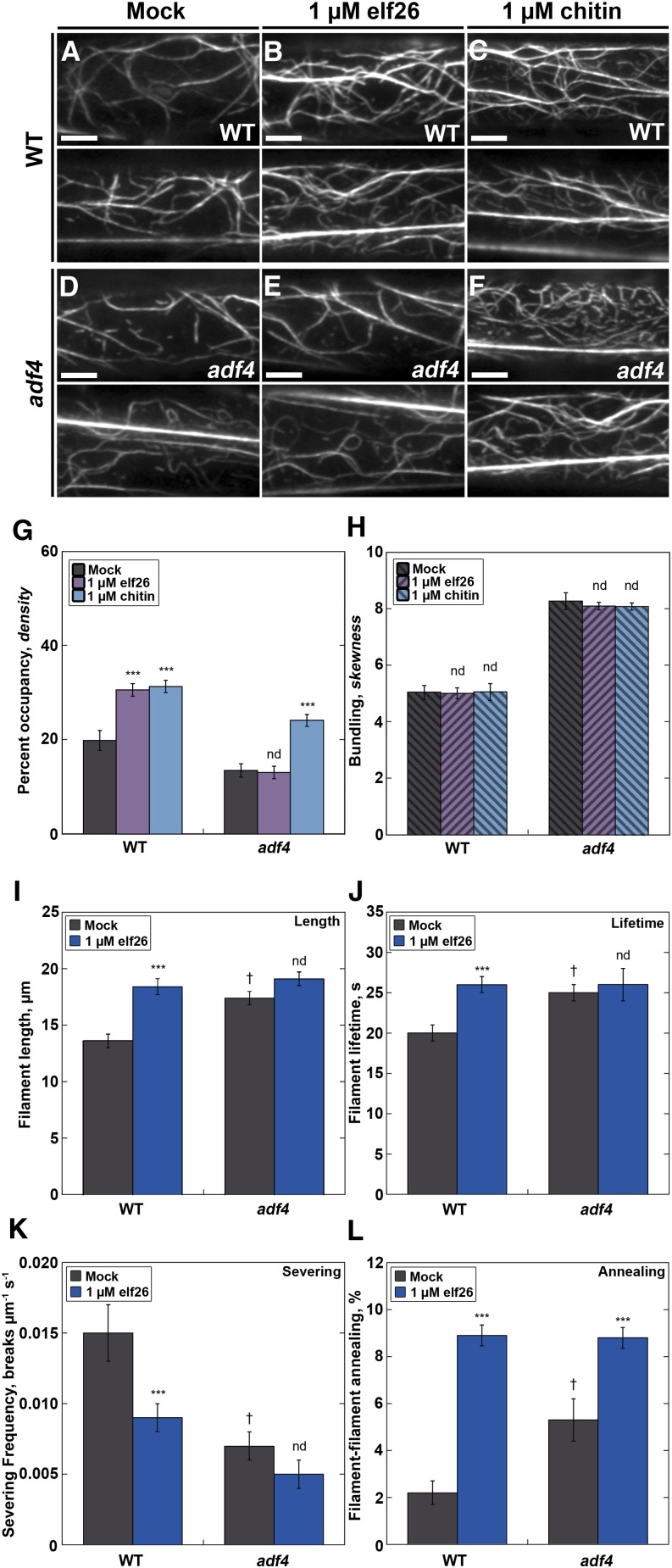
To evaluate the mechanism of MAMP-stimulated changes in actin remodeling, we examined mutants for key actin binding proteins in Arabidopsis. A homozygous adf4 knockout mutant was used previously to implicate ADF4 in effector-triggered immunity. Specifically, adf4 plants treated with P. syringae DC3000 expressing the effector protein AvrPphB showed increased susceptibility to bacterial growth, altered defense gene transcription, and attenuated mitogen-activated protein kinase (MAPK) signaling compared with wild-type Arabidopsis (Tian et al., 2009; Porter et al., 2012). Here, we tested the role of ADF4 in PTI. When wild-type cells were treated with elf26, we observed a response that partially phenocopied a homozygous knockout mutant for adf4 (Supplemental Table 1). Specifically, actin filaments in epidermal cells of the adf4 mutant had significantly increased filament lengths and lifetimes and a 2-fold reduction in severing frequency (Supplemental Table 1), as observed previously (Henty et al., 2011). Overall, it appears that the innate immune response to elf26 involves inhibition of severing activity, which thereby enhances filament stability and lifetimes to increase the abundance of actin filaments in the cortical cytoplasm. Thus, we predict that elf26-induced innate immune signaling works via inhibition or downregulation of ADF4 activity. If this hypothesis is correct, we expect actin organization in the adf4 mutant to be unresponsive or attenuated following elf26 treatment.
To test this prediction, we quantified the actin array architecture and single filament dynamics in the adf4 mutant following a 5-min treatment with elf26 (Figure 4; Supplemental Table 1). We also used a fungal MAMP, chitin, to address the specificity of the actin response because these signaling pathways share the coreceptor protein kinases BAK1 and BIK1 (Chinchilla et al., 2007; Heese et al., 2007). As expected, actin filament levels in wild-type cells were significantly increased after elf26 and chitin treatments compared with mock controls (Figures 4A to 4C and 4G). Actin filament arrays in the mock-treated adf4 mutant (Figure 4D) appeared less abundant and more bundled than the arrays in wild-type epidermal cells (Figure 4A), as reported previously (Henty et al., 2011). Furthermore, the 35S:ADF4;adf4 complementation or rescue line expressing the actin reporter GFP-fABD2 (Henty et al., 2011) restores the actin filament abundance and bundling phenotypes to wild-type levels (Supplemental Figure 3). Importantly, after treatment with elf26, the adf4 mutant did not show an increase in filament abundance (Figures 4E and 4G). However, the actin cytoskeleton in the adf4 mutant still responded to the fungal MAMP chitin with an increase in filament abundance (Figures 4F and 4G), indicating that adf4 is not generally deficient in actin remodeling in response to microbial MAMPs. The ADF4 rescue line responded like the wild type when treated with either elf26 or chitin (Supplemental Figure 3A). The elf26-induced increase in actin filament density in the wild type is quite specific, since there were no significant changes to the extent of filament bundling following either chitin or elf26 treatment in either genotype (Figure 4H).
Figure 4.
The adf4 Mutant Lacks Changes in Actin Filament Architecture and Dynamics following Treatment with elf26.
(A) to (F) Actin filaments in mock-treated epidermal cells from the adf4 mutant (D) appeared to be significantly less abundant and more bundled compared with wild-type cells. Notably, the actin architecture in adf4 cells did not seem to change following treatment with elf26 peptide (E), unlike wild-type cells (B), where actin filaments appeared to be more abundant after MAMP treatment. In response to the fungal MAMP, chitin, cortical actin abundance increased in both the wild type (C) and the adf4 mutant (F). Bars = 5 µm.
(G) Actin filament abundance was measured in epidermal cells at the base of wild-type and adf4 mutant hypocotyls after treatment with MAMPs for 5 min. Wild-type epidermal cells treated with 1 µM elf26 or 1 µM chitin had a significant increase in filament abundance compared with mock-treated seedlings. Actin filament abundance in cells from the adf4 mutant treated with 1 µM elf26 remained unchanged compared with mock-treated controls. However, actin filament abundance in the adf4 mutant was significantly elevated following treatment with chitin.
(H) The extent of actin filament bundling was not altered following treatment with chitin in either wild-type or adf4 hypocotyl epidermal cells. The same images analyzed in (G) were measured for actin filament bundling.
(I) to (K) Several parameters of actin filament turnover do not change in the adf4 mutant following elf26 treatment. The adf4 mutant had significantly enhanced filament lengths (I) and lifetimes (J) as well as a reduction in severing frequency (K) compared with the wild type. Whereas each of these parameters was significantly changed in wild-type seedlings upon treatment with elf26, these do not change in the adf4 mutant.
(L) By contrast, there were significant increases in filament-filament annealing in the adf4 loss-of-function mutant compared with wild-type cells. Following elf26 treatment, both the wild type and the adf4 mutant respond with significantly enhanced filament–filament annealing.
Values given are means ±se (n = 300 cells per treatment and genotype from at least 30 hypocotyls). Asterisks represent significant differences by ANOVA, with Tukey HSD posthoc analysis (nd = not significantly different from genotype-specific mock control; ***P < 0.001; † denotes significant differences between the wild type and adf4). For more details, see Supplemental Table 1.
To investigate whether these changes were specific to a single ADF isovariant, we generated a second homozygous ADF knockout mutant, adf1, expressing the actin reporter GFP-fABD2 (Supplemental Figure 4). We chose ADF1 because it is the most highly expressed ADF in dark-grown hypocotyls (Ma et al., 2005; Henty et al., 2011), is present in the same phylogenetic clade as ADF4 (Ruzicka et al., 2007), its biochemical properties are well characterized (Carlier et al., 1997; Khurana et al., 2010; Zheng et al., 2013), and mutant plants have perturbed actin organization (Dong et al., 2001; Tian et al., 2009). The adf1 knockout mutant had markedly reduced actin filament density in the cortical array of epidermal cells compared with wild-type cells (Supplemental Figure 5). Interestingly, adf1 seedlings failed to respond to either elf26 or chitin treatment with changes in actin architecture (Supplemental Figure 5), further indicating the lack of filament abundance changes in the adf4 mutant are specific to the elf26-stimulated pathway, whereas ADF1 appears to be involved in a general response to diverse MAMPs.
Most single filament parameters central to ADF activity and filament turnover also did not change following treatment of the adf4 mutant with a bacterial MAMP (Supplemental Table 1; Figures 4I to 4K). In particular, actin filament severing frequency was not reduced significantly in the adf4 mutant after elf26 treatment compared with the mock-treated mutant (Figure 4K). Moreover, maximum filament lengths and lifetimes did not increase significantly when adf4 was treated with elf26 (Figures 4I and 4J). However, filament–filament annealing increased nearly 2-fold in adf4 cells treated with elf26, albeit not as dramatically as in wild-type cells (Figure 4L). Collectively, the failure of actin array organization and dynamics to change in adf4 mutant cells treated with a bacterial MAMP indicate that ADF4 is necessary for signaling during innate immunity through the EFR receptor complex to the actin cytoskeleton. However, since the adf4 mutant still responds to the fungal MAMP chitin, there are likely multiple, distinct signaling pathways that require the host actin cytoskeleton during the plant innate immune response. Moreover, there may be additional actin binding proteins contributing to the altered actin dynamics and increased filament abundance because some dynamics parameters like filament–filament annealing still increase following MAMP treatment in the adf4 mutant.
Hallmarks of Innate Immunity Are Perturbed in the adf4 Mutant
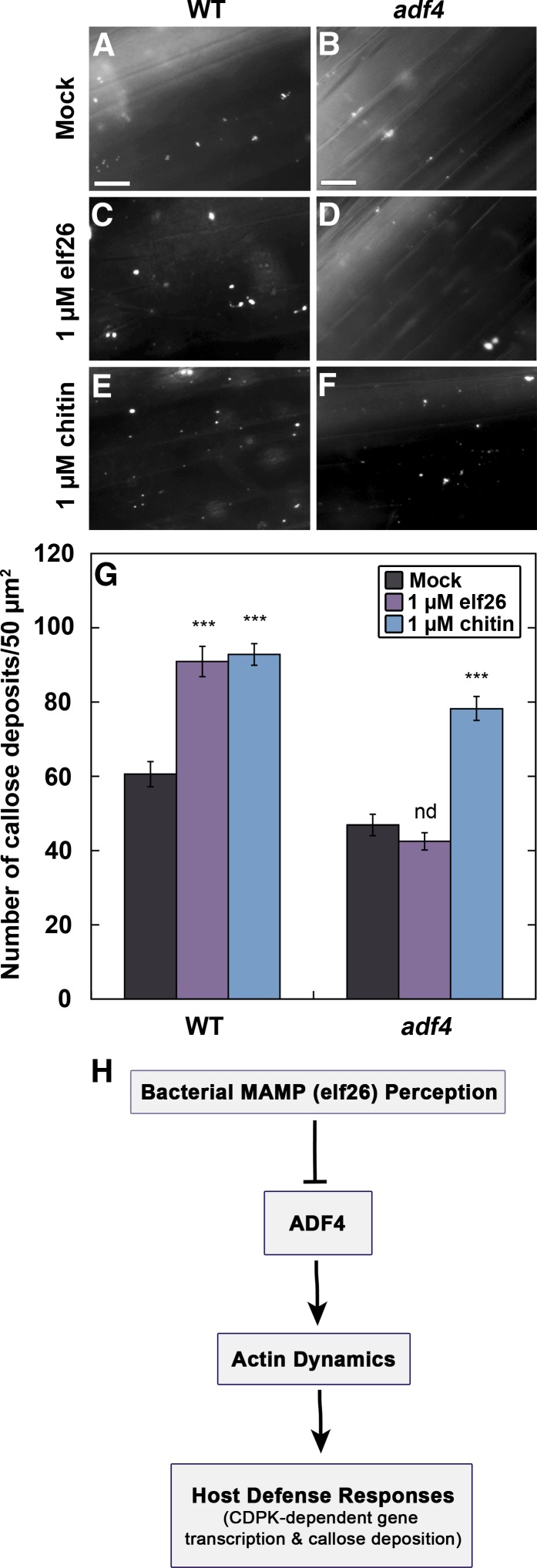
During plant innate immunity, several fundamental cellular responses that might require actin cytoskeleton remodeling have been described, including transcriptional activation, generation of ROS and other antimicrobial compounds, directed vesicle trafficking, and fortification of the cell wall (Day et al., 2011). To test which, if any, of these processes are downstream of ADF4 and actin remodeling, we examined the adf4 mutant following treatment with bacterial and fungal MAMPs. The deposition of the β-1,3-glucan polymer callose fortifies the cell wall hours into the host immune response to bacteria and fungi and requires a functional actin cytoskeleton (Skalamera et al., 1997; Koh et al., 2005). Wild-type hypocotyls responded to both elf26 and chitin treatments with a significant increase in callose deposition (Figures 5C, 5E, and 5G). Notably, the adf4 mutant displays a marked reduction in callose deposits compared with wild-type cells (Figures 5A, 5B, and 5G). Furthermore, the adf4 mutant fails to increase callose deposition in response to elf26 but responds normally to chitin. This provides additional strong evidence for distinct defense signaling pathways that use the actin cytoskeleton during innate immunity. These results confirm that actin array remodeling and ADF4 are necessary for efficient callose deposition during the response to elf26 (Figure 5H).
Figure 5.
Callose Deposition Fails to Occur in the adf4 Mutant following elf26 Treatment.
(A) to (F) Callose deposits in epidermal cells from the base of wild-type hypocotyls appeared to increase following treatment with 1 µM elf26 (C) or 1 µM chitin (E) compared with mock control (A). Callose deposits in the adf4 mutant (B) appeared less abundant than in mock-treated WT cells (A). Furthermore, the number of callose deposits did not appear to increase in adf4 following 1 µM elf26 treatment (D), whereas they did increase following 1 µM chitin treatment (F). Bars = 20 µm.
(G) Quantification of aniline blue-stained spots demonstrates that callose deposition was inhibited in the adf4 mutant following 1 µM elf26 treatment. Values given are means ±se (n = 50 images per treatment, from at least 30 hypocotyls). Asterisks represent significant differences by ANOVA, with Tukey HSD posthoc analysis (nd = not significantly different from mock; ***P < 0.001).
(H) A model describing the role of ADF4 during innate immune signaling.
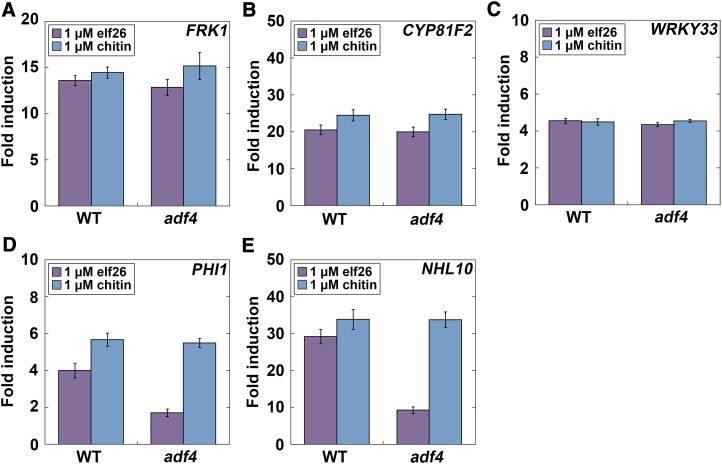
Another feature of innate immune activation is the rapid modulation of transcriptional programming for defense-related target genes through distinct and overlapping signaling pathways, including the MAPK and calcium-dependent protein kinase (CDPK) cascades, which fine-tune multiple aspects of plant immune and stress responses (Boller and Felix, 2009; Boudsocq et al., 2010; Tena et al., 2011; Boudsocq and Sheen, 2013). To define this process, and to investigate whether the adf4 mutant had an altered transcriptional response to MAMP treatment compared with the wild type, we measured gene transcripts by quantitative RT-PCR for the MAPK and CDPK pathways (Figure 6). After 1-h MAMP treatment, target gene transcripts of the MAPK signaling pathway (Backues et al., 2010; Boudsocq et al., 2010; Mao et al., 2011), including the MAPK-specific target gene FRK1 (Figure 6A), the MAPK-dominant gene CYP81F2 (Figure 6B), and a downstream indicator of MAPK signaling, WRKY33 (Figure 6C; Boudsocq et al., 2010), were elevated in both wild-type and adf4 seedlings following treatment with both elf26 and chitin. This suggests that ADF4 and actin remodeling are not upstream of transcriptional activation in the MAPK-dependent pathway and is consistent with previous results showing that FRK1 responded normally to flg22 treatment of light-grown plants (Porter et al., 2012). We also measured target gene transcripts for a CDPK-dependent pathway, including the CDPK-specific gene PHOSPHATE-INDUCED1 (PHI1) and the CDPK-synergistic gene HIN1-like10 (NHL10; Boudsocq et al., 2010), to assess the contribution of early defense transcription (Figures 6D and 6E). Notably, the induction of PHI1 (Figure 6D) and NHL10 (Figure 6E) transcription was markedly attenuated in the adf4 mutant following treatment with elf26 but not with chitin. In summary, these data indicate that ADF4 functions upstream of CDPK-dependent target gene transcription during plant innate immune signaling (Figure 5H).
Figure 6.
Transcriptional Activation in the CDPK Pathway Is Attenuated in adf4 following elf26 Treatment.
Quantitative analysis of marker genes for early defense signaling in the MAPK and CDPK pathways.
(A) to (C) A MAPK-specific reporter gene, FRK1 (A), as well as the MAPK dominant-pathway genes CYP81F2 (B) and WRKY33 (C), were induced in both wild-type and adf4 plants following treatment with elf26 or chitin.
(D) and (E) PHI1, a CDPK-specific response gene (D), and the CDPK-synergistic pathway gene, NHL10 (E), were induced in the wild type following treatment with elf26; however, PHI1 and NHL10 induction was markedly reduced in the adf4 mutant. By contrast, both wild-type and adf4 plants responded with increased PHI1 and NHL10 transcripts following treatment with chitin. Expression of each defense signaling gene and the housekeeping gene GAPD were absent from controls lacking reverse transcriptase (data not shown). Mean values from triplicate biological samples and technical replications are plotted ±se, normalized to GAPD expression and presented as fold induction from mock. Defense gene expression was significantly increased following treatment with either elf26 or chitin on wild-type and the adf4 mutant seedlings compared with mock-treated controls (P < 0.001, ANOVA with Tukey HSD posthoc analysis).
DISCUSSION
Precise regulation of the host actin cytoskeleton is a critical facet of innate immunity in both plants and animals. Here, we established a novel system to dissect, at high spatial and temporal resolution, the changes to actin cytoskeletal organization and turnover that occur during plant innate immune signaling. Within minutes following treatment with a bacterial MAMP, elf26, we observed a significant increase in filament abundance and altered filament dynamics. Aspects of single-filament turnover were also significantly changed, including an increase in filament lengths and lifetimes, a 4-fold increase in filament–filament annealing, and a 2-fold reduction in severing frequency. These changes are similar to the phenotype of an Arabidopsis adf4 knockout mutant (Henty et al., 2011). Importantly, the adf4 mutant failed to undergo actin rearrangements in response to treatment with elf26 but did respond to chitin. Therefore, we infer that ADF4 activity is negatively regulated during innate immune signaling downstream of elf26 perception. Furthermore, callose deposition and CDPK-dependent target gene transcription were markedly reduced in cells of the adf4 mutant after elf26 treatment. These results provide strong genetic and cytological evidence that the inhibition of ADF/cofilin (AC) activity regulates actin dynamics and actin-dependent processes during innate immune signaling (Figure 5H).
The AC family is regarded as a central regulator of actin filament turnover in eukaryotes (Maloney et al., 2008). Phosphorylation on a conserved Ser residue, pH changes, fluxes in phosphoinositide lipids, and interactions with other proteins modulate AC activity (Maloney et al., 2008). Moreover, the stoichiometry of AC to actin dictates its major activities within cells. At low concentrations of AC, filament disassembly is favored, whereas at high concentrations, filament nucleation occurs (Andrianantoandro and Pollard, 2006; Chan et al., 2009). Although AC-facilitated filament disassembly was originally thought to occur through pointed-end depolymerization (Carlier et al., 1997), most disassembly in vivo likely occurs through severing activity (Staiger et al., 2009; Henty et al., 2011). Severing activity mediated by AC also increases the number of free barbed ends; therefore, when there is a large pool of subunits available for assembly, this can result in net actin polymerization. This unconventional role of AC promotes local actin filament assembly and can lead to directed cell motility events (Ghosh et al., 2004).
Investigation of the divergent roles of ADF and cofilin in a mouse model system demonstrate specialized roles for AC during innate immunity (Jönsson et al., 2012). Nonmuscle cofilin is required for antigen presentation, macrophage motility, and polarity, whereas ADF appears to be dispensable for macrophage motility and antigen presentation but contributes to cell shape and polarity (Jönsson et al., 2012). These data suggest cofilin drives dynamic actin rearrangements for receptor availability in mammalian immune signaling cascades. Dynamic actin rearrangements conferred by AC have also been shown to facilitate effector-mediated internalization of bacterial pathogens into host mammalian cells. For example, internalization of Salmonella requires the strict regulation of AC through cycles of phosphorylation and dephosphorylation. To compensate for an increase in actin filaments, AC is transiently activated by the host protein phosphatase, slingshot, and then deactivated by LIM kinase–induced phosphorylation (Dai et al., 2004). Later, AC is recruited to host cell membrane ruffles where it is indirectly activated by the effector protein SopE, through Rho-GTPase Cdc42 activation, to promote Salmonella entry (Dai et al., 2004). By contrast, Listeria and Rickettsia use effector proteins to recruit actin nucleation machinery directly to the bacterial surface from the host cytoplasm and use the mechanical forces generated to drive intercellular motility and to facilitate the propagation of bacteria to neighboring cells (Goley and Welch, 2006). Simple biomimetic systems have recapitulated these polymerization-based motility machines on the surface of polystyrene beads or bacteria in vitro (Loisel et al., 1999; Reymann et al., 2012). In addition to the Arp2/3 complex, the activities of capping protein and AC are required for sustained comet tail motility of beads (Loisel et al., 1999; Reymann et al., 2012). Although two different nucleation systems are used by intracellular pathogens like Rickettsia and Listeria, both require AC activity for actin filament turnover during pathogenesis (Goley and Welch, 2006). By contrast, bacterial phytopathogens do not display intracellular motility and are not engulfed through phagocytosis by plant cells.
Plant cells often respond to diverse microbes and elicitors with increased actin filament abundance or filament bundling (Takemoto and Hardham, 2004; Henty-Ridilla et al., 2013a). Here, we show a rapid production of actin filaments in epidermal cells following application of a bacterial MAMP, elf26, and the fungal MAMP, chitin, on dark-grown hypocotyls. However, additional microbial signals appear to elicit distinct changes to cellular actin arrays. Studies investigating the interaction between plant cells and nonpathogenic, mutualistic bacteria or Nod factor elicitors suggest that actin polymerization occurs in order to promote host–microbe interactions (Crdenas et al., 1998; Cárdenas et al., 2003). Significantly, overexpression of AC or treatment with actin disrupting drugs facilitates the invasion of plant tissues by several fungal and bacterial species (Miklis et al., 2007; Porter et al., 2012; Henty-Ridilla et al., 2013a). In contrast with increased actin abundance during host–microbe interactions, certain signals from pathogenic fungi like the Verticillium dahlia toxin or the bacterial Harpin elicitor can stimulate the dose-dependent destruction of cortical actin filaments (Yuan et al., 2006; Qiao et al., 2010). Moreover, a study with flg22 treatment of tobacco (Nicotiana tabacum) Bright Yellow-2 cells failed to demonstrate any quantitative changes in actin organization, perhaps due to cell type–specific signaling cascades (Guan et al., 2013). Gross rearrangements in actin cytoskeletal architecture often correlate with the altered dynamics of single actin filaments (Henty et al., 2011; Li et al., 2012; Tóth et al., 2012; Henty-Ridilla et al., 2013b). The incessant rearrangement of the cortical actin cytoskeleton has been hypothesized to function as a surveillance mechanism that directly links actin remodeling events, such as those observed during plant innate immunity, to signaling cascades (Staiger et al., 2009; Day et al., 2011). The bacterial effector protein AvrPphB is predicted to target ADF4 through the control of the cognate resistance-gene RESISTANT TO P. SYRINGAE5 to circumvent defense signaling during the plant immune response (Porter et al., 2012). Furthermore, the loss of ADF4 results in a reduction in late defense gene transcription (Porter et al., 2012) and failure to activate effector-triggered immunity (Tian et al., 2009). Here, we show that an increase in actin filament abundance during PTI correlates with a reduction in single filament turnover, which we infer is due to downregulation of ADF4 activity. Genetic ablation of ADF4 abrogates the elf26-induced increase in actin filament abundance, cell wall fortification by callose, and CDPK-dependent target gene transcription. Because all of these responses are normal in the adf4 mutant following treatment with the fungal MAMP chitin, ADF4 is likely part of a distinct pathway used for actin remodeling in response to elf26 perception. Collectively, these results implicate a key ABP in modulating actin dynamics during plant innate immune signaling.
The regulation of actin dynamics in plant cells requires the coordinated activity of numerous ABPs in addition to AC (Smertenko et al., 2010; Staiger et al., 2010). Therefore, besides downregulation of filament turnover, additional mechanisms may also elicit an increase in actin filament abundance during the plant defense response. Perhaps the most efficient way to elicit an increase in actin filament abundance is to stimulate actin polymerization. Indeed, using LatB or cytochalasins, several studies have demonstrated the importance of actin polymerization during the response to beneficial microbes as well as pathogens (Heath et al., 1997; Cárdenas et al., 2006; Hardham et al., 2007; Chang and Nick, 2012; Henty-Ridilla et al., 2013a). An increase in the availability of filament plus ends through a reduction in barbed-end capping also results in enhanced filament–filament annealing, as well as an increase in maximum filament lengths and lifetimes (Li et al., 2012). Such a mechanism could contribute to the increase in actin abundance during innate immune signaling and may explain why filament–filament annealing still changed following elf26 treatment of the adf4 mutant. Moreover, the cellular signals that occur within seconds or minutes of MAMP perception could regulate both AC and other ABPs (Chinchilla et al., 2007; Heese et al., 2007; Boller and Felix, 2009; Zipfel and Robatzek, 2010; Segonzac and Zipfel, 2011). The rapid increase in filament abundance is likely not due to the acidification of the cytosol that occurs within seconds of pathogen perception (Day et al., 2011) because the activity of AC is enhanced at decreased pH (Allwood et al., 2002). However, the downregulation of AC activity by phosphorylation, increased ROS levels, or fluxes in phosphoinositide lipids could all result in an increase in actin filament abundance (Maloney et al., 2008). The inactivation of plant AC during innate immunity most likely occurs by the phosphorylation of Ser-6. Upon stimulation with a MAMP, both nonphosphorylatable ADF4-S6A and the phosphomimic ADF4-S6D localize to the nucleus, and this localization correlates with the reduction of defense gene transcripts (Porter et al., 2012). Notably, the LIM or TESK family kinases that directly phosphorylate AC in many eukaryotic systems do not occur in plants (Bernard, 2007). Consequently, AC activity is likely regulated through other kinases, such as the CDPKs, which can phosphorylate AC in plant and animal systems in vitro (Allwood et al., 2001; Bernard, 2007; Maloney et al., 2008). Unfortunately, the kinase responsible for the phosphorylation of AC in plant cells has not been identified (Allwood et al., 2001; Tian et al., 2009; Porter et al., 2012).
The exact purpose for an increase in actin filament abundance during plant immunity remains to be fully addressed. In response to both pathogenic and nonpathogenic fungi, a focal increase in actin filaments and bundles occurs under the site of attempted fungal penetration (Takemoto and Hardham, 2004; Hardham et al., 2007), presumably to serve as tracks for the deposition of callose, secretion of Golgi-derived antimicrobial products, and vectorial vesicle delivery. Here, we showed that the adf4 mutant has reduced callose levels and elf26 treatment does not stimulate additional callose deposition. This, and previous pharmacological data, demonstrate that actin dynamics are an important facet of cell wall fortification during the response to microbes (Heath et al., 1997; Skalamera et al., 1997). Callose deposition often takes hours to accumulate in the host cell wall during pathogenesis. Although cells treated with bacteria or MAMPs do not elicit a strong focal response but also require wall fortification, actin is likely required for additional cellular processes during plant innate immunity. Based on work from mammalian and yeast cells, a rapid increase in actin filament abundance can be predicted to facilitate the endocytosis of PRRs. However, the mechanisms controlling the dynamic rearrangements of the actin cytoskeleton during endocytosis in plant cells are unclear. Pharmacological and colocalization studies imply that the actin-myosin system is required for FLS2 receptor internalization and/or vesicle trafficking (Robatzek et al., 2006; Beck et al., 2012). By contrast, the inhibition of clathrin-mediated endocytosis in a Rho-GTPase mutant correlates with an accumulation of actin filaments (Nagawa et al., 2012). Upon treatment with LatB, actin filaments are disrupted and normal clathrin-mediated endocytosis is restored (Nagawa et al., 2012). Impaired plant growth and development has been linked to the constitutive activation of immune responses mediated by the FLS2 and EFR signaling pathways in the stomatal cytokinesis-defective1 mutant (Korasick et al., 2010). Notably, scd1 mutants display attenuated immune responses to MAMPs, including reduced ROS levels and reduced seedling growth inhibition, but enhanced resistance to bacterial pathogens and normal accumulation of defense gene transcripts (Korasick et al., 2010). Components of the FLS2 (Henty-Ridilla et al., 2013a) and EFR receptor complexes (this work) are required for an increase in actin filament abundance following MAMP treatments. Broadly speaking, our data demonstrate that AC is a target of bacterial MAMP (elf26) signaling and a key regulator of actin filament dynamics, cell wall fortification, and CDPK-dependent gene transcription during the plant innate immune response. Further genetic dissection of these actin responses at high spatial and temporal resolution will facilitate the dissection of actin-based processes during plant pathogenesis.
METHODS
Plant Material and Growth
The homozygous efr-1 mutant (SALK_044334; Alonso et al., 2003; Zipfel et al., 2004) and the homozygous adf1 mutant (SALK_144459) of Arabidopsis thaliana were transformed with a binary vector for the actin reporter GFP-fABD2 (Sheahan et al., 2004) using the floral dip method (Zhang et al., 2006). Transformed plants were screened on 0.5× Murashige and Skoog medium containing kanamycin and with fluorescence microscopy. In this study, T3 plants were used for actin architecture analysis. Wild-type plants (Columbia-0) as well as the adf4 (GARLIC_823_A11.b.1b.Lb3Fa), fls2 (SALK_062054), bak1-4 (SALK_116202), and bik1 (SALK_005291) homozygous mutants expressing GFP-fABD2 were prepared and characterized previously (Staiger et al., 2009; Henty et al., 2011; Henty-Ridilla et al., 2013a).
Arabidopsis seeds were surface sterilized and stratified for 3 d at 4°C on agar plates containing 0.5× Murashige and Skoog medium supplemented with 1% (w/v) Suc. Stratified seeds were exposed to 4 h of light, and the plates were wrapped in three layers of aluminum foil and transferred to a growth chamber. Seedlings were germinated and grown in the dark at 21°C for 5 d. Plants used for measuring bacterial growth were sown into soil and grown under long-day conditions (16 h light/8 h dark) for 24 d.
Quantitative Analyses of Cortical Actin Filament Architecture
Actin filament abundance (density) and the extent of filament bundling (skewness) were measured as described previously (Higaki et al., 2010; Henty et al., 2011; Li et al., 2012). Briefly, VAEM was used to collect single optical sections from the cortical cytoplasm of 5-d-old hypocotyl epidermal cells expressing GFP-fABD2 (Staiger et al., 2009). VAEM was performed using a TIRF illuminator mounted on an IX-71 microscope equipped with a ×60 1.45–numerical aperture PlanApo TIRF objective (Olympus). Illumination was from a solid-state 50-mW laser (Intelligent Imaging Innovations) attenuated to 10% power. The 488-nm laser emission was captured with an electron multiplying charge-coupled device camera (ORCA-EM C9100-12; Hamamatsu Photonics). The microscope platform was operated and images collected with Slidebook software (version 5.5.0; Intelligent Imaging Innovations). A fixed exposure and gain were selected so that single actin filaments could be seen but higher order filament structures were not intensity saturated. Micrographs were cropped and analyzed in Fiji (http://fiji.sc/wiki/index.php/downloads) for the percentage of occupancy of GFP signal (density) or the difference from a Gaussian distribution of pixel intensities (skewness) in a given micrograph (Higaki et al., 2010). Because VAEM generates high-contrast, low-background images, there was no need to skeletonize images or to apply a Gaussian blur or a high-band-pass filter to the data (Henty et al., 2011; Li et al., 2012). At least 300 images of hypocotyl epidermal cells per treatment or genotype, from at least 30 individual seedlings, were collected and analyzed.
Time-Lapse Imaging of Actin Filament Dynamics
The cortical actin cytoskeleton in epidermal cells from 5-d-old dark-grown hypocotyls expressing GFP-fABD2 was imaged by time-lapse VAEM as described previously (Staiger et al., 2009). The MAMP peptides, flg22 (Meindl et al., 2000) or elf26 (Kunze et al., 2004), both from NeoBioSci, as well as chitin (C9752; Sigma-Aldrich), were diluted in 1× PBS at various concentrations, and 5-d-old seedlings were mounted in the presence of MAMPs for no more than 15 min. Filament severing frequency, maximum filament length, filament lifetime, filament origin, and elongation rates were measured as described previously, as was tracking the individual behavior of newly created filament ends following severing, regrowth, and annealing (Staiger et al., 2009; Henty et al., 2011; Li et al., 2012).
Quantification of Callose Deposition
Callose deposits in hypocotyl epidermal cells were measured as described previously (Kim et al., 2009) with the following changes: Hypocotyls were soaked in 1 µM MAMP for 16 h prior to aniline blue staining, and epidermal cells were viewed with epifluorescence microscopy. Hypocotyls were not cleared with lacto-phenol prior to staining. The mean number of callose spots present in micrographs from the base of the hypocotyl was counted for at least 50 hypocotyls per treatment. Epifluorescence microscopy was performed using a Nikon E600 equipped with a ×40 0.75–numerical aperture PlanFluor objective. Illumination was from a 100-W Hg lamp, and light was filtered through a 360/340-nm filter. Images were captured with a charge-coupled device camera (ORCA-ER C4742-95; Hamamatsu Photonics) and Metamorph software (version 4.6r9).
Real-Time Quantitative PCR
Five-day-old dark-grown seedlings soaked with MAMPs for 1 h were flash frozen in liquid nitrogen and ground to a fine powder. RNA isolation was performed with TRIzol according to the manufacturer’s instructions (Invitrogen). Two-step quantitative RT-PCR was performed using 2× SYBR Green master mix (Qiagen), normalized to GAPD transcript levels, and analyzed with Excel software as described previously (Henty et al., 2011). Gene-specific primers are described in Supplemental Table 2.
Statistical Analyses
Mean values and standard errors were calculated using Microsoft Excel (version 14.2.2). Statistical significance was assessed by ANOVA with Tukey honestly significant difference (HSD) posthoc analysis or Student’s t test, and histograms were plotted with KaleidaGraph (version 4.1.3b1; Synergy Software).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: ADF4, At5g59890; FLS2, At5g46330; EFR-1, At5g20480; BAK1-4, At4g33430; BIK1, At2g39660; and ADF1, At3g4610.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Dose-Dependent Changes in Cortical Actin Arrays Are Elicited by elf26 Treatment.
Supplemental Figure 2. FLAGELLIN SENSING2 Is Not Expressed in Dark-Grown Arabidopsis Seedlings.
Supplemental Figure 3. A 35S:ADF4;adf4 Rescue Line Responds to elf26 Treatment.
Supplemental Figure 4. The Homozygous adf1 Mutant Is a Knockout Line.
Supplemental Figure 5. The adf1 Mutant Fails to Respond to elf26 or Chitin Treatment.
Supplemental Table 1. Actin Dynamics Parameters from Mock and elf26-Treated adf4 Hypocotyl Epidermal Cells.
Supplemental Table 2. Gene-Specific Primers Used for Real-Time Quantitative PCR.
Supplementary Material
Acknowledgments
We thank members of the Staiger lab as well as Daoguo Zhou (Purdue University), Bruce Goode (Brandeis University), and the Antje Heese lab (University of Missouri) for valuable feedback during the course of this study. Seed stocks used in this study were generously provided by Tesfaye Mengiste (Purdue University), Ping He (Texas A&M University), Silke Robatzek (The Sainsbury Laboratory), and the ABRC (Ohio State University). We also thank Hongbing Luo for the excellent care and maintenance of plant materials. This research was supported by a collaborative grant from the U.S. National Science Foundation–Arabidopsis 2010 Program (IOS-1021185 and IOS-1021044) to C.J.S. and B.D., respectively.
AUTHOR CONTRIBUTIONS
J.L.H.-R. and C.J.S. conceptualized and designed experiments. J.L. and B.D. provided valuable input. J.L.H.-R. and J.L. performed experiments and data analysis. J.L.H.-R. and C.J.S. wrote the article.
Glossary
- TLR
toll-like receptor
- MAMP
microbe-associated molecular pattern
- DC
dendritic cell
- PTI
pattern-triggered immunity
- ROS
reactive oxygen species
- LatB
latrunculin B
- VAEM
Variable-angle epifluorescence microscopy
- PRR
pattern recognition receptor
- MAPK
mitogen-activated protein kinase
- CDPK
calcium-dependent protein kinase
- HSD
honestly significant difference
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Allwood E.G., Anthony R.G., Smertenko A.P., Reichelt S., Drøbak B.K., Doonan J.H., Weeds A.G., Hussey P.J. (2002). Regulation of the pollen-specific actin-depolymerizing factor LlADF1. Plant Cell 14: 2915–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allwood E.G., Smertenko A.P., Hussey P.J. (2001). Phosphorylation of plant actin-depolymerising factor by calmodulin-like domain protein kinase. FEBS Lett. 499: 97–100 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Andrianantoandro E., Pollard T.D. (2006). Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24: 13–23 [DOI] [PubMed] [Google Scholar]
- Backues S.K., Korasick D.A., Heese A., Bednarek S.Y. (2010). The Arabidopsis dynamin-related protein2 family is essential for gametophyte development. Plant Cell 22: 3218–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Zhou J., Faulkner C., MacLean D., Robatzek S. (2012). Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 24: 4205–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard O. (2007). Lim kinases, regulators of actin dynamics. Int. J. Biochem. Cell Biol. 39: 1071–1076 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Boujemaa-Paterski R., Henty J.L., Khurana P., Staiger C.J. (2010). Actin dynamics in plant cells: A team effort from multiple proteins orchestrates this very fast-paced game. Curr. Opin. Plant Biol. 13: 714–723 [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Willmann M.R., McCormack M., Lee H., Shan L., He P., Bush J., Cheng S.-H., Sheen J. (2010). Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M., Sheen J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18: 30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas L., McKenna S.T., Kunkel J.G., Hepler P.K. (2006). NAD(P)H oscillates in pollen tubes and is correlated with tip growth. Plant Physiol. 142: 1460–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas L., Thomas-Oates J.E., Nava N., López-Lara I.M., Hepler P.K., Quinto C. (2003). The role of nod factor substituents in actin cytoskeleton rearrangements in Phaseolus vulgaris. Mol. Plant Microbe Interact. 16: 326–334 [DOI] [PubMed] [Google Scholar]
- Carlier M.F., Laurent V., Santolini J., Melki R., Didry D., Xia G.X., Hong Y., Chua N.H., Pantaloni D. (1997). Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: Implication in actin-based motility. J. Cell Biol. 136: 1307–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crdenas L., Vidali L., Domnguez J., Prez H., Snchez F., Hepler P.K., Quinto C. (1998). Rearrangement of actin microfilaments in plant root hairs responding to rhizobium etli nodulation signals. Plant Physiol. 116: 871–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C., Beltzner C.C., Pollard T.D. (2009). Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr. Biol. 19: 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang X., Nick P. (2012). Defence signalling triggered by Flg22 and Harpin is integrated into a different stilbene output in Vitis cells. PLoS ONE 7: e40446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D.G., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Dai S., Sarmiere P.D., Wiggan O., Bamburg J.R., Zhou D. (2004). Efficient Salmonella entry requires activity cycles of host ADF and cofilin. Cell. Microbiol. 6: 459–471 [DOI] [PubMed] [Google Scholar]
- Day B., Henty J.L., Porter K.J., Staiger C.J. (2011). The pathogen-actin connection: A platform for defense signaling in plants. Annu. Rev. Phytopathol. 49: 483–506 [DOI] [PubMed] [Google Scholar]
- Dong C.H., Xia G.X., Hong Y., Ramachandran S., Kost B., Chua N.H. (2001). ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13: 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M., Song X.Y., Mouneimne G., Sidani M., Lawrence D.S., Condeelis J.S. (2004). Cofilin promotes actin polymerization and defines the direction of cell motility. Science 304: 743–746 [DOI] [PubMed] [Google Scholar]
- Goley E.D., Welch M.D. (2006). The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7: 713–726 [DOI] [PubMed] [Google Scholar]
- Gordón-Alonso M., Veiga E., Sánchez-Madrid F. (2010). Actin dynamics at the immunological synapse. Cell Health Cytoskelet. 2: 33–47 [Google Scholar]
- Granucci F., Vizzardelli C., Pavelka N., Feau S., Persico M., Virzi E., Rescigno M., Moro G., Ricciardi-Castagnoli P. (2001). Inducible IL-2 production by dendritic cells revealed by global gene expression analysis. Nat. Immunol. 2: 882–888 [DOI] [PubMed] [Google Scholar]
- Guan X., Buchholz G., Nick P. (2013). The cytoskeleton is disrupted by the bacterial effector HrpZ, but not by the bacterial PAMP flg22, in tobacco BY-2 cells. J. Exp. Bot. 64: 1805–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardham A.R., Jones D.A., Takemoto D. (2007). Cytoskeleton and cell wall function in penetration resistance. Curr. Opin. Plant Biol. 10: 342–348 [DOI] [PubMed] [Google Scholar]
- Heath M.C., Nimchuk Z.L., Xu H. (1997). Plant nuclear migrations as indicators of critical interactions between resistant or susceptible cowpea epidermal cells and invasion hyphae of the cowpea rust fungus. New Phytol. 135: 689–700 [Google Scholar]
- Heese A., Hann D.R., Gimenez-Ibanez S., Jones A.M.E., He K., Li J., Schroeder J.I., Peck S.C., Rathjen J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henty J.L., Bledsoe S.W., Khurana P., Meagher R.B., Day B., Blanchoin L., Staiger C.J. (2011). Arabidopsis actin depolymerizing factor4 modulates the stochastic dynamic behavior of actin filaments in the cortical array of epidermal cells. Plant Cell 23: 3711–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henty-Ridilla J.L., Li J., Blanchoin L., Staiger C.J. (2013b). Actin dynamics in the cortical array of plant cells. Curr. Opin. Plant Biol. 16: 678–687 [DOI] [PubMed] [Google Scholar]
- Henty-Ridilla J.L., Shimono M., Li J., Chang J.H., Day B., Staiger C.J. (2013a). The plant actin cytoskeleton responds to signals from microbe-associated molecular patterns. PLoS Pathog. 9: e1003290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T., Kutsuna N., Sano T., Kondo N., Hasezawa S. (2010). Quantification and cluster analysis of actin cytoskeletal structures in plant cells: role of actin bundling in stomatal movement during diurnal cycles in Arabidopsis guard cells. Plant J. 61: 156–165 [DOI] [PubMed] [Google Scholar]
- Huang Q., Liu D., Majewski P., Schulte L.C., Korn J.M., Young R.A., Lander E.S., Hacohen N. (2001). The plasticity of dendritic cell responses to pathogens and their components. Science 294: 870–875 [DOI] [PubMed] [Google Scholar]
- Irving A.T., Wang D., Vasilevski O., Latchoumanin O., Kozer N., Clayton A.H., Szczepny A., Morimoto H., Xu D., Williams B.R., Sadler A.J. (2012). Regulation of actin dynamics by protein kinase R control of gelsolin enforces basal innate immune defense. Immunity 36: 795–806 [DOI] [PubMed] [Google Scholar]
- Janeway C.A., Jr, Medzhitov R. (2002). Innate immune recognition. Annu. Rev. Immunol. 20: 197–216 [DOI] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jönsson F., Gurniak C.B., Fleischer B., Kirfel G., Witke W. (2012). Immunological responses and actin dynamics in macrophages are controlled by N-cofilin but are independent from ADF. PLoS ONE 7: e36034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana P., Henty J.L., Huang S., Staiger A.M., Blanchoin L., Staiger C.J. (2010). Arabidopsis VILLIN1 and VILLIN3 have overlapping and distinct activities in actin bundle formation and turnover. Plant Cell 22: 2727–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.G., Geng X., Lee S.Y., Mackey D. (2009). The Pseudomonas syringae type III effector AvrRpm1 induces significant defenses by activating the Arabidopsis nucleotide-binding leucine-rich repeat protein RPS2. Plant J. 57: 645–653 [DOI] [PubMed] [Google Scholar]
- Koh S., André A., Edwards H., Ehrhardt D., Somerville S. (2005). Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J. 44: 516–529 [DOI] [PubMed] [Google Scholar]
- Korasick D.A., McMichael C., Walker K.A., Anderson J.C., Bednarek S.Y., Heese A. (2010). Novel functions of Stomatal Cytokinesis-Defective 1 (SCD1) in innate immune responses against bacteria. J. Biol. Chem. 285: 23342–23350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G., Zipfel C., Robatzek S., Niehaus K., Boller T., Felix G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Henty-Ridilla J.L., Huang S., Wang X., Blanchoin L., Staiger C.J. (2012). Capping protein modulates the dynamic behavior of actin filaments in response to phosphatidic acid in Arabidopsis. Plant Cell 24: 3742–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel T.P., Boujemaa R., Pantaloni D., Carlier M.-F. (1999). Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature 401: 613–616 [DOI] [PubMed] [Google Scholar]
- Ma L., Sun N., Liu X., Jiao Y., Zhao H., Deng X.W. (2005). Organ-specific expression of Arabidopsis genome during development. Plant Physiol. 138: 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney, M.T., Kinley, A.W., Pak, C.W., and Bamburg, J.R. (2008). ADF/cofilin, actin dynamics, and disease. In Actin-Binding Proteins and Disease, C.G. Remedios and D. Chhabra, eds (New York: Springer), pp. 83–187. [Google Scholar]
- Mao G., Meng X., Liu Y., Zheng Z., Chen Z., Zhang S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzinger P. (2002). The danger model: A renewed sense of self. Science 296: 301–305 [DOI] [PubMed] [Google Scholar]
- Meindl T., Boller T., Felix G. (2000). The bacterial elicitor flagellin activates its receptor in tomato cells according to the address-message concept. Plant Cell 12: 1783–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklis M., Consonni C., Bhat R.A., Lipka V., Schulze-Lefert P., Panstruga R. (2007). Barley MLO modulates actin-dependent and actin-independent antifungal defense pathways at the cell periphery. Plant Physiol. 144: 1132–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyahara A., Richens J., Starker C., Morieri G., Smith L., Long S., Downie J.A., Oldroyd G.E. (2010). Conservation in function of a SCAR/WAVE component during infection thread and root hair growth in Medicago truncatula. Mol. Plant Microbe Interact. 23: 1553–1562 [DOI] [PubMed] [Google Scholar]
- Nagawa S., Xu T., Lin D., Dhonukshe P., Zhang X., Friml J., Scheres B., Fu Y., Yang Z. (2012). ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biol. 10: e1001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Cooper J.A. (2009). Actin, a central player in cell shape and movement. Science 326: 1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter K., Shimono M., Tian M., Day B. (2012). Arabidopsis Actin-Depolymerizing Factor-4 links pathogen perception, defense activation and transcription to cytoskeletal dynamics. PLoS Pathog. 8: e1003006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao F., Chang X.-L., Nick P. (2010). The cytoskeleton enhances gene expression in the response to the Harpin elicitor in grapevine. J. Exp. Bot. 61: 4021–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymann A.-C., Boujemaa-Paterski R., Martiel J.-L., Guérin C., Cao W., Chin H.F., De La Cruz E.M., Théry M., Blanchoin L. (2012). Actin network architecture can determine myosin motor activity. Science 336: 1310–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S., Chinchilla D., Boller T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka D.R., Kandasamy M.K., McKinney E.C., Burgos-Rivera B., Meagher R.B. (2007). The ancient subclasses of Arabidopsis Actin Depolymerizing Factor genes exhibit novel and differential expression. Plant J. 52: 460–472 [DOI] [PubMed] [Google Scholar]
- Segonzac C., Zipfel C. (2011). Activation of plant pattern-recognition receptors by bacteria. Curr. Opin. Microbiol. 14: 54–61 [DOI] [PubMed] [Google Scholar]
- Sheahan M.B., Staiger C.J., Rose R.J., McCurdy D.W. (2004). A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 136: 3968–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalamera D., Jibodh S., Heath M.C. (1997). Callose deposition during the interaction between cowpea (Vigna unguiculata) and the monokaryotic stage of the cowpea rust fungus (Uromyces vignae). New Phytol. 136: 511–524 [DOI] [PubMed] [Google Scholar]
- Smertenko A.P., Deeks M.J., Hussey P.J. (2010). Strategies of actin reorganisation in plant cells. J. Cell Sci. 123: 3019–3028 [DOI] [PubMed] [Google Scholar]
- Staiger C.J., Poulter N.S., Henty J.L., Franklin-Tong V.E., Blanchoin L. (2010). Regulation of actin dynamics by actin-binding proteins in pollen. J. Exp. Bot. 61: 1969–1986 [DOI] [PubMed] [Google Scholar]
- Staiger C.J., Sheahan M.B., Khurana P., Wang X., McCurdy D.W., Blanchoin L. (2009). Actin filament dynamics are dominated by rapid growth and severing activity in the Arabidopsis cortical array. J. Cell Biol. 184: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart L.M., Paquette N., Boyer L. (2013). Effector-triggered versus pattern-triggered immunity: How animals sense pathogens. Nat. Rev. Immunol. 13: 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto D., Hardham A.R. (2004). The cytoskeleton as a regulator and target of biotic interactions in plants. Plant Physiol. 136: 3864–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena G., Boudsocq M., Sheen J. (2011). Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 14: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M., Chaudhry F., Ruzicka D.R., Meagher R.B., Staiger C.J., Day B. (2009). Arabidopsis actin-depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB. Plant Physiol. 150: 815–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth R., Gerding-Reimers C., Deeks M.J., Menninger S., Gallegos R.M., Tonaco I.A.N., Hübel K., Hussey P.J., Waldmann H., Coupland G. (2012). Prieurianin/endosidin 1 is an actin-stabilizing small molecule identified from a chemical genetic screen for circadian clock effectors in Arabidopsis thaliana. Plant J. 71: 338–352 [DOI] [PubMed] [Google Scholar]
- Tsuda K., Katagiri F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13: 459–465 [DOI] [PubMed] [Google Scholar]
- West M.A., Wallin R.P.A., Matthews S.P., Svensson H.G., Zaru R., Ljunggren H.-G., Prescott A.R., Watts C. (2004). Enhanced dendritic cell antigen capture via toll-like receptor-induced actin remodeling. Science 305: 1153–1157 [DOI] [PubMed] [Google Scholar]
- Yokota K., et al. (2009). Rearrangement of actin cytoskeleton mediates invasion of Lotus japonicus roots by Mesorhizobium loti. Plant Cell 21: 267–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H.-Y., Yao L.-L., Jia Z.-Q., Li Y., Li Y.-Z. (2006). Verticillium dahliae toxin induced alterations of cytoskeletons and nucleoli in Arabidopsis thaliana suspension cells. Protoplasma 229: 75–82 [DOI] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.-S., Niu Q.-W., Chua N.-H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Xie Y., Jiang Y., Qu X., Huang S. (2013). Arabidopsis actin-depolymerizing factor7 severs actin filaments and regulates actin cable turnover to promote normal pollen tube growth. Plant Cell 25: 3405–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D.G., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S. (2010). Pathogen-associated molecular pattern-triggered immunity: Veni, vidi...? Plant Physiol. 154: 551–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D., Felix G., Boller T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.