FtsH is a ubiquitous protease that mediates the degradation of membrane proteins. The chloroplast FtsH is known for its involvement in repairing photosystem II damaged by light. This work reports the characterization of an ftsh mutant in Chlamydomonas and demonstrates the wider role of FtsH in thylakoid membrane protein maintenance under light as well as nutrient stresses.
Abstract
FtsH is the major thylakoid membrane protease found in organisms performing oxygenic photosynthesis. Here, we show that FtsH from Chlamydomonas reinhardtii forms heterooligomers comprising two subunits, FtsH1 and FtsH2. We characterized this protease using FtsH mutants that we identified through a genetic suppressor approach that restored phototrophic growth of mutants originally defective for cytochrome b6f accumulation. We thus extended the spectrum of FtsH substrates in the thylakoid membranes beyond photosystem II, showing the susceptibility of cytochrome b6f complexes (and proteins involved in the ci heme binding pathway to cytochrome b6) to FtsH. We then show how FtsH is involved in the response of C. reinhardtii to macronutrient stress. Upon phosphorus starvation, photosynthesis inactivation results from an FtsH-sensitive photoinhibition process. In contrast, we identified an FtsH-dependent loss of photosystem II and cytochrome b6f complexes in darkness upon sulfur deprivation. The D1 fragmentation pattern observed in the latter condition was similar to that observed in photoinhibitory conditions, which points to a similar degradation pathway in these two widely different environmental conditions. Our experiments thus provide extensive evidence that FtsH plays a major role in the quality control of thylakoid membrane proteins and in the response of C. reinhardtii to light and macronutrient stress.
INTRODUCTION
Photosynthesis allows the conversion of light energy, captured by chlorophyll–protein complexes, into reducing power (NADPH) and chemical energy (ATP). In oxygenic photosynthesis, the photoinduced reduction of NADP+ is performed by a photosynthetic electron transfer chain, which joins three major oligomeric proteins embedded in the thylakoid membranes: photosystem II (PSII), cytochrome b6f complex, and photosystem I (PSI). The assembly, degradation and repair of these protein complexes require some coordination in the expression of numerous subunits encoded either in the chloroplast or in the nucleus, to which numerous cofactors such as chlorophylls, carotenoids, hemes, and iron-sulfur clusters must be added. The traffic of these proteins and of their molecular cofactors to their proper destination requires the action of a diverse set of chaperones, assembly factors, and proteases that ensure the proper biogenesis and recycling of these heterooligomeric complexes.
In recent years, the field of chloroplast protease studies has drawn increasing interest because of their role in the response to oxidative stress that results from the reaction of molecular oxygen with radical species, both of which are produced during illumination of the photosynthetic apparatus. Genomic and proteomic studies provided extensive identification of a well-defined set of chloroplast proteases of bacterial origin (reviewed in Sokolenko et al., 2002; Adam et al., 2006; Sakamoto, 2006; Huesgen et al., 2009), most of which are encoded by nuclear genes, with the exception of the catalytic subunit ClpP1 of the Clp protease, which is chloroplast encoded. Together with Clp, the Deg and FtsH proteases are the major proteolytic enzymes whose activities have been implicated in the regulation of biogenesis and the repair of photosynthetic proteins. A variety of studies proposed a role of these proteases in PSII repair upon photoinhibition (Lindahl et al., 2000; Majeran et al., 2001; Bailey et al., 2002; Silva et al., 2003; Kapri-Pardes et al., 2007; Kato and Sakamoto, 2009; Kato et al., 2012). However, we still have limited knowledge of the diversity of their substrates and regulatory functions.
The thylakoid membrane–anchored ATP-dependent protease FtsH is involved in the processive degradation of stroma-exposed thylakoid proteins, both in Arabidopsis thaliana and the cyanobacterium Synechocystis (reviewed in Lindahl et al., 1996; Narberhaus et al., 2009; Rodrigues et al., 2011). FtsH is a member of the large and diverse AAA+ (for ATPase associated with diverse cellular activities) protein family (Neuwald et al., 1999). It contains an ATPase domain with Walker A and B motifs and a second region of homology (SRH) with a protease domain displaying a zinc binding motif. FtsH is the only membrane-anchored and essential ATP-dependent protease in Escherichia coli, and it participates in protein quality control for only a few known membrane-bound and cytoplasmic substrates (Suzuki et al., 1997; Herman et al., 2003; Ito and Akiyama, 2005; Narberhaus et al., 2009). It has been suggested based on study in E. coli that ATPase activity is required for pulling out substrate proteins from the membrane and pushing them into the internal pore of the FtsH ring structure for proteolysis (Ito and Akiyama, 2005). While FtsH is encoded by a unique gene in most prokaryotes, multiple isoforms are found in plants, algae, and cyanobacteria (see the phylogenetic tree of FtsH homologs in Supplemental Figure 1). Characterization of the functional contribution of these isoforms has only started and combines in vivo and in vitro approaches. Four FtsH homologs (FtsH1 to FtsH4) are found in Synechocystis sp PCC 6803: FtsH3 copurifies with FtsH2 or FtsH1 in heterohexamers (Boehm et al., 2012). In Arabidopsis, the 12 nucleus-encoded FtsH family members (FtsH1 to FtsH12) are distributed between mitochondria and chloroplasts; in addition, five Arabidopsis FtsH-like proteins (FtsHi1 to FtsHi5) targeted to the chloroplast envelope lack the conserved zinc binding motif HisGluXxxXxxHis and are presumably inactive as protease (reviewed in Wagner et al., 2012; see also Arabidopsis proteome databases [Sun et al., 2009; Ferro et al., 2010]). Four isoforms have been identified in Arabidopsis thylakoid membranes: two of type A (FtsH1 and FtsH5) and two of type B (FtsH2 and FtsH8; Friso et al., 2004). The thylakoid enzyme is made of an active heterohexameric ring structure containing type A and B subunits whose accumulation is concerted, with a proposed stoichiometry of two type A and four type B subunits (Zaltsman et al., 2005; Moldavski et al., 2012) or three type A and three type B subunits (Yu et al., 2004; Boehm et al., 2012). Arabidopsis type A and type B subunits use different membrane insertion pathways (Rodrigues et al., 2011).
Here, we isolated two mutants in the gene encoding the thylakoid-embedded FtsH1 protein in the unicellular green alga Chlamydomonas reinhardtii. These mutants were recovered in a genetic screen to identify proteases that would target cytochrome b6f complex to degradation. The cytochrome b6f complex differs from bc1 by an additional ci heme located at the quinone reduction site (Kurisu et al., 2003; Stroebel et al., 2003). In recent years, we characterized cofactor assembly to complex C subunit B (CCB) factors involved in the covalent binding of ci heme to Cys-35 of cytochrome b6 in C. reinhardtii (Kuras et al., 1997, 2007) and Arabidopsis (Lezhneva et al., 2008). Mutants lacking ci heme cannot grow photoautotrophically because they show too little accumulation of a dysfunctional b6f complex (Saint-Marcoux et al., 2009). This observation provided the basis of a screen for suppressor mutations that affected FtsH and restored higher accumulation of dysfunctional b6f complexes to a level sufficient to sustain phototrophic growth.
Here, we show that the thylakoid-bound FtsH protease from C. reinhardtii is mainly found in heterooligomers, larger than 1 MD, comprising two subunits: FtsH1 and FtsH2. Mutation of an Arg residue required for ATP hydrolysis in FtsH1 hampers the integration of the whole FtsH protease in these large supercomplexes and impairs the processive degradation of photosynthetic membrane proteins, especially when they are difficult to unfold. In particular, we investigate the role of FtsH in PSII repair upon C. reinhardtii photoinhibition, described by others in cyanobacteria and plant chloroplasts, but we extend its function to the regulation of PSII accumulation upon macronutrient stress as well as to the quality control of neosynthesized cytochrome b6f complexes.
RESULTS
A Genetic Suppressor Approach to Obtaining Protease Mutants in C. reinhardtii
In a previous study (de Vitry et al., 2004), we had shown that ccb mutants of C. reinhardtii, which are nonphototrophic as a result of their inability to bind ci heme to apocytochrome b6, accumulate very low amounts of b6f complex subunits. However, Blue Native (BN)-PAGE showed that these remaining subunits still assembled into oligomeric complexes corresponding to cytochrome b6f dimers, similar to those found in the wild-type strain. In addition, we gathered preliminary evidence that these b6f complexes retained some electron transfer properties, as detected by their fluorescence induction kinetics, which pointed to some reoxidation of the plastoquinol pool under continuous illumination (Saint-Marcoux et al., 2009).
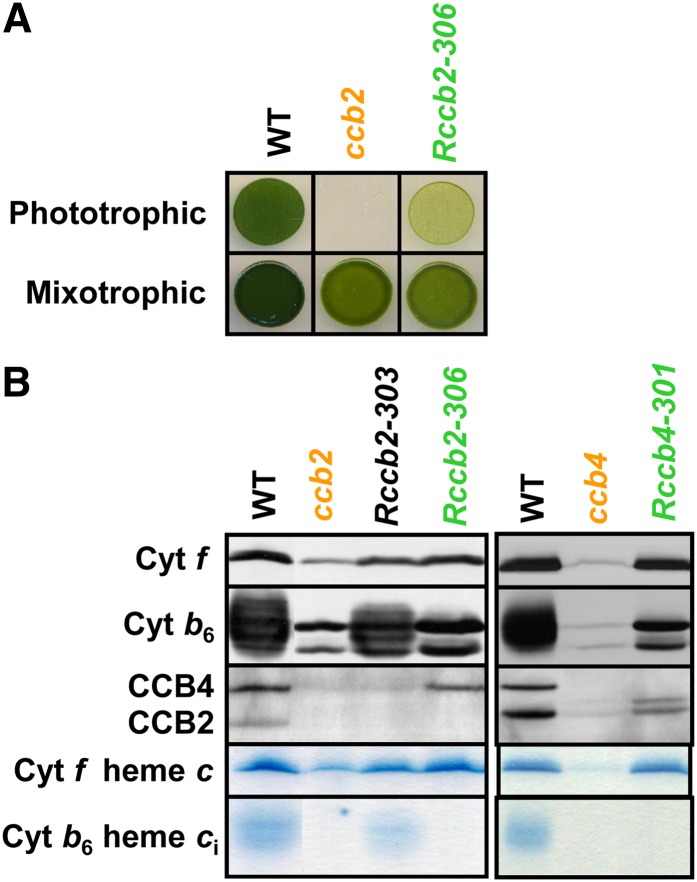
In a search for phototrophic suppressors, we performed a UV light mutagenesis of nuclear ccb mutants and selected revertants for their ability to grow on minimal medium (Figure 1A). Some of these, as illustrated by strain Rccb2-303 (Figure 1B), showed wild-type levels of b6f complexes together with a restoration of heme ci binding, as viewed by the recovery of a heme-stained cytochrome b6 upon electrophoresis, whereas others, as exemplified by Rccb2-306 and Rccb4-301 in Figure 1B, still lacked ci heme, despite a wild-type accumulation of cytochrome b6f subunits. The latter strains were promising candidates for being protease mutants unable to degrade b6f complexes lacking heme ci. In further support of this hypothesis, Figure 1B shows that Rccb2-306 also recovered wild-type levels of CCB4, which was hardly detectable in the original ccb2 mutant, owing to its concerted accumulation with CCB2 (Saint-Marcoux et al., 2009), and reciprocally, Rccb4-301 recovered some CCB2. We note that CCB2 is detected as a doublet in the absence of CCB4, as we observed previously (Saint-Marcoux et al., 2009).
Figure 1.
Revertants Recovered the Wild-Type Level of b6f Complexes.
(A) Phototrophic growth on minimal medium and mixotrophic growth control on acetate medium at 10 µE m−2 s−1.
(B) Total cell proteins of the wild type, ccb2 and ccb4 mutants, and Rccb2-303, Rccb2-306, and Rccb4-301 revertants grown in acetate medium at 6 µE m−2 s−1 were separated by SDS/urea-PAGE and analyzed by immunodetection with antibodies against cytochrome b6f subunits (cytochrome f and cytochrome b6) and CCB factors involved in the covalent binding of ci heme to cytochrome b6 (CCB2 and CCB4) and by heme peroxidase activity detection using tetramethylbenzamidine.
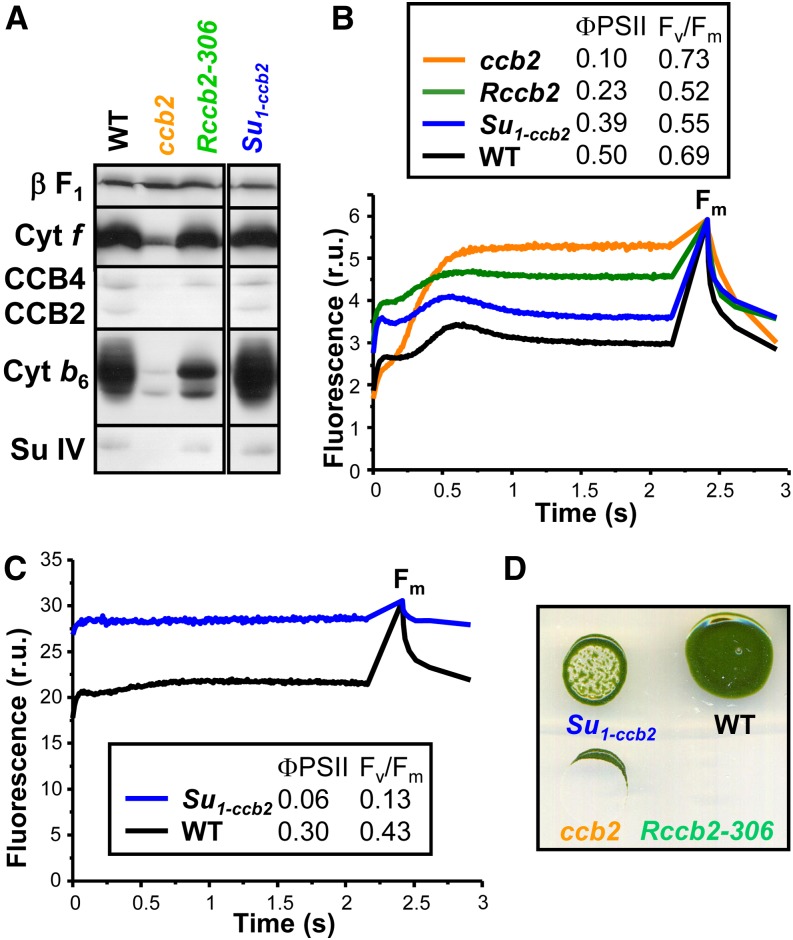
In a backcross between Rccb2-306 and the original ccb2 mutant, we observed only parental ditype tetrads with a Mendelian segregation of cytochrome b6f accumulation and phototrophy, which indicated that the revertant phenotype was due to a single suppressor mutation in the nuclear genome (Supplemental Figure 2). The cross between revertant Rccb2-306 and the wild-type strain yielded 2 parental ditype and 10 tetratype tetrads, indicative of an extragenic suppressor mutation, unlinked to the CCB2 gene. We isolated solely the suppressor named Su1-ccb2 from a tetratype tetrad and characterized its phenotype (Figure 2). The suppressor effect of Su1-ccb2 was confirmed by crossing it back to ccb2 (Supplemental Figure 3). Figure 2A shows that the contents of cytochrome b6f subunits and of CCB2/4 proteins in the suppressor Su1-ccb2 were undistinguishable from those in the wild-type strain. The photosynthetic electron transfer properties were examined in vivo on dark-adapted cells by fluorescence induction kinetics under continuous illumination, giving initial and steady state fluorescence in the light, followed by a saturating pulse to determine maximal fluorescence (Fm). These values allow calculation of the maximum quantum efficiency of PSII primary photochemistry, (Fm − initial fluorescence in the light)/Fm, denoted Fv/Fm, and the quantum yield of PSII photochemistry, (Fm − steady state fluorescence in the light)/Fm, denoted ΦPSII (Genty et al., 1989; Baker, 2008). The ccb2 mutant could be identified by its fluorescence yield, which rose continuously to a stationary fluorescence level close to Fm. The double mutant Rccb2 displayed fluorescence kinetics and ΦPSII intermediate between those in ccb2 and wild-type strains. When Su1-ccb2 was grown under low light, Fv/Fm and ΦPSII were decreased yet comparable to the wild-type strain, indicative of an efficient photosynthetic electron transfer (Figure 2B). However, when grown at higher light intensity, the suppressor mutant Su1-ccb2 underwent a considerable loss in Fv/Fm and ΦPSII, indicative of a nearly complete loss in PSII activity (Figure 2C). Its growth under high light was heavily compromised even under mixotrophic conditions (Figure 2D), an observation that led us to conclude that the suppressor mutation conferred a highly photosensitive phenotype to the Su1-ccb2 strain, a feature that we used for map-based cloning and molecular identification of the suppressor mutation.
Figure 2.
The Suppressor Su1-ccb2 Is Photosensitive.
(A) Total proteins of cells grown at 6 μE m−2 s−1 separated by SDS/urea-PAGE and analyzed by immunodetection with antibodies against cytochrome b6f subunits (cytochrome f, cytochrome b6, and subunit IV) and CCB factors (CCB2 and CCB4), with the ATP synthase β-subunit (β F1) as a loading control.
(B) Fluorescence induction kinetics in relative units (r.u.) at low actinic light of dark-adapted cells followed by a saturating pulse to determine Fm. Strains were grown at 6 μE m−2 s−1.
(C) Fluorescence kinetics. Strains were grown at 6 μE m−2 s−1 followed by 16 h at 200 μE m−2 s−1.
(D) Mixotrophic growth of the strains at 200 μE m−2 s−1.
Map-Based Cloning of the Point Mutation ftsh1-1
The above Su1-ccb2 phenotype did not offer a selection scheme for identification of the mutation by genetic complementation. Since it was obtained by UV light mutagenesis, it did not allow for the possibility to identify DNA-flanking regions as after insertional mutagenesis. As an alternative method, we used map-based cloning that relies on crosses of C. reinhardtii with the interfertile species Chlamydomonas grossii (also known as S1-D2 or CC-2290), which has a suitable profusion of genomic polymorphisms to permit the identification of the region bearing a mutation (Rymarquis et al., 2005). In each of 127 tetrads, we took one progeny carrying the Su1-ccb2 mutation for linkage analysis to various markers. We localized the suppressor mutation on chromosome 12 in the vicinity of the PSR1 and BDF1 markers (Supplemental Figure 4). Next, we amplified and sequenced several candidate genes in this region and found a mutation in the gene encoding the protease FtsH1. The Su1-ccb2 mutation is a CC-to-TT modification leading to a substitution of Gly419GGCArg420CGC into Gly419GGTCys420TGC. In the following, the Su1-ccb2 strain, which bears this ftsH1-R420C substitution, will be referred to as ftsh1-1. Similarly, the revertant Rccb4-301 harbored an ftsH1-T382I mutation (ftsh1-2) with a Thr382ACC into Ile382ATT substitution. Both Su1-ccb2 and Rccb4-301 displayed a wild-type FTSH2 sequence that encodes the other thylakoid-targeted subunit of the FtsH protease (see below). The mutated Arg and Thr residues are well conserved among FtsH sequences from various organisms and are located, respectively, in or before the SRH domain of the FtsH protein (Figures 3A and 3B). Several mutations of the conserved Arg-420 in E. coli were shown to compromise ATP hydrolysis and thereby the protease activity of FtsH homohexamers (Karata et al., 1999, 2001).
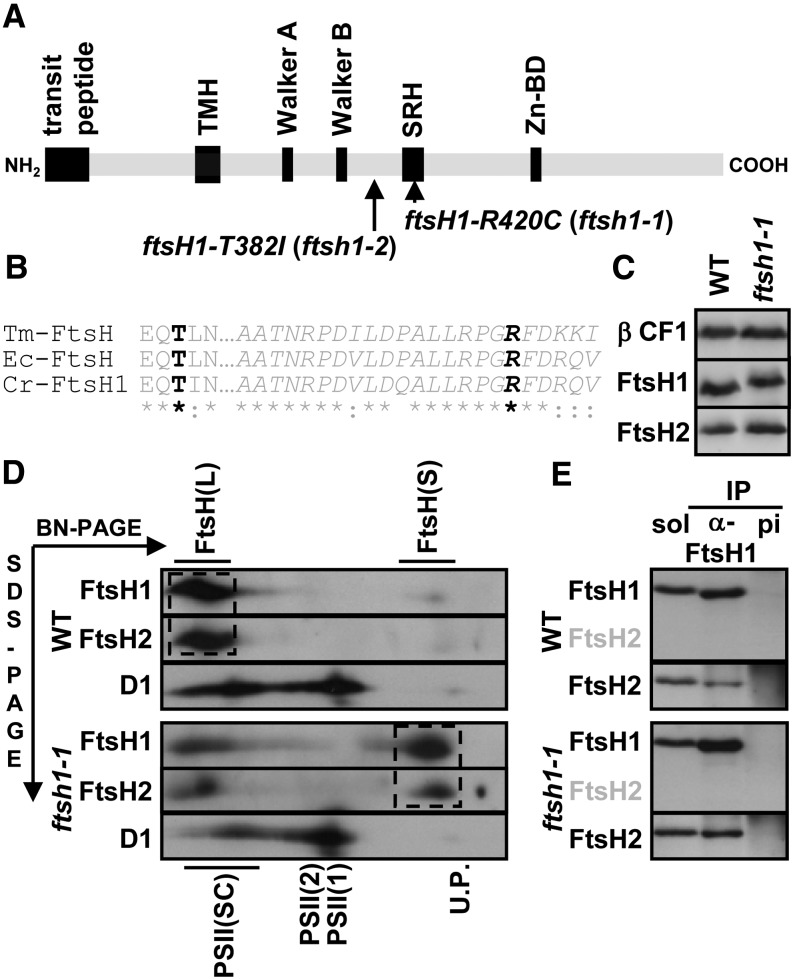
Figure 3.
Mutant ftsH1-R420C Accumulates Wild-Type Levels of FtsH1 and FtsH2 but with Impaired Oligomerization.
(A) Schematic representation of ATP-dependent zinc metalloprotease FtsH1 with the transit peptide, one thylakoid transmembrane helix (TMH), ATPase Walker A and B motifs, the SRH containing conserved Arg residues required for ATP hydrolysis (one of which is substituted to a Cys residue in the ftsH1-R420C [ftsh1-1] mutant of C. reinhardtii), and the zinc binding domain (Zn-BD).
(B) Alignment of the SRH (in italics) from T. maritima FtsH (Tm-FtsH), E. coli FtsH (Ec-FtsH), and C. reinhardtii FtsH1 (Cr-FtsH1), indicating the conservation of the Arg and Thr residues (in black) substituted in the ftsh1-1 and ftsh1-2 mutants.
(C) Mutant ftsH1-R420C accumulates wild-type levels of FtsH1 and FtsH2. Total cell proteins were separated on an 8% polyacrylamide gel in the presence of 8 M urea and analyzed by immunodetection with specific antibodies against FtsH1, FtsH2, and the ATP synthase β-subunit (β CF1) as a loading control.
(D) FtsH complexes after BN-PAGE of digitonin-solubilized membranes from cells grown at 6 μE m−2 s−1. Digitonin-solubilized proteins were separated in the first dimension by BN-PAGE (3 to 12%) and in the second dimension as in (C) and blotted onto a polyvinylidene difluoride membrane. The blot was sequentially probed with antibodies specific for FtsH2, FtsH1, and D1. FtsH1 and FtsH2 migrated mainly and similarly in two populations, the smaller FtsH(S) and the larger FtsH(L) proteins. FtsH(L) is abundant in the wild type and FtsH(S) in ftsh1-1 (dashed boxes). The positions of unassembled proteins (U.P.), monomers [PSII(1)], dimers [PSII(2)], and supercomplexes [PSII(SC)] of PSII are indicated and used as molecular mass markers.
(E) Coimmunoprecipitation analysis of FtsH1 and FtsH2 interactions. Digitonin-solubilized membranes (prepared as for BN-PAGE) from the wild-type and ftsh1-1 strains were incubated with protein A–Sepharose beads coupled to antibodies against a peptide of FtsH1 (α-FtsH1) or against preimmune serum (pi). Aliquots of digitonin-solubilized membranes (sol) and immunoprecipitates (IP) were separated as in (C) and analyzed by immunodetection with antibodies against FtsH1 (not cross-reacting with FtsH2, as indicated with gray letters) and FtsH2.
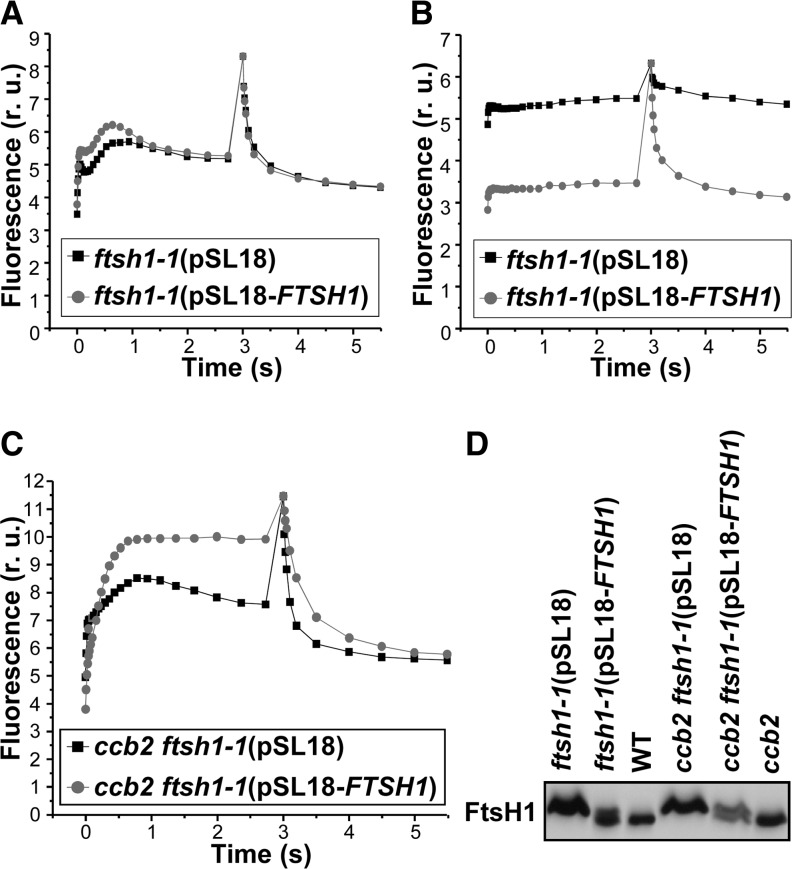
The ftsh1-1 mutation is recessive, as demonstrated by the low accumulation of b6f complex subunits observed in vegetative diploids from the cross ftsh1-1 ccb2 arg7 × FTSH1 ccb2 arg2 (Supplemental Figure 5). Transformation of the ftsh1-1 and ccb2 ftsh1-1 strains with the vector pSL18 containing FTSH1 genomic DNA (pSL18-FTSH1) under the control of the PSAD promoter (see Methods) showed that FTSH1 can complement the loss of PSII activity upon high-light treatment (Figures 4A and 4B) and restore a ccb2 fluorescence phenotype (Figure 4C), respectively. Transformation of these strains with an empty vector (pSL18) did not change their fluorescence kinetics, demonstrating that the FTSH1 gene alone was sufficient for complementation.
Figure 4.
Complementation of ftsh1-1 and ccb2 ftsh1-1 Strains with FTSH1 Genomic DNA Restores Wild-Type FtsH1 Phenotypes.
(A) Fluorescence induction kinetics in relative units (r.u.) at low actinic light of dark-adapted cells followed by a saturating pulse to determine Fm. Strains were grown at 6 μE m−2 s−1.
(B) Fluorescence kinetics. Strains were grown at 6 μE m−2 s−1 followed by 19 h at 200 μE m−2 s−1.
(C) Fluorescence kinetics. Strains were grown at 6 μE m−2 s−1.
(D) Total cell proteins were separated on an 8% polyacrylamide gel in the presence of 8 M urea and analyzed by immunodetection with specific antibodies against FtsH1. Strains ftsh1-1 and ccb2 ftsh1-1 were complemented with FTSH1 genomic DNA (pSL18-FTSH1) or as a control with empty pSL18 vector (pSL18).
ftsh1-1 Still Allows Accumulation of the FtsH1-R420C Mutated Protein but Largely Impairs Its Oligomerization in High Molecular Mass Complexes
Thylakoid FtsH proteases form various heterohexamers in cyanobacteria (Boehm et al., 2012) and Arabidopsis (Sakamoto et al., 2003; Yu et al., 2004; Zaltsman et al., 2005; Moldavski et al., 2012). Whereas four isoforms of thylakoid FtsH were identified in Arabidopsis (Zaltsman et al., 2005), only two thylakoid isoforms, FtsH1 and FtsH2, have been identified by proteomic studies in C. reinhardtii (Allmer et al., 2006). We raised specific antibodies against peptides from each of these isoforms (see Methods). The immunoblots of Figure 3C show that neither the accumulation of FtsH1 nor that of FtsH2 was compromised in the ftsh1-1 mutant. However, the mutated FtsH1-R420C protein migrates slightly higher than the wild-type FtsH1. We used these electrophoresis conditions, which separate the wild-type FtsH1 protein from the mutated one, to immunodetect their coexpression as a doublet in strains comprising the ftsh1-1 mutation complemented with pSL18-FTSH1 (Figure 4D).
To examine whether the ftsh1-1 mutation altered the state of oligomerization of the thylakoid FtsH protease, we combined a mild digitonin solubilization of the thylakoid membranes with two-dimensional BN/SDS-PAGE to identify oligomeric forms of FtsH1 and FtsH2 in C. reinhardtii wild-type and ftsh1-1 strains (Figure 3D). The two isoforms, FtsH1 and FtsH2, showed the same migration pattern, being mainly found in either of two populations, the smaller FtsH(S) and the larger FtsH(L) proteins. FtsH(L) is localized in the 1-MD region, as evidenced by its comigration with D1-containing PSII supercomplexes (Heinemeyer et al., 2004).
The presence of FtsH in PSII complexes was previously reported in cyanobacteria and Arabidopsis and proposed to allow immediate access to damaged D1 and thereby efficient D1 degradation (Kashino et al., 2002; Yoshioka et al., 2010). However, we could not coimmunoprecipitate PSII subunits with our antibody to FtsH1 (Supplemental Figure 6), which suggests either that these high molecular mass complexes merely comigrate in BN-PAGE or that they offer limited accessibility of the FtsH1 antigenic peptide.
FtsH(S) was found in the 150- to 200-kD region and is likely to correspond to an FtsH1/FtsH2 heterodimer. In stark contrast to the wild type, which displayed most of the FtsH1 and FtsH2 isoforms in the FtsH(L) population, up to approximately two-thirds of FtsH1 and FtsH2 were found in the FtsH(S) population in the ftsh1-1 mutant. Notably, the PSII supercomplex level was decreased in the ftsh1-1 mutant, in line with data on the Arabidopsis FtsH2 (var2) mutant (Kato et al., 2009). The similar distribution patterns of FtsH1 and FtsH2 suggested an interaction between the two within heterooligomers, which was confirmed by coimmunoprecipitation with purified antibodies against a peptide of FtsH1 that does not cross-react with FtsH2 (Figure 3E). In both the wild-type strain and the ftsh1-1 mutant, FtsH2 was pulled down together with FtsH1. Therefore, the phenotype of the ftsh1-1 mutant results from the expression of a defective enzyme that is still made of FtsH1/FtsH2 heterooligomers. However, the ftsh1-1 mutation prevents the integration of most of the FtsH1/FtsH2 heterodimers in larger supercomplexes.
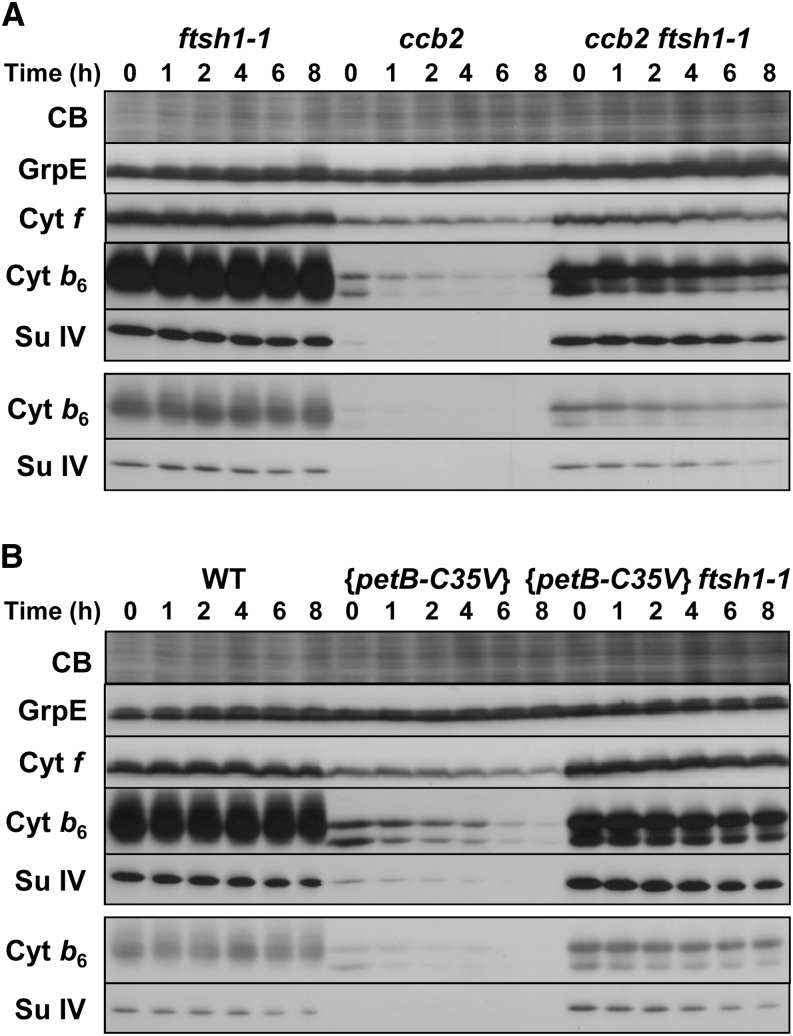
The Accumulation and Lifetime of Cytochrome b6f Complexes Lacking ci Heme Are Highly Increased in the ftsh1-1 Context
The suppressor strategy that led us to the identification of the ftsh1-1 mutant suggested that the assembled b6f complex is a substrate for the FtsH protease. We combined, by genetic crosses, the ftsh1-1 mutation with the {petB-C35V} mutation, which prevents covalent binding of heme ci to cytochrome b6 and leads to a low accumulation level of cytochrome b6f complexes (de Vitry et al., 2004). As was the case for the ccb2 ftsh1-1 mutant, the double mutant {petB-C35V} ftsh1-1 now accumulated wild-type levels of b6f complex subunits (compared with lanes 0 of the single and double mutants in Figure 5, right side). The accumulation of cytochrome b6f subunits in mutants defective for ci binding in the ftsh1-1 mutated context is due to a decrease in their rate of degradation, as also shown in Figure 5. After blocking protein synthesis in the chloroplast with chloramphenicol at time 0, we observed (over the 8-h time span of the immunochase experiment) that the lifetime of cytochrome b6 and subunit IV showed a considerable increase when the ftsh1-1 mutation was introduced into strains containing a mutation that impedes the binding of ci heme, as exemplified by the ccb2 and {petB-C35V} mutations (Figures 5A and 5B). This result demonstrates that the higher accumulation of b6f complexes in the double mutants is due to their lower rate of degradation. We attribute the limited degradation of b6f complexes still occurring in the double mutants to some residual FtsH activity and/or to other proteases (see Discussion).
Figure 5.
Accumulation and Lifetime of b6f Complexes Lacking ci Heme Are Highly Increased in the ftsh1-1 Context.
(A) Immunochase of ftsh1-1, ccb2, and ccb2 ftsh1-1 mutants. Immunochase was performed in the presence of 100 μg mL−1 chloramphenicol, an inhibitor of the translation of chloroplast genes. Total cell proteins were loaded at the chlorophyll constant, separated by SDS/urea-PAGE, and blotted onto a polyvinylidene difluoride membrane. The blot was stained with Coomassie blue (CB) and analyzed by enhanced chemiluminescence immunodetection with antibodies against cytochrome b6f subunits and the nucleotide exchange factor GrpE.
(B) Immunochase of the wild type and the {petB-C35V} and {petB-C35V} ftsh1-1 mutants in the same conditions as in (A).
Note that for better comparison of the content in cytochrome b6f subunits between mutants, shorter times of exposure of the immunoblots are shown at the bottom part of each panel.
The ftsh1-1 Mutation Increases PSII Sensitivity to Photoinhibition, Prevents PSII Repair, and Leads to the Accumulation of D1 Fragments
Photoinhibition, a common feature of light stress in plants, algae, and cyanobacteria, occurs when the rate of light-induced damage to PSII exceeds the repair capacity. The repair of PSII involves partial disassembly of the damaged complex, selective proteolytic degradation and replacement of the damaged subunit (predominantly the D1 reaction center subunit) by a de novo synthesized copy, and reassembly. In plants and cyanobacteria, FtsH regulates PSII repair, through its ability to digest photooxidatively damaged D1 proteins, either alone in cyanobacteria or in combination with Deg proteases in plants (reviewed in Kato and Sakamoto, 2009; Nixon et al., 2010). The role of FtsH during algal photoinhibition is unclear, and the role of FtsH and the possible contribution of chloroplast Deg proteases to this process has not been confirmed experimentally. The ftsh1-1 mutant enabled us to address these issues.
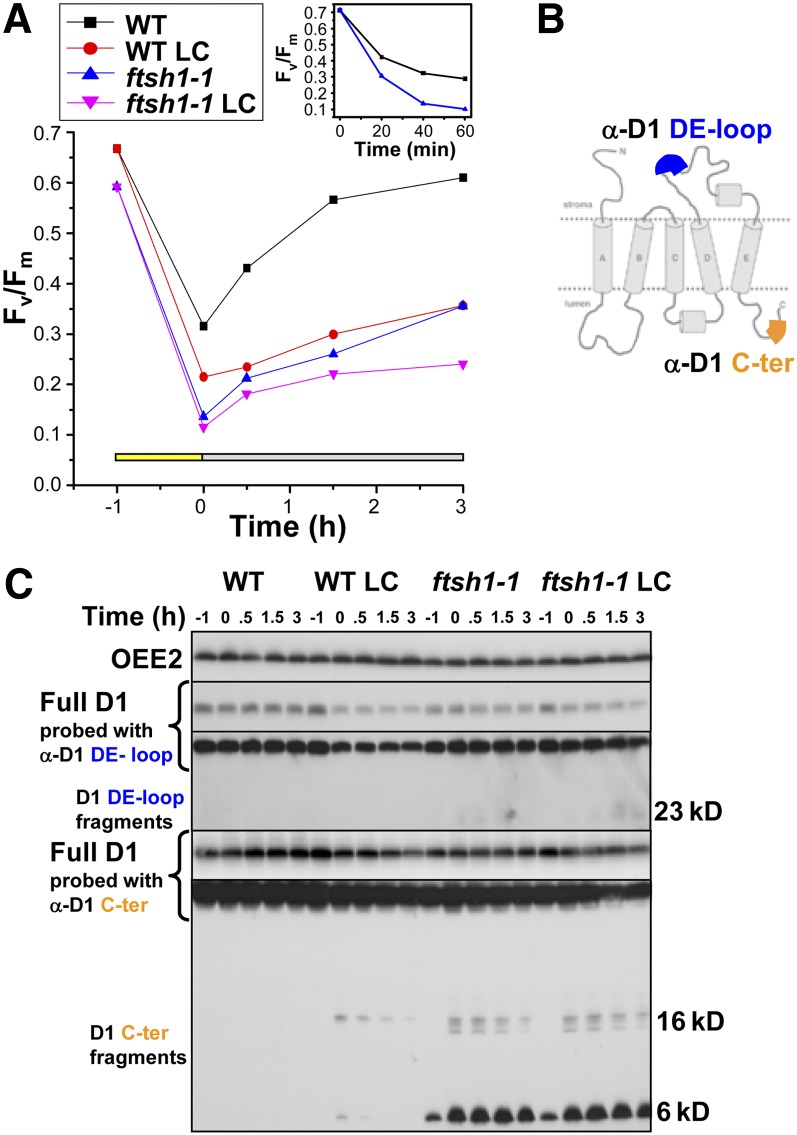
To explore further the strong decrease in Fv/Fm upon higher light exposure and the photosensitive growth phenotype of the ftsh1-1 mutant observed in Figures 2C and 2D, we monitored PSII activity by measuring the Fv/Fm during a mild photoinhibition treatment (250 μE m−2 s−1) and a subsequent recovery under low light (6 μE m−2 s−1). The addition of inhibitors of chloroplast translation, lincomycin and chloramphenicol (LC), prevents D1 synthesis and permits the discrimination of photodamage from repair. As shown in Figure 6A, the ftsh1-1 mutation dramatically increased PSII sensitivity to a mild photoinhibition: over a 1-h illumination period with photoinhibitory light, the loss in variable fluorescence was more extensive and developed faster (Figure 6A, inset) in the mutant than in the wild type, and most of the functional recovery was prevented in a subsequent low-light illumination period, irrespective of inhibitor addition. The means and sd of PSII activity recovery after 1.5 h were 63% ± 12% for the wild type and 19% ± 8% for the ftsh1-1 mutant (three independent experiments). This lack of recovery is similar to what was observed in the wild-type strain when D1 synthesis was inhibited and thus delineates the important role of FtsH in the PSII repair cycle during algal photoinhibition.
Figure 6.
ftsh1-1 Increases PSII Sensitivity to Photoinhibition, Prevents PSII Repair, and Leads to the Accumulation of D1 Fragments.
(A) Time course of photoinhibition at 250 μE m−2 s−1 and recovery at 6 μE m−2 s−1 in wild-type and ftsh1-1 strains in the absence or presence of inhibitors of the translation of chloroplast genes (500 μg mL−1 lincomycin and 100 μg mL−1 chloramphenicol [LC]) as monitored by PSII activity (Fv/Fm). The inset shows kinetics of the PSII activity loss upon photoinhibition in an independent experiment.
(B) D1 model with the localization of the D1 DE loop and C-terminal peptides recognized by antibody (adapted from Kapri-Pardes et al., 2007).
(C) Time course of photoinhibition and recovery in wild-type and ftsh1-1 strains as monitored by levels of D1 and D1 fragments immunodetected with D1 DE loop and C-terminal antibodies using the PSII OEE2 extrinsic subunit as a loading control. Total cell proteins were separated by SDS/urea-PAGE.
Using antibodies against the DE loop or the C terminus of D1 (Figure 6B), we probed the fate of full-length D1 protein and the accumulation of D1 fragments in the same experiment, using longer exposure times of the immunoblots for proper detection of fragments (Figure 6C). Full-length D1 did not decrease during photoinhibition and recovery in the wild type or ftsh1-1 unless translation inhibitors were added during the experiment (LC condition). In the latter condition, the loss of D1 was more extensive in the wild type than in the mutant, as viewed when probed with either of the two antibodies. However, D1 fragments accumulated mainly in the ftsh1-1 mutant. They were detected around 23 kD with the anti-D1 DE loop (Figures 7B and 8B) and in the 16- to 20-kD and 6- to 10-kD regions with the anti-D1 C terminus. We note that the 6- to 10-kD D1 fragments were particularly abundant in ftsh1-1 and even could be well detected before photoinhibitory treatment (Figure 6C). We quantified the content in D1 upon photoinhibition by chemiluminescence using the D1 C-terminal antibody and OEE2 as a loading control when repair was prevented by the addition of translation inhibitors (LC condition). The fraction of full-length D1 degraded over 4 h was 53% in the wild type and 35% in ftsh1-1. When using the D1 DE loop antibody, we observed a similar increase in degradation of full-length D1 in the wild type as compared with the ftsh1-1 mutant, with an additional 17% of full-length D1 being degraded in the wild type only. C-terminal epitopes recovered in D1 fragments amounted to 20% of total initial D1 (full-length plus fragments) in the ftsh1-1 mutant, which indicates that D1 fragmentation was actively occurring in these experimental conditions. Thus, the defective activity of the FtsH protease impairs the degradation of damaged D1 and leads to some accumulation of D1 fragments. We note that the total amount of antibody-recognized C-terminal epitopes, borne by D1 full-length protein and fragments, decreased by 20% in the mutant in the LC condition, suggesting either that some degradation of the D1 C-terminal fragments still occurs or that there is some processive degradation of D1 that does not result in the production of D1 C-terminal fragments. A residual FtsH activity in the mutant may account for either of these two processes.
Figure 7.
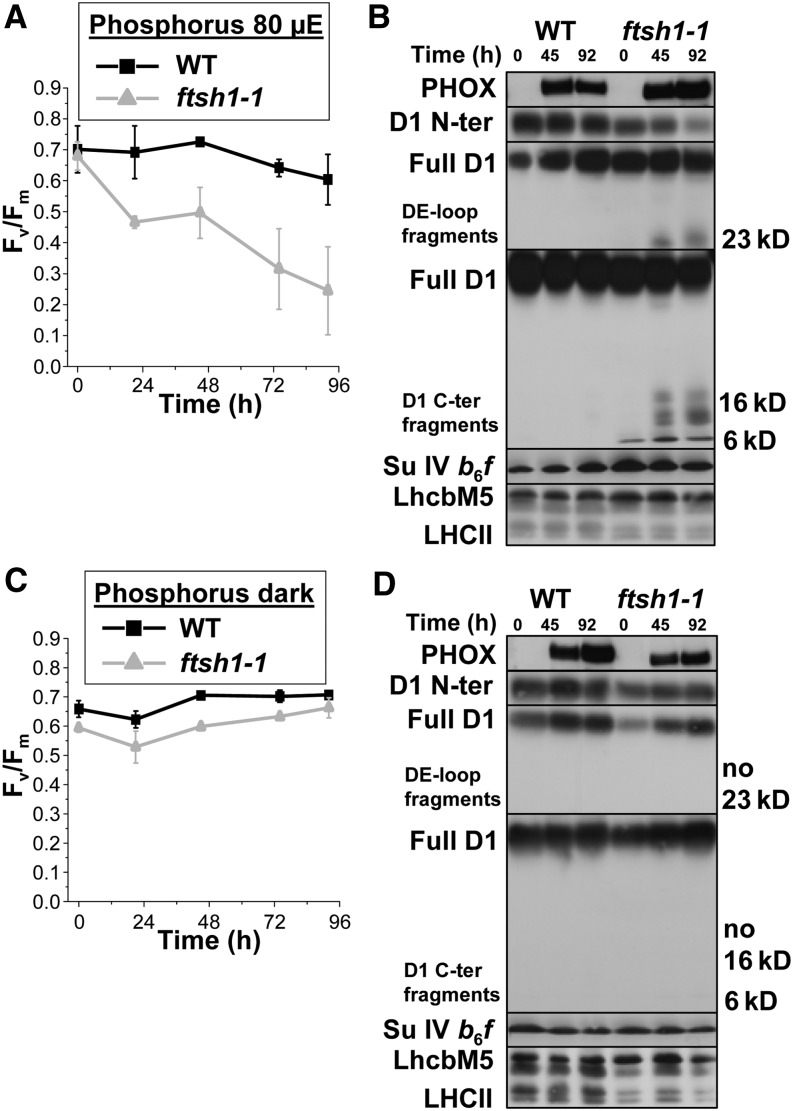
PSII upon Phosphorus Starvation in Wild-Type and ftsh1-1 Strains.
(A) Time course of phosphorus starvation under 80 μE m−2 s−1 as monitored by PSII activity detection.
(B) Time course of phosphorus starvation under 80 μE m−2 s−1 as monitored by immunodetection of inducible PHOX (a marker of the cell response to phosphorus deprivation), full D1 and D1 fragments with D1 DE loop and C-terminal antibodies, cytochrome b6f subunit IV, and light-harvesting LhcbM5. Total cell proteins were separated on a 7.5 to 15% polyacrylamide gel in the presence of 8 M urea.
(C) Time course of phosphorus starvation in the dark as monitored by PSII activity detection.
(D) Time course of phosphorus starvation in the dark as monitored by immunodetection.
Means and sd of two independent experiments for each starvation condition are given.
Figure 8.
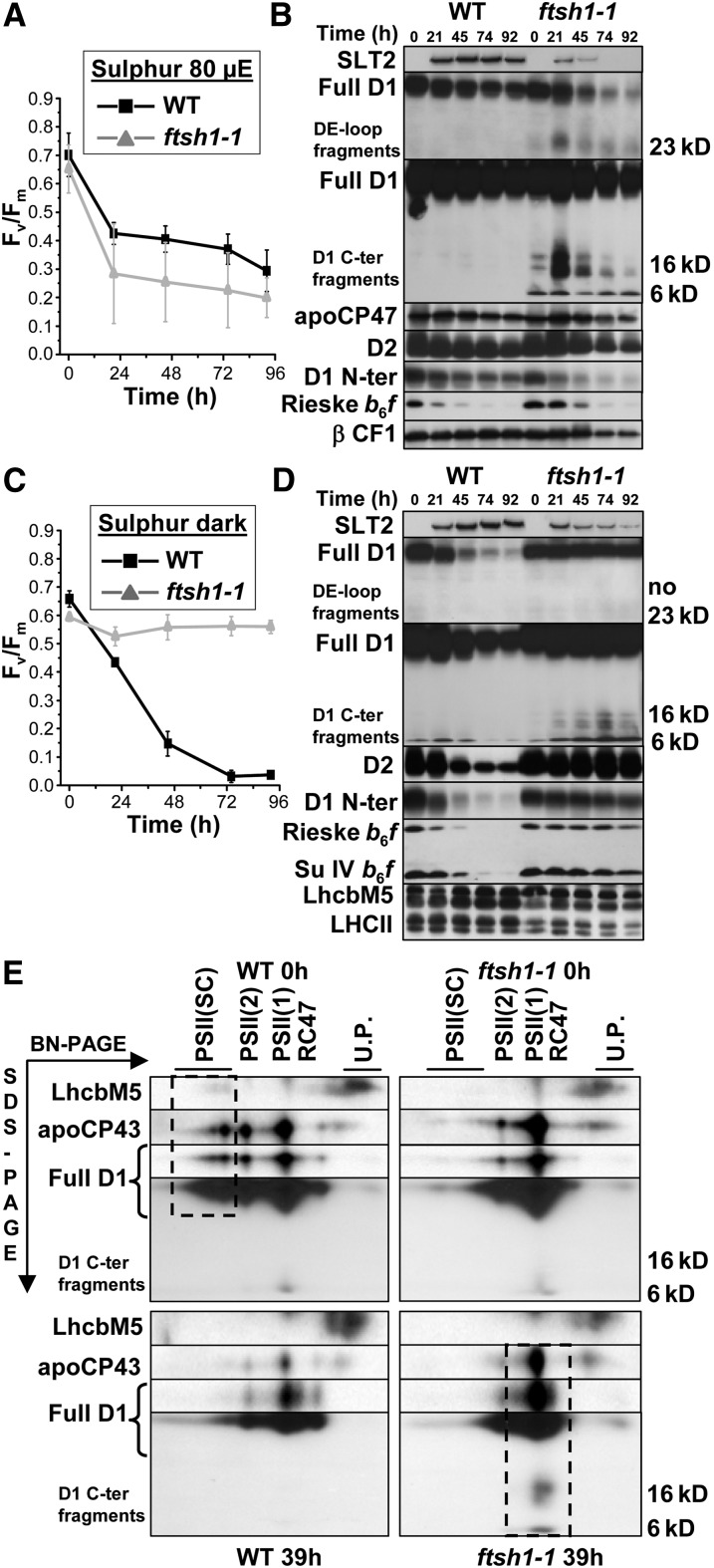
PSII and Cytochrome b6f upon Sulfur Starvation in Wild-Type and ftsh1-1 Strains.
(A) Time course of sulfur starvation under 80 μE m−2 s−1 as monitored by PSII activity detection.
(B) Time course of sulfur starvation under 80 μE m−2 s−1 as monitored by immunodetection of a cell response marker to sulfur deprivation (sulfate transporter SLT2) and of PSII subunits (D1, D2, and CP47), cytochrome b6f subunits (IV and Rieske), and light-harvesting LhcbM5. Total cell proteins were separated as in Figure 7.
(C) Time course of sulfur starvation in the dark as monitored by PSII activity detection.
(D) Time course of sulfur starvation in the dark as monitored by immunodetection.
(E) PSII complexes after BN-PAGE of dodecyl-maltoside–solubilized membranes at 0 and 39 h of sulfur deprivation under 80 μE m−2 s−1 were separated by two-dimensional BN (3 to 12%)/SDS-PAGE (12% in the presence of 8 M urea), blotted onto a polyvinylidene difluoride membrane, and immunodetected. The positions of unassembled proteins (U.P.), a reaction center core lacking CP43 (RC47), and monomers [PSII(1)], dimers [PSII(2)], and supercomplexes [PSII(SC)] of PSII are indicated. At 0 h, supercomplexes are more abundant in the wild-type than in ftsh1-1; at 39 h, fragmented D1 was found assembled in monomeric PSII and RC47 complexes in the ftsh1-1 mutant (dashed boxes).
Means and sd of two independent experiments for each starvation condition are given.
PSII Inactivation upon Phosphorus Starvation Is the Result of Enhanced Photoinhibition
Interestingly, impaired PSII activity also has been reported when C. reinhardtii is subjected to macronutrient stress, namely upon phosphorus and sulfur starvation (Wykoff et al., 1998; Zhang et al., 2002). In both cases, a loss of PSII proteins upon starvation was found responsible for PSII inactivation. The fact that the light intensity used in these experiments was ∼80 μE m−2 s−1 suggested to us that a photoinhibitory effect might have developed, since macronutrient deprivation would lower the photosynthetic ability to fix carbon. If such was the case, we predicted that the ftsh1-1 mutant, being unable to repair damaged D1, would be more sensitive than the wild type to these nutrient stresses and that PSII inactivation would only occur in the light.
We first subjected the ftsh1-1 mutant to phosphorus starvation for 96 h either under illumination of 80 μE m−2 s−1 or in the dark. The cell response marker to phosphorus deprivation, alkaline phosphatase (PHOX; Hallmann, 1999; Moseley et al., 2006), was induced in the light as well as in the dark in both the wild-type and ftsh1-1 strains (top of Figures 7B and 7D). At 80 μE m−2 s−1, the ftsh1-1 mutant showed a more rapid and much larger loss in PSII activity than the wild type, as determined by their respective changes in Fv/Fm (Figure 7A) and a net decrease in the content of full-length D1 in the mutant at the late stage of starvation, probed with the D1 N-terminal antibody (Figure 7B, top). We then overexposed immunoblots reacted with D1 antibodies to the DE loop and C terminus to examine the pattern of accumulation of D1 fragments upon phosphorus starvation under 80 μE m−2 s−1 in the ftsh1-1 mutated context. This pattern was similar to the one observed upon photoinhibition, showing an increased accumulation of D1 fragments, around 23 kD and in the 16- to 20-kD and 6- to 10-kD regions (Figure 7B). When phosphorus starvation was performed in the dark, both the ftsh1-1 and wild-type strains preserved PSII activity over a period of 96 h (Figure 7C), and neither of them showed any accumulation of D1 fragments (Figure 7D). These results clearly demonstrate that compromised photosynthetic activity upon phosphorus starvation merely results from enhanced photoinhibition.
Compromised Photosynthetic Activity upon Sulfur Starvation under Illumination Is Due to Both Photoinhibition and Degradation of b6f Complexes
When the ftsh1-1 mutant was starved of sulfur under illumination of 80 μE m−2 s−1 over 96 h, it showed a larger PSII inactivation than the wild type (Figure 8A). This observation suggests, here again, an efficient FtsH-dependent repair process upon sulfur starvation of C. reinhardtii wild-type cells, similar to the repair upon photoinhibition. Indeed, the decrease in D1 content during sulfur starvation was far more pronounced in the mutant than in the wild type (Figure 8B).
However, there were marked differences with regard to phosphorus starvation or a regular photoinhibition process: the cytochrome b6f complex, as probed with an antibody against the Rieske protein subunit, was now targeted to degradation in the two strains, and the drastic degradation of full-length D1 in the ftsh1-1 mutant was accompanied by a transient overaccumulation of D1 fragments at 21 h of starvation, which were subsequently disposed of. Whereas at a late stage of sulfur starvation there was a general decrease in protein content in the mutant, as shown by the decreased content in the cell response marker to sulfur deprivation, SLT2 (Pootakham et al., 2010), and the β-subunit of CF1 (Figure 8B), we noted that the other subunits of the PSII core, D2 and CP47, better resisted degradation than D1. This result raised the question of whether D2 and CP47 were in a disassembled state or perhaps still assembled with fragmented D1 in the ftsh1-1 mutant. We assayed this hypothesis by two-dimensional BN/SDS-PAGE of dodecyl-maltoside–solubilized membranes isolated after 39 h of sulfur starvation (Figure 8E, bottom). We found that fragmented D1 remained assembled within PSII monomers and reaction center cores lacking CP43 (RC47) in the ftsh1-1 mutant. This finding is in line with what has been reported for the Arabidopsis FtsH2 (var2) mutant after 1 h of extreme high-light exposure at 2500 μE m−2 s−1 (Kato et al., 2012). We note that, before starvation, PSII supercomplexes were far more abundant in the wild type than in the ftsh1-1 strain (Figure 8E, top), as also was suggested by the analysis of digitonin-solubilized membranes (Figure 3D).
We also examined the behavior of the wild type and the ftsh1-1 mutant when sulfur starvation was performed in photoautotrophic conditions, under an illumination of 80 μE m−2 s−1 in the absence of the carbon source acetate. Both strains showed a delayed loss in PSII activity and downstream electron transfer (Supplemental Figures 7A to 7D). They also showed a retarded induction of the sulfate transporter SLT2, probably due to their slower growth and lower sulfur consumption in minimal medium (Supplemental Figures 7E and 7F).
The ftsh1-1 Mutant Conserves Most PSII and b6f Complexes upon Sulfur Starvation in the Dark, in Contrast to the Wild-Type Strain
To further understand to what extent photoinhibitory processes contributed to the loss of PSII upon sulfur starvation, we performed the same starvation experiments in complete darkness. To our surprise, wild-type cells showed a more dramatic inactivation of PSII (Figure 8C), which was now accompanied by a net loss of D1 and D2 proteins. Thus, the repair process that was efficient in the light (Figures 8A and 8B) was either heavily compromised in the dark or unable to make up for higher rates of PSII degradation (Figures 8C and 8D). That the process of PSII degradation in the dark was widely different from the one operating under illumination was demonstrated by the behavior of the ftsh1-1 mutant, which preserved fully active PSII when starved of sulfur in the dark (Figures 8C and 8D). SLT2 was induced upon sulfur depletion in the dark in the two strains, indicating that the differences in PSII degradation should not be attributed to an absence of signaling of sulfur deficiency in the ftsh1-1 mutant. The cytochrome b6f complexes were degraded in the wild type starved of sulfur in the dark, as they were under 80 μE m−2 s−1, but this degradation in the dark was now largely FtsH dependent, since the ftsh1-1 mutant showed very limited cytochrome b6f degradation in contrast to its behavior in the light (Figures 8B and 8D).
The accumulation pattern of D1 fragments upon sulfur starvation of the ftsh1-1 mutant in the dark also differed from the one observed upon photoinhibition, with an absence of 23-kD fragments (Figure 8D). This is consistent with the previously characterized light-induced production of a 23-kD D1 fragment as a substrate of the thylakoid FtsH protease (Lindahl et al., 2000). Importantly, the 6- to 10-kD and 16- to 20-kD fragments still accumulated in the dark. We also note that sulfur starvation induced extensive accumulation of D1/D2 oligomers of high molecular weight in the dark but not in the light in the ftsh1-1 mutated context only (Supplemental Figure 8).
Finally, we tested whether increased expression in some other major chloroplast proteases would have occurred upon FtsH inactivation, which would explain the more pronounced PSII degradation in the ftsh1-1 mutant upon sulfur deprivation in mixotrophic conditions. We used antibodies against FtsH1/FtsH2, stromal ClpP1, lumenal Deg5, and stromal membrane–associated SppA1 to probe the content of major chloroplast proteases in wild-type and ftsh1-1 cells starved of sulfur in the presence or absence of acetate (Supplemental Figures 7E and 7F). The patterns of accumulation were rather similar for the two strains, indicating that there was no upregulation of the probed proteases in the FtsH-defective mutant. Sulfur starvation caused a rapid loss in Deg5 and ClpP1 in the two strains and some processing of SppA1 in mixotrophic conditions only. FtsH and SppA1 were better preserved than Deg5 and ClpP1 when sulfur starvation was performed in the absence of acetate, especially in the ftsh1-1 mutant (Supplemental Figure 7F).
The ftsh1-1 Mutation Has a Limited, if Any, Ability to Prevent the Degradation of Unassembled Subunits from the Cytochrome b6f Complex or the PSII Protein Complex
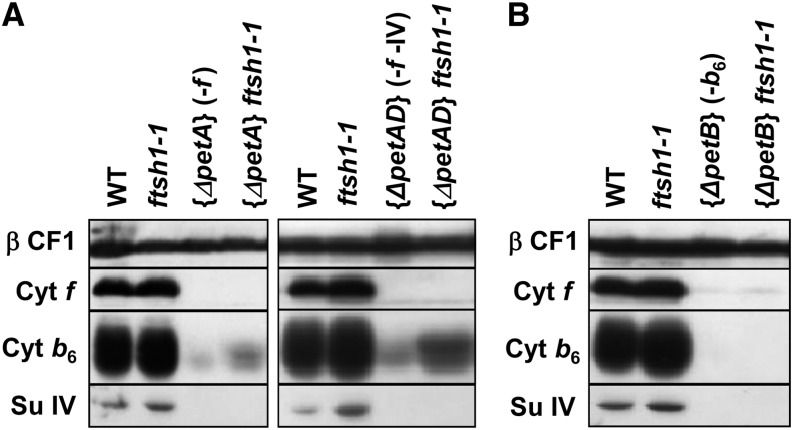
A concerted accumulation of PSII subunits that make up an active PSII protein has been demonstrated to result from a posttranslational regulation leading to the proteolytic disposal of unassembled subunit D2 and CP43 and a downregulation of the translation of subunit D1 or CP47, which are control by epistasy of synthesis (CES) subunits (de Vitry et al., 1989; Minai et al., 2006). We crossed the ftsh1-1 nuclear mutant to chloroplast mutants with a deleted PsbC (CP43) gene or harboring a premature stop codon in PsbA that leads to the expression of a truncated and labile D1. We found no evidence that the mutation ftsh1-1 rescued the accumulation of the other PSII subunits when PSII assembly was compromised in these mutants (Supplemental Figure 9). To assess whether this protease also determines the lifetime of unassembled subunits of the cytochrome b6f complex, most of which are subjected to rapid degradation, with the exception of cytochrome f, which is a CES subunit (Kuras and Wollman, 1994; Choquet et al., 1998), we tested whether the mutation ftsh1-1 rescues the accumulation of cytochrome b6 in the absence of cytochrome f and vice versa. To this end, we crossed the nuclear ftsh1-1 mutant with chloroplast mutants lacking either the petA gene, encoding cytochrome f, the petA and petD genes, the latter encoding subunit IV, or the petB gene, encoding cytochrome b6. The accumulation of unassembled cytochrome b6 showed a limited but noticeable increase in an FtsH-defective context (Figure 9A), whereas unassembled cytochrome f, whose decreased accumulation is due to a decreased translation initiation according to the CES process, proved, as expected, insensitive to the status of the FtsH protease (Figure 9B). In these experiments, the level of accumulation of subunit IV in the absence of its assembly partners remained below detectable levels.
Figure 9.
The Accumulation of Cytochrome b6 Lacking Assembly Partners Shows a Limited but Noticeable Increase in an FtsH-Defective Context.
(A) Double mutants obtained by crosses of the nuclear ftsh1-1 mutant with chloroplast mutants deleted for either the petA gene, encoding cytochrome f, or the petA and petD genes, the latter encoding subunit IV, accumulated more cytochrome b6 when cytochrome f was missing than the {ΔpetA} and {ΔpetAD} mutants. Total cell proteins were separated by SDS/urea-PAGE and analyzed by immunodetection with antibodies against cytochrome b6f subunits (cytochrome f, cytochrome b6, and subunit IV) and the ATP synthase β-subunit (β CF1) as a loading control.
(B) Unassembled cytochrome f when cytochrome b6 is missing, as in the {ΔpetB} mutant context, was insensitive to the presence of the ftsh1-1 mutation.
DISCUSSION
The Thylakoid FtsH Protease of C. reinhardtii Forms Heterooligomers Comprising FtsH1 and FtsH2
The C. reinhardtii nuclear genome encodes six members of the FtsH family distributed between the mitochondria (Atteia et al., 2009) and the chloroplast and three FtsH-like proteins (FHL1 to FHL3) that retain a complete AAA+ domain but lack the zinc binding motif (reviewed in Schroda and Vallon, 2009; see phylogenetic tree of FtsH proteins in Supplemental Figure 1). Only two isoforms have been identified in C. reinhardtii thylakoid membranes: FtsH1 (type A) and FtsH2 (type B; Allmer et al., 2006). Our two-dimensional BN/SDS-PAGE and coimmunoprecipitation experiments demonstrate that FtsH1 also forms heterocomplexes with FtsH2 in C. reinhardtii (Figures 3D and 3E). FtsH(L), however, has a much larger apparent molecular mass than expected for a heterohexamer (∼410 kD, to which should be added a digitonin contribution that has a micellar mass of ∼70 kD). Such large FtsH-containing complexes, around or above 1 MD, were previously observed in E. coli, yeast mitochondria, and Arabidopsis mitochondria, where they also include prohibitin-like proteins whose function remains unclear (Steglich et al., 1999; Saikawa et al., 2004; Piechota et al., 2010). Similar high molecular mass FtsH-containing complexes were described in Synechocystis (Boehm et al., 2012) with a transient or detergent-sensitive association with prohibitin-like proteins (Boehm et al., 2009). Thus, the thylakoid enzyme from C. reinhardtii shows a similar but simpler subunit composition than its counterpart from Arabidopsis, which assembles four distinct FtsH isoforms. We cannot exclude that some FtsH1 and/or FtsH2 may form homohexamer complexes, although homomeric structures are thought to be less likely to exist since they are less stable, as modeled by Moldavski et al. (2012).
The ftsh1-1 Mutation Largely Impairs FtsH Olimerization and Should Affect ATP Hydrolysis in the C. reinhardtii FtsH Protease
Here, we generated, by a genetic suppressor approach, an FtsH mutant in C. reinhardtii, which we named ftsh1-1. It carries a mutation in the FtsH1 gene that substitutes a well-conserved Arg, Arg-420, in the SRH domain by a Cys residue (Figures 3A and 3B). Arg-420 was first proposed to form an Arg finger (Karata et al., 1999), which should coordinate the γ-phosphate of a bound ATP and contribute to its hydrolysis through stabilization of the transition state of the reaction, as described in GTPase-activating proteins (Ahmadian et al., 1997). Indeed, several mutations of the two SRH-conserved Arg residues in E. coli were shown to compromise ATP hydrolysis and thereby protease activity (Karata et al., 1999, 2001). Arg-420 from C. reinhardtii corresponds to the second SRH-conserved Arg, Arg-315, in E. coli, which was proposed to be located at hydrogen bonding distance of the γ-phosphate of bound ATP to the adjacent subunit in the dimeric crystal structure of the apoFtsH AAA domain of E. coli (Krzywda et al., 2002; Ogura et al., 2004). However, in the hexameric crystal structures of ADP-FtsH lacking the transmembrane domain, it is the other conserved Arg, FtsH1-R417 in C. reinhardtii, that was proposed to form an Arg finger in the thermophilic bacterium Aquifex aeolicus (Suno et al., 2006), while the conserved Arg residue corresponding to C. reinhardtii FtsH1-R420 is localized at the intersubunit interface, where it could contribute to the oligomerization of the enzyme by its participation in a salt bridge with a conserved Asp residue from the same subunit in the thermophilic bacterium Thermotoga maritima (Bieniossek et al., 2006).
Indeed, the ftsh1-1 mutant still displays wild-type amounts of the two FtsH isoforms (Figure 3C) with an altered oligomerization pattern (Figure 3D). We observed, however, a well-preserved interaction between FtsH1 and FtsH2, as shown by coimmunoprecipitation (Figure 3E). The striking increase in the proportion of FtsH1 and FtsH2 found in FtsH(S) in the ftsh1-1 mutant (Figure 3D) is consistent with an accumulation of FtsH1/FtsH2 heterodimers due to the location of the mutation at the intersubunit interface. This Arg/Cys amino acid substitution would loosen the formation of the FtsH hexamers and hamper their integration with partner proteins, or substrates, in larger heterooligomers, resulting in a higher yield of FtsH heterodimers. As the ClpX hexameric unfoldase of the AAA+ protein family was shown to retain some function even if only one subunit could hydrolyze ATP (Martin et al., 2005), ATP hydrolysis by wild-type FtsH2 subunits in ftsh1-1 heterocomplexes may allow some digestive power. We conclude that these changes in oligomerization and probably also in ATP hydrolysis deeply affect the protease and unfoldase activities, leading to a lower quality control of partially assembled or misassembled complexes that are particularly difficult to unfold.
Degradation of Unassembled Subunits from the Cytochrome b6f Complex or the PSII Protein Complex Still Occurs with High Efficiency in the ftsh1-1 Mutated Context
The above conclusion that the ftsh1-1 mutation may prevent the unfolding of substrates for degradation rather than proteolysis itself is consistent with its very limited effect on the accumulation level of unassembled subunits from the cytochrome b6f complex and the PSII complex. For instance, D2 and CP43 showed no increased accumulation when the ftsh1-1 mutation was introduced in a strain showing premature termination of D1 that prevents PSII core assembly (Supplemental Figure 9B), thus leading to proteolytic disposal of these subunits (Minai et al., 2006). Similarly, subunit IV was still efficiently degraded in the ftsh1-1 mutated context whether cytochrome f or cytochrome b6 was missing (Figure 9). We noted, however, some increase in the accumulation of unassembled cytochrome b6 when the activity of FtsH was compromised, which suggests that unfolding of holocytochrome b6 may facilitate its degradation (Figure 9A). However, the ftsh1-1 mutant did not show any increase in the accumulation of the partially assembled PSII subcomplex D2/D1/CP47 in a mutant lacking CP43 (Supplemental Figure 9A), which is at variance with the effect of a knockout of the FtsH gene (slr0228) in Synechocystis, which stabilized various PSII assembly intermediates in PSII mutants lacking CP43 or impaired in the assembly or stability of the manganese cluster (Komenda et al., 2006, 2010). Although we cannot fully exclude a residual FtsH activity that would degrade these unassembled, or partly assembled, subunits in the ftsh1-1 mutated context, we tentatively attribute these differences to the lower protein quality control in cyanobacterial thylakoid membranes when compared with C. reinhardtii chloroplasts (Wollman et al., 1999). In particular, crippled PSII complexes are less efficiently degraded in Synechocystis (de Vitry et al., 1989; Vermaas, 1998) than in photosynthetic eukaryotes. These observations suggest the presence of an as yet unidentified additional housekeeping protease that degrades these subunits in the thylakoid membranes of C. reinhardtii.
FtsH Is Involved in Quality Control and the Regulation of Accumulation of Cytochrome b6f Complexes and CCB Proteins
The first suggestion of a cytochrome b6f subunit degraded by FtsH was the unassembled Rieske iron-sulfur protein imported into isolated chloroplasts in vitro (Ostersetzer and Adam, 1997). The involvement of FtsH in cytochrome b6f degradation in vivo is demonstrated here with the identification of the ftsh1-1 mutant by the present genetic suppressor strategy. In marked contrast to the continued degradation of the cytochrome b6f subunits when a subunit is absent (Figure 9), a diversity of labile ci heme–deficient cytochrome b6f complexes proved resistant to degradation in the ftsh1-1 mutated context. They ranged from site-directed substitution of the Cys involved in the binding of the ci heme (Figure 5) to site-directed substitution of the His ligand of the bh heme (Malnoë et al., 2011) and to mutants in the CCB heme binding pathway (Figure 2). These observations demonstrate that FtsH operates as a major quality control protease that recognizes altered conformations in cytochrome b6f assemblies to trigger their degradation through the combined action of its unfoldase and proteolytic activities. A similar situation may prevail in Saccharomyces cerevisiae mitochondria, where FtsH proteases with the catalytic site facing the matrix (m-FtsH) Yta10/Yta12 have been described as involved in maturase-dependent intron splicing of cytochrome b transcripts. However, when using an intronless mitochondrial genome where cytochrome b synthesis was not affected, m-FtsH was shown to contribute to posttranslational bc1 complex assembly and degradation (Guzélin et al., 1996; Arlt et al., 1998; Van Dyck and Langer, 1999). As we report here for sulfur starvation (Figures 8B and 8D) and elsewhere for nitrogen starvation (Bulté and Wollman, 1992; Wei et al., 2013), FtsH has a prominent role in regulating the accumulation level of cytochrome b6f complexes upon these macronutrient stresses in low light or in the dark.
We also found that degradation of unassembled CCB4 in the absence of CCB2 (and vice versa), both of which are transmembrane proteins involved in the ci heme binding pathway to cytochrome b6 (Saint-Marcoux et al., 2009), proved to be regulated by FtsH (Figure 1B), in line with our report elsewhere (Wei et al., 2013) of a global role for FtsH in the degradation of photosynthetic complexes and their assembly factors.
The ClpP protease also has been demonstrated to play a role in cytochrome b6f degradation in C. reinhardtii (Majeran et al., 2000), as does Deg1 on cytochrome b6 in Arabidopsis (Zienkiewicz et al., 2012). However, Clp and FtsH may have distinct actions on the degradation of cytochrome b6f complexes, since a Clp-deficient mutant was unable to protect b6f complexes from degradation in mutants lacking ci heme (Majeran et al., 2000), while the ftsh1-1 mutation did so (Figure 5).
Here, we observed a cytochrome b6f degradation process that was selectively activated in sulfur-deficient conditions. Cytochrome b6f was degraded in the light but not in darkness in the ftsh1-1 mutant, suggesting the action of some additional protease upon illumination. Steady state levels of the chloroplast proteases that we probed, including ClpP1 and Deg5, were rather reduced upon sulfur starvation in the light, but we cannot rule out that some proteases might have better access to cytochrome b6f or be activated by oligomerization in the light in these starvation conditions.
FtsH1 Regulates the Degradation of PSII upon Photoinhibition and Phosphorus and Sulfur Starvation
In cyanobacteria, FtsH has mainly been studied for its role in D1 degradation (reviewed in Nixon et al., 2010; Komenda et al., 2012) and the removal of other damaged or unassembled PSII proteins (Komenda et al., 2006, 2010; Boehm et al., 2011). In one study, FtsH was also shown to contribute to the regulation of PSI amounts (Mann et al., 2000). The mechanism for D1 protein degradation is not fully elucidated, but it is known to involve the reaction of damaging reactive oxygen species, partial disassembly of the PSII complex to form the RC47 complex, and the action of several proteases, notably FtsH, that selectively remove damaged D1 (reviewed in Edelman and Mattoo, 2008; Komenda et al., 2012). The best-documented function of FtsH in thylakoid membranes is in this PSII repair cycle (reviewed in Kato and Sakamoto, 2009). A model for the photoinhibition of cyanobacteria has been proposed in which damaged D1 is removed by FtsH heterohexamers, with degradation proceeding from the N terminus of D1 in a highly processive manner without the requirement of any other proteases (reviewed in Nixon et al., 2010). In particular, photoinhibition studies with the ΔhtrA/hhoA/hhoB triple mutant, defective for the three genes encoding Deg proteases in Synechocystis sp PCC6803, showed that Deg proteases are not required for the removal of damaged D1 protein during the PSII repair cycle when exposed to high light intensity (Barker et al., 2006). These results are in contrast to several photoinhibition studies performed with Arabidopsis showing that PSII protein turnover required both the involvement of FtsH and Deg/Htr proteases (Itzhaki et al., 1998; Kapri-Pardes et al., 2007; Sun et al., 2007). High-light treatment was reported to increase the expression of Deg/Htr and FtsH types of proteases in Arabidopsis (Sinvany-Villalobo et al., 2004), to raise the level of FtsH hexamerization and unstacking of thylakoids (reviewed in Yoshioka and Yamamoto, 2011), and to activate the lumenal Deg protease, based on in vitro experiments showing its hexamerization upon acidification (Kley et al., 2011). Thus, a model for D1 degradation in the chloroplast was proposed in which Deg proteases act as endoproteases fragmenting D1 and FtsH proteases act as exoproteases processively degrading D1 and its fragments (reviewed in Kato and Sakamoto, 2009). The reason why cyanobacterial FtsH would be capable of processive degradation of the whole D1 protein from the stromal side, while the ability of its homolog in the chloroplast is far reduced, may rest in the difference in subunit composition of FtsH from the two types of organisms as well as from the embedment of chloroplast D1 in grana membranes, where it has increased interaction with transmembrane antenna proteins and lumenal oxygen-evolving enhancer subunits (for further discussion, see Edelman and Mattoo, 2008).
Here, in agreement with previous photoinhibition studies from vascular plant chloroplasts and cyanobacteria, we showed that the ftsh1-1 mutation increased PSII sensitivity to photoinhibition and impaired proteolytic degradation and replacement of D1 by a de novo synthesized copy (Figure 6). There is a striking similarity in the fate of D1 in the ftsh1-1 mutant when subjected to light stress or phosphorus starvation under moderate illumination, both of which lead to PSII inactivation in C. reinhardtii (Wykoff et al., 1998; Zhang et al., 2002). Here, we have demonstrated that, indeed, compromised photosynthetic activity upon phosphorus starvation was mainly due to enhanced photoinhibition (Figure 7). In contrast, PSII inactivation upon sulfur starvation cannot be attributed solely to photoinhibition, since it develops even more strongly when sulfur starvation is performed in complete darkness. Since PSII inactivation and D1 degradation in the ftsh1-1 mutant are markedly increased upon sulfur starvation in the light but fully prevented when performed in darkness (Figure 8), one could consider a light-induced upregulation of ClpP when FtsH activity is defective, as recently demonstrated (Kato et al., 2012). Although we did not detect more ClpP in ftsh1-1 than in the wild-type strain in these conditions (Supplemental Figure 7E), we cannot exclude an upregulation of ClpP activity through changes in its state of oligomerization, substrate accessibility, or ATP availability.
FtsH1 Regulates the Degradation of PSII by Removing D1 Fragments
A clear consequence of the inactivation of FtsH in the ftsh1-1 mutant was the detection of D1 fragments upon photoinhibition, which suggests that primary endoproteolytic cuts occurred in the mutant but that their processive degradation by FtsH was inhibited (Figure 6C). According to what has been proposed for plant chloroplasts, there could be a joint action of Deg and FtsH proteases in the degradation and repair of D1 upon photoinhibition of C. reinhardtii. Indeed, the set of chloroplast Deg proteases is similar in Arabidopsis (lumenal Deg1, Deg5, and Deg8 and stromal Deg2, Deg6, Deg7, Deg9, and Deg16) and in C. reinhardtii (lumenal Deg1A, Deg1B, Deg1C, Deg5, and Deg8 and stromal Deg2, Deg7, and Deg9; Huesgen et al., 2009; Schroda and Vallon, 2009; Sun et al., 2010). The model of Kato and Sakamoto (2009), derived from studies with Arabidopsis, predicts the production of Deg-dependent fragments whose apparent molecular mass and epitope content match nicely to those we detected with the two distinct D1 antibodies we used (Supplemental Figure 10).
The physiological significance of the Deg-dependent D1 degradation pathway has been under debate (for discussion, see Edelman and Mattoo, 2008; Komenda et al., 2012). Is it synergistic with FtsH, or is it an escape pathway that supplements the FtsH-mediated one to accelerate D1 degradation under photoinhibitory conditions? We conclude that in our experimental conditions, when the function of FtsH is impaired, the Deg pathway would be rather active, with 20% of the C-terminal epitopes of D1 being found in its fragments (Figure 6C).
We note, however, that most of these fragments accumulated to a limited extent. This could be due to a negative feedback inhibition of the Deg proteases by the accumulation of these early D1 degradation products whose stroma-exposed domains are not processed by FtsH. This is consistent with the observation that the only degradation product that accumulates extensively in the ftsh1-1 mutant is in the ∼6-kD region, where the C-terminal fragment of D1, which is lumen localized, should be found. This fragment differs from all the others in that it should never have access to the stroma-located proteolytic chamber of FtsH in a wild-type context and, therefore, should not be degraded by FtsH. Its increased production in the ftsh1-1 mutant, as compared with the wild type, probably results from the slower digestion of D1 products by FtsH, thus releasing more 6-kD fragments in the lumen.
A conspicuous feature of the various PSII degradation processes upon macronutrient starvation, whether it be phosphorus or sulfur starvation under moderate illumination or sulfur starvation in the dark, is the production of D1 fragments in the ftsh1-1 mutated context. With the exception of the 23-kD fragment, which is detected only upon illumination and serves as a diagnostic of photoinhibition, as suggested previously (Lindahl et al., 2000), the D1 fragmentation patterns upon macronutrient stress (Figures 7B, 8B, and 8D) are very similar to one resulting from photoinhibition (Figure 6C), with a group of fragments accumulating in the 16- to 20-kD and 6- to 10-kD regions. These observations strongly suggest a contribution of the Deg protease family to the degradation of D1 upon nutrient stress as they do upon photoinhibition. Indeed, Degs were also proposed to contribute to D1 degradation under stress conditions by cleaving cross-links to neighboring proteins (Komenda et al., 2012). A modest expression increase in lumenal Deg1C in C. reinhardtii has been found upon phosphorus and sulfur starvation (Zhang et al., 2004; Moseley et al., 2006), and a 3-fold increase of the transcript level of stromal Deg2 upon sulfur starvation in the light in the wild-type strain was also observed (González-Ballester et al., 2010). Here, however, we detected a net decrease of the amount of chloroplast proteases upon sulfur depletion in the presence of acetate (Supplemental Figure 7E). In the latter experimental conditions, D1 fragments transiently overaccumulate in the ftsh1-1 mutant (Figure 8B), which supports a degradation pathway in which Deg proteases produce a first set of early endoproteolytic cuts that can be continued only once the resulting D1 fragments are themselves degraded by processive proteases such as FtsH.
It is of note that light-induced oxidative damage to D1 should not be considered as a prerequisite for the activation of its degradation by the combined effects of FtsH and Deg proteases, since D1 degradation yields the same 16- to 20-kD and 6- to 10-kD fragments when operating in the dark in sulfur stress conditions (Figure 8D). However, the activation of the Deg proteases in the lumen has been shown to be triggered by the high ΔpH formed in the light (Kley et al., 2011). How they are activated in the dark remains unclear unless a high proton gradient is present during sulfur starvation in the dark, an issue that needs to be addressed in the future.
Enhanced Degradation of PSII upon Sulfur Starvation in the Dark Is Fully FtsH Dependent
That PSII degradation was dramatically enhanced when sulfur starvation was performed in the dark was unexpected (Figure 8D). In this case, PSII degradation became a fully FtsH-dependent process. The rationale for this targeted degradation of PSII in darkness upon sulfur starvation remains unclear. It is not a consequence of functional damage to PSII complexes, since the ftsh1-1 mutant, which does not degrade PSII, displays a fluorescence pattern with a high Fv/Fm typical of active PSII centers (Figure 8C). It should not be aimed either at scavenging sulfur, since most of the sulfur-containing cofactors for photosynthesis are found within PSI and not in association with PSII. Interestingly, we noted a spectacular increase in the accumulation of D1/D2 oligomers in the ftsh1-1 mutant relative to the wild type when deprived of sulfur in the dark (Supplemental Figure 8). Similar cross-linking products or adducts had been reported previously either during a light stress or in the dark when a heat or oxidative stress was applied (reviewed in Yamamoto et al., 2008). The accumulation of these oligomers in the dark during a nutrient stress is highly unusual and raises the question of how they are formed. These oligomers may be the privileged substrate for PSII degradation by FtsH upon sulfur stress in the dark. Being resistant to other proteases, they might be a form of active PSII, which is worthy of further investigation.
The mechanism of activation of this degradation pathway is unknown and might be related to sulfur stress–induced expression of chaperones. Indeed, it has been shown in E. coli that chaperones can facilitate degradation by FtsH (Rodriguez et al., 2008). The chloroplast DnaJ chaperone Atg8, induced in darkness and stabilized by FtsH (Chen et al., 2011), is thus a good candidate for being involved in the dark-activated and FtsH-dependent degradation of PSII upon sulfur deprivation.
METHODS
Phylogenetic Analysis
Multiple sequence alignment was performed using ClustalW2.1 (Larkin et al., 2007) with default parameters followed by manual editing. After removing ambiguously aligned N- and C-terminal regions, 639 sites were retained for the phylogenetic analysis (Supplemental Data Set 1). The best-fit amino acid substitution model was determined using ProtTest2.4 (Abascal et al., 2005). The best-fit model was the Le-Gascuel model (Le and Gascuel, 2008) supplemented with a γ distribution for rate variations among sites (LG+G). The tree was reconstructed by the maximum likelihood method based on the LG+G model using RaxML7.2.6 (Stamatakis, 2006). Statistical support for nodes was assessed with 1000 bootstrap replicates.
Chlamydomonas reinhardtii Strains, Growth Conditions, and Genetic Methods
C. reinhardtii mutants have been described previously and were as follows: ccb, {petB-C35V}, {petB-H202Q}, {petB-H187G}, {ΔpetB}, {ΔpetA}, {Fud34}, {Fud7}, {D1TR}, arg7, and arg2 (Eversole, 1956; Matagne, 1978; de Vitry et al., 1989, 2004; Kuras and Wollman, 1994; Kuras et al., 1997, 2007; Minai et al., 2006). Cells were grown at 25°C in Tris-acetate-phosphate (TAP) medium or, when indicated, in minimal medium lacking acetate, pH 7.2, under dim light at 6 μE m−2 s−1 and collected during the exponential phase at 2 × 106 cells mL−1. Plasmid pΔpetA-D was constructed by ligating the ScaI-PstI fragment of 4296 bp of plasmid pΔpetA (Kuras and Wollman, 1994) bearing the ΔpetA deletion with the ScaI-PstI fragment of 2134 bp of plasmid pKfΔpetD (Choquet et al., 2003) bearing the ΔpetD deletion and containing the aadA cassette conferring spectinomycin resistance. Plasmid pΔpetA-D was transformed by the biolistic method into C. reinhardtii wild-type strain (Boynton et al., 1988; Kuras and Wollman, 1994). Nonphotosynthetic {ΔpetAD} transformants were screened in the dark on TAP-agar-spectinomycin medium, and homoplasmy was checked by RNA gel blotting for the absence of PETA and PETD mRNA as described (Drapier et al. 2002) with probes derived from coding sequences (Eberhard et al., 2002). UV light mutagenesis on ccb strains to obtain phototrophic revertants was performed as described (Li et al., 1996). Plasmid pSL18-FTSH1 was constructed by ligating the NdeI/XbaI-digested fragment of 3.5 kb (partially digested to avoid cutting the NdeI site in FtsH1g) of the amplified PCR-coding phase of FtsH1 genomic DNA, adding the NdeI site before and the XbaI site after the coding sequence, with the NdeI/XbaI-digested fragment of 6069 bp of vector pSL18 (Depège et al., 2003) carrying an expression cassette composed of the PSAD promoter and polyadenylation site and a paromomycin resistance cassette. Plasmids pSL18-FTSH1 and pSL18 were transformed by the electroporation method (Shimogawara et al., 1998) into C. reinhardtii ftsh1-1 and ftsh1-1 ccb2 mutants. Transformants were screened on TAP-agar-paromomycin medium in the presence of 10 μg mL−1 paromomycin and analyzed by fluorescence; the expression of wild-type FtsH1 was checked by immunodetection. Crosses were performed and vegetative diploids were isolated according to published protocols (Harris, 1989).
Map-Based Cloning of Su1-ccb2
The C. reinhardtii Su1-ccb2 mutant was crossed with the interfertile species Chlamydomonas grossii, which shows suitable profusion of genomic polymorphism (Rymarquis et al., 2005). In each of 127 tetrads, we took one progeny carrying the Su1-ccb2 mutation, as identified by its photosensitivity, for linkage analysis to various markers by a PCR method. The primers were used in a single reaction and generated PCR products that differ in size for the C. reinhardtii or the C. grossii allele and were assayed directly on an agarose gel to distinguish alleles.
PCR Detection of the Wild-Type FTSH1 Allele
DNA templates were purified from a pin point of C. reinhardtii cells resuspended in 10 μL of water to which was added 10 μL of ethanol and 80 μL of 5% Chelex 100 molecular biology–grade resin (Bio-Rad) and incubated for 8 min at 95°C. One-microliter samples were used in 10-μL reactions performed in a thermocycler (PTC-200; MJ Research) in a PCR buffer containing 1 M betaine, high-fidelity buffer, and polymerase (PCR extender system; 5PRIME). The FtsH1-WTS allele-specific primer 5′-CTGCTGCGCCCCGGCC-3′ was used for the amplification of wild-type FTSH1 genomic DNA with reverse FtsH1-A3 primer 5′-CGCCCCTTATGCAGTGCG-3′. At the stringent annealing temperature of 65°C, these primers allowed the detection of the wild-type FTSH1 allele by specific amplification on genomic DNA of an FtsH1-WTS/FtsH1-A3 product of 277 bp (Supplemental Figure 11).
Chlorophyll Fluorescence Analysis
Fluorescence kinetics were performed with a home-built fluorometer and fluorescence camera (Johnson et al., 2009) by using exponential phase cultures maintained in aerobic conditions by vigorous agitation in the dark before the experiments. Fluorescence was triggered by continuous actinic light at 590 nm. Fm values were determined in the presence of 10 μM DCMU, an inhibitor of the PSII acceptor side, or by a 250-ms saturating flash delivered between two 150-μs probe pulses, delivered 300 ms apart. Means and sd of two independent experiments for each starvation condition are given.
Protein Isolation and Immunoblot Analysis
For polypeptide analysis, cells were resuspended in 200 mM 1,4-DTT and 200 mM Na2CO3 and solubilized in the presence of 2% SDS at 100°C for 50 s. Polypeptides were separated by SDS/urea-PAGE on 12 to 18% SDS-polyacrylamide gels in the presence of 8 M urea (de Vitry et al., 1989), except when indicated differently. Heme peroxidase activity was detected by using 3,3′,5,5′-tetramethylbenzidine (Thomas et al., 1976) or on blots by using chemiluminescence (de Vitry et al., 2004). Proteins were electrotransferred onto Immobilon NC membranes (Westran S membranes; GE Healthcare) in a semidry blotting apparatus at 0.8 mA cm−2 for 45 min. Rabbit-specific antibodies against peptides of FtsH1 (DFGRSKSKFQEVPET) or FtsH2 (QVSVDLPDQKGRLEI) were produced and purified by peptide affinity by Eurogentec. For immunodetection by the chemiluminescence method (GE Healthcare), we used antisera against cytochrome b6, cytochrome f, cytochrome b6f subunit IV, cytochrome b6f iron-sulfur Rieske protein, PSII extrinsic subunits OEE2 and OEE3, SLT2 (Agrisera AS07 244), and GrpE at a 1:10,000 dilution; ATP synthase CF1 and F1 β-subunit at a 1:100,000 dilution; LhcbM5 (LHCII) at a 1:20,000 dilution; PSII subunits CP43, CP47, and D2 (Agrisera AS06 146) and ClpP1 at a 1:5000 dilution; D1 C terminus (Agrisera AS05 084) at a 1:100,000 dilution; PHOX and D1 DE loop (Agrisera AS10 704) at a 1:8000 dilution; D1 N terminus (Agrisera AS11 1786) at a 1:5000 dilution; FtsH1 of Arabidopsis thaliana at a 1:80,000 dilution; FtsH1 and FtsH2 of C. reinhardtii at a 1:5000 dilution; Deg5 (Agrisera AS10 703) at a 1:2500 dilution; and CCB2 and CCB4 at a 1:300 dilution.
BN-PAGE and Immunoprecipitations
Membranes were fractionated from sonicated cell lysates (Saint-Marcoux et al., 2009). All steps up to the second dimension analysis were performed at 4°C. Prepared membranes were resuspended in 50 mM Bis-Tris, pH 7.0, 750 mM aminocaproic acid, 0.5 mM EDTA, supplemented with 1× complete EDTA-free protease inhibitor mixture (Roche) at a final chlorophyll concentration of 0.75 mg mL−1. For PSII complex fractionation (Figure 8E), membranes were solubilized by 3% (w/v) freshly prepared n-dodecyl-β-d-maltoside for 10 min on ice. Supernatant was collected after centrifugation in a TLA-100 rotor (Beckmann Coulter) at 279,000g for 10 min. For FtsH complex fractionation (Figure 3D), membranes were solubilized by 4% (w/v) freshly prepared digitonin for 30 min at 4°C, and supernatant was collected after centrifugation at 16,110g for 30 min. For each sample, 10 μL of supernatant (corresponding to 5 μg of chlorophyll) complemented to 0.5% Coomassie Brilliant Blue G 250 was loaded on a precast 3 to 12% NativePAGE Bis-Tris gel (Invitrogen). Electrophoresis was performed with cathode buffer (50 mM Tricine, 15 mM Bis-Tris, pH 7.0, and 0.04% Coomassie Brilliant Blue G 250) and anode buffer (50 mM Bis-Tris, pH 7.0) for 20 min at 50 V and then 2 h at 150 V. For the denaturing second dimension analysis, strips cut from the first dimension gel were incubated at room temperature for 15 min in 2% SDS, 66 mM Na2CO3, and 0.67% β-mercaptoethanol and applied on 12% (Figure 8E) or 8% (Figure 3D) SDS-polyacrylamide gels containing 8 M urea for separating PSII subunits and FtsH subunits, respectively. After electrophoresis, protein content was detected by immunoblotting. For immunoprecipitation (Figure 3E), antibodies and preimmune serum were coupled to protein A–Sepharose beads (Schroda et al., 2001). The coupled beads were washed twice in 50 mM Bis-Tris, pH 7.0, 750 mM aminocaproic acid, and 0.5 mM EDTA with 0.25% digitonin and incubated with digitonin-solubilized membranes (prepared as for BN-PAGE) for 1 h at 4°C. Afterward, the coupled beads were washed three times in the previous solution followed by two washes in 10 mM Tris-HCl, pH 7.5. Immunoprecipitated proteins were recovered by boiling the beads in 2% SDS and 12% Suc for 50 s. After centrifugation, the supernatant was complemented to 0.1 M DTT and 0.1 M Na2CO3 and analyzed by SDS-PAGE. Protein gel blots of immunoprecipitated samples were detected using horseradish peroxidase–conjugated protein A instead of horseradish peroxidase–conjugated secondary antibodies to decrease the background (Lal et al., 2005).
Photoinhibition
Strains to be compared were diluted from precultures to equal cell densities and cultured for an additional 15 h before photoinhibition (Schroda et al., 1999). Cells at 2 × 106 cells mL−1 were transferred to 500-mL Erlenmeyer flasks, and these were placed onto equally illuminated spots of a rotary shaker below LEDs. Inhibitors of the translation of chloroplast genes (500 μg mL−1 lincomycin and 100 μg mL−1 chloramphenicol) were added or not just before photoinhibition. Cells were photoinhibited at 23°C for 1 h at 250 μE m−2 s−1. During the light treatment, the temperature of the cultures remained constant. Recovery of photosynthesis was allowed to take place at 6 μE m−2 s−1.
Nutrient Starvation
Precultures were grown at 25°C in TAP medium, pH 7.2, at 6 μE m−2 s−1 or in the dark for nutrient starvations, respectively, in the light or in the dark. For phosphorus starvation, cells of precultures in the exponential phase were washed twice in TAP phosphorus-deprived medium, resuspended at 5 × 105 cells mL−1, illuminated at 80 μE m−2 s−1 or maintained in the dark, washed again, and resuspended to 106 cells mL−1 after 2 d (Wykoff et al., 1998). For sulfur starvation, cells of precultures in the exponential phase were washed twice in TAP sulfur-deprived medium, resuspended at 5 × 105 cells mL−1, and illuminated at 80 μE m−2 s−1 or maintained in the dark (Wykoff et al., 1998; Zhang et al., 2002). For comparison of sulfur starvation at 80 μE m−2 s−1 in the presence and absence of acetate, precultures were grown at 25°C in TAP medium at 6 μE m−2 s−1 followed by 15 h at 80 μE m−2 s−1 before sulfur starvation. Cells of precultures in the exponential phase were collected, washed twice in TAP sulfur-deprived medium or in minimum sulfur-deprived medium, resuspended at 5 × 105 cells mL−1, and maintained at 80 μE m−2 s−1.
Accession Numbers
Sequence data from this article can be found in GenBank/Swiss-Prot database under the following accession numbers: Cr-FtsH1, EDP05335.1; Ec-FtsH, AAA23813.1; and Tm-FtsH, Q9WZ49.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Unrooted Phylogenetic Tree of FtsH Proteins.
Supplemental Figure 2. Backcross of Rccb2-306 mt− with ccb2 mt+ Yields Only Parental Ditype Tetrads, Showing That the Suppressor Mutation Is Monogenic and Nuclear.
Supplemental Figure 3. Backcross of Su1-ccb2 with ccb2 Confirms the Suppressor Effect.
Supplemental Figure 4. Map-Based Cloning of the Suppressor.
Supplemental Figure 5. Mutation ftsh1-1 Is Recessive in Vegetative Diploids.
Supplemental Figure 6. PSII Subunit D1 Did Not Coimmunoprecipitate with the Antibody against the FtsH1 Peptide.
Supplemental Figure 7. Cell Response Is Delayed and Thylakoid Proteases Are Preserved upon Sulfur Starvation When Cell Viability Solely Depends on Photosynthesis.
Supplemental Figure 8. D1/D2 Oligomers Accumulate Only in the ftsh1-1 Mutant upon Sulfur Starvation in the Dark.
Supplemental Figure 9. ftsh1-1 Does Not Increase the Accumulation of PSII Integral Subunits Lacking Assembly Partners.
Supplemental Figure 10. D1 Fragments.
Supplemental Figure 11. PCR Test Used to Discriminate between ftsh1-1 and Wild-Type FTSH1 Alleles.
Supplemental Data Set 1. FtsH Sequence Alignment of the Sites Retained for the Phylogenetic Analysis.
Supplementary Material
Acknowledgments
We thank Dominique Drapier and Alix Boulouis for sharing their expertise in map-based cloning, Yves Choquet for assistance with {ΔpetAD} mutant construction, Stéphane Lemaire for the pSL18 plasmid, Karim Mouzannar for help in pSL18-FTSH1 construction, Adrien Quiles, Palak Garg, and Sarah Pousset for assistance with mutant characterization, Olivier Vallon and Nicolas Tourasse for help in FtsH phylogeny, and Arthur Grossman, Kevin Redding, Michael Schroda, Yuichiro Takahashi, and Francesca Zito for PHOX, PsaA, GrpE, LhcbM5, and subunit IV antibodies, respectively. This work was funded by the Agence Nationale de la Recherche (projects ANR-07-BLAN-0114, ANR-12-BSV8-0011-01, and ANR-11-LABX-0011), the European Union (project 245070 FP7-KBBE-2009-3 SUNBIOPATH), and the Unité Mixte de Recherche 7141/Centre National de la Recherche Scientifique/Université Paris 6.
AUTHOR CONTRIBUTIONS
A.M., J.G.-B., and C.d.V. isolated C. reinhardtii ccb2 revertant strains and did the genetic and molecular analysis. A.M., F.W., F.-A.W., and C.d.V. performed biochemical and physiological analyses of mutant ftsh1-1 and of strains combining the ftsh1-1 mutation with assembly mutants of photosynthetic complexes. All authors contributed to designing the study, analyzing the data, and writing the article. C.d.V. coordinated the project.
Glossary
- PSII
photosystem II
- PSI
photosystem I
- SRH
second region of homology
- BN
Blue Native
- Fm
maximal fluorescence
- Fv/Fm
maximum quantum efficiency of photosystem II primary photochemistry
- ΦPSII
quantum yield of photosystem II photochemistry
- LC
lincomycin and chloramphenicol
- PHOX
alkaline phosphatase
- CES
control by epistasy of synthesis
- TAP
Tris-acetate-phosphate
Footnotes
Online version contains Web-only data.
References
- Abascal F., Zardoya R., Posada D. (2005). ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105 [DOI] [PubMed] [Google Scholar]
- Adam Z., Rudella A., van Wijk K.J. (2006). Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr. Opin. Plant Biol. 9: 234–240 [DOI] [PubMed] [Google Scholar]
- Ahmadian M.R., Stege P., Scheffzek K., Wittinghofer A. (1997). Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat. Struct. Biol. 4: 686–689 [DOI] [PubMed] [Google Scholar]
- Allmer J., Naumann B., Markert C., Zhang M., Hippler M. (2006). Mass spectrometric genomic data mining: Novel insights into bioenergetic pathways in Chlamydomonas reinhardtii. Proteomics 6: 6207–6220 [DOI] [PubMed] [Google Scholar]
- Arlt H., Steglich G., Perryman R., Guiard B., Neupert W., Langer T. (1998). The formation of respiratory chain complexes in mitochondria is under the proteolytic control of the m-AAA protease. EMBO J. 17: 4837–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atteia A., et al. (2009). A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the alpha-proteobacterial mitochondrial ancestor. Mol. Biol. Evol. 26: 1533–1548 [DOI] [PubMed] [Google Scholar]
- Bailey S., Thompson E., Nixon P.J., Horton P., Mullineaux C.W., Robinson C., Mann N.H. (2002). A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J. Biol. Chem. 277: 2006–2011 [DOI] [PubMed] [Google Scholar]
- Baker N.R. (2008). Chlorophyll fluorescence: A probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59: 89–113 [DOI] [PubMed] [Google Scholar]
- Barker M., de Vries R., Nield J., Komenda J., Nixon P.J. (2006). The deg proteases protect Synechocystis sp. PCC 6803 during heat and light stresses but are not essential for removal of damaged D1 protein during the photosystem two repair cycle. J. Biol. Chem. 281: 30347–30355 [DOI] [PubMed] [Google Scholar]
- Bieniossek C., Schalch T., Bumann M., Meister M., Meier R., Baumann U. (2006). The molecular architecture of the metalloprotease FtsH. Proc. Natl. Acad. Sci. USA 103: 3066–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M., Nield J., Zhang P., Aro E.M., Komenda J., Nixon P.J. (2009). Structural and mutational analysis of band 7 proteins in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 191: 6425–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M., Romero E., Reisinger V., Yu J., Komenda J., Eichacker L.A., Dekker J.P., Nixon P.J. (2011). Investigating the early stages of photosystem II assembly in Synechocystis sp. PCC 6803: Isolation of CP47 and CP43 complexes. J. Biol. Chem. 286: 14812–14819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M., Yu J., Krynicka V., Barker M., Tichy M., Komenda J., Nixon P.J., Nield J. (2012). Subunit organization of a Synechocystis hetero-oligomeric thylakoid FtsH complex involved in photosystem II repair. Plant Cell 24: 3669–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton J.E., Gillham N.W., Harris E.H., Hosler J.P., Johnson A.M., Jones A.R., Randolph-Anderson B.L., Robertson D., Klein T.M., Shark K.B., Sanford J.C. (1988). Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 240: 1534–1538 [DOI] [PubMed] [Google Scholar]
- Bulté L., Wollman F.A. (1992). Evidence for a selective destabilization of an integral membrane protein, the cytochrome b6/f complex, during gametogenesis in Chlamydomonas reinhardtii. Eur. J. Biochem. 204: 327–336 [DOI] [PubMed] [Google Scholar]
- Chen K.M., Piippo M., Holmström M., Nurmi M., Pakula E., Suorsa M., Aro E.M. (2011). A chloroplast-targeted DnaJ protein AtJ8 is negatively regulated by light and has rapid turnover in darkness. J. Plant Physiol. 168: 1780–1783 [DOI] [PubMed] [Google Scholar]
- Choquet Y., Zito F., Wostrikoff K., Wollman F.A. (2003). Cytochrome f translation in Chlamydomonas chloroplast is autoregulated by its carboxyl-terminal domain. Plant Cell 15: 1443–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y., Stern D.B., Wostrikoff K., Kuras R., Girard-Bascou J., Wollman F.A. (1998). Translation of cytochrome f is autoregulated through the 5′ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc. Natl. Acad. Sci. USA 95: 4380–4385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depège N., Bellafiore S., Rochaix J.D. (2003). Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299: 1572–1575 [DOI] [PubMed] [Google Scholar]
- de Vitry C., Desbois A., Redeker V., Zito F., Wollman F.A. (2004). Biochemical and spectroscopic characterization of the covalent binding of heme to cytochrome b6. Biochemistry 43: 3956–3968 [DOI] [PubMed] [Google Scholar]
- de Vitry C., Olive J., Drapier D., Recouvreur M., Wollman F.A. (1989). Posttranslational events leading to the assembly of photosystem II protein complex: A study using photosynthesis mutants from Chlamydomonas reinhardtii. J. Cell Biol. 109: 991–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapier D., Girard-Bascou J., Stern D.B., Wollman F.A. (2002). A dominant nuclear mutation in Chlamydomonas identifies a factor controlling chloroplast mRNA stability by acting on the coding region of the atpA transcript. Plant J. 31: 687–697 [DOI] [PubMed] [Google Scholar]
- Eberhard S., Drapier D., Wollman F.A. (2002). Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 31: 149–160 [DOI] [PubMed] [Google Scholar]
- Edelman M., Mattoo A.K. (2008). D1-protein dynamics in photosystem II: The lingering enigma. Photosynth. Res. 98: 609–620 [DOI] [PubMed] [Google Scholar]
- Eversole R.A. (1956). Biochemical mutants of Chlamydomonas reinhardtii. Am. J. Bot. 43: 404–407 [Google Scholar]
- Ferro M., et al. (2010). AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell. Proteomics 9: 1063–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G., Giacomelli L., Ytterberg A.J., Peltier J.B., Rudella A., Sun Q., Wijk K.J. (2004). In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: New proteins, new functions, and a plastid proteome database. Plant Cell 16: 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genty B., Briantais J.M., Baker N.R. (1989). The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 990: 87–92 [Google Scholar]
- González-Ballester D., Casero D., Cokus S., Pellegrini M., Merchant S.S., Grossman A.R. (2010). RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell 22: 2058–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzélin E., Rep M., Grivell L.A. (1996). Afg3p, a mitochondrial ATP-dependent metalloprotease, is involved in degradation of mitochondrially-encoded Cox1, Cox3, Cob, Su6, Su8 and Su9 subunits of the inner membrane complexes III, IV and V. FEBS Lett. 381: 42–46 [DOI] [PubMed] [Google Scholar]
- Hallmann A. (1999). Enzymes in the extracellular matrix of Volvox: An inducible, calcium-dependent phosphatase with a modular composition. J. Biol. Chem. 274: 1691–1697 [DOI] [PubMed] [Google Scholar]
- Harris, E.H. (1989). The Chlamydomonas Source Book. (San Diego, CA: Academic Press). [Google Scholar]
- Heinemeyer J., Eubel H., Wehmhöner D., Jänsch L., Braun H.P. (2004). Proteomic approach to characterize the supramolecular organization of photosystems in higher plants. Phytochemistry 65: 1683–1692 [DOI] [PubMed] [Google Scholar]
- Herman C., Prakash S., Lu C.Z., Matouschek A., Gross C.A. (2003). Lack of a robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol. Cell 11: 659–669 [DOI] [PubMed] [Google Scholar]
- Huesgen P.F., Schuhmann H., Adamska I. (2009). Deg/HtrA proteases as components of a network for photosystem II quality control in chloroplasts and cyanobacteria. Res. Microbiol. 160: 726–732 [DOI] [PubMed] [Google Scholar]
- Ito K., Akiyama Y. (2005). Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59: 211–231 [DOI] [PubMed] [Google Scholar]
- Itzhaki H., Naveh L., Lindahl M., Cook M., Adam Z. (1998). Identification and characterization of DegP, a serine protease associated with the luminal side of the thylakoid membrane. J. Biol. Chem. 273: 7094–7098 [DOI] [PubMed] [Google Scholar]
- Johnson X., Vandystadt G., Bujaldon S., Wollman F.A., Dubois R., Roussel P., Alric J., Béal D. (2009). A new setup for in vivo fluorescence imaging of photosynthetic activity. Photosynth. Res. 102: 85–93 [DOI] [PubMed] [Google Scholar]
- Kapri-Pardes E., Naveh L., Adam Z. (2007). The thylakoid lumen protease Deg1 is involved in the repair of photosystem II from photoinhibition in Arabidopsis. Plant Cell 19: 1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karata K., Verma C.S., Wilkinson A.J., Ogura T. (2001). Probing the mechanism of ATP hydrolysis and substrate translocation in the AAA protease FtsH by modelling and mutagenesis. Mol. Microbiol. 39: 890–903 [DOI] [PubMed] [Google Scholar]
- Karata K., Inagawa T., Wilkinson A.J., Tatsuta T., Ogura T. (1999). Dissecting the role of a conserved motif (the second region of homology) in the AAA family of ATPases. Site-directed mutagenesis of the ATP-dependent protease FtsH. J. Biol. Chem. 274: 26225–26232 [DOI] [PubMed] [Google Scholar]
- Kashino Y., Lauber W.M., Carroll J.A., Wang Q., Whitmarsh J., Satoh K., Pakrasi H.B. (2002). Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41: 8004–8012 [DOI] [PubMed] [Google Scholar]
- Kato Y., Sakamoto W. (2009). Protein quality control in chloroplasts: A current model of D1 protein degradation in the photosystem II repair cycle. J. Biochem. 146: 463–469 [DOI] [PubMed] [Google Scholar]
- Kato Y., Sun X., Zhang L., Sakamoto W. (2012). Cooperative D1 degradation in the photosystem II repair mediated by chloroplastic proteases in Arabidopsis. Plant Physiol. 159: 1428–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Miura E., Ido K., Ifuku K., Sakamoto W. (2009). The variegated mutants lacking chloroplastic FtsHs are defective in D1 degradation and accumulate reactive oxygen species. Plant Physiol. 151: 1790–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kley J., Schmidt B., Boyanov B., Stolt-Bergner P.C., Kirk R., Ehrmann M., Knopf R.R., Naveh L., Adam Z., Clausen T. (2011). Structural adaptation of the plant protease Deg1 to repair photosystem II during light exposure. Nat. Struct. Mol. Biol. 18: 728–731 [DOI] [PubMed] [Google Scholar]
- Komenda J., Sobotka R., Nixon P.J. (2012). Assembling and maintaining the photosystem II complex in chloroplasts and cyanobacteria. Curr. Opin. Plant Biol. 15: 245–251 [DOI] [PubMed] [Google Scholar]
- Komenda J., Knoppová J., Krynická V., Nixon P.J., Tichý M. (2010). Role of FtsH2 in the repair of photosystem II in mutants of the cyanobacterium Synechocystis PCC 6803 with impaired assembly or stability of the CaMn(4) cluster. Biochim. Biophys. Acta 1797: 566–575 [DOI] [PubMed] [Google Scholar]
- Komenda J., Barker M., Kuviková S., de Vries R., Mullineaux C.W., Tichy M., Nixon P.J. (2006). The FtsH protease slr0228 is important for quality control of photosystem II in the thylakoid membrane of Synechocystis sp. PCC 6803. J. Biol. Chem. 281: 1145–1151 [DOI] [PubMed] [Google Scholar]
- Krzywda S., Brzozowski A.M., Verma C., Karata K., Ogura T., Wilkinson A.J. (2002). The crystal structure of the AAA domain of the ATP-dependent protease FtsH of Escherichia coli at 1.5 A resolution. Structure 10: 1073–1083 [DOI] [PubMed] [Google Scholar]
- Kuras R., Wollman F.A. (1994). The assembly of cytochrome b6/f complexes: An approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J. 13: 1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras R., Saint-Marcoux D., Wollman F.A., de Vitry C. (2007). A specific c-type cytochrome maturation system is required for oxygenic photosynthesis. Proc. Natl. Acad. Sci. USA 104: 9906–9910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras R., de Vitry C., Choquet Y., Girard-Bascou J., Culler D., Büschlen S., Merchant S., Wollman F.A. (1997). Molecular genetic identification of a pathway for heme binding to cytochrome b6. J. Biol. Chem. 272: 32427–32435 [DOI] [PubMed] [Google Scholar]
- Kurisu G., Zhang H., Smith J.L., Cramer W.A. (2003). Structure of the cytochrome b6f complex of oxygenic photosynthesis: Tuning the cavity. Science 302: 1009–1014 [DOI] [PubMed] [Google Scholar]
- Lal A., Haynes S.R., Gorospe M. (2005). Clean western blot signals from immunoprecipitated samples. Mol. Cell. Probes 19: 385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Le S.Q., Gascuel O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25: 1307–1320 [DOI] [PubMed] [Google Scholar]
- Lezhneva L., Kuras R., Ephritikhine G., de Vitry C. (2008). A novel pathway of cytochrome c biogenesis is involved in the assembly of the cytochrome b6f complex in Arabidopsis chloroplasts. J. Biol. Chem. 283: 24608–24616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.H., Quinn J., Culler D., Girard-Bascou J., Merchant S. (1996). Molecular genetic analysis of plastocyanin biosynthesis in Chlamydomonas reinhardtii. J. Biol. Chem. 271: 31283–31289 [DOI] [PubMed] [Google Scholar]
- Lindahl M., Spetea C., Hundal T., Oppenheim A.B., Adam Z., Andersson B. (2000). The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 12: 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M., Tabak S., Cseke L., Pichersky E., Andersson B., Adam Z. (1996). Identification, characterization, and molecular cloning of a homologue of the bacterial FtsH protease in chloroplasts of higher plants. J. Biol. Chem. 271: 29329–29334 [DOI] [PubMed] [Google Scholar]
- Majeran W., Wollman F.A., Vallon O. (2000). Evidence for a role of ClpP in the degradation of the chloroplast cytochrome b6f complex. Plant Cell 12: 137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W., Olive J., Drapier D., Vallon O., Wollman F.A. (2001). The light sensitivity of ATP synthase mutants of Chlamydomonas reinhardtii. Plant Physiol. 126: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnoë A., Wollman F.A., de Vitry C., Rappaport F. (2011). Photosynthetic growth despite a broken Q-cycle. Nat Commun 2: 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann N.H., Novac N., Mullineaux C.W., Newman J., Bailey S., Robinson C. (2000). Involvement of an FtsH homologue in the assembly of functional photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 479: 72–77 [DOI] [PubMed] [Google Scholar]
- Martin A., Baker T.A., Sauer R.T. (2005). Rebuilt AAA+ motors reveal operating principles for ATP-fuelled machines. Nature 437: 1115–1120 [DOI] [PubMed] [Google Scholar]
- Matagne R.F. (1978). Fine structure of the arg-7 cistron in Chlamydomonas reinhardtii. Complementation between arg-7 mutants defective in argininosuccinate lyase. Mol. Gen. Genet. 160: 95–99 [PubMed] [Google Scholar]
- Minai L., Wostrikoff K., Wollman F.A., Choquet Y. (2006). Chloroplast biogenesis of photosystem II cores involves a series of assembly-controlled steps that regulate translation. Plant Cell 18: 159–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldavski O., Levin-Kravets O., Ziv T., Adam Z., Prag G. (2012). The hetero-hexameric nature of a chloroplast AAA+ FtsH protease contributes to its thermodynamic stability. PLoS ONE 7: e36008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.L., Chang C.W., Grossman A.R. (2006). Genome-based approaches to understanding phosphorus deprivation responses and PSR1 control in Chlamydomonas reinhardtii. Eukaryot. Cell 5: 26–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narberhaus F., Obrist M., Führer F., Langklotz S. (2009). Degradation of cytoplasmic substrates by FtsH, a membrane-anchored protease with many talents. Res. Microbiol. 160: 652–659 [DOI] [PubMed] [Google Scholar]
- Neuwald A.F., Aravind L., Spouge J.L., Koonin E.V. (1999). AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9: 27–43 [PubMed] [Google Scholar]
- Nixon P.J., Michoux F., Yu J., Boehm M., Komenda J. (2010). Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. (Lond.) 106: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T., Whiteheart S.W., Wilkinson A.J. (2004). Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J. Struct. Biol. 146: 106–112 [DOI] [PubMed] [Google Scholar]
- Ostersetzer O., Adam Z. (1997). Light-stimulated degradation of an unassembled Rieske FeS protein by a thylakoid-bound protease: The possible role of the FtsH protease. Plant Cell 9: 957–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechota J., Kolodziejczak M., Juszczak I., Sakamoto W., Janska H. (2010). Identification and characterization of high molecular weight complexes formed by matrix AAA proteases and prohibitins in mitochondria of Arabidopsis thaliana. J. Biol. Chem. 285: 12512–12521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pootakham W., Gonzalez-Ballester D., Grossman A.R. (2010). Identification and regulation of plasma membrane sulfate transporters in Chlamydomonas. Plant Physiol. 153: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues R.A., Silva-Filho M.C., Cline K. (2011). FtsH2 and FtsH5: Two homologous subunits use different integration mechanisms leading to the same thylakoid multimeric complex. Plant J. 65: 600–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez F., Arsène-Ploetze F., Rist W., Rüdiger S., Schneider-Mergener J., Mayer M.P., Bukau B. (2008). Molecular basis for regulation of the heat shock transcription factor sigma32 by the DnaK and DnaJ chaperones. Mol. Cell 32: 347–358 [DOI] [PubMed] [Google Scholar]
- Rymarquis L.A., Handley J.M., Thomas M., Stern D.B. (2005). Beyond complementation. Map-based cloning in Chlamydomonas reinhardtii. Plant Physiol. 137: 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikawa N., Akiyama Y., Ito K. (2004). FtsH exists as an exceptionally large complex containing HflKC in the plasma membrane of Escherichia coli. J. Struct. Biol. 146: 123–129 [DOI] [PubMed] [Google Scholar]
- Saint-Marcoux D., Wollman F.A., de Vitry C. (2009). Biogenesis of cytochrome b6 in photosynthetic membranes. J. Cell Biol. 185: 1195–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W. (2006). Protein degradation machineries in plastids. Annu. Rev. Plant Biol. 57: 599–621 [DOI] [PubMed] [Google Scholar]
- Sakamoto W., Zaltsman A., Adam Z., Takahashi Y. (2003). Coordinated regulation and complex formation of YELLOW VARIEGATED1 and YELLOW VARIEGATED2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15: 2843–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda, M., and Vallon, O. (2009). Chaperones and proteases. In The Chlamydomonas Sourcebook: Organellar and Metabolic Processes, D. Stern, ed (Ithaca, NY: Academic Press), pp. 671–729. [Google Scholar]
- Schroda M., Vallon O., Wollman F.A., Beck C.F. (1999). A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11: 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M., Vallon O., Whitelegge J.P., Beck C.F., Wollman F.A. (2001). The chloroplastic GrpE homolog of Chlamydomonas: Two isoforms generated by differential splicing. Plant Cell 13: 2823–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawara K., Fujiwara S., Grossman A., Usuda H. (1998). High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148: 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva P., Thompson E., Bailey S., Kruse O., Mullineaux C.W., Robinson C., Mann N.H., Nixon P.J. (2003). FtsH is involved in the early stages of repair of photosystem II in Synechocystis sp PCC 6803. Plant Cell 15: 2152–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinvany-Villalobo G., Davydov O., Ben-Ari G., Zaltsman A., Raskind A., Adam Z. (2004). Expression in multigene families. Analysis of chloroplast and mitochondrial proteases. Plant Physiol. 135: 1336–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolenko A., Pojidaeva E., Zinchenko V., Panichkin V., Glaser V.M., Herrmann R.G., Shestakov S.V. (2002). The gene complement for proteolysis in the cyanobacterium Synechocystis sp. PCC 6803 and Arabidopsis thaliana chloroplasts. Curr. Genet. 41: 291–310 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Steglich G., Neupert W., Langer T. (1999). Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol. Cell. Biol. 19: 3435–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroebel D., Choquet Y., Popot J.L., Picot D. (2003). An atypical haem in the cytochrome b6f complex. Nature 426: 413–418 [DOI] [PubMed] [Google Scholar]
- Sun Q., Zybailov B., Majeran W., Friso G., Olinares P.D., van Wijk K.J. (2009). PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res. 37: D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Peng L., Guo J., Chi W., Ma J., Lu C., Zhang L. (2007). Formation of DEG5 and DEG8 complexes and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. Plant Cell 19: 1347–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Fu T., Chen N., Guo J., Ma J., Zou M., Lu C., Zhang L. (2010). The stromal chloroplast Deg7 protease participates in the repair of photosystem II after photoinhibition in Arabidopsis. Plant Physiol. 152: 1263–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suno R., Niwa H., Tsuchiya D., Zhang X., Yoshida M., Morikawa K. (2006). Structure of the whole cytosolic region of ATP-dependent protease FtsH. Mol. Cell 22: 575–585 [DOI] [PubMed] [Google Scholar]
- Suzuki C.K., Rep M., van Dijl J.M., Suda K., Grivell L.A., Schatz G. (1997). ATP-dependent proteases that also chaperone protein biogenesis. Trends Biochem. Sci. 22: 118–123 [DOI] [PubMed] [Google Scholar]
- Thomas P.E., Ryan D., Levin W. (1976). An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75: 168–176 [DOI] [PubMed] [Google Scholar]
- Van Dyck L., Langer T. (1999). ATP-dependent proteases controlling mitochondrial function in the yeast Saccharomyces cerevisiae. Cell. Mol. Life Sci. 56: 825–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaas W.F. (1998). Gene modifications and mutation mapping to study the function of photosystem II. Methods Enzymol. 297: 293–310 [DOI] [PubMed] [Google Scholar]
- Wagner R., Aigner H., Funk C. (2012). FtsH proteases located in the plant chloroplast. Physiol. Plant. 145: 203–214 [DOI] [PubMed] [Google Scholar]
- Wei L., Derrien B., Gautier A., Houille-Vernes L., Boulouis A., Saint-Marcoux D., Malnoë A., Rappaport F., de Vitry C., Vallon O., Choquet Y., Wollman F.-A. (2014). Nitric oxide–triggered remodeling of chloroplast bioenergetics and thylakoid proteins upon nitrogen starvation in Chlamydomonas reinhardtii. Plant Cell 26: 353–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman F.A., Minai L., Nechushtai R. (1999). The biogenesis and assembly of photosynthetic proteins in thylakoid membranes1. Biochim. Biophys. Acta 1411: 21–85 [DOI] [PubMed] [Google Scholar]
- Wykoff D.D., Davies J.P., Melis A., Grossman A.R. (1998). The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol. 117: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Aminaka R., Yoshioka M., Khatoon M., Komayama K., Takenaka D., Yamashita A., Nijo N., Inagawa K., Morita N., Sasaki T., Yamamoto Y. (2008). Quality control of photosystem II: Impact of light and heat stresses. Photosynth. Res. 98: 589–608 [DOI] [PubMed] [Google Scholar]
- Yoshioka M., Yamamoto Y. (2011). Quality control of photosystem II: Where and how does the degradation of the D1 protein by FtsH proteases start under light stress? Facts and hypotheses. J. Photochem. Photobiol. B 104: 229–235 [DOI] [PubMed] [Google Scholar]
- Yoshioka M., Nakayama Y., Yoshida M., Ohashi K., Morita N., Kobayashi H., Yamamoto Y. (2010). Quality control of photosystem II: FtsH hexamers are localized near photosystem II at grana for the swift repair of damage. J. Biol. Chem. 285: 41972–41981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Park S., Rodermel S.R. (2004). The Arabidopsis FtsH metalloprotease gene family: Interchangeability of subunits in chloroplast oligomeric complexes. Plant J. 37: 864–876 [DOI] [PubMed] [Google Scholar]
- Zaltsman A., Ori N., Adam Z. (2005). Two types of FtsH protease subunits are required for chloroplast biogenesis and photosystem II repair in Arabidopsis. Plant Cell 17: 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Happe T., Melis A. (2002). Biochemical and morphological characterization of sulfur-deprived and H2-producing Chlamydomonas reinhardtii (green alga). Planta 214: 552–561 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Shrager J., Jain M., Chang C.W., Vallon O., Grossman A.R. (2004). Insights into the survival of Chlamydomonas reinhardtii during sulfur starvation based on microarray analysis of gene expression. Eukaryot. Cell 3: 1331–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zienkiewicz M., Ferenc A., Wasilewska W., Romanowska E. (2012). High light stimulates Deg1-dependent cleavage of the minor LHCII antenna proteins CP26 and CP29 and the PsbS protein in Arabidopsis thaliana. Planta 235: 279–288 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.