This work reveals aspects of the properties and function of the phospholipid cardiolipin (CL). In Arabidopsis, CL mediates mitochondrial fission at least in part by stabilizing the higher order protein complexes of dynamin-related protein DRP3, a major organelle division factor. CL also protects plants from stresses that induce programmed cell death.
Abstract
Mitochondria are essential and dynamic organelles in eukaryotes. Cardiolipin (CL) is a key phospholipid in mitochondrial membranes, playing important roles in maintaining the functional integrity and dynamics of mitochondria in animals and yeasts. However, CL’s role in plants is just beginning to be elucidated. In this study, we used Arabidopsis thaliana to examine the subcellular distribution of CL and CARDIOLIPIN SYNTHASE (CLS) and analyzed loss-of-function cls mutants for defects in mitochondrial morphogenesis and stress response. We show that CL localizes to mitochondria and is enriched at specific domains, and CLS targets to the inner membrane of mitochondria with its C terminus in the intermembrane space. Furthermore, cls mutants exhibit significantly impaired growth as well as altered structural integrity and morphogenesis of mitochondria. In contrast to animals and yeasts, in which CL’s effect on mitochondrial fusion is more profound, Arabidopsis CL plays a dominant role in mitochondrial fission and exerts this function, at least in part, through stabilizing the protein complex of the major mitochondrial fission factor, DYNAMIN-RELATED PROTEIN3. CL also plays a role in plant responses to heat and extended darkness, stresses that induce programmed cell death. Our study has uncovered conserved and plant-specific aspects of CL biology in mitochondrial dynamics and the organism response to environmental stresses.
INTRODUCTION
Mitochondria are essential and dynamic organelles that house diverse biochemical pathways for energy production, metabolism, and signaling (Jacoby et al., 2012). To maintain optimal biochemical activities, mitochondria remodel their morphology and function in response to developmental and environmental cues and use continual fission and fusion as an efficient quality control mechanism. Mitochondrial fission and fusion are two topologically opposite but molecularly similar processes that involve comparable steps of lipid bilayer remodeling, such as local disruption, bending, and bridging (Palmer et al., 2011). Thus, it is not surprising that dynamin-related proteins (DRPs) are at the core of both mitochondrial fusion and fission machines (Chan, 2012). In contrast to typical animal and yeast cells, in which mitochondria are highly tubular and interconnected (Chan, 2012), normal plant cells contain fragmented and discrete mitochondria, indicating the dominance of mitochondrial fission in plants as opposed to fusion in many other eukaryotes. Arabidopsis thaliana possesses conserved and plant-specific mitochondrial fission factors, including the homologous DRP3A and DRP3B proteins that play dual roles in mitochondrial and peroxisomal fission (Hu et al., 2012). However, neither organelle fusion DRPs nor other components of the mitochondrial fusion apparatus have been identified in plants. Plant mitochondria do fuse (Logan, 2010), which may indicate the existence of a weak and plant-specific mitochondrial fusion machinery.
Cardiolipin (CL) is a key component of both prokaryotic and eukaryotic membranes, with unique structure and functions (Lewis and McElhaney, 2009). It is an anionic phospholipid with a dimeric structure and contains a triple glycerol backbone and four acyl groups, most of which are highly unsaturated. CL’s acyl chain composition is quite simple. For example, almost all acyl chains in human heart CL are composed of 18-carbon fatty acids, 80% of them being linoleic acid [18:2 (n-6); Schlame et al., 2005]. This feature is rather unusual, because most other phospholipids, including CL’s precursor phosphatidylglycerol (PG), do not show an obvious preference for specific fatty acids. Although it is unknown how the acyl chain preference is determined in CL biosynthesis, the simple composition of acyl chains in CL apparently leads to a high degree of structural uniformity and molecular symmetry, which may facilitate the assembly of organized membrane domains and enhance the stability of mitochondrial membrane protein homodimers (Schlame et al., 2005).
In bacteria, cardiolipin-enriched membrane domains (CMDs) localize at the negatively curved regions of the cell membrane, such as the poles of Escherichia coli (Renner and Weibel, 2011) and the septa of Bacillus subtilis undergoing spore formation, where decreasing CL levels cause a reduced rate of sporulation (Kawai et al., 2004). CMDs also colocalize with E. coli cell division proteins like Division IVA and Minicell D (Lenarcic et al., 2009; Renner and Weibel, 2012) and facilitate the function of the cell fission protein Fission Protein B in B. subtilis (Doan et al., 2013). In eukaryotes, although the distribution pattern of CL in mitochondria has not been fully characterized, biochemical analysis revealed that in yeasts, ∼75% of total CL localizes in the mitochondrial inner membrane and ∼25% is in the outer membrane (Osman et al., 2011).
In yeasts and animals, CL promotes mitochondrial fusion by affecting the enzymatic processing and/or aggregation of fusion DRPs (DeVay et al., 2009; Ban et al., 2010; Joshi et al., 2012). CL tends to concentrate in curved membrane regions, promoting the formation of hexagonal membrane structures, which can be the intermediate state of double membranes in fission and fusion (Ortiz et al., 1999). However, under reduced CL levels caused by deletion of a positive regulator of CL, yeast mitochondria are more fragmented, indicating a stronger effect of CL on mitochondrial fusion (Tamura et al., 2009). In addition, CL contributes to the bioenergetics and biogenesis of mitochondria in animals and yeasts by enhancing the organization, stabilization, and/or activity of several protein complexes involved in respiration and protein import (Osman et al., 2011). CL also regulates the early events of apoptotic programmed cell death (PCD) in animals by providing an activating platform for Bcl-2 family proteins that are required for inducing outer membrane permeabilization, which leads to the release of mitochondrial cytochrome c and other proapoptotic molecules (Lutter et al., 2000; Kuwana et al., 2002; Kagan et al., 2005; Gonzalvez and Gottlieb, 2007; Montessuit et al., 2010). In humans, changes in CL levels can cause various pathological conditions, including Barth syndrome, aging, and heart failure (Chicco and Sparagna, 2007).
A previous study in the yeast Pichia pastoris discovered CL to constitute 2 to 4% of the phospholipids in peroxisomal membranes (Wriessnegger et al., 2007), raising the possibility that CL may also be involved in the dynamics and function of peroxisomes, essential organelles linked to mitochondria through metabolism and shared fission DRPs (Schrader and Yoon, 2007; Hu et al., 2012). This finding, together with the distinct morphology of plant mitochondria from that of animals and yeasts, prompted us to investigate CL’s role in the dynamics of mitochondria and possibly other organelles in plants. In addition, plant PCD is a highly regulated cell death process that shares some morphological hallmarks with apoptosis (Gadjev et al., 2008). However, the plant PCD network is poorly characterized at the molecular level. To this end, it was important to investigate CL’s role in plant PCD as well.
Recent studies of the Arabidopsis CARDIOLIPIN SYNTHASE (CLS) T-DNA insertion mutants demonstrated defects of the mutants in embryo development, growth and reproduction, mitochondrial ultrastructure and respiratory function, and protoplast response to UV light and heat shock (Katayama and Wada, 2012; Pineau et al., 2013). In this study, we investigated the role of CL in organelle dynamics and explored CL’s function in protecting plants from stresses at the whole-organism level. Subcellular distribution of CL and the targeting and membrane topology of CLS were determined, and Arabidopsis CLS T-DNA mutant and artificial microRNA (amiRNA) lines were characterized for defects in mitochondrial morphology and plant response to stresses that induce PCD. We demonstrate that Arabidopsis CL mediates mitochondrial fission at least in part by stabilizing the higher order protein complexes of a key organelle division factor, DRP3, and plays a protective role in the plant response to stresses that induce PCD.
RESULTS
Mitochondrial Localization of CL and CLS in Plant Cells
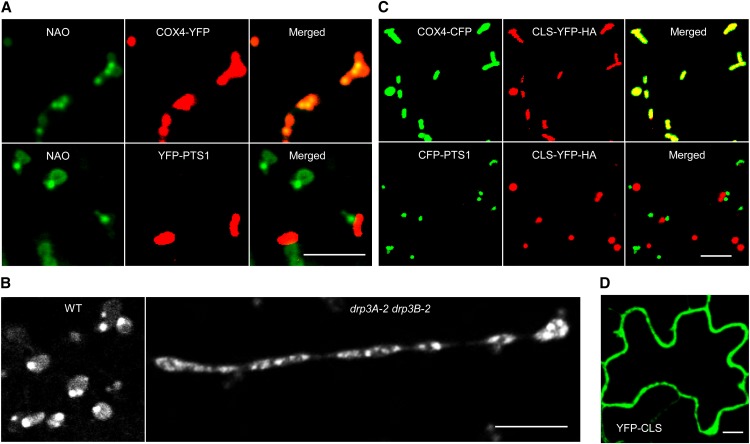
To understand the role of CL in plant organelle dynamics, we first determined the subcellular distribution of CL and explored the possibility that it may also be present in other organelles such as peroxisomes. Arabidopsis seedlings containing a mitochondrial marker (Saccharomyces cerevisiae CYTOCHROME C OXIDASE4-YELLOW FLUORESCENT PROTEIN [COX4-YFP]; Nelson et al., 2007) or a peroxisomal marker (YFP-PEROXISOMAL TARGETING SIGNAL1 [PTS1]; Fan et al., 2005) were stained with the CL-specific fluorescent dye 10-N-nonyl-acridine orange (NAO; Mileykovskaya and Dowhan, 2000; Kawai et al., 2004; Kaewsuya et al., 2007; Renner and Weibel, 2011), which gives consistent and specific staining patterns under our experimental conditions. Confocal laser scanning microscopic analysis showed that, whereas peroxisomes were negatively stained for NAO, all fluorescent signals colocalized with the mitochondrial marker (Figure 1A). Somewhat similar to the polar distribution of CL on bacterial cell membranes (Kawai et al., 2004; Renner and Weibel, 2011), CL was particularly enriched at specific domains in mitochondria, a pattern that was obvious in both the wild type and the drp3A-2 drp3B-2 double mutant that frequently contained extremely elongated mitochondria (Zhang and Hu, 2009; Aung and Hu, 2012; Figures 1A and 1B). We concluded that, in plants, CL localizes primarily, if not exclusively, to mitochondria and is enriched at specific mitochondrial regions, the CMDs.
Figure 1.
Subcellular Localization of CL and CLS in Plant Cells.
(A) Confocal images showing the localization of CL to mitochondria in Arabidopsis. Leaf epidermal cells of 2-week-old T3 plants expressing COX4-YFP or YFP-PTS1 were stained with the CL-specific dye NAO. Bar = 5 μm.
(B) Confocal images showing CMDs on mitochondria in leaf epidermal cells from 2-week-old wild-type and drp3 double mutant plants stained with NAO. Bar = 5 μm.
(C) CLS localizes to mitochondria but not peroxisomes. Two-week-old T2 lines expressing CLS-YFP-HA and COX4-CFP or CLS-YFP-HA and CFP-PTS1 were used for analysis. Bar = 5 μm.
(D) Cytosolic diffusion of YFP-CLS transiently expressed in tobacco leaf epidermal cells. Bar = 10 μm.
To further examine the distribution and biosynthesis of CL in plants, we analyzed the subcellular localization of CLS, which is conserved in diverse eukaryotic species (Supplemental Figure 1A and Supplemental Data Set 1). Arabidopsis has a single-copy CLS gene (At4g04870), and its protein is capable of catalyzing CL synthesis from cytidinediphosphate-diacylglycerol and PG in E. coli; CLSN terminus-GFP was localized to mitochondria in onion (Allium cepa) epidermis (Katayama et al., 2004; Nowicki et al., 2005). To get a complete view of CLS localization, full-length CLS fused with a C-terminal YFP-HEMAGGLUTININ (HA) tag was expressed under the control of the 35S constitutive promoter in Arabidopsis plants containing the organelle marker COX4-CYAN FLUORESCENT PROTEIN (CFP) or CFP-PTS1 (Aung and Hu, 2011). Confocal microscopic analysis showed that CLS-YFP-HA overlapped completely with the mitochondrial marker but not the peroxisomal marker (Figure 1C). By contrast, CLS fused to an N-terminal YFP (YFP-CLS) was diffuse in the cytosol when expressed in tobacco (Nicotiana tabacum) leaves (Figure 1D), proving that the N terminus is vital for organelle targeting of CLS. Immunoblot analysis of proteins from tobacco plants expressing the cytosolic YFP-CLS protein or the mitochondrial CLS-YFP-HA protein detected the former as a single band, while the latter appeared as two bands, one of which was slightly smaller than the full-length protein (Supplemental Figure 2A). After fractionation by several centrifugation steps, only the smaller band was detectable in the pellet fraction enriched in mitochondria (Supplemental Figure 2B). These data suggested that an N-terminal signal peptide was cleaved from the CLS precursor of the mitochondria-localized CLS-YFP-HA, which is consistent with the target peptide processing mechanism for many mitochondrial inner membrane proteins (Teixeira and Glaser, 2013).
Although the catalytic domain is well conserved, the N-terminal region of plant CLS homologs is divergent from those in nonplant CLS sequences (Supplemental Figures 1B and 1C). To dissect this plant-specific region, we made a series of CLS deletion constructs (Supplemental Figure 3A) and transiently expressed the 35S-driven truncated CLS-YFP proteins together with COX4-CFP in tobacco. Confocal microscopic analysis revealed that, as predicted, mitochondrial targeting signals reside at the N terminus, as the C terminus of CLS (CLS141-341) mistargeted the fusion protein to compartments such as the plasma membrane, endoplasmic reticulum (ER), and Golgi (Supplemental Figures 3B and 3C). Interestingly, two separate regions at the N terminus seemed to contain mitochondrial targeting signals. Signal 1 (amino acids 1 to 20) was sufficient to target the protein to mitochondria. However, when signal 1 was deleted, signal 2 (amino acids 42 to 140) could direct the protein to both mitochondria and chloroplasts (Supplemental Figure 3B). Since both signals are fairly well conserved in plant CLS sequences (Supplemental Figure 1C), they may represent plant-specific targeting mechanisms for CLS and possibly some other proteins as well, although further investigations will be needed to clearly address this aspect of targeting in planta.
Membrane Association and Topology of CLS
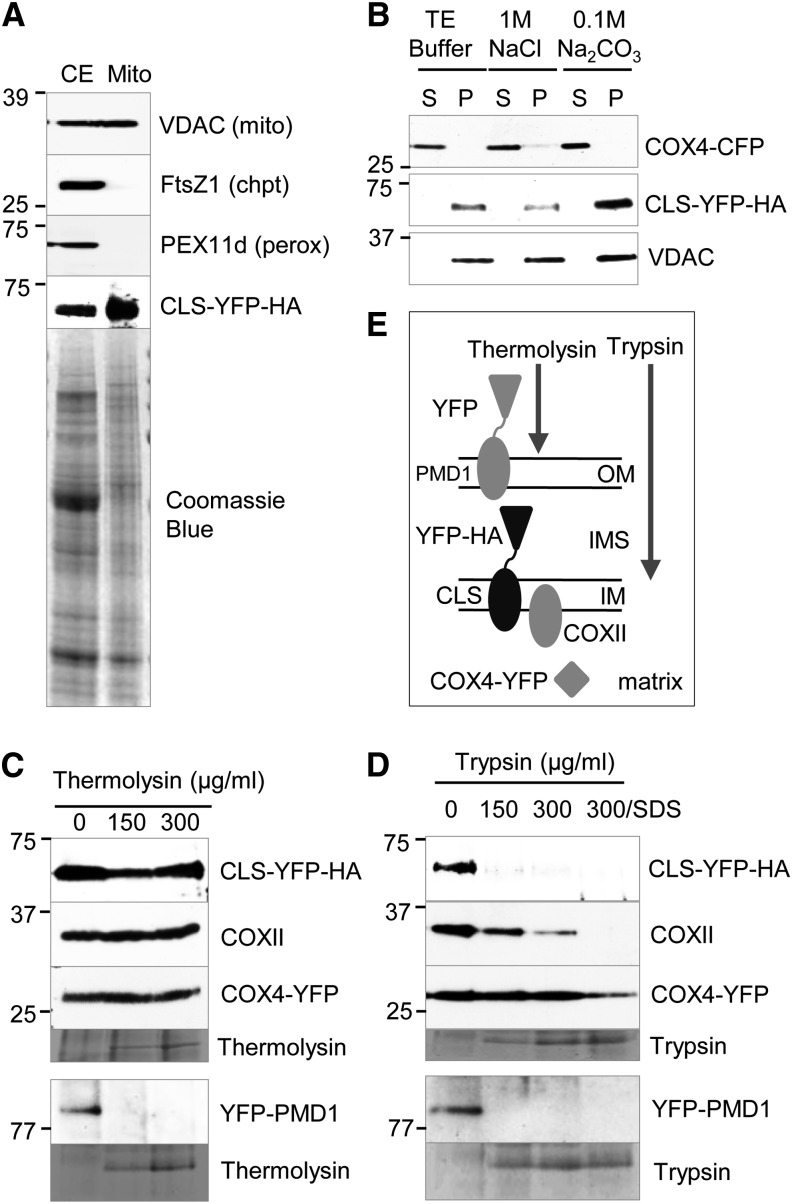
In animals and yeasts, CL biosynthesis occurs in the mitochondrial inner membrane (Osman et al., 2011), but the membrane topology of the key enzyme CLS was unknown. To address this issue, mitochondria were isolated from transgenic plants coexpressing CLS-YFP-HA and COX4-YFP, and the purity of mitochondria was determined by immunoblot analysis using organelle-specific antibodies (Figure 2A). The presence of the CLS-YFP-HA fusion protein in mitochondria was confirmed by its detection with the GFP antibody (Figure 2A). After treatments with Tris-EDTA, high concentrations of NaCl, and Na2CO3, CLS-YFP-HA was detected in the pellet after each treatment, suggesting that CLS is indeed a mitochondrial integral membrane protein (Figure 2B).
Figure 2.
Analyses of Mitochondrial Membrane Association and Topology of CLS.
(A) Immunoblot analysis to assess the purity of mitochondria isolated from leaf tissue of Arabidopsis transgenic plants. Organelle-specific antibodies used were against plant VDAC (mitochondria), FtsZ1 (chloroplasts), and PEX11d (peroxisomes). The presence of CLS-YFP-HA was confirmed by detection with the GFP antibody. CE, Crude extract. Protein molecular mass markers in kilodaltons are indicated for the immunoblots.
(B) Membrane association of CLS. Purified mitochondria were treated with Tris-EDTA (10 mM Tricine and 1 mM EDTA, pH 7.5), NaCl, or Na2CO3 (pH 11.0) and later separated into soluble (S) and pellet (P) fractions by centrifugation. Immunoblot analyses were performed on the proteins using α-GFP (for CFP and YFP fusions) and α-VDAC antibodies. COX4-YFP and VDAC were controls for matrix and membrane proteins, respectively. Protein size markers in kilodaltons are indicated for the immunoblots.
(C) and (D) Protease protection assays to determine the membrane topology of CLS. Antibodies used were α-GFP and α-COXII. Mitochondria used for the detection of YFP-PMD1 were isolated independently from Arabidopsis plants expressing 35Spro:YFP-PMD1 (Aung and Hu, 2011). Protein markers in kilodaltons are indicated for the immunoblots.
(E) Illustration of the digestive ability of thermolysin and trypsin. PMD1, COXII, and COX4-YFP were controls for distinct subcompartments of mitochondria. IM, inner membrane; OM, outer membrane.
We further dissected the localization and topology of CLS by performing protease protection assays with thermolysin, which degrades proteins on the surface of the organelles, and trypsin, which can access the intermembrane space. For the controls, the outer membrane protein YFP-PEROXISOME AND MITOCHONDRIAL DIVISION FACTOR1 (PMD1) was digestible by both proteases, whereas the inner membrane protein COXII and the matrix protein COX4-YFP were protected from both enzymes (Figures 2C and 2D). CLS-YFP-HA was resistant to thermolysin but digestible by trypsin (Figures 2C and 2D), suggestive of a topology in which CLS is anchored to the mitochondrial inner membrane with its C terminus facing the intermembrane space (Figure 2E).
Disruption of CLS Leads to Defects in Mitochondrial Structure and Fission
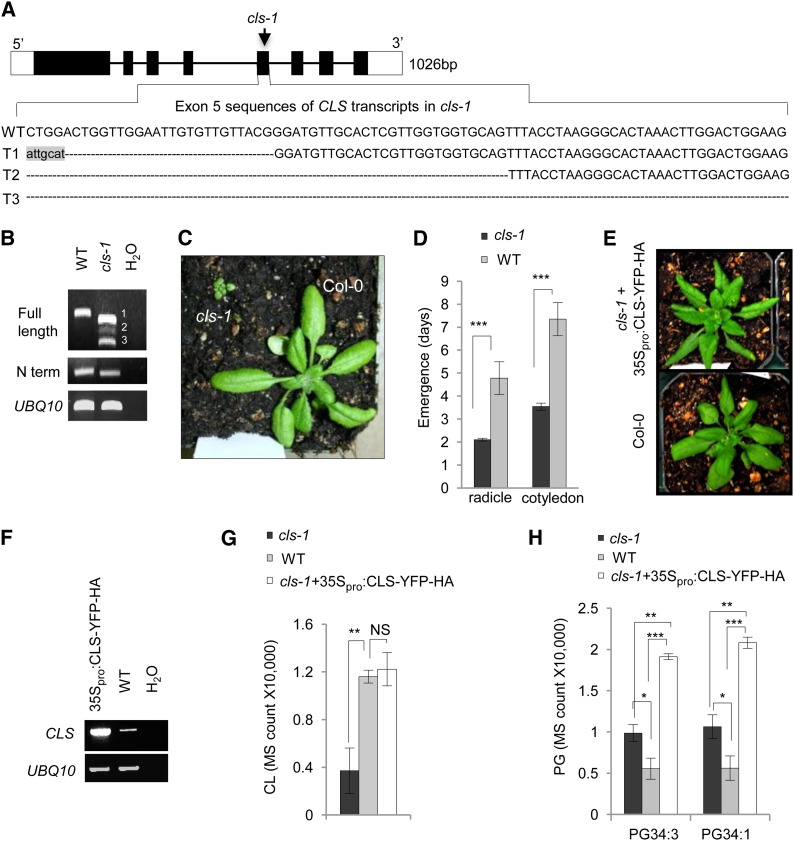
The localization pattern of NAO-stained CMDs shared some similarities with those of the mitochondrial/peroxisomal fission proteins DRP3A and DRP3B (i.e., at the tips and fission sites of the organelles; Arimura and Tsutsumi, 2002; Arimura et al., 2004; Mano et al., 2004; Zhang and Hu, 2009), suggesting a possible role of CL in plant mitochondrial division. To test this prediction, we characterized the cls-1 (SALK_049840) mutant, which has a T-DNA insertion in exon 5 (Figure 3A). A previous report of the same cls mutant allele described extreme retardation in embryo development and plant growth and low fertility of the mutant (Katayama and Wada, 2012), yet the molecular basis of the mutation was not fully characterized. RT-PCR analysis revealed that cls-1 contained three truncated CLS transcripts (Figure 3B). Sequencing of the RT-PCR products showed that transcripts 1 and 2 each retained a partial sequence of exon 5 and were still in frame, and thus may encode truncated proteins lacking part of the catalytic domain, whereas transcript 3 was missing the entire exon 5 and was out of frame (Figure 3A). Consistent with the previously reported growth phenotypes of cls T-DNA insertion mutants (Katayama and Wada, 2012; Pineau et al., 2013), cls-1 was severely dwarfed (Figure 3C; Supplemental Figure 4A) and produced very small siliques that contained nonviable seeds (Supplemental Figure 4B). The homozygous cls-1 seeds produced by heterozygous plants were significantly delayed in germination (Figure 3D). These phenotypes are in agreement with the expression pattern of CLS, which shows an elevation during seed development and peaks in dry seeds, followed by a quick decrease to the basal level after germination (Supplemental Figures 5A and 5B). The mutant phenotypes could be rescued by 35Spro:CLS-YFP-HA (Figures 3E and 3F; Supplemental Figure 4C), confirming the essential role of CL in plant development and the proper function of the CLS-YFP-HA fusion protein.
Figure 3.
Molecular, Phenotypic, and Biochemical Analyses of the cls-1 Mutant and Rescued Lines.
(A) Schematics of the Arabidopsis CLS gene and exon 5 sequences in the three CLS transcripts (T1 to T3) in cls-1 as a result of the T-DNA insertion. White boxes, untranslated regions; black boxes, exons; lines, introns. Dashed lines indicate deletions from exon 5 in the three transcripts in cls-1. Nucleic acids in lowercase letters are foreign sequences introduced by the T-DNA insertion.
(B) RT-PCR analysis of RNA from 4-week-old cls-1. N term, 500 bp of the N terminus of the CLS cDNA. UBIQUITIN10 (UBQ10) was used as a loading control. Bands 1 to 3 represents transcripts 1 to 3.
(C) Three-week-old wild-type and cls-1 plants.
(D) Delayed germination of cls-1 seeds produced by heterozygotes. Average numbers of days required for the appearance of radicles and the two green cotyledons were quantified. Error bars indicate sd; n = 5. ***P < 0.001 in Student’s t test.
(E) Three-week-old Col-0 and a cls-1 plant rescued by 35Spro:CLS-YFP-HA.
(F) RT-PCR analysis of CLS mRNA in the CLS overexpression line shown in (E). UBQ10 was used as the loading control.
(G) and (H) LC-MS analysis of CL (G) and PG (H) levels in the wild type, cls-1, and the complemented cls-1 line (cls-1+35Spro:CLS-YFP-HA). Mass spectrometry counts normalized to total protein content were used for quantifications. Error bars indicate sd; n = 4. NS, not significant (P > 0.05); **P < 0.01, ***P < 0.001. Total CL was used in (G), and two major PG species were used in (H).
[See online article for color version of this figure.]
Liquid chromatography followed by mass spectrometry analysis (LC-MS) of lipids from leaf tissue revealed an ∼70% reduction of CL in cls-1, which was restored by overexpression of CLS-YFP-HA (Figure 3G). In cls-1, there was an ∼2-fold accumulation of the two major species of PG, substrates for CLS; however, PG levels in the complemented cls-1 lines were even higher than in the mutant (Figure 3H), possibly reflecting the complex regulation of phospholipid levels in the CLS overexpression lines.
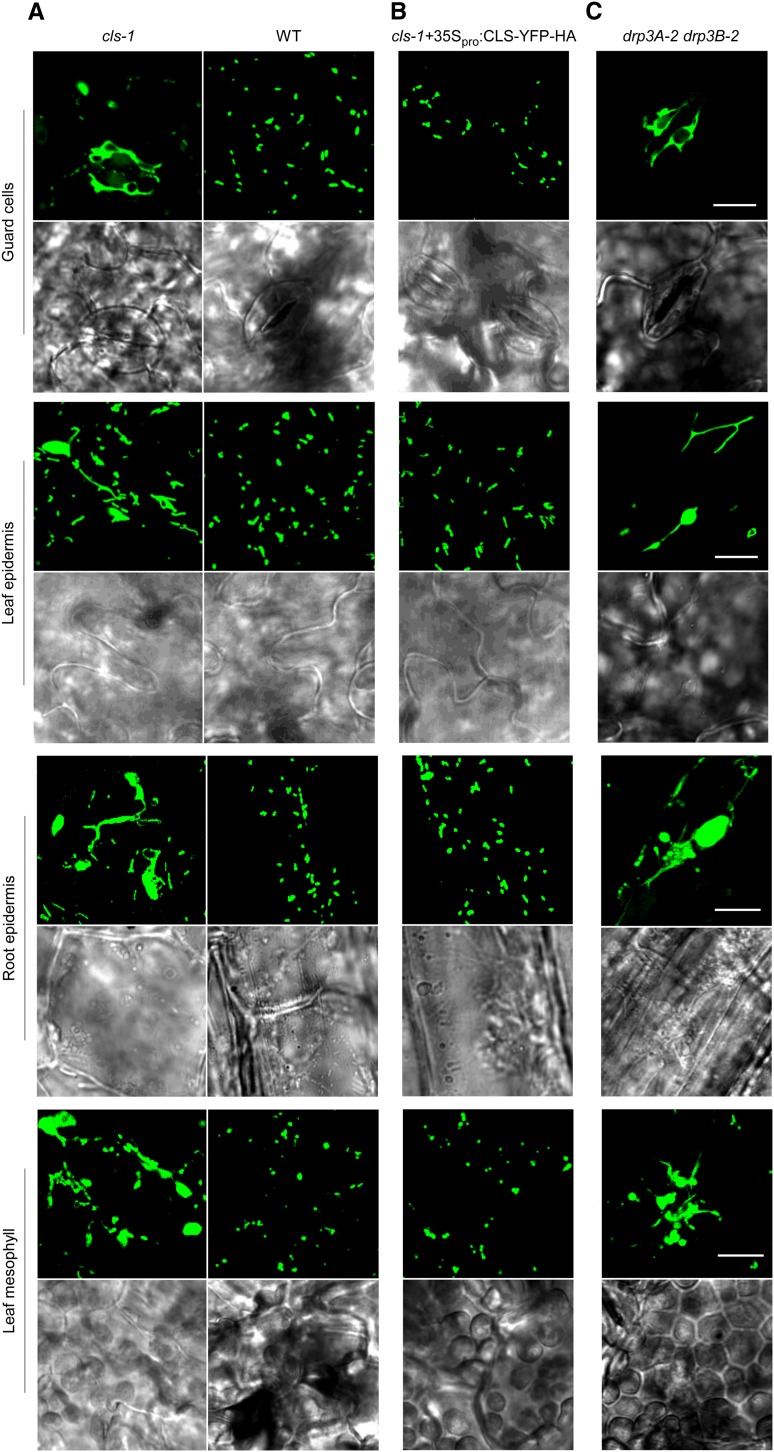
To investigate CL’s role in plant mitochondrial dynamics, COX4-YFP was transformed into cls-1. Confocal microscopy of various cell types in transgenic plants showed that in cls-1, mitochondria were remarkably elongated and enlarged, and these phenotypes could be largely rescued by CLS-YFP-HA (Figures 4A and 4B). Similar to the cls-1 mutant, the drp3A-2 drp3B-2 double mutant (Zhang and Hu, 2009; Aung and Hu, 2012) also displayed significantly elongated and frequently enlarged mitochondria (Figure 4C), suggesting that both mitochondrial elongation and enlargement are direct consequences of the fission defect caused by the absence of DRP3 functions. Consistent with the lack of detection of CL in peroxisomes, peroxisome morphology exhibited no obvious changes in cls-1 (Supplemental Figure 4D). Based on these observations, we concluded that, opposite to what occurs in animals and yeasts, reduction of CL levels in plants has a much stronger effect on mitochondrial fission than fusion. It is less likely that CL inhibits mitochondrial fusion in plants, because CL has the intrinsic property of promoting membrane curvature and hexagonal structure, an intermediate step required for the fission and fusion processes (Osman et al., 2011).
Figure 4.
Mitochondrial Morphology in cls-1.
Confocal images show mitochondrial morphologies in different cell types in the wild type and cls-1 (A), cls-1 complemented by 35Spro:CLS-YFP-HA (B), and the drp3 double mutant (C). All lines are expressing COX4-YFP. Bars = 10 μm.
[See online article for color version of this figure.]
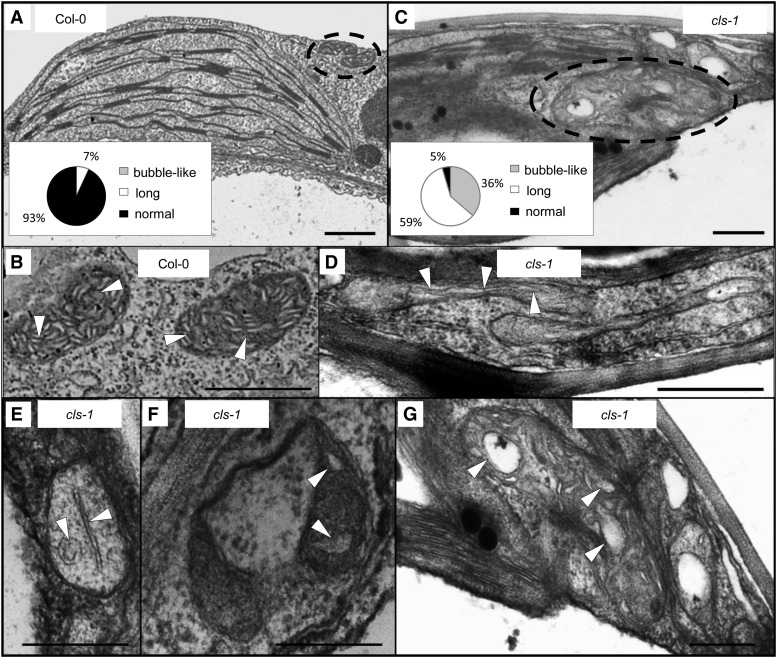
Transmission electron microscopy (TEM) further revealed that, in addition to the abnormal mitochondrial shape, the inner mitochondrial membrane structure in cls-1 was also strongly altered (Figure 5). In contrast to the wild type (Figures 5A and 5B), most mitochondria in the mutant contained markedly long or enlarged/bubble-like cristae structures and decreased abundance of cristae (Figures 5C to 5G), supporting the crucial role of CL in maintaining the structural integrity of mitochondria. The disruption of cristae structure may also cause mitochondrial swelling/enlargement, as these two morphological changes are often found to occur concurrently in isolated mitochondria and chemical-treated cell lines and under disease conditions (Malamed, 1965; Myron and Connelly, 1971; Faller, 1978; Mannella et al., 2001; Sun et al., 2007; Magdalan et al., 2009). Taken together, our results demonstrated that CL plays a positive role in mitochondrial morphogenesis, including in fission of the organelle and cristae formation in the inner membrane.
Figure 5.
TEM Analysis of Mitochondrial Structure from 4-Week-Old Wild-Type and cls-1 Leaves.
(A) and (C) show large areas in the cell, where dotted circles indicate individual mitochondria and insets are quantifications of mitochondrial cristae structure with different morphologies. (B) and (D) to (G) show enlarged views of individual mitochondria, with arrowheads indicating mitochondrial inner membrane and cristae structure. (G) is a magnified view of a part of (C). Data for the quantification were collected from 29 mitochondria from the wild type and 22 mitochondria from the mutant. Long cristae are defined as >0.4 µm in length, and bubble-like cristae are defined as bubble-shaped and >0.2 µm in diameter. Bars = 1 µm.
CL Regulates Mitochondrial Fission through DRP3 Proteins
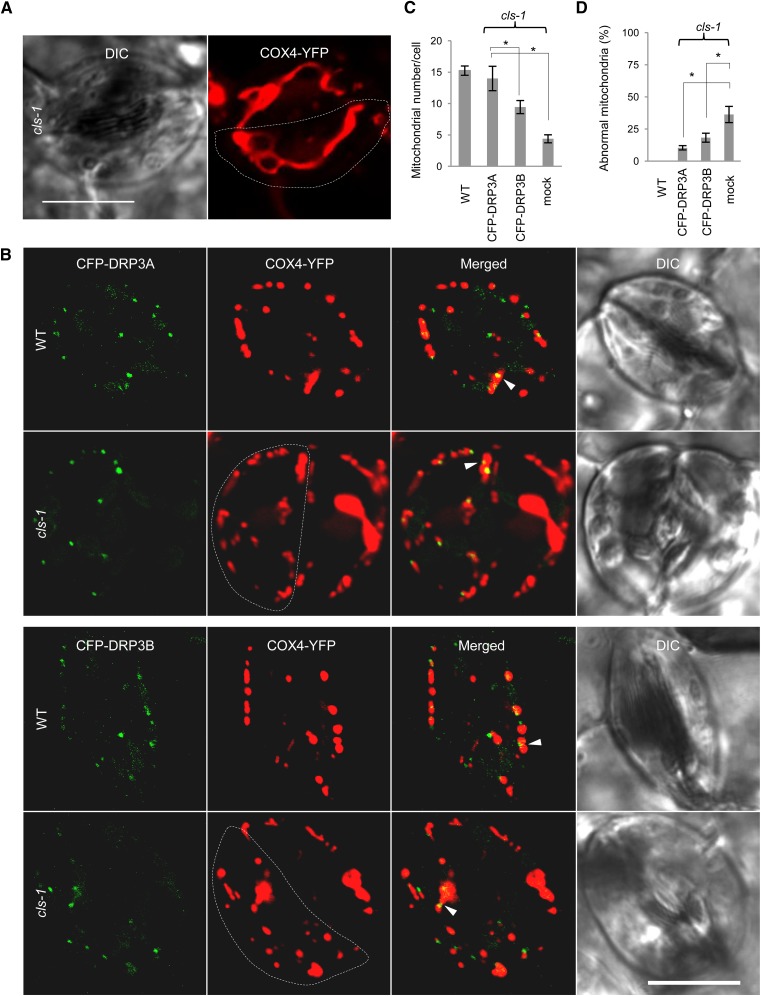
The similarities between cls-1 and drp3 mutants in mitochondrial fission defects suggested that, in plants, CL promotes mitochondrial fission and may exert this function via key mitochondrial division factors, such as DRP3. To elucidate the link between CL and DRP3, we expressed 35Spro:CFP-DRP3 in COX4-YFP–containing cls-1 to see whether the mitochondrial fission defects could be partially rescued. Compared with the cls-1 control (Figure 6A), cls-1 lines expressing 35Spro:CFP-DRP3 contained more fragmented mitochondria, decreased levels of elongated and enlarged mitochondria, and an increased number of mitochondria per cell close to that of the wild type (Figures 6B to 6D). This result provided genetic evidence that DRP3 acts downstream from CL in mitochondrial fission. It is not surprising that CFP-DRP3 overexpression could not fully complement the cls-1 defect in mitochondrial morphology and plant development to the wild-type levels, as CL is a membrane structural component that is involved in multiple aspects of mitochondrial morphogenesis besides DRP3-mediated fission.
Figure 6.
Partial Complementation of the Mitochondrial Morphological Phenotype in cls-1 by DRP3 Proteins.
(A) Mitochondrial morphology in cls-1 guard cells. Dashed lines delineate single guard cells. DIC, differential interference contrast. Bar = 10 μm.
(B) Mitochondrial morphology in wild-type and cls-1 guard cells expressing 35Spro:CFP-DRP3A or 35Spro:CFP-DRP3B. Arrowheads point to some mitochondrion-associated DRP3 proteins. Dashed lines delineate single guard cells. Bar = 10 μm.
(C) and (D) Quantification of the rescue of mitochondrial fission defects in cls-1 by CFP-DRP3 overexpression in comparison with the wild type. Independent mitochondria per guard cell were quantified. Abnormal mitochondria include enlarged and elongated mitochondria, where enlarged mitochondria have diameters >2 μm and elongated mitochondria have lengths >4 μm. Error bars indicate se; n = 6 (number of cells analyzed). *P < 0.05 in Student’s t test.
In animals and yeasts, CL physically binds to fusion and fission DRPs (DeVay et al., 2009; Ban et al., 2010; Montessuit et al., 2010). DRP3 and the human mitochondrial and peroxisomal fission DRP, DRP1, belong to the same DRP subclade, in which most members have dual functions in the division of mitochondria and peroxisomes (Miyagishima et al., 2008). Human DRP1 interacts with CL and stimulates oligomerization of the Bcl-2 family protein, Bax, in the initiation of apoptosis; Arg-247, a positively charged residue exposed on the surface of DRP1, was shown to mediate this interaction (Montessuit et al., 2010). Sequence alignment of fission DRPs from diverse species showed that this Arg is conserved in mitochondrial fission DRPs but not in Arabidopsis ARC5 (DRP5B; Supplemental Figure 6), which is localized to chlooplasts and peroxisomes (Gao et al., 2003; Zhang and Hu, 2010). Comparison of the protein tertiary structure of the GTPase domain revealed structural similarities between human DRP1 and Arabidopsis DRP3s, including the surface-exposed region in which the conserved Arg is located (Supplemental Figure 7A). These observations strongly suggested that, in Arabidopsis, CL also interacts with DRP3 and the conserved Arg on DRP3 is likely required for this interaction.
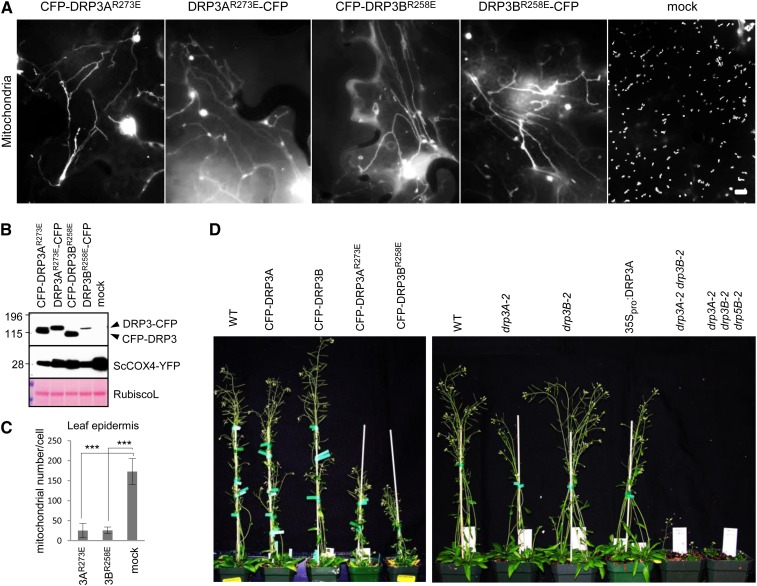
We then tested the hypothesis that the conserved Arg-273 on DRP3A and Arg-258 on DRP3B, which presumably mediate the interaction with CL, also affect DRP3 function in mitochondrial fission. CFP fusions with DRP3AR273E and DRP3BR258E, in which the conserved CL-interacting residue Arg was substituted by a negatively charged residue, Glu, were generated. Tobacco cells transiently coexpressing DRP3R→E with N- or C-terminal CFP and COX4-YFP showed strong inhibition of mitochondrial fission and, as a result, dramatic reduction of the total number of mitochondria per cell, which is in stark contrast to cells expressing wild-type DRP3s (Figures 7A to 7C; Supplemental Figure 7B). As expected, no morphological changes were observed for peroxisomes when coexpressing the mutant DRP3 proteins and the peroxisomal marker (Supplemental Figures 7C and 7D). Similar to the phenotype caused by DRP3R→E in wild-type tobacco cells, CFP-DRP3AR273E transiently expressed in Arabidopsis leaves of drp3A-2 seedlings by the Fast Agro–mediated Seedling Transformation method (FAST; Li et al., 2009) strongly enhanced the mitochondrial fission defect in drp3A-2 to the level of the drp3A-2 drp3B-2 double mutant (Supplemental Figure 8A). Transgenic plants expressing CFP-DRP3R→E showed similar mitochondrial phenotypes to those in tobacco and were dwarf (Figure 7D). These data together strongly support the idea that the proper function of DRP3 in mitochondrial fission is CL dependent.
Figure 7.
DRP3 Function in Mitochondrial Fission Is Dependent on Interaction with CL.
(A) Epifluorescence images of tobacco leaf epidermis coexpressing CFP-DRP3R→E (or DRP3R→E-CFP) and the mitochondrial marker COX4-YFP. Bar = 10 μm.
(B) Immunoblot analysis of the expression of DRP3 fusion proteins in crude protein extracts from tobacco leaves coexpressing CFP-DRP3 (or DRP3-CFP) and COX4-YFP. Proteins were detected by the GFP antibody. The large subunit of Rubisco (RubiscoL) stained by Ponceau S is shown as a protein loading control. Protein molecular mass markers in kilodaltons are indicated for the immunoblots.
(C) Quantification of the number of independent mitochondria in cells expressing CFP-DRP3AR273E or CFP-DRP3BR258E. Error bars indicate sd; n = 5. ***P < 0.001.
(D) Six-week-old plants overexpressing CFP-DRP3 or CFP-DRP3R→E and various DRP3 loss- and gain-of-function lines.
[See online article for color version of this figure.]
Previous studies showed that residues Lys-38, Ser-39, and Thr-59 of human DRP1 are important for protein function; K38A and S39N reduced GTP binding, and T59A decreased GTP hydrolysis. When ectopically expressed, these mutant forms of hDRP1 led to fission inhibition and thus dramatic elongation of mitochondria (Smirnova et al., 2001). Overexpression of DRP1K38A caused mild peroxisome elongation, but when co-overexpressed with the peroxisomal elongation factor PEX11β, DRP1K38A strongly inhibited peroxisome fission (Koch et al., 2003). Furthermore, overexpression of DRP1S39N and DRP1T59A resulted in reduced abundance of peroxisomes and, for DRP1S39N, peroxisome elongation as well (Li and Gould, 2003). These three residues are conserved in Arabidopsis organelle division DRPs; for example, Lys-72, Ser-73, and Thr-93 on DRP3A are conserved with hDRP1 Lys-38, Ser-39, and Thr-59, respectively (Supplemental Figure 6). To compare the inhibitory effects of DRP3AR273E on mitochondrial division with those caused by CFP-DRP3AK72A, CFP-DRP3AS73N, and CFP-DRP3AT93A, we made 35Spro:CFP-DRP3A constructs that contained K72A, S73N, and T93A, respectively. When transiently expressed in Arabidopsis drp3A-2 plants and tobacco leaves, CFP-DRP3AK72A, CFP-DRP3AS73N, and CFP-DRP3AT93A led to alterations in mitochondrial morphology comparable to those caused by CFP-DRP3AR273E (Supplemental Figures 8A and 8C). By contrast, the morphology or abundance of peroxisomes did not show obvious changes (Supplemental Figures 8B and 8D). Furthermore, CFP-DRP3AR273E, but not CFP-DRP3AK72A, CFP-DRP3AS73N, or CFP-DRP3AT93A, complemented the peroxisome phenotype in drp3A-2 (Supplemental Figure 8B), suggesting that disrupting the CL-DRP3 interaction does not interfere with DRP3’s role in peroxisome fission, yet reducing DRP3’s GTPase activity would abolish its function in the division of both peroxisomes and mitochondria. In summary, our data reinforce the idea that CL is much more important to DRP3’s role in mitochondrial fission than peroxisome fission.
CL Stabilizes the Mitochondrion-Associated DRP3 Protein Complex
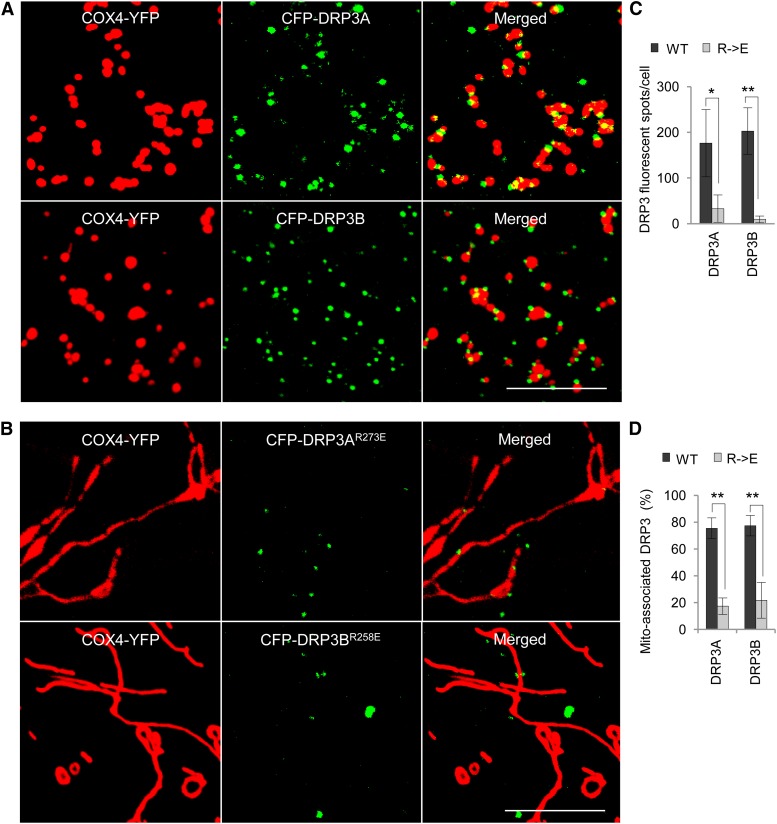
To further determine the effect of CL on DRP3 at the mechanistic level, we analyzed the localization of the DRP3 proteins in mitochondria in tobacco plants coexpressing various forms of CFP-DRP3 and COX4-YFP. Wild-type CFP-DRP3 proteins were found to be frequently associated with mitochondria (Figure 8A). Although some CFP-DRP3AR273E and CFP-DRP3BR258E proteins still localized to the massively elongated mitochondria, the total number of fluorescent spots per cell and the percentage of mitochondrion-associated fluorescent spots were significantly reduced (Figures 8B to 8D). These data suggested that CL is likely required for the stabilization of DRP3A and DRP3B proteins/complexes on mitochondria.
Figure 8.
Association of CFP-DRP3 and CFP-DRP3R→E Proteins with Mitochondria.
(A) and (B) Confocal analysis of the association of CFP-DRP3 and CFP-DRP3R→E with mitochondria in tobacco leaf epidermis expressing COX4-YFP. Bars = 10 μm.
(C) and (D) Quantification of the number of CFP-DRP3 fluorescent spots and their mitochondrial association. Error bars indicate sd; n = 4. *P < 0.05, **P < 0.01.
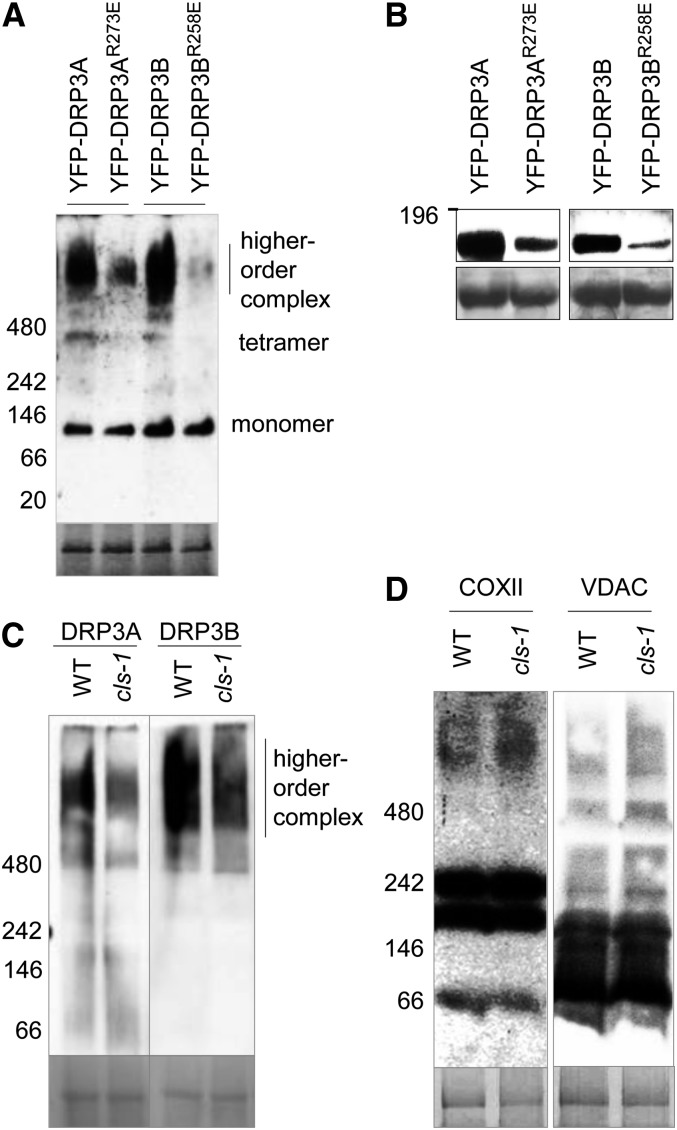
To detect the formation of the DRP3 protein complexes, we performed blue native (BN)-PAGE of proteins from tobacco leaves expressing YFP-DRP3 or YFP-DRP3R→E, followed by immunoblot analysis. YFP-DRP3R→E monomers remained at a similar level to that of the wild-type DRP3, but the level of the higher order protein complexes was greatly reduced (Figure 9A). Consistent with this result, the total protein levels of YFP-DRP3R→E were also reduced (Figure 9B). To examine the possibility that the reduction of DRP3R→E higher order complexes was due to impaired DRP3 self-interacting ability, coimmunoprecipitation was performed with HA- and YFP-tagged wild-type and mutant DRP3 proteins expressed in tobacco leaves. No obvious defects in self-interaction were observed for any of the DRP3 pairs (Supplemental Figure 9A), indicating that CL’s role in DRP3 function was likely to stabilize the DRP3 oligomers.
Figure 9.
CL Stabilizes DRP3 Protein Complexes.
(A) BN-PAGE followed by immunoblot analysis to detect the level of oligomerization of DRP3 proteins overexpressed in tobacco leaves. Crude protein extracts were analyzed, and the α-GFP antibody was used to detect YFP-DRP3 proteins. Coomassie blue staining of Rubisco is shown at bottom as the loading control.
(B) SDS-PAGE followed by immunoblot analysis of DRP3 fusion proteins analyzed in (A), detected by α-GFP antibodies. The loading control was the large subunit of Rubisco stained by Ponceau S.
(C) and (D) BN-PAGE followed by immunoblot analysis to detect the level of complex formation for DRP3 (C) and COXII and VDAC proteins (D) in cls-1. Crude protein extracts from leaves were used, and endogenous DRP3A, DRP3B, COXII, and VDAC proteins were detected by their respective antibodies. Coomassie blue staining of Rubisco is shown as the loading control.
Protein molecular mass markers in kilodaltons are indicated on all blots.
To further examine the view that CL can stabilize DRP3 oligomers, we analyzed the formation of the endogenous DRP3 complex in the cls-1 mutant, using BN-PAGE followed by immunoblot analysis with DRP3 peptide antibodies. In our previous study, these antibodies had been able to detect the DRP3 higher order complexes but not monomers, possibly due to the low level of endogenous DRP3 monomers (Aung and Hu, 2012). As expected, there was a reduction in the level of the DRP3 higher order complexes in cls-1 (Figure 9C), whereas no obvious decrease was noted for the DRP3 transcripts in the mutant (Supplemental Figure 9B). To further determine the specificity of this stabilization effect, we analyzed the complex formation of endogenous COXII (mitochondrial inner membrane) and voltage-dependent anion-selective channel (VDAC; mitochondrial outer membrane) proteins. The complex patterns of these two proteins on the BN-PAGE gel did not show significant differences between the wild type and cls-1 (Figure 9D), suggesting that the reduction of the DRP3 complex in cls-1 is fairly specific. The total protein levels of endogenous DRP3 and COXII were also analyzed by SDS-PAGE followed by immunoblots. In comparison with COXII, the total protein levels of DRP3 were slightly decreased in cls-1 (Supplemental Figure 9C). These results together led us to the conclusion that CL can stabilize the mitochondrion-associated DRP3 protein complex.
CL Plays a Role in Plant Responses to PCD-Inducing Stresses
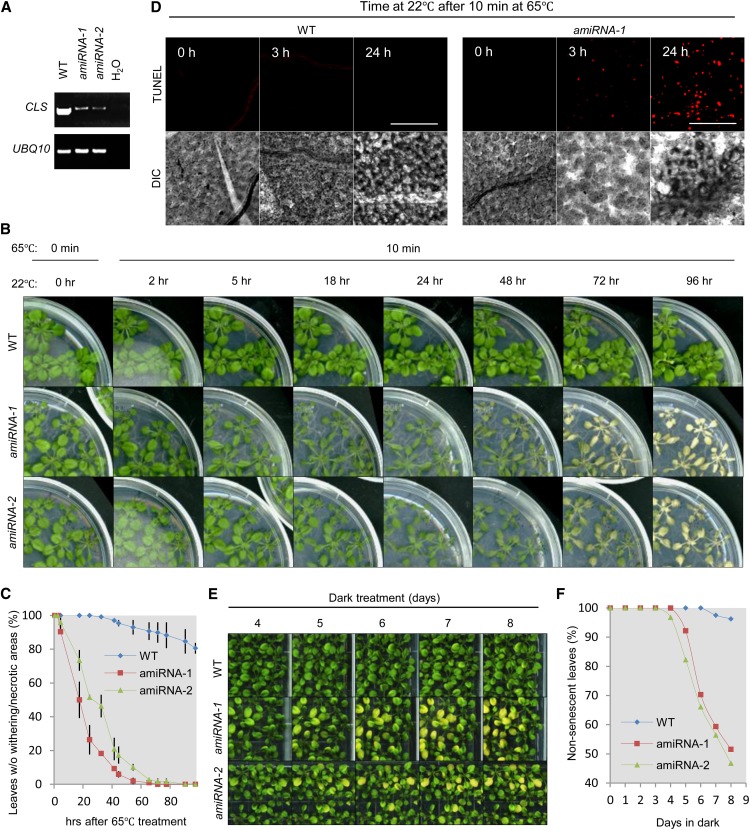
The significant role of CL in mammalian apoptosis prompted us to explore the function of CL in plant PCD, a process that shares morphological and biochemical features with apoptosis (see Introduction). In fact, UV-C light and heat shock treatments had been found to cause increased cell death in protoplasts from a cls mutant harboring an allele different from cls-1, supporting CL’s protective role in plant stress response (Pineau et al., 2013). To address this question in whole plants, we first treated wild-type and CL-deficient plants with heat, one of the most effective PCD stimuli in plants (Gadjev et al., 2008). The cls-1 mutant was weak and severely inhibited in growth; therefore, it was not suitable for physiological analysis. As an alternative, we generated amiRNA lines and selected two lines in which the levels of the CLS transcripts were significantly reduced (Figure 10A). These CLS amiRNA lines did not show growth defects but exhibited obvious changes in mitochondrial morphology (Supplemental Figure 10A).
Figure 10.
CLS-Deficient Plants Are More Susceptible to Heat Stress and Prolonged Darkness.
(A) RT-PCR analysis of CLS mRNA levels in 4-week-old plants. UBQ10 was the internal control.
(B) Appearance of 3-week-old Arabidopsis plants treated with 65°C heat for 10 min and left to recover at 22°C for varied lengths of time.
(C) Quantitative analysis of the tolerance of wild-type and CLS-deficient lines to heat stress. The percentage of healthy leaves showing no chlorosis or withering was calculated. Error bars indicate se; n = 2.
(D) TUNEL staining of fragmented DNA in leaf epidermal cells from wild-type and CLS amiRNA-1 lines treated with heat for the indicated times. Bars = 100 μm.
(E) Accelerated induction of senescence in dark-treated CLS-deficient lines. Three-week-old seedlings were subjected to dark treatment for 8 d, during which the plates were brought to light for imaging before being placed back in the dark.
(F) Quantitative analysis of the number of senescent leaves in wild-type and CLS-deficient lines shown in (E). The percentage of healthy leaves showing no visible signs of senescence was calculated.
To determine the optimal length of time for the heat treatment, we first placed wild-type Columbia-0 (Col-0) plants at 65°C and analyzed genomic DNA fragmentation, a feature of late-stage PCD (Collins et al., 1997). DNA fragmentation was detected after 30 min of heat treatment (Supplemental Figure 10B). As CL presumably exerts its function in the early stage of PCD, we selected 10 min at 65°C as the PCD-inducing condition and analyzed responses in both wild-type and CLS-deficient seedlings after the treatment. Whereas wild-type Col-0 did not show strong differences in appearance, CLS-deficient lines displayed leaf chlorosis and shriveling and even plant death (Figures 10B and 10C), despite their lack of apparent growth deficiencies under normal conditions (see plants in Figure 10B at 0 h). To detect the level of PCD in the plants, we used the terminal deoxynucleotidyl transferase–mediated deoxy-UTP nick end labeling (TUNEL) assay to stain fragmented DNA in the wild type and the amiRNA-1 line, which exhibited stronger chlorosis than amiRNA-2 in response to heat (Figure 10B). Three hours after the heat treatment, a significant level of DNA fragmentation was detected in amiRNA-1, whereas wild-type plants stained negative for TUNEL (Figure 10D), indicating a much earlier burst of PCD in the CLS-deficient line. Plants were also treated with a longer and milder heat stress: 37°C for 24 h. Accelerated formation of chlorotic lesions was evident in amiRNA lines, again indicating decreased stress tolerance of these plants (Supplemental Figures 10C and 10D).
To determine whether CL plays a specific role in the heat stress response or is broadly involved in PCD, we applied another commonly known PCD-inducing stimulus, prolonged darkness (Gadjev et al., 2008). Unlike the wild-type plants, CLS-deficient lines started to demonstrate signs of senescence after 4 to 5 d in the dark (Figures 10E and 10F). Our results from the heat and extended dark treatments and the TUNEL assays together provided strong evidence that CL plays a protective role in plants against PCD-inducing stresses.
DISCUSSION
New and Plant-Specific Aspects of CL Biology Uncovered
Previous studies using nonplant model organisms revealed CL’s pivotal roles in mitochondrial dynamics and integrity and human health (Houtkooper and Vaz, 2008). CL’s role in plants is just beginning to be revealed. CL is essential for normal plant growth and development, as shown by the two previous studies (Katayama and Wada, 2012; Pineau et al., 2013) and this study. Thus, the functions of CL in mitochondrial electron transport, ATP production, and protein import are very likely to be conserved among eukaryotes. In this study, we also discovered aspects of CL biology that were previously uncharacterized or are unique to plants.
First, we observed a distinct pattern of CMDs in a significant portion of plant mitochondria, which is similar to the pattern shown for CL on bacterial cell membranes (Mileykovskaya and Dowhan, 2000; Kawai et al., 2004) but has not been reported in animal or yeast cells. However, CMDs have been defined in mammals biochemically as detergent-resistant membrane fractions (Sorice et al., 2009). That fluorescence microscopy detected CMDs only in plants and bacteria suggests that the number of CMDs might be lower in other eukaryotic systems.
Second, we showed that the N terminus of Arabidopsis CLS consists of two peptides with mitochondrial targeting signals, signal 1 and signal 2, which are conserved in plant CLS proteins but divergent from CLSs from nonplant species, suggesting a possible plant-specific targeting mechanism for this protein. It is possible that signal 1 is a mitochondrial matrix targeting signal that directs the precursor protein partially into the matrix and that signal 2 targets the protein to the mitochondrial intermembrane space (IMS) after signal 1 is cleaved off. Together, the N-terminal (putative) transmembrane domain and signal 2 ensure that CLS resides in the inner membrane with the correct topology. Signal 2 by itself leads the fusion proteins to both mitochondria and chloroplasts, suggesting the importance of signal 1 for initial mitochondrial targeting. At this point, we do not completely exclude the possibility that targeting of the protein to both organelles by signal 2 was an artifact in our experiment; further investigations are needed to address this possibility.
Third, we determined the topology of CLS, which resides in the mitochondrial inner membrane with the C terminus facing the IMS. This suggests that the C-terminal catalytic domain might be in the IMS. However, a previous study using rat liver suggested that CL is synthesized on the matrix side of the mitochondrial inner membrane (Schlame and Haldar, 1993); thus, it is unlikely that the catalytic domain is solely in the IMS. Since several transmembrane domains are predicted in the C-terminal region of CLS, we predict that the enzymatic domain of CLS may pass the inner membrane.
More importantly, we showed a specific role of CL in mitochondrial fission through the dynamin-related protein DRP3 and in protecting plants from some PCD-inducing stresses (see below).
CL-Mediated Mitochondrial Fission and Fusion
The contrasting morphology of mitochondria in plant versus yeast and animal cells suggests highly unbalanced mitochondrial dynamics leaning toward fission in plants, as opposed to animals and yeasts. Consistent with this notion, plant mitochondria exhibit impaired fission upon CL deficiency, which is linked at least in part to CL’s ability to stabilize DRP3 higher order complexes. Taking into consideration the positive impact of CL on fusion DRP functions in yeasts and animals, we predict that CL plays equally critical roles in mitochondrial fission and fusion. The ultimate outcome of CL deficiency in mitochondrial dynamics may be determined by the relative strengths of the antagonistic fusion and fission machineries in an organism. As such, when CL is deficient, the impairment of the dominant machinery is more pronounced, resulting in the shift of mitochondrial dynamics toward fission in animals and yeasts and toward fusion in plants.
In this study, we showed the similarities in mitochondrial morphological phenotypes caused by the loss of DRP3 and CLS, partial rescue of the mitochondrial phenotype in cls-1 by overexpressing DRP3, and stabilization of DRP3 oligomers by CL, suggesting a direct role of CL in mitochondrial fission through DRP3. In yeasts and mammals, CL is an important structural component of the mitochondrial membrane involved in the organization, stabilization, and/or activity of several protein complexes required for respiration, protein import, and fusion (see Introduction). Reduction in the level of respiratory chain complexes was also shown for an Arabidopsis cls mutant (Pineau et al., 2013). However, CL’s role in mitochondria does not appear to be nonselective, as shown by the lack of reduction in the levels of the mitochondrial membrane protein complexes COXII and VDAC in cls-1. CL tends to be enriched in specific membrane domains and promotes the formation of hexagonal membrane structure, an intermediate step of the fission and fusion processes (see Introduction). It will be interesting to determine in the future whether CL is also important for the function of other proteins involved in mitochondrial division/morphogenesis, such as FISSION1, a conserved protein that presumably recruits DRP3 to both mitochondria and peroxisomes (Scott et al., 2006; Lingard et al., 2008; Zhang and Hu, 2008, 2009); ELONATED MITOCHONDRIA1, a plant-specific factor that recruits DRP3A to mitochondria (Arimura et al., 2008); and PMD1, another plant-specific factor that regulates mitochondrial and peroxisome morphogenesis in a DRP3-independent manner (Aung and Hu, 2011).
We cannot completely rule out the possibility that significantly increased fusion of mitochondria may also contribute to the observed phenotype in cls mutants. However, CL has been shown by multiple studies to play a positive role in the mitochondrial fusion process in nonplant systems. This is achieved not only by its role in stabilizing the oligomerization and enzymatic processing of the fusion DRPs but also by a structural role in promoting membrane curvature and hexagonal structure (see Introduction). Therefore, although mitochondrial fusion machinery has not been discovered in plants and thus it is difficult to address this question directly at present, it is unlikely that CL plays an inhibitory role in mitochondrial fusion in plants.
Different Membrane Lipid Compositions of Mitochondria and Peroxisomes May Contribute to the Distinct Modes of DRP3 Functions in These Organelles
DRP higher order complexes form a ring structure that encircles the organelle fission site to mediate membrane fission (Chan, 2012). However, whether and how the same DRP functions in distinct organelles in different manners is still an open question. In this study, we showed that DRP3R→E mutant proteins conferred a dominant negative effect on mitochondrial fission when overexpressed, possibly by competing with wild-type mitochondrial DRP3 proteins and, as a result, significantly reducing the interaction between DRP3 and CL needed for DRP3 complex stabilization.
Ectopically expressed dominant negative forms of human DRP1 with impaired GTP hydrolysis or GTP binding lead to dramatic morphological changes to mitochondria and, to some extent, peroxisomes (Thoms and Erdmann, 2005). In our study, transient expression of CFP-DRP3A containing changes of the corresponding conserved amino acids (i.e., CFP-DRP3AK72A, CFP-DRP3AS73N, and CFP-DRP3AT93A) also caused strong inhibition of mitochondrial fission but virtually no changes to the number or morphology of peroxisomes (Supplemental Figure 8D). It is possible that the endogenous DRP3 and/or DRP5B in plants retained enough competence to divide peroxisomes even in the presence of ectopically expressed mutant DRP3 proteins. Nonetheless, CFP-DRP3AR273E, but not the three GTPase activity–reduced dominant negative forms of DRP3A, attenuated the peroxisome elongation phenotype in drp3A-2, supporting the conclusion that CL is not as essential to peroxisome fission as to mitochondrial fission. It is well known that membrane lipid composition can modulate membrane-associated protein functions (Marsh, 2008; Vögler et al., 2008). Thus, the mitochondrion-specific presence of CL and CMDs may reflect a mechanism for DRP3 proteins to distinguish mitochondria from peroxisomes.
CL May Link Mitochondrial Fission to PCD
In apoptosis, CL on the outer membrane provides an anchor and activating platform for CASPASE8 as well as BID, a factor required for activating Bax and Bak, proteins that induce outer membrane permeabilization and the release of cytochrome c (Lutter et al., 2000; Kuwana et al., 2002; Gonzalvez and Gottlieb, 2007; Montessuit et al., 2010). CL also directly contributes to cytochrome c release. Prior to apoptosis, CL on the outer leaflet of the mitochondrial inner membrane binds to cytochrome c to retain it in the cristae, and stress signals induce the peroxidation of CL by cytochrome c, which presumably allows for remodeling of the cristae and the subsequent release of cytochrome c (Kagan et al., 2005; Gonzalvez and Gottlieb, 2007).
Multiple studies in nonplant systems have linked mitochondrial fission to the onset of PCD. For example, inhibition of human DRP1 activity reduces apoptotic mitochondrial fission and cytochrome c release (Frank et al., 2001). Ectopic expression of Caenorhabditis elegans DRP1 induces both mitochondrial fission and cell death (Jagasia et al., 2005), and the S. cerevisiae fission DRP, Dnm1, was shown to promote cell death (Fannjiang et al., 2004). CL and CMDs are believed to provide an activating platform for the PCD pathway by recruiting/stabilizing Bcl-2 family proteins required for the release of cytochrome c and other pro-PCD molecules (Gonzalvez and Gottlieb, 2007). It is possible that during PCD, CL’s interactions with fission DRP and Bcl-2 family proteins are both strengthened, leading to increased mitochondrial scission and the rupture of mitochondrial outer membranes, thus promoting the initiation of PCD.
Plant PCD is involved in embryogenesis, tissue differentiation, reproductive organ formation, pollination, and senescence and serves as a response mechanism to some abiotic and biotic stresses (Lord and Gunawardena, 2012). It shares several morphological and biochemical hallmarks with animal apoptotic PCD, such as an increased number of vesicles, cytoplasmic condensation, plasma membrane shrinkage, DNA fragmentation, and cytochrome c release (Gadjev et al., 2008). Although the well-characterized protein machinery that mediates animal apoptosis does not seem to be conserved in plants, plants possess caspase-like activities and Bcl-like proteins (Lord and Gunawardena, 2012). It would be of great interest to focus future investigations on the role of CL and DRP3 proteins in plant PCD. Our finding that CLS-deficient plants have reduced tolerance of heat stress and prolonged darkness treatment is a first step in this direction.
METHODS
Plant Material and Transformation
Arabidopsis thaliana plants were grown at 22°C with 70% humidity and 70 to 80 μmol m−2 s−1 white light for 14 h per day. Col-0 was used as the wild type. Seeds for the T-DNA insertion mutant cls-1 (SALK_049840) were provided by the ABRC, and homozygous mutants were identified by PCR of genomic DNA. Peroxisome marker lines CFP-PTS1 and YFP-PTS1 were generated in our previous study (Fan et al., 2005). Mitochondrial marker lines COX4-YFP and COX4-CFP were generated in a previous study (Nelson et al., 2007) and obtained from the ABRC. Transgenic Arabidopsis plants were generated via a floral dipping method (Clough and Bent, 1998) using Agrobacterium tumefaciens strain GV3101 (pMP90). To screen for transformants with BASTA, T1 plants were grown on soil and sprayed at 7 and 9 d after germination with 0.1% (v/v) BASTA (Finale; Farnam) with 0.05% (v/v) Silwet L-77. For other transformants, T1 seeds were plated on half-strength Linsmaier and Skoog medium containing 50 μg/mL kanamycin or 50 μg/mL hygromycin. Transient gene expression in tobacco (Nicotiana tabacum) plants was performed as described previously (Aung and Hu, 2011). Information on the primers and vectors used is provided in Supplemental Tables 1 and 2.
Gene Cloning, Plasmid Construction, and RT-PCR
The coding sequences of specific genes were amplified with Gateway-compatible primers (Supplemental Table 1) by Phusion polymerase (New England Biolabs) using cDNA made from total RNA of Col-0 plants. The PCR products were cloned into pDonor207 vector and various destination vectors (Supplemental Table 2) via a standard Gateway cloning system (Invitrogen; http://www.invitrogen.com/). To generate point mutations of DRP3AR273E, DRP3BR258E, DRP3AK72A, DRP3AS73N, and DRP3AT93A, overlapping PCR was performed (http://gfp.stanford.edu/protocol/index5.html) with primers containing the mutation sites (Supplemental Table 1).
To clone the amiRNA construct, CLS (5′-TTACACACCCGATAAGAACCC-3′) was designed by WMD3-Web MicroRNA Designer (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi). Overlapping PCR was used to clone the amiRNA according to the WMD3-Web MicroRNA Designer (http://wmd3.weigelworld.org/downloads/Cloning_of_artificial_microRNAs.pdf). The precursor microRNA was amplified by Gateway-compatible primers (Supplemental Table 1) and cloned into pEarley100 (Supplemental Table 2).
To analyze gene expression level in plants, RT-PCR was performed as described previously (Aung and Hu, 2011).
Protein Preparation and Immunoblot Analysis
For crude plant protein extraction, 50 mg fresh tissue from Arabidopsis or tobacco plants was ground with plastic pestles using liquid nitrogen and 500 μL of extraction buffer (60 mM Tris-HCl, pH 8.8, 2% SDS, 2.5% glycerol, 0.13 mM EDTA, pH 8.0, and 1× protease inhibitor cocktail complete from Roche). The tissue lysates were vortexed for 30 s, heated at 70°C for 10 min, and centrifuged at 13,000g twice for 5 min at room temperature. The supernatants were then transferred to new tubes. Supernatant (15 μL) mixed with 5 μL of 4× lithium dodecyl sulfate (LDS) sample buffer was loaded onto SDS-PAGE. To prepare mitochondrial proteins for SDS-PAGE analysis, 15 μL of purified mitochondria (see below) mixed with 5 μL of 4× LDS sample buffer was heated at 70°C for 10 min before being loaded onto SDS-PAGE.
To prepare crude native protein extracts, 100 mg fresh weight of Arabidopsis or tobacco leaves was homogenized with a pestle in liquid nitrogen. Total native proteins were isolated using a NativePAGE Sample Prep kit (Invitrogen). Four hundred microliters of 1× Tris-buffered saline buffer containing 2% Triton X-100 was added to the homogenized samples before the mixture was incubated on ice for 30 min and later centrifuged at 17,000g two times (15 min each) at 4°C. Twenty-five microliters of supernatant was transferred to a new tube, mixed with 3 μL of 5% Coomassie Brilliant Blue G 250, and then separated on a 4 to 16% NativePAGE gel according to the product manual.
After separation on the gel, proteins were transferred to a polyvinylidene difluoride membrane. For native gels, the polyvinylidene difluoride membrane was fixed by 8% acetic acid, rinsed, air dried, and washed with methanol, as described in the product manual. The membrane was blocked with 5% milk in 1× Tris-buffered saline plus Tween (TBST; 50 mM Tris base, 150 mM NaCl, and 0.05% Tween 20, pH 8.0) for 1 h at room temperature and then incubated for 1 h at room temperature with primary antibody prepared in the blocking buffer as follows: 1:20,000 for α-GFP (Abcam), 1:5000 for α-ACTIN (Sigma-Aldrich), 1:1000 for α-PEX11d (Orth et al., 2007), 1:5000 for α-VDAC (Reumann et al., 2009), 1:3000 for α-FtsZ1 (Schmitz et al., 2009; provided by Kathy Osteryoung, Michigan State University), 1:250 for α-COXII (generated by the former lab of Lee McIntosh at Michigan State University and used in a previous study [Moellering and Benning, 2010]), 1:2000 for α-HA (Cell Signaling), and 1:400 for α-DRP3A and α-DRP3B (Aung and Hu, 2012). The probed membrane was washed with 1× TBST three times for 5 min each time, followed by incubation with the secondary antibody (i.e., 1:20,000 goat anti-rabbit IgG or 1:20,000 goat anti-mouse IgG; Millipore) at room temperature, washed four times with 1× TBST for 10 min each time, and then visualized with SuperSignal West Dura Extended Duration Substrate (Pierce Biotechnology).
NAO Staining and Microscopy
For NAO staining, an NAO (Invitrogen) stock solution of 2 mg/mL in 100% ethanol was prepared. Ten-day-old seedlings were incubated with 10 µg/mL NAO in distilled, deionized water (diluted from the stock) in the dark for 10 min at room temperature. A leaf from the treated seedlings was used for fluorescence microscopy analysis using a Zeiss Axio Imager upright epifluorescence microscope and an Olympus Fluoview FV1000 confocal laser scanning microscope.
For confocal microscopy, NAO was excited with a 458-nm laser and detected at 575 to 620 nm. CFP, YFP, and chlorophyll autofluorescence were excited with 458-, 515-, and 515-nm lasers, respectively, and detected at 465 to 490 nm, 505 to 555 nm, and 655 to 755 nm. Fluorescent mitochondrial, Golgi, and ER makers used were from a previously study (Nelson et al., 2007). The mitochondrial marker ScCOX4-YFP is a fusion of the first 29 amino acids of the yeast Saccharomyces cerevisiae COX4 and YFP. The Golgi marker GmMan1-CFP contains the first 49 amino acids of soybean (Glycine max) α-1,2-MANNOSIDASE I, which includes the cytoplasmic tail and transmembrane domain. The ER marker AtWAK2-CFP contains the signal peptide of Arabidopsis WALL-ASSOCIATED KINASE2 at the N terminus of the CFP and the ER retention signal His-Asp-Glu-Leu at the C terminus of CFP.
To observe the ultrastructure of organelles in mesophyll cells, 4-week-old wild-type and cls-1 Arabidopsis plants were subjected to TEM analysis using methods described previously (Fan et al., 2005; Aung and Hu, 2011).
Total Lipid Extraction
Lipid extraction was performed following a modified protocol from the Kansas Lipidomics Research Center (http://www.k-state.edu/lipid/lipidomics/index.htm). Approximately 50 mg of Arabidopsis leaf tissue was collected and quickly immersed in 3 mL of 75°C (preheated) isopropanol with 0.01% butylated hydroxytoluene (Sigma-Aldrich; B1378) for 15 min. After adding 1.5 mL of chloroform and 0.6 mL of water, the mixture was briefly vortexed and then incubated 1 h at room temperature with agitation. After the liquid portion was transferred to a new glass tube, 4 mL of chloroform:methanol (2:1) with 0.01% butylated hydroxytoluene was added to the tube with the leaf tissue and agitated for 30 min at room temperature. This step was repeated until the tissue turned white. Then, 1 mL of 1 M aqueous KCl was added to the combined lipid extract. After a brief vortex, the sample was centrifuged at 1000g for 5 min. After the upper phase was discarded, 2 mL of water was added. After another brief vortex, the mixture was centrifuged again at 1000g for 5 min. After discarding the upper phase, the organic extract was evaporated to dryness with blowing nitrogen in a 37°C water bath. Finally, the dried residue was dissolved in 1 mL of chloroform and the tube was filled with nitrogen to protect the sample from oxidation.
LC-MS Analysis
Analyses of lipids were performed using a Waters LCT Premier mass spectrometer interfaced to Shimadzu LC-20 HPLC pumps and a Shimadzu SIL-5000 autosampler. A 10-μL aliquot of each reconstituted lipid extract was injected onto a Supelco Ascentis Express C18 column (2.1 × 50 mm, 2.7-µm particles), and gradient elution was performed using total flow rates of 0.4 mL/min and gradients based on solvent A (10 mM aqueous ammonium formate, pH 2.7) and solvent B (acetonitrile:2-propanol, 1:2 [v/v]) as follows (A/B): 0 to 4 min (90/10), linear gradient to 20/80 at 15 min, linear gradient to 1/99 at 20 min and held at these conditions until 28 min, at which time the solvent composition was returned to the initial condition. Mass spectra were acquired using W-mode ion optics (mass resolution at full-width half-maximum = 9000) using negative-ion mode electrospray ionization over m/z 100 to 2000. Mass spectra were acquired in five quasisimultaneous functions using aperture 1 potential settings of 20, 35, 50, 65, and 80 V to generate nonselective ion fragmentation using a protocol known as multiplexed collision-induced dissociation (Gu et al., 2010; Stagliano et al., 2010) that provides additional evidence of lipid structure from fragment ion masses. Levels of CL were quantified by integration (Waters QuanLynx software) of the extracted ion chromatogram for m/z 1401.98, corresponding to [M-H]− of the most abundant form of CL (with two 16-carbon and two 18-carbon fatty acids and a total of three double bonds). Amounts of other forms of CL relative to the major form remained constant across all wild-type and mutant extracts. An external standard of 1′,3′-bis[1,2-dioleoyl-sn-glycero-3-phospho]-sn-glycerol (Avanti) was used for method development and quantitative analysis.
Coimmunoprecipitation
Tobacco leaves (∼1 g fresh weight) transiently expressing the YFP-DRP3 and HA-DRP3 fusion proteins were homogenized in 4 mL of RIPA buffer (Thermo) with 1× complete protease inhibitor cocktail (Roche) and lysed on a rotator for 1 h at 4°C. After the lysed tissue was centrifuged at 13,000g for 10 min, the supernatant was incubated with 20 μL of agarose-conjugated anti-GFP (MBL) on a rotator at 4°C for 1 h to pull down the YFP fusion proteins. The agarose beads were spun down at 3000g for 15 s and washed three times with RIPA buffer. YFP-interacting proteins were eluted by adding 1× NuPAGE LDS sample buffer (Invitrogen) and heating at 75°C for 10 min. The eluted proteins were subjected to immunoblot analysis as described above.
Mitochondrial Isolation and Determination of Protein Membrane Association and Topology
Mitochondria were isolated from rosette leaves of 4-week-old transgenic plants expressing 35Spro:YFP-PMD1 or 35Spro:CLS-YFP-HA and the mitochondrial marker COX4-YFP using a previously published protocol (Aung and Hu, 2011). The purity of mitochondrial preparations was determined using immunoblot analyses and organelle-specific antibodies as described above. Mitochondrial membrane association was tested using previously described methods (Orth et al., 2007).
To determine the topology of CLS on the mitochondrial membrane, protease protection assays were employed using the proteases thermolysin and trypsin. For thermolysin treatment, 200 μL of purified mitochondria was treated with 0, 150, or 300 μg/mL thermolysin in an incubation buffer containing 50 mM HEPES-NaOH, pH 7.5, 0.33 M sorbitol, and 0.5 mM CaCl2. The reactions were performed at 4°C for 1 h and stopped by incubating on ice for 5 min with 5 mM EDTA. For trypsin treatment, 200 μL of purified mitochondria was treated with 0, 150, or 300 μg/mL trypsin in an incubation buffer containing 50 mM HEPES-NaOH, pH 7.5, and 0.33 M sorbitol. As a control, mitochondria were also treated with 1% SDS and 300 μg/mL trypsin, in which SDS served to permeabilize the membrane to cause the loss of protection of inner membrane proteins against trypsin. The reactions were performed at 4°C for 30 min and stopped by incubating on ice for 5 min with 0.5 mM phenylmethylsulfonyl fluoride. Immunoblot analysis was then performed as described above.
TUNEL Assay
Whole Arabidopsis seedlings were fixed overnight in 4% paraformaldehyde in PBS, pH 7.4, and washed with PBS for 10 min. Samples were then permeabilized by immersion in 0.1% (v/v) Triton X-100 in 0.1% sodium citrate for 2 min on ice, washed with PBS for 10 min, and incubated with 4% pectinase and 2% cellulase for 30 min. Treated samples were washed with PBS for 10 min before being used for the TUNEL assay with protocols provided by the manufacturer (Roche).
Statistical Analysis
Quantitative analysis results are presented as means ± sd (or se) from repeated experiments as indicated in the figure legends. Pairwise Student’s t test was used to analyze statistical significance.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Arabidopsis CLS (AT4G04870), DRP3A (At4g33650), and DRP3B (At2g14120).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Comparison of CLS Proteins from Different Species.
Supplemental Figure 2. CLS-YFP-HA Is Processed upon Mitochondrial Targeting.
Supplemental Figure 3. Analysis of Organelle Targeting Signals on CLS.
Supplemental Figure 4. Further Characterization of cls-1.
Supplemental Figure 5. Expression Profile of the Arabidopsis CLS Gene.
Supplemental Figure 6. Alignment of the N Terminus (Containing the GTPase Domain) of Mitochondrial/Peroxisome Division DRPs from Various Species.
Supplemental Figure 7. Functional Association of CL and DRP3 in Mitochondrial Fission.
Supplemental Figure 8. Analysis of the Roles of a Few Conserved Residues in Mitochondrial and Peroxisomal Fission.
Supplemental Figure 9. Analyses of DRP3R→E Protein Self-Interaction in Tobacco and DRP3 Transcript and Protein Levels in cls-1.
Supplemental Figure 10. Mitochondrial Phenotypes of CLS AmiRNA Lines and Characterization of CL’s Role in Plant Response to PCD-Inducing Stresses.
Supplemental Table 1. Primers Used in This Study.
Supplemental Table 2. Vectors Used in This Study.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis of CLS Proteins Shown in Supplemental Figure 1A.
Supplementary Material
Acknowledgments
We thank Christoph Benning for comments on the article, Weili Yang, Henrik Tjellström, Min Zhang, and John Froehlich for technical assistance, Melinda Frame and Alicia Pastor for their help with confocal microscopy and TEM analyses, and Tsuyoshi Nakagawa for providing the pGWB544 and pGWB545 vectors. This work was supported by grants to J.H. from the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (Grant DE-FG02-91ER20021) and the National Science Foundation (Grant MCB 1330441) and to A.D.J. by Michigan AgBioResearch.
AUTHOR CONTRIBUTIONS
R.P. designed and performed most experiments and data analysis. A.D.J. designed the LC-MS protocols and performed LC-MS analyses, including data processing. J.H. participated in the designing of experiments and data interpretation. R.P., A.D.J., and J.H. cowrote the article.
Glossary
- CL
cardiolipin
- PG
phosphatidylglycerol
- CMD
cardiolipin-enriched membrane domain
- DRP
dynamin-related protein
- PCD
programmed cell death
- NAO
10-N-nonyl-acridine orange
- ER
endoplasmic reticulum
- LC-MS
liquid chromatography–mass spectrometry
- TEM
transmission electron microscopy
- BN
blue native
- VDAC
voltage-dependent anion-selective channel
- amiRNA
artificial microRNA
- Col-0
Columbia-0
- TUNEL
deoxynucleotidyl transferase–mediated deoxy-UTP nick end labeling
- IMS
intermembrane space
- LDS
lithium dodecyl sulfate
- TBST
Tris-buffered saline plus Tween
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Arimura S., Tsutsumi N. (2002). A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc. Natl. Acad. Sci. USA 99: 5727–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura S., Aida G.P., Fujimoto M., Nakazono M., Tsutsumi N. (2004). Arabidopsis dynamin-like protein 2a (ADL2a), like ADL2b, is involved in plant mitochondrial division. Plant Cell Physiol. 45: 236–242 [DOI] [PubMed] [Google Scholar]
- Arimura S., Fujimoto M., Doniwa Y., Kadoya N., Nakazono M., Sakamoto W., Tsutsumi N. (2008). Arabidopsis ELONGATED MITOCHONDRIA1 is required for localization of DYNAMIN-RELATED PROTEIN3A to mitochondrial fission sites. Plant Cell 20: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung K., Hu J. (2011). The Arabidopsis tail-anchored protein PEROXISOMAL AND MITOCHONDRIAL DIVISION FACTOR1 is involved in the morphogenesis and proliferation of peroxisomes and mitochondria. Plant Cell 23: 4446–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung K., Hu J. (2012). Differential roles of Arabidopsis dynamin-related proteins DRP3A, DRP3B, and DRP5B in organelle division. J. Integr. Plant Biol. 54: 921–931 [DOI] [PubMed] [Google Scholar]
- Ban T., Heymann J.A., Song Z., Hinshaw J.E., Chan D.C. (2010). OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation. Hum. Mol. Genet. 19: 2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D.C. (2012). Fusion and fission: Interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 46: 265–287 [DOI] [PubMed] [Google Scholar]
- Chicco A.J., Sparagna G.C. (2007). Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am. J. Physiol. Cell Physiol. 292: C33–C44 [DOI] [PubMed] [Google Scholar]
- Clough S., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collins J.A., Schandi C.A., Young K.K., Vesely J., Willingham M.C. (1997). Major DNA fragmentation is a late event in apoptosis. J. Histochem. Cytochem. 45: 923–934 [DOI] [PubMed] [Google Scholar]
- DeVay R.M., Dominguez-Ramirez L., Lackner L.L., Hoppins S., Stahlberg H., Nunnari J. (2009). Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J. Cell Biol. 186: 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T., Coleman J., Marquis K.A., Meeske A.J., Burton B.M., Karatekin E., Rudner D.Z. (2013). FisB mediates membrane fission during sporulation in Bacillus subtilis. Genes Dev. 27: 322–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faller A. (1978). [Energetic swelling and lysis of mitochondria]. In German. Acta Anat. (Basel) 100: 573–581 [PubMed] [Google Scholar]
- Fan J., Quan S., Orth T., Awai C., Chory J., Hu J. (2005). The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol. 139: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fannjiang Y., Cheng W.C., Lee S.J., Qi B., Pevsner J., McCaffery J.M., Hill R.B., Basañez G., Hardwick J.M. (2004). Mitochondrial fission proteins regulate programmed cell death in yeast. Genes Dev. 18: 2785–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S., Gaume B., Bergmann-Leitner E.S., Leitner W.W., Robert E.G., Catez F., Smith C.L., Youle R.J. (2001). The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell 1: 515–525 [DOI] [PubMed] [Google Scholar]
- Gadjev I., Stone J.M., Gechev T.S. (2008). Programmed cell death in plants: New insights into redox regulation and the role of hydrogen peroxide. Int. Rev. Cell Mol. Biol. 270: 87–144 [DOI] [PubMed] [Google Scholar]
- Gao H., Kadirjan-Kalbach D., Froehlich J.E., Osteryoung K.W. (2003). ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl. Acad. Sci. USA 100: 4328–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalvez F., Gottlieb E. (2007). Cardiolipin: Setting the beat of apoptosis. Apoptosis 12: 877–885 [DOI] [PubMed] [Google Scholar]
- Gu L., Jones A.D., Last R.L. (2010). Broad connections in the Arabidopsis seed metabolic network revealed by metabolite profiling of an amino acid catabolism mutant. Plant J. 61: 579–590 [DOI] [PubMed] [Google Scholar]
- Houtkooper R.H., Vaz F.M. (2008). Cardiolipin, the heart of mitochondrial metabolism. Cell. Mol. Life Sci. 65: 2493–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Baker A., Bartel B., Linka N., Mullen R.T., Reumann S., Zolman B.K. (2012). Plant peroxisomes: Biogenesis and function. Plant Cell 24: 2279–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby R.P., Li L., Huang S., Pong Lee C., Millar A.H., Taylor N.L. (2012). Mitochondrial composition, function and stress response in plants. J. Integr. Plant Biol. 54: 887–906 [DOI] [PubMed] [Google Scholar]
- Jagasia R., Grote P., Westermann B., Conradt B. (2005). DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans. Nature 433: 754–760 [DOI] [PubMed] [Google Scholar]
- Joshi A.S., Thompson M.N., Fei N., Hüttemann M., Greenberg M.L. (2012). Cardiolipin and mitochondrial phosphatidylethanolamine have overlapping functions in mitochondrial fusion in Saccharomyces cerevisiae. J. Biol. Chem. 287: 17589–17597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewsuya P., Danielson N.D., Ekhterae D. (2007). Fluorescent determination of cardiolipin using 10-N-nonyl acridine orange. Anal. Bioanal. Chem. 387: 2775–2782 [DOI] [PubMed] [Google Scholar]
- Kagan V.E., et al. (2005). Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat. Chem. Biol. 1: 223–232 [DOI] [PubMed] [Google Scholar]
- Katayama K., Wada H. (2012). T-DNA insertion in the CLS gene for cardiolipin synthase affects development of Arabidopsis thaliana. Cytologia (Tokyo) 7: 123–129 [Google Scholar]
- Katayama K., Sakurai I., Wada H. (2004). Identification of an Arabidopsis thaliana gene for cardiolipin synthase located in mitochondria. FEBS Lett. 577: 193–198 [DOI] [PubMed] [Google Scholar]
- Kawai F., Shoda M., Harashima R., Sadaie Y., Hara H., Matsumoto K. (2004). Cardiolipin domains in Bacillus subtilis Marburg membranes. J. Bacteriol. 186: 1475–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A., Thiemann M., Grabenbauer M., Yoon Y., McNiven M.A., Schrader M. (2003). Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278: 8597–8605 [DOI] [PubMed] [Google Scholar]
- Kuwana T., Mackey M.R., Perkins G., Ellisman M.H., Latterich M., Schneiter R., Green D.R., Newmeyer D.D. (2002). Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111: 331–342 [DOI] [PubMed] [Google Scholar]
- Lenarcic R., Halbedel S., Visser L., Shaw M., Wu L.J., Errington J., Marenduzzo D., Hamoen L.W. (2009). Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 28: 2272–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis R.N., McElhaney R.N. (2009). The physicochemical properties of cardiolipin bilayers and cardiolipin-containing lipid membranes. Biochim. Biophys. Acta 1788: 2069–2079 [DOI] [PubMed] [Google Scholar]
- Li J.F., Park E., von Arnim A.G., Nebenführ A. (2009). The FAST technique: A simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 5: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Gould S.J. (2003). The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J. Biol. Chem. 278: 17012–17020 [DOI] [PubMed] [Google Scholar]
- Lingard M.J., Gidda S.K., Bingham S., Rothstein S.J., Mullen R.T., Trelease R.N. (2008). Arabidopsis PEROXIN11c-e, FISSION1b, and DYNAMIN-RELATED PROTEIN3A cooperate in cell cycle-associated replication of peroxisomes. Plant Cell 20: 1567–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan D.C. (2010). The dynamic plant chondriome. Semin. Cell Dev. Biol. 21: 550–557 [DOI] [PubMed] [Google Scholar]
- Lord C.E., Gunawardena A.H. (2012). Programmed cell death in C. elegans, mammals and plants. Eur. J. Cell Biol. 91: 603–613 [DOI] [PubMed] [Google Scholar]
- Lutter M., Fang M., Luo X., Nishijima M., Xie X., Wang X. (2000). Cardiolipin provides specificity for targeting of tBid to mitochondria. Nat. Cell Biol. 2: 754–761 [DOI] [PubMed] [Google Scholar]
- Magdalan J., Ostrowska A., Podhorska-Okołów M., Piotrowska A., Izykowska I., Nowak M., Dolińska-Krajewska B., Zabel M., Szelag A., Dziegiel P. (2009). Early morphological and functional alterations in canine hepatocytes due to alpha-amanitin, a major toxin of Amanita phalloides. Arch. Toxicol. 83: 55–60 [DOI] [PubMed] [Google Scholar]
- Malamed S. (1965). Structural changes during swelling of isolated rat mitochondria. Z. Zellforsch. 65: 10–15 [Google Scholar]
- Mannella C.A., Pfeiffer D.R., Bradshaw P.C., Moraru I.I., Slepchenko B., Loew L.M., Hsieh C.E., Buttle K., Marko M. (2001). Topology of the mitochondrial inner membrane: Dynamics and bioenergetic implications. IUBMB Life 52: 93–100 [DOI] [PubMed] [Google Scholar]
- Mano S., Nakamori C., Kondo M., Hayashi M., Nishimura M. (2004). An Arabidopsis dynamin-related protein, DRP3A, controls both peroxisomal and mitochondrial division. Plant J. 38: 487–498 [DOI] [PubMed] [Google Scholar]
- Marsh D. (2008). Protein modulation of lipids, and vice-versa, in membranes. Biochim. Biophys. Acta 1778: 1545–1575 [DOI] [PubMed] [Google Scholar]
- Mileykovskaya E., Dowhan W. (2000). Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 182: 1172–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima S.Y., Kuwayama H., Urushihara H., Nakanishi H. (2008). Evolutionary linkage between eukaryotic cytokinesis and chloroplast division by dynamin proteins. Proc. Natl. Acad. Sci. USA 105: 15202–15207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering E.R., Benning C. (2010). Phosphate regulation of lipid biosynthesis in Arabidopsis is independent of the mitochondrial outer membrane DGS1 complex. Plant Physiol. 152: 1951–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montessuit S., Somasekharan S.P., Terrones O., Lucken-Ardjomande S., Herzig S., Schwarzenbacher R., Manstein D.J., Bossy-Wetzel E., Basañez G., Meda P., Martinou J.C. (2010). Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell 142: 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myron D.R., Connelly J.L. (1971). The morphology of the swelling process in rat liver mitochondria. J. Cell Biol. 48: 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nowicki M., Müller F., Frentzen M. (2005). Cardiolipin synthase of Arabidopsis thaliana. FEBS Lett. 579: 2161–2165 [DOI] [PubMed] [Google Scholar]
- Orth T., Reumann S., Zhang X., Fan J., Wenzel D., Quan S., Hu J. (2007). The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis. Plant Cell 19: 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz A., Killian J.A., Verkleij A.J., Wilschut J. (1999). Membrane fusion and the lamellar-to-inverted-hexagonal phase transition in cardiolipin vesicle systems induced by divalent cations. Biophys. J. 77: 2003–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C., Voelker D.R., Langer T. (2011). Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 192: 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C.S., Osellame L.D., Stojanovski D., Ryan M.T. (2011). The regulation of mitochondrial morphology: Intricate mechanisms and dynamic machinery. Cell. Signal. 23: 1534–1545 [DOI] [PubMed] [Google Scholar]
- Pineau B., Bourge M., Marion J., Mauve C., Gilard F., Maneta-Peyret L., Moreau P., Satiat-Jeunemaître B., Brown S.C., De Paepe R., Danon A. (2013). The importance of cardiolipin synthase for mitochondrial ultrastructure, respiratory function, plant development, and stress responses in Arabidopsis. Plant Cell 25: 4195–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner L.D., Weibel D.B. (2011). Cardiolipin microdomains localize to negatively curved regions of Escherichia coli membranes. Proc. Natl. Acad. Sci. USA 108: 6264–6269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner L.D., Weibel D.B. (2012). MinD and MinE interact with anionic phospholipids and regulate division plane formation in Escherichia coli. J. Biol. Chem. 287: 38835–38844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Quan S., Aung K., Yang P., Manandhar-Shrestha K., Holbrook D., Linka N., Switzenberg R., Wilkerson C.G., Weber A.P., Olsen L.J., Hu J. (2009). In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol. 150: 125–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlame M., Haldar D. (1993). Cardiolipin is synthesized on the matrix side of the inner membrane in rat liver mitochondria. J. Biol. Chem. 268: 74–79 [PubMed] [Google Scholar]
- Schlame M., Ren M., Xu Y., Greenberg M.L., Haller I. (2005). Molecular symmetry in mitochondrial cardiolipins. Chem. Phys. Lipids 138: 38–49 [DOI] [PubMed] [Google Scholar]
- Schmitz A.J., Glynn J.M., Olson B.J., Stokes K.D., Osteryoung K.W. (2009). Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development. Mol. Plant 2: 1211–1222 [DOI] [PubMed] [Google Scholar]
- Schrader M., Yoon Y. (2007). Mitochondria and peroxisomes: Are the ‘big brother’ and the ‘little sister’ closer than assumed? Bioessays 29: 1105–1114 [DOI] [PubMed] [Google Scholar]
- Scott I., Tobin A.K., Logan D.C. (2006). BIGYIN, an orthologue of human and yeast FIS1 genes functions in the control of mitochondrial size and number in Arabidopsis thaliana. J. Exp. Bot. 57: 1275–1280 [DOI] [PubMed] [Google Scholar]
- Smirnova E., Griparic L., Shurland D.L., van der Bliek A.M. (2001). Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12: 2245–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorice M., Manganelli V., Matarrese P., Tinari A., Misasi R., Malorni W., Garofalo T. (2009). Cardiolipin-enriched raft-like microdomains are essential activating platforms for apoptotic signals on mitochondria. FEBS Lett. 583: 2447–2450 [DOI] [PubMed] [Google Scholar]
- Stagliano M.C., DeKeyser J.G., Omiecinski C.J., Jones A.D. (2010). Bioassay-directed fractionation for discovery of bioactive neutral lipids guided by relative mass defect filtering and multiplexed collision-induced dissociation. Rapid Commun. Mass Spectrom. 24: 3578–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M.G., Williams J., Munoz-Pinedo C., Perkins G.A., Brown J.M., Ellisman M.H., Green D.R., Frey T.G. (2007). Correlated three-dimensional light and electron microscopy reveals transformation of mitochondria during apoptosis. Nat. Cell Biol. 9: 1057–1065 [DOI] [PubMed] [Google Scholar]
- Tamura Y., Endo T., Iijima M., Sesaki H. (2009). Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J. Cell Biol. 185: 1029–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira P.F., Glaser E. (2013). Processing peptidases in mitochondria and chloroplasts. Biochim. Biophys. Acta 1833: 360–370 [DOI] [PubMed] [Google Scholar]
- Thoms S., Erdmann R. (2005). Dynamin-related proteins and Pex11 proteins in peroxisome division and proliferation. FEBS J. 272: 5169–5181 [DOI] [PubMed] [Google Scholar]
- Vögler O., Barceló J.M., Ribas C., Escribá P.V. (2008). Membrane interactions of G proteins and other related proteins. Biochim. Biophys. Acta 1778: 1640–1652 [DOI] [PubMed] [Google Scholar]
- Wriessnegger T., Gübitz G., Leitner E., Ingolic E., Cregg J., de la Cruz B.J., Daum G. (2007). Lipid composition of peroxisomes from the yeast Pichia pastoris grown on different carbon sources. Biochim. Biophys. Acta 1771: 455–461 [DOI] [PubMed] [Google Scholar]
- Zhang X., Hu J. (2009). Two small protein families, DYNAMIN-RELATED PROTEIN3 and FISSION1, are required for peroxisome fission in Arabidopsis. Plant J. 57: 146–159 [DOI] [PubMed] [Google Scholar]
- Zhang X., Hu J. (2010). The Arabidopsis chloroplast division protein DYNAMIN-RELATED PROTEIN5B also mediates peroxisome division. Plant Cell 22: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.C., Hu J.P. (2008). FISSION1A and FISSION1B proteins mediate the fission of peroxisomes and mitochondria in Arabidopsis. Mol. Plant 1: 1036–1047 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.