Dense vesicles (DVs) are known to be key carriers for sorting of storage proteins to protein storage vacuoles (PSVs) in seed plants. This work characterizes the kelch-repeat protein GLUTELIN PRECURSOR ACCUMULATION3, which forms a functional protein complex with the small GTPase Rab5a via its guanine exchange factor, VPS9a, to regulate directional trafficking of DVs to PSVs in rice endosperm.
Abstract
In seed plants, a major pathway for sorting of storage proteins to the protein storage vacuole (PSV) depends on the Golgi-derived dense vesicles (DVs). However, the molecular mechanisms regulating the directional trafficking of DVs to PSVs remain largely elusive. Here, we report the functional characterization of the rice (Oryza sativa) glutelin precursor accumulation3 (gpa3) mutant, which exhibits a floury endosperm phenotype and accumulates excess proglutelins in dry seeds. Cytological and immunocytochemistry studies revealed that in the gpa3 mutant, numerous proglutelin-containing DVs are misrouted to the plasma membrane and, via membrane fusion, release their contents into the apoplast to form a new structure named the paramural body. Positional cloning of GPA3 revealed that it encodes a plant-specific kelch-repeat protein that is localized to the trans-Golgi networks, DVs, and PSVs in the developing endosperm. In vitro and in vivo experiments verified that GPA3 directly interacts with the rice Rab5a-guanine exchange factor VPS9a and forms a regulatory complex with Rab5a via VPS9a. Furthermore, our genetic data support the notion that GPA3 acts synergistically with Rab5a and VPS9a to regulate DV-mediated post-Golgi traffic in rice. Our findings provide insights into the molecular mechanisms regulating the plant-specific PSV pathway and expand our knowledge of vesicular trafficking in eukaryotes.
INTRODUCTION
Vacuoles, one of the landmarks of plant cells, perform diverse functions, especially in the degradation and storage of proteins and secondary metabolites. Plant cells contain two functionally and morphologically distinct vacuoles: the lytic vacuole (LV) and the protein storage vacuole (PSV; Rogers, 2008). To perform their duties as the LV or PSV, these organelles need to receive their respective protein cargos via vacuolar branch routes. Most vacuolar proteins start with synthesis from the endoplasmic reticulum (ER) and then to the Golgi apparatus for further sorting to post-Golgi compartments (e.g., the trans-Golgi network [TGN] and prevacuolar compartment [PVC]) before reaching the LV or PSV (Robinson et al., 2005, 2008; Hwang, 2008).
Previous studies suggested that the TGN serves as a major sorting station for biosynthetic cargos in plants (Lam et al., 2007a; Robinson et al., 2008). Cytological evidence from pea (Pisum sativum) and Arabidopsis thaliana show that sorting of storage proteins occurs early in the cis-cisternae of the Golgi apparatus (Hillmer et al., 2001; Otegui et al., 2006), where two types of putative vacuolar sorting determinants (VSRs), the BP-80 family members and the receptor homology region transmembrane domain RING H2 MOTIF PROTEIN1 (RMR1), most probably act as receptors for soluble storage proteins (Robinson et al., 2005; Shen et al., 2011). Their interactions may form a condensation nucleus that facilitates subsequent storage protein aggregation at the periphery of the Golgi cis-cisternae, which continuously traverses the Golgi apparatus and buds off from the TGN as dense vesicles (DVs; 100 to 200 nm in diameter; Robinson et al., 2005). Characterized by electron-dense contents, the DVs are enclosed by a single membrane but lack visible protein coats on their cytosolic surface (Hohl et al., 1996; Otegui et al., 2006; Wang et al., 2012). Thus, the DV defines a unique carrier for trafficking of storage proteins to the PSV. Growing evidence is showing that post-Golgi trafficking of storage proteins to the PSV also requires the retromer components (MAG1/VPS29, VPS35, and SNXs) involved in the recycling of VSRs (Shimada et al., 2003, 2006; Yamazaki et al., 2008; Zouhar et al., 2009; Pourcher et al., 2010), the Rab family of small GTPases as well as their common guanine exchange factor (GEF) for specifying vesicular trafficking (Wang et al., 2010; Ebine et al., 2011; Liu et al., 2013), and the SNARE complex for mediating membrane fusion between post-Golgi compartments (Ebine et al., 2008). However, plant-specific regulators required for these processes largely remain to be identified.
Rice (Oryza sativa), a staple food for ∼60% of the world’s population, has been a primary model plant for molecular genetic and functional studies of crops. Three major types of storage proteins coexist in rice endosperm: glutelins, prolamins, and α-globulins (Takemoto et al., 2002). Among them, prolamins directly aggregate within the ER lumen and form the spherical protein body I (PBI; Li et al., 1993). By contrast, the most abundant glutelins are synthesized as 57-kD proglutelins at the ER and later follow the DV-mediated transport route to the PSV (Takemoto et al., 2002), where cleavage of proglutelins into mature acidic and basic subunits occurs, and eventually form the irregularly shaped protein body II (PBII) together with α-globulins (Washida et al., 2012). Additionally, there have been reports of precursor-accumulating (PAC) vesicles (200 to 400 nm in diameter) in the PSV pathway, suggesting the existence of a direct ER-to-PSV route for the transport of proglutelins and α-globulins in rice endosperm (Hara-Nishimura et al., 1998; Takahashi et al., 2005).
Previous studies have identified eight independent rice 57H mutants that accumulate high levels of 57-kD proglutelins. These mutants were named endosperm storage protein (esp2) and glutelin precursor (glup1 to glup7; Ueda et al., 2010). Among them, ESP2 encodes PDI1-1 (for PROTEIN DISULFIDE ISOMERASE-LIKE1-1), involved in proglutelin maturation in the ER (Takemoto et al., 2002). GLUP4 and GLUP6 encode the small GTPase Rab5a and its GEF, VPS9a, respectively, and they were shown to work together in regulating post-Golgi trafficking of proglutelins and α-globulins (Fukuda et al., 2011, 2013), while GLUP3 codes for a vacuolar processing enzyme, which proteolytically processes proglutelins into acidic and basic subunits within the PSV (Kumamaru et al., 2010). We previously reported three 57H mutants named W379, glutelin precursor accumulation1 (gpa1), and gpa2, which are allelic to glup3, glup4, and glup6, respectively (Wang et al., 2009, 2010; Liu et al., 2013). Despite these significant advances, our understanding of the molecular mechanisms regulating storage protein sorting in rice still remains limited and fragmented. It is expected that molecular cloning and characterization of additional 57H mutants should help elucidate the regulatory mechanisms of vacuolar storage protein sorting in rice and other cereal plants.
In this article, we report the functional characterization of the rice mutant gpa3 that accumulates excess amounts of proglutelins in dry seeds. Cytological and immunocytochemistry studies revealed that in the gpa3 mutant, large amounts of proglutelins are misrouted to the apoplast via fusion between DVs and the plasma membrane (PM) to form a new structure named the paramural body (PMB) containing proglutelins, α-globulins, aquaporin TIP3, and abnormally deposited cell wall components. We show that GPA3 encodes a plant-specific kelch-repeat protein that is localized to the TGN, DV, and PBII in developing rice endosperm. Our combined molecular, genetic, and cytological data support the notion that GPA3 forms a regulatory complex with Rab5a through physical interaction with VPS9a and that these factors acts synergistically in regulating DV-mediated post-Golgi trafficking of major storage proteins in the rice endosperm during grain filling and maturation.
RESULTS
Phenotypic Characterization of the gpa3 Mutant
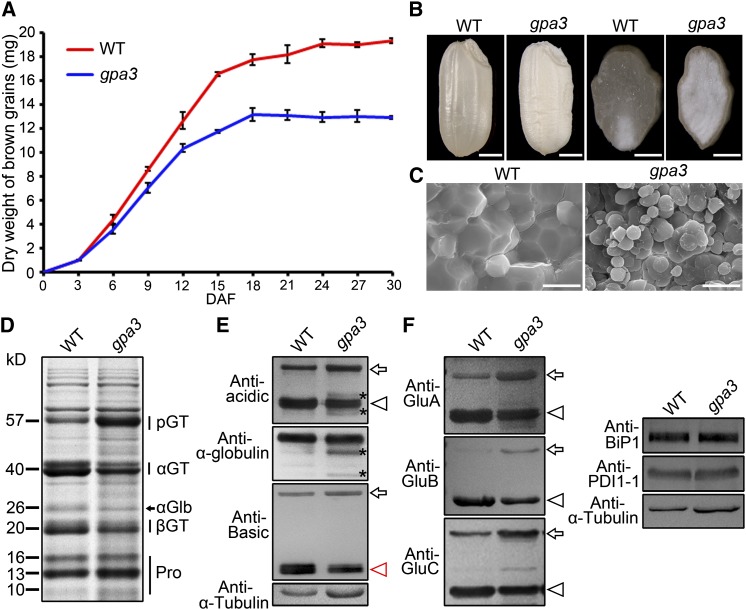
To better dissect the molecular machineries regulating proglutelin trafficking in rice, we isolated another 57H mutant named gpa3. The gpa3 plants exhibited no visible abnormalities before the grain-filling stage, such as plant height, tiller number, and heading date. After fertilization, the gpa3 mutant exhibited a markedly slower grain-filling rate (Figure 1A) and eventually produced a shrunken and floury endosperm (Figure 1B), which resulted in an ∼30% reduction in grain weight (Supplemental Table 1). Scanning electron microscopy analysis revealed that the wild-type endosperm was filled with densely packed, polyhedral starch granules, but endosperm of the gpa3 mutant was packed with round, irregularly arranged compound starch granules (Figure 1C). Further analysis showed that, compared with the wild type, amylose content was reduced ∼22% in the gpa3 mutant, whereas its protein and lipid contents increased ∼5 and 88%, respectively (Supplemental Table 1). Thus, the mutation in GPA3 causes a pleiotropic defect in storage substance accumulation in the rice endosperm.
Figure 1.
Phenotypic Analyses of the gpa3 Mutant.
(A) Grain-filling process in the wild type and the gpa3 mutant. Values are means ± sd (n = 3).
(B) Transverse sections of representative wild-type and gpa3 mutant dry seeds. Bars = 1 mm.
(C) Scanning electron microscopy images of transverse sections of wild-type and gpa3 mutant grains. Bars = 10 μm.
(D) SDS-PAGE storage protein profiles of dry seeds from the wild type and the gpa3 mutant. pGT, proglutelins; αGT, glutelin acidic subunits; αGlb, α-globulin; βGT, glutelin basic subunits; Pro, prolamins.
(E) Immunoblot analysis of storage proteins from dry seeds of the wild type and the gpa3 mutant, using antiglutelin and anti-α-globulin specific antibodies. Possible partial degradation products of glutelins and α-globulins are labeled with asterisks.
(F) Immunoblot analysis of the glutelin subfamily proteins (GluA, GluB, and GluC) and the molecular chaperones BiP1 and PDI1-1.
Arrows denote the 57-kD proglutelins, while arrowheads indicate the glutelin acidic subunits (black) and basic subunits (red). Antitubulin antibodies were used as a loading control in (E) and (F).
SDS-PAGE and immunoblot analyses revealed that the accumulation of prolamins was comparable in the dry seeds of the wild type and the gpa3 mutant. However, the gpa3 mutant seeds exhibited increased accumulation of 57-kD proglutelins, accompanied by reduced accumulation of the mature acidic and basic subunits as well as α-globulins, compared with wild-type seeds (Figures 1D and 1E; Supplemental Figure 1). In addition, immunoblot analysis using isoform-specific antibodies revealed increased accumulation of proglutelins for all glutelin subfamilies (GluA, GluB, and GluC; Takemoto et al., 2002), accompanied by decreased accumulation of their respective acidic subunits in the gpa3 mutant (Figure 1F), suggesting that a defect in a regulatory factor rather than glutelin structural genes may be responsible for the gpa3 mutant phenotypes. Furthermore, time-course analysis revealed that the defects in storage protein accumulation were visible in endosperm 9 d after flowering (DAF) and became evident in endosperm 12 DAF and thereafter (Supplemental Figure 2).
To test whether gpa3 is defective in storage protein sorting at the ER level or in post-Golgi trafficking, we compared the levels of ER lumen BINDING PROTEIN1 (BiP1) and PDI1-1, two molecular chaperones known to be induced in 57H rice mutants defective in maturation and export of proglutelins from the ER (Ueda et al., 2010). Immunoblot analysis showed that the levels of BiP1 and PDI1-1 were comparable in the gpa3 mutant and the wild type (Figure 1F). Furthermore, ultrastructural studies with samples prepared using a high-pressure frozen/freeze substitution (HPF) procedure revealed no obvious differences in the morphology of ER and Golgi between gpa3 and wild-type endosperm (Supplemental Figure 3). These results collectively suggest that the gpa3 mutant is most likely defective in post-Golgi trafficking of storage proteins.
Abnormal Deposition of Glutelins and α-Globulins in the gpa3 Mutant
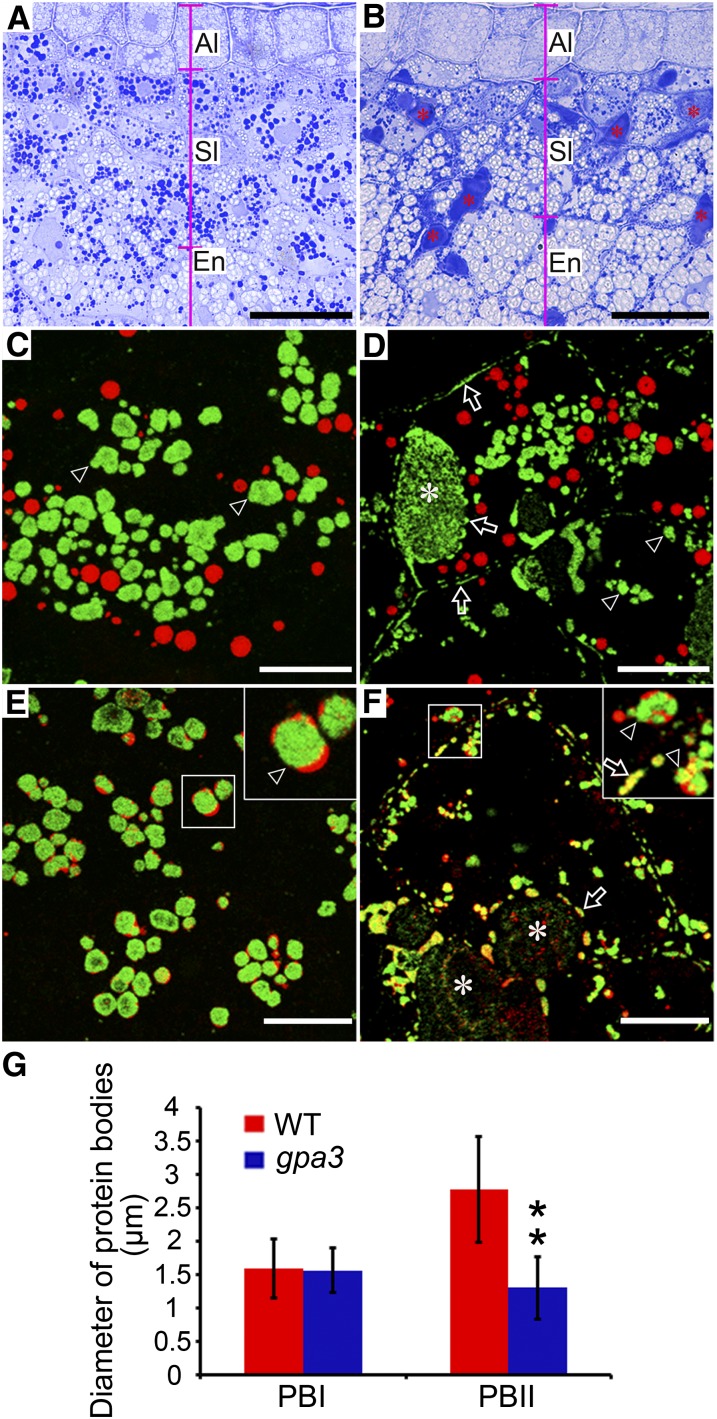
To obtain direct evidence for the missorting of storage proteins in the gpa3 mutant, we prepared semithin sections (0.5 μm thickness) of wild-type and gpa3 developing grains (9 DAF). As shown in Figures 2A and 2B, Coomassie blue staining showed that the storage proteins were most abundant in the subaleurone layer of endosperm in both the wild type and the gpa3 mutant. Notably, we observed numerous abnormal protein-filled structures (average section area ± sd: 56.6 ± 45.5 μm2, n = 69) in the gpa3 mutant but not in the wild type (Figure 2B, asterisks). Similar structures were observed in the gpa1/glup4/rab5a mutant, and they are referred to as vesicle-filled structures (Wang et al., 2010) or PMBs (Fukuda et al., 2011). In this work, we also refer to these abnormal protein-filled structures as PMBs. Indirect immunofluorescence experiments using specific antibodies against various storage proteins showed that the PMBs contained not only glutelins but also α-globulins. Moreover, we observed numerous glutelin- and α-globulin–containing protein granules distributed near the cell periphery in the gpa3 mutant endosperm but not in wild-type endosperm (Figures 2C to 2F). Furthermore, the average sizes of PBIIs and PSVs were evidently smaller in the gpa3 mutant, although the sizes of the spherical PBIs showed no evident differences between the gpa3 mutant and the wild type (Figure 2G; Supplemental Figure 4). As in the wild type (Figure 2E), α-globulins were sequestered mainly in the periphery of PBIIs surrounding glutelins, but they overlapped with glutelins in those protein granules in the gpa3 mutant (Figure 2F). These observations suggest that both glutelins and α-globulins are missorted to the PMB and cell periphery, resulting in smaller PBIIs and PSVs in the gpa3 mutant.
Figure 2.
Light and Immunofluorescence Microscopy Images of Protein Bodies in the Subaleurone Cells of the Wild Type and the gpa3 Mutant.
(A) and (B) Light microscopy observation of wild-type (A) and gpa3 mutant (B) grain sections stained with Coomassie blue. Note the numerous PMB structures (asterisks) present in the gpa3 mutant. Al, aleurone layers; Sl, subaleurone layers; En, starchy endosperm. Bars = 25 μm.
(C) to (F) Immunofluorescence microscopy images of storage proteins in wild-type ([C] and [E]) and gpa3 mutant ([D] and [F]) grains. Secondary antibodies conjugated with Alexa fluor 488 (green) and Alexa fluor 555 (red) were used to trace the antigens recognized by the antiglutelin and antiprolamin antibodies, respectively, in (C) and (D). Similar reactions were performed with anti-α-globulin antibodies instead of antiprolamin antibodies in (E) and (F). The insets represent magnified images of the selected areas in (E) and (F). Arrows indicate the protein granules lying along the cell periphery and PMB structures (asterisks), while arrowheads indicate the PBIIs. Bars = 10 μm.
(G) Measurement of the diameters of PBIs and PBIIs. Values are means ± sd. **P < 0.01 (n > 300, Student’s t test).
Abnormal Deposition of Callose and Cell Wall Components in PMBs
In our immunofluorescence experiment, we observed that the rice grain cell wall displayed strong autofluorescence under UV light or a 405-nm laser. We then used this feature to mark cell walls in our study. In wild-type grain, the aleurone cells had much thicker cell walls than the endosperm cells (Supplemental Figure 5A). Interestingly, such a difference disappeared in the gpa3 mutant due to thickening of the endosperm cell wall, especially in those PMB-containing cells (Supplemental Figure 5B). In addition, we found that the peripheral regions of the PMBs also displayed the cell wall characteristic autofluorescence signals (Supplemental Figures 5C to 5E), implying that PMBs may also contain cell wall components. To test this, we performed immunofluorescence experiments using three representative cell wall antibodies against callose, (1,3;1,4)-β-glucan, and pectin. In developing wild-type subaleurone cells, callose was deposited as dotted structures at the plasmodesmata (Supplemental Figure 6A), as reported previously (Chen and Kim, 2009). In contrast, (1,3;1,4)-β-glucan and pectin were deposited evenly in the cell wall (Supplemental Figures 6C and 6E). In the gpa3 mutant, we observed abnormal deposition of callose, (1,3;1,4)-β-glucan, and pectin in the PMBs, giving the large PMBs a stratified appearance (Supplemental Figures 6B, 6D, and 6F).
Mistargeting of DVs Leads to the Formation of PMBs
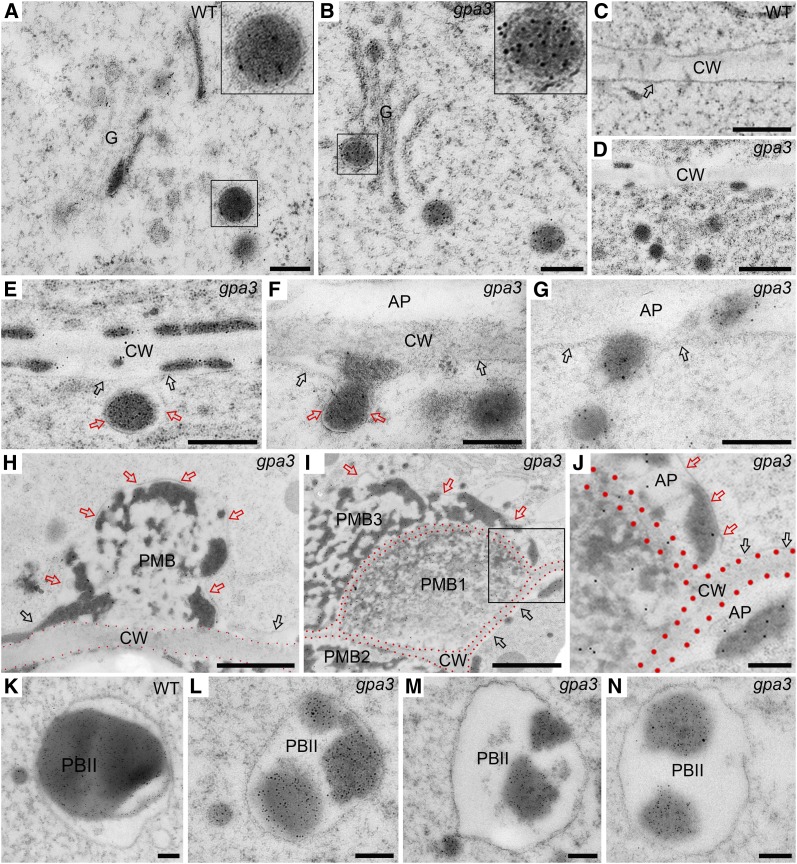
To further dissect the origin and formation of PMBs, we performed immunogold labeling studies of glutelins with ultrathin sections prepared from high-pressure frozen/freeze-substituted subaleurone cells at various developmental stages. As in the wild type (Figure 3A), glutelin-containing single membrane–enclosed DVs can normally bud off from the Golgi in the gpa3 mutant (Figure 3B), and the sizes of DVs near the Golgi were comparable in the wild type and the gpa3 mutant (mean ± sd: 153.09 ± 33.95 nm [n = 67] in the wild type versus 156.13 ± 33.26 nm [n = 337] in the gpa3 mutant; P > 0.05, Student’s t test). Notably, we observed that numerous DVs (mean ± sd: 169.72 ± 37.18 nm [n = 126]) accumulated near the cell wall in the gpa3 mutant but not in the wild type (Figures 3C and 3D). More strikingly, we observed that these single membrane–enclosed DVs could fuse directly with the PM and expel their contents into the apoplast space, leading to the formation of small PMBs (Figures 3E to 3H). Continuous fusion of DVs with small PMBs led to the formation of larger and more complex PMBs, which were divided into several parts by cell wall components (Figure 3I; Supplemental Figures 6D and 6E). In support of this notion, we observed some DVs accumulated as clusters outside the outermost PMB structure, suggesting that these DVs might be en route to fuse with the PMB envelope (Figure 3I). Similarly, we also observed the colocalization of storage proteins and another DV cargo, TIP3 (a PSV marker protein; Hinz et al., 1999; Hillmer et al., 2001), in PMBs in PTIP3:TIP3-GFP transgenic plants in the gpa3 mutant background (Supplemental Figure 7). Together, these observations suggest that mistargeting of DVs to the PM and releasing of their contents via DV-PM fusion give rise to the formation of PMBs.
Figure 3.
Immunoelectron Microscopy Localization of Glutelins Depicting the Biogenesis of PMBs in the gpa3 Mutant.
Ultrathin sections of developing wild-type and gpa3 subaleurone cells were prepared using an HPF procedure, and developing wild-type and gpa3 subaleurone cells were labeled with antiglutelin primary antibodies and gold colloid–conjugated secondary antibodies. Gold particles were observed in the Golgi (G), DVs, protein granules in the apoplast (AP), PMBs, and PBIIs. Black arrows indicate the PM, while red arrows indicate the envelope of the PMB. Red dots outline the cell wall (CW) region. Bars in (A) to (C), (E) to (G), and (J) to (N) = 200 nm; bars in (D) = 500 nm; bars in (H) and (I) = 1 mm.
(A) and (B) Electron micrographs showing that, as in the wild type (A), DVs can normally bud off from the Golgi in the gpa3 mutant (B). Note that DVs are enclosed with a single membrane (insets). The insets represent the magnified images of the selected DVs.
(C) and (D) General overview of cell periphery in developing wild-type (C) and gpa3 (D) subaleurone cells. Note that numerous glutelin-containing DVs accumulate near the PM.
(E) to (G) Electron micrographs showing that single membrane (red arrows)–enclosed DVs can fuse with the PM (black arrows) and expel their contents into the apoplast.
(H) to (J) Electron micrographs showing young (H) and mature (I) PMB structures. Note that the young PMB has a single membrane envelope (red arrows), which is physically connected with the PM (black arrows). (J) represents the magnified image of the selected area in (I).
(K) to (N) Electron micrographs showing that DVs can fuse with PSVs to form the PBIIs in the wild type (K) and the gpa3 mutant ([L] to [N]), but the PBII appears smaller in the gpa3 mutant.
In addition to the formation of PMBs, we also observed that the sizes of DVs near the PBIIs were comparable in the wild type and the gpa3 mutant (mean ± sd: 174.93 ± 35.13 nm [n = 26] in the wild type versus 165.26 ± 30.59 nm [n = 152] in the gpa3 mutant; P > 0.05, Student’s t test), but the sizes of PBIIs appeared evidently smaller in the gpa3 mutant (mean ± sd: 3.93 ± 2.55 μm2 [n = 171] in the wild type versus 1.17 ± 0.92 μm2 [n = 355] in the gpa3 mutant; P < 0.01, Student’s t test). These observations collectively suggest that in the gpa3 mutant, the sorting route of DVs to the PSV is severely blocked, resulting in smaller PBIIs and the formation of complex PMBs.
GPA3 Encodes a Plant-Specific Kelch-Repeat Protein
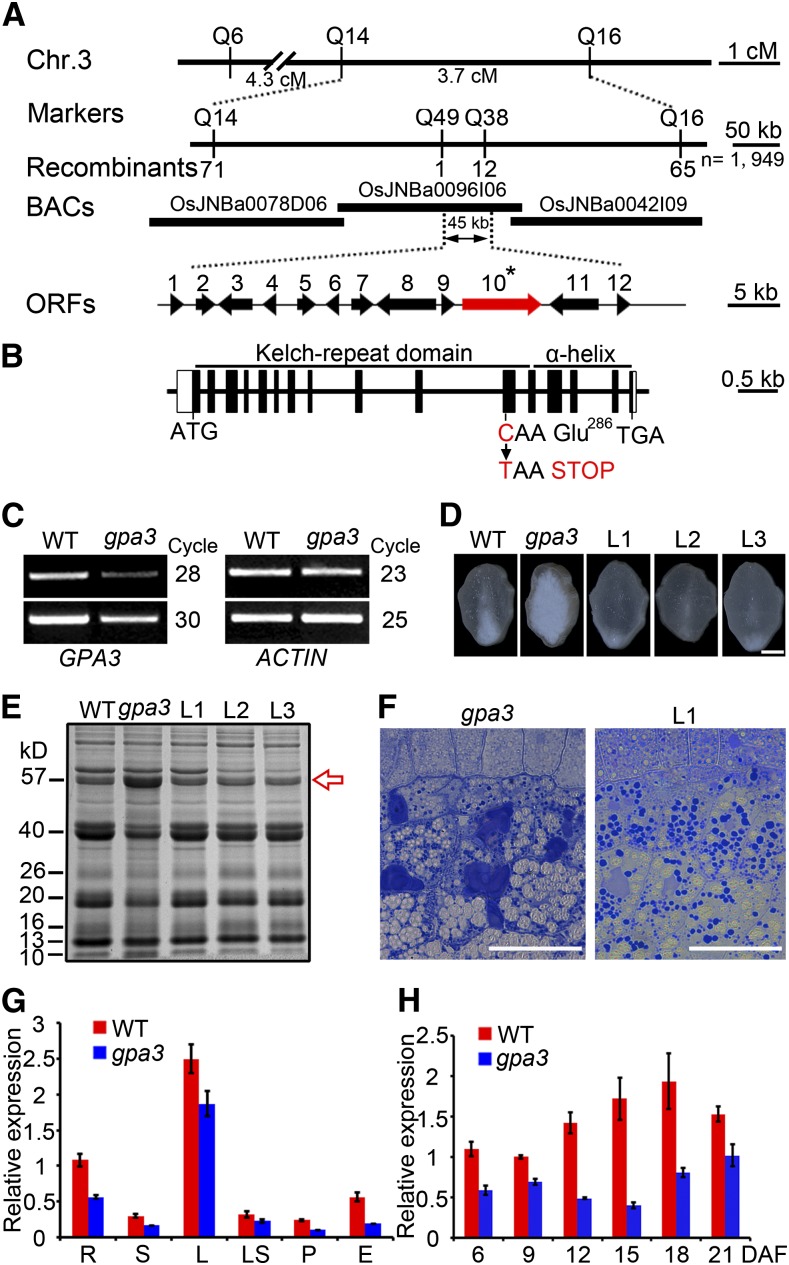
The gpa3 mutant was isolated from a 60Co-irradiated population of indica var Baifeng B. Genetic analysis revealed that the mutant phenotypes were inherited as a single nuclear recessive mutation (Supplemental Table 2). A map-based cloning approach was used to isolate the GPA3 gene. We first crossed the gpa3 mutant with the japonica var 02428 to generate an F2 mapping population. The gpa3 locus was initially placed in an 8.0-centimorgan interval between the insertion/deletion markers Q6 and Q16, on the long arm of rice chromosome 3, based on a dozen recessive gpa3 individuals. By screening a mapping population of 1949 F2 homozygous gpa3 background lines, the gpa3 locus was mapped to a 45-kb genomic region flanked by the markers Q49 (derived cleaved-amplified polymorphic sequence) and Q38 (simple sequence repeat) on the BAC clone OsJNBa0096I06 (Figure 4A). Annotation analysis of this 45-kb genomic DNA segment identified 12 putative open reading frames (Figure 4A). Sequence analysis revealed a single nucleotide substitution of cytimidine (C) to thymidine (T) in the 11th exon of Os03g0835800, which encodes a kelch-repeat protein. This point mutation caused Glu-286 replacement by a premature termination stop codon (Figure 4B). Further expression analysis showed that of the 12 candidate open reading frames, only the expression of Os03g0835800 (harboring the C-to-T single-base mutation) was downregulated in the gpa3 mutant (Figure 4C).
Figure 4.
Positional Cloning and Expression Analysis of GPA3.
(A) Fine mapping of the GPA3 locus. The molecular markers and the number of recombinants are shown. cM, centimorgan; ORF, open reading frame.
(B) Exon/intron structure and mutation site of GPA3. The GPA3 gene comprises 16 exons (closed boxes) and 15 introns (open boxes). ATG and TGA represent the start and stop codons, respectively.
(C) RT-PCR assay showing that expression of the candidate GPA3 gene is reduced in the gpa3 mutant compared with the wild type.
(D) to (F) A 10.4-kb wild-type genomic segment of GPA3 completely rescues the grain appearance (D), the storage protein composition pattern (E), and the storage protein sorting defects (F) of the gpa3 mutant. L1 to L3 denote grains from three different T1 transgenic lines. The red arrow indicates the 57-kD proglutelins. Bar in (D) = 1 mm; bars in (F) = 50 μm.
(G) Real-time RT-PCR assay showing that GPA3 is expressed in various organs examined, with highest accumulation in the leaf. R, root; S, stem; L, leaf; LS, leaf sheath; P, panicle; E, endosperm.
(H) Real-time RT-PCR assay showing that GPA3 is expressed throughout endosperm development.
α-Tubulin was used as an internal control in real-time RT-PCR. For each RNA sample, three technical replicates were performed. Representative results from two biological replicates are shown. Values are means ± sd.
The kelch motif is a type of ancient and evolutionarily widespread protein segment of 44 to 56 amino acids in length that was first identified in the Drosophila kelch ORF1 protein (Xue and Cooley, 1993). It occurs typically as four to seven repeats that form a tertiary structure called the β-propeller. To test whether the kelch-repeat gene corresponded to the candidate GPA3 gene, a 10.4-kb wild-type genomic fragment spanning the putative coding sequence and regulatory region of Os03g0835800 was introduced into the gpa3 homozygous mutant. Four positive GPA3 transgenic lines were obtained, and they all showed complete rescue of the mutant phenotypes, including the floury endosperm appearance, storage protein composition, and storage protein deposit pattern in the subaleurone cells (Figures 4D to 4F), thus confirming that Os03g0835800 indeed represents the GPA3 gene.
The GPA3 gene was predicted to encode a protein composed of 501 amino acid residues, with the N-terminal (amino acids 1 to 343) and C-terminal (amino acids 344 to 501) regions containing six kelch-repeat motifs and an α-helical domain, respectively. The former was predicted to fold into a β-propeller (Supplemental Figures 8A and 8B), while the latter showed no homology with any proteins of known function. The mutation in the gpa3 mutant completely removed the α-helical domain and led to an incomplete kelch-repeat domain that lost three β-strands of the first mixed kelch-repeat motif and the last β-strand of the sixth kelch-repeat motif. BLAST search showed that GPA3 was a single-copy gene in the rice genome. Homologous genes of GPA3 can be found in other plant genomes, such as maize (Zea mays), barley (Hordeum vulgare), cypress (Taxodium distichum), castor bean (Ricinus communis), poplar (Populus spp), soybean (Glycine max), grape (Vitis vinifera), and Arabidopsis, but not in animals or yeast (Supplemental Figure 8C).
To test whether mutation in gpa3 affected storage protein synthesis, representative storage protein members were examined temporally during endosperm development at the transcriptional level. The results indicated that expression of neither the glutelin members nor α-globulin and prolamin genes showed evident differences between the gpa3 mutant and the wild type (Supplemental Figure 9), suggesting that defects in the post-Golgi trafficking of storage proteins did not significantly affect storage protein gene expression. Although the phenotypes of the gpa3 mutant were restricted to endosperm, GPA3 expression was detected in all tissues examined, including root, stem, leaf, leaf sheath, panicle, and endosperm, with highest accumulation in the leaf (Figure 4G). During endosperm development, the expression of GPA3 was low at an early stage, then peaked at ∼18 DAF, and slowly decreased after 21 DAF (Figure 4H). Together, we concluded that GPA3 encodes a plant-specific kelch-repeat protein and that it is broadly expressed in multiple organs and tissues.
GPA3 Protein Is Localized to Post-Golgi Compartments
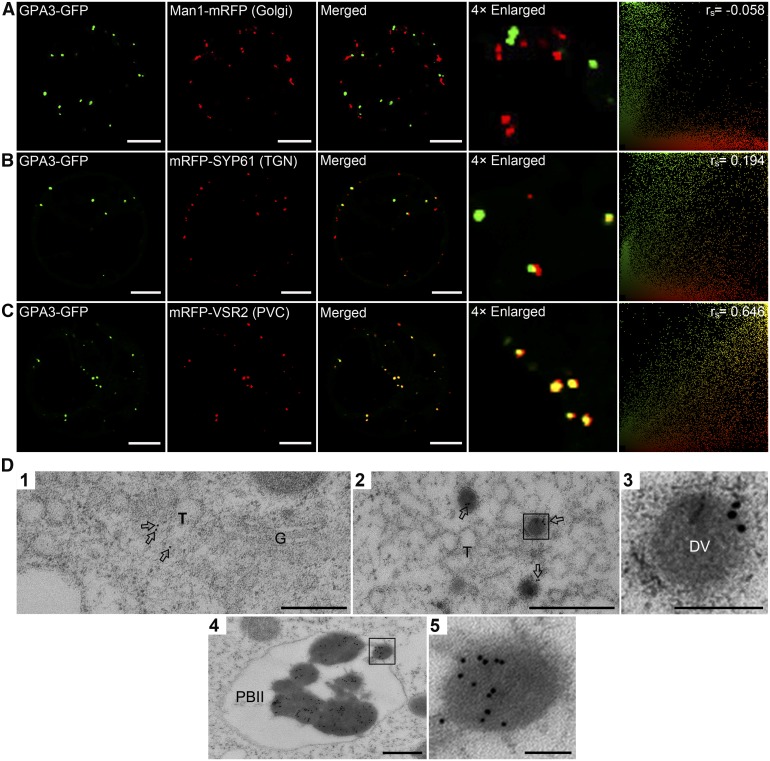
To determine the subcellular localization of the GPA3 protein, we transiently expressed P35S:GPA3-GFP in Arabidopsis protoplasts. As expected, GFP itself was distributed evenly in the cytoplasm and the nucleus, whereas the GPA3-GFP fusion protein was mainly localized to the cytoplasm and to punctate compartments in the cytosol (Supplemental Figures 10A and 10B). Moreover, we observed colocalization of various combinations of GPA3 fused with either fluorescent protein (GFP or mCherry) or a Flag tag at the N terminus or C terminus in Arabidopsis protoplasts (Supplemental Figures 10C to 10E). To determine the nature of these punctate compartments, we coexpressed GPA3-GFP and fluorescent marker proteins characteristic for the Golgi apparatus (Man1-mRFP; Tse et al., 2004), the TGN (mRFP-SYP61; Lam et al., 2007b), and the PVC (mRFP-VSR2; Miao et al., 2006). As shown in Figures 5A to 5C, the punctate compartments of GPA3-GFP were obviously distinct from the Golgi but partially overlapped with the TGN and PVC. Furthermore, correlation analysis using the Pearson-Spearman correlation (PSC) plugin for ImageJ (French et al., 2008) revealed strong correlation between GPA3-GFP and the PVC marker (rs = 0.646), but the correlation between GPA3-GFP and the TGN marker appeared to be weaker (rs = 0.194).
Figure 5.
Subcellular Localization of GPA3 in Arabidopsis Protoplasts and Developing Subaleurone Cells.
(A) to (C) Confocal microscopy images showing that GPA3-GFP is localized as punctate signals in the cytosol and its distribution is obviously distinct from the marker for Golgi (Man1-mRFP [A]) but partially overlaps with the markers for TGN (mRFP-SYP61 [B]) and PVC (mRFP-VSR2 [C]). PSC coefficients (rs) between GPA3-GFP and each marker are shown in the right panels. Bars = 10 μm.
(D) Immunoelectron microscopy localization of GPA3 in developing subaleurone cells. Ultrathin sections prepared using HPF subaleurone cells of transgenic plants expressing P35S:GPA3-GFP were labeled with anti-GFP antibodies, where gold particles (arrows) are found in the TGN (T; panel 1), DVs (panels 2 and 3), and PBIIs (panels 4 and 5). Panels 3 and 5 are the magnified images of selected areas in panels 2 and 4, respectively. G, Golgi. Bars in panels 1, 2, and 4 = 500 nm; bars in panels 3 and 5 = 100 nm.
To verify the subcellular localization of GPA3 in vivo, we generated P35S:GPA3-GFP transgenic rice lines, and they all exhibited the wild-type phenotypes (Supplemental Figure 11), indicating that GPA3-GFP represents a functional fusion protein. Fluorescence microscopy observation revealed that GPA3-GFP displayed a similar localization pattern in the transgenic root cells to that observed in the Arabidopsis protoplasts (Supplemental Figure 10G). Furthermore, immunoelectron microscopy analysis with anti-GFP antibodies revealed that GPA3-GFP was targeted to the TGN, DVs, and PBIIs (Figure 5D). Together, these data indicated that GPA3 is localized to various post-Golgi compartments irrespective of the cell types.
GPA3 Forms a Protein Complex with Rab5a and Its GEF VPS9a
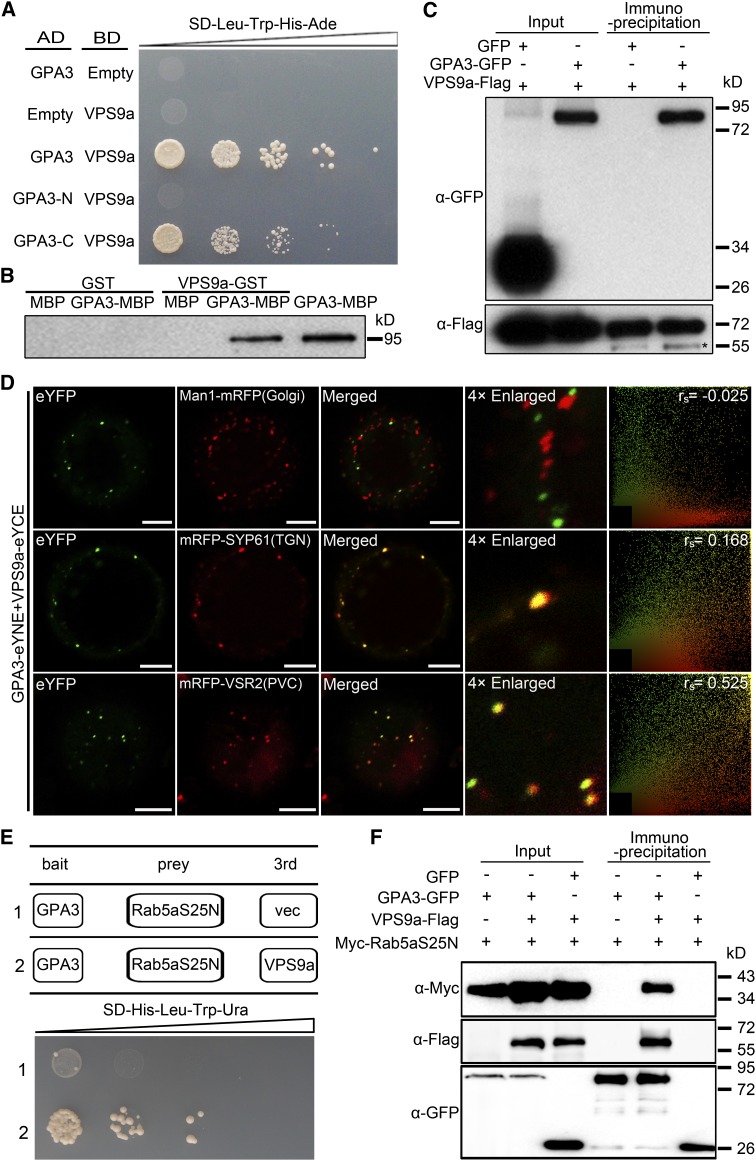
To dissect the molecular mechanisms of GPA3 in regulating the post-Golgi trafficking of storage proteins, we tested possible protein–protein interaction between GPA3 and a group of selected storage proteins using the yeast two-hybrid (Y2H) assay. The results indicated that neither GluA2 nor α-globulin interacted with GPA3 in the Y2H assay (Supplemental Figure 12), suggesting that GPA3 does not directly function in storage protein recognition. We then tested possible interaction between GPA3 and two post-Golgi trafficking regulators recently identified in our laboratory, GPA1/RAB5a (Wang et al., 2010) and its GEF VPS9a encoded by GPA2 (Liu et al., 2013). The Y2H assay showed that GPA3 indeed interacted with VPS9a but not with Rab5a or its GDP-bound derivative form Rab5aS25N (Figure 6A; Supplemental Figure 12). By contrast, we found that VPS9a physically interacted with Rab5aS25N but not with the wild-type Rab5a (Supplemental Figure 12). Furthermore, deletion analysis revealed that GPA3-N failed to interact with VPS9a but GPA3-C displayed a specific interaction with VPS9a (Figure 6A), indicating that the C-terminal 216 amino acids lost in the gpa3 mutant played an essential role in mediating the interaction between GPA3 and VPS9a. In addition, an in vitro pull-down assay confirmed that VPS9a-GST, but not GST itself, pulled down the GPA3-MBP fusion protein (Figure 6B). Furthermore, a coimmunoprecipitation (CoIP) experiment confirmed the in vivo interaction of GPA3 and VPS9a in leaf epidermal cells of Nicotiana benthamiana (Figure 6C).
Figure 6.
GPA3 Physically Interacts with Rice Rab5a via Its GEF VPS9a.
(A) Y2H assay showing that both the full-length GPA3 and GPA3-C (containing the C-terminal 216 amino acids of GPA3) interact with VPS9a, while GPA3-N (denotes the truncated gpa3 protein in the gpa3 mutant, which loses the C-terminal 216 amino acids of GPA3) fails to interact with VPS9a.
(B) In vitro GST pull-down assay showing that VPS9a-GST, but not GST itself, pulls down GPA3-MBP.
(C) In vivo CoIP assay showing that GPA3-GFP can be coimmunoprecipitated in the total leaf extract of N. benthamiana with anti-Flag agarose beads. The left two lanes show the protein immunoblots of an input control with anti-GFP and anti-Flag antibodies. The right two lanes show the results of CoIP. The asterisk indicates the IgG heavy chain.
(D) BiFC assay showing that GPA3 can interact with VPS9a in TGN and PVC in leaf epidermal cells of N. benthamiana. PSC coefficients (rs) between eYFP and each marker are shown in the right panel. Bars = 10 μm.
(E) Yeast three-hybrid assay showing that GPA3 interacts with Rab5aS25N in the presence of VPS9a. Ura, uracil.
(F) In vivo CoIP assay showing that Myc-Rab5aS25N can be coimmunoprecipitated in the presence of the VPS9a-Flag fusion in the total leaf extract of N. benthamiana with anti-GFP agarose beads.
To visualize the interaction between GPA3 and VPS9a, we performed a bimolecular fluorescence complementation (BiFC) assay using leaf epidermal cells of N. benthamiana. When GPA3-GFP and VPS9a-GFP were separately expressed in N. benthamiana leaves, GPA3-GFP displayed a similar localization pattern to that observed in the Arabidopsis protoplasts and stable transgenic rice plants (Supplemental Figure 13B), whereas the VPS9a-GFP fusion showed a typical cytosolic localization pattern (Supplemental Figure 13C). Strikingly, when GPA3-eYNE was coexpressed with VPS9a-eYCE, strong eYFP signal was detected in the TGN and PVC (Figure 6D; Supplemental Figure 14), suggesting that GPA3 may recruit VPS9a into these punctate compartments via direct protein–protein interaction. In support of this view, a yeast three-hybrid experiment using VPS9a as the bridge molecule showed that GPA3 interacted with Rab5aS25N (Rab5a’s GDP-fixed mutant form) in the presence of VPS9a (Figure 6E). Furthermore, an in vivo CoIP assay confirmed that Myc-Rab5aS25N can be coimmunoprecipitated by GPA3-GFP in the presence of the VPS9a-Flag fusion in the total leaf extract of N. benthamiana with anti-GFP agarose (Figure 6F).
GPA3 Functions Synergistically with Rab5a and VPS9a in Storage Protein Trafficking to the PSV in Rice Endosperm
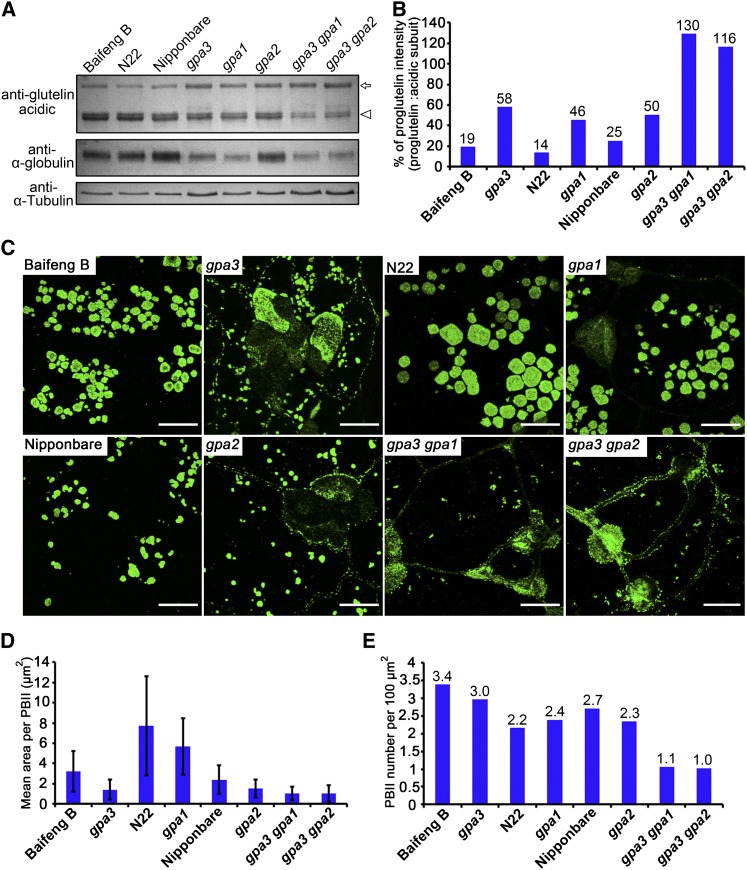
To further investigate the genetic interaction between GPA3, GPA1/Rab5a, and GPA2/VPS9a, we generated the gpa3 gpa1 and gpa3 gpa2 double mutants and determined their proglutelin intensity. Although gpa1/rab5a and gpa2/vps9a exhibited weaker proglutelin accumulation phenotypes than gpa3, either the gpa1/rab5a or gpa2/vps9a mutation significantly enhanced proglutelin accumulation in the gpa3 background (Figures 7A and 7B), suggesting that GPA3, GPA1, and GPA2 act synergistically in regulating proglutelin transport. Consistent with this notion, immunofluorescence experiments revealed a significant reduction in the number of PBIIs in the gpa3 gpa1 and gpa3 gpa2 double mutants, although the sizes of PBIIs were only slightly reduced in the double mutants in comparison with the gpa3 parental mutant (Figures 7C to 7E). Together, these results suggest that GPA3 functions synergistically with GPA1/Rab5a and GPA2/VPS9a to regulate proglutelin transport to PBIIs.
Figure 7.
GPA3, GPA1/Rab5a, and GPA2/VPS9a Act Synergistically to Regulate Storage Protein Trafficking in Rice Endosperm.
(A) Immunoblot analysis of storage proteins in dry seeds using antiglutelin acidic subunit (top) and anti-α-globulin (middle) antibodies showing increased accumulation of 57-kD proglutelins (arrow), accompanied by reduced accumulation of acidic subunits (arrowhead) and α-globulins in various single and double mutants, compared with their respective wild-type variety. Antitubulin antibodies were used as a loading control (bottom).
(B) Quantitative comparison of the proglutelin intensity (proglutelins/acidic subunits) in the wild type, single mutants, and double mutants.
(C) Morphology comparison of PBIIs from subaleurone cells among the wild type, single mutants, and double mutants. Secondary antibodies conjugated with Alexa fluor 488 (green) were used to visualize the reaction between glutelins and antiglutelin acidic subunit antibodies. Bars = 10 μm.
(D) and (E) Quantitative comparison of PBII area (D) and number (E) in the wild type, single mutants, and double mutants. Values are means ± sd (n > 500, Student’s t test).
DISCUSSION
gpa3 Is Defective in Post-Golgi Trafficking of Storage Proteins with Distinct Features
Mutants that abnormally accumulate storage protein precursors are ideal genetic resources for dissecting the molecular mechanisms underlying the PSV sorting pathways. Previous studies have identified eight 57H mutants in rice, including esp2 and glup1 to glup7 (Ueda et al., 2010). Compared with these previously reported rice 57H mutants, the gpa3 mutant displayed several distinct features. First, gpa3 accumulated normal levels of PDI1-1 and BiP1, whereas those mutants that have defects in ER export of storage proteins usually exhibited elevated levels of these chaperones, such as esp2/pdi1-1 (Takemoto et al., 2002), glup1, glup2, and glup7 (Ueda et al., 2010). Second, despite the fact that these mutants all accumulate increased levels of proglutelins, there are distinct differences in their subcellular characteristics due to lesions in different trafficking processes. For example, the esp2/pdi1-1 mutant forms small PBIs, W379/vpe1 develops round PBIIs (Wang et al., 2009), while gpa3 contains normal PBIs but small PBIIs. The observed gpa3 mutant phenotypes and subcellular defects are most reminiscent of the recently reported glup4/gpa1/rab5a and glup6/gpa2/vps9a mutants (Wang et al., 2010; Fukuda et al., 2011, 2013; Liu et al., 2013). However, the defects in gpa3 appeared to be more severe than those observed in glup4/gpa1/rab5a and glup6/gpa2/vps9a (Figure 7). Together, these data suggest that GPA3 represents a regulator of post-Golgi vesicular traffic essential for vacuolar protein sorting in rice endosperm.
Mistargeting of DVs to the PM Leads to PMB Formation
Previous morphological and cytological studies have generated a whole body of evidence showing that storage proteins are sorted via a vesicle-mediated trafficking pathway to PSVs in seed plants (Robinson et al., 2005; Vitale and Hinz, 2005). This notion is further supported by the identification of several Arabidopsis mutants defective in post-Golgi trafficking of storage protein to PSVs, such as atvsr1, vps29, vps35, kam2, and vps9a (Shimada et al., 2003, 2006; Tamura et al., 2007; Yamazaki et al., 2008; Ebine et al., 2011). A conspicuous phenotype for those Arabidopsis mutants is partial mistargeting of storage protein precursors to the extracellular space, as observed in the rice mutants defective in DV-mediated post-Golgi trafficking of storage proteins, such as gpa1/rab5a, gpa2/vps9a, and gpa3 (Wang et al., 2010; Liu et al., 2013). In addition, there were also reports that multivesicular bodies (MVBs) and PAC vesicles are possibly involved in storage protein sorting to the PSVs in dicotyledonous plants (Hara-Nishimura et al., 1998; Robinson et al., 1998; Otegui et al., 2006). However, the role of MVBs and PAC vesicles in storage protein sorting in monocotyledonous plants remains unclear.
In this study, our immunoelectron microscopy analysis with ultrathin sections prepared using the HPF method showed direct fusion of proglutelin-containing DVs with the PSV in both the wild type and the gpa3 mutant (Figures 3K to 3N), but we did not detect classical MVBs or PAC-like vesicles near the PSVs in either wild-type or gpa3 subaleurone cells, despite extensive efforts. However, we found that sorting of DVs to the PSV was severely blocked in the gpa3 mutant, in which numerous proglutelin-filled DVs were missorted to the PM, and via membrane fusion, releasing their contents to the apoplast to form PMBs (Figure 3). In addition, our light and confocal microscopy analyses showed that both glutelins and α-globulins were missorted to the PMB and cell periphery, resulting in smaller PBIIs and PSVs in the gpa3 mutant (Figure 2). Moreover, the PMBs contained not only storage proteins but also large amounts of callose, and the PMB was divided into different districts by other cell wall components, such as (1,3;1,4)-β-glucan and pectin (Supplemental Figure 6). These results collectively indicate that mistargeting of DV to the PM and releasing of their contents via DV-PM fusion leads to the formation of PMBs. Interestingly, a similar DV-PM fusion phenotype was also observed in wortmannin (a specific inhibitor of phosphoinositide 3-kinases)-treated mung bean (Vigna radiata) cotyledons (Wang et al., 2012), indicating that the target protein (phosphatidylinositol 3-kinase) of wortmannin may play a similar role to GPA3 in regulating post-Golgi trafficking of DVs to the PSV.
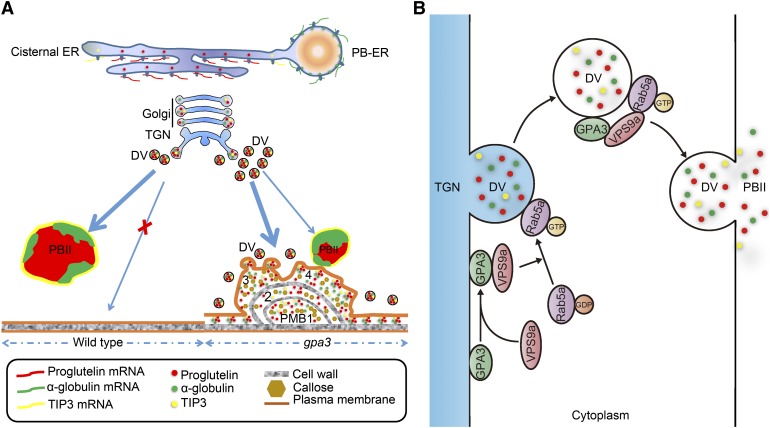
It is known that callose synthesis and deposition can be quickly induced by biotic stresses such as plasmolysis in plants (Chen and Kim, 2009). The uploading of mistargeted DV cargos into the apoplast in the gpa3 mutant may represent a physiologically similar process to plasmolysis. We speculate that such processes may trigger abnormal callose synthesis at the plasmodesmata, and continuous entry of DV cargos will further stimulate callose synthesis and its misdeposition, resulting in larger and complex PMBs. In addition, these processes may also stimulate the abnormal synthesis and deposition of other cell wall components, such as (1,3;1,4)-β-glucan and pectin, leading to the formation of compartmentalized PMB structures (Figure 8A).
Figure 8.
Working Model Depicting the Function of GPA3 in Vacuolar Protein Sorting in Rice Endosperm.
In the wild type, proglutelins and α-globulins are separately synthesized in the cisternal ER and protein body ER, and both of them converge in the Golgi apparatus, where they are packaged together into DVs, and then directly targeted to the PSV (PBII). The PSV marker protein TIP3 follows the same route to the PSV (PBII) with proglutelins. In the gpa3 mutant, directional targeting of DVs to the PSV (PBII) is severely blocked. Therefore, numerous DVs are mistargeted to the PM, and via membrane fusion, releasing their contents into the apoplast to form PMBs, in which massive amounts of cargo proteins, including proglutelins, α-globulins, and TIP3, as well as cell wall components abnormally accumulate (A). It can be speculated that GPA3 most likely functions as an adaptor protein to recruit VPS9a (Rab5a-GEF) and facilitate the interaction between Rab5a and VPS9a on the TGN and DVs, which is required for proper membrane fusion and/or directional targeting of DVs to the PSV (PBII [B]).
GPA3 Acts Synergistically with GLUP4/GPA1/Rab5a and GLUP6/GPA2/VPS9a in Regulating Post-Golgi Trafficking in the Rice Endosperm
In eukaryotic cells, Rabs are considered to be coordinators of vesicular trafficking, and they play an essential role in several distinct vesicular trafficking steps, including vesicle formation, mobility, docking, and fusion with the target membrane (Stenmark, 2009). It has been reported that loss of function of rice GLUP4/GPA1/Rab5a results in partial secretion of proglutelins and α-globulins to the extracellular space (Wang et al., 2010; Fukuda et al., 2011). Recently, rice VPS9a has also been identified to be the GEF of Rab5a, and its loss-of-function mutant gpa2/glup6 displays similar but more severe phenotypic defects than glup4/gpa1/rab5a (Fukuda et al., 2013; Liu et al., 2013). These observations support a role for Rab5a and VPS9a in the transport of proglutelins from the Golgi apparatus to PSV. The rice GPA1/Rab5a protein has been localized to the Golgi apparatus, the Golgi-derived DVs, and the PBIIs in developing rice endosperm (Fukuda et al., 2011).
In this work, we showed that GPA3 encodes a plant specific kelch-repeat protein. Homologous genes of GPA3 can be found in other plant genomes, such as maize, barley, cypress, castor bean, poplar, soybean, grape, and Arabidopsis, but not in animals or yeast (Supplemental Figure 8C), suggesting that GPA3 and its homologs may play a unique role specific in seed plants. Subcellular localization studies showed that the GPA3-GFP fusion protein is localized to the TGN and PVC in Arabidopsis protoplasts (Figures 5A to 5C). Furthermore, immunoelectron microscopy studies showed that GPA3 is localized to the TGN, Golgi-localized DVs, and PBIIs in the subaleurone cells of P35S:GPA3-GFP transgenic plants (Figure 5D). The similar subcellular defects of the gpa3 mutant with gpa1/rab5a and gpa2/vps9a mutants and their subcellular localization patterns suggest that GPA3 may act in the same genetic pathway to regulate DV-mediated post-Golgi trafficking in rice endosperm. This notion was also supported by the observed physical interaction between GPA3 and GLUP6/GPA2/VPS9a. Moreover, we found that in the presence of VPS9a, GPA3 can indirectly interact with Rab5aS25N (the GDP-fixed form of Rab5a; Figure 6). These observations suggest that GPA3 forms a regulatory protein complex with Rab5aS25N via VPS9a. More strikingly, our BiFC result indicated that GPA3 may recruit VPS9a into the TGN and PVCs via their physical interaction in N. benthamiana leaf cells (Figure 6). Furthermore, our genetic interactions indicated that GPA3, Rab5a, and VPS9a acted in a synergistic manner to regulate the DV-mediated post-Golgi trafficking of storage proteins to PSV (Figure 7). Based on these observations, we envisaged that GPA3 may serve as an adaptor protein that helps to recruit the Rab5a-VPS9a (GEF) complex and facilitate their physical interaction and mediate directional targeting and/or membrane fusion en route to the PSV (Figure 8B).
Consistent with the above proposition, it was previously reported that p40, a kelch-repeat protein in human, formed a complex with Rab9 to regulate the recycling of mannose-6-phosphate receptors from PVC to the TGN (Diaz et al., 1997), suggesting a role for kelch-repeat proteins in regulating retrograde post-Golgi trafficking. However, plants have no direct counterpart of Rab9; thus, it seems unlikely that GPA3 and its homologs can exert a similar function to that of p40 to drive the TGN docking of retromer-coated vesicles carrying VSRs in plants. In addition, VPS9 domain–containing GEFs in mammals and yeast usually contain an extra domain other than the VPS9 domain itself. These domains play essential roles in signal transduction, such as the CUE domain in yeast VPS9p, SH2, and the Ras-associated domain in human Rin1 (Carney et al., 2006). However, the plant VPS9 homologs lack such known domains but contain a C-terminal region of unknown function. Therefore, it is possible that GPA3 and its homologs may perform a similar regulatory role to that provided by the extra domain in mammals and yeast VPS9. The identification and functional characterization of GPA3 suggest that plants have evolved unique molecular machinery and mechanisms to regulate Rab5-based vesicular trafficking during the directional sorting of vacuolar storage proteins to PSV.
METHODS
Plant Materials and Growth Conditions
The gpa3 mutant was identified from a 60Co-irradiated M2 population of the indica rice (Oryza sativa) var Baifeng B. Reciprocal crosses between Baifeng B and gpa3 were used for genetic analysis. An F2 population was generated from a cross between the gpa3 mutant and the japonica var 02428. The rab5a/gpa1 mutant was isolated from a 60Co-irradiated mutant pool of the indica rice var N22 (Wang et al., 2010). The vps9a/gpa2 mutant was obtained from in vitro tissue culture of the japonica var Nipponbare (Liu et al., 2013). The gpa3 gpa1 and gpa3 gpa2 double mutants were isolated from the F2 populations of their respective crosses by phenotype observation and verified by genotyping with the primer pairs Q4041-F/Q4041-R, T5390-F/T5390-R, and Q25-F/Q25-R (Supplemental Table 3). All plants were grown in paddy fields during the normal growing seasons or in a greenhouse at the Chinese Academy of Agricultural Sciences, in Beijing.
Measurement of Starch, Lipid, and Total Protein Contents
Rice grains were processed using a dehuller and ground into fine flour with a miller. Amylose, lipid, and total protein contents were subsequently measured according to a previous report (Han et al., 2012).
Generation of Antibodies
Polyclonal antibodies against TIP3, the glutelin acidic subunits, and α-globulins were raised in rabbits as described elsewhere (Takahashi et al., 2004; Wang et al., 2010). Polyclonal antibodies against the 13-kD prolamins, GluA, GluB, and GluC, were produced in rabbits according to a previous report (Takagi et al., 2006). Monoclonal antibodies against the glutelin acidic subunits, glutelin basic subunits, α-globulins, BiP1, and PDI1-1 were raised in mouse (Abmart). Monoclonal antibodies against callose (1,3-β-glucan), (1,3;1,4)-β-glucan, and pectin (JIM7) were purchased from Biosupplies Australia and the University of Georgia. Antitubulin antibodies and anti-GFP antibodies were purchased from Sigma-Aldrich.
Protein Extraction from Rice Seeds and Immunoblot Analysis
Total protein extraction and immunoblot assay were performed as described previously (Wang et al., 2010). The intensities of protein gel blot bands were determined using Bio-Rad Quantity One imaging software.
Scanning Electron Microscopy and Transmission Electron Microscopy
Scanning electron microscopy, HPF, and subsequent immunogold labeling experiments were performed as described previously (Wang et al., 2010, 2012).
Light and Immunofluorescence Microscopy
Semithin (0.5 μm thickness) sections were stained in 1% (w/v) Coomassie Brilliant Blue R 250 in 25% (v/v) isopropanol and 10% (v/v) acetic acid, followed by destaining in 10% (v/v) acetic acid, and then subjected to light microscopy observation (Leica CTR 5000).
For immunofluorescence analysis, sections were blocked in TBST (10 mM Tris-HCl, 150 mM NaCl, and 0.05% Tween 20, pH 7.4) containing 3% BSA for 1 h at 37°C and then incubated with the primary antibodies (1:100 dilution) for 1 h at 37°C in TBST containing 1% BSA. After washing three times with TBST, the sections were incubated with the Alexa fluor 488 (green) or Alexa fluor 555 (red) conjugated secondary antibodies (Invitrogen) for 1 h at 37°C and then washed three times in TBST, followed by confocal imaging with a Leica TCS SP5 laser scanning confocal microscope. Double staining was performed in the same way as described above. For the preparation of thick sections (60 μm thickness), freshly harvested seeds (12 DAF) were cross-sectioned with a Leica VT 1200S vibratome, and then the sections were immediately immersed in MTSB (50 mM PIPES-KOH, pH 6.9, 10 mM EGTA, 10 mM MgSO4, 1% DMSO, and 0.1% Triton X-100) prior to fixation for 30 min in freshly prepared 4% paraformaldehyde in MTSB. For cell wall labeling, the sections were directly stained using Calcofluor white (Sigma-Aldrich) at room temperature for 5 min, followed by washing three times with PBS and then observed with a Leica stereo fluorescence microscope. For PBI staining, thick sections were incubated with 1 μM rhodamine (Sigma-Aldrich) for 30 min at 37°C. For immunofluorescence staining, sections (60 μm thickness) were further digested with 1% cellulase as described elsewhere (Satoh-Cruz et al., 2010). The remaining procedures were performed in the same way as described for immunofluorescence staining of thin sections.
Map-Based Cloning
To map the GPA3 locus, floury seeds were first selected from the F2 population of gpa3 and the japonica var 02428. The embryoless halves were ground into flour for total protein extraction, and the remaining halves were placed in ordered arrays using 96-well PCR plates. Seeds with floury and high-proglutelin phenotypes were identified as recessive gpa3 individuals, and the corresponding embryo-containing halves were used for germination for subsequent genomic DNA extraction. Newly developed molecular markers are shown in Supplemental Table 3.
Vector Construction and Rice Transformation
For genetic complementation, a 10.4-kb genomic fragment spanning the entire coding region, 2000-bp upstream sequence, and 500-bp downstream sequence was recombined into the pCAMBIA1305 vector (SmaI site) to generate the pCAMBIA1305-GPA3 construct using an Infusion cloning kit (Clontech). Subsequently, the construct was introduced into the Agrobacterium tumefaciens strain EHA105 and used to infect calli of recessive gpa3 lines isolated from a gpa3/02428 F3 population.
For subcellular localization of GPA3 in rice, full-length GPA3 was amplified from the wild-type cDNA and cloned into the pCAMBIA1305-GFP vector (generated by insertion of a SacI-SalI fragment containing the GFP expression cassette of pAN580 into the pCAMBIA1305 vector at the SacI-SalI sites) to generate the binary vector pCAMBIA1305-GPA3GFP (XbaI-BamHI sites). The empty plasmid pCAMBIA1305-GFP and pCAMBIA1305-GPA3GFP were individually introduced into the EHA105 strain and used to infect calli of the japonica var Kitaake and recessive gpa3 lines isolated from a gpa3/02428 F3 population, respectively. Transgenic roots were analyzed by a Leica TCS SP5 laser scanning confocal microscope.
For localization analysis of TIP3 protein, we made a translational fusion between the full-length TIP3 and GFP driven by TIP3’s native promoter (PTIP3:TIP3-GFP) according to a previous report (Onda et al., 2009). The construct was introduced into the EHA105 strain and used to infect calli of the indica var N22 and recessive gpa3 lines isolated from a gpa3/02428 F3 population. Immunolocalization of glutelins in transgenic lines expressing TIP3-GFP was performed as described above. The primer sets used here are listed in Supplemental Table 3.
RNA Extraction and RT-PCR
Total RNA was extracted from various tissues using a plant RNA extraction kit (Tiangen) according to the manufacturer’s instructions. RNA was reverse transcribed to cDNA using a reverse transcription kit (SuperScript II; TaKaRa). For RT-PCR analysis, specific primer pairs for GPA3, storage protein genes, and the internal reference gene Actin are shown in Supplemental Table 3. To ensure that amplification was in the logarithmic phase for PCR products, RT-PCR was performed to amplify gene transcripts with low cycles, and the PCR bands were separated on a 1% agarose gel containing ethidium bromide, followed by imaging with the Bio-Rad Gel Doc XR+ system.
Real-time PCR analysis was performed using the ABI7900HT real-time system with the SYBR Premix Ex Taq kit (TaKaRa). The real-time RT-PCR experiments were done with two independent groups of RNA samples. Each RNA sample was extracted from a pool of tissues collected from at least three individual plants. For each sample, quantification was made in triplicate. The specificity of the PCR amplification was ensured by detecting the melt curves at the end of each reaction. All primer sequences used for PCR are listed in Supplemental Table 3.
Sequence Analysis and Prediction of Protein Structure
Gene prediction was performed using the Rice Genome Automated Annotation System (http://ricegaas.dna.affrc.go.jp/). Sequence analysis of GPA3 was performed using the Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de/). Homologs of GPA3 were identified using the BLASTP search program of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). The secondary structure of GPA3 was predicted using the PSIPRED Protein Sequence Analysis Workbench (http://bioinf.cs.ucl.ac.uk/psipred/). Amino acid sequence alignment of kelch-repeat motifs of GPA3 was performed according to a previous report (Adams et al., 2000). The three-dimensional model of the GPA3 kelch domain was constructed on the basis of its similarity with the kelch domain of human KLHL12 (Protein Data Bank code 2vpj chain A) using the SWISS-MODEL program (Bordoli et al., 2009). Multiple sequence alignments were conducted with the ClustalW method using BioEdit software.
Transient Expression Analysis in Arabidopsis thaliana and Nicotiana benthamiana
Wild-type GPA3 was amplified and inserted into the pAN series of transient expression vectors (http://www.bio.utk.edu/cellbiol/markers/vectors.htm) to produce the GPA3-GFP (XbaI-BamHI sites), GFP-GPA3 (BglII-NotI sites), GPA3-Flag (XbaI-BamHI sites), and GPA3-mCherry (XbaI-BamHI sites) fusions. A 3×Flag tag was inserted into XbaI-BglII–digested pAN580 to give the pAN580-Flag vector for immunofluorescence staining studies. Both GPA3-GFP and GPA3-Flag were introduced into Arabidopsis mesophyll protoplasts by a polyethylene glycol–mediated transformation method (Park et al., 2005). Subsequent immunohistochemistry was performed as described previously (Park et al., 2005). The transient expression using protoplasts derived from Arabidopsis suspension cells was performed following a well-established protocol (Miao and Jiang, 2007). At least 15 independent protoplasts were analyzed with the PSC plugin for ImageJ to quantify the colocalization of GPA3-GFP and each marker (French et al., 2008).
For transient expression analysis in N. benthamiana, the binary vector pCAMBIA1305-GPA3GFP was constructed as described above, while the binary vectors pCAMBIA1305-VPS9aGFP and pEGAD-GFPRab5a were generated by cloning the coding sequences of VPS9a (XbaI-BamHI) and Rab5a (EcoRI-BamHI) into the pCAMBIA1305-GFP vector and pEGAD, respectively. Constructs were introduced into the Agrobacterium strain EHA105 and then used to infiltrate N. benthamiana leaves, as described previously (Waadt and Kudla, 2008). N. benthamiana protoplasts were isolated using the same method used with Arabidopsis (Park et al., 2005). Confocal imaging analysis was performed using a Leica TCS SP5 laser scanning confocal microscope.
Protein–Protein Interaction Analysis
For the Y2H assay, the coding sequences of GPA3, GluA2, α-globulin, Rab5a, and VPS9a were amplified using gene-specific primer sets. Full-length GPA3 was ligated into pGADT7 to form the GPA3-AD plasmid, while full-length rice GluA2, α-globulin, Rab5a, Rab5aS25N, and VPS9a were cloned into pGBKT7 to generate the GluA2-BD, α-globulin-BD, Rab5a-BD, Rab5aS25N-BD, and VPS9a-BD plasmids, respectively (see Supplemental Table 3 for primer sequences and cloning sites). Yeast transformation, screening for positive clones, and subsequent reporter gene assays were performed according to the manufacturer’s instructions (Clontech).
For in vitro pull-down assay, full-length GPA3 and VPS9a were inserted into pMAL-c2X and pGEX4T-1 vectors to generate the GPA3-MBP and VPS9a-GST plasmids, respectively (see Supplemental Table 3 for primer sets and cloning sites). Proteins including fusions and empty tags were expressed in Escherichia coli BL21 cells (TransGen) and were then purified using the corresponding resin (amylose resin for MBP purification [New England Biolabs] and GST binding resin for GST purification [Merck]). GST or VPS9a-GST coupled beads were used to capture MBP or GPA3-MBP. The pull-down analyses were performed as described (Miernyk and Thelen, 2008) and detected with horseradish peroxidase–conjugated anti-MBP monoclonal antibodies (1:2000; New England Biolabs).
For the BiFC assay, GPA3 and VPS9a were cloned into the binary BiFC vectors pSPYNE173 and pSPYCE (M) to produce the GPA3-eYNE and VPS9a-eYCE plasmids, respectively (see Supplemental Table 3 for primers and cloning sites; Waadt et al., 2008). Constructs including empty vectors were introduced into the Agrobacterium strain EHA105 and then used to infiltrate N. benthamiana leaves, as described above, followed by confocal imaging analysis at various time points after infiltration using a Leica TCS SP5 laser scanning confocal microscope. The excitation and detection wavelengths for eYFP were 514 and 527 nm, respectively. For coexpression analysis of the BiFC vector set (GPA3-eYNE and VPS9a-eYCE) and various Golgi or post-Golgi fluorescent marker vectors, protoplasts were isolated from infiltrated leaves of N. benthamiana as described above, followed by confocal imaging analysis using a Leica TCS SP5 laser scanning confocal microscope. At least 15 independent protoplasts were analyzed with the PSC plugin for ImageJ to quantify colocalization of the BiFC punctate signals and each marker (French et al., 2008).
For the CoIP assay, coding sequences of GPA3, VPS9a, and Rab5aS25N were introduced into the binary vectors pCAMBIA1305-GFP, pCAMBIA1300-221-Flag, and pCAMBIA1300-221-myc, respectively, and then transformed into the Agrobacterium strain EHA105. The subsequent steps were performed as reported (Liu et al., 2010). The anti-Flag agarose (Sigma-Aldrich) or anti-GFP agarose (MBL) was used to immunoprecipitate the VPS9a or GPA3 complex, and the complex was detected by anti-Flag (1:2000 dilution; Sigma-Aldrich), anti-GFP (1:2000 dilution; Sigma-Aldrich), and anti-myc (1:2000 dilution; Sigma-Aldrich) antibodies. The horseradish peroxidase–conjugated anti-mouse and anti-rabbit IgGs (1:5000 dilution; MBL) were used as the secondary antibodies. The immunoblot was detected with ECL Plus reagent (GE Healthcare), followed by visualization with x-ray autography.
For the yeast three-hybrid assay, the coding sequences of GPA3, Rab5aS25N, and VPS9a were amplified using gene-specific primer sets. Full-length Rab5aS25N was ligated into pGADT7 to form the Rab5aS25N-AD (EcoRI site) plasmid, and full-length GPA3 and VPS9a were cloned into pGBKT7 and pYES2 (Invitrogen) to generate the GPA3-BD (EcoRI site) and HA-tagged VPS9a (BamHI site) plasmids, respectively. These three plasmids were simultaneously transformed into the yeast Gold strain, positive clones were screened on three dropout plates (SD-Leu-Trp-Ura), and then positive clones were transferred to four dropout plates (SD-His-Leu-Trp) to grow.
Accession Numbers
Genes reported in this article can be found in the GenBank/EMBL databases under the following accession numbers: GPA3 (Os03g0835800, AK070689); Rab5a (Os12g0631100, AY029301); VPS9a (Os03g0262900, ABF95105); GluA2 (Os10g0400200, AK107314); α-globulin (Os05g0499100, AK242943); TIP3 (Os10g0492600, AK111931); Actin (Os03g0718100, AK100267); and Tubulin (Os03g0234200, AC103891).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Evaluation of the Specificity of the Storage Protein Monoclonal Antibodies Developed in This Work.
Supplemental Figure 2. Time-Course Analysis of Storage Proteins during Wild-Type and gpa3 Endosperm Development.
Supplemental Figure 3. The Morphology of ER and Golgi in Developing Wild-Type and gpa3 Subaleurone Cells.
Supplemental Figure 4. gpa3 Forms Smaller PSVs in Comparison with the Wild Type.
Supplemental Figure 5. Abnormal Deposition of Cell Wall–Like Materials in the gpa3 Subaleurone Cells.
Supplemental Figure 6. Abnormal Accumulation of Cell Wall Components in the PMBs.
Supplemental Figure 7. Accumulation of TIP3 and Glutelins to the Apoplast in gpa3 Subaleurone Cells.
Supplemental Figure 8. Structural and Sequence Analyses of GPA3.
Supplemental Figure 9. Expression Analysis of Representative Storage Protein Genes at Various Developmental Stages of Endosperm.
Supplemental Figure 10. Subcellular Localization of GPA3 in Arabidopsis Protoplasts and Complemented Transgenic Rice Root Cells.
Supplemental Figure 11. Complementation of gpa3 Mutant Phenotypes by 35S Promoter–Driven GPA3-GFP.
Supplemental Figure 12. Y2H Assay between GPA3 and Related Proteins.
Supplemental Figure 13. Subcellular Localization of GPA3 and VPS9a in Leaf Epidermal Cells of N. benthamiana.
Supplemental Figure 14. BiFC Assay of GPA3 and VPS9a in Leaf Epidermal Cells of N. benthamiana.
Supplemental Table 1. Characteristics of Wild-Type and gpa3 Grains
Supplemental Table 2. Segregation of Mutant Phenotypes in Reciprocal Crosses between the Wild Type and the gpa3 Mutant
Supplemental Table 3. List of Primer Pairs Used in This Study
Supplementary Material
Acknowledgments
We thank Liwen Jiang (Chinese University of Hong Kong) for kind help with protein subcellular localization experiments and Meiqin Liu (Beijing Forestry University) for assisting in operating the Leica SP5 laser scanning confocal microscope. This work was supported by grants from the National Natural Science Foundation of China (Grants 31330054 and 31371598), a project from the Ministry of Agriculture of China for Transgenic Research (Grants 2013ZX08001-006, 2009ZX08009-114B, and 2013ZX08009-003), and the Jiangsu 333 Project (Grant BRA2012126).
AUTHOR CONTRIBUTIONS
J.W., H.W., and Y.R. designed the research. Y.R., Y.-H.W., F.L., and X.H. screened the mutant materials. Y.D. and Y.-H.W. performed HPF. Y.D., F.L., and Y.R. performed immunogold labeling experiments. Y.R., K.Z., F.L., F.Z., Y.W., K.L., and Y.-H.W. performed all other experiments. L.G., W.M., X.Z., X.G., F.W., Z.C., J.-L.W., C.L., Q.L., L.J., C.W., and Y.B. provided technical assistance. J.W., H.W., and Y.R. analyzed the data and wrote the article.
Glossary
- LV
lytic vacuole
- PSV
protein storage vacuole
- ER
endoplasmic reticulum
- TGN
trans-Golgi network
- PVC
prevacuolar compartment
- VSR
vacuolar sorting determinant
- DV
dense vesicle
- GEF
guanine exchange factor
- PAC
precursor-accumulating
- PBI
protein body I
- PBII
protein body II
- PMB
paramural body
- DAF
days after flowering
- HPF
high-pressure frozen/freeze substitution
- PM
plasma membrane
- PSC
Pearson-Spearman correlation
- Y2H
yeast two-hybrid
- CoIP
coimmunoprecipitation
- BiFC
bimolecular fluorescence complementation
- MVB
multivesicular body
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Adams J., Kelso R., Cooley L. (2000). The kelch repeat superfamily of proteins: Propellers of cell function. Trends Cell Biol. 10: 17–24 [DOI] [PubMed] [Google Scholar]
- Bordoli L., Kiefer F., Arnold K., Benkert P., Battey J., Schwede T. (2009). Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 4: 1–13 [DOI] [PubMed] [Google Scholar]
- Carney D.S., Davies B.A., Horazdovsky B.F. (2006). Vps9 domain-containing proteins: Activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 16: 27–35 [DOI] [PubMed] [Google Scholar]
- Chen X.Y., Kim J.Y. (2009). Callose synthesis in higher plants. Plant Signal. Behav. 4: 489–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz E., Schimmöller F., Pfeffer S.R. (1997). A novel Rab9 effector required for endosome-to-TGN transport. J. Cell Biol. 138: 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K., Okatani Y., Uemura T., Goh T., Shoda K., Niihama M., Morita M.T., Spitzer C., Otegui M.S., Nakano A., Ueda T. (2008). A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell 20: 3006–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K., et al. (2011). A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 13: 853–859 [DOI] [PubMed] [Google Scholar]
- French A.P., Mills S., Swarup R., Bennett M.J., Pridmore T.P. (2008). Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat. Protoc. 3: 619–628 [DOI] [PubMed] [Google Scholar]
- Fukuda M., Satoh-Cruz M., Wen L., Crofts A.J., Sugino A., Washida H., Okita T.W., Ogawa M., Kawagoe Y., Maeshima M., Kumamaru T. (2011). The small GTPase Rab5a is essential for intracellular transport of proglutelin from the Golgi apparatus to the protein storage vacuole and endosomal membrane organization in developing rice endosperm. Plant Physiol. 157: 632–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., et al. (2013). A guanine nucleotide exchange factor for Rab5 proteins is essential for intracellular transport of the proglutelin from the Golgi apparatus to the protein storage vacuole in rice endosperm. Plant Physiol. 162: 663–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., et al. (2012). The failure to express a protein disulphide isomerase-like protein results in a floury endosperm and an endoplasmic reticulum stress response in rice. J. Exp. Bot. 63: 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura I., Shimada T., Hatano K., Takeuchi Y., Nishimura M. (1998). Transport of storage proteins to protein storage vacuoles is mediated by large precursor-accumulating vesicles. Plant Cell 10: 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmer S., Movafeghi A., Robinson D.G., Hinz G. (2001). Vacuolar storage proteins are sorted in the cis-cisternae of the pea cotyledon Golgi apparatus. J. Cell Biol. 152: 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz G., Hillmer S., Baumer M., Hohl I.I. (1999). Vacuolar storage proteins and the putative vacuolar sorting receptor BP-80 exit the Golgi apparatus of developing pea cotyledons in different transport vesicles. Plant Cell 11: 1509–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl I., Robinson D.G., Chrispeels M.J., Hinz G. (1996). Transport of storage proteins to the vacuole is mediated by vesicles without a clathrin coat. J. Cell Sci. 109: 2539–2550 [DOI] [PubMed] [Google Scholar]
- Hwang I. (2008). Sorting and anterograde trafficking at the Golgi apparatus. Plant Physiol. 148: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamaru T., Uemura Y., Inoue Y., Takemoto Y., Siddiqui S.U., Ogawa M., Hara-Nishimura I., Satoh H. (2010). Vacuolar processing enzyme plays an essential role in the crystalline structure of glutelin in rice seed. Plant Cell Physiol. 51: 38–46 [DOI] [PubMed] [Google Scholar]
- Lam S.K., Tse Y.C., Robinson D.G., Jiang L. (2007a). Tracking down the elusive early endosome. Trends Plant Sci. 12: 497–505 [DOI] [PubMed] [Google Scholar]
- Lam S.K., Siu C.L., Hillmer S., Jang S., An G., Robinson D.G., Jiang L. (2007b). Rice SCAMP1 defines clathrin-coated, trans-Golgi-located tubular-vesicular structures as an early endosome in tobacco BY-2 cells. Plant Cell 19: 296–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wu Y., Zhang D.Z., Gillikin J.W., Boston R.S., Franceschi V.R., Okita T.W. (1993). Rice prolamine protein body biogenesis: A BiP-mediated process. Science 262: 1054–1056 [DOI] [PubMed] [Google Scholar]
- Liu F., et al. (2013). OsVPS9A functions cooperatively with OsRAB5A to regulate post-Golgi dense vesicle-mediated storage protein trafficking to the protein storage vacuole in rice endosperm cells. Mol. Plant 6: 1918–1932 [DOI] [PubMed] [Google Scholar]
- Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., Xie Q. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61: 893–903 [DOI] [PubMed] [Google Scholar]
- Miao Y., Jiang L. (2007). Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat. Protoc. 2: 2348–2353 [DOI] [PubMed] [Google Scholar]
- Miao Y., Yan P.K., Kim H., Hwang I., Jiang L. (2006). Localization of green fluorescent protein fusions with the seven Arabidopsis vacuolar sorting receptors to prevacuolar compartments in tobacco BY-2 cells. Plant Physiol. 142: 945–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk J.A., Thelen J.J. (2008). Biochemical approaches for discovering protein-protein interactions. Plant J. 53: 597–609 [DOI] [PubMed] [Google Scholar]
- Onda Y., Kumamaru T., Kawagoe Y. (2009). ER membrane-localized oxidoreductase Ero1 is required for disulfide bond formation in the rice endosperm. Proc. Natl. Acad. Sci. USA 106: 14156–14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui M.S., Herder R., Schulze J., Jung R., Staehelin L.A. (2006). The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies. Plant Cell 18: 2567–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M., Lee D., Lee G.J., Hwang I. (2005). AtRMR1 functions as a cargo receptor for protein trafficking to the protein storage vacuole. J. Cell Biol. 170: 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcher M., Santambrogio M., Thazar N., Thierry A.M., Fobis-Loisy I., Miège C., Jaillais Y., Gaude T. (2010). Analyses of sorting nexins reveal distinct retromer-subcomplex functions in development and protein sorting in Arabidopsis thaliana. Plant Cell 22: 3980–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.G., Jiang L., Schumacher K. (2008). The endosomal system of plants: Charting new and familiar territories. Plant Physiol. 147: 1482–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.G., Oliviusson P., Hinz G. (2005). Protein sorting to the storage vacuoles of plants: A critical appraisal. Traffic 6: 615–625 [DOI] [PubMed] [Google Scholar]
- Robinson D.G., Baumer M., Hinz G., Hohl I. (1998). Vesicle transfer of storage proteins to the vacuole: The role of the Golgi apparatus and multivesicular bodies. J. Plant Physiol. 152: 659–667 [Google Scholar]
- Rogers J.C. (2008). Multiple vacuoles in plant cells. Plant Physiol. 146: 1024–1025, author reply 1026–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Cruz M., Crofts A.J., Takemoto-Kuno Y., Sugino A., Washida H., Crofts N., Okita T.W., Ogawa M., Satoh H., Kumamaru T. (2010). Protein disulfide isomerase like 1-1 participates in the maturation of proglutelin within the endoplasmic reticulum in rice endosperm. Plant Cell Physiol. 51: 1581–1593 [DOI] [PubMed] [Google Scholar]
- Shen Y., Wang J., Ding Y., Lo S.W., Gouzerh G., Neuhaus J.M., Jiang L. (2011). The rice RMR1 associates with a distinct prevacuolar compartment for the protein storage vacuole pathway. Mol. Plant 4: 854–868 [DOI] [PubMed] [Google Scholar]
- Shimada T., Fuji K., Tamura K., Kondo M., Nishimura M., Hara-Nishimura I. (2003). Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100: 16095–16100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Koumoto Y., Li L., Yamazaki M., Kondo M., Nishimura M., Hara-Nishimura I. (2006). AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol. 47: 1187–1194 [DOI] [PubMed] [Google Scholar]
- Stenmark H. (2009). Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10: 513–525 [DOI] [PubMed] [Google Scholar]
- Takagi H., Hirose S., Yasuda H., Takaiwa F. (2006). Biochemical safety evaluation of transgenic rice seeds expressing T cell epitopes of Japanese cedar pollen allergens. J. Agric. Food Chem. 54: 9901–9905 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Rai M., Kitagawa T., Morita S., Masumura T., Tanaka K. (2004). Differential localization of tonoplast intrinsic proteins on the membrane of protein body type II and aleurone grain in rice seeds. Biosci. Biotechnol. Biochem. 68: 1728–1736 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Saito Y., Kitagawa T., Morita S., Masumura T., Tanaka K. (2005). A novel vesicle derived directly from endoplasmic reticulum is involved in the transport of vacuolar storage proteins in rice endosperm. Plant Cell Physiol. 46: 245–249 [DOI] [PubMed] [Google Scholar]
- Takemoto Y., Coughlan S.J., Okita T.W., Satoh H., Ogawa M., Kumamaru T. (2002). The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol. 128: 1212–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Takahashi H., Kunieda T., Fuji K., Shimada T., Hara-Nishimura I. (2007). Arabidopsis KAM2/GRV2 is required for proper endosome formation and functions in vacuolar sorting and determination of the embryo growth axis. Plant Cell 19: 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse Y.C., Mo B., Hillmer S., Zhao M., Lo S.W., Robinson D.G., Jiang L. (2004). Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16: 672–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y., Satoh-Cruz M., Matsusaka H., Takemoto-Kuno Y., Fukuda M., Okita T.W., Ogawa M., Satoh H., Kumamaru T. (2010). Gene-gene interactions between mutants that accumulate abnormally high amounts of proglutelin in rice seed. Breed. Sci. 60: 568–574 [Google Scholar]
- Vitale A., Hinz G. (2005). Sorting of proteins to storage vacuoles: How many mechanisms? Trends Plant Sci. 10: 316–323 [DOI] [PubMed] [Google Scholar]
- Waadt R., Kudla J. (2008). In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). CSH Protoc. 2008: t4995. [DOI] [PubMed] [Google Scholar]
- Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Wang J., Tse Y.C., Hinz G., Robinson D.G., Jiang L. (2012). Storage globulins pass through the Golgi apparatus and multivesicular bodies in the absence of dense vesicle formation during early stages of cotyledon development in mung bean. J. Exp. Bot. 63: 1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhu S., Liu S., Jiang L., Chen L., Ren Y., Han X., Liu F., Ji S., Liu X., Wan J. (2009). The vacuolar processing enzyme OsVPE1 is required for efficient glutelin processing in rice. Plant J. 58: 606–617 [DOI] [PubMed] [Google Scholar]
- Wang Y., et al. (2010). OsRab5a regulates endomembrane organization and storage protein trafficking in rice endosperm cells. Plant J. 64: 812–824 [DOI] [PubMed] [Google Scholar]
- Washida H., Sugino A., Doroshenk K.A., Satoh-Cruz M., Nagamine A., Katsube-Tanaka T., Ogawa M., Kumamaru T., Satoh H., Okita T.W. (2012). RNA targeting to a specific ER sub-domain is required for efficient transport and packaging of α-globulins to the protein storage vacuole in developing rice endosperm. Plant J. 70: 471–479 [DOI] [PubMed] [Google Scholar]
- Xue F., Cooley L. (1993). kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell 72: 681–693 [DOI] [PubMed] [Google Scholar]
- Yamazaki M., Shimada T., Takahashi H., Tamura K., Kondo M., Nishimura M., Hara-Nishimura I. (2008). Arabidopsis VPS35, a retromer component, is required for vacuolar protein sorting and involved in plant growth and leaf senescence. Plant Cell Physiol. 49: 142–156 [DOI] [PubMed] [Google Scholar]
- Zouhar J., Rojo E., Bassham D.C. (2009). AtVPS45 is a positive regulator of the SYP41/SYP61/VTI12 SNARE complex involved in trafficking of vacuolar cargo. Plant Physiol. 149: 1668–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.