Elucidating the mechanism of a plant’s responses to phosphate deficiency will help to breed plants that can utilize environmental phosphate more efficiently. Here, we show that the level of the phosphate transporter PT2 is regulated by NITROGEN LIMITATION ADAPTATION and PHOSPHATE2 in a phosphate-responsive manner.
Abstract
The NITROGEN LIMITATION ADAPTION (NLA) gene was initially shown to function in nitrogen limitation responses; however, recent work shows that the nla mutant hyperaccumulates Pi, phenocopying the Pi signaling mutant pho2. PHO2 encodes a putative E2 conjugase, UBC24. Here, we show that NLA is an E3 ligase that specifically requires UBC24 for polyubiquitination in Arabidopsis thaliana. Among five members of the Pht1 Pi-transporter family tested, NLA associates only with PT2 (Pht1;4). The NLA-UBC24 pair mediates polyubiquitination of PT2 but not PT1. Posttranslational decay of PT2 at high Pi is blocked in pho2 and inhibited by MG132, indicating the requirement of UBC24 and 26S proteasomes. Consistent with NLA/UBC24 function, induced NLA expression causes a UBC24-dependent decrease in PT2 levels. Confocal microscopy of fusion proteins revealed an NLA/PT2 interaction at the plasma membrane. Collectively, these results show that under Pi-replete conditions, NLA and UBC24 target the PT2 transporter for destruction. During the Pi deprivation response, NLA and PHO2 transcripts are cleaved by miR399 and miR827, respectively, and our results suggest that this downregulation relieves the posttranslational repression of PT2, allowing it to accumulate and participate in Pi uptake. Our work provides additional molecular details describing Pi signaling/homeostasis regulation by identifying NLA and UBC24 as partners and PT2 as one of their downstream targets.
INTRODUCTION
Pi is one of the six macronutrients required for normal plant development and reproductive growth. It is one of the three major constituents of synthetic fertilizers used world-wide to boost crop yield. Although fertilizer application has clearly increased crop productivity by 30 to 50%, its manufacturing is an energy-intensive process. Moreover, overapplication of fertilizers leads to runoff and pollution of waterways, causing a host of environmental problems. One possible approach to reducing the application of fertilizers in agriculture is to optimize their efficient use by crop plants (Holford, 1997; Raghothama, 1999; Yang and Finnegan, 2010; Secco et al., 2012).
Efforts were initiated more than a decade ago to investigate the molecular responses of Pi limitation in an attempt to optimize Pi use efficiency by plants. Genetic screens in Arabidopsis thaliana yielded several genes that play key roles in Pi acquisition and transport and also in Pi starvation responses. Prominent among these genes are PHO1 and PHO2. PHO1 encodes a membrane-bound protein with SPX and EXS domains, which is implicated in Pi loading into xylem vessels (Poirier et al., 1991; Hamburger et al., 2002; Wang et al., 2004). Based on sequence homology, PHO2 is likely to be an E2 ubiquitin conjugase named UBC24; however, neither its enzyme activity nor its cognate E3 partner has been confirmed. Genetic and physiological experiments have provided evidence that PHO2 is an important central negative regulator of the Pi response pathway. Mutant pho2 plants appear to mount a constitutive Pi response, with shoots accumulating high Pi levels under normal conditions. PHO2 transcription does not appear to be regulated by Pi, but its mRNA is targeted for cleavage by miR399 in a Pi-regulated manner. Upon Pi deprivation, miR399 levels are elevated, resulting in a downregulation of PHO2 mRNA levels (Aung et al., 2006; Bari et al., 2006; Liu et al., 2010; Rouached et al., 2010; Liu et al., 2012). If PHO2 is indeed an E2 conjugase, it is reasonable to assume that this enzyme may be involved in destabilizing a number of downstream proteins via ubiquitin-mediated proteolysis. Therefore, reduction in PHO2 expression should relieve proteolysis of downstream targets, allowing the latter to accumulate to sufficiently high levels to initiate Pi starvation responses.
Although the pho2 mutant was isolated and characterized almost two decades ago (Delhaize and Randall, 1995), little is known about the molecular mode of action of PHO2, and its downstream targets are unclear. Liu et al. (2012) hypothesized that PHO2 might function as a chimeric E2-E3 enzyme that directly degrades target proteins. On the other hand, a typical ubiquitin E2 conjugase would bind to a partner E3 ligase, but not directly to a substrate, with the E3 ligase conferring substrate specificity. Identification of the cognate E3 ligase would provide the opportunity to advance our knowledge of the interacting target proteins and clarify how the Pi response circuit is negatively regulated by PHO2. Among previously identified Pi-responsive genes, none encodes an E3 ligase.
As a first step toward identifying the cognate E3 for PHO2, we conducted a literature survey on molecular responses to nutritional stresses in Arabidopsis. We found that a central positive regulator for nitrogen adaptability in Arabidopsis is a RING protein called NITROGEN LIMITATION ADAPTION (NLA). Mutant plants deficient in NLA displayed earlier onset of senescence compared with wild-type plants under nitrogen-limiting conditions (Peng et al., 2007b). Two observations provide circumstantial evidence that the NLA gene may be related to Pi responses. First, nla mutant plants behave similarly to wild-type plants when grown under low Pi or challenged with abiotic stresses, but under normal conditions, nla shoots accumulate high Pi levels (Kant et al., 2011). The phenotype of high shoot Pi levels in the nla mutant mimics that of the pho2 mutant, strongly suggesting that the two genes may function in the same pathway. Second, Pi deprivation elevates the expression level of miR827, which targets the 5′-untranslated region of NLA mRNA for cleavage and downregulation (Kant et al., 2011). This scenario is reminiscent of the regulation of PHO2 mRNA by Pi-inducible miR399 and suggests that NLA may be somehow involved in Pi starvation responses.
Several groups of transporters are implicated in Pi uptake from soil, its subsequent partition among intracellular endomembrane compartments, and Pi distribution between cells/organs. A major Pi transporter family is the Pht1 family, which has nine members in Arabidopsis (Rausch and Bucher, 2002). Given the large number of Pht1 family members, it is not surprising that these transporters may have overlapping functions. The best-studied family members are Pht1;1 and Pht1;4, both of which are high-affinity transporters involved in Pi acquisition from soil. Analysis of single and double mutants shows that Pht1;1 and Pht1;4 are responsible for ∼50% of Pi uptake under Pi-limiting conditions, but other transporters, such as Pht1;2 and Pht1;3, also play a role (Misson et al., 2004; Shin et al., 2004). More recently, Remy et al. (2012) provided evidence that Pht1;8 and Pht1;9 are also involved in Pi uptake. Pht1;5, on the other hand, plays a key role in Pi partition between roots and shoots (Smith et al., 2011; Remy et al., 2012). The functions of the remaining two Pht1 family members (Pht1;6 and Pht1;7) are presently unknown.
Little is known about how Pi deprivation can lead to increased Pi uptake from soil; however, transcriptional regulation of the Pht1 gene family members is an important contributing factor. Under Pi starvation conditions, Pht1;1 (hereafter called PT1), Pht1;2, Pht1;3, and Pht1.4 (hereafter called PT2) transcript levels are induced, and this induction is likely mediated by the myb transcription factor PHR1 (Nilsson et al., 2007). Consistent with this hypothesis, the imperfect palindromic sequence (GNATATNC), which serves as a binding site for PHR1 dimers, is found on the promoters of these four Pht1 genes. The elevated transcript levels of the four Pht1 family members, and presumably a corresponding increase in their protein levels, can explain in part the increased Pi uptake. On the other hand, the hyperaccumulation of Pi in the pho2 mutant under Pi-replete conditions suggests possible regulation of Pi transporters at the posttranslational level as well. It is unclear whether the regulation by UBC24 is direct or indirect.
In this work, we show that NLA can act as an E3 ubiquitin ligase for PHO2 (hereafter termed UBC24 when referring to its molecular function) in the regulation of Pi homeostasis in Arabidopsis. Consistent with their shared functions in mediating ubiquitination, NLA interacts with UBC24 in vitro and in vivo, and the E2/E3 pair together polyubiquitinates the high-affinity Pi transporter PT2 and targets it for destruction. In addition to explaining the mechanism of Pi homeostasis regulation by PHO2, our work uncovers an intimate crosstalk between Pi and nitrogen response pathways at the level of regulated proteolysis.
RESULTS
NLA Homodimerizes via Its RING-Containing Region
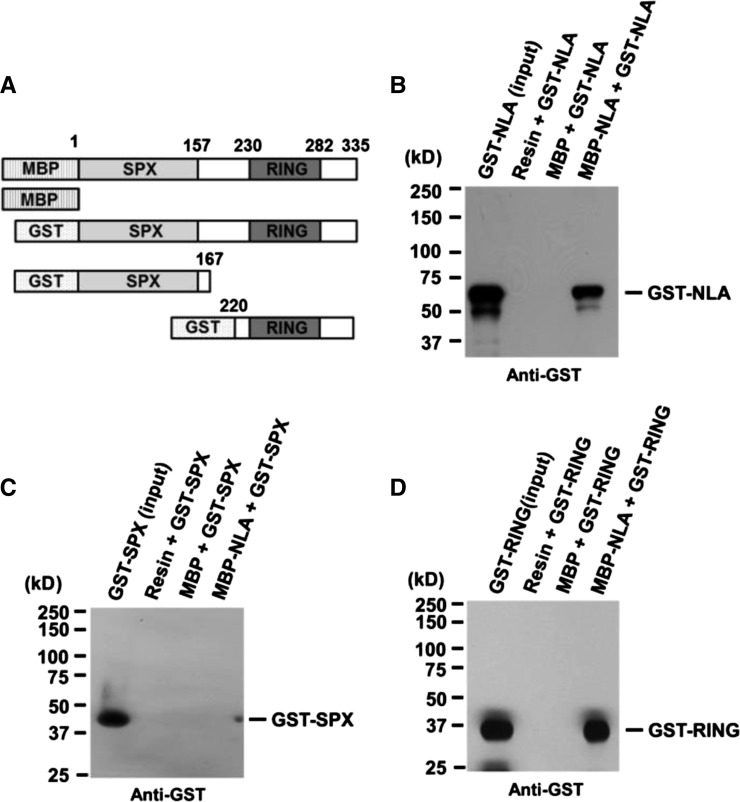
NLA is a RING motif protein belonging to the RING HCa type (Stone et al., 2005). Among the RING motif proteins, NLA is unusual in that its RING domain is located at the C terminus (Figure 1A). All RING motif proteins reported to date form dimers, and this dimerization is required to activate E3 ligase activity (Xie et al., 2002; Seo et al., 2003). To assess if NLA also dimerizes, we tested the ability of purified NLA to self-associate, using full-length protein, its N-terminal fragment (SPX; amino acids 1 to 167), or its C-terminal fragment (RING; amino acids 220 to 335) (Figure 1). We found that full-length NLA was able to interact with the C-terminal but not the N-terminal fragment. These results indicate that NLA can homodimerize via its C-terminal region, which includes the RING motif.
Figure 1.
NLA Forms Homodimers via Its RING Finger Domain.
(A) Schematic diagrams of full-length NLA and its deletion derivatives used to test for NLA homodimerization. MBP or MBP-NLA was used as a bait protein and GST-NLA (amino acids 1 to 335), GST-SPX (amino acids 1 to 270), or GST-RING (amino acids 220 to 335) was used as a prey protein.
(B) In vitro pull-down assay of MBP-NLA and GST-NLA. Purified GST-NLA protein as prey was mixed with MBP or MBP-NLA as bait.
(C) In vitro pull-down assay of MBP-NLA and GST-SPX.
(D) In vitro pull-down assay of MBP-NLA and GST-RING. For (B) to (D), the reaction mix contained 1 μg each of prey and bait proteins. Pulled-down protein was analyzed by protein gel blot using anti-GST antibody.
PHO2 Is a Cognate E2 for the NLA E3 Ligase
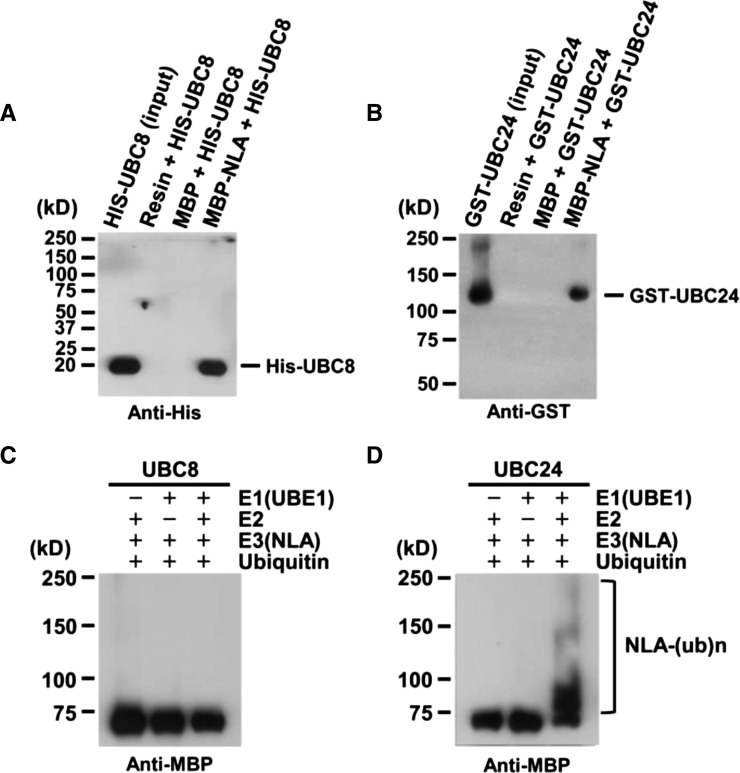
Given that many RING domain proteins function as E3 ligases, we deduced that NLA would also be functioning as an E3; however, in their activity screen of a large number of Arabidopsis RING family proteins, Stone et al. (2005) failed to detect any E3 activity associated with NLA purified from Escherichia coli extracts. As with other E3 ligases, it is possible that NLA may require a specific E2 for autoubiquitination. Peng et al. (2007b) have previously reported that NLA interacts with UBC8 in yeast (Saccharomyces cerevisiae) as well as in plant cells. Using in vitro pull-down assays, we confirmed the interaction between purified NLA and Arabidopsis UBC8 (Figure 2A). Nevertheless, this interaction did not promote autoubiquitination of NLA in vitro (Figure 2C).
Figure 2.
UBC24 (PHO2) Is a Specific E2 Conjugase for NLA.
(A) In vitro pull-down assay of His-UBC8 with MBP-NLA full-length. His-UBC8 as prey was mixed with MBP or MBP-NLA as bait. His-UBC8 was analyzed by protein gel blot using anti-His antibody.
(B) In vitro pull-down assay of GST-UBC24 with MBP-NLA full-length. GST-UBC24 as prey was mixed with MBP or MBP-NLA as bait. GST-UBC24 was analyzed by protein gel blot using anti-GST antibody.
(C) UBC8 does not support NLA self-ubiquitination.
(D) UBC24 supports NLA autoubiquitination.
For (C) and (D), the reaction mix contained 25 ng UBE1 (rabbit E1), 100 ng UBC8 or UBC24 (E2), 500 ng MBP-NLA (E3), and 8 μg His-Ubiquitin. After the reaction, polyubiquitinated NLA was analyzed by protein gel blot using anti-MBP antibody.
The function of NLA was first characterized through analysis of its deficient Arabidopsis mutant nla, which displayed earlier onset of senescence than the wild type in nitrogen-deficient medium (Peng et al., 2007b). Further analysis showed that nla mutant plants accumulated more Pi compared with wild-type plants (Kant et al., 2011). This overaccumulation of Pi phenocopies the pho2 mutant. Given that PHO2 encodes UBC24, an E2 enzyme (Aung et al., 2006; Bari et al., 2006; Liu et al., 2010; Rouached et al., 2010; Liu et al., 2012), we hypothesized that NLA may impose its function on the Pi signaling pathway as an E3 ligase and use UBC24 as the cognate E2. To test this hypothesis, we used yeast two-hybrid assays to examine possible interaction between NLA and UBC24. UBC8, which has been previously reported to interact with NLA (Peng et al., 2007b), was used as a positive control. Supplemental Figure 1 shows that NLA was able to interact not only with UBC8 but also with UBC24. These interactions in yeast were confirmed by in vitro pull-down assays (Figure 2B).
Next, we tested whether UBC24 could promote NLA autoubiquitination. We performed in vitro ubiquitination assays using purified UBC24. In contrast with UBC8, UBC24 was very active in mediating NLA autoubiquitination in vitro (Figure 2D). These results indicate that binding of an E2 enzyme to an E3 ligase does not necessarily imply that the E2 enzyme is able to support E3-mediated ubiquitination.
PT2 Is a Substrate of NLA and UBC24 in Vitro
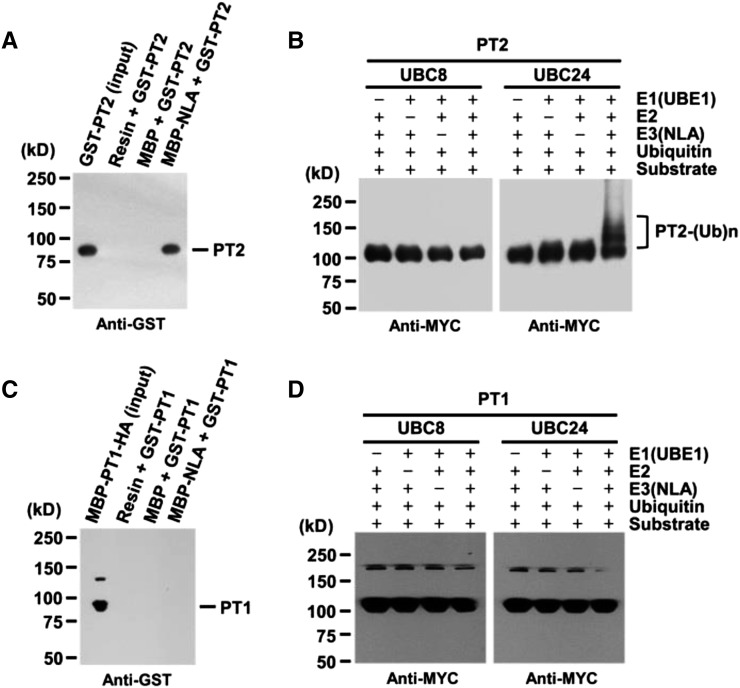
The observation that NLA interacted with UBC24 in vitro and the fact that, under Pi-sufficient conditions, both nla and pho2 mutant plants accumulated higher Pi levels than wild-type plants suggest that the E2/E3 pair may function together to degrade downstream components such as high-affinity Pi transporters (Aung et al., 2006; Bari et al., 2006; Peng et al., 2007a). In nla or pho2 mutants (Aung et al., 2006; Bari et al., 2006; Peng et al., 2007a), one or more Pi transporters may be more stable, leading to an increased uptake and accumulation of Pi in mutant plants. To explore this possibility, we first examined possible interactions between NLA and five different Pi transporters (PHT1;1/PT1, PHT1;4/PT2, PHT1;3/PT4, PHT1;5/PHT5, and PHT1;6/PHT6) (Supplemental Figure 2) by yeast two-hybrid assays. Of these five Pi transporters, only PHT1;4, a high-affinity Pi transporter (henceforth referred to as PT2), interacted with NLA. This interaction was further confirmed by in vitro pull-down experiments using purified proteins. Figures 3A and 3B show that NLA interacted with PT2 but not PT1.
Figure 3.
UBC24 E2 Conjugase Is Required for PT2 Polyubiquitination by NLA E3 Ligase.
(A) and (C) In vitro pull-down assay of PT1 (C) and PT2 (A) with NLA. MBP or MBP-NLA as bait and GST-PT1 or GST-PT2 as prey were mixed. GST-PT1 or GST-PT2 was analyzed by protein gel blots using anti-GST antibody.
(B) and (D) In vitro ubiquitination of PT1 (D) and PT2 (B). The reaction mix contained 25 ng UBE1 (rabbit E1), 100 ng UBC8 or UBC24 (E2), 500 ng MBP-NLA (E3), 100 ng of substrate (GST-PT1-MYC or GST-PT2-MYC), and 8 μg His-Ubiquitin.
After the reaction, polyubiquitinated GST-PT1-MYC or GST-PT2-MYC was analyzed by protein gel blot using anti-MYC antibody.
Given that NLA binds to PT2, we wanted to test whether PT2 may be a substrate of NLA and its functional E2 conjugase, UBC24. To this end, we performed in vitro ubiquitination assays using NLA as an E3 ligase, UBC24 as an E2 conjugase, and PT2 as a substrate. Figures 3C and 3D show that indeed PT2 but not PT1 was polyubiquitinated by NLA in a reaction dependent on E1, E2, and E3 enzymes. Control experiments showed that UBC8 was inactive in this assay (Figures 3C and 3D).
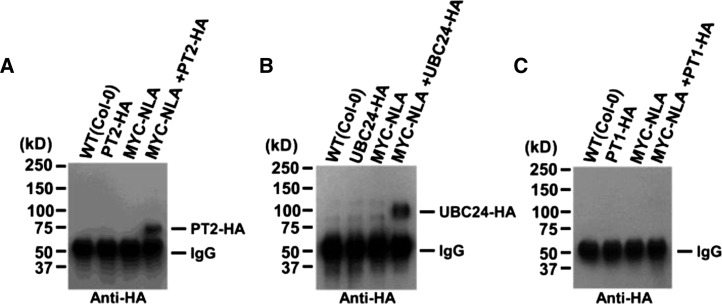
PT2/UBC24/NLA Interactions in Vivo
To provide physiological relevance to the in vitro interaction results, we tested for NLA-UBC24 and NLA-PT2 associations in vivo using extracts prepared from double transgenic plants expressing tagged proteins. Figure 4 shows that NLA displayed specific binding to UBC24 and PT2 but not to PT1.
Figure 4.
NLA Interacts with PT2 and UBC24 in Arabidopsis.
NLA interaction with PT1 (C), PT2 (A), and UBC24 (B) in vivo was detected by coimmunoprecipitation with full-length NLA, PT1, and PT2 in double transgenic plants (35S:MYC-NLA/35S:PT1-HA, 35S:MYC-NLA/35S:PT2-HA, and 35S:MYC-NLA/35S:PHO2-HA). Pulled-down proteins were analyzed by protein gel blots using anti-HA antibody. PT1 was used as a negative control.
Pi Concentrations Regulate Posttranslational Stability of PT2
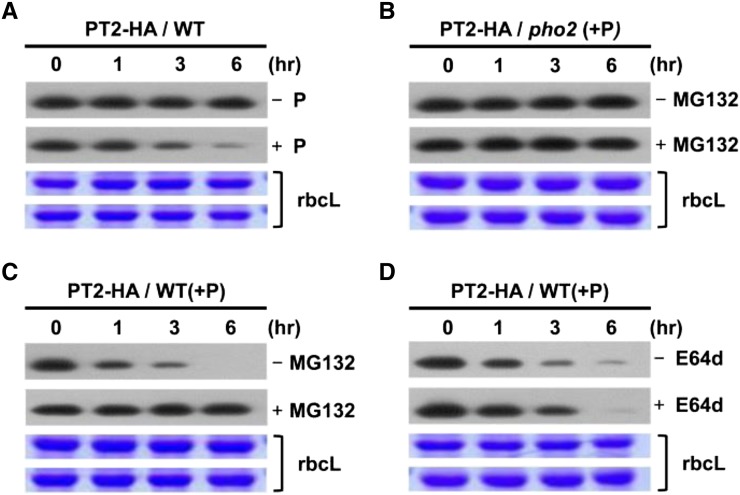
It is known that upon initiation of Pi limitation, miR399 is greatly induced to mediate cleavage of PHO2 mRNAs (Aung et al., 2006; Bari et al., 2006). We surmised that if PT2 is an in vivo target of UBC24, reduced PHO2 expression would result in PT2 protein accumulation. We hypothesized that upon readdition of Pi, miR399 levels would be downregulated because of the attenuation of Pi starvation response. Consequently, PHO2 mRNA levels would recover, and newly synthesized UBC24 protein would be able to mediate PT2 degradation at a posttranslational level. To confirm the validity of the above scenario, we examined the effect of Pi on PT2 protein levels in the absence of new protein synthesis. We grew plants at 1.75 mM Pi and added additional Pi to the medium to a final concentration of 3.5 mM. Plants were treated with cycloheximide (CHX) to block new protein synthesis, and PT2 levels were determined in a time-course experiment. Figure 5A shows that Pi addition triggered rapid posttranslational PT2 degradation, which decayed with a half-life of between 1 and 3 h. Control experiments with PT1 showed the relative stability of this Pi translocator under the same conditions. The decay of PT2 required UBC24 because it was blocked by the pho2 mutation (Figure 5B). Moreover, the decrease in PT2 levels was arrested by the 26S proteasome inhibitor MG132 (Figure 5C) but insensitive to E74d, a vacuolar Cys protease inhibitor (Figure 5D). Taken together, our results show that the posttranslational degradation of PT2 is mediated by the UBC24 E2 conjugase and that the polyubiquitinated PT2 proteins are degraded via 26S proteasomes.
Figure 5.
Pi Destabilizes PT2 Protein in Vivo.
(A) Wild-type transgenic plants expressing 35S:PT2 in 1 mM CHX were treated with (+) or without (−) 3.5 mM Pi.
(B) pho2 mutant plants expressing 35S:PT2 in 3.5 mM Pi and 1 mM CHX were treated with (+) or without (−) MG132.
(C) Wild-type transgenic plants expressing 35S:PT2 in 3.5 mM Pi and 1 mM CHX were treated with (+) or without (−) 50 μM MG132 (proteasome inhibitor). Plants were treated with 3.5 mM Pi and 1 mM CHX.
(D) Wild-type transgenic plants expressing 35S:PT2 in 3.5 mM Pi and 1 mM CHX were treated with or without 50 μM E64d (Cys protease inhibitor).
For (A) to (D), samples were taken at various time points, and PT2-HA protein levels were analyzed by protein gel blots using anti-HA antibody. Stained gel bands of the large subunit of Rubisco, rbcL, were used as a loading control.
NLA and UBC24 Mediate PT2 Degradation in Vivo
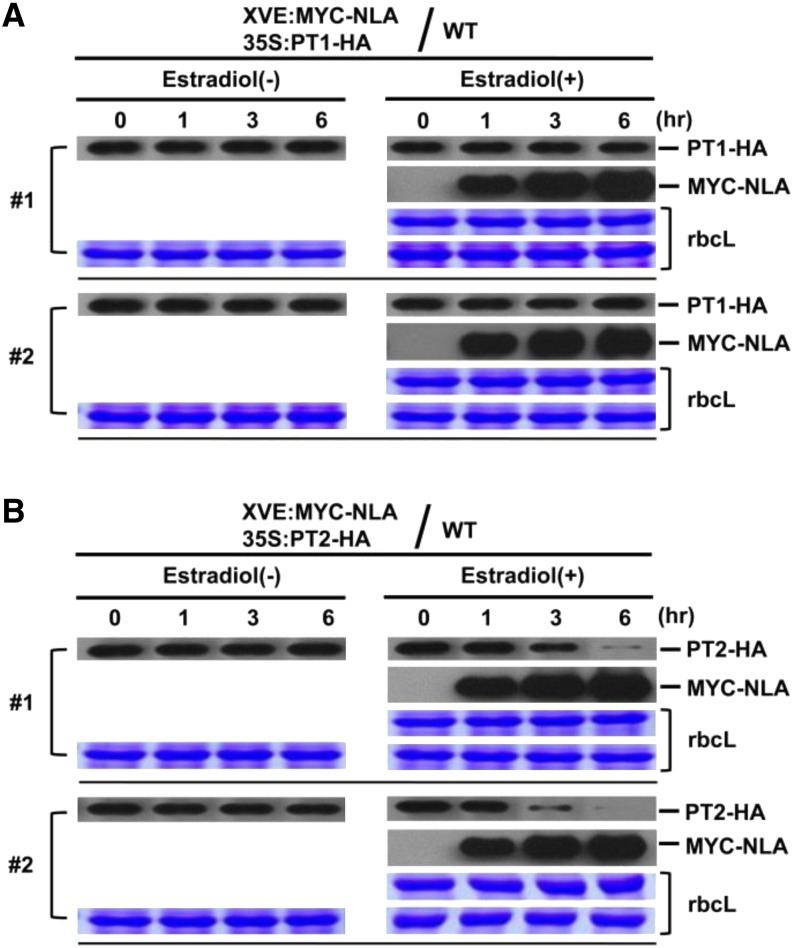
The requirement of UBC24 for PT2 degradation under Pi-sufficient conditions raises the issue of whether the NLA E3 ligase is involved in this process. We generated transgenic plants carrying inducible XVE:NLA and 35S:PT2 to investigate the effect of increasing NLA levels on PT2 levels in vivo. Figure 6 shows that induced expression of NLA resulted in a decrease in PT2 levels in a time-dependent manner. Similar results were obtained in two independent transgenic lines. By contrast, induced expression of NLA had no effect on the level of PT1, which did not bind to NLA and served as a convenient negative control. Quantitative RT-PCR experiments verified that there was no significant change in either PT1 or PT2 transcript levels upon NLA induction (Supplemental Figure 3).
Figure 6.
NLA Targets PT2 but Not PT1 for Degradation in Vivo.
(A) Protein levels of PT1 were determined in wild-type transgenic plants carrying two transgenes, XVE:MYC-NLA (inducible) and 35S:PT1-HA. Plants were either treated or not with β-estradiol to induce NLA expression. Samples were taken at 0, 1, 3, and 6 h after induction, and the expression levels of PT1-HA and MYC-NLA were analyzed by protein gel blots. Stained gel bands of rbcL were used as a loading control. Two independent transgenic lines (no. 1 and 2) were analyzed.
(B) Wild-type transgenic plants carrying two transgenes, XVE:MYC-NLA (inducible) and 35S:PT2-HA, were used. Plants were treated with or without β-estradiol to induce the NLA E3 ubiquitin ligase. Proteins levels were analyzed by protein gel blots using anti-HA antibody. Stained gel bands of rbcL were used as a loading control. Two independent transgenic lines (no. 1 and 2) were analyzed.
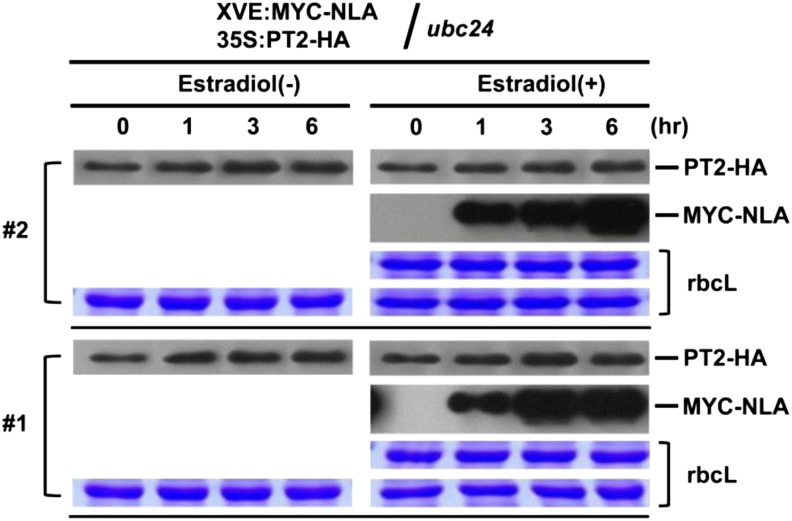
To see if PT2 degradation in vivo is dependent on UBC24, we transformed the pho2 mutant with XVE:NLA and 35S:PT2. In contrast with the results of the wild type, induced expression of NLA in the pho2 mutant background had no effect on PT2 levels in two independent transgenic lines (Figure 7). These results, along with those described earlier (Figure 5), establish the requirement of UBC24 for NLA-mediated PT2 decay.
Figure 7.
NLA-Mediated Degradation of PT2 Is Dependent on PHO2 in Vivo.
pho2 mutant transgenic plants carrying two transgenes, XVE:MYC-NLA (inducible) and 35S:PT2-HA, were used. Plants were treated with or without β-estradiol to induce the NLA E3 ubiquitin ligase. Proteins levels were analyzed by protein gel blots using anti-HA antibody. Stained gel bands of rbcL were used as a loading control. Two independent transgenic lines (no. 1 and 2) were analyzed.
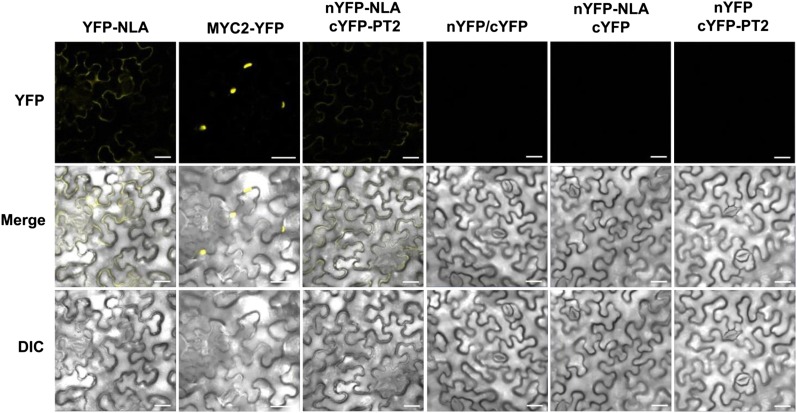
Colocalization of NLA and PT2 on Plasma Membranes
We first investigated subcellular localization of yellow florescent protein (YFP)–NLA using the nuclear protein MYC2-YFP as a control. Confirming previous reports (Lorenzo et al., 2004; Chini et al., 2009), MYC2-YFP was indeed localized to the nucleus, whereas in parallel experiments, YFP-NLA was localized to plasma membranes (Figure 8). To see if the plasma membrane–localized NLA would interact with PT2, we used the bimolecular fluorescence complementation (BiFC) system (Hu et al., 2002; Kerppola, 2008) in which the YFP fluorescence would be reconstituted only if the two proteins would interact. Figure 8 shows that coexpression of 35S:nYFP-NLA and 35S:cYFP-PT2 reconstituted a clear YFP signal along plasma membranes. No signal was detected with 35S:nYFP/35S:cYFP, 35S:nYFP-NLA/35S:cYFP, and 35S:nYFP/35S:cYFP-PT2, all of which served as negative controls.
Figure 8.
Interaction of NLA and PT2 on Plasma Membranes.
Fluorescent fusion genes (YFP-NLA, MYC2-YFP, nYFP, nYFP-NLA, cYFP, and cYFP-PT2) were transiently expressed in N. benthamiana leaves infiltrated with agrobacterial cultures. Confocal images were collected 3 days after infiltration. Experiments were repeated twice, and ∼25 to 30 cells were examined in each experiment. Bars = 20 μm.
DISCUSSION
NLA Displays E3 Ubiquitin Ligase Activity with PHO2 as Its E2 Conjugase
The nla and pho2 mutants with lesions in nitrogen limitation adaptation and Pi starvation response, respectively, were recovered in independent genetic screens (Aung et al., 2006; Bari et al., 2006; Peng et al., 2007a). Notwithstanding their independent origins, these two mutants share a similar phenotype in their hyperaccumulation of Pi under Pi-replete conditions, suggesting a genetic relationship between the two genes in Pi response. Here, we provide biochemical and molecular evidence that NLA encodes an E3 ubiquitin ligase and PHO2 encodes a cognate E2 conjugase for NLA. This claim is supported by two lines of evidence: (1) NLA associated with UBC24 in vitro and in vivo; and (2) NLA displayed autoubiquitination activity in vitro using UBC24 but not UBC8 as the E2 conjugase.
The Arabidopsis genome encodes ∼40 ubiquitin conjugases (UBCs) but >1300 E3 ligases, including those with RING motifs, plant U-box motif, and F-box proteins (Smalle and Vierstra, 2004). The overwhelming ratio (>35) between E3s and E2s suggests one UBC is likely to service multiple E3 ligases. In this regard, it would not be surprising for PHO2 (UBC24) to have other E3 partners dedicated to the modification/destruction of other target proteins. Whether these other E3 ligases are also involved in the Pi deprivation response requires further studies. On the other hand, notwithstanding the large number of E3 ligases compared with the limited number of E2 conjugases, an E3 ligase may also select different E2 partners to execute cell type–specific or other specialized functions. In the case of NLA, Peng et al. (2007b) have previously shown binding of NLA to UBC8, an observation confirmed in this work. Nevertheless, UBC8 was unable to support NLA autoubiquitination activity in vitro (Kraft et al., 2005). Given this, it is possible that ubiquitination mediated by the NLA-UBC8 pair may have other functional consequences not related to protein destabilization (e.g., change of cellular location of ubiquitinated target protein). The targets of the NLA-UBC8 pair remain to be identified.
PT2 Is a Target of NLA-UBC24
Using yeast two-hybrid assays, we tested five (PHT1;1/PT1, PHT1;4/PT2, PHT1;3/PT4, PHT1;5/PHT5, and PHT1;6/PHT6) of the nine members of the Pht1 transporter family for possible interactions with NLA, and only PT2 (Pht1;4) was found to interact. This interaction was shown to be direct by in vitro pull-down assays as well as to occur in plants via in vivo coimmunoprecipitation experiments. More importantly, we showed that PT2 can be polyubiquitinated by NLA in vitro and the in vitro ubiquitination reaction specifically required UBC24 because UBC8 was inactive. Moreover, PT1, which did not bind to NLA, was not modified under the same assay conditions.
Consistent with its role in PT2 degradation, we showed that YFP-NLA localizes to plasma membranes of tobacco (Nicotiana tabacum) leaf cells (Figure 8). Moreover, using the BiFC system (Hu et al., 2002; Kerppola, 2008), we found that the plasma membrane-localized NLA interacts with PT2, providing additional evidence for in vivo interaction of the two proteins. Peng et al. (2007b) have previously reported nuclear localization of NLA in onion (Allium sepa) skin cells and detected NLA/UBC8 complex in nucleus speckles. Although NLA can interact with UBC8 in vivo, we have shown that UBC8 is unable to support ubiquitination of PT2 (or PT1) mediated by UBC24 (PHO2); therefore, the nuclear localization reported by Peng et al. (2007b) is not relevant to the set of results reported here. The differences between our results and those of Peng et al. (2007b) could be due to the use of different cell type (onion skin cells versus tobacco leaf cells) and two different types of plants (onion versus tobacco). It should not be surprising that NLA may have functions other than in Pi/nitrogen responses and that the NLA-UBC9 connection may have a biological function different from the NLA-UBC24 pair.
In eukaryotes, proteins targeted for destruction are first polyubiquitinated before being degraded by 26S proteasomes. The physiological relevance of the in vitro ubiquitination of PT2 by the NLA-UBC24 pair was validated by transgenic plant experiments. Using wild-type transgenic plants carrying two different transgenes, we showed that induced NLA expression led to a clear decrease in PT2 but not PT1 protein levels. By contrast, in the pho2 mutant background, no change in PT2 levels was seen upon induced expression of NLA. Together, these results provide strong evidence that PT2 is targeted by NLA/UBC24 for destruction in vivo. These results are not consistent with the hypothesis that UBC24 functions as a chimeric E2-E3 enzyme (Liu et al., 2012).
While this work was being reviewed, Huang et al. (2013) used comparative proteomics and specific antibodies to show that PHT1;1/2/3 and PHT1;4 levels are higher in roots of pho2 mutant compared with the wild type in Pi-sufficient media. Moreover, the half-lives of these Pi transporters are longer in the mutant compared with the wild type. In a related study, Lin et al. (2013) reported the regulation of the same four Pi transporters by NLA and PHO2. Whereas the approaches used by Huang et al. (2013) and Lin et al. (2013) are different from those used in this work, the conclusions are substantially similar. There is agreement that PHT1;4 (PT4) is targeted by NLA. Whereas we demonstrated a direct involvement of UBC24 (PHO2) in the targeted destruction of PT2 by NLA, Lin et al. (2013) suggested that PHO2 acts synergistically but sequentially with NLA. Lin et al. (2013) and Huang et al. (2013) also used a specific but nondiscriminating antibody to the three transporters to demonstrate regulation of PHT1;1/2/3 levels by NLA and UBC24. In this work, we showed that PHT1;1 and PHT1;3 do not interact with NLA. Therefore, our results considered together with theirs would suggest that the changes in protein levels seen by Lin et al. (2013) and Huang et al. (2013) can be mainly attributed to PHT1;2.
Posttranslational Regulation of PT2 by Pi Concentration
The regulation of PT2 by NLA-UBC24 at the level of protein stability raises the question of posttranslational regulation of this transporter by Pi concentration in the medium. Here, we showed that PT2 is stable at a low Pi concentration (1.75 mM) but becomes destabilized at a high Pi concentration (3.5 mM). This Pi-triggered degradation is of physiological significance, as it is abolished in the pho2 mutant which, in contrast with the wild type, accumulates high levels of Pi under Pi-replete conditions. The arrest of PT2 degradation by MG132 but not E64d indicates that ubiquitinated PT2 protein is destroyed by 26S proteasomes rather than vacuolar Cys proteases. It is possible that modifications of other Pi response–related proteins by UBC24 may lead to a different fate. Liu et al. (2012) have recently reported that PHO2 colocalizes with PHO1 on Arabidopsis endomembranes and assists PHO1 degradation via vacuolar proteolysis. In this system, it is assumed that there is a direct ubiquitination of PHO1 by PHO2 without the involvement of any E3 ligase. The authors suggested that under conditions of Pi sufficiency, ubiquitinated PHO1 on endoplasmic reticulum or Golgi membranes may be redirected for vacuolar destruction.
Our biochemical results on the posttranslational degradation of PT2 by NLA are further supported by subcellular localization experiments. We found that NLA interacts with PT2 under Pi-sufficient but not Pi-deficient conditions. Peng et al. (2007b) found that NLA was localized to the nucleus in cells of onion epidermal cell. It is possible that upon Pi starvation, NLA dissociates from the plasma membrane and migrates to the nucleus, thereby allowing recovery of PT2 proteins to transport Pi into cells.
Our results add considerable molecular details to the Pi signaling pathway and Pi homeostasis regulation. It was previously unclear whether NLA was involved in the Pi starvation response. Here, we have identified this E3 ligase as an upstream component of UBC24/PHO2 and the PT2 Pi transporter as one of the downstream targets of UBC24. Under Pi-sufficient conditions, NLA, together with UBC24, polyubiquitinates PT2 and maintains the PT2 protein at a low level, thus limiting Pi uptake. Upon Pi starvation, miR399 and miR827 are induced and these two miRNAs mediate cleavage of PHO2 mRNA and NLA mRNA, respectively. The reduced expression of NLA and PHO2 allows accumulation of PT2, which plays a role in Pi uptake.
Whereas our results clarified certain key aspects of Pi signaling and Pi homeostasis regulation, many related issues still remain to be addressed. For example, given the functional redundancy of the Pht1 family members, it would be important to understand how other family members (e.g., PT1) are regulated during the Pi response. The identification of NLA as a partner of PHO2 suggests intimate crosstalk between the nitrogen-limitation response and the Pi deprivation response pathways. Elucidation of the mechanism of crosstalk of signaling pathways will improve our understanding of how plants regulate endogenous cellular processes when challenged with a limiting nutrient supply.
METHODS
Plant Materials, Culture Conditions, and Preparation of Plant Samples
Plant materials used were wild-type Arabidopsis thaliana (Columbia-0 [Col-0]), pho2 T-DNA insertion mutant (SALK_026788C), and wild-type (Col-0) transgenic plants carrying the following transgenes: 35S:MYC-NLA, 35S:PT1-HA, 35S:PT2-HA, 35S:PHO2-HA, XVE:MYC-NLA, 35S:PT1-HA/XVE:MYC-NLA, and 35S:PT2-HA/XVE:MYC-NLA. All plants were grown either on soil or in a growth chamber at 22°C under 16 h light/8 h dark on 0.8% agar media containing Murashige and Skoog salts, 0.5 g/L MES, and 10 g/L Suc. For chemical treatments, plants were incubated in basal MGRL hydroponic medium or a Pi-depleted medium containing 1.75 mM MES, pH 5.8, instead of 1.75 mM sodium Pi (pH 5.8) (Woo et al., 2012).
Preparation of Constructs
The Arabidopsis NLA cDNA encoding full-length protein was amplified by PCR, cloned in pENTR/D vector (Invitrogen) and then transferred into pMBP-DC (Jang et al., 2007) by recombination using the LR Clonase enzyme (Invitrogen). cDNAs encoding full-length Arabidopsis PT1, PT2, UBC8, and PHO2 were inserted by the Gateway Cloning System into pGEX4T-1 (Amersham Biosciences) to give GST-PT1-HA, GST-PT1-MYC, GST-PT2-HA, GST-PT2-MYC, GST-PHO2, and into EcoRI and XhoI of pET28a (Novagen) to give HIS-UBC8. To investigate possible homodimerization of NLA, cDNA and fragments encoding full-length NLA, SPX domain (amino acids 1 to 167), and RING domain (amino acids 220 to 335) were inserted by the Gateway Cloning System into pGEX-4T-1 (Amersham Biosciences) to generate GST-NLA, GST-SPX, and GST-RING, respectively.
To probe for in vivo protein–protein interaction, cDNAs encoding PT1, PT2, UBC24 (PHO2), and NLA were inserted into pBA002 (Kost et al., 1998) or pER8 (Zuo et al., 2000) to produce 35S:MYC-NLA, 35S:PT1-HA, 35S:PT2-HA, 35S:PHO2-HA, and XVE:MYC-NLA. Transgenic plants expressing combinations of these transgenes were used for coimmunoprecipitation experiments. To investigate subcellular localization of NLA and PT2, cDNAs encoding NLA and PT2 were inserted into pBA002 to produce 35S:YFP-NLA, 35S:MYC2-YFP (Lorenzo et al., 2004; Chini et al., 2009), 35S:nYFP-NLA, and 35S:cYFP-PT (Hu et al., 2002; Kerppola, 2008). Agrobacterial cultures carrying each of these plasmids were used for transient expression in leaves of Nicotiana benthamiana (Liu et al., 2012).
Yeast Two-Hybrid Assays
Yeast two-hybrid assays were performed using the GAL4-based two-hybrid system (Clontech). Full-length PHT1;1/PT1, PHT1;4/PT2, PHT1;3/PT4, PHT1;5/PHT5, PHT1;6/PHT6, UBC8, and PHO2 cDNA fragments were cloned into pGAD424 (Clontech) to generate constructs carrying the activating comain. Full-length NLA cDNA was cloned into pGBT8 (Clontech) to generate a construct carrying the binding domain. All constructs were transformed into yeast strain AH109 by the lithium acetate method (Elble, 1992). Transformed yeast cells were grown on Leu−/Trp− minimal medium and were plated onto Leu−/Trp−/, Leu−/Trp−/His− minimal medium and Leu−/Trp−/His− minimal medium containing 20 mM 3-amino-1,2,4-triazole to test for possible interactions between NLA and candidate proteins.
In Vitro and in Vivo Pull-Down of APT1, PT2, and UBC24 with NLA
Constructs MBP-NLA-HA, GST-PT1-MYC, GST-PT2-MYC, and GST-PHO2 were transformed into Escherichia coli BL21/DE3(pLysS) cells. Transformed cells were treated with 1 mM isopropyl-β-d-thiogalactoside for 12 h under 22°C to induce expression of fusion proteins. To determine possible interactions of NLA with PT1, PT2, and UBC24 (PHO2), both 1 μg of MBP-NLA and 1 μg of GST-PT1-MYC, GST-PT2-MYC, or GST-UBC24 (PHO2) were added to 1 mL of binding buffer (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1% Triton X-100, 0.2% glycerol, and 0.5 mM β-mercaptoethanol). After incubation at 25°C for 2 h, the reaction mixtures were incubated with amylose resin for 2 h before being washed four times with buffer (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, and 1% Triton X-100). Absorbed proteins were analyzed by SDS-PAGE and detected by protein gel blot using an anti-GST antibody at a 1:5000 dilution (Santa Cruz Biotechnology). To determine in vivo interactions between NLA and PT1 or PT2 or UBC24, (PHO2) single and double transgenic plants carrying the transgenes 35S:HA-PT1, 35S:HA-PT2, 35S:PHO2-HA, 35S:MYC-NLA, 35S:HA-PT1/XVE:MYC-NLA, 35S:HA-PT2/XVE:MYC-NLA, and 35S:PHO2-HA/XVE:MYC-NLA were used. Constructs for single and double transgenic plants were transformed into Agrobacterium tumefaciens strain ABI and infiltrated into the wild type (Col-0) using the floral dip method (Clough and Bent, 1998; Zhang et al., 2006).
In Vitro Ubiquitination Assays of PT1 and PT2 and Self-Ubiquitination of NLA
The reaction mix of 30 μL contained 50 mM Tris, pH 7.5, 50 mM MgCl2, 20 µM ZnCl2, 20 mM ATP, 100 ng of purified GST-PT1-MYC and GST-PT2-MYC, 500 ng of purified MBP-NLA (E3), 25 ng UBE1 (rabbit E1; BostonBiochem), 100 ng HIS-UBC8 or GST-UBC24 (E2), and 8 μg of HIS-ubiquitin (Sigma-Aldrich). After incubation in a 30°C water bath for 2 h, the reaction mixtures were separated on an 8% SDS-polyacrylamide gel, and substrates (GST-PT1-MYC and GST-PT2-MYC) were detected by protein gel blot analysis using anti-MYC antibody at a 1:4000 dilution (Santa Cruz Biotechnology). In vitro self-ubiquitination of NLA was performed using the same reaction conditions but without substrate. Self-ubiquitinated MBP-NLA was detected by protein gel blot using anti-MBP antibody at a 1:5000 dilution (Santa Cruz Biotechnology).
Effects of NLA Expression on PT1 and P2 Protein Stability
Double transgenic plants expressing 35S:PT1-HA/XVE:MYC-NLA and 35S:PT2-HA/XVE:MYC-NLA in the wild type (Col-0) and ubc24 mutant background were generated. Seedlings (10 d old) were treated with or without 25 μM β-estradiol (Sigma-Aldrich), and samples were taken at 0, 1, 3, and 6 h. Two independent transgenic lines were analyzed.
The 35S:PT2-HA overexpression plants were used to examine phosphate transporter (PT2) protein stability with or without 3.5 mM Pi. Seedlings (10 d old) grown in hydroponic medium (with or without 3.5 mM Pi) were treated with 1 mM CHX (Sigma-Aldrich) to block new protein synthesis, and samples were collected at 0, 1, 3, and 6 h. In experiments using 26S proteasome inhibitor or Cys protease inhibitor, 35S:PT2-HA plants were incubated in hydroponic culture medium containing 3.5 mM Pi with 1 mM CHX and with/without 50 μM MG132 or 50 μM E64d. Protein samples were analyzed by protein gel blots using anti-HA at a 1:4000 dilution (Santa Cruz Biotechnology) antibody for the tagged proteins.
RNA Extraction and Quantitative RT-PCR Analysis
Total RNA was isolated from two independent lines (10 d old) of two double transgenic plant (35S:PT1-HA/XVE:MYC-NLA and 35S:PT2-HA/XVE:MYC-NLA) using Qiagen RNeasy mini kits including DNase treatment according to the manufacturer’s instructions. Reverse transcriptional reaction was performed using Ready-To-Go You-Prime first-strand beads (GE Healthcare) from 1 µg of total RNA according to the manufacturer’s instructions. Quantitative PCR for 100 ng of single-strand cDNA was performed using the SYBR Primix Ex Taq system (TaKaRa) on the CFS96 real-time system (Bio-Rad) with three sets of primers: 5′-TGATGATCTTGTGCTCTGTCG-3′ and 5′-ATGACACCCTTGGCTTCGT-3′ for PT1 (Lei et al., 2011), 5′-CGAAGCTCCTCGGTCGTAT-3′ and 5′-GGAGAGTCCCAGGCTTTTGT-33′ for PT2 (Lei et al., 2011), and 5′-AGTGGTCGTACAACCGGTATTGT-3′ and 5′-GATGGCATGAGGAAGAGAGAAAC-3′ for ACT2. All quantitataive PCR data show average value (se) with n = 3 independent biological samples.
Infiltration of Agrobacterial Cultures into N. benthamiana Leaves and Fluorescence Confocal Microscopy of Infiltrated Cells
Leaves of two-week-old N. benthamiana plants were infiltrated with agrobacterial cultures according to Liu et al. (2012), and samples were analyzed 3 d later. Fluorescent signals were observed using a LSM 780 laser scanning confocal microscope (Zeiss) with excitation-emission wavelengths of 500/20 to 535/30 nm for YFP (Kerppola, 2006).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: NLA, At1g02860;UBC8, At5g41700;UBC24(PHO2), At2g33770; PHT1;1, At5g43350;PHT1;2, At5g43370;PHT1;3, At5g43360;PHT1;5, At2g32830;PHT1;6, At5g43340;MYC2, At1g32640; and ACT2, At3g18780.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. NLA Interacts with UBC24 in Yeast.
Supplemental Figure 2. Yeast Two-Hybrid Assays for Possible Interactions between NLA and Five Different Pi Transporters.
Supplemental Figure 3. qPCR Analysis of PT1 and PT2 Transcript Levels in Samples Shown in Figure 6.
Supplementary Material
Acknowledgments
This work was supported in part by the Cooperative Research Program for Agricultural Science and Technology Development (PJ906910), Rural Development Administration, Republic of Korea. We thank Michelle Kean and Maria Cruz de Carvalho for reading the article and members of the laboratory for discussion.
AUTHOR CONTRIBUTIONS
B.S.P. and N.-H.C. designed the experiments. B.S.P. performed all experiments except the BiFC experiment, which was done with the assistance of J.S.S. B.S.P. and N.-H.C. wrote the article.
Glossary
- CHX
cycloheximide
- BiFC
bimolecular fluorescence complementation
- Col-0
Columbia-0
Footnotes
Online version contains Web-only data.
References
- Aung K., Lin S.I., Wu C.C., Huang Y.T., Su C.L., Chiou T.J. (2006). pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R., Datt Pant B., Stitt M., Scheible W.R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Chico J.M., Fernández-Calvo P., Solano R. (2009). The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 59: 77–87 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Delhaize E., Randall P.J. (1995). Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol. 107: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elble R. (1992). A simple and efficient procedure for transformation of yeasts. Biotechniques 13: 18–20 [PubMed] [Google Scholar]
- Hamburger D., Rezzonico E., MacDonald-Comber Petétot J., Somerville C., Poirier Y. (2002). Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holford I.C.R. (1997). Soil phosphorus: Its measurement, and its uptake by plants. Aust. J. Soil Res. 35: 227–239 [Google Scholar]
- Hu C.D., Chinenov Y., Kerppola T.K. (2002). Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Huang T.K., et al. (2013). Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 25: 4044–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.C., Yang S.W., Yang J.Y., Chua N.H. (2007). Independent and interdependent functions of LAF1 and HFR1 in phytochrome A signaling. Genes Dev. 21: 2100–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S., Peng M.S., Rothstein S.J. (2011). Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet. 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola T.K. (2006). Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 1: 1278–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola T.K. (2008). Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37: 465–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B., Spielhofer P., Chua N.H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16: 393–401 [DOI] [PubMed] [Google Scholar]
- Kraft E., Stone S.L., Ma L., Su N., Gao Y., Lau O.S., Deng X.W., Callis J. (2005). Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 139: 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M.G., Liu Y.D., Zhang B.C., Zhao Y.T., Wang X.J., Zhou Y.H., Raghothama K.G., Liu D. (2011). Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiol. 156: 1116–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W.Y., Huang T.K., Chiou T.J. (2013). NITROGEN LIMITATION ADAPTATION, a target of MicroRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 25: 4061–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.Q., Allan D.L., Vance C.P. (2010). Systemic signaling and local sensing of phosphate in common bean: Cross-talk between photosynthate and microRNA399. Mol. Plant 3: 428–437 [DOI] [PubMed] [Google Scholar]
- Liu T.Y., Huang T.K., Tseng C.Y., Lai Y.S., Lin S.I., Lin W.Y., Chen J.W., Chiou T.J. (2012). PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24: 2168–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J., Thibaud M.C., Bechtold N., Raghothama K., Nussaume L. (2004). Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol. Biol. 55: 727–741 [DOI] [PubMed] [Google Scholar]
- Nilsson L., Müller R., Nielsen T.H. (2007). Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ. 30: 1499–1512 [DOI] [PubMed] [Google Scholar]
- Peng M., Bi Y.M., Zhu T., Rothstein S.J. (2007a). Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Mol. Biol. 65: 775–797 [DOI] [PubMed] [Google Scholar]
- Peng M., Hannam C., Gu H., Bi Y.M., Rothstein S.J. (2007b). A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J. 50: 320–337 [DOI] [PubMed] [Google Scholar]
- Poirier Y., Thoma S., Somerville C., Schiefelbein J. (1991). Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 97: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama K.G. (1999). Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Rausch C., Bucher M. (2002). Molecular mechanisms of phosphate transport in plants. Planta 216: 23–37 [DOI] [PubMed] [Google Scholar]
- Remy E., Cabrito T.R., Batista R.A., Teixeira M.C., Sá-Correia I., Duque P. (2012). The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol. 195: 356–371 [DOI] [PubMed] [Google Scholar]
- Rouached H., Arpat A.B., Poirier Y. (2010). Regulation of phosphate starvation responses in plants: Signaling players and cross-talks. Mol. Plant 3: 288–299 [DOI] [PubMed] [Google Scholar]
- Secco D., Wang C., Arpat B.A., Wang Z.Y., Poirier Y., Tyerman S.D., Wu P., Shou H.X., Whelan J. (2012). The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 193: 842–851 [DOI] [PubMed] [Google Scholar]
- Seo H.S., Yang J.Y., Ishikawa M., Bolle C., Ballesteros M.L., Chua N.H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Shin H., Shin H.S., Dewbre G.R., Harrison M.J. (2004). Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Smalle J., Vierstra R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Smith A.P., Nagarajan V.K., Raghothama K.G. (2011). Arabidopsis Pht1;5 plays an integral role in phosphate homeostasis. Plant Signal. Behav. 6: 1676–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Hauksdóttir H., Troy A., Herschleb J., Kraft E., Callis J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ribot C., Rezzonico E., Poirier Y. (2004). Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 135: 400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J., MacPherson C.R., Liu J., Wang H., Kiba T., Hannah M.A., Wang X.J., Bajic V.B., Chua N.H. (2012). The response and recovery of the Arabidopsis thaliana transcriptome to phosphate starvation. BMC Plant Biol. 12: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Guo H.S., Dallman G., Fang S., Weissman A.M., Chua N.H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170 [DOI] [PubMed] [Google Scholar]
- Yang X.J., Finnegan P.M. (2010). Regulation of phosphate starvation responses in higher plants. Ann. Bot. (Lond.) 105: 513–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.S., Niu Q.W., Chua N.H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646 [DOI] [PubMed] [Google Scholar]
- Zuo J.R., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.