Abstract
The circadian clock is a cellular time-keeper mechanism that regulates biological rhythms with a period of ∼24 h. The circadian rhythms in metabolism, physiology, and development are synchronized by environmental cues such as light and temperature. In plants, proper matching of the internal circadian time with the external environment confers fitness advantages on plant survival and propagation. Accordingly, plants have evolved elaborated regulatory mechanisms that precisely control the circadian oscillations. Transcriptional feedback regulation of several clock components has been well characterized over the past years. However, the importance of additional regulatory mechanisms such as chromatin remodeling, protein complexes, protein phosphorylation, and stability is only starting to emerge. The multiple layers of circadian regulation enable plants to properly synchronize with the environmental cycles and to fine-tune the circadian oscillations. This review focuses on the diverse posttranslational events that regulate circadian clock function. We discuss the mechanistic insights explaining how plants articulate a high degree of complexity in their regulatory networks to maintain circadian homeostasis and to generate highly precise waveforms of circadian expression and activity.
INTRODUCTION
The circadian clock is a cellular time-keeper mechanism able to perceive external synchronizing inputs to generate endogenous rhythmic outputs with a period of ∼24 h. In many plant species, synchronization of the clock with the environment confers fitness advantages by controlling key essential processes, such as photosynthetic activity, hypocotyl elongation, and the floral transition (Doyle et al., 2002; Green et al., 2002; Imaizumi et al., 2003; Dodd et al., 2005; Zhang et al., 2008; Niwa et al., 2009; Resco et al., 2009; Yerushalmi and Green, 2009; Nusinow et al., 2011). A large fraction of the plant transcriptome is clock controlled, suggesting that the circadian clock globally modulates diverse signals and metabolic pathways that mediate development and environmental adaptation responses (Nagel and Kay, 2012).
The transcriptional regulation of several clock components has been well characterized at a molecular level over the past years (reviewed in Carré and Veflingstad, 2013). Multiple intertwined regulatory networks define the basic architecture of the Arabidopsis thaliana circadian clock. Two single MYB transcription factors, CIRCADIAN CLOCK-ASSOCIATED1 (CCA1) (Wang and Tobin, 1998) and LATE ELONGATED HYPOCOTYL (LHY) (Schaffer et al., 1998), and TIMING OF CAB EXPRESSION1/ PSEUDO-RESPONSE REGULATOR1 (TOC1/PRR1) (Strayer et al., 2000; Makino et al., 2002) comprise a central regulatory module (Alabadí et al., 2001). CCA1 and LHY repress TOC1 expression that in turn represses the transcription of CCA1 and LHY (Gendron et al., 2012; Huang et al., 2012). This regulatory module is interlocked with a morning loop and an evening loop (Locke et al., 2006). In the morning loop, members of the PRR family (PRR5, PRR7, and PRR9) bind to promoters of CCA1 and LHY and repress their expression (Nakamichi et al., 2010). CCA1 and LHY in turn promote the expression of PRR7 and PRR9 by direct association with their promoters (Farré et al., 2005). The reciprocal regulation between TOC1 and GIGANTEA (GI) together with the recently identified evening complex (EC) comprise the evening loop (Locke et al., 2006). The EC is composed of EARLY FLOWERING3 (ELF3), ELF4, and LUX ARRYTHMO (LUX)/PHYTOCLOCK1 and acts at dusk as a transcriptional repressor of PRR9 expression (Helfer et al., 2011; Nusinow et al., 2011). Further connections between the different loops are exemplified by the widespread repressing function of TOC1, regulating nearly all of the components of the morning and evening loops (Huang et al., 2012).
The complex network of transcriptional regulators at the core of the clock underscores the role of transcriptional regulation as a central regulatory mechanism for circadian oscillation. However, emerging evidence reinforces the notion that circadian clock components are further regulated by additional regulatory mechanisms (Más and Yanovsky, 2009). In this review, we summarize some of the recent advances on the role of chromatin remodeling and posttranslational clock protein modification as key regulatory mechanisms controlling the circadian function in Arabidopsis. Many excellent recent reviews (Harmer, 2009; Adams and Carré, 2011; Sanchez et al., 2011; Nagel and Kay, 2012; Haydon et al., 2013; Kinmonth-Schultz et al., 2013) cover in detail other particular aspects of clock organization and function that are not addressed in this review.
POSTTRANSLATIONAL REGULATION
Ubiquitination and Degradation
Covalent attachment of ubiquitin is a common mechanism of modulating protein stability. The ubiquitination process that leads to protein degradation is mediated by the sequential action of three enzymes: ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin-ligase (E3). The fact that ∼5% of Arabidopsis genes are involved in ubiquitination underscores the significance of this regulatory process in plants (Mazzucotelli et al., 2006; Lee and Kim, 2011; Sadanandom et al., 2012).
To date, two E3 ligases and three F-box proteins have been characterized as circadian clock regulators in Arabidopsis: the E3s CONSTITUTIVE PHOTOMORPHOGENESIS1 (COP1) and SINAT5 and the F-box proteins ZEITLUPE (ZTL), FLAVIN BINDING, KELCH REPEAT AND F-BOX1 (FKF1), and LOV KELCH PROTEIN2 (LKP2) (Yu et al., 2008; Baudry et al., 2010; Park et al., 2010). The ZTL, FKF1, and LKP2 proteins contain three specific domains: a blue light–absorbing PAS domain (Per-ARNT-Sim/LOV [for light, oxygen, or voltage]), which binds the flavin mononucleotide chromophore (Ito et al., 2012), an F-box domain with E3 ligase activity as a component of the SKP-Cullin-Rbx-F-box (SCF) complex, and a Kelch domain responsible for interactions with substrates. ZTL, FKF1, and LKP2 contribute to the ubiquitin-mediated clock protein degradation by conferring substrate specificity to the SCF E3 ubiquitin ligase complexes (Ito et al., 2012).
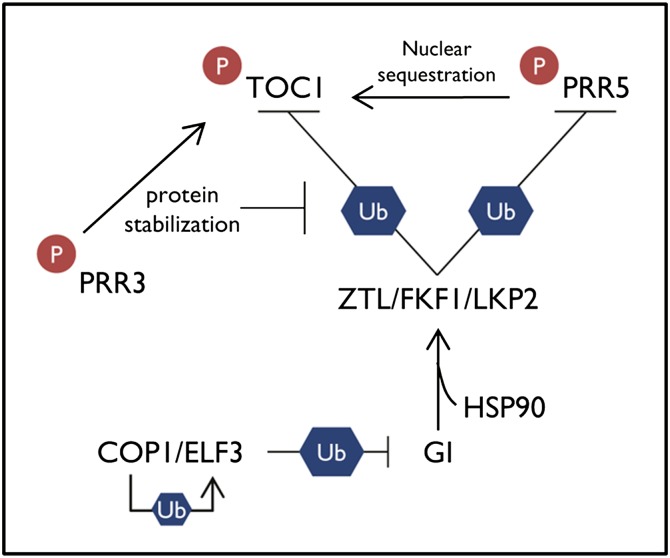
ZTL assembles into a functional SCF complex by interacting with the SKP1 homolog ARABIDOPSIS SKP-LIKE PROTEIN1, with CULLIN1, and with the RING finger protein RBX1 (Han et al., 2004; Harmon et al., 2008). ZTL regulation of circadian period is accomplished via regulation of TOC1 and PRR5 stability. The LOV domain of ZTL directly interacts with TOC1 and PRR5 through their pseudo-receiver domain and mediates the dark-dependent protein degradation by the 26S proteasome (Más et al., 2003; Kiba et al., 2007). Genetic analyses further demonstrated the physiological relevance of these interactions. The long period phenotype of ztl mutants is abolished in the absence of a functional TOC1 (Más et al., 2003); likewise, the phenotypes of ztl mutant are also suppressed by the prr5 mutation (Kiba et al., 2007). Notably, toc1 prr5 double mutants phenocopy transgenic plants overexpressing ZTL, which is consistent with the ZTL-dependent regulation of TOC1 and PRR5 protein stability (Ito et al., 2008). Recent studies have shown that FKF1 and LKP2 are also involved in proper circadian oscillation. While fkf1-deficient mutants have no obvious alterations in circadian period, the fkf1 mutation enhances the long period phenotype of ztl mutants (Baudry et al., 2010). Moreover, the ztl fkf1 lkp2 triple mutants further lengthen the circadian period relative to the ztl fkf1 double mutant. Consistently, FKF1 and LKP2 also interact with and degrade TOC1 and PRR5 (Wang et al., 2010), thus contributing along with ZTL to their protein oscillation (Figure 1).
Figure 1.
Phosphorylation Modulates TOC1 Protein Stability.
Phosphorylation of TOC1 enhances the interaction with ZTL, which leads to proteasomal degradation. ZTL also targets PRR5 for degradation. Phosphorylation also favors TOC1 stabilization both by PRR3-mediated competitive inhibition of the proteasomal degradation and by PRR5 nuclear sequestration. ZTL is stabilized by interaction with GI and HSP90. GI is regulated by the COP1-ELF3 complex. P, phosphorylation; Ub, ubiquitination.
Interestingly, ZTL is also targeted by proteasomal degradation (Kim et al., 2003). ZTL mRNA accumulation does not oscillate, but the protein cycles with significant variation in amplitude. The rhythmic oscillation of ZTL protein is controlled by phase-specific degradation through the proteasome (Kim et al., 2003). This proteasome-dependent degradation of ZTL is antagonized by the flowering regulator and clock component GI. ZTL interacts with GI in a blue light–dependent manner and cooperatively enhances stability of both proteins (Kim et al., 2007). Therefore, the cyclic accumulation of GI protein facilitates the rhythmic regulation of ZTL turnover and the subsequent control in the oscillations of TOC1 and PRR5 (Figure 1). Regulation of ZTL maturation into its active conformation and assembly into a functional SCF E3 ligase is also modulated by the chaperone protein HEAT SHOCK PROTEIN90 (HSP90). HSP90 physically interacts with ZTL and protects it from denaturation and aggregation (Kim et al., 2011). Inactivation of HSP90 diminishes ZTL protein accumulation and lengthens the circadian period, most likely through accumulation of TOC1 protein (Kim et al., 2011). Indeed, TOC1 and PRR5, the proteolytic targets of ZTL, are more stable in plants with defects in HSP90 function. This chaperone protein may contribute to maintaining an intact structure of the F-box protein, thus ensuring clock-controlled proteome homeostasis (Kim et al., 2011) (Figure 1).
Degradation and regulation of clock protein stability might be dependent on COP1 function. COP1 is an E3 ubiquitin ligase previously shown to be involved in the degradation of components of the light signaling pathway associated with photomorphogenesis and floral transition. The COP1-deficient mutants also display shortened circadian oscillations (Yu et al., 2008), most likely as the result of the interaction of COP1 with ELF3, which undergoes ubiquitination and proteasome-dependent degradation in a COP1-dependent manner. Notably, the ELF3 degradation does not lead to an antagonistic functional relationship with COP1 (Yu et al., 2008) but allows the recruitment of newly synthesized ELF3 to further enhance the extent of ELF3 function. COP1 and ELF3 appear to act together to control the stability of GI. COP1 and ELF3 accumulate at night and promote GI destabilization through the proteasome pathway (Figure 1). ELF3 appears to act as a substrate adaptor that facilitates the interaction of COP1 and GI, and the subsequent COP1-mediated degradation of ELF3 controls the extent of ELF3 function (Yu et al., 2008). Thus, the time-dependent interaction of COP1 and ELF3 might be key to ensuring a precise shaping of GI protein accumulation.
Another component involved in clock protein degradation is the light signaling mediator DE-ETIOLATED1 (DET1), which regulates LHY protein stability (Song and Carré, 2005). Although LHY undergoes proteasome-dependent proteolysis by the E3 ligase SINAT5, DET1 suppresses turnover by physical interaction with LHY (Song and Carré, 2005; Park et al., 2010). In the det1-1 mutant, LHY degradation is accelerated, which is concomitant with a short circadian period of gene expression, a phenotype that is similar to the one observed in lhy loss-of-function mutants (Song and Carré, 2005).
Deubiquitinating enzymes counteract the functions of the F-box proteins. The Arabidopsis genome contains 27 predicted ubiquitin-specific proteases (UBPs), with a role in a variety of cellular signaling pathways (Doelling et al., 2001, 2007). In particular, UBP12 and UBP13, which are circadian-regulated genes, play a role in the control of circadian period. Consistently, the rhythmic oscillation of LHY and TOC1 transcripts is shortened in ubp12-deficient and ubp12 ubp13 double mutants (Cui et al., 2013). Taken together, these studies show that dynamic and reversible modulation of ubiquitin attachment to clock proteins fine-tunes circadian oscillation and facilitates daylength measurement.
Phosphorylation
Phosphorylation is a fundamental regulatory mechanism by which protein activity is dynamically regulated mostly through regulation of complex formation, protein turnover, and nuclear localization (Budde and Chollet, 1988). The expression of a considerable number of genes encoding kinases and phosphatases is under the control of the circadian clock (Kusakina and Dodd, 2012). The first studies connecting phosphorylation with the Arabidopsis circadian system came from a yeast two-hybrid screening that identified the Ser/Thr protein kinase CK2 (formerly CASEIN KINASE2) as an interacting partner of CCA1 (Sugano et al., 1998). In Arabidopsis, the CK2 holoenzyme comprises two catalytic α-subunits and two regulatory β-subunits, forming a α2β2 tetramer (Pinna, 2002; Salinas et al., 2006). While the α-subunits have catalytic activity and are critical for phosphorylation, the β-subunits enhance the catalytic activity and define substrate specificity (Sugano et al., 1998). Thus, the Arabidopsis CK2 β-subunits interact with and facilitate the phosphorylation of CCA1 and LHY (Sugano et al., 1998, 1999; Daniel et al., 2004).
Several lines of evidence suggest that CK2-mediated phosphorylation antagonistically regulates CCA1 transcriptional activity (Portolés and Más, 2010). The dephosphorylated CCA1 protein is preferentially bound to the promoters of its target clock genes. Consistently, the Arabidopsis ckα1α2α3 triple mutants, which have both reduced CK2 kinase activity and CCA1 phosphorylation, lengthen the circadian period in a similar fashion to that observed in CCA1-overexpressing plants (Lu et al., 2011b). By contrast, transgenic plants overexpressing either CK2 β-SUBUNIT3 (CKB3) or CKB4, which show enhanced CK2 activity, exhibit a shortened period of expression similar to the phenotype of cca1 mutant plants (Sugano et al., 1998, 1999; Perales et al., 2006). The phosphorylation state of CK2 might be important in the modulation of CCA1 and LHY phosphorylation. Indeed, the regulatory subunit CKB4 is also phosphorylated and the CKB4 hyperphosphorylated isoforms are more susceptible to ubiquitination and degradation through the proteasome pathway (Perales et al., 2006). Degradation of CKB4 preferentially occurs during the day and is under the control of the circadian clock (Perales et al., 2006). These results are in agreement with a previous observation showing that CK2 activity is reduced during the light period (Hardtke et al., 2000).
Insights about the biological relevance of CK2 and CCA1 interaction in clock function were provided in a recent study. The study shows that CK2 phosphorylation does not affect CCA1 protein accumulation or subcellular localization but interferes with CCA1 binding activity to the promoters of the oscillator genes. High temperature enhances both CCA1 binding and CK2 phosphorylation. This parallel regulation in opposite directions generates a balance that contributes to maintaining a stable period across a physiological range of temperatures, a clock property known as temperature compensation. Therefore, two counterbalanced and temperature-dependent activities (CCA1 and CK2) underlie, at least in part, the mechanism behind clock temperature compensation in Arabidopsis (Portolés and Más, 2010).
TOC1 and other PRRs are also phosphorylated in a time of day–dependent manner, although the specific kinases responsible for this phosphorylation remain elusive (Fujiwara et al., 2008). Overall, phosphorylation of PRRs affects their protein–protein interactive networks and makes the proteins more susceptible to degradation (Fujiwara et al., 2008). This is illustrated by the interaction of TOC1 and PRR5 with ZTL, whereby the binding affinity of TOC1 and PRR5 to ZTL is enhanced following TOC1 and PRR5 phosphorylation (Fujiwara et al., 2008). On the other hand, TOC1 phosphorylation also contributes to its stabilization. Phosphorylation of both TOC1 and PRR3 also boosts their interaction (Fujiwara et al., 2008). As PRR3 and ZTL interact with TOC1 through the same region at the TOC1 N terminus, PRR3 competes with ZTL for the interaction with TOC1 and thereby relieves TOC1 from the ZTL-dependent degradation (Para et al., 2007; Fujiwara et al., 2008). PRR5 interacts with TOC1 regardless of their phosphorylation status but the interaction favors the phosphorylation of TOC1 (Wang et al., 2010). Phosphorylation of TOC1 triggers its nuclear localization (Wang et al., 2010), preventing the cytoplasmic ZTL-dependent degradation (Kim et al., 2007). Altogether, the studies indicate a dual effect of TOC1 phosphorylation on its stability. Phosphorylation of TOC1 not only facilitates its protein degradation by enhancing the interaction with ZTL but also stabilizes it through competitive inhibition by PRR3 and nuclear sequestration by PRR5 (Figure 1). Complete characterization of this dual mode of regulation of TOC1 protein phosphorylation is still lacking.
Protein–Protein Interaction
Protein–protein interaction networks are also critical for regulation of circadian clock function. Dynamic dimer formation of central clock components is an important way to ensure proper circadian oscillation. For instance, CCA1 and LHY contain a single MYB DNA binding domain, but at least two MYB domains are required for DNA binding (Jin and Martin, 1999). Hence, CCA1 and LHY form homo and heterodimers in the nucleus (Lu et al., 2009; Yakir et al., 2009). Although the molecular and biochemical functions of the dimers have not yet been described in detail, it seems likely that the interactions may affect nuclear localization, transcriptional activity, DNA binding affinity and specificity, and protein complex stability, which clearly diversify their regulatory schemes at the basis of their circadian function.
The protein–protein interaction network buildup from CCA1 and LHY further extends the repertoires of their circadian control. In addition to the role in the ubiquitination pathway, DET1 also acts as a transcriptional corepressor together with CCA1 and LHY (Lau et al., 2011). By interacting with CCA1 and LHY, DET1 localizes to the promoters of their target genes and represses their expression (Lau et al., 2011). Consistently, binding of DET1 to gene promoters is substantially diminished in cca1 lhy double mutant plants. The det1-1 mutant shows no alterations in CCA1 and LHY protein abundance but displays a shortened period of TOC1 and GI expression, a similar phenotype observed in cca1 and lhy loss-of-function mutants. Furthermore, the circadian phenotypes of CCA1-ox transgenic plants are compromised in CCA1-ox/det1-1 plants, indicating that DET1 is required for proper function of CCA1.
CCA1 also bolsters circadian oscillation of clock output expression through additional protein–protein interaction networks. Indeed, CCA1 interacts with key regulators of light signaling, such as ELONGATED HYPOCOTYL5 (HY5), FAR RED-IMPAIRED RESPONSE1 (FAR1), and FAR RED-ELONGATED HYPOCOTYL3 (FHY3), which provide important crosstalk points between the clock and the light signaling pathways (Andronis et al., 2008; Li et al., 2011). Notably, CCA1 synergistically increases the DNA binding activity of HY5 on the LHCB1*1 promoter (Andronis et al., 2008), while CCA1 disrupts the transcriptional activating function of FHY3, HY5, and FAR1 on the ELF4 promoter through inhibition of their DNA binding in a time of day–specific manner (Li et al., 2011). Further experiments are required to decipher the mechanistic and molecular insights behind the differential modulation of transcriptional activity by CCA1.
At the core of the evening oscillator, ELF3, ELF4, and LUX are known to be important for sustaining circadian oscillation. It has been demonstrated that the three proteins function as transcriptional repressors through the formation of the EC that is diurnally regulated and peaks at dusk (Nusinow et al., 2011; Herrero et al., 2012). LUX is a bona fide DNA binding transcription factor, while ELF3 and ELF4 are plant-specific nuclear proteins with no known functional domains. ELF3 seems to provide the basic platform of the complex and interacts independently both with ELF4 and LUX (Nusinow et al., 2011; Herrero et al., 2012). The EC is associated with the promoters of PRR9, PIF4, and PIF5 (Helfer et al., 2011; Nusinow et al., 2011), which is relevant for circadian rhythms and phase-dependent gating of growth and development.
A recent study has shown that TOC1 is associated with the promoters of nearly all the oscillator genes to repress their expression (Huang et al., 2012). TOC1 directly binds to DNA through its conserved CCT domain (for CONSTANS, CONSTANS-like, TOC1) (Gendron et al., 2012). The regulation specificity of TOC1 to target genes seems to be determined by time-of-day interactions with other regulatory proteins. For example, regulation of CCA1 expression might be facilitated by TOC1 interaction with CHE, a transcription factor from the TCP (for TEOSINTE BRANCHED1, CYCLOIDEA, and PCFs) family (Pruneda-Paz et al., 2009). CHE directly binds to CCA1 promoter through the TCP binding site and represses its expression. CCA1 and LHY reciprocally regulate CHE expression, forming a transcriptional feedback loop (Pruneda-Paz et al., 2009).
In addition to its interaction with TOC1 and PRR5, ZTL also interacts with the putative transcription factor EARLY BIRD (EBI). EBI-deficient mutant plants show defects in circadian rhythmicity with an advanced phase in the expression of clock genes, a period of shortening, and an early flowering (Johansson et al., 2011). Notably, the interaction of EBI with ZTL does not lead to EBI protein degradation; rather, ZTL regulates the transcriptional activity of EBI in a time-dependent manner. Thus, different modes of action characterize ZTL role on the circadian clock.
Subcellular Compartmentalization
Some clock components are localized both in the nucleus and in the cytoplasm. The subcellular compartmentalization of clock proteins might provide an efficient way to regulate circadian oscillation. Subcellular compartmentalization as a circadian regulatory event is also complemented with other regulatory mechanisms, as exemplified by the PRR5–TOC1 interaction (Wang et al., 2010).
The function of some clock factors such as ELF4 seems to be related to the translocation of several clock components to the nucleus. Indeed, on one hand, ELF4 recruits ELF3 in the nucleus, which leads to nuclear accumulation and nuclear body formation (Chow et al., 2012; Herrero et al., 2012). Concomitantly, ELF4 interaction with ELF3 facilitates their transcriptional repressive action as a component of EC (Herrero et al., 2012). On the other hand, ELF4 is also involved in the nuclear compartmentalization of GI. GI is expressed both in the nucleus and in the cytoplasm, but its nuclear localization is modulated in part by ELF4 (Kim et al., 2013b). In the nucleus, GI forms nuclear bodies, and ELF4 is required for this process. The ELF4 sequestration of GI from the nucleoplasm provides a mechanism for retaining GI activity without exhaustion.
The biological relevance of the nucleocytoplasmic distribution of GI has been further investigated. Nuclear and cytoplasmic localization of GI have different roles in regulating LHY expression (Kim et al., 2013a). Nuclear GI activates LHY expression, whereas cytoplasmic GI delays the induction kinetics of LHY, forming an incoherent feed-forward loop. Notably, robust rhythms of LHY expression require the coordinated action of nuclear and cytoplasmic GI, which demonstrate that spatial partitioning is a regulatory event that enhances the robustness of the clock.
In addition, the dynamic interaction of clock components with nuclear proteins may facilitate their nuclear sequestration. For example, the LKP2 protein is clearly transported to the nucleus and forms nuclear bodies when coexpressed with the flowering-related components CONSTANS or CONSTANS-LIKE1 (Fukamatsu et al., 2005). Although there are only few examples, further studies would confirm whether this regulatory scheme is important for clock control.
POSTTRANSLATIONAL MODIFICATIONS OF HISTONES
Chromatin architecture modulates the accessibility of transcriptional regulatory proteins and thereby dynamically alters gene expression in response to developmental and environmental cues (Pfluger and Wagner, 2007). Multiple chemical and reversible modifications regulate chromatin activity and function. The modifications include, among others, DNA methylation/demethylation by DNA methyltransferases and methylcytosine DNA glycosylases, histone acetylation/deacetylation by histone acetyltransferases (HATs) and histone deacetylases (HDACs), histone methylation/demethylation by histone methyltransferases and histone demethylases, and histone variant exchange (Pandey et al., 2002; Pfluger and Wagner, 2007; Chen et al., 2011). According to structural and functional analyses, a number of chromatin remodeling factors have been identified in Arabidopsis. Some of them have been shown to be implicated in plant growth and development, floral transition, cellular differentiation, and genomic imprinting (Chaudhury and Berger, 2001; Baroux et al., 2002; Berger and Gaudin, 2003; Jarillo et al., 2009; Zografos and Sung, 2012).
Recent studies have also shown that changes in chromatin architecture also modulate circadian function (Table 1). Indeed, the pattern of histone acetylation at the TOC1 promoter follows a circadian oscillation that is closely associated with TOC1 rhythmic expression (Perales and Más, 2007). At dawn, TOC1 expression is repressed, and this repression is concomitant with CCA1 binding to the TOC1 promoter. Circadian-regulated binding of CCA1 antagonizes H3 acetylation most likely by blocking HAT accessibility (Stratmann and Más, 2008). As CCA1 binding decreases during the day, a yet to be identified HAT is recruited to the TOC1 promoter and thereby TOC1 is transcriptionally derepressed. The declining phase of TOC1 is facilitated by HDAC activities at the light-to-dark transition (Perales and Más, 2007). The HDACs lead to histone H3 hypoacetylation that favors a repressive chromatin state at the TOC1 promoter.
Table 1. Posttranslational Regulation of Arabidopsis Clock Proteins.
| Core Clock Component | Epigenetic Regulators | Phosphorylation | Ubiquitination Degradation | Protein–Protein Interaction |
|---|---|---|---|---|
| CCA1 | SDG2/ATXR3, JMJ30/JMJD5, TPL-HDA6 | CK2 | CK2, CCA1, LHY, DET1, FHY3 | |
| LHY | SDG2/ATXR3, JMJ30/JMJD5, TPL-HDA6 | CK2 | SINAT5 | CK2, CCA1, LHY, DET1 |
| TOC1 | CCA1, LCL5, SDG2/ATXR3 | Unknown | ZTL, FKF1, LKP2 | PRR3, PRR5, CHE, ZTL, FKF1, LKP2 |
| PRR3 | Unknown | TOC1 | ||
| PRR5 | Unknown | ZTL, FKF1, LKP2 | TOC1, ZTL, FKF1, LKP2, TPL | |
| PRR7 | SDG2/ATXR3 | TPL | ||
| PRR9 | SDG2/ATXR3 | TPL | ||
| GI | COP1 | ELF3, COP1, HSP90, ZTL, FKF1, LKP2 | ||
| CHE | TOC1 | |||
| ELF3 | ELF4, LUX, COP1, GI | |||
| ELF4 | ELF3, LUX | |||
| LUX | SDG2/ATXR3 | ELF3, ELF4 |
The molecular components responsible for the different posttranslational regulatory mechanisms at the core of the clock are listed. In some instances, the specific molecular components responsible for the modification remain to be discovered (unknown).
Dynamic changes in chromatin structure at the TOC1 promoter are further regulated by another morning-expressed MYB transcription factor, REVEILLE8/LHY-CCA1-LIKE5 (RVE8/LCL5) (Farinas and Mas, 2011). Despite their structural similarities, the molecular function of CCA1 and RVE8/LCL5 markedly differs. Although both RVE8/LCL5 and CCA1 bind to the TOC1 promoter, CCA1 favors histone hypoacetylation, while RVE8/LCL5 leads to H3 hyperacetylation at the TOC1 promoter. Overexpression of RVE8/LCL5 results in a short circadian period with an advanced rising phase of TOC1 expression, which coincides with increased H3 acetylation (Farinas and Mas, 2011). The opposite phenotypes for period, phase, and histone acetylation are observed in rve8/lcl5 loss-of-function mutant. These results indicate that RVE8/LCL5 acts during the rising phase of TOC1 by facilitating histone acetylation and thus counterbalancing the repressing activity of CCA1 (Farinas and Mas, 2011). Following TOC1 peak of expression, the relevant HDAC activities at the TOC1 promoter interfere with and antagonize RVE8/LCL5 function, thus contributing to the formation of repressive chromatin structures that lead to the declining phase of TOC1.
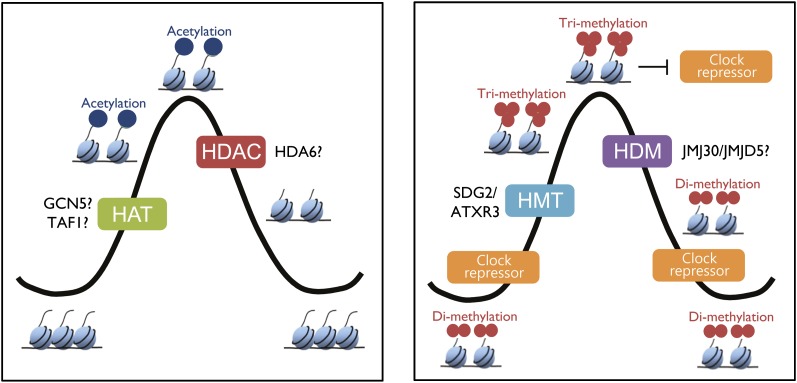
Regulation of circadian expression by oscillating histone marks are not exclusive of TOC1 but also pervades other oscillator genes (Hemmes et al., 2012; Malapeira et al., 2012; Song and Noh, 2012). Indeed, histone acetylation (H3K56ac and H3K9/14ac) and methylation (H3K4me3) closely correlate with the rhythmic expression of LHY, CCA1, and TOC1 (Hemmes et al., 2012; Malapeira et al., 2012; Song and Noh, 2012) as well as PRR9, PRR7, and LUX (Malapeira et al., 2012). As inferred by their rhythmic phase and by the results obtained following treatment with specific inhibitors, histone acetylation (H3ac) and methylation (H3K4me3) seem to be active marks promoting the rhythmic activation of the oscillator genes (Malapeira et al., 2012). However, histone acetylation and methylation are not fully redundant activating marks. Histone acetylation contributes to the circadian peak of expression, while H3K4me3 regulates clock repressor binding, ensuring a proper timing and duration of gene activation (Malapeira et al., 2012) (Figure 2).
Figure 2.
Epigenetic Regulation at the Core of the Arabidopsis Circadian Oscillator.
H3K56ac, H3K9/14ac, H3K4me3, and H3K4me2 are representative oscillating epigenetic marks that correlate with and contribute to the rhythmic expression of the core clock genes. The timing of histone acetylation regulates gene expression by influencing transcription factor accessibility, whereas histone trimethylation antagonizes clock repressor binding. The molecular components responsible for the reversible histone acetylation and demethylation are not known. SDG2/ATXR3 is responsible for histone trimethylation at the core of the Arabidopsis circadian clock. HMT, histone methyltransferase; HDM, histone demethylase.
The molecular components responsible for histone modifications are just beginning to emerge. For instance, H3K4me3 accumulation at the oscillator gene promoters is regulated by the HMT SET DOMAIN GROUP2/ARABIDOPSIS TRITHORAX-RELATED3 (SDG2/ATXR3) (Malapeira et al., 2012). The SDG2/ATXR3-deficient mutants globally decrease H3K4me3 in the Arabidopsis genome. Decreased H3K4me3 accumulation correlates with reduced oscillator gene expression in sdg2/atxr3 mutant plants (Malapeira et al., 2012). Consistent with the role of H3K4me3 regulating repressor activity, in sdg2/atxr3 mutant plants, the timing of clock repressor binding is affected. Altogether, these results support a direct function of histone marks in fine-tuning the shape of the circadian waveforms at the core of the clock.
Regarding the possible components involved in histone acetylation-deacetylation, it was recently shown that the TOPLESS/TOPLESS RELATED PROTEIN (TPL/TPR) members of the Groucho/Tup1 family interact with the PRRs and repress transcription. TPL also associates with HISTONE DEACETYLASE6 (HDA6) to repress circadian gene expression (Wang et al., 2013). HATs involved in photomorphogenesis, such as TATA BINDING PROTEIN-ASSCIATED FACTOR1 and GENERAL CONTROL NONREPRESSED5, are plausible candidates controlling chromatin-dependent circadian clock oscillation (Stratmann and Más, 2008) (Figure 2). Further studies are required to examine these hypotheses.
Another component recently found to be involved in regulating histone marks is the E3 ligase HISTONE MONOUBIQUITINATION1 (HUB1) (Himanen et al., 2012b). This protein controls histone H2B monoubiquitination, a modification that does not entail protein degradation. H2B monoubiquitination is associated with H3K4me3 accumulation at the gene coding regions (Sridhar et al., 2007; Cao et al., 2008) to facilitate transcriptional elongation. Transcriptomic analysis of HUB1 misexpressing lines showed that a number of circadian clock genes are targets of HUB1 in Arabidopsis. The amplitude of circadian gene expression is affected in the hub1-1 mutant plants. This alteration coincides with reduced monoubiquitination of histone H2B at their coding regions (Himanen et al., 2012b) and with altered plant fitness (Himanen et al., 2012a).
Jumonji C domain–containing proteins that are known as histone demethylases are also involved in circadian control. The expression of JMJ30/JMJD5 displays a robust circadian regulation with a peak at dusk (Jones et al., 2010; Lu et al., 2011a). The core oscillators CCA1 and LHY repress JMJ30/JMJD5 expression by directly binding to its promoter. In turn, JMJ30/JMJD5 promotes expression of CCA1 and LHY, presumably through histone demethylase activity (Jones et al., 2010). Consistently, jmj30/jmjd5 loss-of-function mutants shortened the circadian period in the expression of clock genes. The Arabidopsis and human JMJ30/JMJD5 orthologs rescue the circadian phenotypes of the mutants in the reciprocal organism (Jones et al., 2010; Jones and Harmer, 2011), which suggests a common function of JMJ30/JMJD5 in plants and animals.
CONCLUDING REMARKS
The circadian clock enables plants to match biological processes with the most appropriate time of day, thus conferring a fitness advantage. Multiple regulatory layers underlie both the fine-tuning of circadian oscillation and synchronization of internal physiology with the changing environment. Core clock genes are regulated through a diverse array of mechanisms, which ensure fully functional activities connected with plant physiology and development. However, we are still far from a comprehensive view of the higher order architectural regulation underlying the circadian interactive networks. Identification of novel clock components and their actual biochemical function will have a substantial impact in the circadian research field. The complex layers of regulation also involve a broad range of connections between internal and external signals that once incorporated into the circadian system provide important nodes of crosstalk with other relevant plant pathways. Given that circadian rhythms govern many physiological processes in plants, future research in this area would contribute to precisely define the maps of physiology and metabolism in Arabidopsis.
Acknowledgments
Research in the laboratory of P.J.S. is supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (NRF-2013R1A1A1004831). Research in P.M.’s lab is supported by the Ramón Areces Foundation, the Spanish Ministry of Science and Innovation, and the European Young Investigator Award through the European Science Foundation.
AUTHOR CONTRIBUTIONS
Both authors contributed equally to writing the article.
References
- Adams S., Carré I.A. (2011). Downstream of the plant circadian clock: Output pathways for the control of physiology and development. Essays Biochem. 49: 53–69 [DOI] [PubMed] [Google Scholar]
- Andronis C., Barak S., Knowles S.M., Sugano S., Tobin E.M. (2008). The clock protein CCA1 and the bZIP transcription factor HY5 physically interact to regulate gene expression in Arabidopsis. Mol. Plant 1: 58–67 [DOI] [PubMed] [Google Scholar]
- Alabadí D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Baroux C., Spillane C., Grossniklaus U. (2002). Genomic imprinting during seed development. Adv. Genet. 46: 165–214 [DOI] [PubMed] [Google Scholar]
- Baudry A., Ito S., Song Y.H., Strait A.A., Kiba T., Lu S., Henriques R., Pruneda-Paz J.L., Chua N.H., Tobin E.M., Kay S.A., Imaizumi T. (2010). F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22: 606–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F., Gaudin V. (2003). Chromatin dynamics and Arabidopsis development. Chromosome Res. 11: 277–304 [DOI] [PubMed] [Google Scholar]
- Budde R.J.A., Chollet R. (1988). Regulation of enzyme activity in plants by reversible phosphorylation. Physiol. Plant. 72: 435–439 [Google Scholar]
- Cao Y., Dai Y., Cui S., Ma L. (2008). Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 20: 2586–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carré I., Veflingstad S.R. (2013). Emerging design principles in the Arabidopsis circadian clock. Semin. Cell Dev. Biol. 24: 393–398 [DOI] [PubMed] [Google Scholar]
- Chaudhury A.M., Berger F. (2001). Maternal control of seed development. Semin. Cell Dev. Biol. 12: 381–386 [DOI] [PubMed] [Google Scholar]
- Chen X., Hu Y., Zhou D.X. (2011). Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochim. Biophys. Acta 1809: 421–426 [DOI] [PubMed] [Google Scholar]
- Chow B.Y., Helfer A., Nusinow D.A., Kay S.A. (2012). ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signal. Behav. 7: 170–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Lu F., Li Y., Xue Y., Kang Y., Zhang S., Qiu Q., Cui X., Zheng S., Liu B., Xu X., Cao X. (2013). Ubiquitin-specific proteases UBP12 and UBP13 act in circadian clock and photoperiodic flowering regulation in Arabidopsis. Plant Physiol. 162: 897–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel X., Sugano S., Tobin E.M. (2004). CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A.N., Salathia N., Hall A., Kévei E., Tóth R., Nagy F., Hibberd J.M., Millar A.J., Webb A.A. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633 [DOI] [PubMed] [Google Scholar]
- Doelling J.H., Phillips A.R., Soyler-Ogretim G., Wise J., Chandler J., Callis J., Otegui M.S., Vierstra R.D. (2007). The ubiquitin-specific protease subfamily UBP3/UBP4 is essential for pollen development and transmission in Arabidopsis. Plant Physiol. 145: 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J.H., Yan N., Kurepa J., Walker J., Vierstra R.D. (2001). The ubiquitin-specific protease UBP14 is essential for early embryo development in Arabidopsis thaliana. Plant J. 27: 393–405 [DOI] [PubMed] [Google Scholar]
- Doyle M.R., Davis S.J., Bastow R.M., McWatters H.G., Kozma-Bognár L., Nagy F., Millar A.J., Amasino R.M. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77 [DOI] [PubMed] [Google Scholar]
- Farinas B., Mas P. (2011). Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 66: 318–329 [DOI] [PubMed] [Google Scholar]
- Farré E.M., Harmer S.L., Harmon F.G., Yanovsky M.J., Kay S.A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Fujiwara S., Wang L., Han L., Suh S.S., Salomé P.A., McClung C.R., Somers D.E. (2008). Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 283: 23073–23083 [DOI] [PubMed] [Google Scholar]
- Fukamatsu Y., Mitsui S., Yasuhara M., Tokioka Y., Ihara N., Fujita S., Kiyosue T. (2005). Identification of LOV KELCH PROTEIN2 (LKP2)-interacting factors that can recruit LKP2 to nuclear bodies. Plant Cell Physiol. 46: 1340–1349 [DOI] [PubMed] [Google Scholar]
- Gendron J.M., Pruneda-Paz J.L., Doherty C.J., Gross A.M., Kang S.E., Kay S.A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.M., Tingay S., Wang Z.Y., Tobin E.M. (2002). Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 129: 576–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Mason M., Risseeuw E.P., Crosby W.L., Somers D.E. (2004). Formation of an SCF(ZTL) complex is required for proper regulation of circadian timing. Plant J. 40: 291–301 [DOI] [PubMed] [Google Scholar]
- Hardtke C.S., Gohda K., Osterlund M.T., Oyama T., Okada K., Deng X.W. (2000). HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19: 4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S.L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Harmon F., Imaizumi T., Gray W.M. (2008). CUL1 regulates TOC1 protein stability in the Arabidopsis circadian clock. Plant J. 55: 568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon M.J., Hearn T.J., Bell L.J., Hannah M.A., Webb A.A. (2013). Metabolic regulation of circadian clocks. Semin. Cell Dev. Biol. 24: 414–421 [DOI] [PubMed] [Google Scholar]
- Helfer A., Nusinow D.A., Chow B.Y., Gehrke A.R., Bulyk M.L., Kay S.A. (2011). LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 21: 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmes H., Henriques R., Jang I.C., Kim S., Chua N.H. (2012). Circadian clock regulates dynamic chromatin modifications associated with Arabidopsis CCA1/LHY and TOC1 transcriptional rhythms. Plant Cell Physiol. 53: 2016–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E., et al. (2012). EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K., Boccardi T.M., De Rycke R., Odeny O.P., Van Lijsebettens M. (2012a). Is HUB1 a hub for plant fitness? Plant Signal. Behav. 7: 1537–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K., Woloszynska M., Boccardi T.M., De Groeve S., Nelissen H., Bruno L., Vuylsteke M., Van Lijsebettens M. (2012b). Histone H2B monoubiquitination is required to reach maximal transcript levels of circadian clock genes in Arabidopsis. Plant J. 72: 249–260 [DOI] [PubMed] [Google Scholar]
- Huang W., Pérez-García P., Pokhilko A., Millar A.J., Antoshechkin I., Riechmann J.L., Mas P. (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79 [DOI] [PubMed] [Google Scholar]
- Imaizumi T., Tran H.G., Swartz T.E., Briggs W.R., Kay S.A. (2003). FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426: 302–306 [DOI] [PubMed] [Google Scholar]
- Ito S., Niwa Y., Nakamichi N., Kawamura H., Yamashino T., Mizuno T. (2008). Insight into missing genetic links between two evening-expressed pseudo-response regulator genes TOC1 and PRR5 in the circadian clock-controlled circuitry in Arabidopsis thaliana. Plant Cell Physiol. 49: 201–213 [DOI] [PubMed] [Google Scholar]
- Ito S., Song Y.H., Imaizumi T. (2012). LOV domain-containing F-box proteins: Light-dependent protein degradation modules in Arabidopsis. Mol. Plant 5: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo J.A., Piñeiro M., Cubas P., Martínez-Zapater J.M. (2009). Chromatin remodeling in plant development. Int. J. Dev. Biol. 53: 1581–1596 [DOI] [PubMed] [Google Scholar]
- Jin H., Martin C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41: 577–585 [DOI] [PubMed] [Google Scholar]
- Johansson M., McWatters H.G., Bakó L., Takata N., Gyula P., Hall A., Somers D.E., Millar A.J., Eriksson M.E. (2011). Partners in time: EARLY BIRD associates with ZEITLUPE and regulates the speed of the Arabidopsis clock. Plant Physiol. 155: 2108–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.A., Covington M.F., DiTacchio L., Vollmers C., Panda S., Harmer S.L. (2010). Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc. Natl. Acad. Sci. USA 107: 21623–21628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.A., Harmer S. (2011). JMJD5 Functions in concert with TOC1 in the Arabidopsis circadian system. Plant Signal. Behav. 6: 445–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T., Henriques R., Sakakibara H., Chua N.H. (2007). Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19: 2516–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.S., Kim W.Y., Fujiwara S., Kim J., Cha J.Y., Park J.H., Lee S.Y., Somers D.E. (2011). HSP90 functions in the circadian clock through stabilization of the client F-box protein ZEITLUPE. Proc. Natl. Acad. Sci. USA 108: 16843–16848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.Y., Fujiwara S., Suh S.S., Kim J., Kim Y., Han L., David K., Putterill J., Nam H.G., Somers D.E. (2007). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360 [DOI] [PubMed] [Google Scholar]
- Kim W.Y., Geng R., Somers D.E. (2003). Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc. Natl. Acad. Sci. USA 100: 4933–4938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Han S., Yeom M., Kim H., Lim J., Cha J.Y., Kim W.Y., Somers D.E., Putterill J., Nam H.G., Hwang D. (2013a). Balanced nucleocytosolic partitioning defines a spatial network to coordinate circadian physiology in plants. Dev. Cell 26: 73–85 [DOI] [PubMed] [Google Scholar]
- Kim Y., Lim J., Yeom M., Kim H., Kim J., Wang L., Kim W.Y., Somers D.E., Nam H.G. (2013b). ELF4 regulates GIGANTEA chromatin access through subnuclear sequestration. Cell Rep. 3: 671–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinmonth-Schultz H.A., Golembeski G.S., Imaizumi T. (2013). Circadian clock-regulated physiological outputs: Dynamic responses in nature. Semin. Cell Dev. Biol. 24: 407–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakina J., Dodd A.N. (2012). Phosphorylation in the plant circadian system. Trends Plant Sci. 17: 575–583 [DOI] [PubMed] [Google Scholar]
- Lau O.S., Huang X., Charron J.B., Lee J.H., Li G., Deng X.W. (2011). Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol. Cell 43: 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Kim W.T. (2011). Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol. Cells 31: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., et al. (2011). Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat. Cell Biol. 13: 616–622 [DOI] [PubMed] [Google Scholar]
- Locke J.C., Kozma-Bognár L., Gould P.D., Fehér B., Kevei E., Nagy F., Turner M.S., Hall A., Millar A.J. (2006). Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol. Syst. Biol. 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.X., Knowles S.M., Andronis C., Ong M.S., Tobin E.M. (2009). CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 150: 834–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.X., Knowles S.M., Webb C.J., Celaya R.B., Cha C., Siu J.P., Tobin E.M. (2011a). The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 155: 906–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.X., Liu H., Knowles S.M., Li J., Ma L., Tobin E.M., Lin C. (2011b). A role for protein kinase casein kinase2 α-subunits in the Arabidopsis circadian clock. Plant Physiol. 157: 1537–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Matsushika A., Kojima M., Yamashino T., Mizuno T. (2002). The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol. 43: 58–69 [DOI] [PubMed] [Google Scholar]
- Malapeira J., Khaitova L.C., Mas P. (2012). Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 109: 21540–21545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P., Kim W.Y., Somers D.E., Kay S.A. (2003). Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- Más P., Yanovsky M.J. (2009). Time for circadian rhythms: Plants get synchronized. Curr. Opin. Plant Biol. 12: 574–579 [DOI] [PubMed] [Google Scholar]
- Mazzucotelli E., Belloni S., Marone D., De Leonardis A., Guerra D., Di Fonzo N., Cattivelli L., Mastrangelo A. (2006). The e3 ubiquitin ligase gene family in plants: regulation by degradation. Curr. Genomics 7: 509–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel D.H., Kay S.A. (2012). Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 22: R648–R657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kiba T., Henriques R., Mizuno T., Chua N.H., Sakakibara H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y., Yamashino T., Mizuno T. (2009). The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol. 50: 838–854 [DOI] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farré E.M., Kay S.A. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R., Müller A., Napoli C.A., Selinger D.A., Pikaard C.S., Richards E.J., Bender J., Mount D.W., Jorgensen R.A. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30: 5036–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Para A., Farré E.M., Imaizumi T., Pruneda-Paz J.L., Harmon F.G., Kay S.A. (2007). PRR3 Is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19: 3462–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B.S., Eo H.J., Jang I.C., Kang H.G., Song J.T., Seo H.S. (2010). Ubiquitination of LHY by SINAT5 regulates flowering time and is inhibited by DET1. Biochem. Biophys. Res. Commun. 398: 242–246 [DOI] [PubMed] [Google Scholar]
- Perales M., Más P. (2007). A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 19: 2111–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M., Portolés S., Más P. (2006). The proteasome-dependent degradation of CKB4 is regulated by the Arabidopsis biological clock. Plant J. 46: 849–860 [DOI] [PubMed] [Google Scholar]
- Pfluger J., Wagner D. (2007). Histone modifications and dynamic regulation of genome accessibility in plants. Curr. Opin. Plant Biol. 10: 645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinna L.A. (2002). Protein kinase CK2: A challenge to canons. J. Cell Sci. 115: 3873–3878 [DOI] [PubMed] [Google Scholar]
- Portolés S., Más P. (2010). The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet. 6: e1001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Para A., Kay S.A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resco V., Hartwell J., Hall A. (2009). Ecological implications of plants ability to tell the time. Ecol. Lett. 12: 583–592 [DOI] [PubMed] [Google Scholar]
- Sadanandom A., Bailey M., Ewan R., Lee J., Nelis S. (2012). The ubiquitin-proteasome system: central modifier of plant signalling. New Phytol. 196: 13–28 [DOI] [PubMed] [Google Scholar]
- Salinas P., Fuentes D., Vidal E., Jordana X., Echeverria M., Holuigue L. (2006). An extensive survey of CK2 alpha and beta subunits in Arabidopsis: Multiple isoforms exhibit differential subcellular localization. Plant Cell Physiol. 47: 1295–1308 [DOI] [PubMed] [Google Scholar]
- Sanchez S.E., Petrillo E., Kornblihtt A.R., Yanovsky M.J. (2011). Alternative splicing at the right time. RNA Biol. 8: 954–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R., Ramsay N., Samach A., Corden S., Putterill J., Carré I.A., Coupland G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Song H.R., Carré I.A. (2005). DET1 regulates the proteasomal degradation of LHY, a component of the Arabidopsis circadian clock. Plant Mol. Biol. 57: 761–771 [DOI] [PubMed] [Google Scholar]
- Song H.R., Noh Y.S. (2012). Rhythmic oscillation of histone acetylation and methylation at the Arabidopsis central clock loci. Mol. Cells 34: 279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar V.V., Kapoor A., Zhang K., Zhu J., Zhou T., Hasegawa P.M., Bressan R.A., Zhu J.K. (2007). Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 447: 735–738 [DOI] [PubMed] [Google Scholar]
- Stratmann T., Más P. (2008). Chromatin, photoperiod and the Arabidopsis circadian clock: A question of time. Semin. Cell Dev. Biol. 19: 554–559 [DOI] [PubMed] [Google Scholar]
- Strayer C., Oyama T., Schultz T.F., Raman R., Somers D.E., Más P., Panda S., Kreps J.A., Kay S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Sugano S., Andronis C., Green R.M., Wang Z.Y., Tobin E.M. (1998). Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc. Natl. Acad. Sci. USA 95: 11020–11025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S., Andronis C., Ong M.S., Green R.M., Tobin E.M. (1999). The protein kinase CK2 is involved in regulation of circadian rhythms in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 12362–12366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Fujiwara S., Somers D.E. (2010). PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 29: 1903–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Kim J., Somers D.E. (2013). Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. USA 110: 761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Tobin E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Yakir E., Hilman D., Kron I., Hassidim M., Melamed-Book N., Green R.M. (2009). Posttranslational regulation of CIRCADIAN CLOCK ASSOCIATED1 in the circadian oscillator of Arabidopsis. Plant Physiol. 150: 844–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerushalmi S., Green R.M. (2009). Evidence for the adaptive significance of circadian rhythms. Ecol. Lett. 12: 970–981 [DOI] [PubMed] [Google Scholar]
- Yu J.W., et al. (2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 32: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Li H., Li R., Hu R., Fan C., Chen F., Wang Z., Liu X., Fu Y., Lin C. (2008). Association of the circadian rhythmic expression of GmCRY1a with a latitudinal cline in photoperiodic flowering of soybean. Proc. Natl. Acad. Sci. USA 105: 21028–21033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zografos B.R., Sung S. (2012). Vernalization-mediated chromatin changes. J. Exp. Bot. 63: 4343–4348 [DOI] [PubMed] [Google Scholar]