Abstract
Background: Atherosclerotic vascular disease (AVD), a leading cause of morbidity and mortality, is increasing in prevalence in the developing world. We describe an approach to establish a biorepository linked to medical records with the eventual goal of facilitating discovery of biomarkers for AVD. Methods: The Vascular Disease Biorepository at Mayo Clinic was established to archive DNA, plasma, and serum from patients with suspected AVD. AVD phenotypes, relevant risk factors and comorbid conditions were ascertained by electronic medical record (EMR)-based electronic algorithms that included diagnosis and procedure codes, laboratory data and text searches to ascertain medication use. Results: Up to December 2012, 8800 patients referred for vascular ultrasound examination and non-invasive lower extremity arterial evaluation were approached, of whom 5268 consented. The mean age of the initial 2182 patients recruited was 70.4 ± 11.2 years, 62.6% were men and 97.6% were whites. The prevalences of AVD phenotypes were: carotid artery stenosis 48%, abdominal aortic aneurysm 21% and peripheral arterial disease 38%. Positive predictive values for electronic phenotyping algorithms were>0.90 for cases (and>0.95 for controls) for each AVD phenotype, using manual review of the EMR as the gold standard. The prevalences of risk factors and comorbidities were as follows: hypertension 78%, diabetes 29%, dyslipidemia 73%, smoking 70%, coronary heart disease 37%, heart failure 12%, cerebrovascular disease 20% and chronic kidney disease 19%. Conclusions: Our study demonstrates the feasibility of establishing a biorepository of plasma, serum and DNA, with relatively rapid annotation of clinical variables using EMR-based algorithms.
Keywords: atherosclerotic vascular disease, biorepository, electronic medical records, electronic phenotyping
Background
Atherosclerotic vascular disease (AVD) is a leading cause of mortality and morbidity worldwide. 1 Several circulating biomarkers and genetic variants have been reported to be associated with AVD in cohorts of European ancestry. 2 As AVD becomes increasingly prevalent in developing countries, there is an urgent need to identify biomarkers for early identification, prognostication and new drug development in diverse ethnic groups. Although significant progress has been made in identifying novel risk factors of coronary heart disease, little is known about genetic susceptibility variants and circulating biomarkers for peripheral vascular diseases1 – a group of diverse diseases characterized by atherosclerotic lesions in carotid arteries, aorta and lower extremity arteries. As a step towards identifying novel genetic and circulating biomarkers, we describe our approach to create an electronic medical record (EMR)-linked vascular disease-specific biorepository of DNA, plasma and serum. The biorepository includes patients with carotid artery stenosis (CAS), abdominal aortic aneurysm (AAA), and peripheral arterial disease (PAD), with linkage of biospecimens to clinical characteristics.
The EMR archives billing information, laboratory and imaging results, medications, and clinical documentation, thereby serving as a resource for genotype-phenotype association studies. A key issue we attempted to address was the feasibility and accuracy of capturing relevant clinical data using EMR-based phenotyping algorithms. Such algorithms have the potential to cost-effectively and efficiently ascertain phenotypes and relevant clinical covariates for conducting genomic studies 3–5 , whereas traditional manual review of medical records to ascertain clinical covariates can be time-consuming and expensive.
Methods
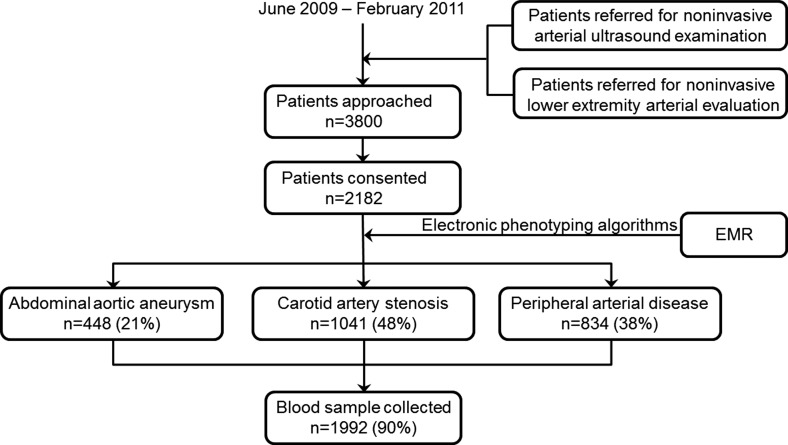
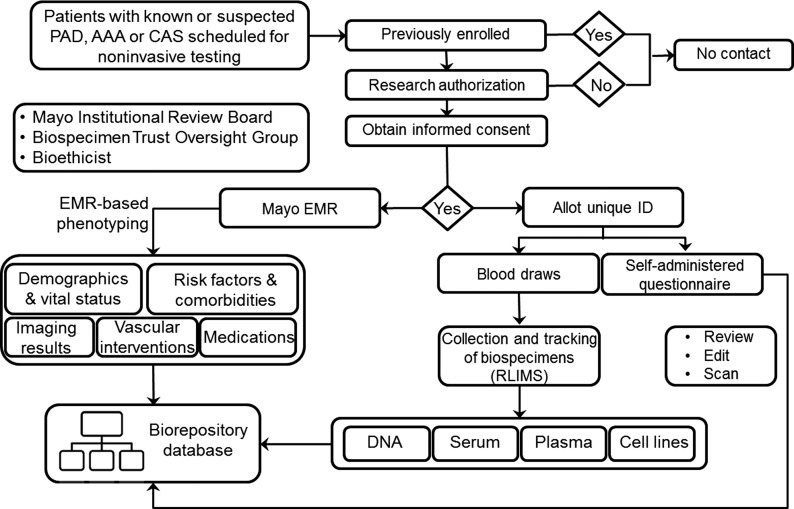
The study protocol was approved by the Institutional Review Board of the Mayo Clinic. Enrollment of patients and collection of biospecimens started in June 2009 and is still ongoing. The recruitment process is summarized in Figure 1 and the project infrastructure is depicted in Figure 2.
Figure 1. .
Pattern of recruitment for the Vascular Disease Biorepository. EMR = electronic medical record.
Figure 2. .
Overview of the Vascular Disease Biorepository. RLIMS = research laboratory information management systems; CAS = carotid artery stenosis; AAA = abdominal aortic aneurysm; PAD = peripheral arterial disease.
Participant recruitment
Consecutive adult patients with known or suspected CAS, AAA, or PAD, referred for non-invasive vascular ultrasound or lower extremity arterial evaluation, were approached for participation in the biorepository. Definitions of three AVD phenotypes for the study are listed in Table 1. All potential participants were checked against records of patients already enrolled or those who had refused research consent (Figure 1). The informed consent document (see supplement) conformed to the guidelines regarding Bioethics Resources and human subject research on National Institutes of Health Web (http://nih.gov/sigs/bioethics), and International Society of Biological and Environmental Biorepositories Web (http://www.isber.org).
Table 1 .
Criteria for ascertaining atherosclerotic vascular disease phenotypes.
| Carotid artery stenosis |
| (1) ≥ 40% stenosis in internal carotid artery/bulb (peak systolic velocity ≥ 150 cm/second) on either side evaluated with Doppler; OR 2) at least moderate atheromatous plaque in any of the following locations: common carotid artery, bulb, bifurcation or internal carotid artery of either side, or postoperative change of carotid endarterectomy or presence of stent in either side demonstrated by conventional, computed tomography or magnetic resonance angiography; OR 3) any procedure reports of carotid endarterectomy or stenting. |
| Abdominal aortic aneurysm |
| (1) Distal, infrarenal or juxtarenal abdominal aortic anteroposterior diameter ≥ 3 cm, measured with ultrasound, conventional or computed tomography or magnetic resonance angiography, or evidence of abdominal aortic aneurysm repair on imaging; OR 2) any procedure reports of open or endovascular abdominal aortic aneurysm repair; OR 3) abdominal aortic aneurysm documented in physician's note. |
| Peripheral artery disease |
| (1) Rest or 1 min post exercise ankle-brachial index (ABI) ≤ 0.9 or rest ABI ≥ 1.4 or lower extremity systolic BP ≥ 255 mm Hg in either leg; OR 2) at least moderate stenosis in lower extremity arteries in either side (distal to abdominal aortic bifurcation) on imaging; OR 3) postoperative change of lower extremity angioplasty, stenting, open vascular bypass or amputation on imaging or reports of these procedures for lower extremity arterial occlusive disease. |
The study coordinator described the objectives of the study, the risks and potential benefits of participation in the study, and the storage and future use of the samples. The consent form had separate check-off boxes seeking consent for biospecimens to be re-used or shared with collaborating investigators. Lack of immediate benefit for health, the potential to improve risk stratification of AVD, and the right to withdraw from the study any time after consent were specified. A questionnaire on sociodemographic information, cardiovascular health history, physical activity, lifestyle, past medical history and family history, was given to each participant at the time of consent. Once returned, the barcoded questionnaire was reviewed, scanned and added to the study database.
Collection, aliquoting, and storage of peripheral blood samples
Blood was collected in the fasting state whenever possible and the time from blood draw to storage was limited to < 1 h to minimize sample degradation; 52 ml of peripheral blood were drawn into the appropriate collection tubes, labeled with a Mayo-generated barcode ID number, and sent through a pneumatic tube system to the Biospecimens Accessioning and Processing (BAP) laboratory. Blood was centrifuged and EDTA plasma and serum were aliquoted into 0.5 ml tubes and stored in − 80°C freezers. DNA was extracted by Gentra AutoPure chemistries (Gentra systems Inc., Minneapolis, MN) from 5 ml of whole blood contained in EDTA and quantified by ultraviolet absorbance and quality control by 260/280 optical density ratio. In a subset of patients, lymphocytes were cryopreserved for future (see supplement).
Laboratory Information Management System
An in-house software system, Research Laboratory Information Management System (RLIMS), was used to record and monitor sample processing. All tubes and plates that contained an individual's samples were barcode-labeled by patient numbering program (PNP) and entered into the RLIMS. The program contains demographic information and an assigned study number to de-identify participants enrolled and track each one after recruitment. The unique number assigned to each subject is in no way related to his or her identity. The current PNP is web-based and contains a security layer and a logging mechanism for tracking by RLIMS. RLIMS allots unique IDs for all barcoded biospecimens including input (sample tubes) and output (DNA/plasma/white blood cell) tubes. Based on barcoding, RLIMS records the time biospecimens were received, the time of the DNA extraction and the quality of DNA. All pipetting was performed by robotic workstations that incorporate barcode scanners to track the transfer of the biological material from tube to tube, tube to plate, and plate to plate. Extensive integrity checks were made within the tracking system to reduce the risk for error.
Annotation of biospecimens with phenotype data
Broadly, there are two types of data in the EMR: codified data that can be abstracted directly including billing codes, demographics, and laboratory data; and narrative data in free-text format that can be mined by text searches using natural language processing (NLP). Electronic phenotyping algorithms were used to obtain patient characteristics including AVD phenotypes, conventional risk factors, comorbidities, and medication use. A federated warehouse of patient data – the Mayo Clinic Life Sciences Trust, derived from EMR data sources throughout the Mayo Clinic, was used to obtain relevant demographic and clinical data. It accommodates most EMR contents for>7 million patients, including highly annotated, full-text clinical notes, laboratory tests, diagnostic findings, demographics, and related clinical data. Since 1999, all medical records have been entered in this integrated EMR system. Billing codes, including International Classification of Disease (ICD) diagnosis and procedure codes version 9-CM, and Current Procedural Terminology (CPT) codes version 4, were used to obtain diagnoses and procedure information from Mayo's billing systems.
Demographics
Birth date, gender, race/ethnicity, and current residency, were mined directly from the EMR. The categories of self-reported race were “American Indian/Alaskan Native,” “Asian/Pacific Islander,” “Black,” “choose not to disclose,” “Native Hawaiian,” “other,” “Unknown,” and “white.” Current residency information included city, state, and zip code where patient currently resides. The geographic distribution of the enrolled patients was ascertained by zip codes.
Conventional risk factors for vascular disease
Hypertension, diabetes, dyslipidemia, and smoking status were ascertained by electronic phenotyping algorithms as previously described. 3 These algorithms were constructed based on laboratory test values, medications, and ICD-9-CM diagnosis codes. The time window for ICD-9-CM codes to ascertain relevant clinical covariates was any time before and up to 6 months after the enrollment, and for laboratory data, one year around enrollment. Plasma glucose, hemoglobin A1c, total and high-density lipoprotein cholesterol and triglyceride levels were extracted from the laboratory database. Resting systolic and diastolic blood pressure (BP) values were mined as structured observations from the vital signs section. Hypoglycemic agents or insulin, lipid-lowering and anti-hypertensive medications were ascertained by NLP from the current medications, admission medications and dismissal medications sections in clinical notes. Hypertension was defined as either systolic BP ≥ 140 mmHg or diastolic BP ≥ 90 mmHg at two serial measurements within 3 months closest to the enrollment, or a prior diagnosis of hypertension with use of antihypertensive medication. Diabetes was defined as fasting blood glucose ≥ 126 mg/dL, random glucose ≥ 200 mg/dL, hemoglobin A1c ≥ 6.5%, or a prior diagnosis with oral hypoglycemic or insulin therapy. Dyslipidemia was defined as total cholesterol ≥ 220 mg/dL, or high-density lipoprotein cholesterol ≤ 40 mg/dL in men or ≤ 45 mg/dL in women, triglycerides ≥ 200 mg/dL, or the use of lipid-lowing medications. Smoking status was ascertained by NLP as described previously 6 and smokers were defined as either current or past smokers.
Comorbid conditions
We used the following ICD-9-CM diagnosis and procedure codes to identify comorbid conditions: cerebrovascular disease: 433.xx – 434.xx (cerebral infarction), 435.x (cerebral ischemia), 436 – 437.x (vascular disorders of the entire brain); heart failure: 428.0, 428.1, 428.21–22, 428.4x and codes given as primary or secondary diagnosis; coronary heart disease: 410.xx (acute myocardial infarction), 412 (old myocardial infarction), 413.xx (angina), 414.0x and 414.2x (chronic ischemic heart disease), procedure codes 36.0x (percutaneous coronary intervention) and 36.1x (by-pass surgery); chronic kidney disease 585.x (chronic kidney disease stage I – IV), 586 (end stage renal disease), 588.x (renal failure). Fifty patients were randomly selected to validate algorithms for risk factors and comorbid conditions. Manual medical record review was used as gold standard to generate a positive predictive value (PPV) for each algorithm.
AVD phenotypes
We used ICD-9-CM and CPT-4 codes to ascertain the three AVD phenotypes of interest: CAS, AAA, and PAD (Table 2). The algorithms were developed to identify cases and controls for each phenotype. To identify AVD phenotype with high specificity, we required that the relevant diagnosis codes had to be present at least twice in the EMR. For controls, we required that the relevant diagnosis codes had to be absent in the EMR. PPV was calculated to assess the accuracy of each algorithm to ascertain cases and controls. Manual review of random samples was used to improve the algorithm (criteria listed in Table 1) and repeated to obtain a PPV>90% for cases and controls. We reviewed 50 cases and 50 controls for each phenotype at each step of algorithm development and for final validation. Finalized algorithms were run in the entire dataset and a separate dataset of random samples from the Mayo Phase I eMERGE (electronic MEdical Records and GEnomics) cohort to test the performance of the phenotyping algorithms. The Mayo eMERGE study cohort consists of 1687 patients with PAD and 1725 controls recruited from non-invasive vascular laboratory and stress electrocardiography laboratory respectively, as previously described. 3 Accuracy of algorithms to ascertain vascular intervention or surgeries was validated by manually reviewing a random set of 25 cases and 25 non-cases for each phenotype.
Table 2 .
Algorithms to ascertain atherosclerotic vascular disease cases and controls.
| ICD-9-CM codes to ascertain cases | ICD-9-CM codes to rule out controls | |
| Carotid artery stenosis | 433.1, 433.10, 433.11 – Occlusion and stenosis of carotid artery | 433.xx – occlusion and stenosis of precerebral arteries |
| Abdominal aortic aneurysm | 441.3-4, 441.6-7 – abdominal and thoracoabdominal aneurysm | 441.xx – aortic aneurysm and dissection |
| Peripheral arterial disease | 440.21–24 – atherosclerotic limb with claudication, rest pain, ulcer or gangrene | 440.20–24, 440.0, 440.4, 443.9 – atherosclerotic extremity, peripheral vascular disease |
| Vascular procedures | ||
| Carotid stenting or endarterectomy | ICD-9-CM codes: 38.12 and 00.62; CPT-4: 35,301 and 0075 T | |
| Abdominal aortic aneurysm repair | Open vascular repair: ICD-9-CM codes: 38.44, 39.52, 38.34, 38.64, 38.4, 38.6; CPT-4 codes: 33,877; Endovascular repair: CPT-4 codes: 34,800-05: | |
| Lower extremity revascularization or surgery | Open vascular bypass: ICD-9-CM codes: 38.08, 38.18, 38.48, 38.48, 39.25; Angioplasty with or without stenting: ICD-9-CM codes: 39.50, 39.90; CPT-4 codes: 73,725, 75,635, 75,716; Major amputation: ICD-9-CM codes: 84.13–84.17 | |
CPT-4: current procedural terminology codes version 4; ICD-9-CM: international classification disease codes version 9-CM.
Results
From June, 2009 to December 2012, 8800 patients scheduled for vascular ultrasound or lower extremity arterial evaluation in the Gonda Vascular Center were approached, of whom 5268 consented. Demographics and clinical characteristics for the initial 2182 participants are summarized in Table 3.
Table 3 .
Demographics and clinical characteristics.
| Variables | n = 2182 |
| Age (years) | 70.4 ± 11.2 |
| Men | 1367 (62.7%) |
| Non-Hispanic white ethnicity | 2029 (97.6%) |
| Upper midwest residency* | 1856 (84.9%) |
| Atherosclerotic vascular disease phenotype | |
| Carotid artery disease | 1041 (48%) |
| Carotid endarterectomy/carotid stenting | 521 (24%) |
| Abdominal aortic aneurysm | 448 (21%) |
| Repair of abdominal aortic aneurysm | 190 (9%) |
| Peripheral arterial disease | 834 (38%) |
| Lower extremity revascularization/amputation | 224 (10%) |
| Conventional risk factors | |
| Hypertension | 1712 (78%) |
| Diabetes | 632 (29%) |
| Dyslipidemia | 1585 (73%) |
| Ever Smoking | 1538 (70%) |
| Comorbid conditions | |
| Coronary artery disease | 841 (37%) |
| Coronary revascularization | 513 (24%) |
| Heart failure | 255 (12%) |
| Cerebrovascular disease | 427 (20%) |
| Chronic kidney disease | 418 (19%) |
*Upper Midwest includes the following states: Minnesota, Iowa, Illinois, Wisconsin, Michigan, North and South Dakota.
Demographics and clinical characteristics
Our study population (Table 3) was predominantly white (97.6%) and 62.7% were men, with mean age 70.43 ± 11.21 years. All participants were U.S. residents, with 85% from the Upper Midwest. We manually reviewed the “patient-provided information summary” section in the EMR for 50 patients. No mismatches for sex, race and address information were noted between EMR mined data and manually reviewed data.
The most prevalent risk factor was hypertension (78%), followed by dyslipidemia (73%). More than one third (37%) of the participants had coronary heart disease, one fifth (20%) had heart failure and chronic kidney disease (19%) respectively. The PPVs for algorithms were: hypertension 0.96, dyslipidemia 1.00, diabetes 0.98 and smoking 0.90, cerebrovascular disease 0.90, heart failure 0.88, coronary heart disease 0.90 and chronic kidney disease 1.00.
Vascular disease phenotypes
The most common vascular disease in our biorepository was CAS (48%), followed by PAD (38%) and AAA (21%). History of carotid endarterectomy or carotid stenting was present in half of the patients with CAS, history of aneurysm repair was present in 48% of patients with AAA and history of lower extremity revascularization or amputation was present in 34% of patients with PAD. More than 40% of patients had atherosclerotic disease in two or more vascular beds. To get the final PPVs for vascular disease phenotypes, we manually reviewed patients detected by algorithms as cases and controls for each phenotype. The causes of false positives for the final algorithms were ascertained in 50 cases and 50 controls and listed in Table 4. For cases, false positives were due to: 1) codes for a specific phenotype given at the time of non-invasive testing or clinical evaluations even though results were normal subsequently; 2) mild disease not meeting the criteria we used in the present study. For controls, false positives were due to: 1) lack of specific codes for a subphenotype, such as poorly compressible arteries in the case of PAD; 2) codes assigned in error. The accuracy of the algorithms to ascertain history of carotid stenting or endarterectomy in patients with CAS or to ascertain history of aneurysm repair in patients with AAA was 100%. The accuracy of procedure codes to identify vascular interventions in patients with PAD was 98%, with 1 patient who underwent renal artery stenting procedure detected as PAD case and 1 patient with superior femoral artery stenting detected as PAD control by the algorithm. To test the specificity of the EMR-based algorithms, we validated these in random samples from the Mayo eMERGE phase I cohort, in which 49% of the patients have PAD. The PPVs of cases were lower for CAS and AAA, higher for PAD. The PPVs were similar for controls for each vascular disease (Table 4). The false positives for comorbid conditions mainly resulted from billing codes assigned at the time of non-invasive testing.
Table 4 .
Accuracy of electronic phenotyping algorithms – comparison of EMR-based algorithms to manual medical record review in cases (n = 50) and controls (n = 50) in each dataset.
| PPV (VDB dataset) | PPV (Validation dataset) | Causes of false positives | |
| Carotid artery stenosis | |||
| Cases | 0.94 | 0.90 | mild atherosclerotic plaque or stenosis < 40%; |
| Controls | 0.98 | 0.98 | abnormal carotid ultrasound with codes assigned for cerebrovascular disease, not for carotid artery stenosis |
| Abdominal aortic aneurysm | |||
| Cases | 0.96 | 0.94 | ectasia of abdominal aorta diameter < 30 mm |
| Controls | 1.0 | 1.0 | |
| Peripheral arterial disease | |||
| Cases | 0.92 | 0.98 | lower extremity aneurysm; diabetic neuropathy or non-ischemic ulcer only |
| Controls | 0.96 | 0.98 | noncompressible artery in the lower extremity |
Discussion
AVD is the leading cause of death globally despite the development of effective therapies. 7 Changes in lifestyle due to urbanization, industrialization, and longer life expectancy are some of the factors leading to increase in prevalence of cardiovascular disease in developing countries. 8 Varying genetic susceptibility as well as novel circulating biomarkers may help explain some of the disparities in the prevalence of AVD globally. However, to date, most of the attention has focused on coronary heart disease in whites, whereas other AVD phenotypes and ethnic groups remain relatively understudied. To reduce the global burden of AVD, there is a need to identify novel biomarkers for early detection and prognostication, especially in patients of non-European ancestry. We describe the creation of a biorepository of DNA, serum and plasma from patients with AVD encountered in clinical practice. The biorepository was annotated with demographic information, AVD phenotypes, conventional risk factors and comorbidities by using electronic phenotyping algorithms.
The need for biomarker studies of cardiovascular diseases has led to the establishment of biorepositories in several countries, predominantly in the developed world. The Generation Scotland project (n = 15,000) enrolled participants from Scotland's population to identify genetic variants accounting for variations in quantitative traits underlying heart disease, diabetes and mental disease. 9 The UK Biobank (n = 500,000) aims to investigate the association of common complex diseases including stroke or coronary heart disease with genetic and lifestyle factors by recruiting volunteers aged 40–69 years and following them through linked population-level health related medical records. 10 deCode Genetics leverages Iceland's genealogy data and medical records to investigate genetic and molecular causes of common diseases including myocardial infarction and aneurysmal disease 11 . Recently, disease-focused biorepositories have been initiated to study the association of genetic variants with atherosclerosis. 12–14
Electronic phenotyping
To maximize the value of a biorepository, collection of clinical information should not be limited to characteristics for the specific disease, but should include laboratory and imaging reports, treatments, medications, and past medical history as well. 15 Abstracting clinical data from medical records by manual review can be time-consuming and costly. Electronic phenotyping has several advantages over the classic abstraction approach, including rapid and inexpensive generation of large case-control cohorts. 16 An example of EMR-coupled biorepositories is the eMERGE (electronic Medical Records and Genomics) Network, an NHGRI-supported consortium of five institutions, including Mayo Clinic, to explore the potential of DNA repositories linked to EMR for genomic studies. 17 Other examples include BioVU, the Vanderbilt DNA databank 16 and the Marshfield Clinic's Personalized Medicine Research Project (PMRP), a population-based DNA biobank. 18
Accurate ascertainment of phenotypes depends on the approach to establish the diagnoses. Using ICD-9-CM codes alone to ascertain cardiovascular risk factors such as hypertension or diabetes from the EMR is sensitive but not accurate. 19 Combining diagnosis codes, medications, laboratory data, and text searches using NLP may increase accuracy. 16,20 We used a similar approach, including codified data such as billing codes and laboratory results, and narrative data in physician notes, to ascertain risk factors and increase accuracy of electronic algorithms. We validated the accuracy of algorithms in a separate dataset and found similar PPVs for cases and controls. We found high PPVs of electronic phenotyping algorithms based on manual review of the EMR; supporting the view that EMR-based phenotyping could be used instead of traditional manual abstraction. However, billing codes do not provide information on the location of disease in a particular vascular bed and may lead to a significant number of false positives. We have previously demonstrated that the use of text searches by NLP to ascertain PAD in radiology reports 21 can provide information on the extent and location of atherosclerosis.
Ethical and psychosocial issues
Advances in bioinformatics allow the merging of datasets from different centers, for data sharing, and re-analysis in the future. However, this raises ethical and psychosocial issues, such as whether the initial informed consent allows the use of biospecimens for secondary research and the potential aggregation of data into different databases, using and sharing existing databases, and best approaches to avoid participant identifiability. Additional ethical and psychosocial issues that are unique to a particular ethnic group/geographic location may also need to be addressed. To ensure that procedures conform to what has been established during the informed consent process, different approaches have been used as described above. Phenotypic information needs to be used and stored in a manner that protects patients' confidentiality. For example, a redacted version of the data would be created for those who are eligible and wish to use it. The Mayo Institutional Review Board, a Biospecimen Trust Oversight Group and involvement of bioethicists in our study allow rapid adaptation to issues evoked by policy changes and scientific advancement.
Limitations
Billing codes to ascertain relevant covariates and comorbidities are easily available at a relatively low cost, but systematic misclassification and exclusion of conditions or procedures not pertinent to reimbursement are potential limitations to their use. 22 The availability of phenotypes in the EMR may be affected by whether a patient gets care at one or multiple medical institutions. The relatively high prevalences of vascular diseases and related risk factors may have inflated PPVs for our algorithms. Using NLP to conduct more comprehensive and specific free-text search in radiology and procedure reports will increase precision and generalizability of the electronic phenotyping algorithms. Obtaining data for environmental factors such as physical activity or diet from the EMR is difficult, limiting the ability to study gene-environment interactions. EMRs are not in widespread use yet in developing countries. However, study questionnaires could serve as an alternative means of obtaining information on covariates needed to conduct biomarker and genetic studies.
Conclusion
In summary, we describe methodology for establishing a biorepository of plasma, serum and DNA from patients with AVD and demonstrate the use of electronic phenotyping algorithms to annotate such a biorepository with relevant covariates. These methods may inform the establishment of similar biorepositories in different geographic regions of the world, facilitating the identification and validation of novel biomarkers of AVD in diverse ethnic groups.
Authors' contributions
IJK conceived of the study and participated in its design and helped to draft the manuscript. ZY participated in the design of the study and drafted the manuscript. FSK participated in the study design. All authors read and approved the final manuscript.
Acknowledgement
This work was funded by a Marriott Award in Individualized Medicine to I.J.K.
References
- [1].Ding K, Kullo IJ. Genome-wide association studies for atherosclerotic vascular disease and its risk factors. Circ Cardiovasc Genet. 2009;2(1):63–72. doi: 10.1161/CIRCGENETICS.108.816751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hochholzer W, Morrow DA, Giugliano RP. Novel biomarkers in cardiovascular disease: update 2010. Am Heart J. 2010;160(4):583–594. doi: 10.1016/j.ahj.2010.06.010. [DOI] [PubMed] [Google Scholar]
- [3].Kullo IJ, Fan J, Pathak J, Savova GK, Ali Z, Chute CG. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J Am Med Inform Assoc. 2011;17(5):568–574. doi: 10.1136/jamia.2010.004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Manolio TA. Collaborative genome-wide association studies of diverse diseases: programs of the NHGRI's office of population genomics. Pharmacogenomics. 2009;10(2):235–241. doi: 10.2217/14622416.10.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rzhetsky A, Wajngurt D, Park N, Zheng T. Probing genetic overlap among complex human phenotypes. Proc Natl Acad Sci USA. 2007;104(28):11,694–699. doi: 10.1073/pnas.0704820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Savova GK, Ogren PV, Duffy PH, Buntrock JD, Chute CG. Mayo clinic NLP system for patient smoking status identification. J Am Med Inform Assoc. 2008;15(1):25–28. doi: 10.1197/jamia.M2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Roger VL, Go AS, Lloyd-Jones DM, Adams RJ, Berry JD, Brown TM, Carnethon MR, Dai S, de Simone G, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Greenlund KJ, Hailpern SM, Heit JA, Ho PM, Howard VJ, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Makuc DM, Marcus GM, Marelli A, Matchar DB, McDermott MM, Meigs JB, Moy CS, Mozaffarian D, Mussolino ME, Nichol G, Paynter NP, Rosamond WD, Sorlie PD, Stafford RS, Turan TN, Turner MB, Wong ND, Wylie-Rosett J. Heart disease and stroke statistics–2011 update: a report from the american heart association. Circulation. 2011;123(4):e18–e209. doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Perret F, Bovet P, Shamlaye C, Paccaud F, Kappenberger L. High prevalence of peripheral atherosclerosis in a rapidly developing country. Atherosclerosis. 2000;153(1):9–21. doi: 10.1016/s0021-9150(00)00376-2. [DOI] [PubMed] [Google Scholar]
- [9].Smith BH, Campbell H, Blackwood D, Connell J, Connor M, Deary IJ, Dominiczak AF, Fitzpatrick B, Ford I, Jackson C, Haddow G, Kerr S, Lindsay R, McGilchrist M, Morton R, Murray G, Palmer CN, Pell JP, Ralston SH, St Clair D, Sullivan F, Wolf R, Wright A, Porteous D, Morris AD. Generation Scotland: the Scottish family health study; a new resource for researching genes and heritability. BMC Med Genet. 2006;7:74. doi: 10.1186/1471-2350-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Palmer LJ. UK Biobank: bank on it. Lancet. 2007;369(9578):1980–1982. doi: 10.1016/S0140-6736(07)60924-6. [DOI] [PubMed] [Google Scholar]
- [11].Helgadottir A, Thorleifsson G, Magnusson KP, Grétarsdottir S, Steinthorsdottir V, Manolescu A, Jones GT, Rinkel GJ, Blankensteijn JD, Ronkainen A, Jäääskeläinen JE, Kyo Y, Lenk GM, Sakalihasan N, Kostulas K, Gottsäter A, Flex A, Stefansson H, Hansen T, Andersen G, Weinsheimer S, Borch-Johnsen K, Jorgensen T, Shah SH, Quyyumi AA, Granger CB, Reilly MP, Austin H, Levey AI, Vaccarino V, Palsdottir E, Walters GB, Jonsdottir T, Snorradottir S, Magnusdottir D, Gudmundsson G, Ferrell RE, Sveinbjornsdottir S, Hernesniemi J, Niemelä M, Limet R, Andersen K, Sigurdsson G, Benediktsson R, Verhoeven EL, Teijink JA, Grobbee DE, Rader DJ, Collier DA, Pedersen O, Pola R, Hillert J, Lindblad B, Valdimarsson EM, Magnadottir HB, Wijmenga C, Tromp G, Baas AF, Ruigrok YM, van Rij AM, Kuivaniemi H, Powell JT, Matthiasson SE, Gulcher JR, Thorgeirsson G, Kong A, Thorsteinsdottir U, Stefansson K. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40(2):217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- [12].Plant SR, Samsa GP, Shah SH, Goldstein LB. Exploration of a hypothesized independent association of a common 9p21.3 gene variant and ischemic stroke in patients with and without angiographic coronary artery disease. Cerebrovasc Dis. 2011;31(2):117–122. doi: 10.1159/000321510. [DOI] [PubMed] [Google Scholar]
- [13].Shah SH, Bain JR, Muehlbauer MJ, Stevens RD, Crosslin DR, Haynes C, Dungan J, Newby LK, Hauser ER, Ginsburg GS, Newgard CB, Kraus WE. Association of a peripheral blood metabolic profile with coronary artery disease and risk of subsequent cardiovascular events. Circ Cardiovasc Genet. 2010;3(2):207–214. doi: 10.1161/CIRCGENETICS.109.852814. [DOI] [PubMed] [Google Scholar]
- [14].Koontz JI, Haithcock D, Cumbea V, Waldron A, Stricker K, Hughes A, Nilsson K, Sun A, Piccini JP, Kraus WE, Pitt GS, Shah SH, Hranitzky P. Rationale and design of the Duke Electrophysiology Genetic and Genomic Studies (EPGEN) biorepository. Am Heart J. 2009;158(5):719–725. doi: 10.1016/j.ahj.2009.08.011. [DOI] [PubMed] [Google Scholar]
- [15].Collins FS. The case for a US prospective cohort study of genes and environment. Nature. 2004;429(6990):475–477. doi: 10.1038/nature02628. [DOI] [PubMed] [Google Scholar]
- [16].Ritchie MD, Denny JC, Crawford DC, Ramirez AH, Weiner JB, Pulley JM, Basford MA, Brown-Gentry K, Balser JR, Masys DR, Haines JL, Roden DM. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86(4):560–572. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, Li R, Masys DR, Ritchie MD, Roden DM, Struewing JP, Wolf WA. The eMERGE network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wilke RA, Berg RL, Peissig P, Kitchner T, Sijercic B, McCarty CA, McCarty DJ. Use of an electronic medical record for the identification of research subjects with diabetes mellitus. Clin Med Res. 2007;5(1):1–7. doi: 10.3121/cmr.2007.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Birman-Deych E, Waterman AD, Yan Y, Nilasena DS, Radford MJ, Gage BF. Accuracy of ICD-9-CM codes for identifying cardiovascular and stroke risk factors. Med Care. 2005;43(5):480–485. doi: 10.1097/01.mlr.0000160417.39497.a9. [DOI] [PubMed] [Google Scholar]
- [20].Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, Crawford D. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26(9):1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Savova GK, Fan J, Ye Z, Murphy SP, Zheng J, Chute CG, Kullo IJ. Discovering peripheral arterial disease cases from radiology notes using natural language processing. AMIA Annu Symp Proc. 2010;2010:722–726. [PMC free article] [PubMed] [Google Scholar]
- [22].Tirschwell DL, Longstreth WT., Jr. Validating administrative data in stroke research. Stroke. 2002;33(10):2465–2470. doi: 10.1161/01.str.0000032240.28636.bd. [DOI] [PubMed] [Google Scholar]