Introduction
The zebrafish Danio rerio, a tropical freshwater fish, belongs to the family of cyprinidae, which in the last 30 years has developed into a very popular model organism for studies of embryonic development and human diseases. Initially the zebrafish species has been selected on the basis of its small size of approximately 3–5 cm, its transparency during development and its high fertility, qualities first identified by George Stresinger, the founding father of zebrafish research. 1 The ability to house thousands of small fishes and the ease of screening mutations in the translucent embryos made it feasible to perform large-scale forward genetic screens in a vertebrate model organism. The abundance of eggs obtained, approximately 200 eggs per female per week, is ideal for genetic and statistical analysis. The mutagenesis screens performed in the early 1990s have led to the identification of genes important in vertebrate organogenesis in an unbiased fashion. 2,3 Many of the isolated mutants have now been fully characterized and the mutated genes mapped, as the zebrafish genome sequencing completes. The knowledge derived has led to a better understanding of the underlying genetic networks governing vertebrate development. More sophisticated phenotype-based screens have since been developed to screen for mutations in defined biological processes. 4
The development of the zebrafish is very fast and by 24 hours post fertilization (hpf), the fertilized egg has developed into an embryo that has established most organ primordia including the nervous system and a contracting heart tube. This is followed by the commencement of blood circulation. The embryo hatches after about 2 days post fertilization (dpf) and reaches the larval stage at 3 dpf when most internal organs have matured. At 30 dpf, the zebrafish metamorphose and after 90 days has reached adulthood.
Zebrafish are easily maintained and are relatively economical in comparison to mammalian species. About 5 adult fishes are kept in 1 liter and therefore a small laboratory aquarium facility can easily house more than 1000 animals. Non-invasive imaging can follow the entire phase of embryonic development while the embryo is kept in a petri dish, which allows cell biology to be performed in an intact organism. 5,6 In fact, by introducing pigments and scale mutations that renders the fish transparent, tumors formation can be visualized and thus enable transplantation experiments to be performed in adult zebrafish. 7 The small size of the embryo allows it to be placed into 384 well plates for small molecule screenings. 8 There are however also drawbacks that restrict the utility of the zebrafish. These include the currently limited biological resources for this model organism in comparison to the mouse. For example antibodies used in mammals often do not cross-react in the zebrafish limiting the utilization of this model organism for protein biochemistry. Likewise during teleost evolution, genome duplication took place, 9 which resulted in the presence of many duplicated genes. On one hand, this complicates genetic analysis but it allows the examination of multiple functions of a gene in separate mutants since the duplicated genes display subfunctionalization. 10 The lack of embryonic stem cells, which is required for reverse genetic approaches such as the generation of knockout strains, has hampered the progress to utilize this organism in biomedical research. Nonetheless, recent technological advances have greatly improved this situation. 11–13
In this review, we will outline the knowledge of cardiac development in the zebrafish. Moreover, the current state of knowledge in studying development and function of the cardiac conduction system in zebrafish will be discussed. The zebrafish as a novel disease model in cardiac hypertrophy and heart failure will be introduced. We will also discuss the role of the zebrafish as a premier model to study cardiac regeneration. Since no single review is able to cover all aspects of this rapidly growing field, the reader is also referred to some other excellent reviews in the field, which have been recently published. 14–17
Heart development
Apart from sharing well-conserved genetic pathways that govern heart formation, zebrafish heart development proceeds through very similar steps as amniotes, and begins with the specification of precardiac cells in the anterior lateral plate mesoderm followed by fusion of the bilateral heart fields at the embryonic midline (Table 1). The mechanism of this seemingly simple morphogenetic event at the very early stages of embryonic development became better understood largely due to the powerful genetic amenability and high-resolution bioimaging of zebrafish embryos as illustrated below. 18–20
Table 1 .
Milestones of early heart development shared by different organisms. (modified from 57 ).
| Cardiac Events | Human | Mouse | Zebrafish |
| Migration of precardiac cells from epiblast | 15–16 days | 7 dpc (primitive streak) | 50% epiboly (5.5 hpf) |
| First evident assembly of myocardial plate | 18 days | 7 dpc (late primitive streak) | 8–10 somites (∼13 hpf) |
| Generation of single heart tube initiated | 22 days (4–10 somites) | 8 dpc (5–10 somites) | 20 somites (∼19 hpf) |
| Tubular heart starts contraction | 23 days | 8.5 dpc (8–10 somites) | 26 somites (22 hpf) |
| Looping | 23 days | 8.5 dpc | 33 hpf |
| Endocardial cushions form | 28 days (30–38 somites) | 9.5 dpc | 48 hpf |
In the zebrafish, cardiac progenitors are specified as early as 5 hpf, 21 just before gastrulation, a developmental phase lasting nearly 5 hours, which results in the formation of the three primary germ layers: ectoderm, mesoderm and the endoderm. Fusion of the bilateral heart fields by 16 hpf produces a cardiac cone, which expresses cardiac differentiation markers such as the sarcomeric genes tropomyosin and troponin. 22 The cardiac cone then extends anteriorly and is gradually remodeled into a primitive heart tube.
By 24 hpf, the early heart tube consists of an outer myocardial and an inner endocardial layer separated by extracellular matrix, known as the “cardiac jelly”. Shortly after its formation, the linear heart tube is capable of contracting rhythmically in a peristaltic manner with a conduction velocity of 1 mms− 1. 23
The heart tube loops at 33 hpf, into an S-shaped configuration, displacing the ventricle to the right of the atrium. The transition from slow peristaltic wave to sequential chamber contraction by 36 hpf suggests the onset of cardiac conduction system (CCS) function.
Fate mapping studies have shown that chamber identities are established as early as 5 hpf, 21 attributing to differential gene expression that governs later differences in size, morphology and contractile properties conferred by different sets of sarcomeric proteins (atrial myosin heavy chain, amhc, in the atrium versus ventricular myosin heavy chain, vmhc, in the ventricle). By 2 dpf, the ventricular chamber balloons into a natriuretic peptide precursor A (nppa) –labeled outer curvature (OC) and an inner curvature (IC), which forms the future apex and base of the mature heart respectively. Myocardial cells of the OC are elongated, bigger and conduct electrical activity thrice faster than cells located at the IC. 23,24
Heart muscle growth
As the heart develops, the atrium remains a thin layer of myocardial cells whereas the ventricle thickens to form trabeculae (myocardial projections), which are necessary to generate sufficient contractile force to propel circulation in the growing embryo. At the same time myofibrillogenesis is completed whereby thin and thick filaments, M line, and Z discs are assembled into uniform arrays of cardiac sarcomeres. 25 Studies in mice have shown that trabeculation of the ventricular myocardium is dependent on endocardial-derived signals such as neuregulin, BMP10, and Brg1 (a chromatin remodeling protein) as well as a supportive microenvironment provided by the cardiac jelly. 26–29 The requirement for the neuregulin signaling pathway in trabeculation is resonated in the zebrafish where embryos mutated in erbB2 encoding the neuregulin receptor fail to undergo trabeculation and hence show aberrant electrical conduction in the ventricular myocardium. 30
Starting with 150 cardiac myocytes at 24 hpf, the cell number has doubled at 3 dpf. However the embryonic heart does not rely on cell proliferation, which is negligible at this stage in development. Addition of cells at both arterial and venous poles of the heart followed by waves of cardiac myocyte differentiation is found to be the major mechanism for increasing cell numbers. 31
The adult zebrafish heart grows to a size with a ventricular length of 1 mm or larger, its atrium is lined internally with pectinate muscle and the ventricle consists of a compact layer enveloping the inner trabeculae, which in contrast to the mammalian heart persists into adulthood (Fig. 1). To investigate the complexity of ventricular wall formation, Gupta and Poss recently employed a highly sophisticated multicolor lineage tracing tool, termed Brainbow, which allows after modification and optimization of the technique, to label approximately 20 cardiac myocytes with different colors at embryonic stage and to trace their descendants in the adult heart. 32 These experiments not only uncovered an “inside-out” mechanism for the formation of the outer compact layer of the heart where the source of surface myocardium originates from inner muscle fibers, but also has led to the identification of the amazing properties of a few ‘clonally dominant’ cardiac myocytes, which display proliferative ability in a stem cell-like manner to give rise to the entire heart wall. Hence, further investigation into the properties of this specific cardiac myocyte population, in particular its expression profile, which underlies its unique abilities, is necessary and may provide an insight to rebuild the damaged heart.
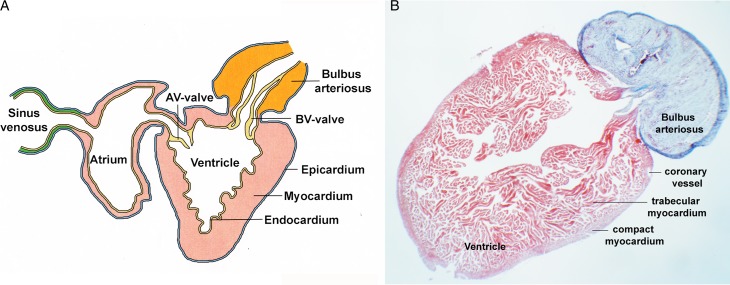
Figure 1. .
Anatomy of the adult zebrafish heart. (A) Schematic representation of the adult zebrafish heart. (B) Trichrome stained histological section through the ventricle and bulbus arteriosus, which labels myocardium in red whereas nonmuscle tissue and extracellular matrix in blue. Note the trabecular layer, which is extensive in the adult zebrafish heart, while the compact layer makes only a minor fractor of the total mass of the ventricular wall. Rarely coronary vessels are observed in the compact layer mocardium.
Endocardium and valves
Cardiac valves in the form of leaflets develop between the chambers to ensure unidirectional blood flow. 33,34 Valve formation is initiated by the restriction of bmp4, tbx2b, and vcana expression to the AV myocardium at 37 hpf 35,36 followed by the endocardial expression of notch1b, has2 and neuregulin. 37,38 These changes in gene expression are accompanied by changes in endocardial cell morphology. The endocardial cells are initially cuboidal in shape and by 72 hpf form primitive valve leaflets, which serve to block retrograde blood flow. 34 In zebrafish, the process of valve maturation, which also involves epithelial–mesenchymal transition (EMT) proceeds into the larval stages of development and is completed by 16 dpf. 39 In amniotes, endocardial cells also participate in septum formation in the atria, ventricles and outflow tract (OFT). The significance of proper valve formation, illustrated in congenital valve defects and adult onset congenital valve disease (CVD), renders it one of the most extensively studied aspects of heart development. Studies in vertebrate embryos and in vitro with the help of EMT collagen gel assays have demonstrated the involvement of multiple signaling pathways, including VEGF, 40 Notch, 41,42 ErbB, 43,44 Bmp, 45,46 TGFβ, 47 Wnt 48 and NFAT 49 (for reviews 50,51 ).
Zebrafish mutants with defective heart valves usually display symptoms of blood regurgitation, which is used as an indicator in a forward genetic screen for valvular defects. 52 The importance of extracellular matrix, heparan sulfate proteoglycan production, in valve specification is highlighted in the Jekyll mutant that encodes UDP-glucose dehydrogenase, 35 which has also been recently linked to CVD in humans. 53 Another ECM protein, nephronectin, regulates AV valve differentiation acting through a Bmp4-Has2 pathway. 54 In addition, zebrafish mutants defective in WNT signaling pathway components – axin (mbl), adenomatous polyposis coli (apc) and glycogen synthase kinase 3 (gsk3) do not form proper valves. 48–55
The endocardium, which is connected to the vasculature, is constantly exposed to hemodynamic forces. Experimental changes of cardiac hemodynamics have a strong impact on the formation and growth of the valve leaflets. 52 With the help of high speed video imaging and blood flow pattern analysis, Vermot et al. demonstrated recently the requirement of reverse blood flow in patterning the atrioventricular valve in addition to the identification of endothelium-derived klf2a acting as a sensor of hemodynamic force and possibly as an indicator of defective valve formation. 56 Blood flow also has its impact on cardiac trabeculation, myocyte growth and chamber maturation, most likely via the endocardium. 57–59
In the mature heart two valves are present, the atrioventricular (AV) and the bulboventricular (BV) valves (Fig. 1). 60 Some significant morphological differences exist between the valvular apparatus in fish and the mammalian heart. For example, the AV valve lacks papillary muscles and chordae tendinae. Moreover, the BV valve, homologous to the aortic valve, has only two leaflets instead of three, which is found in the mammalian heart. 60 The outflow tract in the zebrafish also shows a significant divergence form the mammalian heart in that is has an additional chamber-like structure, the bulbus arteriosus (BA), which is highly distensible due to the presence of large amounts of elastic fibers (Fig. 1). The BA is believed to be a vascular adaptation, which is required to accommodate the short distance between the BA and the gill apparatus and to maintain constant blood perfusion. 61,62
The endocardium also plays a non-cell autonomous role in myocardial wall formation. The zebrafish mutants heart of glass (heg), santa (san) and valentine (vtn) display a dilatation of the ventricular chamber during early development. 63,64 While the cell number is maintained, the ventricular myocardium is unable to develop into a multilayered wall. As a consequence, the chambers are massively dilated and dysfunctional. Heg1, which is the gene mutated in heart of glass, encodes a transmembrane protein that directly associates with the proteins Ccm1 and Ccm2, which are defective in the santa and valentine mutants and also display ventricular dilatation due to insufficient wall formation. 64,65 CCM1, CCM2 and CCM3 have been genetically linked to a common neurological disease, cerebral cavernous malformation, which causes a dilatation of blood vessels in the brain and ultimately causes cerebral hemorrhage. 65 The CCM proteins form with Heg1 a plasma membrane-associated complex that affects endothelial cell-cell-interaction through the modulation of the actin cytoskeleton. 66 Presently, it is unclear how defective endocardial cell-cell interaction affects ventricular wall formation, but secretion of an endocardium-derived signaling molecule is a likely scenario. In the liver, the CCM complex in the vascular endothelium affects apicobasal polarity of hepatocytes in a non-cell autonomous manner 67 and it can be envisioned that a similar activity is required at least transiently in the early heart. It will be interesting to find out whether such a signaling pathway is still active in the adult myocardium and possibly involved in the pathogenesis of dilated cardiomyopathy.
Secondary heart field
Recent advances in understanding heart development in mouse and chick embryos has led to the identification of two different sources of cells that give rise to the early embryonic heart. A first heart field lineage contributes mainly to the linear heart tube and subsequently develops into the left ventricle, whereas the second heart field lineage develops into the right ventricle and both the outflow and inflow tract of the heart. 68–70 The second heart field cells are located dorsal to the primitive heart tube in the pharyngeal mesoderm. Mutants of genes that are expressed in the second heart field, such as Isl1, are missing outflow tract, right ventricle and most of the atria. 71 Interestingly, second heart field cells also give rise to a subset of craniofacial muscles. 72,73
The presence of only two non-septated heart chambers and a primitive OFT prompts the question of the presence of the second heart field in the zebrafish. Recent work verified that there exist secondary heart field-like cells that contribute to specific lineages of the outflow tract and distal ventricular myocardium, which acts under latent TGFβ?signaling. 74 Interestingly, isl1, which is essential for second heart field development in the mouse is dispensable for zebrafish OFT development. 31
Epicardium and neural crest cells
In amniotes, epicardial cells are derived from a transient structure, the proepicardium (PE) located adjacent to the sinus venosus at the venous pole; they migrate to the surface of the myocardium, proliferate rapidly, and envelop most of the heart except the OFT, which is covered by epicardium from an arterial source. 75 It has been shown that morphogenetic signaling from the epicardium is necessary for proper heart morphogenesis as well as development of coronary vasculature and cardiac fibroblasts (reviewed in 76–78 ). Only a few marker genes, namely wt1, tbx18 and tcf21 have been linked to the formation of the PE and its transformation into the epicardium. As a result, the epicardium remains the least understood cell lineage of the heart. In zebrafish, the presence of the PE and its derivative, the epicardium, has been documented, 11,79,80 but very little research has been done. This is partly due to the lack of useful live markers. Tbx5 and Bmp signaling in PE specification, which has been previously demonstrated both in the chicken and mouse embryo, are also essential for PE development in the zebrafish. 79 In contrast, zebrafish PE induction does not require signals from the liver primordium as proposed in higher vertebrates. 79,81 Epicardium–derived cells (EPDC) were initially reported to also contribute to the myocardial lineage. 82 However, it was subsequently demonstrated to be due to the endogenous expression of epicardial marker genes in a subset of cardiac myocytes. 83 Similarly in zebrafish, the non-epicardial expression of wt1 and tbx18 rendered these genes unsuitable for lineage studies. 84
The cardiac neural crest cells (NCC) is another extra-cardiac cell population, which in the amniote embryo migrates into the forming pharyngeal arches and is involved in re-patterning the initially bilaterally symmetrical pharyngeal arch arteries to form the asymmetric great arteries of the thorax. 85 The NCC will form the smooth muscle tunica media of the arteries. A subpopulation of the cardiac NCC in the caudal pharyngeal arches migrates into the cardiac outflow tract and forms the aorticopulmonary septum dividing the common arterial outflow into the aorta and pulmonary trunk. 86 A combination of lineage tracing and ablation experiments have demonstrated Wnt-dependent contribution of cardiac neural crest to myocardium in all regions of the embryonic fish heart 87–89 ; this is in contrast to higher vertebrates, where neural crest cells contribute mostly to the OFT and do not appear to invade deeply into the developing heart tube. However, further analysis of the functional importance of the NCC in zebrafish heart development has yet to be performed.
Mutant screening, transgenics and drugs
In the 1990s, the first large-scale chemical mutagenesis screens using ENU in Tübingen and Boston has generated hundreds of mutants. 2,3 Mark Fishman, a cardiologist who initiated the Boston screen envisioned the strength of the zebrafish as model to study cardiovascular diseases. As described throughout the review, many heart mutants are derived from these two screens. 57
Due to a lack of embryonic stem cells in the zebrafish, targeted knockout by genetic recombination has not been possible. Hence, the zebrafish community has been using antisense technology mainly morpholinos, which are modified oligonucleotides that block translation or mRNA splicing leading to a knockdown of specific genes of interest. 90 However, some morpholinos can cause undesirable nonspecific side effects such as apoptosis, edema of the fourth brain ventricle and the pericardial sac. Careful experimental design that includes stringent controls and rescue experiments is required when using morpholinos. 91,92
Regardless, the effect of morpholinos is short-lived and therefore analysis of gene function is confined to the embryonic and early larval stages. Attempts to generate mutants include targeting induced local lesions in genomes (TILLING) and insertional mutagenesis. Tilling is slow and tedious, however it has generated a significant number of mutations in important genes. 93 Insertional mutagenesis, which aims to create mutants by insertion of a foreign DNA into the zebrafish genome, has been carried out using a number of different constructs with varying results. Large-scale screens were performed with a mouse retroviral vector 94 and transposons. Tol2 transposon 95,96 and the sleeping beauty transposon 97 have been used in enhancer and gene trap screenings. The use of Tol2 transposon to mutagenize genes has not been successful, but hundreds of transgenic marker lines have been produced instead that serve as markers for studying various biological process or organ structures (Fig. 2). 11,95,98 A promising novel approach involves the use of an in vivo protein-trap mutagenesis system, which can be used to assess spatiotemporal protein expression dynamics and gene function. 99
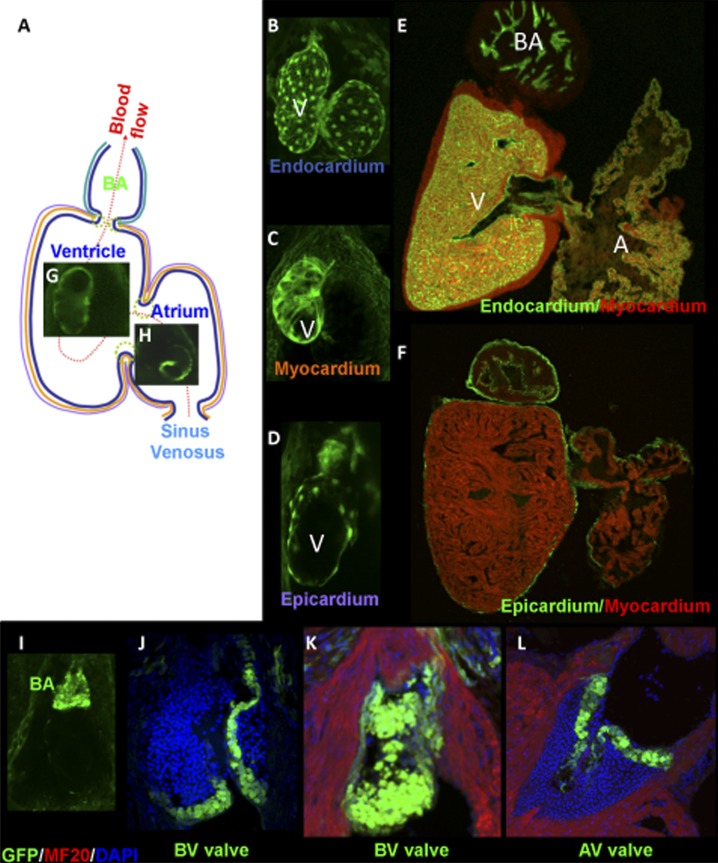
Figure 2. .
Transgenic labeling of tissues and cell types in the zebrafish heart using GFP reporter genes. (A) The different cell types of the zebrafish heart are GFP-labeled by transposon mediated insertional transgenesis. (A) Schematic depiction of a zebrafish heart indicating the route of blood flow from the sinus venous, through the atrium, ventricle and exiting from the bulbus arteriosus. GFP expression present in the three cardiac layers - (B,E) endocardium, (C) myocardium and (D, F) epicardium as well as examples of transgenes with a region-specific expression pattern including the (G) ventricle, (H) atrium, (I) bulbus arteriosus and (J-L) valves. Expression patterns are shown in the (B-D, I) embryonic, or (E-F,J-L) adult heart. Abbreviations: A, atrium; AV, atrioventricular valve; BA, bulbus arteriosus; BV, bulboventricular valve; V, ventricle. From 11 with permission.
The plan to saturate the zebrafish genome or to generate a mutant for every gene is becoming possible with the arrival of zinc-finger based mutagenesis tools - zinc finger technology 100 and TALEN (transcription activator-like effector nucleases). 101 Zinc-finger nucleases (ZFNs) induce a targeted double-strand break in the genome that is repaired to generate small insertions and deletions. 102 The major drawback for the use of ZFNs is the cost involved and the requirement of laborious screening efforts. TALEN however seems to be a technology with potential to become the method of choice in creating mutants of any gene of interest. TALEN proteins have a simpler mode of DNA base recognition than zinc fingers, with an individual base recognized by only two amino acids in an individual TALE repeat unit. 101 Due to this property, gene-specific TALENs are rapidly assembled even for small laboratories, thus making it likely that this technology will find wide distribution. Initial reports suggest that mutagenized genes are germline transmitted, and therefore novel mutant lines are rapidly and efficiently produced. 103 A recent report suggests that not only indel mutations are possible but also site-directed integration of loxP sites, which is a prerequisite for Cre-mediated conditional mutagenesis. Likewise, the DNA repair following TALEN nuclease treatment can also be modified so that single point mutations can be introduced in any gene, which will be useful to model gene mutations associated with cardiovascular disease in the zebrafish model. 104
Another important area, in which the zebrafish model may find its use, is chemical genetics, i.e. the search for novel cardiovascular drugs. 105 The zebrafish larvae can be kept in 384 well plates and chemical compounds are directly added to its aquatic environment, and are, therefore, readily absorbed avoiding the need for time-consuming injections. Zebrafish embryos can be maintained for a few days in 96-well plates with several embryos per well or singly in 384 well plates. Compounds can be added and the plates can be screened either by eye or with the help of high-throughput video techniques. Numerous chemical libraries exist, from small collections of characterized compounds to larger libraries consisting of tens of thousands of compounds of uncharacterized function. In a recent report, larval zebrafish were used to screen chemical compounds that modulate heart failure-specific gene expression patterns. 106 The authors made use of the promoter of the zebrafish nppb gene, which appears to respond to pathological signaling pathways in a very similar way as in mammals. The long QT (LQT) phenotype, which develops in the zebrafish kcnh2 mutant, was utilized in a large-scale screening approach to search for chemical compounds with a LQT suppressive ability. 107 A screen for molecules that enhances FGF signaling resulted in the identification of a novel compound, which increased the size of the embryonic zebrafish heart. 108 This compound may have the potential to enhance cardiac regeneration in vivo or to improve the efficiency of generating cardiac myocytes from stem cell cultures. These few examples demonstrate the power of the zebrafish model for chemical genetic approaches.
Cardiac Conduction System (CCS)
Even with only a two-chambered heart, a functional CCS is necessary to initiate, maintain and coordinate heart rhythm so that synchronized contraction can take place. A large-scale screen for gene mutations affecting the CCS was recently conducted which helped to define four stages of CCS development in zebrafish and furthermore lead to the identification of 17 mutants with defects in one of these stages. 109 Stage 1 occurs between 20–24 hpf, when a linear activation wave travels across the heart tube from the sinus venosus to the OFT; in stage 2 (36–48 hpf), a significant AV conduction delay develops; in stage 3 (72–96 hpf), an immature fast conduction network develops within the ventricle; and in stage 4, this fast conduction network fully matures resulting in appearance of an apex-to-base activation pattern. 109
The presence of the pacemaker in zebrafish had been enigmatic for many years with only two publications describing a mutant, slow mo, with defective pacemaker current. 110,111 However, pacemaker activity is clearly present in fishes and its function is similar to that of mammals. 112 By recording action potentials from the hearts of a variety of fishes, Saito (1969) concluded that the pacemaker site is at the sinoatrial valve. 113 Consistent with this, a recent study showed that knockdown of shox2 resulted in pronounced sinus bradycardia. 114 Finally, with a combination of optogenetics, which utilizes transgenic expression of light-controlled ion channels and light sheet microscopy, the cardiac pacemaker of the zebrafish has been localized to a few cells in the sinoatrial region. 115 The Isl1 genes in zebrafish, human and mouse heart is expressed in sinoatrial pacemaker cells. 116 While in mammals Isl1 labels a subset of sinoatrial node cells, in zebrafish, isl1 expression demarcates the entire pacemaker population, which forms a ring-like structure at the border between the sinus venosus and the atrium. 116 The isl1-positive cells also express tbx2b and are characterized by the presence of primitive action with a pacemaker potential.
The AV conduction system is responsible for delaying the electrical impulse between the chambers so as to achieve coordinated contractions. By calcium mapping with calcium green dyes, a retardation of calcium waves at the AV junction is observed at 40 hpf in the zebrafish. Interestingly, the specification of zebrafish AV conduction is mediated by the endocardial signals notch1b and neuregulin. 117 In addition, foxn4, which drives the expression of tbx2b, is involved in AV canal specification and mutants of both genes display AV conduction defects. 36 Recently, one member of the Popeye domain containing gene family, popdc2 has been shown to exhibit AV conduction abnormalities, which reflects its role in the regulation of the cardiac conduction system (Fig. 3). 118
Figure 3. .
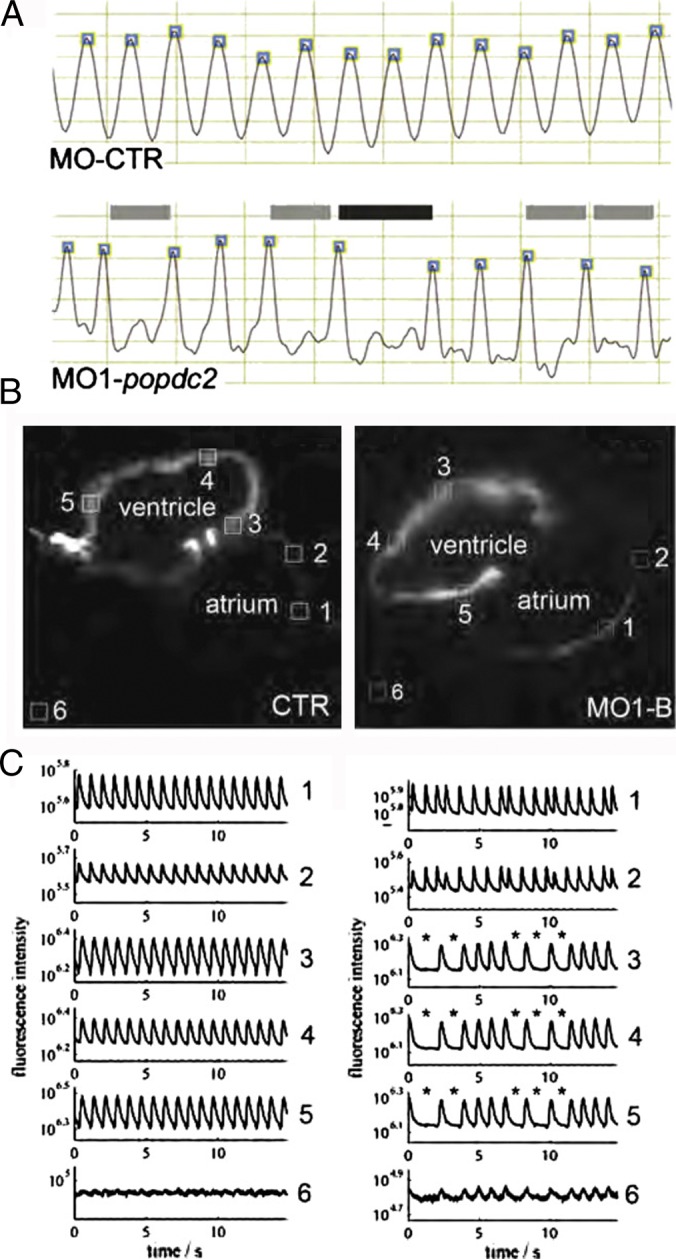
Analysis of cardiac arrhythmias in zebrafish popdc2 morphants. (A) Ventricular contractions are regular in control animals (CTR) but display 2:1 and 3:1 secondary AV block in popdc2 morphants (MO1). (B, C) Optical mapping of calcium waves in control and morphant hearts reveal aberrant calcium waves in Popdc2 morphants. From 111 with permission.
The apex-to-base ventricular activation in both embryonic and adult zebrafish has been optically mapped by individual groups. 109,119 This electrical activity is most likely transmitted by the gap junction protein Cx40, which immunoreactivity has been detected in the embryo at 4 dpf, supporting the presence of a fast CCS at this stage in development. 109 In the absence of morphologically defined His-Purkinje bundles in zebrafish, it has been proposed that the impulse travels through ventricular trabeculae, which are the evolutionary precursors of Purkinje fibers in amniotes. In fact, the CCS does not mature in a mutant that is devoid of trabeculae. 30 Studies in embryonic zebrafish with an aberrant cardiac conduction revealed the importance of cardiac conduction in influencing chamber morphogenesis. 120 The authors furthermore uncovered a novel connexin gene, cx46, in conduction of electrical impulse which results in CCS defects in mutant mouse that resemble human heart failure patients.
Cardiac electrophysiology
Despite the small size of the zebrafish embryonic heart (100–150 μm), an electrocardiogram (ECG) of the zebrafish heart can be recorded using micropipette electrodes from as early as 5 dpf with apparent P and R waves that reflect atrial and ventricular depolarization, respectively. 121 As the larvae develop further and the heart matures, the ECG pattern changes accordingly with the emergence of T waves at 2 weeks post fertilization followed by the shortening of QRS interval and finally distinct P waves, QRS complexes, and T waves, which are detectable by 35 dpf. 122,123 The surprisingly close resemblance of the zebrafish ECG morphology to that of the human heart probably reflects the crucial importance of electrical activity for governing the heartbeat and is therefore evolutionary conserved. The relative simple design of the zebrafish heart, containing only the essential elements of the cardiac conduction system, offers an advantage to study fundamental questions of cardiac electrophysiology.
The fish heart beats at a rate of 120–180 beats per minute (bpm), which is closer to the human heart rate (60–100 bpm) than the mouse (300–600 bpm). As a result, the cardiac repolarization phase, a diagnostic feature for possible arrhythmias represented by the length of the corrected QT (QTc) interval, is more similar between human (300–450 msec) and zebrafish (300–440 msec), than mouse (83–96 msec). 124,125 Furthermore, both the human and the zebrafish generate a repolarizing current during the cardiac action potential (AP) using a similar rapidly activating delayed rectifier potassium current (I Kr), which is in contrast to the mouse that uses a different set of potassium channels. 126 The similarity in cardiac repolarization between humans and mice is also evident in the phenotypes of the breakdance and reggae mutants, which are defective in the ERG potassium channel and exhibiting either long QT syndrome or short QT syndrome. 127–129 In fact, QT syndrome mutations have been successfully modeled in the zebrafish but not in mice. 125
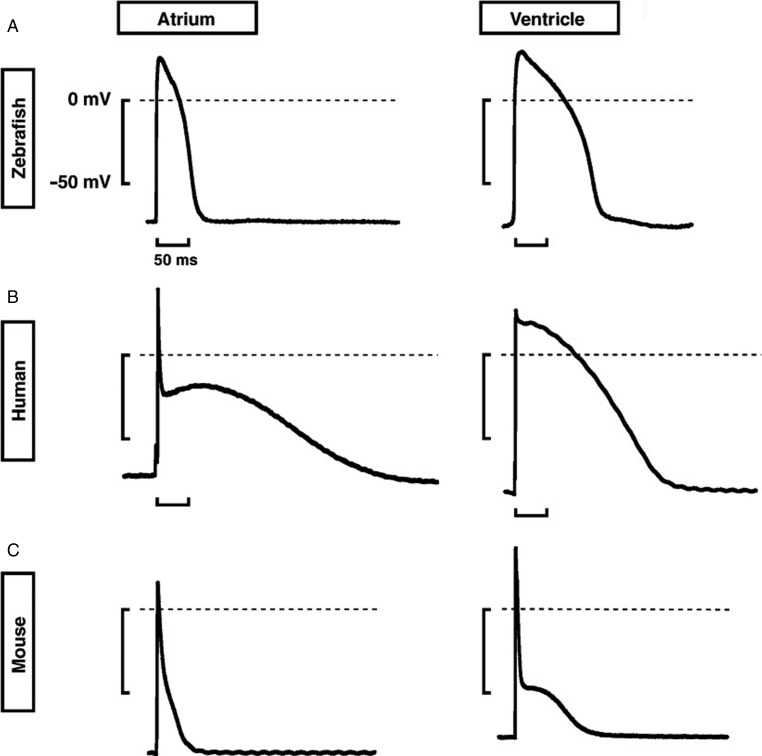
Specific ion channels, mainly gating sodium, potassium and/or calcium ions generate unique currents that shape the different phases of the cardiac AP. Knowledge of the set of ion channels that contribute to the zebrafish cardiac AP are derived from fish mutants defective in ion channels as well as with the help of pharmacological experiments. 126 By detailed comparison of the AP between human, mouse and zebrafish cardiac myocytes, it was found that the AP parameters - AP amplitude and the resting membrane potential were highly similar between zebrafish and humans, whereas the fish has a lower maximum depolarization velocity (dV/dtmax) (Fig. 4). This is possibly due to less competent or fewer fast sodium channel that drives INA. The AP profile of both atrial and ventricular adult zebrafish cardiac myocytes are comparable to that of the humans, especially the long plateau phase 2 and the resulting action potential duration (APD) except for the lack of the sharp early repolarization phase 1, which is probably due to a non-functional I ks current in zebrafish cardiac myocytes. 119 Sodium channels blocker (tetrodotoxin, TTX) decelerate the AP upstroke velocity confirming the role of INA in establishing the steep slope of the rapid depolarization phase 0 of the AP. Interestingly zebrafish cardiac myocytes are 10–100x more sensitive to TTX than mammalian cardiac myocytes due to the presence of an aromatic amino acid at a critical position, which is thought to increase the TTX sensitivity. 130 The L-type calcium channel (LTCC) contributes to the AP plateau phase indicated by the result that LTCC blocker, nifedipine, shortens both atrial and ventricular AP. Moreover, the LTCC mutant island beat embryos exhibit an absence of the QRS complex causing atrial fibrillation and a silent ventricle. 131 Unexpectedly, apart from LTCC, functional T-type calcium channel (TTCC) also contribute to the calcium current in adult zebrafish cardiac myocytes, 126 which can be considered as an indication of an immature state of the adult zebrafish heart in comparison to mammals, where the TTCC is only present in the embryonic heart and in pacemaker tissue.
Figure 4. .
Atrial and ventricular action potentials in the zebrafish, human and mouse. Representative shapes of action potentials in (A) zebrafish, (B) human, and (C) mouse. Note the similarity between the ventricular action potentials in humans and zebrafish. From 119 with permission.
Although this has not yet been confirmed, the zebrafish heart most likely employs a similar pacemaking mechanism as mammals and probably depends on the activity of the hyperpolarization-activated cyclic nucleotide-gated channel. Unfortunately, the slow mo mutation, which has a defective pacemaker current, has not been molecularly characterized until now. 110,111
In terms of establishing ion homeostasis, cardiac myocytes utilize the sodium/calcium exchanger (Ncx1) and calcium ATPase at the plasma membrane (PMCA) and sarcoplasmic reticulum (SERCA) to regulate the cytosolic calcium concentration and the Na/K-ATPase for maintaining a proper sodium/potassium gradient. The mouse Ncx1 mutant is embryonic lethal due to a vascular function of the Ncx1 gene whereas a cardiac-specific Ncx1 null mutant reaches adulthood with cardiac myocytes exhibiting normal calcium transients. 132,133 In contrast, the cardiac-specific ncx1 mutant (tremblor, tre) displays atrial fibrillation and arrhythmia whereas the ventricle is noncontractile due to calcium overload. 134,135 The difference in phenotype severity of the mutants is most likely based on differences in the calcium homeostasis and genetic buffering mechanisms in these species.
One interesting observation is that some channel proteins do have non-electrical related functions in early embryonic development. The heart and mind (had), which is defective in the Na/K-ATPase α1B1 isoform, causes severe abnormalities in primitive heart tube extension, cardiac myocyte differentiation and compromised cardiac function. These phenotypic anomalies are found to be direct consequences of defective pump activity resulting in ionic imbalances that affect the maintenance of proper myocardial cell junctions in the early embryonic heart. 136,137 The morpholino-mediated knockdown of the voltage-gated sodium channel scn5a caused a reduction in the expression of the cardiac transcription factors and had an negative impact on proliferation and differentiation of cardiac myocyte resulting in abnormal cardiac morphogenesis. 138 These phenotypes were not reproduced by the administration of TTX, suggesting a novel non-electrogenic function of scn5a.
Cardiomyopathy
Zebrafish has an established track record in uncovering the molecular mechanisms of human cardiovascular diseases (CVD), which includes prevalent forms of cardiomyopathy - dilated cardiomyopathy (DCM) and hypertrophic cardiomyopathy (HCM). It was observed that some mutants, generated from forward genetic screens, exhibit a lack of blood circulation together with pericardial edema. By high-speed video imaging of the embryonic heart, standard non-invasive assessments of cardiac performance were made and revealed compromised cardiac contractility, shortening fraction (sometimes presented in M mode), stroke volume and/or cardiac output. Positional cloning of these mutants showed that many of the mutated genes were cardiac sarcomeric genes, which have been previously implicated in human cardiomyopathy such as titin (ttn), 139 tropomyosin (tpm4), 140 troponin 2 (tnnt2), 141 myosin light chain genes (cmlc1, myl7) 142–144 and myosin heavy chain (myh6). 145 Moreover, these mutants also exhibit myofibrillar disarray, one of the hallmarks of cardiomyopathy as revealed by ultrastructural examination of sarcomeric organization. 146
Cardiac myofibrillogenesis in the zebrafish has been characterized but not completely understood. Non-striated actin filaments are the first to assemble at the plasma membrane. Sarcomeric myosin independently assembles into thick filaments before being integrated into the thin filament network. 25 Next, M-lines of fixed width and Z-discs, which elongate from Z bodies, are recruited to the contractile units. Phenotypic differences of different mutations of sarcomeric genes also provide an opportunity to understand their unique and specific roles in myofibrillogenesis. This becomes apparent in the case of titin isoforms and has also been observed for the alkali and regulatory myosin light chains, which have distinct roles in the process of myofibril assembly. 143,147 The zebrafish is also instrumental to unravel the function of novel myofibrillar genes. Kindlin2, an integrin-binding protein first identified in Caenorhabditis elegans localizes to the intercalated disc. Morpholino-mediated knockdown abolishes sarcomere assembly at the plasma membrane and causes contractile dysfunction. 148,149 Myozap, another intercalated disc protein, which directly interacts with desmoplakin and zonula occludens-1 participates in the Rho-signaling pathway and activates serum response factor dependent transcription. 150 Consistent with its role in myofibrillar-nuclear communication is the fact that the knockdown of Myozap in zebrafish causes a severe cardiomyopathy-like phenotype. Chap is another newly identified Z-Disks proteins and the loss-of-function phenotype includes severely impaired heart function. 151
The efficient and cost-effective zebrafish system is also used to study the physiological roles of mutations found in patients with heart failure such as in TNNT2 146 and especially non-sarcomeric genes, including transcriptional coactivator EYA4 152 and RNA-binding protein RBM24 (Fig. 5). 153 Cardiac myosin light chain kinase (mlck) was identified in a microarray comparison of healthy and failing human hearts. Its crucial role in cardiac function is suggested by its morphants, which develop a dilated cardiac ventricle with immature sarcomere organization. 154 Nexilin, a novel Z disk protein, functions to stabilize the Z-disks in zebrafish and is a candidate in causing DCM in patients. 155
Figure 5. .
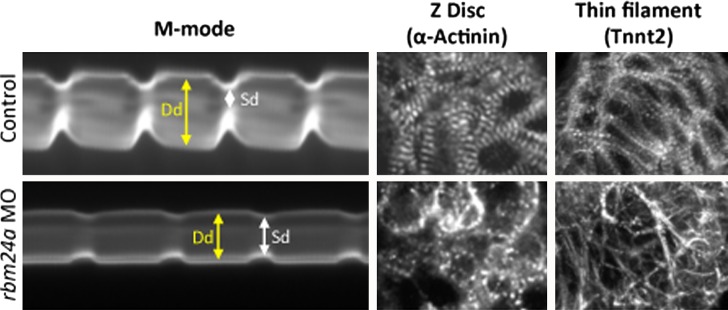
Contractile dysfunction in the zebrafish heart. Morpholino-mediated knockdownn of the gene encoding the RNA-binding protein Rbm24a causes contractile dysfunction as evidenced by M mode analysis (left panels) which shows significant difference between end-diastolic (Dd) and end-systolic (Sd) diameter of the rbm24a morphant (lower panel) as compared to control embryo (upper panel). Furthermore, sarcomeric defects are observed in the thin filament (visualized by anti-Tnnt2 immunohistochemistry) and Z-Disc (visualized by anti-α-Actinin immunohistochemistry) that most likely cause the contractile dysfunction. From 146 with permission.
Interestingly, more evidence has substantiated the effect of cardiac mechanical stretch sensing on regulation of heart function hence resulting in cardiomyopathy. In zebrafish, the first clue comes from the positional cloning of the contractility-deficient mutant main squeeze (msq), which fails to express stretch-responsive genes. 156 The extracellular matrix protein integrin-linked kinase (ilk) is mutated in msq. ILK, found to localize to Z-Disks interacts with integrin and β-parvin together with many other proteins forming a complex at the cell membrane, which acts as a mechanosensor and signals into cardiac myocytes. PINCH, which is also part of the mechanosensor complex, has been shown to regulate contraction in the zebrafish heart. 157 More importantly, human patients with DCM were also found to bear mutations in ILK or laminin-alpha4 (Lama4), yet another of ILK interaction partner. 156,158 Collectively, these genetic and biochemistry data emphasize the importance of the mechanosensor complex in the regulation of cardiac function. In addition, a M-band protein, myomasp (Myosin-interacting, M-band-associated stress-responsive protein) has been shown to regulate stretch responsive genes and hence cardiac stretch sensing via an alternative pathway that involves serum response factor (SRF)-dependent signaling. 159 To investigate myocardial function in greater detail, the micromechanical properties of zebrafish myofibril has been evaluated and proven to be a feasible contractile model where functional studies at subcellular level can be carried out. 160 This might serve as another step forward in reaching a detailed molecular understanding of the etiology of cardiomyopathy. On another note, Werdich et al. tested the stretch response of embryonic zebrafish heart exploring the feasibility of using the zebrafish as novel model to study the mechano-electrical feedback of the heart. 161
One main limitation of the zebrafish embryonic models of cardiomyopathy is that it does not go through the progressive stages of cardiac remodeling as in patients due to the fact that so far most of the studies utilized zebrafish embryos or larvae. Hence, developing an adult cardiomyopathy model in zebrafish, which more closely mimics the human pathological process would be an important improvement. Recently, two adult models of cardiomyopathy have indeed been developed. The first is anemia-induced either by a band3 mutant that inflicts chronic stress in the heart or through the use of the anemia-inducing drug phenylhydrazine. 162 Encouragingly, the mutant hearts displayed myocyte hypertrophy and hyperplasia as well as several features of human cardiomyopathy, comprising myocyte disarray, fetal gene expression reactivation, and severe arrhythmia. The second model was generated by cardiomyopathy-inducing doxorubicin (DOX) injection. 163 These two zebrafish cardiomyopathy models used in combination with a target of rapamycin (ztor) mutant fish, served to provide genetic evidence for a cardioprotective function of target of rapamycin (TOR) signaling.
Heart regeneration
The zebrafish heart demonstrates the surprising capability of myocardial regeneration within months after experimental induction of myocardial injury. Experimental procedures to induce myocardial regeneration include 20% ventricular resection, 164,165 or cryocautherization, which induces localized injury of 25% of myocardium 166,167 or diphtheria toxin A chain (DTA)-induced cytoxicity that kills up to 60% of cardiac myocyte. 168 Apart from demonstrating a similar capacity for cardiac regeneration in neonatal mice, 169 this trait of regeneration is absent or possibly turned off in adult mammals. This prompted the zebrafish to become the key model for elucidating the molecular machinery of heart regeneration, which would fuel the development of regenerative medicine for human cardiovascular diseases. Newts and some other lower vertebrates also exhibit a similar level of cardiac regenerative capability 170,171 but are not as favorable to work on due to practical reasons relating to the lack of genetic infrastructure and tools as compared to the zebrafish model.
Within hours of ventricular resection, the endocardium undergoes extensive changes in morphology and gene expression, most notably the upregulation of raldh2, which is a key enzyme, involved in the synthesis of retinoic acid (RA) and promotes cardiac myocyte proliferation. 172 The stimulation and prolongation of retinoic acid signaling is supported by epicardial cells and it is thought that these combined efforts of the endocardium and epicardium, may serve as the major trigger of cardiac myocyte proliferation.
The activated epicardial cells (3 days post amputation) invade the wound site in the zebrafish heart and give rise to coronary vessels via epithelial–mesenchymal transition (EMT), regulated by snail2 and twist1b. 173 This process is reminiscent of coronary arteriogenesis during embryonic heart development, which is regulated by FGF and PDGF signaling. 174–176 FGFRs (fgfr2 and fgfr4) in the epicardial cells respond to their ligand, fgf17b, which is transcribed in the myocardium, to promote epicardial EMT and further neovascularisation. 173 This process when inhibited will results in a block in regeneration. A hint for an involvement of PDGF signaling stems from an earlier transcriptome analysis of regenerated heart tissues, which identified an upregulation of pdgfa. 177 Blocking PDGF using an inhibitor demonstrates the requirement of epicardial-derived pdgfrb for epicardial proliferation and coronary vessel formation. In experiments where zebrafish epicardial cells are cultured on fibrin gel in vitro, PDGF is found to induce stress fiber formation and loss of cell–cell contacts, mediated by Rho-associated protein kinase, ROCK. 178 Cardiac revascularization, inherent to the zebrafish, may account for the difference in regeneration abilities, between fish and mammals. Therefore, elucidation of the process of epicardial EMT and subsequent neovascularisation may help to shed light on new developments for regenerative medicine.
The origin of the new myocardium that replaced the injured myocardium has been a case of speculation and epicardial cells with their ability to undergo cell type switch is suspected of contributing to new myocardium. However, the studies described above indicate that the epicardium does not contribute to new cardiac myocytes, but instead serves as a source of coronary vasculature, which is essential for the growth of new cardiac myocytes. Most recently, Kikuchi et al. confirmed that tcf21positive epicardial cells are restricted to non-myocardial cell fates by Cre-based fate-mapping experiments. 84
The quest for the origin of new myocardium has lead to several genetic cell lineage studies that finally revealed that the source is not cardiac progenitors but pre-existing cardiac myocytes. 179,180 In response to injury, these cardiac myocytes undergo dedifferentiation and is accompanied by sarcomeric disassembly and aberrant electrical phenotypes including a reduction in conduction velocity and breaking of the depolarization wavefront, which are signs of functional uncoupling of cardiac myocytes. Dedifferentiated cardiac myocytes can then re-enter the cell cycle and proliferate to supply new functional cardiac myocytes. This intriguing finding suggests the possibility of harnessing the potential of pre-existing cardiac myocytes in failing human hearts.
Cardiac myocyte proliferation is thus an important step, and if blocked, as in the cell cycle mutants mitotic checkpoint kinase (mps1) and polo-like kinase 1 (plk1) 181,182 or by overexpression of the microRNA miR-133, 183 cardiac regeneration is retarded. miR-133 is one of the microRNAs, which is downregulated during the course of cardiac regeneration. Similarly, if injured zebrafish heart is blocked from entering into the cell cycle by activating the well-known mammalian cell cycle inhibitors p38α MAPK, the ability of cardiac regeneration is compromised. 182 It is exciting to note that the zebrafish heart employs similar molecular mechanisms for the regulation of cardiac myocyte proliferation as in mammals, substantiating the relevance of the zebrafish model to develop regenerative therapies for the human heart.
Recently, the question of how proliferating cardiac myocytes actually reach the wounded myocardium was addressed. Expression screening using the regenerating zebrafish heart identified the genes cxcl12a, encoding a chemokine ligand (also known as sdfa1), and its receptor cxcrb4 to be expressed during heart regeneration. 184 The CXCL12-CXCR4 system is known to regulate directed cell migration during embryonic development. 185 Interestingly, cxcrb4 was expressed in proliferating cardiac myocytes, while the ligand was found in activated epicardium. Loss-of-function experiments clearly demonstrated that blocking Cxcr4 function only affects the migration of proliferating cardiac myocytes to the wounded area, while other aspects like the level of cell proliferation or gene expression were not affected. Thus an epicardium-mediated guidance of proliferating cardiac myocytes via CXCL12-CXCR4 signaling to the wounded area is essential for zebrafish heart regeneration.
As mentioned earlier, cryoinjury was also applied as technique to cause damage to zebrafish cardiac cells. 166,186 This method appears to be superior to ventricular resection, since it better resembles myocardial infarction in patients, which is characterized by irreversible replacement of damaged myocardium by a fibrotic collagenous scar that lacks the functional contractile properties of healthy myocardial cells. Despite scar formation, the teleost is able to rapidly regenerate its injured heart by elimination of the scar through apoptosis and triggering proliferation of cardiac myocytes. In fact, it has been recognized that transient scar formation is advantageous for cardiac regeneration serving as a transitional support required by the dynamic beating heart, before being removed and the space is populated by cardiac myocytes. 187 Chablais and Jaźwińska further elucidated that Smad3-dependent TGFβ/activin signaling mediates multiple important steps: inflammatory response of infiltrating immune cells during cardiac cells apoptosis; ECM production by fibrotic cells to form scar and subsequent cardiac myocyte proliferation. The coordination of this highly complex process remains to be clarified.
In a short span of ten years since the discovery of the regenerative potential of the zebrafish heart, our understanding of this process has greatly advanced and further research promises more answers and mechanistic insights into the amazing property of the zebrafish heart, which potentially will also lead to novel therapeutic options to treat heart failure patients.
Future prospects
The zebrafish has seen a surprisingly rapid and extremely successful career as a model organism in cardiovascular research. As an affordable in vivo model, the zebrafish will continue to be used to assess candidate genes (from microarray or genome wide association studies) that may have a cardiac phenotype. There are only a few areas in cardiovascular research in which the potential of the zebrafish model has not yet been tested. The challenge for the zebrafish community will be to model human cardiovascular pathology in this organism even closer than it is now. Recent technical improvements allow the generation of mutants for any gene. Thus it is likely that in the not too far future, mutants for nearly every gene will be available. Genetic interactions between genes, gene-environment interactions, or the role of modifier genes, which are important determinants of the phenotype severity of a particular mutation, can be analyzed at ease and with significant lower costs than in mice. There is exciting progress in the increasingly sophisticated means of genetic manipulation in zebrafish that parallels that of mice. Certainly the zebrafish will not be able to fully substitute the mouse, however it will reduce the number of experiments that need to be performed in mammals. Moreover, recent improvements in mass screening approaches such as chemical genetics will be a prime area for the zebrafish model. In conclusion, with well-constructed thoughtful experimental designs, a bright future is assured for the small but mighty zebrafish model.
Acknowledgements
The drawing of the adult zebrafish heart and the histological section are the courtesy of Dr. Jan Schlüter, Harefield Heart Science Centre, which is herewith gratefully acknowledged. Work in the authors' lab is funded through the MRC (MR/J010383/1) and the Magdi Yacoub Institute.
References
- [1].Grunwald DJ, Streisinger G. Induction of recessive lethal and specific locus mutations in the zebrafish with ethyl nitrosourea. Genet Res. 1992;59:103–116. doi: 10.1017/s0016672300030317. [DOI] [PubMed] [Google Scholar]
- [2].Haffter P, Nusslein-Volhard C. Large scale genetics in a small vertebrate, the zebrafish. Int J Dev Biol. 1996;40:221–227. [PubMed] [Google Scholar]
- [3].Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, Belak J, Boggs C. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- [4].Milan DJ, Kim AM, Winterfield JR, Jones IL, Pfeufer A, Sanna S, Arking DE, Amsterdam AH, Sabeh KM, Mably JD, Rosenbaum DS, Peterson RT, Chakravarti A, Kaab S, Roden DM, MacRae CA. Drug-sensitized zebrafish screen identifies multiple genes, including GINS3, as regulators of myocardial repolarization. Circulation. 2009;120:553–559. doi: 10.1161/CIRCULATIONAHA.108.821082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Beis D, Stainier DY. In vivo cell biology: following the zebrafish trend. Trends Cell Biol. 2006;16:105–112. doi: 10.1016/j.tcb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- [6].Distel M, Jennifer CH, Koster RW. In vivo cell biology using Gal4-mediated multicolor subcellular labeling in zebrafish. Commun Integr Biol. 2011;4:336–339. doi: 10.4161/cib.4.3.15037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].White RM, Sessa A, Burke C, Bowman T, LeBlanc J, Ceol C, Bourque C, Dovey M, Goessling W, Burns CE, Zon LI. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ni TT, Rellinger EJ, Mukherjee A, Xie S, Stephens L, Thorne CA, Kim K, Hu J, Lee E, Marnett L, Hatzopoulos AK, Zhong TP. Discovering small molecules that promote cardiomyocyte generation by modulating Wnt signaling. Chem Biol. 2011;18:1658–1668. doi: 10.1016/j.chembiol.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kleinjan DA, Bancewicz RM, Gautier P, Dahm R, Schonthaler HB, Damante G, Seawright A, Hever AM, Yeyati PL, van Heyningen V, Coutinho P. Subfunctionalization of duplicated zebrafish pax6 genes by cis-regulatory divergence. PLoS Genet. 2008;4:e29. doi: 10.1371/journal.pgen.0040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Poon KL, Liebling M, Kondrychyn I, Garcia-Lecea M, Korzh V. Zebrafish cardiac enhancer trap lines: new tools for in vivo studies of cardiovascular development and disease. Dev Dyn. 2010;239:914–926. doi: 10.1002/dvdy.22203. [DOI] [PubMed] [Google Scholar]
- [12].Koga A, Cheah FS, Hamaguchi S, Yeo GH, Chong SS. Germline transgenesis of zebrafish using the medaka Tol1 transposon system. Dev Dyn. 2008;237:2466–2474. doi: 10.1002/dvdy.21688. [DOI] [PubMed] [Google Scholar]
- [13].Kettleborough RN, Bruijn E, Eeden F, Cuppen E, Stemple DL. High-throughput target-selected gene inactivation in zebrafish. Methods Cell Biol. 2011;104:121–127. doi: 10.1016/B978-0-12-374814-0.00006-9. [DOI] [PubMed] [Google Scholar]
- [14].Bakkers J. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc Res. 2011;91:279–288. doi: 10.1093/cvr/cvr098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tu S, Chi NC. Zebrafish models in cardiac development and congenital heart birth defects. Differentiation. 2012;84:4–16. doi: 10.1016/j.diff.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liu J, Stainier DY. Zebrafish in the study of early cardiac development. Circ Res. 2012;110:870–874. doi: 10.1161/CIRCRESAHA.111.246504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Miura GI, Yelon D. A guide to analysis of cardiac phenotypes in the zebrafish embryo. Methods Cell Biol. 2011;101:161–180. doi: 10.1016/B978-0-12-387036-0.00007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kupperman E, An S, Osborne N, Waldron S, Stainier DY. A sphingosine-1-phosphate receptor regulates cell migration during vertebrate heart development. Nature. 2000;406:192–195. doi: 10.1038/35018092. [DOI] [PubMed] [Google Scholar]
- [19].Osborne N, Brand-Arzamendi K, Ober EA, Jin SW, Verkade H, Holtzman NG, Yelon D, Stainier DY. The spinster homolog, two of hearts, is required for sphingosine 1-phosphate signaling in zebrafish. Curr Biol. 2008;18:1882–1888. doi: 10.1016/j.cub.2008.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kawahara A, Nishi T, Hisano Y, Fukui H, Yamaguchi A, Mochizuki N. The sphingolipid transporter spns2 functions in migration of zebrafish myocardial precursors. Science. 2009;323:524–527. doi: 10.1126/science.1167449. [DOI] [PubMed] [Google Scholar]
- [21].Keegan BR, Meyer D, Yelon D. Organization of cardiac chamber progenitors in the zebrafish blastula. Development. 2004;131:3081–3091. doi: 10.1242/dev.01185. [DOI] [PubMed] [Google Scholar]
- [22].Glickman NS, Yelon D. Cardiac development in zebrafish: coordination of form and function. Semin Cell Dev Biol. 2002;13:507–513. doi: 10.1016/s1084952102001040. [DOI] [PubMed] [Google Scholar]
- [23].Panakova D, Werdich AA, Macrae CA. Wnt11 patterns a myocardial electrical gradient through regulation of the L-type Ca(2+) channel. Nature. 2010;466:874–878. doi: 10.1038/nature09249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Auman HJ, Coleman H, Riley HE, Olale F, Tsai HJ, Yelon D. Functional modulation of cardiac form through regionally confined cell shape changes. PLoS Biol. 2007;5:e53. doi: 10.1371/journal.pbio.0050053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Huang W, Zhang R, Xu X. Myofibrillogenesis in the developing zebrafish heart: a functional study of tnnt2. Dev Biol. 2009;331:237–249. doi: 10.1016/j.ydbio.2009.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gassmann M, Casagranda F, Orioli D, Simon H, Lai C, Klein R, Lemke G. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature. 1995;378:390–394. doi: 10.1038/378390a0. [DOI] [PubMed] [Google Scholar]
- [27].Lee K-F, Simon H, Chen H, Bates B, Hung M-C, Hauser C. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature. 1995;378:394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- [28].Chen H, Shi S, Acosta L, Li W, Lu J, Bao S, Chen Z, Yang Z, Schneider MD, Chien KR, Conway SJ, Yoder MC, Haneline LS, Franco D, Shou W. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development. 2004;131:2219–2231. doi: 10.1242/dev.01094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Stankunas K, Hang CT, Tsun ZY, Chen H, Lee NV, Wu JI, Shang C, Bayle JH, Shou W, Iruela-Arispe ML, Chang CP. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev Cell. 2008;14:298–311. doi: 10.1016/j.devcel.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liu J, Bressan M, Hassel D, Huisken J, Staudt D, Kikuchi K, Poss KD, Mikawa T, Stainier DY. A dual role for ErbB2 signaling in cardiac trabeculation. Development. 2010;137:3867–3875. doi: 10.1242/dev.053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].de Pater E, Clijsters L, Marques SR, Lin YF, Garavito-Aguilar ZV, Yelon D, Bakkers J. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development. 2009;136:1633–1641. doi: 10.1242/dev.030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gupta V, Poss KD. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature. 2012;484:479–484. doi: 10.1038/nature11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Stainier DY, Beis D, Jungblut B, Bartman T. Endocardial cushion formation in zebrafish. Cold Spring Harb Symp Quant Biol. 2002;67:49–56. doi: 10.1101/sqb.2002.67.49. [DOI] [PubMed] [Google Scholar]
- [34].Scherz PJ, Huisken J, Sahai-Hernandez P, Stainier DY. High-speed imaging of developing heart valves reveals interplay of morphogenesis and function. Development. 2008;135:1179–1187. doi: 10.1242/dev.010694. [DOI] [PubMed] [Google Scholar]
- [35].Walsh EC, Stainier DY. UDP-glucose dehydrogenase required for cardiac valve formation in zebrafish. Science. 2001;293:1670–1673. doi: 10.1126/science.293.5535.1670. [DOI] [PubMed] [Google Scholar]
- [36].Chi NC, Shaw RM, De Val S, Kang G, Jan LY, Black BL, Stainier DY. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 2008;22:734–739. doi: 10.1101/gad.1629408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Milan DJ, Giokas AC, Serluca FC, Peterson RT, MacRae CA. Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development. 2006;133:1125–1132. doi: 10.1242/dev.02279. [DOI] [PubMed] [Google Scholar]
- [38].Kortschak RD, Tamme R, Lardelli M. Evolutionary analysis of vertebrate Notch genes. Dev Genes Evol. 2001;211:350–354. doi: 10.1007/s004270100159. [DOI] [PubMed] [Google Scholar]
- [39].Martin RT, Bartman T. Analysis of heart valve development in larval zebrafish. Dev Dyn. 2009;238:1796–1802. doi: 10.1002/dvdy.21976. [DOI] [PubMed] [Google Scholar]
- [40].Dor Y, Camenisch TD, Itin A, Fishman GI, McDonald JA, Carmeliet P, Keshet E. A novel role for VEGF in endocardial cushion formation and its potential contribution to congenital heart defects. Development. 2001;128:1531–1538. doi: 10.1242/dev.128.9.1531. [DOI] [PubMed] [Google Scholar]
- [41].Rutenberg JB, Fischer A, Jia H, Gessler M, Zhong TP, Mercola M. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development. 2006;133:4381–4390. doi: 10.1242/dev.02607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Timmerman LA, Grego-Bessa J, Raya A, Bertran E, Perez-Pomares JM, Diez J, Aranda S, Palomo S, McCormick F, Izpisua-Belmonte JC, dela Pompa JL. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18:99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Iwamoto R, Yamazaki S, Asakura M, Takashima S, Hasuwa H, Miyado K, Adachi S, Kitakaze M, Hashimoto K, Raab G, Nanba D, Higashiyama S, Hori M, Klagsbrun M, Mekada E. Heparin-binding EGF-like growth factor and ErbB signaling is essential for heart function. Proc Natl Acad Sci USA. 2003;100:3221–3226. doi: 10.1073/pnas.0537588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Camenisch TD, Schroeder JA, Bradley J, Klewer SE, McDonald JA. Heart-valve mesenchyme formation is dependent on hyaluronan-augmented activation of ErbB2-ErbB3 receptors. Nat Med. 2002;8:850–855. doi: 10.1038/nm742. [DOI] [PubMed] [Google Scholar]
- [45].Sugi Y, Yamamura H, Okagawa H, Markwald RR. Bone morphogenetic protein-2 can mediate myocardial regulation of atrioventricular cushion mesenchymal cell formation in mice. Dev Biol. 2004;269:505–518. doi: 10.1016/j.ydbio.2004.01.045. [DOI] [PubMed] [Google Scholar]
- [46].Ma L, Lu MF, Schwartz RJ, Martin JF. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;132:5601–5611. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- [47].Potts JD, Dagle JM, Walder JA, Weeks DL, Runyan RB. Epithelial-mesenchymal transformation of embryonic cardiac endothelial cells is inhibited by a modified antisense oligodeoxynucleotide to transforming growth factor beta 3. Proc Natl Acad Sci USA. 1991;88:1516–1520. doi: 10.1073/pnas.88.4.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hurlstone AF, Haramis AP, Wienholds E, Begthel H, Korving J, Van Eeden F, Cuppen E, Zivkovic D, Plasterk RH, Clevers H. The Wnt/beta-catenin pathway regulates cardiac valve formation. Nature. 2003;425:633–637. doi: 10.1038/nature02028. [DOI] [PubMed] [Google Scholar]
- [49].Chang CP, Neilson JR, Bayle JH, Gestwicki JE, Kuo A, Stankunas K, Graef IA, Crabtree GR. A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis. Cell. 2004;118:649–663. doi: 10.1016/j.cell.2004.08.010. [DOI] [PubMed] [Google Scholar]
- [50].Lindsey SE, Butcher JT. The cycle of form and function in cardiac valvulogenesis. Aswan Heart Centre Sci Pract Ser. 2011;2:10. [Google Scholar]
- [51].Hitz MP, Brand T, Andelfinger G. Genetic regulation of heart valve development. Clinical implications. Aswan Heart Centre Sci Pract Ser. 2011;2 [Google Scholar]
- [52].Beis D, Bartman T, Jin SW, Scott IC, D'Amico LA, Ober EA, Verkade H, Frantsve J, Field HA, Wehman A, Baier H, Tallafuss A, Bally-Cuif L, Chen JN, Stainier DY, Jungblut B. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132:4193–4204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- [53].Hyde AS, Farmer E, Easley KE, van Lammeren K, Christoffels VM, Barycki JJ, Bakkers J, Simpson MA. UDP-glucose dehydrogenase polymorphisms from patients with congenital heart valve defects disrupt enzyme stability and quaternary assembly. J Biol Chem. 2012;287:32708–32716. doi: 10.1074/jbc.M112.395202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Patra C, Diehl F, Ferrazzi F, van Amerongen MJ, Novoyatleva T, Schaefer L, Muhlfeld C, Jungblut B, Engel FB. Nephronectin regulates atrioventricular canal differentiation via Bmp4-Has2 signaling in zebrafish. Development. 2011;138:4499–4509. doi: 10.1242/dev.067454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lee HC, Tsai JN, Liao PY, Tsai WY, Lin KY, Chuang CC, Sun CK, Chang WC, Tsai HJ. Glycogen synthase kinase 3 alpha and 3 beta have distinct functions during cardiogenesis of zebrafish embryo. BMC Dev Biol. 2007;7:93. doi: 10.1186/1471-213X-7-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Vermot J, Gallego Llamas J, Fraulob V, Niederreither K, Chambon P, Dolle P. Retinoic acid controls the bilateral symmetry of somite formation in the mouse embryo. Science. 2005;308:563–566. doi: 10.1126/science.1108363. [DOI] [PubMed] [Google Scholar]
- [57].Fishman M, Chien K. Fashioning the vertebrate heart: earliest embryonic decisions. Development. 1997;124:2099–2117. doi: 10.1242/dev.124.11.2099. [DOI] [PubMed] [Google Scholar]
- [58].Peshkovsky C, Totong R, Yelon D. Dependence of cardiac trabeculation on neuregulin signaling and blood flow in zebrafish. Dev Dyn. 2011;240:446–456. doi: 10.1002/dvdy.22526. [DOI] [PubMed] [Google Scholar]
- [59].Lin YF, Swinburne I, Yelon D. Multiple influences of blood flow on cardiomyocyte hypertrophy in the embryonic zebrafish heart. Dev Biol. 2012;362:242–253. doi: 10.1016/j.ydbio.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hu N, Sedmera D, Yost HJ, Clark EB. Structure and function of the developing zebrafish heart. Anat Rec. 2000;260:148–157. doi: 10.1002/1097-0185(20001001)260:2<148::AID-AR50>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- [61].Grimes AC, Duran AC, Sans-Coma V, Hami D, Santoro MM, Torres M. Phylogeny informs ontogeny: a proposed common theme in the arterial pole of the vertebrate heart. Evol Dev. 2010;12:552–567. doi: 10.1111/j.1525-142X.2010.00441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Grimes AC, Stadt HA, Shepherd IT, Kirby ML. Solving an enigma: arterial pole development in the zebrafish heart. Dev Biol. 2006;290:265–276. doi: 10.1016/j.ydbio.2005.11.042. [DOI] [PubMed] [Google Scholar]
- [63].Mably JD, Mohideen MAPK, Burns CG, Chen JN, Fishman MC. Heart of glass regulates the concentric growth of the heart in zebrafish. Curr Biol. 2003;13:2138–2147. doi: 10.1016/j.cub.2003.11.055. [DOI] [PubMed] [Google Scholar]
- [64].Mably JD, Chuang LP, Serluca FC, Mohideen MAPK, Chen JN, Fishman MC. Santa and valentine pattern concentric growth of the cardiac myocardium in the zebrafish. Development. 2006;133:3139–3146. doi: 10.1242/dev.02469. [DOI] [PubMed] [Google Scholar]
- [65].Kleaveland B, Zheng X, Liu JJ, Blum J, Tung JT, Zou Z, Sweeney SM, Chen M, Lu MM, Zhou D, Kitajewski J, Affolter M, Ginsberg MH, Kahn ML. Regulation of the cardiovascular development and integrity by the heart of glass-cerbral cavernous malformation protein pathway. Nat Med. 2006;15:169–176. doi: 10.1038/nm.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zheng X, Xu C, Di Lorenzo A, Kleaveland B, Zou Z, Seiler C, Chen M, Cheng L, Xiao J, Je J, Pack MA, Sessa WC, Kahn ML. CCM3 signaling through sterile 20-like kinases plays an essential role during zebrafish cardiovascular development and cerebral cavernous malformations. J Clin Invest. 2010;120:2795–2804. doi: 10.1172/JCI39679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Sakaguchi TF, Sadler KC, Cosnier C, Stainier DYI. Endothelial signals modulate hepatocyte apicobasal polarization in zebrafish. Curr Biol. 2008;18:1565–1571. doi: 10.1016/j.cub.2008.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- [69].Dyer LA, Kirby ML. The role of secondary heart field in cardiac development. Dev Biol. 2009;336:137–144. doi: 10.1016/j.ydbio.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kelly RG. The second heart field. Curr Top Dev Biol. 2012;100:33–65. doi: 10.1016/B978-0-12-387786-4.00002-6. [DOI] [PubMed] [Google Scholar]
- [71].Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Tzahor E. Heart and craniofacial muscle development: a new developmental theme of distinct myogenic fields. Dev Biol. 2009;327:273–279. doi: 10.1016/j.ydbio.2008.12.035. [DOI] [PubMed] [Google Scholar]
- [73].Lescroart F, Kelly RG, Le Garrec JF, Nicolas JF, Meilhac SM, Buckingham M. Clonal analysis reveals common lineage relationships between head muscles and second heart field derivatives in the mouse embryo. Development. 2010;137:3269–3279. doi: 10.1242/dev.050674. [DOI] [PubMed] [Google Scholar]
- [74].Zhou Y, Cashman TJ, Nevis KR, Obregon P, Carney SA, Liu Y, Gu A, Mosimann C, Sondalle S, Peterson RE, Heideman W, Burns CE, Burns CG. Latent TGF-beta binding protein 3 identifies a second heart field in zebrafish. Nature. 2011;474:645–648. doi: 10.1038/nature10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Manner J, Perez-Pomares JM, Macias D, Munoz-Chapuli R. The origin, formation and developmental significance of the epicardium: a review. Cells Tissues Organs. 2001;169:89–103. doi: 10.1159/000047867. [DOI] [PubMed] [Google Scholar]
- [76].Schlueter J, Brand T. Origin and fates of the proepicardium. Aswan Heart Centre Sci Pract Ser. 2011;2:11. [Google Scholar]
- [77].Schlueter J, Brand T. Epicardial progenitor cells in cardiac development and regeneration. J Cardiovasc Transl Res. 2012;5(5):641–653. doi: 10.1007/s12265-012-9377-4. [DOI] [PubMed] [Google Scholar]
- [78].Mikawa T, Brand T. Epicardial lineage: origins and fates. In: Harvey RP, Rosenthal N, editors. Heart Development and Regeneration. Vol. 1. Academic Press; 2010. pp. 325–344. [Google Scholar]
- [79].Liu J, Stainier DY. Tbx5 and Bmp signaling are essential for proepicardium specification in zebrafish. Circ Res. 2010;106:1818–1828. doi: 10.1161/CIRCRESAHA.110.217950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Serluca FC. Development of the proepicardial organ in the zebrafish. Dev Biol. 2008;315:18–27. doi: 10.1016/j.ydbio.2007.10.007. [DOI] [PubMed] [Google Scholar]
- [81].Ishii Y, Langberg JD, Hurtado R, Lee S, Mikawa T. Induction of proepicardial marker gene expression by the liver bud. Development. 2007;134:3627–3637. doi: 10.1242/dev.005280. [DOI] [PubMed] [Google Scholar]
- [82].Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Rudat C, Kispert A. Wt1 and epicardial fate mapping. Circ Res. 2012;111:165–169. doi: 10.1161/CIRCRESAHA.112.273946. [DOI] [PubMed] [Google Scholar]
- [84].Kikuchi K, Gupta V, Wang J, Holdway JE, Wills AA, Fang Y, Poss KD. tcf21+epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011;138:2895–2902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Keyte A, Hutson MR. The neural crest in cardiac congenital anomalies. Differentiation. 2012;84:25–40. doi: 10.1016/j.diff.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Kirby ML, Waldo KL. Neural crest and cardiovascular patterning. Circ Res. 1995;77:211–215. doi: 10.1161/01.res.77.2.211. [DOI] [PubMed] [Google Scholar]
- [87].Sun X, Zhang R, Lin X, Xu X. Wnt3a regulates the development of cardiac neural crest cells by modulating expression of cysteine-rich intestinal protein 2 in rhombomere 6. Circ Res. 2008;102:831–839. doi: 10.1161/CIRCRESAHA.107.166488. [DOI] [PubMed] [Google Scholar]
- [88].Li YX, Zdanowicz M, Young L, Kumiski D, Leatherbury L, Kirby ML. Cardiac neural crest in zebrafish embryos contributes to myocardial cell lineage and early heart function. Dev Dyn. 2003;226:540–550. doi: 10.1002/dvdy.10264. [DOI] [PubMed] [Google Scholar]
- [89].Sato M, Yost HJ. Cardiac neural crest contributes to cardiomyogenesis in zebrafish. Dev Biol. 2003;257:127–139. doi: 10.1016/s0012-1606(03)00037-x. [DOI] [PubMed] [Google Scholar]
- [90].Nasevicius A, Ekker SC. Effective targeted gene ’knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- [91].Bill BR, Petzold AM, Clark KJ, Schimmenti LA, Ekker SC. A primer for morpholino use in zebrafish. Zebrafish. 2009;6:69–77. doi: 10.1089/zeb.2008.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Eisen JS, Smith JC. Controlling morpholino experiments: don't stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- [93].Moens CB, Donn TM, Wolf-Saxon ER, Ma TP. Reverse genetics in zebrafish by TILLING. Brief Funct Genomic Proteomic. 2008;7:454–459. doi: 10.1093/bfgp/eln046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, Lin SY, Nissen RM, Hopkins N. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet. 2002;31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- [95].Parinov S, Kondrichin I, Korzh V, Emelyanov A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev Dyn. 2004;231:449–459. doi: 10.1002/dvdy.20157. [DOI] [PubMed] [Google Scholar]
- [96].Kawakami K. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 2004;77:201–222. doi: 10.1016/s0091-679x(04)77011-9. [DOI] [PubMed] [Google Scholar]
- [97].Balciunas D, Davidson AE, Sivasubbu S, Hermanson SB, Welle Z, Ekker SC. Enhancer trapping in zebrafish using the sleeping beauty transposon. BMC Genomics. 2004;5:62. doi: 10.1186/1471-2164-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Kondrychyn I, Garcia-Lecea M, Emelyanov A, Parinov S, Korzh V. Genome-wide analysis of Tol2 transposon reintegration in zebrafish. BMC Genomics. 2009;10:418. doi: 10.1186/1471-2164-10-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Clark KJ, Balciunas D, Pogoda HM, Ding Y, Westcot SE, Bedell VM, Greenwood TM, Urban MD, Skuster KJ, Petzold AM, Ni J, Nielsen AL, Patowary A, Scaria V, Sivasubbu S, Xu X, Hammerschmidt M, Ekker SC. In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nat Methods. 2011;8:506–515. doi: 10.1038/nmeth.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Zhu C, Smith T, McNulty J, Rayla AL, Lakshmanan A, Siekmann AF, Buffardi M, Meng X, Shin J, Padmanabhan A, Cifuentes D, Giraldez AJ, Look AT, Epstein JA, Lawson ND, Wolfe SA. Evaluation and application of modularly assembled zinc-finger nucleases in zebrafish. Development. 2011;138:4555–4564. doi: 10.1242/dev.066779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- [102].Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, Khayter C, Joung JK, Sander JD, Peterson RT, Yeh JR. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012;40:8001–8010. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug II RG, Tan W, Penheiter SG, Ma AC, Leung AY, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012. doi: 10.1038/nature11537. [in the press] [DOI] [PMC free article] [PubMed]
- [105].Peal DS, Peterson RT, Milan D. Small molecule screening in zebrafish. J Cardiovasc Transl Res. 2010;3:454–460. doi: 10.1007/s12265-010-9212-8. [DOI] [PubMed] [Google Scholar]
- [106].Becker JR, Robinson TY, Sachidanandan C, Kelly AE, Coy S, Peterson RT, MacRae CA. In vivo natriuretic peptide reporter assay identifies chemical modifiers of hypertrophic cardiomyopathy signalling. Cardiovasc Res. 2012;93:463–470. doi: 10.1093/cvr/cvr350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Peal DS, Mills RW, Lynch SN, Mosley JM, Lim E, Ellinor PT, January CT, Peterson RT, Milan DJ. Novel chemical suppressors of long QT syndrome identified by an in vivo functional screen. Circulation. 2011;123:23–30. doi: 10.1161/CIRCULATIONAHA.110.003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Molina G, Vogt A, Bakan A, Dai W, Queiroz de Oliveira P, Znosko W, Smithgall TE, Bahar I, Lazo JS, Day BW, Tsang M. Zebrafish chemical screening reveals an inhibitor of Dusp6 that expands cardiac cell lineages. Nat Chem Biol. 2009;5:680–687. doi: 10.1038/nchembio.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Chi NC, Shaw RM, Jungblut B, Huisken J, Ferrer T, Arnaout R, Scott I, Beis D, Xiao T, Baier H, Jan LY, Tristani-Firouzi M, Stainier DY. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 2008;6:e109. doi: 10.1371/journal.pbio.0060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Baker K, Warren KS, Yellen G, Fishman MC. Defective “pacemaker” current (Ih) in a zebrafish mutant with a slow heart rate. Proc Natl Acad Sci USA. 1997;94:4554–4559. doi: 10.1073/pnas.94.9.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Warren KS, Baker K, Fishman MC. The slow mo mutation reduces pacemaker current and heart rate in adult zebrafish. Am J Physiol Heart Circ Physiol. 2001;281:H1711–H1719. doi: 10.1152/ajpheart.2001.281.4.H1711. [DOI] [PubMed] [Google Scholar]
- [112].Irisawa H. Comparative physiology of the cardiac pacemaker mechanism. Physiol Rev. 1978;58:461–498. doi: 10.1152/physrev.1978.58.2.461. [DOI] [PubMed] [Google Scholar]
- [113].Saito A. Electophysiological studies on the pacemaker of several fish hearts. Zool Mag. 1969;78:291–296. [Google Scholar]
- [114].Blaschke RJ, Hahurij ND, Kuijper S, Just S, Wisse LJ, Deissler K, Maxelon T, Anastassiadis K, Spitzer J, Hardt SE, Scholer H, Feitsma H, Rottbauer W, Blum M, Meijlink F, Rappold G, Gittenberger-de Groot AC. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- [115].Arrenberg AB, Stainier DY, Baier H, Huisken J. Optogenetic control of cardiac function. Science. 2010;330:971–974. doi: 10.1126/science.1195929. [DOI] [PubMed] [Google Scholar]
- [116].Tessadori F, van Weerd JH, Burkhard SB, Verkerk AO, de Pater E, Boukens BJ, Vink A, Christoffels VM, Bakkers J. Identification and functional characterization of cardiac pacemaker cells in zebrafish. PLoS One. 2012;7:e47644. doi: 10.1371/journal.pone.0047644. doi: 10.1371/journal.pone.0047644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Milan DJ, Giokas AC, Serluca FC, Peterson RT, MacRae CA. Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development. 2006;133:1125–1132. doi: 10.1242/dev.02279. [DOI] [PubMed] [Google Scholar]
- [118].Kirchmaier BC, Poon KL, Schwerte T, Huisken J, Winkler C, Jungblut B, Stainier DY, Brand T. The Popeye domain containing 2 (popdc2) gene in zebrafish is required for heart and skeletal muscle development. Dev Biol. 2012;363:438–450. doi: 10.1016/j.ydbio.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Sedmera D, Reckova M, deAlmeida A, Sedmerova M, Biermann M, Volejnik J, Sarre A, Raddatz E, McCarthy RA, Gourdie RG, Thompson RP. Functional and morphological evidence for a ventricular conduction system in zebrafish and Xenopus hearts. Am J Physiol Heart Circ Physiol. 2003;284:H1152–H1160. doi: 10.1152/ajpheart.00870.2002. [DOI] [PubMed] [Google Scholar]
- [120].Chi NC, Bussen M, Brand-Arzamendi K, Ding C, Olgin JE, Shaw RM, Martin GR, Stainier DY. Cardiac conduction is required to preserve cardiac chamber morphology. Proc Natl Acad Sci USA. 2010;107:14662–14667. doi: 10.1073/pnas.0909432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Forouhar AS, Hove JR, Calvert C, Flores J, Jadvar H, Gharib M. Electrocardiographic characterization of embryonic zebrafish. Conf Proc IEEE Eng Med Biol Soc. 2004;5:3615–3617. doi: 10.1109/IEMBS.2004.1404016. [DOI] [PubMed] [Google Scholar]
- [122].Yu F, Huang J, Adlerz K, Jadvar H, Hamdan MH, Chi N, Chen JN, Hsiai TK. Evolving cardiac conduction phenotypes in developing zebrafish larvae: implications to drug sensitivity. Zebrafish. 2010;7:325–331. doi: 10.1089/zeb.2010.0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Milan DJ, Jones IL, Ellinor PT, MacRae CA. In vivo recording of adult zebrafish electrocardiogram and assessment of drug-induced QT prolongation. Am J Physiol Heart Circ Physiol. 2006;291:H269–H273. doi: 10.1152/ajpheart.00960.2005. [DOI] [PubMed] [Google Scholar]
- [124].Wehrens XH, Doevendans PA, Ophuis TJ, Wellens HJ. A comparison of electrocardiographic changes during reperfusion of acute myocardial infarction by thrombolysis or percutaneous transluminal coronary angioplasty. Am Heart J. 2000;139:430–436. doi: 10.1016/s0002-8703(00)90086-3. [DOI] [PubMed] [Google Scholar]
- [125].Leong IU, Skinner JR, Shelling AN, Love DR. Zebrafish as a model for long QT syndrome: the evidence and the means of manipulating zebrafish gene expression. Acta Physiol. 2010;199:257–276. doi: 10.1111/j.1748-1716.2010.02111.x. [DOI] [PubMed] [Google Scholar]
- [126].Nemtsas P, Wettwer E, Christ T, Weidinger G, Ravens U. Adult zebrafish heart as a model for human heart? An electrophysiological study. J Mol Cell Cardiol. 2010;48:161–171. doi: 10.1016/j.yjmcc.2009.08.034. [DOI] [PubMed] [Google Scholar]
- [127].Kopp R, Schwerte T, Pelster B. Cardiac performance in the zebrafish breakdance mutant. J Exp Biol. 2005;208:2123–2134. doi: 10.1242/jeb.01620. [DOI] [PubMed] [Google Scholar]
- [128].Arnaout R, Ferrer T, Huisken J, Spitzer K, Stainier DY, Tristani-Firouzi M, Chi NC. Zebrafish model for human long QT syndrome. Proc Natl Acad Sci USA. 2007;104:11316–11321. doi: 10.1073/pnas.0702724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Hassel D, Scholz EP, Trano N, Friedrich O, Just S, Meder B, Weiss DL, Zitron E, Marquart S, Vogel B, Karle CA, Seemann G, Fishman MC, Katus HA, Rottbauer W. Deficient zebrafish ether-a-go-go-related gene channel gating causes short-QT syndrome in zebrafish reggae mutants. Circulation. 2008;117:866–875. doi: 10.1161/CIRCULATIONAHA.107.752220. [DOI] [PubMed] [Google Scholar]
- [130].Haverinen J, Hassinen M, Vornanen M. Fish cardiac sodium channels are tetrodotoxin sensitive. Acta Physiol. 2007;191:197–204. doi: 10.1111/j.1748-1716.2007.01734.x. [DOI] [PubMed] [Google Scholar]
- [131].Rottbauer W, Baker K, Wo ZG, Mohideen MA, Cantiello HF, Fishman MC. Growth and function of the embryonic heart depend upon the cardiac-specific L-type calcium channel alpha1 subunit. Dev Cell. 2001;1:265–275. doi: 10.1016/s1534-5807(01)00023-5. [DOI] [PubMed] [Google Scholar]
- [132].Cho CH, Kim SS, Jeong MJ, Lee CO, Shin HS. The Na+–Ca2+ exchanger is essential for embryonic heart development in mice. Mol Cells. 2000;10:712–722. doi: 10.1007/s10059-000-0712-2. [DOI] [PubMed] [Google Scholar]
- [133].Henderson SA, Goldhaber JI, So JM, Han T, Motter C, Ngo A, Chantawansri C, Ritter MR, Friedlander M, Nicoll DA, Frank JS, Jordan MC, Roos KP, Ross RS, Philipson KD. Functional adult myocardium in the absence of Na+–Ca2+ exchange: cardiac-specific knockout of NCX1. Circ Res. 2004;95:604–611. doi: 10.1161/01.RES.0000142316.08250.68. [DOI] [PubMed] [Google Scholar]
- [134].Ebert AM, Hume GL, Warren KS, Cook NP, Burns CG, Mohideen MA, Siegal G, Yelon D, Fishman MC, Garrity DM. Calcium extrusion is critical for cardiac morphogenesis and rhythm in embryonic zebrafish hearts. Proc Natl Acad Sci USA. 2005;102:17705–17710. doi: 10.1073/pnas.0502683102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Langenbacher AD, Dong Y, Shu X, Choi J, Nicoll DA, Goldhaber JI, Philipson KD, Chen JN. Mutation in sodium–calcium exchanger 1 (NCX1) causes cardiac fibrillation in zebrafish. Proc Natl Acad Sci USA. 2005;102:17699–17704. doi: 10.1073/pnas.0502679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Shu X, Cheng K, Patel N, Chen F, Joseph E, Tsai HJ, Chen JN. Na,K-ATPase is essential for embryonic heart development in the zebrafish. Development. 2003;130:6165–6173. doi: 10.1242/dev.00844. [DOI] [PubMed] [Google Scholar]
- [137].Cibrian-Uhalte E, Langenbacher A, Shu X, Chen JN, Abdelilah-Seyfried S. Involvement of zebrafish Na+, K+ATPase in myocardial cell junction maintenance. J Cell Biol. 2007;176:223–230. doi: 10.1083/jcb.200606116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Chopra SS, Stroud DM, Watanabe H, Bennett JS, Burns CG, Wells KS, Yang T, Zhong TP, Roden DM. Voltage-gated sodium channels are required for heart development in zebrafish. Circ Res. 2010;106:1342–1350. doi: 10.1161/CIRCRESAHA.109.213132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Xu X, Meiler SE, Zhong TP, Mohideen M, Crossley DA, Burggren WW, Fishman MC. Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat Genet. 2002;30:205–209. doi: 10.1038/ng816. [DOI] [PubMed] [Google Scholar]
- [140].Zhao L, Zhao X, Tian T, Lu Q, Skrbo-Larssen N, Wu D, Kuang Z, Zheng X, Han Y, Yang S, Zhang C, Meng A. Heart-specific isoform of tropomyosin4 is essential for heartbeat in zebrafish embryos. Cardiovasc Res. 2008;80:200–208. doi: 10.1093/cvr/cvn177. [DOI] [PubMed] [Google Scholar]
- [141].Sehnert AJ, Huq A, Weinstein BM, Walker C, Fishman M, Stainier DY. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat Genet. 2002;31:106–110. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- [142].Rottbauer W, Wessels G, Dahme T, Just S, Trano N, Hassel D, Burns CG, Katus HA. Cardiac myosin light chain-2: a novel essential component of thick-myofilament assembly and contractility of the heart. Circ Res. 2006;99:323–331. doi: 10.1161/01.RES.0000234807.16034.fe. [DOI] [PubMed] [Google Scholar]
- [143].Chen Z, Huang W, Dahme T, Rottbauer W, Ackerman MJ, Xu X. Depletion of zebrafish essential and regulatory myosin light chains reduces cardiac function through distinct mechanisms. Cardiovasc Res. 2008;79:97–108. doi: 10.1093/cvr/cvn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Meder B, Laufer C, Hassel D, Just S, Marquart S, Vogel B, Hess A, Fishman MC, Katus HA, Rottbauer W. A single serine in the carboxyl terminus of cardiac essential myosin light chain-1 controls cardiomyocyte contractility in vivo. Circ Res. 2009;104:650–659. doi: 10.1161/CIRCRESAHA.108.186676. [DOI] [PubMed] [Google Scholar]
- [145].Berdougo E, Coleman H, Lee DH, Stainier DY, Yelon D. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development. 2003;130:6121–6129. doi: 10.1242/dev.00838. [DOI] [PubMed] [Google Scholar]
- [146].Becker JR, Deo RC, Werdich AA, Panakova D, Coy S, MacRae CA. Human cardiomyopathy mutations induce myocyte hyperplasia and activate hypertrophic pathways during cardiogenesis in zebrafish. Dis Model Mech. 2011;4:400–410. doi: 10.1242/dmm.006148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Seeley M, Huang W, Chen Z, Wolff WO, Lin X, Xu X. Depletion of zebrafish titin reduces cardiac contractility by disrupting the assembly of Z-discs and A-bands. Circ Res. 2007;100:238–245. doi: 10.1161/01.RES.0000255758.69821.b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Dowling JJ, Gibbs E, Russell M, Goldman D, Minarcik J, Golden JA, Feldman EL. Kindlin-2 is an essential component of intercalated discs and is required for vertebrate cardiac structure and function. Circ Res. 2008;102:423–431. doi: 10.1161/CIRCRESAHA.107.161489. [DOI] [PubMed] [Google Scholar]
- [149].Dowling JJ, Vreede AP, Kim S, Golden J, Feldman EL. Kindlin-2 is required for myocyte elongation and is essential for myogenesis. BMC Cell Biol. 2008;9:36. doi: 10.1186/1471-2121-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Seeger TS, Frank D, Rohr C, Will R, Just S, Grund C, Lyon R, Luedde M, Koegl M, Sheikh F, Rottbauer W, Franke WW, Katus HA, Olson EN, Frey N. Myozap, a novel intercalated disc protein, activates serum response factor-dependent signaling and is required to maintain cardiac function in vivo. Circ Res. 2010;106:880–890. doi: 10.1161/CIRCRESAHA.109.213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Beqqali A, Monshouwer-Kloots J, Monteiro R, Welling M, Bakkers J, Ehler E, Verkleij A, Mummery C, Passier R. CHAP is a newly identified Z-disc protein essential for heart and skeletal muscle function. J Cell Sci. 2010;123:1141–1150. doi: 10.1242/jcs.063859. [DOI] [PubMed] [Google Scholar]
- [152].Schonberger J, Wang L, Shin JT, Kim SD, Depreux FF, Zhu H, Zon L, Pizard A, Kim JB, Macrae CA, Mungall AJ, Seidman JG, Seidman CE. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat Genet. 2005;37:418–422. doi: 10.1038/ng1527. [DOI] [PubMed] [Google Scholar]
- [153].Poon KL, Tan KT, Wei YY, Ng CP, Colman A, Korzh V, Xu XQ. RNA-binding protein RBM24 is required for sarcomere assembly and heart contractility. Cardiovasc Res. 2012;94:418–427. doi: 10.1093/cvr/cvs095. [DOI] [PubMed] [Google Scholar]
- [154].Seguchi O, Takashima S, Yamazaki S, Asakura M, Asano Y, Shintani Y, Wakeno M, Minamino T, Kondo H, Furukawa H, Nakamaru K, Naito A, Takahashi T, Ohtsuka T, Kawakami K, Isomura T, Kitamura S, Tomoike H, Mochizuki N, Kitakaze M. A cardiac myosin light chain kinase regulates sarcomere assembly in the vertebrate heart. J Clin Invest. 2007;117:2812–2824. doi: 10.1172/JCI30804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Hassel D, Dahme T, Erdmann J, Meder B, Huge A, Stoll M, Just S, Hess A, Ehlermann P, Weichenhan D, Grimmler M, Liptau H, Hetzer R, Regitz-Zagrosek V, Fischer C, Nurnberg P, Schunkert H, Katus HA, Rottbauer W. Nexilin mutations destabilize cardiac Z-disks and lead to dilated cardiomyopathy. Nat Med. 2009;15:1281–1288. doi: 10.1038/nm.2037. [DOI] [PubMed] [Google Scholar]
- [156].Bendig G, Grimmler M, Huttner IG, Wessels G, Dahme T, Just S, Trano N, Katus HA, Fishman MC, Rottbauer W. Integrin-linked kinase, a novel component of the cardiac mechanical stretch sensor, controls contractility in the zebrafish heart. Genes Dev. 2006;20:2361–2372. doi: 10.1101/gad.1448306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Meder B, Huttner IG, Sedaghat-Hamedani F, Just S, Dahme T, Frese KS, Vogel B, Kohler D, Kloos W, Rudloff J, Marquart S, Katus HA, Rottbauer W. PINCH proteins regulate cardiac contractility by modulating integrin-linked kinase-protein kinase B signaling. Mol Cell Biol. 2011;31:3424–3435. doi: 10.1128/MCB.05269-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Knoll R, Postel R, Wang J, Kratzner R, Hennecke G, Vacaru AM, Vakeel P, Schubert C, Murthy K, Rana BK, Kube D, Knoll G, Schafer K, Hayashi T, Holm T, Kimura A, Schork N, Toliat MR, Nurnberg P, Schultheiss HP, Schaper W, Schaper J, Bos E, Den Hertog J, van Eeden FJ, Peters PJ, Hasenfuss G, Chien KR, Bakkers J. Laminin-alpha4 and integrin-linked kinase mutations cause human cardiomyopathy via simultaneous defects in cardiomyocytes and endothelial cells. Circulation. 2007;116:515–525. doi: 10.1161/CIRCULATIONAHA.107.689984. [DOI] [PubMed] [Google Scholar]
- [159].Will RD, Eden M, Just S, Hansen A, Eder A, Frank D, Kuhn C, Seeger TS, Oehl U, Wiemann S, Korn B, Koegl M, Rottbauer W, Eschenhagen T, Katus HA, Frey N. Myomasp/LRRC39, a heart- and muscle-specific protein, is a novel component of the sarcomeric M-band and is involved in stretch sensing. Circ Res. 2010;107:1253–1264. doi: 10.1161/CIRCRESAHA.110.222372. [DOI] [PubMed] [Google Scholar]
- [160].Iorga B, Neacsu CD, Neiss WF, Wagener R, Paulsson M, Stehle R, Pfitzer G. Micromechanical function of myofibrils isolated from skeletal and cardiac muscles of the zebrafish. J Gen Physiol. 2011;137:255–270. doi: 10.1085/jgp.201010568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Werdich AA, Brzezinski A, Jeyaraj D, Ficker E, Wan X, McDermott BM, Sabeh MK, Macrae CA, Rosenbaum DS. The zebrafish as a novel animal model to study the molecular mechanisms of mechano-electrical feedback in the heart. Prog Biophys Mol Biol. 2012;110(2–3):154–165. doi: 10.1016/j.pbiomolbio.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Sun X, Hoage T, Bai P, Ding Y, Chen Z, Zhang R, Huang W, Jahangir A, Paw B, Li YG, Xu X. Cardiac hypertrophy involves both myocyte hypertrophy and hyperplasia in anemic zebrafish. PLoS One. 2009;4:e6596. doi: 10.1371/journal.pone.0006596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Ding Y, Sun X, Huang W, Hoage T, Redfield M, Kushwaha S, Sivasubbu S, Lin X, Ekker S, Xu X. Haploinsufficiency of target of rapamycin attenuates cardiomyopathies in adult zebrafish. Circ Res. 2011;109:658–669. doi: 10.1161/CIRCRESAHA.111.248260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- [165].Raya A, Koth CM, Buscher D, Kawakami Y, Itoh T, Raya RM, Sternik G, Tsai HJ, Rodriguez-Esteban C, Izpisua-Belmonte JC. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc Natl Acad Sci USA. 2003;100(Suppl 1):11889–11895. doi: 10.1073/pnas.1834204100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Chablais F, Veit J, Rainer G, Jazwinska A. The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Dev Biol. 2011;11:21. doi: 10.1186/1471-213X-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Schnabel K, Wu CC, Kurth T, Weidinger G. Regeneration of Cryoinjury induced necrotic heart lesions in zebrafish is associated with epicardial activation and cardiomyocyte proliferation. PLoS One. 2011;6:e18503. doi: 10.1371/journal.pone.0018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Wang J, Panakova D, Kikuchi K, Holdway JE, Gemberling M, Burris JS, Singh SP, Dickson AL, Lin YF, Sabeh MK, Werdich AA, Yelon D, Macrae CA, Poss KD. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–3430. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Laube F, Heister M, Scholz C, Borchardt T, Braun T. Re-programming of newt cardiomyocytes is induced by tissue regeneration. J Cell Sci. 2006;119:4719–4729. doi: 10.1242/jcs.03252. [DOI] [PubMed] [Google Scholar]
- [171].Oberpriller JO, Oberpriller JC. Response of the adult newt ventricle to injury. J Exp Zool. 1974;187:249–253. doi: 10.1002/jez.1401870208. [DOI] [PubMed] [Google Scholar]
- [172].Kikuchi K, Holdway JE, Major RJ, Blum N, Dahn RD, Begemann G, Poss KD. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell. 2011;20:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- [174].Tomanek RJ, Hansen HK, Christensen LP. Temporally expressed PDGF and FGF-2 regulate embryonic coronary artery formation and growth. Arterioscler Thromb Vasc Biol. 2008;28:1237–1243. doi: 10.1161/ATVBAHA.108.166454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Lavine KJ, White AC, Park C, Smith CS, Choi K, Long F, Hui CC, Ornitz DM. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes Dev. 2006;20:1651–1666. doi: 10.1101/gad.1411406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Smith CL, Baek ST, Sung CY, Tallquist MD. Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling. Circ Res. 2011;108:e15–e26. doi: 10.1161/CIRCRESAHA.110.235531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Lien CL, Schebesta M, Makino S, Weber GJ, Keating MT. Gene expression analysis of zebrafish heart regeneration. PLoS Biol. 2006;4:e260. doi: 10.1371/journal.pbio.0040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Kim J, Wu Q, Zhang Y, Wiens KM, Huang Y, Rubin N, Shimada H, Handin RI, Chao MY, Tuan TL, Starnes VA, Lien CL. PDGF signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. Proc Natl Acad Sci USA. 2010;107:17206–17210. doi: 10.1073/pnas.0915016107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Poss KD, Nechiporuk A, Hillam AM, Johnson SL, Keating MT. Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development. 2002;129:5141–5149. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- [182].Jopling C, Sune G, Morera C, Izpisua Belmonte JC. p38alpha MAPK regulates myocardial regeneration in zebrafish. Cell Cycle. 2012;11:1195–1201. doi: 10.4161/cc.11.6.19637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Yin VP, Lepilina A, Smith A, Poss KD. Regulation of zebrafish heart regeneration by miR-133. Dev Biol. 2012;365:319–327. doi: 10.1016/j.ydbio.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Itou J, Oishi I, Kawakami H, Glass TJ, Richter J, Johnson A, Lund TC, Kawakami Y. Migration of cardiomyocytes is essential for heart regeneration in zebrafish. Development. 2012;139:4133–4142. doi: 10.1242/dev.079756. [DOI] [PubMed] [Google Scholar]
- [185].Raz E, Mahabaleshwar H. Chemokine signaling in embryonic cell migration: a fisheye view. Development. 2009;136:1223–1239. doi: 10.1242/dev.022418. [DOI] [PubMed] [Google Scholar]
- [186].Gonzalez-Rosa JM, Martin V, Peralta M, Torres M, Mercader N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development. 2011;138(9):1663–1674. doi: 10.1242/dev.060897. [DOI] [PubMed] [Google Scholar]
- [187].Chablais F, Jazwinska A. The regenerative capacity of the zebrafish heart is dependent on TGFbeta signaling. Development. 2012;139:1921–1930. doi: 10.1242/dev.078543. [DOI] [PubMed] [Google Scholar]