Abstract
Metastatic disease is the main cause of cancer-related mortality due to almost universal therapeutic resistance. Despite its high clinical relevance our knowledge of how cancer cell populations change during metastatic progression is limited. Here we investigated intratumor genetic and phenotypic heterogeneity during metastatic progression of breast cancer. We analyzed cellular genotypes and phenotypes at the single cell level by performing immuno-FISH in intact tissue sections of distant metastatic tumors from rapid autopsy cases and from primary tumors and matched lymph node metastases collected prior to systemic therapy. We calculated Shannon index of intratumor diversity in all cancer cells and within phneotypically distinct cell populations. We found that the extent of intratumor genetic diversity was similar regardless of the chromosomal region analyzed, implying that it may reflect an inherent property of the tumors. We observed that genetic diversity was highest in distant metastases and was generally concordant across lesions within the same patient, whereas treatment-naïve primary tumors and matched lymph node metastases were frequently genetically more divergent. In contrast, cellular phenotypes were more discordant between distant metastases than primary tumors and matched lymph node metastases. Diversity for 8q24 was consistently higher in HER2+ tumors compared to other subtypes and in metastases of triple negative tumors relative to primary sites. We conclude that our integrative method that couples ecologic models with experimental data in human tissue samples, could be used for the improved prognostication of cancer patients and for the design of more effective therapies for progressive disease.
Major findings
By defining quantitative measures of intratumor cellular genetic and phenotypic heterogeneity in primary and metastatic breast tumors and by assessing tumor topology, we determined that distant metastatic tumors are the most diverse, which can explain the frequent therapy-resistance of advanced stage disease.
Keywords: breast cancer, metastasis, diversity, genetic, lymph node
INTRODUCTION
Metastatic dissemination and the growth of tumors at distant sites is a key step of tumor progression that is responsible for most cancer-related deaths. The accurate prediction of which patient will develop metastatic disease and the prevention and treatment of metastatic lesions remain major challenges largely due to the relative scarcity of studies of distant metastases. Difficulties associated with tissue acquisition, especially repeated sampling of multiple lesions during disease progression, and lack of faithful models of metastatic disease hamper progress in this area. However, a detailed molecular understanding of metastatic tumors is a prerequisite for the development of more effective cancer therapies.
In breast cancer, the risk of distant metastasis and the preferred sites for these lesions strongly correlate with tumor subtype (1). Luminal estrogen receptor positive (ER+) tumors tend to have low probability of metastatic spread and preferentially form bone metastases. In contrast, Her2+ and triple negative (negative for estrogen and progesterone receptors and HER2) tumors have higher propensity for metastatic progression and form visceral and brain metastases.
Metastatic spread traditionally thought to be a late event in tumorigenesis that occurs after substantial tumor growth at the primary site (2). However, recent data in model organisms (3) and in cancer patients (4) suggest that tumor cells may disseminate early, leading to parallel progression of primary and disseminated tumors (5), although clinically relevant distant metastases are still detected relatively late. A limited number of prior studies have analyzed the genetic profiles of primary and metastatic lesions in breast and other carcinomas and in general found a large extent of clonal relatedness between lesions (6–8). However, almost all of these studies used bulk tissue samples that do not allow for detailed characterization of clonal composition, and very few compared multiple lesions in the same patient.
Besides genetic alterations, the presence of cancer cells with more mesenchymal, stem cell-like features has been associated with increased risk of metastatic disease (9, 10); yet distant metastases are largely composed of more differentiated epithelial cells implying sequential EMT (epithelial to mesenchymal transition) followed by MET (mesenchymal to epithelial transition) during dissemination and metastatic growth, respectively. “Self-seeding” of cancer cells among multiple lesions within the same patient may also contribute to heterogeneity both within and among tumors (11). Here we describe the combined analysis of genetic and phenotypic heterogeneity in breast cancer distant and lymph node metastases at the single cell level.
QUICK GUIDE TO EQUATIONS AND ASSUMPTIONS
| Equation 1 |
where pi represents the proportion of individuals belonging to the ith type or species when there are n types in total. This quantity is known as the Shannon index of diversity or Shannon entropy. This index has been widely used in the ecological literature (12). It was originally proposed by Claude Shannon to quantify the entropy (uncertainty or information content) in strings of text (13). His idea was that the more different letters there are, and the more equal their proportional abundances in the string of text, the more difficult it is to correctly predict which letter will be the next one in the string. The Shannon entropy quantifies the uncertainty associated with this prediction. In ecology, pi represents the proportion of individuals belonging to the ith species in the dataset of interest. Then the Shannon entropy quantifies the uncertainty in predicting the species identity of an individual that is taken at random from the dataset.
| Equation 2 |
where pi represents the proportion of individuals belonging to the ith type or species when there are n types in total. This quantity is known as Simpson’s index of diversity. The Simpson index was introduced in 1949 by Edward H. Simpson to measure the degree of concentration when individuals are classified into types (14). The square root of the index had already been introduced in 1945 by the economist Albert O. Hirschman (15). The measure equals the probability that two entities taken at random from the dataset of interest represent the same type. It also equals the weighted arithmetic mean of the proportional abundances pi of the types of interest.
MATERIALS AND METHODS
Human breast cancer samples
Formalin-fixed paraffin embedded human primary tumors and metastases from breast cancer patients were obtained from Johns Hopkins University School of Medicine using protocols approved by the Institutional Review Board. Samples were de-identified prior to analysis. Tumor histology and expression of standard biomarkers (ER, PR, and HER2) were evaluated at the time of diagnosis according to ASCO/CAP guidelines (16). Subtype definitions in this study were as follows: luminal A (ER+ and/or PR+, HER2−), luminal B (ER+ and/or PR+, HER2+), HER2+ (ER−, PR−, HER2+), and triple negative (ER−, PR−, HER2−). In total, we analyzed 11 patients with distant metastases and 12 patients with matched primary tumor and lymph node metastases.
Multicolor immunoFISH
The detection of the copy number gain for 1q32.1, 8q24.13, 10p13, 11q13.2, 12p13.1, 16p13.3, and 17q21 (including the genes NUAK2, NSMCE2, ITGA8, CCND1, H2AFJ, MPFL, and HER2 respectively) or the centromeric region of each chromosome was performed using whole sections of formalin fixed paraffin embedded (FFPE) human breast cancer tissue or breast cancer metastasis. The tissues were dewaxed in xylene and hydrated in a series of ethanol. After heat-induced antigen retrieval overnight at 70°C in citrate buffer (pH 6), the digestion with pepsin was performed in a slide warmer at 37°C for 10–20 minutes depending on the sample. The immunostaining was performed at room temperature and sequentially to avoid cross-reaction between antibodies as follows: CD44 (Neomarkers, clone 156-3C11, mouse monoclonal IgG2) for 1 hour, biotin conjugated Rabbit anti-Mouse IgG2a (Life Technologies Cat#61-0240) for 30 minutes, CD24 (NeoMarkers, clone SN3b, mouse monoclonal IgM) for 1 hour, Streptavidin Pacific Blue conjugated (Life Technologies Cat#S-11222) and Alexa Fluor 647 Goat anti-mouse IgM (Life Technologies Cat# A-21238). The samples were then fixed in Carnoy’s for 10 minutes and dehydrated in a series of ethanol. The probes (BAC probes) for the detection of 8q, 11q, 16p, 12p, 10p, and 17q were labeled with SpectrumOrange (Vysis), and the probe for the detection of 1q with SpectrumGreen (Vysis) using a Nick Translation (Abbot Molecular) according to the manufacturer’s recommendations, mixed with the corresponding centromeric probe for each chromosome (CEP probes) (Vysis), diluted in hybridization buffer and applied to each sample. The denaturalization was performed in a slide warmer at 75°C for several minutes depending on the sample, and then the slides were incubated in a humid chamber for 20 hours at 37°C. Finally, the samples were washed with different stringent SCC buffers, air-dried and protected for long storage with ProLong Gold (Life Technologies). Different immunofluorescence images from multiple areas of each sample were acquired with a Nikon Ti microscope attached to a Yokogawa spinning-disk confocal unit, 60× plan apo objective, and OrcaER camera controlled by Andor iQ software.
Inference of frequencies for cell phenotypes
The frequency of each phenotypically distinct cancer cell subpopulation (i.e., CD44+CD24−, CD44+CD24+, CD44−CD24+, and CD44−CD24−) was calculated by counting an average of 300 cells in each sample.
Statistical analyses
Genetic diversity was determined essentially as described (17), but we calculated diversity indices based on copy number counts for (1) BAC, (2) chromosome specific centromeric, (3) ratio of BAC/CEP counts, and (4) unique BAC and CEP count combinations. Statistical differences in primary tumor vs. lymph node metastasis or between two different metastatic lesions were calculated through 100,000 iterations of bootstrapping the BAC and CEP counts from the larger cell population and comparing the mean counts of each bootstrap repetition against the mean count of the smaller cell population. Statistical differences in the BAC and CEP counts between adjacent cells in two different metastatic sites were calculated through 100,000 iterations of bootstrapping the absolute difference in BAC and CEP counts of adjacent cells from the larger cell population and comparing the mean absolute difference in counts of each bootstrap repetition against the mean absolute difference in count of the smaller cell population. Statistical differences in BAC and CEP counts were evaluated using the achieved significance level (ASL) method (18). This amounts to using 100,000 iterations of bootstrapping the BAC and CEP counts from the larger cell population and comparing the mean counts of each bootstrap repetition against the mean count of the smaller cell population. Statistical differences in the BAC and CEP counts between adjacent cells were calculated through 100,000 iterations of bootstrapping the absolute difference in BAC and CEP counts of adjacent cells from the larger cell population and comparing the mean absolute difference in counts of each bootstrap repetition against the mean absolute difference in count of the smaller cell population. Statistical differences in the BAC and CEP counts between adjacent cells were also calculated using ASL, through 100,000 iterations of bootstrapping the absolute difference in BAC and CEP counts of adjacent cells from the larger cell population and comparing the mean absolute difference in counts of each bootstrap repetition against the mean absolute difference in count of the smaller cell population.
Topology analysis
For the analysis of the topological distribution of cellular subsets, 3×3 images (corresponding to 71,678 μm2 area) were obtained using 60× plan apo objective with 5% overlap between areas in order to be able to assemble them into one montage. The loci for the BAC and CEP probes were automatically detected using previously described algorithms (19). Cellular phenotype was determined manually based on immunofluorescence as described above. Both tumor and stromal cells were analyzed, and both the signals and the coordinates for each cell were recorded; however further analysis was restricted to solely non-stromal tumor cells. For each patient, we determined the distribution of BAC and CEP counts both across all cell phenotype and independently for each phenotype. Statistical differences in the distribution of BAC and CEP counts differences between two different metastatic sites were then determined through bootstrapping. Additionally, to assess the spatial distribution of different cell phenotypes and the topological genomic diversity we also focused on neighboring cells, which we defined as cells for which the shortest distance between cell boundaries is smaller than 10% of the average cell radius in each analyzed field of view. Significance of the differences between neighboring cells was also determined through bootstrapping. The fraction of homotypic neighbors was calculated by counting the number of pairs of neighboring cells where both cells were the same phenotype and dividing by the total number of pairs of neighboring cells. To assess the significance of this fraction independently between two different metastatic sites, we used permutation testing. We determined the fraction of homotypic neighbors in the actual sample and compared this fraction against the fraction of homotypic neighbors in 100,000 randomized ensembles in which the cell phenotypes were randomly shuffled. The null hypothesis of this permutation test is that the pattern of homotypic neighbors seen in the sample falls within the distribution expected under random migration. A two-tailed p-value was used to determine whether the fraction detected fell outside the expected range.
RESULTS
Genetic and phenotypic diversity between distant metastases
To explore genetic heterogeneity in metastatic lesions from the same patient, we first performed SNP (Single Nucleotide Polymorphism) array analysis of paired distant metastases from 11 rapid autopsies of breast cancer patients (20) (Supplementary Table S1). Overall, we detected a relatively small degree of copy number divergence between two lesions from the same patient (data not shown), potentially due to the inability of SNP arrays to detect subclonal populations within tumors when using bulk tissue samples. Thus, to obtain a more detailed picture of the subclonal structure of metastatic lesions, we performed iFISH (combined immunofluorescence and FISH) (17) to assess genetic and phenotypic variability within tumors at the single cell level (Fig. 1A and Supplementary Table S1). Genetic heterogeneity was determined by evaluating copy number variation for chromosomal regions commonly gained in each of the three major breast tumor subtypes (i.e., luminal, HER2+ and triple negative tumors) (21) and corresponding centromeric probes. A probe for 8q24.13 was used in all tumors, for 11q13.2 and 16p13.3 in luminal, for 1q32.1 and 17q21 in HER2+, and for 12p13.1 and 10p13 in triple negative subtypes. Phenotypic heterogeneity was evaluated by staining for CD44 and CD24 cell surface markers, which identify cells with more luminal epithelial and mesenchymal features, respectively, that have different biological properties relevant to metastasis, including invasiveness and angiogenic potential (9, 22–26). We used hematoxylin-eosine (H&E) staining to identify tumor cell-enriched areas and morphologic features to discriminate between normal and neoplastic cells. We also used autofluorescence to define tissue architecture on the FISH images and neoplastic cells were also identifiable based on the presence of copy number gain.
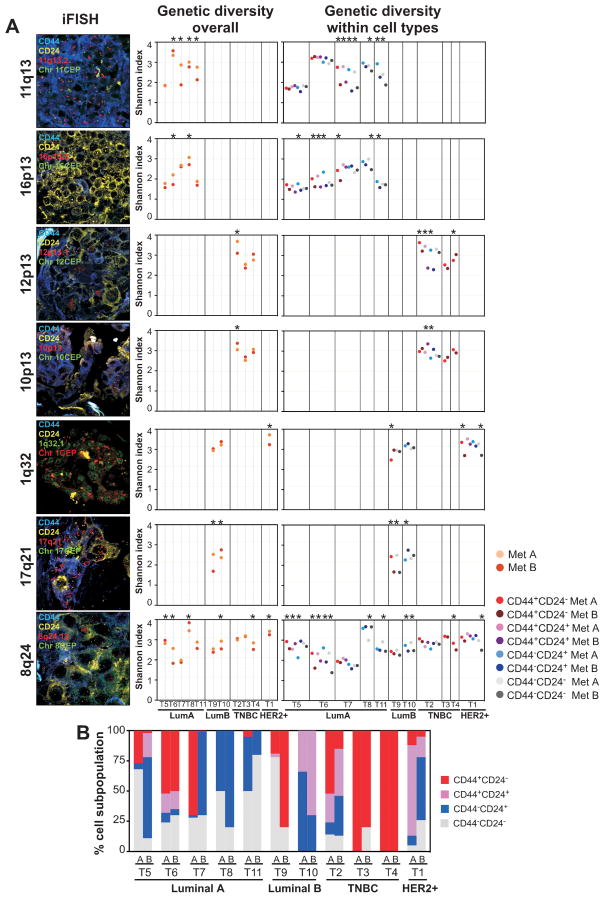
Figure 1. Genetic diversity in distant metastases in the same patient.
A, representative images of iFISH for the indicated probes and markers. Dot plots depict Shannon diversity indices calculated based on unique BAC and CEP counts in all cancer cells combined (overall) and in phenotypically distinct tumor cell subpopulations. Dots represent distinct metastatic lesions or phenotypically distinct tumor cell subpopulations within lesions. Asterisks indicate significant (p<0.05, statistical methodology described in Supplementary methods online) differences. Details of tissue samples and Shannon index of diversity calculations are listed in Supplementary Tables S1 and S3–S4, respectively. B, bar graphs depict the relative frequencies of CD44+CD24−, CD44+CD24+, CD44−CD24+, and CD44−CD24− cells in different metastases (A and B) for a given (T1–T11) patient. TNBC - Triple Negative Breast Cancer.
Chromosomal region-specific BAC (Bacterial Artificial Chromosome) and CEP (Centromeric Probe) signals were counted in ~100 individual cells in each of the four phenotypically distinct tumor cell populations (i.e., CD44+CD24−, CD44+CD24+, CD44−CD24+, and CD44−CD24− cells) (Supplementary Table S2). Overall assessment of copy number differences within phenotypically distinct cell populations in metastatic lesions revealed divergent copy number gain for multiple genomic loci in most cases (Supplementary Fig. S1); this feature was also apparent in the relative changes of unique cancer cells visualized by Kernel density and Whittaker plots (Supplementary Fig. S2) (12, 17).
Next, we calculated the Shannon and Simpson indices of diversity (12) in four different ways based on measures of (1) copy number of the BAC probe, (2) copy number of the CEP probe, (3) the ratio of BAC to CEP counts, and (4) individual copy number of both BAC and CEP probes in each cell (unique counts). Overall, each of the four different calculations displayed similar relative differences among tumors, but as expected, diversity indices were highest based on unique counts (Supplementary Table S3). Measuring BAC probe and BAC to CEP ratio provides information on copy number gain of a specific locus, CEP probe counts alone report the degree of aneuploidy, whereas unique BAC and CEP counts provide combined information on both. Thus, to assess genetic diversity due to both copy number gain and aneuploidy, we subsequently used unique counts for all analyses unless otherwise indicated.
Overall, genetic diversity as measured by the Shannon index was significantly different between two distant metastases in the same patient for most genomic loci analyzed and in almost all cases (Fig. 1A and Supplementary Table S3). Assessment of genetic diversity within phenotypically distinct cell subpopulations provided similar results (Fig. 1A and Supplementary Table S4). However, in several cases, the differences in genetic diversity between metastatic lesions were significant only for certain loci and in specific cell populations; in some cases only one cell population showed differences and some loci were divergent only in one cell subpopulation. These results potentially reflect the order of genetic events during tumor evolution (27) or selection of a particular cell population by local microenvironmental forces. The use of Simpson’s index led to similar results (Supplementary Table S3 and S4).
To determine whether differences in cellular phenotypes contributed to the observed cell type-specific genetic diversity within and between metastatic lesions, we analyzed the relative frequencies of the four distinct cell subpopulations identified using CD24 and CD44 cell surface markers within tumors. Correlating with prior results from our (28) and other labs (29), the frequencies of the four different cell types displayed tumor subtype-specific differences, with CD44+CD24− and CD44−CD24+ cells being more common in triple negative and in luminal tumors, respectively (Fig. 1B). Metastatic lesions within the same patient also displayed substantial differences in the relative frequencies of the four cell types, with the exception of two TNBC cases whose metastases were almost entirely composed of CD44+CD24− cells. Interestingly, TNBC patients had the shortest time interval from diagnosis to death implying rapid emergence and growth of distant metastases, which could potentially explain the higher similarity both for cell types and genotypes between lesions within the same patient. These results emphasize the value of combined genetic and phenotypic characterization of individual cancer cells and highlight the degree of biological heterogeneity between metastatic lesions within the same patient.
Genetic and phenotypic diversity between primary tumors and lymph node metastases
Distant metastases in breast cancer patients are usually detected as recurrences after systemic adjuvant therapy, making it difficult to study the natural course of the disease (30). Indeed, all metastatic patients in our cohort were diagnosed with localized tumors (T1 or T2) and many did not even have lymph node metastases at the time of diagnosis (Supplementary Table S1). Therefore, the observed high degree of genetic and phenotypic heterogeneity between metastatic lesions could be due to selection pressure by the multiple rounds of treatment the patients received. Thus, to investigate potential changes in genetic and phenotypic heterogeneity during the natural progression of breast tumors to metastatic disease, we performed iFISH analysis of primary tumors of different subtypes and matched lymph node metastases (Fig. 2A and Supplementary Tables S1 and S2) from patients who were not exposed to any systemic treatment prior to tissue acquisition.
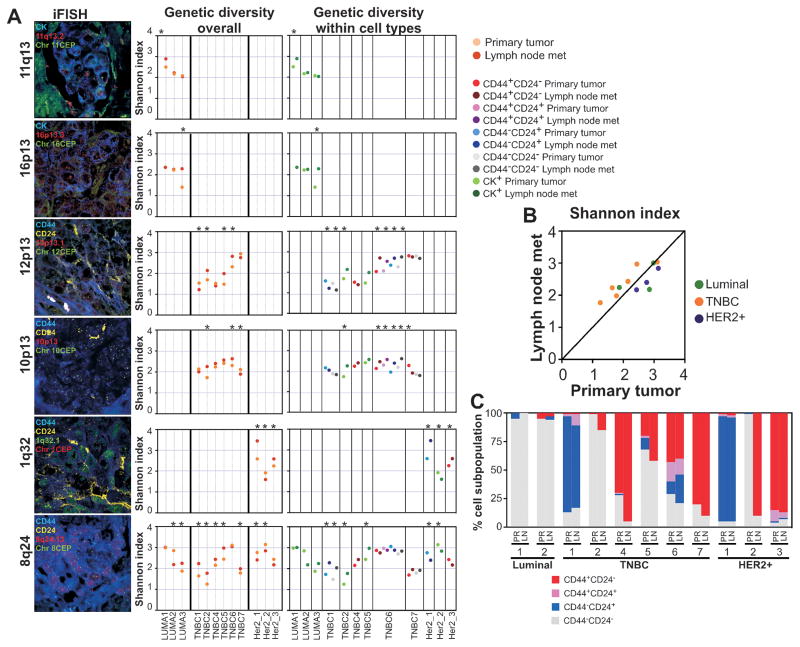
Figure 2. Genetic diversity of matched primary tumors and lymph node metastases.
A, representative images of iFISH for the indicated probes and markers. Dot plots depict Shannon diversity indices calculated based on unique BAC and CEP counts in all cancer cells combined (overall) and in phenotypically distinct tumor cell subpopulations. Dots represent primary tumors and lymph node metastases or phenotypically distinct tumor cell subpopulations within these lesions. Asterisks indicate significant (p<0.05, statistical methodology described in Supplementary methods online) differences. Details of tissue samples and Shannon index of diversity calculations are listed in Supplementary Tables S5–S6. B, Differences in Shannon diversity index between primary tumors and matched lymph node metastases according to breast tumor subtype. C, Bar graphs depict the relative frequencies of CD44+CD24−, CD44+CD24+, CD44−CD24+, and CD44−CD24− cells in matched primary tumors and lymph node metastases. TNBC - Triple Negative Breast Cancer.
In general, the extent of genetic diversity in primary tumors and lymph node metastases was lower and more variable than that observed in distant metastatic lesions, yet the differences in diversity between primary tumors and matched lymph nodes were still statistically significant in almost all cases and for all probes analyzed (Fig. 2A and Supplementary Table S5). Interestingly, TNBCs in general had lower diversity scores for 8q24, the only probe that was analyzed in all tumors, than luminal and HER2+ cases, and it was consistently higher in lymph nodes compared to their matched primaries (Fig. 2B). In contrast, diversity for 8q24 in HER2+ tumors was generally high and it was higher in the primary tumors relative to matched lymph nodes.
The differences in genetic diversity were in general observed for all phenotypically distinct cell populations for most cases and probes, although for a few cases we were not able to assess all four cell types within both primary tumors and lymph node metastases (Fig. 2A and Supplementary Table S6). Thus, in contrast to distant metastatic lesions, we did not observe significant differences in diversity between primary and lymph nodes for certain cell types, potentially indicating the lack of selection for a particular phenotype. Correlating with this hypothesis, we found that the relative frequency of the four phenotypically distinct cell populations was almost identical between lymph node metastases and matched primaries (Fig. 2C).
Differences between distant and lymph node metastases
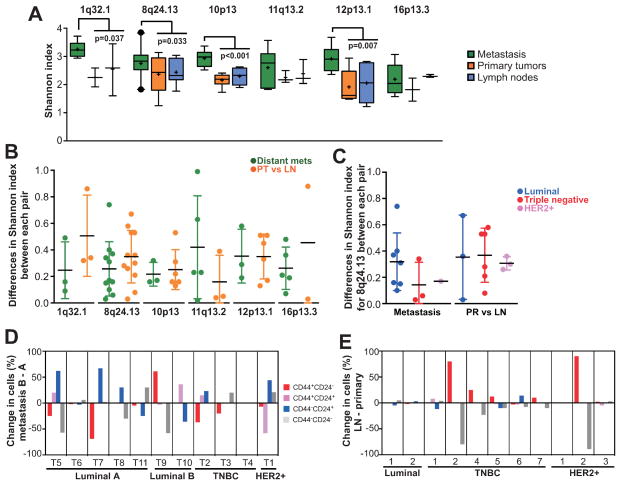
We observed several interesting differences in the genetic and phenotypic diversity of primary tumors, lymph node and distant metastases and differences between lesions in the same patient; these findings could potentially reflect tumor evolution in unperturbed (e.g., no systemic therapy) and perturbed (cancer treatment) environments. Primary tumors and lymph node metastases had significantly lower genetic diversity for almost all chromosomal regions analyzed than distant metastases (Fig. 3A). In contrast, the difference in genetic diversity between a primary tumor and its matched lymph node metastasis was larger for some probes (e.g., 1q32 and 8q24) but smaller for others (e.g., 16p13, 11q13) than that between two distant metastases (Fig. 3B). Triple negative tumors showed the highest differences in diversity for 8q24 between primary tumors and matched lymph nodes, whereas the opposite was observed in distant metastases (Fig. 3C). Importantly, genetic diversity indices were similar for each genomic probe analyzed (Supplementary Table S7), implying that this may reflect an inherent property of the tumor independent of the way of measurement (31).
Figure 3. Differences in diversity between distant and lymph node metastases.
A, box plots depict Shannon diversity indices of primary tumors, and lymph node and distant metastases. Boxes show 25th to 75th percentile whereas whiskers extend to 5th and 95th percentiles. Outliers outside of 5th and 95th percentile are shown as black dots. Significant differences by Mann-Whitney test between two distant metastases within the same patient and primary and lymph node metastasis are shown. B, dot plots showing the differences in Shannon index between each pair of distant metastasis or between each pair of primary and lymph node metastasis for the indicated chromosomal regions. C, differences in Shannon index for 8q24.13 in each tumor subtype. Relative changes in the frequency of each of the indicated cell population is shown in metastases (D) and in matched primary tumors and lymph node metastases (E). Details of tissue samples are listed in Supplementary Table S1. PR - primary tumor, LN - lymph node metastasis, TNBC - Triple Negative Breast Cancer.
Contrary to genetic diversity, phenotypically distinct cell populations were more commonly divergent between two distant metastases than between primary tumor and matched lymph node (Fig. 3D–E). Similar to diversity for 8q24, differences in cellular phenotypes were less pronounced between distant metastatic lesions of TNBCs than primary TNBC and its matched lymph node whereas the opposite trend was observed for luminal tumors.
Topological mapping of genetic and phenotypic diversity in metastasis
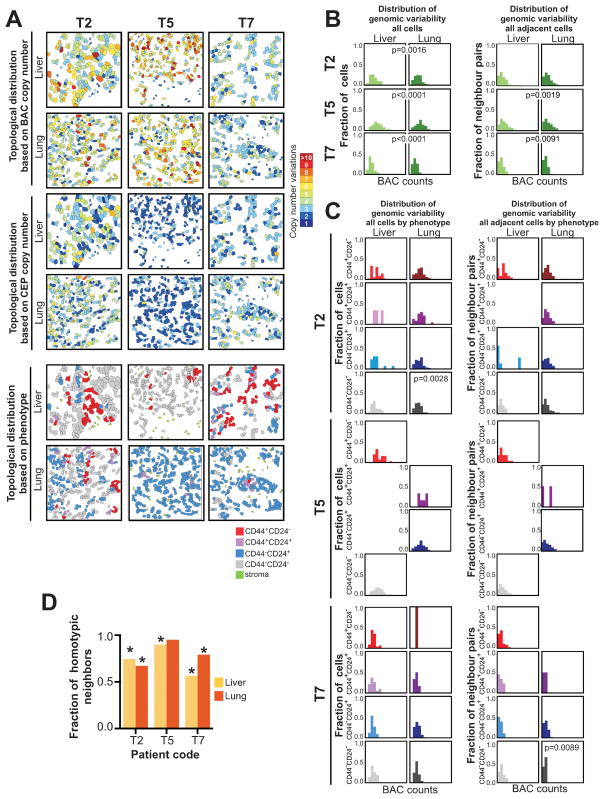
To further explore the impact of local microenvironments on genetic and phenotypic diversity within and between tumors, we analyzed tumor topology - defined as the spatial distribution of genetically and/or phenotypically different cells within tumors - in liver and lung metastases of three patients. Two cases (T5 and T7) were luminal A and one (T2) was triple negative subtype. We generated topology maps depicting individual cancer cells with specific genotypes (copy numbers for 8q24 BAC and chr8 CEP probes) and phenotypes (based on the expression of CD44 and CD24) (Fig. 4A). Next, we calculated the variability for copy number in all cells and in all adjacent cancer cells (Fig. 4B and Supplementary Fig. S3A and S4). Such analysis can potentially discern different modes of tumor evolution. Large variation in all cells but low variation in adjacent cells could indicate the existence of independently evolving spatially coherent clones. In contrast, large variation between spatially adjacent cells could indicate that tumor cells are either rapidly migrating or that mutation rates are extremely high. We detected significant differences in the distribution of genetic variability in all cells and all adjacent cells of liver and lung metastases in cases T5 and T7, whereas in T2, a significant difference was only observed for all cells but not for all adjacent cells (Fig. 4B). These results may potentially indicate the differences in evolution between luminal (T5 and T7) and triple negative (T2) tumors with the latter ones having multiple spatially independently evolving subclones.
Figure 4. Analysis of tumor topology.
A, maps show topologic differences in the distribution of genetically distinct tumor cells based on copy number for 8q24 BAC, chromosome 8 CEP, and cellular phenotype in liver and lung metastases of three breast cancer patients. B, histograms depicting absolute differences in copy numbers for BAC probe counts regardless of cellular phenotype in all cells or in adjacent cells in liver and lung metastases. C, histograms depicting absolute differences in copy numbers for BAC probe counts in all cells of the same phenotype or in adjacent cells of the same phenotype in liver and lung metastases. D, fraction of adjacent cells with the same phenotype in liver and lung metastases. Significance of the differences was determined by calculating the homotypic fraction for 100,000 iterations of permutation testing over randomized cellular phenotypes; asterisks indicate significant (p<0.05) differences.
To determine if the differences in the distribution of genetic variability were present in all or only in some of the phenotypically distinct subpopulations, we also performed similar analyses in each of the four cell types (CD44+CD24−, CD44+CD24+, CD44−CD24+, and CD44−CD24− cells). We observed significant differences in the distribution of genetic variability for all cells only in the CD44−CD24− subpopulation in patient T2, whereas in patient T7, this was significant in all adjacent cells in the CD44−CD24− population (Fig. 4C). We also evaluated differences in the topological distribution of cellular phenotypes and found that the frequency of homotypic interactions (i.e., adjacent cells with the same phenotype) was significantly higher compared to the frequency of heterotypic interactions between two metastatic lesions within the same patient (Fig. 4D). These data suggest that in metastatic lesions, the distribution of genetic variability is fairly even in all of the four cell types analyzed and adjacent cells are more likely to be phenotypically the same but genetically divergent.
These results again highlight the advantage of analyzing tumors in situ at the single cell level as tumor topology can reveal more detailed information on tumor evolution than the assessment of spatially dissociated cancer cells.
DISCUSSION
Tumor evolution culminates in metastatic disease that is almost universally fatal due to the lack of effective therapy. Despite being the main reason of cancer-related mortality, distant metastases are still both rarely sampled and rarely subjected to molecular analyses, especially in patients with multiple metastatic lesions in different organs. Thus, our knowledge of the clonal heterogeneity of multiple lesions within the same patient is limited. Based on the traditional clonal evolution model, cancer metastases are thought to originate from a single clone present in the primary tumor, sometimes at very low frequency (2), but experimental data supporting this model is scarce. Recent high-throughput sequencing studies have attempted to address this issue and identified shared and divergent somatic variants between primary tumors and matched distant metastases in breast and pancreatic carcinomas (6, 32, 33). However, as these studies were performed on bulk tumor samples, the cellular origin and the topologic distribution of the somatic changes could not be determined. Here we have investigated cellular genetic and phenotypic heterogeneity in two different distant metastatic lesions from patients who failed cancer treatment as well as primary tumors and matched lymph node metastases before any systemic therapy. We determined the extent of genetic heterogeneity for chromosomal regions frequently gained in breast cancer and for cellular phenotypes associated with mesenchymal and more differentiated luminal cell features thought to be relevant to metastatic progression and therapeutic resistance. By the combined analysis of genotypes and phenotypes at the single cell level and in intact tissue slices in situ, we obtained a more detailed view of tumor evolution than previously possible. Table 1 lists a brief summary of our major findings.
Table 1.
Brief summary of the major findings.
| Samples | Results |
|---|---|
| Distant metastases | Genetic diversity overall is high especially in TNBCs |
| Genetic diversity indices are similar regardless of the probe used | |
| Two lesions in the same patients have significantly different diversity in some cases and some probes | |
| In some cases genetic diversity differs between the two metastatic lesions only in some cell types | |
| Two lesions in the same patient are frequently phenotypically distinct except in TNBC cases | |
| Matched primary tumors and lymph node metastases | Genetic diversity overall is lower than in distant metastases, especially true in TNBCs |
| Genetic diversity indices are similar regardless of the probe used | |
| Genetic diversity is significantly different in most cases and most probes | |
| Diversity for 8q24 is lower in some primary TNBCs and higher in the lymph node metastasis | |
| Diversity for 8q24 is higher in some primary Her2+ tumors and lower in the lymph node metastasis | |
| Primary and matched lymph node metastasis are phenotypically more similar in most cases |
Assessing diversity using different chromosomal regions commonly yielded different results overall or when also considering specific cellular phenotypes. These differences could be due to many reasons including differences in the (1) acquisition of a particular genetic change during disease progression (i.e., it may reflect the order of events), (2) genomic instability for different loci, (3) therapeutic sensitivity of cancer cells with different copy number gain, and (4) tumor microenvironment – both within tumors and between organs of metastatic sites.
Diversity for 8q24 was found to be significantly different between two distant metastatic lesions within the same patient in most cases both when considering the overall cell population and also when considering distinct cellular phenotypes. The same is true for primary tumors and matched lymph node metastases. The 8q24 chromosomal region harbors several important oncogenes such as C-MYC; however, 8q24 gain is almost always a reflection of gain of the whole 8q arm, thus it is difficult to determine which gene drives the selection for cancer cells with increased copy number for 8q24 (34). Copy number levels of 11q varied most often between distant metastases of luminal tumors, but not between primary tumor and lymph nodes. This amplicon contains several proliferation-related genes (e.g., CCND1), and thus, heterogeneity for this locus may result in differences in proliferation, although several of these genes also influence sensitivity to endocrine therapy (35).
Comparing the extent of diversity among lesions of different progression stages revealed that distant metastatic lesions in general have higher diversity for most loci compared to primary tumors and lymph node metastases. Distant metastases were also more commonly divergent for the relative frequency of cells with different phenotypes, although this was also influenced by tumor subtype. Interestingly we did not see a consistent selection for a particular cellular phenotype during tumor progression; the phenotypic variability among tumors was more a reflection of tumor subtype than stage. Thus, the higher genetic and phenotypic diversity of distant metastatic tumors could be due to the multiple lines of therapy these patients received. It would be necessary to examine matched primary and distant metastatic lesions at the time of diagnosis and before any systemic therapy to differentiate between natural and treatment-induced diversity. Fortunately for patients, breast cancer is very rarely diagnosed at this late stage; thus, the lack of such samples makes such a study difficult to conduct.
We also observed several interesting differences between tumors of different subtypes. Diversity for 8q24 was lower in primary TNBCs and increased in both distant and lymph node metastases whereas the opposite trend was observed for HER2+ cases. TNBCs are thought to metastasize at high frequency and at an earlier stage and TNBC patients who fail treatment tend to have a shorter recurrence-free and overall survival (36). Thus, their metastatic lesions may be less likely to be divergent than those of luminal and Her2+ tumors that typically have a much longer duration of disease. Luminal tumors on the other hand are slower proliferating than TNBCs and HER2+ cases, which may result in lower diversity. The types of treatment given to each subtype may also influence intratumor diversity.
In summary, we found higher genetic and phenotypic diversity in distant metastases compared to primary tumors and lymph node metastases. In contrast, two different metastatic lesions were found to display lower differences in genetic diversity than a primary tumor and its matched lymph node metastasis. Due to difficulties to conduct these types of studies, the cohorts we analyzed were relatively small and the metastatic patients were autopsy cases who failed treatment. Thus, the examination of large cohorts and all stage lesions from the same patient before any systemic therapy would be necessary to conclusively determine the natural evolution of breast tumors.
Supplementary Material
Acknowledgments
The authors thank Lisa Cameron in the DFCI Confocal and Light Microscopy Core Facility for technical assistance, and also thank members of our laboratories for their critical reading of this manuscript and useful discussions.
Notes: Financial support: This work was supported by the National Cancer Institute Physical Sciences-Oncology Centers, U54CA143874 (to A.O.) and U54CA143798 (to F. Michor and M. Gönen), the Cellex foundation (to V. Almendro), and the Breast Cancer Research Foundation (to K. Polyak).
List of abbreviations
- SNP
Single Nucleotide Polymorphism
- FISH
fluorescence in situ hybridization
- BAC
bacterial artificial chromosome
- CEP
centromeric probe
- TNBC
Triple Negative Breast Cancer
Footnotes
Potential conflicts of interest: No potential conflicts of interest were disclosed.
References
- 1.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 3.Weng D, Penzner JH, Song B, Koido S, Calderwood SK, Gong J. Metastasis is an early event in mouse mammary carcinomas and is associated with cells bearing stem cell markers. Breast Cancer Res. 2012;14:R18. doi: 10.1186/bcr3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanger N, Effenberger KE, Riethdorf S, Van Haasteren V, Gauwerky J, Wiegratz I, et al. Disseminated tumor cells in the bone marrow of patients with ductal carcinoma in situ. Int J Cancer. 2011;129:2522–6. doi: 10.1002/ijc.25895. [DOI] [PubMed] [Google Scholar]
- 5.Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009;9:302–12. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 6.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–65. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torres L, Ribeiro FR, Pandis N, Andersen JA, Heim S, Teixeira MR. Intratumor genomic heterogeneity in breast cancer with clonal divergence between primary carcinomas and lymph node metastases. Breast Cancer Res Treat. 2007;102:143–55. doi: 10.1007/s10549-006-9317-6. [DOI] [PubMed] [Google Scholar]
- 9.Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–73. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 10.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–73. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 11.Norton L, Massague J. Is cancer a disease of self-seeding? Nat Med. 2006;12:875–8. doi: 10.1038/nm0806-875. [DOI] [PubMed] [Google Scholar]
- 12.Magurran AE. Measuring biological diversity. Malden: Blackwell; 2004. [Google Scholar]
- 13.Shannon CE. A mathematical theory of communication. The Bell System Technical Journal. 1948;27:379–423. 623–56. [Google Scholar]
- 14.Simpson EH. Measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- 15.Hirschman AO. National power and the structure of foreign trade. University of California Berkeley; 1945. [Google Scholar]
- 16.Hammond ME, Hayes DF, Wolff AC, Mangu PB, Temin S. American society of clinical oncology/college of american pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Journal of oncology practice/American Society of Clinical Oncology. 2010;6:195–7. doi: 10.1200/JOP.777003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park SY, Gönen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. The Journal of Clinical Investigation. 2010;120:636–44. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Efron B, Tibshirani RJ. An introduction to the bootstrap. London: Chapman & Hall; 1993. [Google Scholar]
- 19.Itzkovitz S, Lyubimova A, Blat IC, Maynard M, van Es J, Lees J, et al. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat Cell Biol. 2012;14:106–14. doi: 10.1038/ncb2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singhi AD, Cimino-Mathews A, Jenkins RB, Lan F, Fink SR, Nassar H, et al. MYC gene amplification is often acquired in lethal distant breast cancer metastases of unamplified primary tumors. Mod Pathol. 2011 doi: 10.1038/modpathol.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikolsky Y, Sviridov E, Yao J, Dosymbekov D, Ustyansky V, Kaznacheev V, et al. Genome-wide functional synergy between amplified and mutated genes in human breast cancer. Cancer Res. 2008;68:9532–40. doi: 10.1158/0008-5472.CAN-08-3082. [DOI] [PubMed] [Google Scholar]
- 22.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–26. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 24.Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–9. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 25.Bloushtain-Qimron N, Yao J, Snyder EL, Shipitsin M, Campbell LL, Mani SA, et al. Cell type-specific DNA methylation patterns in the human breast. Proc Natl Acad Sci U S A. 2008;105:14076–81. doi: 10.1073/pnas.0805206105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bloushtain-Qimron N, Yao J, Shipitsin M, Maruyama R, Polyak K. Epigenetic patterns of embryonic and adult stem cells. Cell Cycle. 2009;8:809–17. doi: 10.4161/cc.8.6.7938. [DOI] [PubMed] [Google Scholar]
- 27.Martins FC, De S, Almendro V, Gonen M, Park SY, Blum JL, et al. Evolutionary Pathways in BRCA1-Associated Breast Tumors. Cancer Discov. 2012;2:503–11. doi: 10.1158/2159-8290.CD-11-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SY, Lee HE, Li H, Shipitsin M, Gelman R, Polyak K. Heterogeneity for Stem Cell–Related Markers According to Tumor Subtype and Histologic Stage in Breast Cancer. Clinical Cancer Research. 2010;16:876–87. doi: 10.1158/1078-0432.CCR-09-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honeth G, Bendahl PO, Ringner M, Saal LH, Gruvberger-Saal SK, Lovgren K, et al. The CD44+/CD24− phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10:R53. doi: 10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider C, Fehr MK, Steiner RA, Hagen D, Haller U, Fink D. Frequency and distribution pattern of distant metastases in breast cancer patients at the time of primary presentation. Archives of gynecology and obstetrics. 2003;269:9–12. doi: 10.1007/s00404-002-0445-x. [DOI] [PubMed] [Google Scholar]
- 31.Merlo LM, Shah NA, Li X, Blount PL, Vaughan TL, Reid BJ, et al. A comprehensive survey of clonal diversity measures in Barrett’s esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer Prev Res (Phila) 2010;3:1388–97. doi: 10.1158/1940-6207.CAPR-10-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–13. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 33.Geyer FC, Weigelt B, Natrajan R, Lambros MB, de Biase D, Vatcheva R, et al. Molecular analysis reveals a genetic basis for the phenotypic diversity of metaplastic breast carcinomas. J Pathol. 2010;220:562–73. doi: 10.1002/path.2675. [DOI] [PubMed] [Google Scholar]
- 34.Singhi AD, Cimino-Mathews A, Jenkins RB, Lan F, Fink SR, Nassar H, et al. MYC gene amplification is often acquired in lethal distant breast cancer metastases of unamplified primary tumors. Mod Pathol. 2012;25:378–87. doi: 10.1038/modpathol.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lundgren K, Holm K, Nordenskjold B, Borg A, Landberg G. Gene products of chromosome 11q and their association with CCND1 gene amplification and tamoxifen resistance in premenopausal breast cancer. Breast Cancer Res. 2008;10:R81. doi: 10.1186/bcr2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–48. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.