Abstract
Background
MicroRNAs (miRNAs) are abundant in the circulation and play a central role in diverse biological processes; they may be useful for early diagnosis of hepatocellular carcinoma (HCC).
Methods
We conducted a two-phase, case-control study (20 pairs for the discovery set and 49 pairs for the validation set) to test the hypothesis that genome-wide dysregulation of circulating miRNAs differentiate HCC cases from controls. Taqman low density arrays were used to examine genome-wide miRNA expression for the discovery set, and quantitative RT-PCR was used to validate candidate miRNAs for both discovery and validation sets.
Results
Sixty-six miRNAs were found to be significantly over-expressed in plasma of HCC cases compared to controls after adjusting for false discovery rate (p<0.05). A volcano plot indicated that 7 miRNAs had greater than 2-fold case-control differences with p<0.01. Four significant miRNAs (miR-150, miR-30c, miR-483-5p and miR-520b) detectable in all samples with varied expression levels were further validated in a validation set. MiR-483-5p was statistically significantly over-expressed in HCC cases compared with controls (3.20 vs. 0.82, p<0.0001). HCC risk factors and clinic-pathological characteristics did not influence miR-483-5p expression. The combination of plasma miR-483-5p level and HCV status can significantly differentiate HCC cases from controls with an AUC of 0.908 (p<0.0001). The sensitivity and specificity were, respectively, 75.5% and 89.8%.
Conclusions
These preliminary results suggest the importance of dysregulated circulating miR-483-5p as a potential HCC biomarker.
Impact
Confirmation of aberrant expression of miR-483-5p in a large prospective HCC study will provide support for its application to HCC detection.
Keywords: Genome-wide, circulating miRNAs, miR-483-5p, dysregulation, hepatocellular carcinoma
Introduction
The incidence of hepatocellular carcinoma (HCC) in the United States (US) has doubled over the past 20 years (1,2). HCC prognosis is poor if not diagnosed and treated at an early stage. Current clinical diagnostic approaches for HCC are mainly based on imaging techniques (abdominal ultrasound MRI, contrast-enhanced CT scan) and histology (3,4). Serum α-fetoprotein (AFP) measured in clinically available samples has long been used as an early diagnostic biomarker of HCC, but the sensitivity (39-65%) and specificity (76-94%) are poor (5,6). Therefore, identification of novel and reliable biomarkers in easily accessible clinical biospecimens is extremely important to improve the early detection of HCC. Circulating microRNAs (miRNAs) in cell-free plasma/serum samples have been consistently observed to have high stability and resistance to storage/handling (7-11), suggesting their potential use as diagnostic biomarkers.
miRNAs are a class of small non-coding RNAs that control gene expression by inhibiting translation or inducing cleavage of target messenger RNAs (mRNAs). miRNAs can regulate diverse biological processes including DNA repair, apoptosis, cell proliferation, differentiation and immune function. Aberrant miRNA expression has been associated with a variety of cancers (10,12), including HCC by examining tumor and non-tumor tissues (13-15). Several miRNAs with oncogenic characteristics are significantly up-regulated in HCC tumor tissues compared with non-tumor tissues, such as the miR-17-92 cluster, miR-21, miR-181b, miR-221, miR-222 and miR-500 (16-27). Genome-wide microarray data found over-expression of miR-221 and miR-222 in 50-83% of HCC tissues compared with matched cirrhotic tissue (28-32). Tumor-site specific miRNAs can be extracellularly released into the bloodstream via active secretion from tumor tissues in a protein-bound complex (33) or as membrane-bound vesicles (34). Because of high rates of proliferation and cell lysis in tumors, non-specific passive release of miRNAs is also observed (9). Circulating miRNAs are consistently shown to have high stability due to their protection from RNases (7,9,11), and even under severe conditions, such as boiling, very low or high pH levels, extended storage, and multiple freeze–thaw cycles (7). These data indicate that miRNAs are abundant in circulation, and may be useful for early diagnosis of HCC.
Investigators, using candidate and genome-wide approaches, have found more than 100 miRNAs that were dysregulated in HCC tumor tissue compared to non-tumor tissue (35). But only a small proportion of circulating miRNAs (∼1/10) were aberrantly expressed in the plasma of HCC cases compared to controls, including over-expression of let-7f, miR-122, miR-192, miR-21, miR-221-224, miR-25, miR-26a, miR-27a, miR-375 miR-500, miR-801 and miR-885 (26,36-41) and down-regulation of miR-16, miR-195, miR-199a and miR-92a (27,42). One genome-wide study identified a panel of miRNAs (miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a and miR-801) significantly over-expressed in hepatitis B virus (HBV)-related HCC compared to controls with unknown viral status (40). Interestingly, different expression patterns were observed for miRNAs in plasma and tumor tissue samples. For example, miR-16 (42,43) and miR-92a (27) were over-expressed in HCC tumor tissues but down-regulated in plasma; let-7g (28,41) and miR-122 (28,39) were repressed in tissue but up-regulated in plasma samples. These results indicate that further studies are needed to validate genome-wide circulating miRNAs as biomarkers to improve early diagnosis of HCC. In the current study, we employed a two-phase epidemiological study design to first discover genome-wide circulating miRNAs significantly differentiating HCC cases from controls, and then validating candidate miRNAs in the same discovery set and in a validation set including 49 HCC cases and matched controls.
Materials and Methods
Selection of HCC patients, controls and biospecimens
A hospital-based HCC case-control study is ongoing in Columbia University Medical Center (CUMC), which is approved by the Institutional Review Board. Written informed consent was obtained from each participant. HCC patients and controls in the current study were enrolled between October 2008 and August 2011. Cases were newly diagnosed HCC patients who were treated in the Hepatobiliary Oncology Clinics, CUMC, and examined pathologically. Histologic evaluation of hematoxylin and eosin stained 4 micron thick sections of frozen tissue stored at -20°C assessed for the presence and percent tumor. Tumor stage was determined according to the American Joint Committee on Cancer (AJCC) criteria (44). Separate blocks of non-tumor liver tissues were evaluated with respect to presence (Batts-Ludwig stage of 4) or absence cirrhosis (Batts-Ludwig stage<4). The inclusion criteria for HCC cases were pathologically confirmed diagnosis. Cases were excluded from the current study if they had a history of other cancers.
Controls were recruited from volunteers through the Research Recruitment and Minority Outreach (RRMO) core of Herbert Irving Comprehensive Cancer Center (HICCC). Flyers were placed at strategic locations around CUMC where hospital visitors and employees frequent or were handed out at inreach events at the hospital or at outreach events in the community. Interested participants were directed to contact the trained recruitment staff from the RRMO and given further information about participation. Interested participants were excluded from the control group if diagnosed for any kind of cancer or liver disease. Eligible controls were asked to fill out the same demographic and epidemiological questionnaire as HCC cases. In the current study, controls were matched with HCC cases on age (±5 yrs), gender (male/female) and ethnicities (Caucasian/Hispanic/African-American/Asian).
After completing the questionnaire, all participants were asked to provide a 15 ml blood sample, obtained by a trained phlebotomist. Blood samples were processed by the Biomarkers Shared Resource (BSR) of HICCC according to a standardized protocol (processing blood either the day of collection, normally within 2 hrs, or the next morning for samples drawn after 5 pm). Overnight bloods were kept chilled. Time of collection and processing were recorded in the core database. All blood samples in the current study were collected during the daytime, so the time of phlebotomy was not a confounder. Because a few control samples were processed next day, we compared miRNA profiles for those bloods collected/processed on the same day (16 subjects) and processed the next day (4 subjects) to identify potential confounding. No significant difference was observed for the expression of most miRNAs (747 out of total 750 miRNAs). Only 3 miRNAs (miR-26a, miR-16 and miR-342-3p) were significantly repressed in samples processed the next day (data not shown). These data indicated that no obvious confounding effect exists for different times to blood processing. Samples were stored in -80°C freezers connected to a telephone alarm system. A web-based inventory includes number and volume of aliquots as well as freeze-thaw cycles. In the phase I study, the pre-operative plasma samples from 20 histologically confirmed HCC cases and 20 matched controls as discovery set were screened for genome-wide miRNA expression. In the phase II study, candidate miRNAs were verified in both the 20 pairs of the discovery set, as well as a validation set of 49 HCC cases and 49 matched controls.
Epidemiological and clinicopathological data collection
A short, self-administered epidemiological questionnaire was used to collect information on age, gender, race/ethnicity, place of birth of self and parents, height, weight, education, occupation, active and passive smoking, alcohol consumption and family history of cancer. Information on HBV, hepatitis C virus (HCV) infection and clinicopathological features including AFP levels, anti-viral treatment, cirrhotic status, Milan criteria, Child-Pugh score, tumor stage, tumor size and survival status for HCC cases were obtained from the medical and pathological records.
Laboratory methods
HBV surface antigen (HBsAg) and HCV (anti-HCV) status in plasma samples were determined by ELISA Kits (BioChain Institute Inc., Hayward, CA) if not available from the medical records. Total RNA, including miRNAs was isolated from 250μl plasma using miRNeasy® Mini Kit (Qiagen, Frederick, MA) according to the manufacturer's protocol. TaqMan Low Density Arrays (TLDA, Applied Biosystems, Foster City, CA), including card A and B, were used to measure genome-wide miRNA expression profiles. A three-step process was performed involving a reverse transcription (RT) reaction followed by a pre-amplification reaction and quantitative real-time PCR (qRT-PCR). Cycle threshold (Ct) values were calculated using the SDS2.2.2. The complete set of TLDA data has been deposited in NCBI's Gene Expression Omnibus (GEO) (45) and are available through series accession number GSE50013 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE50013). TaqMan MicroRNA Assays were used for quantification of dysregulated miRNAs identified by TLDA. Real-time PCR reactions were run in duplicate, and the average Ct and standard deviation (SD) were calculated. U6 snRNA was used as an endogenous control to normalize the relative expression of target miRNAs and are presented as 2(-ΔCt) (26). The fold changes of paired samples were determined by the 2(-ΔΔCt) approach using DataAssist™ v2.0 (Applied Biosystems) (46). Because of skewing of miRNA levels, all data were log2 transformed. In a pilot study, we spiked 250μl plasma with 100 fmol of a chemically synthesized cel-miR-39 (Invitrogen, Carlsbad, CA) to normalize the RNA extraction, RT and qRT-PCR processes. The reproducibility of measuring cel-miR-39 was excellent with an overall R2=0.99 (p<0.01). The average inter- and intra-assay CVs were 1.47% and 0.84%, respectively indicating the accuracy of the quantitation.
Statistical analysis
We applied Biometric Research Branch (BRB) array tools to conduct data analysis with the original results transformed to log-scale and adjusted for reference scale (47). A two-sample t-test was used to compare the difference in geometric mean between cases and controls. A volcano plot was constructed for detectable miRNAs (detected in at least two paired samples). We filtered miRNAs that had missing values for more than 20% of samples, and explored significant miRNAs by the univariate test and Benjamini-Hochberg false-discovery rate (FDR) adjustment (p<0.05). We generated heat maps and hierarchical plots to check the clustering of samples based on significant miRNAs.
Four miRNAs over-expressed in HCC and detectable in every sample were selected for further testing by qRT-PCR in the same 20 pair discovery set and a validation set of 49 paired HCC cases and controls. Gene ontology analysis (htpp://www.pantherdb.org/) was performed by the PANTHER classification system to compare potential target genes affected by miRNAs with the NCBI reference (human genome build 36) (48). The binomial test was used to identify significantly enriched pathways, biological processes, molecular functions, cellular components and protein class terms after Bonferroni correction for multiple comparisons with a cutoff of p≤0.05.
miRNA expression levels were compared by genometric means and using the 75th percentile in controls as cutoff points for over-expression in order to increase specificity. A two-sample t-test assuming equal variance was carried out to compare log2 miRNA levels by categorical covariates of HCC risk factors and clinic-pathological characteristics, including gender (male vs. female), HBsAg (negative vs. positive), anti-HCV (negative vs. positive), cigarette smoking (no vs. yes), alcohol drinking (no vs. yes), AFP (<400ng/mL vs. ≥400ng/mL), cirrhosis (no vs. yes), within Milan criteria (yes vs. no), Child-Pugh score (A vs. B), tumor size (<5cm vs. ≥5cm), tumor stage (I-II vs. III-IV) and survival (yes vs. no). Logistic regression was used to construct receiver operating characteristic (ROC) curves by miRNA level adjusted for age and ethnicity (49). The maximum sensitivity and specificity and the area under the curve (AUC) were identified by numerical integration of each ROC curve. A better prediction model was built by fitting miRNAs, HBV and HCV status into the logistic regression model. The stepwise backward model selection was performed to determine the combinations of miRNA and viral status that significantly discriminate HCC patients from controls. A likelihood ratio test p value <0.05 was considered statistically significant. Statistical analyses were completed using Statistical Analysis System 9.0 (SAS Institute, Cary, NC).
Results
The demographic and clinicopathologic characteristics of 20 pairs in the discovery set and the 49 pairs in the validation set are shown in Table 1. The mean ages, gender and ethnicity are similar for HCC cases and controls in both sets. Most cases are HCV infected (55% and 66%), values significantly higher than among matched controls. Twelve and 20% of HCC cases are HBsAg positive; this is significant higher than controls in the discovery set but not the validation set. For the clinical co-variants, there are no statistically significant differences for HCC cases in discovery and validation sets for anti-viral treatment, AFP levels, Milan criteria, Child-Pugh score, tumor stage, cirrhosis, and survival outcome. Only tumor size significantly differs (p=0.047) between the two sets, indicating the need for further subgroup analysis.
Table 1. Demographic and clinico-pathological characteristics of study subjects in the discovery and validation sets.
| Variables | Discovery Set | Validation Set | ||
|---|---|---|---|---|
|
|
|
|||
| HCC Cases (n=20) |
Controls (n=20) |
HCC Cases (n=49) |
Controls (n=49) |
|
| Age at blood collection | ||||
| Mean ± SD (yrs) | 59.1 ± 8.8 | 58.6 ± 9.0 | 61.1 ± 11.7 | 61.5 ± 11.0 |
| < 60 yrs | 11 (55) | 10 (50) | 24 (49) | 23 (47) |
| ≥ 60 yrs | 9 (45) | 10 (50) | 25 (51) | 26 (53) |
| Gender | n (%) | n (%) | n (%) | n (%) |
| Male | 17 (85) | 17 (85) | 41 (84) | 41 (84) |
| Female | 3 (15) | 3 (15) | 8 (16) | 8 (16) |
| Ethnicity | ||||
| Caucasian | 16 (80) | 16 (80) | 32 (65) | 32 (65) |
| Hispanic | 2 (10) | 2 (10) | 11 (23) | 11 (22) |
| African-American | 0 | 0 | 4 (8) | 4 (8) |
| Asian | 2 (10) | 2 (10) | 2 (4) | 2 (4) |
| Viral infection status | ||||
| HBV (−) and HCV (−) | 5 (25) | 19 (95) | 11 (22) | 39 (80) |
| HBV (+) and HCV (−) | 4 (20) ** | 1 (5) | 6 (12) | 7 (14) |
| HBV (−) and HCV (+) | 11 (55) ** | 0 | 32 (66) ** | 1 (2) |
| HBV (+) and HCV (+) | 0 | 0 | 0 | 2 (4) |
| Anti-viral treatment | ||||
| No | 9 (45) | 22 (45) | ||
| Yes | 8 (40) | 15 (31) | ||
| Missing | 3 (15) | 12 (24) | ||
| AFP (ng/mL) | ||||
| <400 | 14 (70) | 32 (65) | ||
| ≥400 | 6 (30) | 15 (31) | ||
| Missing | 0 | 2 (4) | ||
| Cirrhosis | ||||
| Presence | 6 (30) | 18 (37) | ||
| Absence | 5 (25) | 8 (16) | ||
| Missing | 9 (45) | 23 (47) | ||
| Within Milan | ||||
| Yes | 15 (75) | 32 (65) | ||
| No | 4 (20) | 17 (35) | ||
| Missing | 1 (5) | 0 | ||
| Child-Pugh score | ||||
| A | 11 (55) | 34 (69) | ||
| B | 9 (45) | 15 (31) | ||
| Tumor stage | ||||
| I-II | 7 (35) | 19 (39) | ||
| III-IV | 13 (65) | 30 (61) | ||
| Tumor size | ||||
| <5 cm | 5 (25) | 26 (53) | ||
| ≥5 cm | 13 (65) * | 21 (43) | ||
| Missing | 2 (10) | 2 (4) | ||
| Survival status | ||||
| Alive | 10 (50) | 31 (63) | ||
| Deceased | 10 (50) | 17 (35) | ||
| Missing | 0 | 1 (2) | ||
p < 0.05;
p < 0.001;
TLDA data for the 20 pairs in the discovery set showed that 255 miRNAs (34.0% of 750 miRNAs) were detectable in circulation (defined as expression in at least two samples for each group). Analyzing a total of 91 miRNAs with ≥80% detection rates, we found that 66 miRNAs were significantly over-expressed in HCC cases compared to controls after adjusting for false discovery rate (Supplemental Table 1). Hierarchical cluster analysis found that miRNA expression patterns in HCC cases were significantly different from controls, i.e. genome-wide miRNA over-expression was more common in HCC cases compared to controls (Supplemental Figure 1). A volcano plot shows that 7 miRNAs (miR-150, miR-30b, miR-30c, miR-376a, miR-483-5p, miR-520b and miR-720) have over 2-fold case-control differences with a p<0.01 (Figure 1). The log2 fold-changes of these 7 miRNAs between 20 paired HCC cases and controls are shown in Supplemental Figure 2. Over-expression (log2 fold-change>0) is consistently observed for most HCC cases (75-90%). No significantly down-regulated miRNAs were observed.
Figure 1.
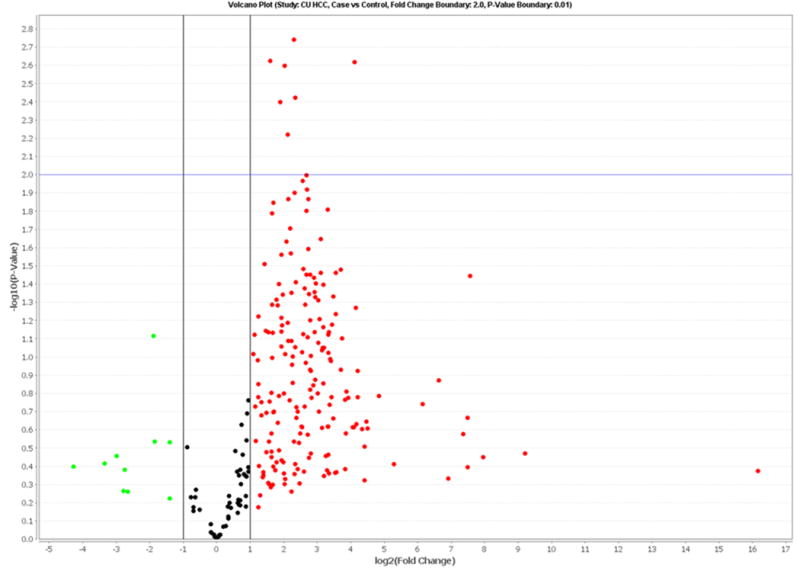
Volcano plot of detectable genome-wide miRNA profiles in differentiating 20 HCC cases from age-, gender- and ethnicity-matched controls. The x-axis shows the log2 fold-change in circulating miRNAs' expression between HCC cases and controls, while the y-axis shows the −log10 of the adjusted p value for each miRNA, representing the strength of the association. Above the dashed line indicates statistically significant (p<0.01) after Bonferroni correction.
There were 36 and 30 miRNAs significantly associated with HBsAg or HCV positive HCC, respectively, compared with viral negative controls at the nominal 0.05 level of the univariate test (Supplemental Table 1). No miRNA remained statistically significant after FDR adjustment, except for miR-1243 among HBsAg positive HCC cases (p=0.014). To examine viral-related miRNAs, we compared expression profiles by viral status (HBsAg or HCV positive) in HCC patients. Seven miRNAs were specifically associated with viral infection, but unrelated with HCC (p<0.05, data not shown).
We further verified 4 miRNAs (miR-150, miR-30c, miR-483-5p and miR-520b) by qRT-PCR in the same 20 pairs of discovery set, and in another validation set including 49 cases and 49 matched controls. The four miRNAs were selected because they were detectable by TLDA with varied expression levels (high, moderate or low) in all subjects. Expression of miR-520b was undetectable for all plasma samples by qRT-PCR, and was omitted in the data analysis. Through PANTHER ontology analysis, we examined the biological characteristics of 1,497 conserved genes potentially targeted by the three detectable miRNAs (miR-150, miR-30c and miR-483-5p). Enriched genes were significantly associated with more than 20 biological pathways, including pathways of Hedgehog, interleukin, TGF-beta, EGF receptor/Wnt signaling, p38 MAPK, Ras, p53, PI3 kinase etc. (Supplemental Table 2). Some of these genes were previously identified as mutated or with copy number changes in HCC (50,51). Our data confirm the broad range of targets for the three miRNAs potentially involved in the pathophysiology of HCC.
The correlations in expression levels (log2 transformed) between TLDA and qRT-PCR assays for the three miRNAs were significant (Supplemental Figure 3). The correlation coefficients were 0.57, 0.60 and 0.79 for miR-30c, miR-150 and miR-483-5p, respectively (p≤0.0001), indicating excellent correlations if the miRNAs were detected by both assays. However, when separately comparing the qRT-PCR data for the three miRNAs between HCC cases and controls in the discovery and validation sets, only miR-483-5p is significantly over-expressed in HCC cases, which is consistent with the result from the TLDA data (Table 2). The fold change in miR-483-5p was 3.8 and 9.4, suggesting the potentially important role of miR-483-5p upregulation in HCC. No consistent expression patterns were observed for miR-30c and miR-150 between the discovery and validation sets.
Table 2. Log2 expression levels of three candidate miRNAs in the discovery and validation sets by TLDA and qRT-PCR assays.
| miRNAs | Methods, Subjects | Log2 expression levels Mean (SD) | Fold change | p | |
|---|---|---|---|---|---|
|
| |||||
| HCC cases | Controls | ||||
| miR-30c | TLDA, 20 pairs | 5.46 (1.97) | 3.05 (2.56) | 5.31 | 0.004 |
| qRT-PCR, 20 pairs | 2.53 (1.88) | 0.93 (3.12) | 3.02 | 0.088 | |
| qRT-PCR, 49 pairs | 0.37 (1.60) | 1.16 (2.11) | 0.58 | 0.038 | |
| miR-150 | TLDA, 20 pairs | 5.18 (2.25) | 3.19 (2.15) | 3.98 | 0.008 |
| qRT-PCR, 20 pairs | 3.17 (1.52) | 1.99 (1.67) | 2.27 | 0.052 | |
| qRT-PCR, 49 pairs | 1.86 (1.49) | 2.55 (1.58) | 0.62 | 0.035 | |
| miR-483-5p | TLDA, 20 pairs | 2.30 (2.52) | -1.74 (2.37) | 16.56 | 9.01E-05 |
| qRT-PCR, 20 pairs | 0.67 (2.02) | -2.47 (2.21) | 9.41 | 0.0004 | |
| qRT-PCR, 49 pairs | 1.68 (1.64) | -0.28 (1.37) | 3.88 | 7.43E-08 | |
To increase the specificity of dysregulated miRNA in detecting HCC, we used the 75th percentile among controls as a cutoff point for over-expression. Only miR-483-5p is significantly associated with increased HCC risk compared to controls in the validation set (OR=9.7, 95%CI: 2.6-35.3, p=0.0006, Table 3) after adjustment for age, gender, ethnicity, HBV and HCV status, which is consistent with the finding in discovery set (Supplemental Table 3). No significant case-control differences were observed for miR-150 and miR-30c in the discovery or validation sets (Table 3 and Supplemental Table 3).
Table 3. Over-expression of plasma miRNAs and HCC prediction in the 49 HCC case-control pairs of the validation set.
| miRNAs over-expression* | HCC, N (%) | Controls, N (%) | OR (95%CI)** | p |
|---|---|---|---|---|
| miR-30c | ||||
| No | 42 (86) | 36 (73) | 1.00 (ref) | |
| Yes | 7 (14) | 13 (27) | 0.23 (0.04-1.43) | 0.116 |
| miR-150 | ||||
| No | 37 (76) | 36 (73) | 1.00 (ref) | |
| Yes | 12 (24) | 13 (27) | 0.72 (0.13-4.05) | 0.712 |
| miR-483-5p | ||||
| No | 14 (29) | 37 (76) | 1.00 (ref) | |
| Yes | 35 (71) | 12 (24) | 9.66 (2.64-35.26) | 0.0006 |
75th percentile in controls is used as the cutoff point for miRNA over-expression
Adjusted for age, gender, ethnicity and viral status (HBV and HCV)
To examine the potential influence of HCC risk factors and clinic-pathological factors on expression of the three miRNAs, we separately conducted subgroup analyses for those covariates in the discovery and validation sets using the qRT-PCR data. Among HCC cases, we found no significant differences in miRNA expression by HBV, HCV, anti-viral treatment, cirrhosis, tumor stage or survival subgroups (Supplemental Tables 4-5). Although differences were seen with respect to certain covariates in one set, no consistent influences were found in both sets. For example, miR-483-5p differs by Child-Pugh score in the discovery but not in validation set; miR-30c and miR-483-5p differ by AFP and tumor size in the validation but not in discovery set. The three miRNAs also display no significant difference in expression by HBV or HCV status among controls (data not shown). These data suggest that the influence of the covariates on miRNAs may be minor, although the small sample size in subgroup analysis limits interpretation of the results.
To assess the predictive accuracy of miRNAs in detecting HCC cases, multivariable logistic regression was used to construct ROC curves in combination with HBV and HCV infection status. Using a stepwise selection model to gradually eliminate non-significant covariates, the best predictive model including miR-483-5p and HCV has a predictive accuracy of 0.908 (p<0.0001), a sensitivity of 75.5% and a specificity of 89.8% (Figure 2). HCV had an AUC of 0.796 (p<0.0001), and either viral infection (HBV, HCV) had an AUC of 0.815 (p<0.0001). MiR-483-5p expression can significantly differentiate HCC cases from controls with an AUC of 0.827 (p<0.0001), which is better than the accuracy of using viral status alone. The sensitivity and specificity for miR-483-5p alone are, respectively, 55.1% (27/49) and 85.7% (42/49) (Supplemental Table 6).
Figure 2.
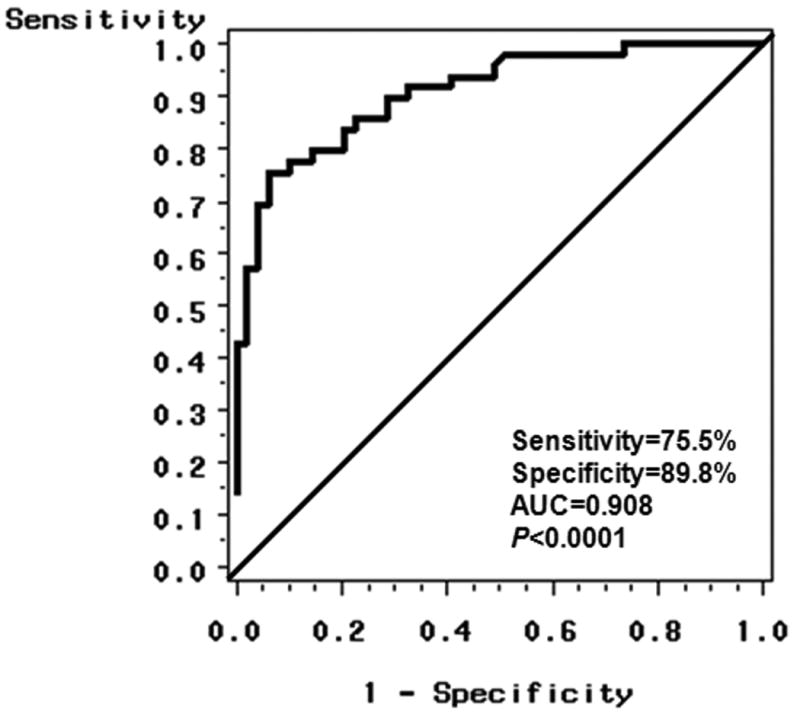
Receiver-operator characteristic (ROC) curve plot of sensitive vs. 1–specificity for miR-483-5p levels and HCV status that can differentiate HCC cases from controls. The area under the curve (AUC) is 0.908 (p<0.0001) for the probability cut-point of 0.50 with a sensitivity of 75.5% (37/49) and a specificity of 89.8% (44/49).
Discussion
We employed low density array to first examine genome-wide expression for 750 miRNAs, and then validated three candidate miRNAs in the same discovery set and a validation set by qRT-PCR. A total of 66 miRNAs were significantly differentially expressed between HCC cases and controls after adjusting for false discovery rate (Supplemental Figure 1, Supplemental Table 1). More importantly, 59 miRNAs were initially identified as circulating biomarkers for HCC, and 7 (miR-19b-1, miR-24, miR-29c, miR-376a, miR-378, miR-520c-3p and miR-92a) were consistent with previous findings. A volcano plot indicated that 7 miRNAs (miR-150, miR-30b, miR-30c, miR-376a, miR-483-5p, miR-520b and miR-720) had over 2-fold case-control differences with an adjusted p value of <0.01 (Figure 1). To our knowledge, no previous study has characterized these miRNAs in HCC. In a validation study, miR-483-5p over-expression was significantly associated with increased HCC risk (OR=6.8, 95%CI: 2.1-22.2, p=0.002, Table 3). Multivariable logistic regression indicated that miR-483-5p expression and HCV status could significantly differentiate HCC cases from controls with an AUC of 0.908 (Figure 2). These data suggest that circulating miR-483-5p may be a useful biomarker for HCC detection.
Limited data are available for miR-483-5p expression in human cancer. Because pre-miR-483-5p maps to intron 2 of IGF2 (20), a gene that is highly expressed in adrenocortical carcinoma (52,53) and pheochromocytomas (54), it is reasonable to assume that miR-483-5p may be co-expressed with its host gene (55). In support of this hypothesis, several previous studies found positive correlations between miR-483 expression and IGF2 mRNA levels in Wilms tumor, colorectal cancer, malignant pheochromocytoma and HCC tissues (52-55). One study also found elevated expression of miR-483-5p in serum from HCC cases (36), which is consistent with our current observation. The pathogenic role and molecular mechanism of action of miR-483-5p in tumorigenesis remain unknown. miR-483-3p was found to function as an anti-apoptotic oncogene in cancer cell lines (HEPG2, liver carcinoma and HCT116, colorectal carcinoma) (53). An in vitro study of adrenocortical carcinoma revealed a growth promoting role for miR-483-5p (56). These data suggest a potential carcinogenic role for miR-483-5p in tumorigenesis.
Despite the promising finding that elevated plasma miR-483-5p can differentiate HCC cases from controls, it is still unknown whether this is due to active secretion from the tumor tissues. This is one of the major limitations in the current study. In an ongoing study, we found that miR-483-3p and miR-483-5p were highly expressed in HCC tumor tissues (2.8-13.9-fold) compared to adjacent non-tumor tissues, consistent with our current finding. From the methodologic point of view, another limitation is the pre-amplification step involved in the discovery phase using the TLDA arrays. The qRT-PCR assays used in the validation approach do not require pre-amplification. This may be one reason for the inconsistent results for miR-30c and miR-520b in the discovery and validation studies. More sensitive assays or protocols to enrich for circulating miRNAs may be required. The cross-sectional study design with no post-surgical plasma samples collected at different time points prevents us from obtaining information on the causal association between aberrant miRNAs and HCC. Therefore, it is imperative to establish longitudinal biospecimen repositories in order to clarify the critical role of circulating miRNAs in the long-term process of hepatocarcinogenesis.
In summary, our study suggests that circulating miR-483-5p may be a potential biomarker for less-invasive detection of HCC. These data support the idea that blood is a promising resource for novel miRNA biomarker discovery in addition to its ability to monitor genetic variation and DNA methylation alterations. Further evaluation of the biological functions of candidate miRNAs in tumorigenesis, such as angiogenesis, chronic inflammation, or cellular proliferation, will provide more evidence to understand the role of miRNAs as HCC biomarkers.
Supplementary Material
Acknowledgments
The authors thank Dr. Victor R. Grann and Kazeem Abdul in the Research Recruitment and Minority Outreach Core, Herbert Irving Comprehensive Cancer Center (HICCC) for recruitment of control subjects for the current study. We would like to thank all subjects who participated by donating blood for this study.
Grant Support: This work was financially supported by NIH grants R03 CA156629 (to J. Shen), R01 ES005116 (to R. Santella), P30 ES009089 (to R. Santella), P30 CA013696 (to R. Santella) and a pilot of NIEHS Center for Environmental Health in Northern Manhattan (to J. Shen).
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of Interest were disclosed.
References
- 1.El Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Benson AB, III, Abrams TA, Ben Josef E, Bloomston PM, Botha JF, Clary BM, et al. NCCN clinical practice guidelines in oncology: hepatobiliary cancers. J Natl Compr Canc Netw. 2009;7:350–91. doi: 10.6004/jnccn.2009.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zinkin NT, Grall F, Bhaskar K, Otu HH, Spentzos D, Kalmowitz B, et al. Serum proteomics and biomarkers in hepatocellular carcinoma and chronic liver disease. Clin Cancer Res. 2008;14:470–7. doi: 10.1158/1078-0432.CCR-07-0586. [DOI] [PubMed] [Google Scholar]
- 5.Collier J, Sherman M. Screening for hepatocellular carcinoma. Hepatology. 1998;27:273–8. doi: 10.1002/hep.510270140. [DOI] [PubMed] [Google Scholar]
- 6.Oka H, Tamori A, Kuroki T, Kobayashi K, Yamamoto S. Prospective study of alpha-fetoprotein in cirrhotic patients monitored for development of hepatocellular carcinoma. Hepatology. 1994;19:61–6. [PubMed] [Google Scholar]
- 7.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 8.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–77. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu AM, Yao TJ, Wang W, Wong KF, Lee NP, Fan ST, et al. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: a retrospective cohort study. BMJ Open. 2012;2:e000825. doi: 10.1136/bmjopen-2012-000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gramantieri L, Fornari F, Callegari E, Sabbioni S, Lanza G, Croce CM, et al. MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med. 2008;12:2189–204. doi: 10.1111/j.1582-4934.2008.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mott JL. MicroRNAs involved in tumor suppressor and oncogene pathways: implications for hepatobiliary neoplasia. Hepatology. 2009;50:630–7. doi: 10.1002/hep.23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munker R, Calin GA. MicroRNA profiling in cancer. Clin Sci (Lond) 2011;121:141–58. doi: 10.1042/CS20110005. [DOI] [PubMed] [Google Scholar]
- 16.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–45. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 17.Connolly E, Melegari M, Landgraf P, Tchaikovskaya T, Tennant BC, Slagle BL, et al. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am J Pathol. 2008;173:856–64. doi: 10.2353/ajpath.2008.080096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Tan W, Neo TW, Aung MO, Wasser S, Lim SG, et al. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci. 2009;100:1234–42. doi: 10.1111/j.1349-7006.2009.01164.x. [DOI] [PubMed] [Google Scholar]
- 19.Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, et al. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–27. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, Patel T, et al. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–61. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–29. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 24.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 25.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–33. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto Y, Kosaka N, Tanaka M, Koizumi F, Kanai Y, Mizutani T, et al. MicroRNA-500 as a potential diagnostic marker for hepatocellular carcinoma. Biomarkers. 2009;14:529–38. doi: 10.3109/13547500903150771. [DOI] [PubMed] [Google Scholar]
- 27.Shigoka M, Tsuchida A, Matsudo T, Nagakawa Y, Saito H, Suzuki Y, et al. Deregulation of miR-92a expression is implicated in hepatocellular carcinoma development. Pathol Int. 2010;60:351–7. doi: 10.1111/j.1440-1827.2010.02526.x. [DOI] [PubMed] [Google Scholar]
- 28.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–9. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 29.Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, et al. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135:257–69. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Wong QW, Ching AK, Chan AW, Choy KW, To KF, Lai PB, et al. MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin Cancer Res. 2010;16:867–75. doi: 10.1158/1078-0432.CCR-09-1840. [DOI] [PubMed] [Google Scholar]
- 31.Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–61. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 32.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:264–9. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nilsson RJ, Balaj L, Hulleman E, van Rijn S, Pegtel DM, Walraven M, et al. Blood platelets contain tumor-derived RNA biomarkers. Blood. 2011;118:3680–3. doi: 10.1182/blood-2011-03-344408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borel F, Konstantinova P, Jansen PL. Diagnostic and therapeutic potential of miRNA signatures in patients with hepatocellular carcinoma. J Hepatol. 2012;56:1371–83. doi: 10.1016/j.jhep.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 36.Gui J, Tian Y, Wen X, Zhang W, Zhang P, Gao J, et al. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin Sci (Lond) 2011;120:183–93. doi: 10.1042/CS20100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Wang Y, Yu W, Chen J, Luo J. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochem Biophys Res Commun. 2011;406:70–3. doi: 10.1016/j.bbrc.2011.01.111. [DOI] [PubMed] [Google Scholar]
- 38.Qi P, Cheng SQ, Wang H, Li N, Chen YF, Gao CF. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS ONE. 2011;6:e28486. doi: 10.1371/journal.pone.0028486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136–42. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- 40.Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29:4781–8. doi: 10.1200/JCO.2011.38.2697. [DOI] [PubMed] [Google Scholar]
- 41.Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70:9798–807. doi: 10.1158/0008-5472.CAN-10-1001. [DOI] [PubMed] [Google Scholar]
- 42.Qu KZ, Zhang K, Li H, Afdhal NH, Albitar M. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J Clin Gastroenterol. 2011;45:355–60. doi: 10.1097/MCG.0b013e3181f18ac2. [DOI] [PubMed] [Google Scholar]
- 43.Huang XH, Wang Q, Chen JS, Fu XH, Chen XL, Chen LZ, et al. Bead-based microarray analysis of microRNA expression in hepatocellular carcinoma: miR-338 is downregulated. Hepatol Res. 2009;39:786–94. doi: 10.1111/j.1872-034X.2009.00502.x. [DOI] [PubMed] [Google Scholar]
- 44.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–4. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 45.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 47.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007;3:11–7. [PMC free article] [PubMed] [Google Scholar]
- 48.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem. 1993;39:561–77. [PubMed] [Google Scholar]
- 50.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–4. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 51.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–8. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patterson EE, Holloway AK, Weng J, Fojo T, Kebebew E. MicroRNA profiling of adrenocortical tumors reveals miR-483 as a marker of malignancy. Cancer. 2011;117:1630–9. doi: 10.1002/cncr.25724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veronese A, Lupini L, Consiglio J, Visone R, Ferracin M, Fornari F, et al. Oncogenic role of miR-483-3p at the IGF2/483 locus. Cancer Res. 2010;70:3140–9. doi: 10.1158/0008-5472.CAN-09-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer-Rochow GY, Jackson NE, Conaglen JV, Whittle DE, Kunnimalaiyaan M, Chen H, et al. MicroRNA profiling of benign and malignant pheochromocytomas identifies novel diagnostic and therapeutic targets. Endocr Relat Cancer. 2010;17:835–46. doi: 10.1677/ERC-10-0142. [DOI] [PubMed] [Google Scholar]
- 55.Ma N, Wang X, Qiao Y, Li F, Hui Y, Zou C, et al. Coexpression of an intronic microRNA and its host gene reveals a potential role for miR-483-5p as an IGF2 partner. Mol Cell Endocrinol. 2011;333:96–101. doi: 10.1016/j.mce.2010.11.027. [DOI] [PubMed] [Google Scholar]
- 56.Ozata DM, Caramuta S, Velazquez-Fernandez D, Akcakaya P, Xie H, Hoog A, et al. The role of microRNA deregulation in the pathogenesis of adrenocortical carcinoma. Endocr Relat Cancer. 2011;18:643–55. doi: 10.1530/ERC-11-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


