Abstract
RNA-directed DNA methylation (RdDM) in Arabidopsis thaliana depends on the upstream synthesis of 24-nucleotide small interfering RNAs (siRNAs) by RNA POLYMERASE IV (Pol IV)1,2 and downstream synthesis of non-coding transcripts by Pol V. Pol V transcripts are thought to interact with siRNAs which then recruit DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) to methylate DNA3-7. The SU(VAR)3-9 homologs SUVH2 and SUVH9 act in this downstream step but the mechanism of their action is unknown8,9. Here we show that genome-wide Pol V association with chromatin redundantly requires, SUVH2 and SUVH9. Although SUVH2 and SUVH9 resemble histone methyltransferases a crystal structure reveals that SUVH9 lacks a peptide-substrate binding cleft and lacks a properly formed S-adenosyl methionine (SAM) binding pocket necessary for normal catalysis, consistent with a lack of methyltransferase activity for these proteins8. SUVH2 and SUVH9 both contain SET- and RING-ASSOCIATED (SRA) domains capable of binding methylated DNA8, suggesting that they function to recruit Pol V through DNA methylation. Consistent with this model, mutation of DNA METHYLTRANSFERASE 1 (MET1) causes loss of DNA methylation, a nearly complete loss of Pol V at its normal locations, and redistribution of Pol V to sites that become hypermethylated. Furthermore, tethering SUVH2 with a zinc finger to an unmethylated site is sufficient to recruit Pol V and establish DNA methylation and gene silencing. These results suggest that Pol V is recruited to DNA methylation through the methyl-DNA binding SUVH2 and SUVH9 proteins, and our mechanistic findings suggest a means for selectively targeting regions of plant genomes for epigenetic silencing.
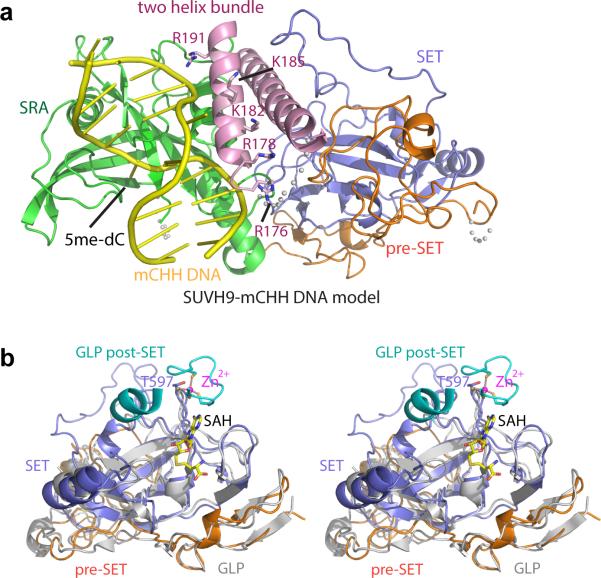
To gain insights into the function of SUVH2/SUVH9, we solved the crystal structure of an N-terminally-truncated SUVH9 construct (residues 134 – 650), which contains all the known functional domains (the SRA, pre-SET, and SET domains) (Fig. 1a, Extended Data 1a and Supplementary Table 1). The structure of SUVH9 is composed of three segments: a two-helix bundle towards the N-terminus (residues 138 - 194), the SRA domain (residues 195 - 379), and the pre-SET/SET domains (residues 380 - 637). There are extensive inter-domain interactions that can stabilize the overall architecture of the protein (Fig 1a and Extended Data Fig. 1b-g).
Figure 1. Crystal structure of SUVH9.
a. Ribbon diagram of the SUVH9 crystal structure containing a two-helix bundle, SRA domain, pre-SET domain, and SET domain colored in pink, green, orange, and blue, respectively. The Zn3Cys9 cluster is highlighted in a ball-and-stick representation and disordered regions are shown with dashed lines.
b. A superposition of SUVH9 SRA domain (in green) and SUVH5 SRA domain (in silver) shows that both proteins adopt a similar fold.
c. Top panel: the crystal structure of human GLP in complex with bound SAH (PDB code: 2IGQ) in a silver ribbon representation. Bottom panel: the SAH binding site in an electrostatic surface representation. The cofactor SAH is shown in a space-filling representation.
d. Top panel: the crystal structure of human GLP in complex with SAH and H3K9me2 peptide (PDB code: 2RFI) in silver ribbon representation. Bottom panel: the peptide binding site in an electrostatic surface representation. The post-SET domain and the acidic loop of the SET domain involved in peptide substrate binding are highlighted in cyan and dark blue, respectively. The bound peptide is shown in a space-filling representation in both panels.
e. Top panel: the crystal structure of SUVH9 in the free state in a color-coded ribbon representation. Bottom panel: an expanded view of the putative SAH binding site in an electrostatic surface representation.
f. Top panel: the crystal structure of SUVH9 in the free state in a color-coded ribbon representation. Bottom panel: an expanded view of the putative peptide-binding site in an electrostatic surface representation. The long insertion loop of the SET domain is highlighted in magenta.
Extended Data Figure 1. Interdomain interactions of SUVH9.
a. Color-coded schematic representation of full length SUVH9 and the N-terminally truncated construct used for crystallization.
b. The hydrophobic interactions and charged interactions within the two-helix bundle shown in two alternate views rotated by 180 degree. Residues involved in inter-helix hydrophobic interactions are highlighted in yellow.
c. The N-terminal part of the first α-helix forms charged and hydrogen bonding interactions with the SRA domain and the SET domain. The interacting residues are shown in stick representation and the hydrogen-bonding interactions are shown with dashed red lines.
d. The C-terminal part of the first α-helix exhibits extensive hydrophobic interactions with the SRA domain and the pre-SET/SET domains. The tip of a long loop from the SET domain covers over the first helix and forms hydrophobic interactions with it.
e. The second α-helix forms some interactions with the SRA domain.
f. The SRA domain forms a hydrophobic core that interacts with the pre-SET/SET domains.
g. A long insertion loop of SUVH9 SET domain (highlighted in magenta) is enriched with hydrophobic residues and forms extensive hydrophobic interactions with the two-helix bundle, the pre-SET and SET domains.
The SRA domain of SUVH9 resembles those of UHRF1 and SUVH510-13 (Fig. 1b). Based on the SUVH5 SRA-mCHH DNA complex structure13, we modeled a mCHH DNA into SUVH9 (Extended Data Fig. 2a). The DNA could be positioned in the nucleic acid-binding cleft of the SRA domain without significant steric clashes and the proposed flipped-out 5mC base readily inserts into the binding pocket of the SRA domain.
Extended Data Figure 2. SUVH9 SRA and pre-SET/SET domains.
a. A model positioning the mCHH DNA to the active site of SUVH9 SRA domain following superposition of the structures of the SUVH5 SRA-mCHH complex [PDB code: 3Q0F) and SUVH9 in the free state. SUVH9 domains are depicted with the same color-coding as in Figure 1a and the modeled DNA is colored in yellow. The DNA fits well into the SRA domain without significant steric clashes. Some surrounding residues on the second α-helix of the two-helix bundle, which can potentially be involved in the binding to the DNA, are highlighted in a stick representation.
b. A stereo view of the superposition of the structure of SUVH9 in the free state and the structure of human GLP catalytic fragment complexed with SAH (PDB code: 2IGQ). The GLP pre-SET and SET domains are colored in silver and its post-SET domain is colored in cyan. The zinc-binding motif of GLP post-SET domain and SET domain, the bound SAH molecule, and the corresponding Thr597 of SUVH9 are highlighted in a stick representation.
Although SUVH9 contains histone methyltransferase pre-SET and SET domains similar to Dim5, G9a, and GLP14-16 (Extended Data Fig. 2b), it exhibits neither detectable histone methyltransferase activity nor binding capacity for the SAM cofactor in vitro8. SUVH9 and SUVH2 lack post-SET domains that are normally critical for cofactor and peptide substrate binding, as well as catalysis (Extended Data Fig. 3). Compared with the structure of human H3K9 methyltransferase GLP (Fig. 1c, d), the putative S-adenosylhomocysteine (SAH)-binding pocket and peptide binding channel of SUVH9 are very open and incompletely formed (Fig. 1e, f) and cannot stably retain either a bound SAH molecule or the peptide substrate, especially in the absence of the stabilizing role of the post-SET domain. In summary, the SUVH9 structure is consistent with the demonstrated ability of SUVH9 to bind to methylated DNA, and supports the view that SUVH9 (and SUVH2) proteins encode inactive methyltransferase homologs.
Extended Data Figure 3. Structure-based sequence alignment of SUVH family proteins from Arabidopsis.
The secondary structural elements of SUVH9 are labeled on the top of the sequence alignment. The domain boundaries are marked on the top and depicted with color-coding as in Figure 1a. Conserved residues involved in the interaction with flipped 5mC base and the DNA backbone available from the published SUVH5-DNA complex structures are highlighted with cyan circles and blue hexagons, respectively. The insertions in the SET domains are highlighted with a purple box. The zinc-coordinating Cys residues are highlighted with black stars in the SET domain and grey stars in the post-SET domain. Two-tyrosine residues that are conserved and normally important for enzymatic activity are highlighted with red dots.
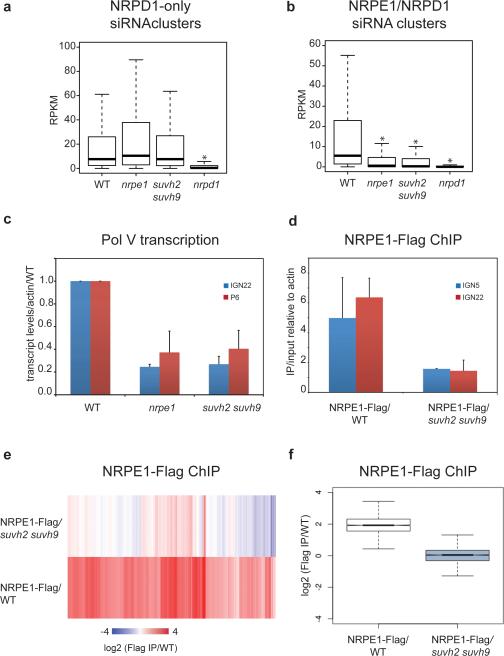
We sought to further characterize the role of SUVH2 and SUVH9 in the RdDM pathway by determining their effect on siRNAs genome-wide. Most siRNAs are eliminated in Pol IV (nrpd1) mutants, while only some siRNAs are reduced in Pol V (nrpe1) mutants17 (Fig 2a). We found that the suvh2 suvh9 double mutant reduced siRNA abundance at siRNAs clusters that were dependent on both Pol IV and Pol V, but not at Pol V-independent clusters (Fig. 2a, b). Thus, the suvh2 suvh9 double mutant siRNA phenotype closely resembles that of a Pol V mutant. We next sought to determine whether SUVH2/9 might be involved in the production of non-coding transcripts by Pol V. At two characterized sites (IGN22 and P6)18,19 we found that suvh2 suvh9 reduced Pol V transcripts to the same extent as nrpe1 (Fig. 2c). Similar results were observed at the AtSN1 locus in suvh2 suvh99. To test if SUVH2/9 are required for Pol V chromatin occupancy, we utilized chromatin immunoprecipitation (ChIP) of a Flag-tagged NRPE1 (the largest subunit of Pol V). We observed only background levels of Pol V binding at IGN5 and IGN22 in suvh2 suvh9 compared to a six-fold enrichment in wild type (WT; Fig. 2d). We further analyzed our ChIPs by next generation sequencing (ChIP-Seq) and found that binding of Pol V at all previously identified sites18 was significantly decreased or eliminated in suvh2 suvh9 (Fig. 2e, f).
Figure 2. SUVH2 and SUVH9 are required for Pol V dependent siRNA production, chromatin binding, and transcription.
a. Boxplot (whiskers extend to ±1.5 interquartile range (IQR)) of RPKM values for 24-nt siRNAs at previously defined siRNA clusters dependent on Pol IV (NRPD1), but not Pol V (NRPE1). 24-nt counts allow for up to 100 identical reads to be counted at any given position. * indicates a significant decrease (P<2.2e-16, Mann-Whitney U test).
b. Similar to plot in (a) for clusters defined as dependent on Pol IV and Pol V.
c. Quantitative reverse transcription PCR (RT-qPCR) of IGN22 and P6 relative to ACTIN7 and normalized to Columbia-0 (WT). Mean +/− standard deviation (SD) of two biological replicas.
d. Quantitative PCR (qPCR) of IGN22 and IGN5 from Flag ChIP shown as enrichment of IP/input relative to ACTIN7 in NRPE1-Flag/WT and NRPE1-Flag/suvh2 suvh9 lines. Mean +/− SD of two biological replicas.
e. Heat map of NRPE1 enrichment at defined NRPE1 sites determined by Flag ChIP-Seq in either NRPE1-Flag/WT or NRPE1-Flag/suvh2 suvh9, with Flag ChIP in WT as negative control.
f. Box plot of NRPE1 enrichment at sites shown in Fig. 2e for NRPE1-Flag/WT and NRPE1-Flag/suvh2 suvh9.
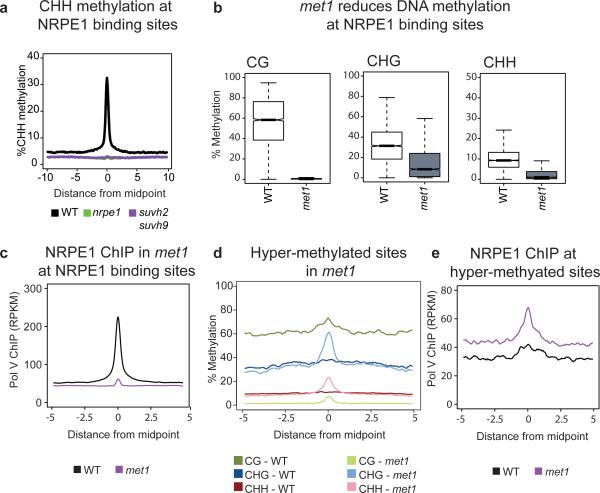
To determine the effect of SUVH2/9 on DNA methylation at defined Pol V binding sites, we utilized whole-genome bisulfite sequencing (BS-Seq). As in nrpe1, CHH methylation at Pol V binding sites was eliminated in suvh2 suvh9 (Fig. 3a). We also analyzed BS-Seq of the single mutants suvh2 and suvh9 to determine if SUVH2/SUVH9 act redundantly at all sites or have non-overlapping sites where they function. We found that suvh2 had a stronger effect than suvh9 at Pol V sites as well as at differentially methylated regions (DMRs) defined in either suvh2 or suvh9 single mutants, or in the suvh2 suvh9 double mutant (Extended Data Fig. 4a-c). These results indicate that SUVH2 and SUVH9 act redundantly throughout the genome to control RdDM.
Figure 3. Pol V binding is dependent on DNA methylation.
a. Metaplot of percent CHH methylation at all defined NRPE1 binding sites as determined by BS-seq in wild type (WT), nrpe1, and suvh2 suvh9.
b. Box plots showing DNA methylation in each cytosine context at defined NRPE1 binding sites in WT and met1.
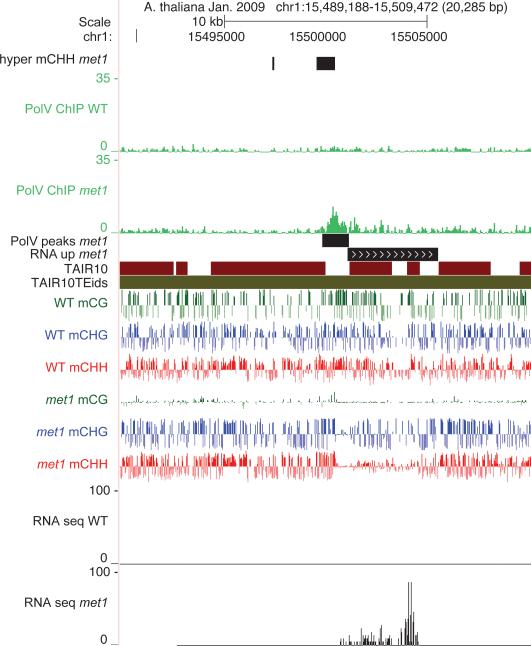
c. Pol V occupancy in met1 is reduced at NRPE1 sites.
d. Metaplot of DNA methylation at sites defined as hyper-methylated in met1.
e. Pol V occupancy in met1 is increased at defined hyper-methylated sites.
Extended Data Figure 4. SUVH2 and SUVH9 act redundantly genome-wide.
a. Metaplots of CHH methylation over DMRs identified in the various SUVH mutants.
b. Metaplots of CHH methylation over Pol V binding sites.
c. Venn diagram detailing the overlaps between CHH hypo-methylated regions in SUVH mutants.
Our results suggest that a reinforcing loop exists between DNA methylation and Pol V binding via SUVH2/9. To further test this model we utilized a mutation in the maintenance methyltransferase MET1 that eliminates CG methylation genome-wide and also reduces CHG and CHH methylation20-22 (Fig. 3b). Using endogenous antibodies to NRPE1, ChIP-seq revealed that Pol V occupancy was virtually eliminated in met1 at sites normally occupied by Pol V (Fig. 3c and Extended Data Figs. 5-7). In contrast, at sites previously identified as gaining methylation in met121, we observed an increase in Pol V binding (Fig. 3d, e, and Extended Data Figs. 8, 9). Furthermore, point mutations in the SRA domains of SUVH2 and SUVH9 were shown to cause a loss of RdDM8. Together, these results suggest that SUVH2/SUVH9 binding to methylated DNA recruits Pol V, thus providing a link between preexisting DNA methylation and the recruitment of further methylation by RdDM.
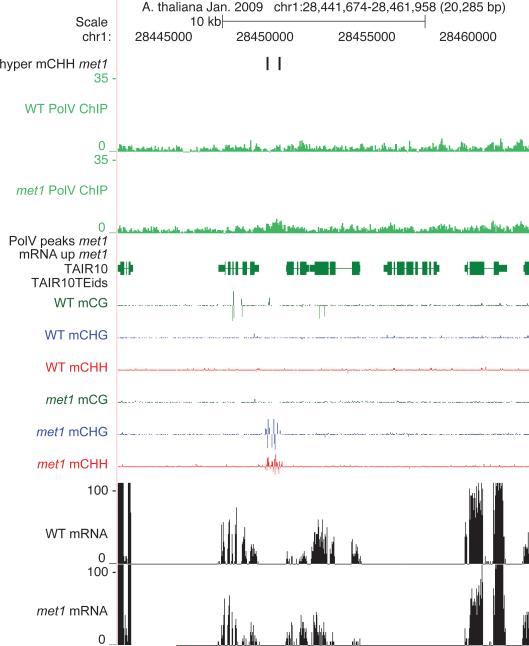
Extended Data Figure 5. Pol V occupancy in WT versus met1.
Chromosome 1 showing Pol V ChIP in WT versus met1 as mapped over TAIR10 (green genes, red TE).
Extended Data Figure 7. Screen shot of Pol V binding in WT versus met1.
Reduction in Pol V binding in a met1 hypomethylated site.
Extended Data Figure 8. Screen shot of Pol V binding at a hyper-CHH methylated site in WT versus met1.
An example of Pol V being redistributed to regions that gain methylation in met1.
Extended Data Figure 9. Pol V binding at hyper-CHH methylated site that is also transcribed.
Strong Pol V binding was detected at regions in the genome that not only retained high levels of non-CG methylation, but also were transcriptionally activated in met1.
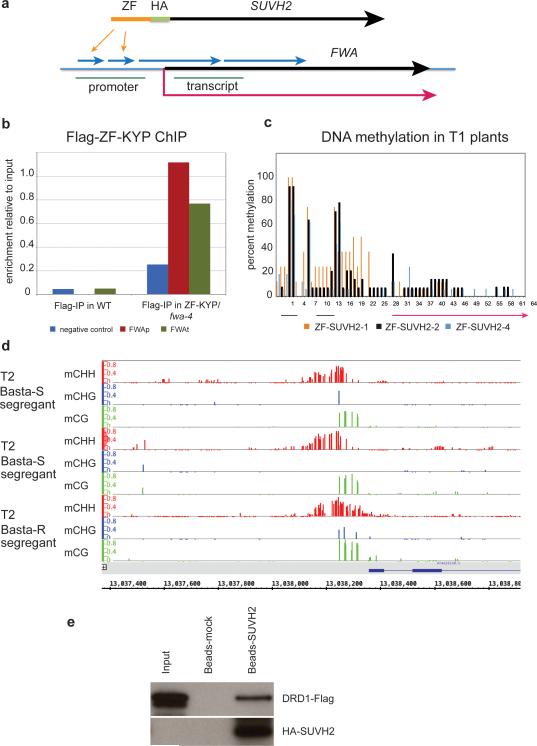
To test directly whether SUVH2/SUVH9 may be sufficient to recruit Pol V, we utilized a zinc finger (ZF) to tether SUVH2 to an unmethylated epiallele of FWA, fwa-4. FWA is normally silenced due to DNA methylation of tandem repeats in its promoter23. FWA epialleles have lost this methylation, leading to ectopic expression of FWA and a heritable late flowering phenotype23,24. Surprisingly, siRNAs are still observed in fwa epialleles but are unable to direct DNA methylation25. We hypothesized that Pol V transcripts may be missing in fwa epialleles and that recruitment of Pol V by SUVH2 might therefore stimulate FWA methylation and silencing. To test this we transformed a ZF-SUVH2 fusion protein construct into the fwa-4 epiallele (Extended Data Fig. 10a). As negative controls, we also transformed an HA-tagged SUVH2 line without the ZF (HA-SUVH2) as well as a construct in which the ZF was fused to KRYPTONITE (KYP/SUVH4) (a SUVH protein not required for RdDM).
Extended Data Figure 10. ZF-SUVH2 construct stably recruits Pol V to FWA through a direct interaction with DRD1.
a. Top: Diagram of SUVH2 with Zn Finger (ZF) inserted immediately before the HA tag. Bottom: Schematic of FWA gene showing the two small and two large repeats (blue arrows), the regions amplified by PCR (promoter and transcript: green lines), the start and direction of transcription (red arrow), and the sites bound by the ZF (indicated by two orange arrows).
b. Flag-ChIP in WT versus ZF-KYP (flag-tagged) showing enrichment at FWA in both the promoter and transcript region (see above).
c. Percent methylation at each C in the FWA repeat region from three individual T1 plants. Percent methylation was determined from 20-25 clones of bisulfite-treated DNA.
d. BS-Seq of FWA from a Basta-resistant line containing the ZF-SUVH2 transgene and two Basta-sensitivie siblings which had lost the ZF-SUVH2 transgene.
e. Pull-down of DRD1-Flag with ZF-SUVH2. Input: DRD1-Flag extract from Arabidopsis; Beads-mock: elution from DRD1-Flag pull-down using HA-magnetic beads pre-bound with Nicotiana benthamiana extract; Beads-ZF-SUVH2: elution from DRD1-Flag pull-down using HA-magnetic beads pre-bound with Nicotiana benthamiana ZF-SUVH2 extract. Top panel: Flag blot; bottom panel: HA blot.
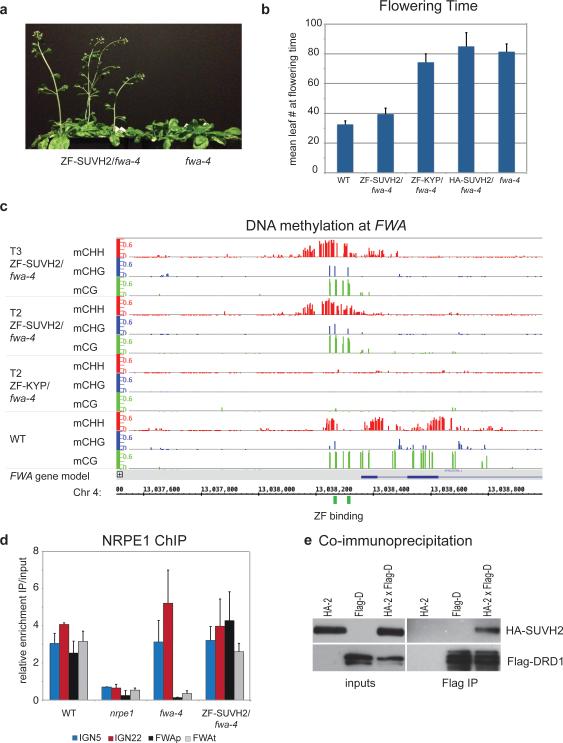
Approximately 75% of the fwa-4 plants transformed with the ZF-SUVH2 (T1 generation) flowered early as compared to the parental fwa-4 line, suggesting silencing of FWA (Fig. 4a). The fwa-4 line transformed with HA-SUVH2 or ZF-KYP flowered at the same time as the fwa-4 parent showing that the effect was specific to the ZF-SUVH2 fusion. Flowering time was measured in the T2 generation confirming these observations (Fig. 4b). The presence of the control ZF-KYP at FWA was shown by ChIP (Extended Data Fig. 10b); however we were unable to detect the ZF-SUVH2 most likely due to its instability or low abundance.
Figure 4. Tethered SUVH2 recruits Pol V through DRD1, resulting in DNA methylation and a late-flowering phenotype.
a. Plants grown side-by-side to illustrate early flowering of ZF-SUVH2 in fwa-4 (T2 plants) compared to fwa-4.
b. Flowering time of Columbia-0 (WT), ZF-SUVH2 in fwa-4, ZF-KYP in fwa-4, HA-SUVH2 in fwa-4, and fwa-4. Flowering time was determined by counting all rosette and cauline leaves up until the terminal flower. The average leaf number and standard deviation of between 20-30 plants was determined. Mean +/− SD.
c. Percent methylation at each cytosine in the FWA repeat region as determined by BS-seq in T2 and T3 ZF-SUVH2/fwa-4 plants compared to T2 ZF-KYP/fwa-4 (unmethylated) and WT (standard methylation pattern). ZF binding sites are shown in green and the FWA gene in blue.
d. NRPE1 ChIP in WT (positive control), nrpe1 mutant (negative control), fwa-4 epiallele, and ZF-SUVH2/fwa-4. qPCR results of two well-characterized NRPE1 binding sites (IGN5 and IGN22) and two regions in FWA (FWAp: promoter; FWAt: transcript) are shown as enrichment of IP/input relative to negative control. Mean +/− SD of two biological replicas.
e. Coimmunoprecipitation of HA-SUVH2 in Arabidopsis using Flag-DRD1. Left panels are inputs from the two parental strains (expressing either HA-SUVH2 (HA-2) or Flag-DRD1 (Flag-D)) and an F2 line expressing both HA-SUVH2 and Flag-DRD1 (HA-2xFlag-d). The right panels show elution from Flag-magnetic beads. Top panels are HA western blots, bottom panels are Flag western blots.
We next utilized bisulfite sequencing to determine if FWA gene silencing was associated with DNA methylation. In wild type, DNA methylation was detected over a large region, while in both fwa-4 and transformants with ZF-KYP or HA-SUVH2, this region was devoid of DNA methylation (see ZF-KYP; Fig. 4c). In three independent ZF-SUVH2 T1 lines, DNA methylation was observed immediately around the Zn finger binding sites in all cytosine sequence contexts (Extended Data Fig. 10c). We analyzed one of the lines (ZF-SUVH2-2) in the T2 and T3 generations using BS-seq and found that methylation extended approximately 150 base pairs in either direction from the binding sites and did not expand significantly between generations (Fig. 4c). FWA methylation and gene silencing were maintained in T2 plants that had segregated away the ZF-SUVH2 transgene (Extended Data Fig. 10d), indicating that targeting by SUVH2 is capable of inducing DNA methylation and gene silencing that can be maintained in the absence of the initial trigger.
To determine whether Pol V was present at FWA, we utilized NRPE1 ChIP. As expected, we found enrichment of Pol V at two known Pol V sites, IGN5 and IGN22, in the wild-type and fwa-4 lines, but not in nrpe1 mutant plants (Fig. 4d). At FWA, we found enrichment of Pol V in the wild type at both the promoter and transcript regions, but not in nrpe1 or fwa-4 (Fig. 4d). However in the ZF-SUVH2-transformed fwa-4 plants we could now see enrichment of Pol V at FWA (Fig. 4d), indicating that SUVH2 is sufficient to recruit Pol V.
To look for a direct interaction between SUVH2/9 and Pol V, we queried several IP-mass spectrometric datasets from purifications of NRPE1-Flag, but failed to find any SUVH2/9 peptides. However, we did identify SUVH2 peptides in two independent mass spectrometric datasets from DRD1 purifications6 (Supplementary Table 2). DRD1 is a component of the DDR complex (also containing DMS3 and RDM1) which interacts with Pol V6 and is required for Pol V occupancy throughout the genome18. The number of SUVH2 peptides observed was lower than those from the DMS3 and RDM1 proteins and also lower than the level of peptides of most Pol V complex components, suggesting that the interaction between SUVH2 and DRD1 is weaker or more transient than the interaction between the DDR components or between DDR and Pol V. To confirm the interaction, we performed co-immunoprecipitation experiments with HA-tagged SUVH2 and Flag-tagged DRD1 in transgenic Arabidopsis plants and were able to detect HA-SUVH2 in a Flag-DRD1 pull-down (Fig. 4e). We were also able to detect Flag-DRD1 in a pull-down using HA-SUVH2 expressed in leaves of Nicotiana benthamiana and purified on HA magnetic beads (Extended Data 10e). These results confirm the IP-mass spectrometry observations and are consistent with a model in which SUVH2 acts indirectly via a transient interaction with DRD1 to recruit Pol V.
Because RdDM is triggered at genomic locations that synthesize both Pol IV-dependent siRNAs and Pol V-dependent non-coding transcripts, it is critical to understand the mechanisms that direct Pol IV and Pol V to chromatin. Furthermore, since RdDM functions to silence transposons that do not necessarily share consensus DNA sequences, a strong prediction is that Pol IV and Pol V will utilize epigenetic information in their targeting. We propose that SUVH2/9 serve as critical components of this targeting system by recruiting Pol V to DNA methylation. We recently showed that recruitment of Pol IV involves SHH1, a protein that binds to histone H3 lysine 9 methylation, a silencing mark also found at RdDM sites26-28. Therefore, the dual marks of DNA methylation and histone H3 lysine 9 methylation appear to act in self-reinforcing loop mechanisms to maintain Pol V and Pol IV at sites of RdDM. These mechanistic findings also suggest a means for selectively targeting DNA methylation by directing both siRNAs and Pol V to overlapping regions of genome, paving the way to chromatin engineering in crop plants.
Similar to other components of RdDM, SUVH2/SUVH9 are required for the establishment of de novo DNA methylation during transformation of an unmethylated FWA gene into Arabidopsis8. An interesting question is how an originally unmethylated DNA is first recognized as an RdDM target if SUVH2/9 DNA methylation-mediated Pol V recruitment cannot initially occur. One possibility is that shortly after the transgene is integrated into the genome, a permissive chromatin environment may allow RNA Pol V (or another polymerase) to produce surveillance transcripts that might serve to initiate RdDM through siRNA-mediated recruitment of DRM2. After this initial round of DNA methylation, SUVH2/9 could then bind methylated DNA and recruit Pol V for subsequent rounds of transcription. In this way, a self-reinforcing loop would be established where preexisting DNA methylation is required for the maintenance of RNA-directed DNA methylation.
Online Methods
Biological materials
All plants used in this study were of the Arabidopsis thaliana Columbia-0 (Col-0) accession, with WT referring to the parental strain. The nrpe1-12 T-DNA (SALK_033852), met1-3, and the suvh2 suvh9 double mutant lines were described previously18,29,8. NRPE1-FLAG transgenic plants30 were crossed to the suvh2 suvh9 double mutant and homozygous F2 plants were identified. The fwa-4 epiallele was isolated from a met1-3 segregating population.
Experimental procedures in Arabidopsis
ChIPs were performed as described previously31 except that ground tissue was cross-linked with formaldehyde in the following buffer: 10 mM HEPES pH 8.0, 1 M Sucrose, 5 mM KCl, 5 mM MgCl2, 5 mM EDTA, and 0.6% Triton X-100. Libraries for NRPE1 ChIP-Seq were generated using the Ovation Ultralow DR Multiplex System (NuGen). Bisulfite sequencing followed by PCR amplification and cloning of FWA fragments was done using EZ DNA Methylation-Gold kit (Zymo Research) as described previously8. BS-Seq libraries were generated as previously reported26 and all libraries were sequenced using the HiSeq 2000 platform following manufacturer instructions (Illumina) at a length of 50 bp.
Co-immunoprecipitation experiments
Nicotiana benthamiana was infiltrated with pLJ322 (ZF-HA-SUVH2 in JP7468) and leaves were collected after 3 days. Leaves that were not infiltrated were used as a negative control. 3 g of tissue was ground in liquid N2 and resuspended in 12 ml of IP buffer (50 mM Tris pH 8.0, 150 mM NaCl, 5 mM MgCl2, 10% glycerol, and 0.1% NP40) with 1 μg/ml pepstatin, 1 mM PMSF, and 1X complete protease inhibitor tab-EDTA (Roche). The extracts were filtered through miracloth and centrifuged for 5 min at 3,000g. 200 μl HA-magnetic beads (MRL) were added and rotated at 4°C for 45 min. After 3 washes with IP buffer, these were then incubated with DRD1-Flag Arabidopsis flower extracts made in the same fashion. Incubation continued for 45 min rotating at 4°C. The beads were washed 3 times with IP buffer and boiled in 60 μl SDS dyes. Western blots were probed with either anti-Flag-HRP or anti-HA-HRP antibodies. The co-IP experiment in Arabidopsis was performed starting with 2 g of flowers from plants expressing HA-SUVH2, Flag-DRD1 or T2 plants expressing both HA-SUVH2 and Flag-DRD1. Extracts were made as described above and 100 ul of Flag-magnetic beads (Sigma) were added and incubated with rotation at 4°C for 45 min. Washes and western blots were performed as described above.
Data analysis
Bisulfite-Seq (BS-Seq), small RNA-Seq, and ChIP-Seq reads were aligned to the TAIR10 version of the Arabidopsis thaliana reference genome using BS-seeker for BS-Seq data and Bowtie for the small RNA-Seq and ChIP-Seq data. For BS-Seq up to 2 mismatches were allowed, whereas for endogenous NRPE1 ChIP and small RNA sequencing only 1 mismatch was allowed. For NRPE1-Flag ChIP, only perfect matches were used and the TAIR8 version of the genome was used to match an earlier report18. For BS-seq and ChIP-seq, only uniquely mapping reads were used. For small RNA-seq, reads were normalized as previously described26.
The small RNA clusters used in Fig. 2a and 2b were previously defined as “pol-iv only” and “shh1/drm2/pol-v” dependent clusters, respectively26.
For NRPE1-Flag ChIP, enrichment over NRPE1 sites18 was normalized to Flag ChIP in Columbia (negative control). For ChIP-Seq using the endogenous Pol V antibody, the read density over Pol V peaks was calculated using the reads per kilobase per million reads (RPKM) metric. For the purposes of Figs. 2e, f, 3a, b, and d, and Extended Data Fig. 4b, Pol V sites were used as previously defined18. For the purposes of Fig. 3e, Pol V enrichment was compared to met1 hypermethylated CHH sites21. To ensure these sites were comparable in genomic character, all sites were expanded to be 1,000 bp in length, defined by extending the borders of sites +/−500 bp from the previously defined midpoint. For met1 hypermethylated CHH sites, we applied a size filter to the previously described sites21 so that we retained only sites larger than 100 bp. This was in an effort to filter out sites of spurious CHH methylation and retain bona fide hypermethylation sites in the met1 mutant.
DMR definitions
Deferentially methylated regions (DMRs) were defined for the suvh2 suvh9, suvh2, and suvh9 libraries by comparing methylation profiles to a suite of wild-type libraries as previously described32.
Zn finger constructs
The FWA promoter region contains two small direct repeats followed by two larger repeats33. We designed a peptide containing six ZFs that would recognize a sequence found in each of two small repeats (CGGAAAGATGTATGGGCT)34,35. The peptide containing 6 Zn fingers was designed as described previously33,34 and cloned into pUC57 with Xho I sites at both ends (Genewiz). This Xho I fragment was excised and inserted into the unique Xho I site located upstream of the HA tag in our pENTR-SUVH2 construct8, which encodes an HA-tagged SUVH2 driven by the endogenous SUVH2 promoter. This ZF-SUVH2 fragment was then recombined into JP7268 and introduced into fwa-4 using agrobacterium-mediated transformation. The ZF was also fused to the N terminus of the KRYPTONITE H3K9 methyltransferase and transformed into fwa-4.
Protein preparation
The N-terminal truncated SUVH9 (residues 134 - 650) was cloned into a pFastBacHT B vector (Invitrogen), which fuses a hexa-histidine tag followed by a TEV cleavage site to the N-terminus of the target gene. The plasmid was transformed into E. coli strain DH10Bac (Invitrogen) to generate the bacmid. Baculovirus was generated by transfecting Sf9 cells with the bacmid following standard Bac-to-Bac protocol (Invitrogen). The harvested virus was subsequently used to infect the suspended Hi5 cell for recombinant protein expression. The recombinant protein was first purified using nickel affinity chromatography column (GE Healthcare). The hexa-histidine tag was cleaved by TEV protease. The target protein was further purified using a Q sepharose column and a Superdex G200 gel filtration column (GE Healthcare). The purified protein was concentrated to 15 mg/ml and stored at −80°C.
Crystallization
Before crystal screening, the SUVH9 protein was mixed with the putative cofactor S-adenosyl-L-homocysteine (SAH) in a molar ratio of 1:3 or with SAH and a histone H3(1-15) peptide in a molar ratio of 1:3:3 at 4°C for 2 hours. Crystallization of the SUVH9 was carried out at 20°C using the hanging drop vapor diffusion method by mixing 1 μl protein sample at a concentration of 11.5 mg/ml and 1 μl reservoir solution and equilibrating against 500 μl reservoir solution. SUVH9 was crystallized in the condition of 0.2 M potassium thiocyanate, 0.1 M Bis-Tris propane, pH 7.5, and 20% PEG 3350 in free form, as well as in the presence of SAH or in the presence of SAH and H3(1-15) peptide. Square-shaped crystals appeared in 2 weeks. All the crystals were dipped into reservoir solution supplemented with 15% glycerol and flash cooled into liquid nitrogen for diffraction data collection. The diffraction data were collected at beamline X29A, National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory (BNL), New York at the zinc peak wavelength. The data were indexed, integrated, and further scaled with the program HKL200036. The statistics of the diffraction data are summarized in Supplementary Table 1.
For investigating the molecular mechanism for recognition of mCHH containing DNA by SUVH9, we have extensively screened SUVH9 in complex with DNAs as a function of length and termini. We used DNA lengths from 8 bp to 20 bp, with a blunt end or with one or two nucleotides overhanging at either 5’ ends or 3’ ends, all of which containing a central mCHH site. Unfortunately, we were not able to get any hits on the SUVH9-DNA complex. The methylated and unmethylated oligos were purchased from Keck Oligonucleotide Synthesis Facility at Yale University and Life Technologies, respectively.
Structure Determination and Refinement
The structure of SUVH9 in the free state was solved using single-wavelength anomalous dispersion method implemented in the program Phenix37. The model building was carried out using the program Coot38 and structural refinement using the program Phenix37. Throughout the refinement, a free R factor was calculated using 5% random chosen reflections. The stereochemistry of the structural model was analyzed using the program Procheck39. The statistics of the refinement and structure models are shown in Supplementary Table 1. Although we have also collected data for the crystals of SUVH9 in the presence of SAH and in the presence of SAH and H3(1-15) peptide, we observed no electron density for either the SAH moiety or the peptide. Our data are indicative that crystals of SUVH9 in presence of SAH and in the presence of SAH and H3(1-15) peptide are indeed the crystal of free form SUVH9. All the molecular graphics were generated with the program Pymol (DeLano Scientific LLC). The sequence alignment was conducted using the program ClustalX240 and illustrated using the ESPript server41.
Supplementary Material
Extended Data Figure 6. Screen shot of Pol V binding in WT versus met1.
An example of reduced Pol V binding in met1 at sites that become hypomethylated.
Acknowledgments
We are grateful to the Dr. Wuxian Shi at National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory (BNL) for support in diffraction data collection. We thank Craig Pikaard for the NRPE1 antibodies and Mahnaz Akhavan for technical assistance. High-throughput sequencing was performed in the UCLA BSCRC BioSequencing Core Facility. This work was supported by the Abby Rockefeller Mauze Trust and the Maloris and STARR foundations to D.J.P., and NIH grant GM60398 to S.E.J. C.J.H. is supported by the Damon Runyon post-doctoral fellowship, S.B. is supported by a post-doctoral fellowship of the Swiss National Science Foundation, S.F. is a Special Fellow of the Leukemia & Lymphoma Society, and X.Z. is supported by Ruth L. Kirschstein National Research Service grant F32GM096483-01. S.E.J. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Author contributions
Protein purification and crystallography were designed and performed by J.D., G.M., and D.J.P., siRNA libraries were made by S.B., whole-genome bisulfite sequencing was performed and analyzed by S.F., L.M.J., C.H., R.K.C., and M.P., Pol V transcription assays were performed by X.Z., ChIP-Seq was performed and analyzed by L.M.J. and C.H., cloning and flowering time experiments were performed by L.M.J., the Zn finger was designed by D.J.S. and S.E.J., pull-down experiments were performed by L.M.J. and S.B., the manuscript was written by L.M.J., J.D., D.J.P., and S.E.J., and S.E.J. participated in all experimental design.
Accession codes
The coordinates and structure factors of SUVH9 have been deposited in the RCSB Protein Data Bank with the accession codes 4NJ5. Sequencing data have been deposited at GEO (GSE52041).
References
- 1.Pelissier T, Wassenegger M. A DNA target of 30 bp is sufficient for RNA-directed DNA methylation. RNA. 2000;6:55–65. doi: 10.1017/s135583820099201x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aufsatz W, Mette MF, van der Winden J, Matzke AJ, Matzke M. RNA-directed DNA methylation in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(Suppl 4):16499–16506. doi: 10.1073/pnas.162371499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Reviews. Genetics. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pontier D, et al. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes & Development. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pikaard CS, Haag JR, Ream T, Wierzbicki AT. Roles of RNA polymerase IV in gene silencing. Trends in Plant Science. 2008;13:390–397. doi: 10.1016/j.tplants.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Law JA, et al. A protein complex required for polymerase V transcripts and RNA-directed DNA methylation in Arabidopsis. Current Biology : CB. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nature Genetics. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson LM, Law JA, Khattar A, Henderson IR, Jacobsen SE. SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genetics. 2008;4 doi: 10.1371/journal.pgen.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhlmann M, Mette MF. Developmentally non-redundant SET domain proteins SUVH2 and SUVH9 are required for transcriptional gene silencing in Arabidopsis thaliana. Plant Molecular Biology. 2012 doi: 10.1007/s11103-012-9934-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hashimoto H, et al. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Avvakumov GV, et al. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 12.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 13.Rajakumara E, et al. A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes & Development. 2011;25:137–152. doi: 10.1101/gad.1980311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, et al. Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. Cell. 2002;111:117–127. doi: 10.1016/s0092-8674(02)00999-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, et al. Structural basis for the product specificity of histone lysine methyltransferases. Molecular Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H, et al. Structural biology of human H3K9 methyltransferases. PloS One. 2010;5:e8570. doi: 10.1371/journal.pone.0008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci U S A. 2008;105:3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong X, et al. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nature Structural & Molecular Biology. 2012;19:870–876. doi: 10.1038/nsmb.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aufsatz W, Mette MF, Matzke AJ, Matzke M. The role of MET1 in RNA-directed de novo and maintenance methylation of CG dinucleotides. Plant Molecular Biology. 2004;54:793–804. doi: 10.1007/s11103-004-0179-1. [DOI] [PubMed] [Google Scholar]
- 21.Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soppe WJ, et al. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Molecular Cell. 2000;6:791–802. doi: 10.1016/s1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- 24.Kakutani T. Genetic characterization of late-flowering traits induced by DNA hypomethylation mutation in Arabidopsis thaliana. The Plant Journal : for Cell and Molecular Biology. 1997;12:1447–1451. doi: 10.1046/j.1365-313x.1997.12061447.x. [DOI] [PubMed] [Google Scholar]
- 25.Chan SW, Zhang X, Bernatavichute YV, Jacobsen SE. Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biology. 2006;4:e363. doi: 10.1371/journal.pbio.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Law JA, et al. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013;498:385–389. doi: 10.1038/nature12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, et al. DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8290–8295. doi: 10.1073/pnas.1300585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Law JA, Vashisht AA, Wohlschlegel JA, Jacobsen SE. SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genetics. 2011;7:e1002195. doi: 10.1371/journal.pgen.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tariq M, et al. Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc Natl Acad Sci U S A. 2003;100:8823–8827. doi: 10.1073/pnas.1432939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Shami M, et al. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes & Development. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PloS One. 2008;3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greenberg MV, et al. Interplay between Active Chromatin Marks and RNA-Directed DNA Methylation in Arabidopsis thaliana. PLoS Genetics. 2013;9:e1003946. doi: 10.1371/journal.pgen.1003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinoshita Y, et al. Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. The Plant Journal : for Cell and Molecular Biology. 2007;49:38–45. doi: 10.1111/j.1365-313X.2006.02936.x. [DOI] [PubMed] [Google Scholar]
- 34.Kolb AF, et al. Site-directed genome modification: nucleic acid and protein modules for targeted integration and gene correction. Trends in Biotechnology. 2005;23:399–406. doi: 10.1016/j.tibtech.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Segal DJ, et al. Evaluation of a modular strategy for the construction of novel polydactyl zinc finger DNA-binding proteins. Biochemistry. 2003;42:2137–2148. doi: 10.1021/bi026806o. [DOI] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica. Section D, Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica. Section D, Biological Crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 40.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 41.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.