Abstract
Glioblastoma contains a hierarchy of stem-like cancer cells, but how this hierarchy is established is unclear. Here, we show that asymmetric Numb localization specifies glioblastoma stem-like cell (GSC) fate in a manner that does not require Notch inhibition. Numb is asymmetrically localized to CD133-hi GSCs. The predominant Numb isoform, Numb4, decreases Notch and promotes a CD133-hi, radial glial-like phenotype. However, upregulation of a novel Numb isoform, Numb4 delta 7 (Numb4d7), increases Notch and AKT activation while nevertheless maintaining CD133-hi fate specification. Numb knockdown increases Notch and promotes growth while favoring a CD133-lo, glial progenitor-like phenotype. We report the novel finding that Numb4 (but not Numb4d7) promotes SCFFbw7 ubiquitin ligase assembly and activation to increase Notch degradation. However, both Numb isoforms decrease epidermal growth factor receptor (EGFR) expression, thereby regulating GSC fate. Small molecule inhibition of EGFR activity phenocopies the effect of Numb on CD133 and Pax6. Clinically, homozygous NUMB deletions and low Numb mRNA expression occur primarily in a subgroup of proneural glioblastomas. Higher Numb expression is found in classical and mesenchymal glioblastomas and correlates with decreased survival. Thus, decreased Numb promotes glioblastoma growth, but the remaining Numb establishes a phenotypically diverse stem-like cell hierarchy that increases tumor aggressiveness and therapeutic resistance.
Introduction
A primary mechanism for generating neural diversity is the asymmetric localization of cell fate determinants. During neural stem cell division, asymmetric segregation of the cell fate determinant, Numb, into daughter cells is thought to promote neuronal differentiation by antagonizing Notch. However, genetic deletion of Numb and its homolog, Numb-like, in double-knockout mice leads to premature neural stem cell depletion, indicating an essential role for Numb in neural stem cell and neural progenitor cell maintenance [1]. Recent evidence suggests that epidermal growth factor receptor (EGFR) activity promotes the transition from neural stem cell to intermediate progenitor cell by upregulating Numb and inhibiting Notch [2]. How Numb promotes the maintenance of neural stem cells while also promoting their differentiation has not been completely resolved.
Numb antagonizes Notch signaling through multiple mechanisms. For example, Numb binds Notch and increases its ubiquitination and degradation through interactions with amyloid precursor protein (APP) or the HECT (homologous to E6-AP carboxy terminus) domain ubiquitin ligase, Itch [3, 4]. Numb also increases endocytic trafficking of the Notch receptor [5]. In addition, Numb may regulate differentiation and proliferation through Notch-independent mechanisms such as stabilizing p53 [6] or promoting the Itch-dependent degradation of the Sonic hedgehog pathway transcription factor, Gli1 [7].
In drosophila and mice, disruption of asymmetric cell division leads to neural overgrowth [8, 9]. In humans, Numb restrains the growth of breast cancer cells by inhibiting Notch and stabilizing p53 [6]. In addition, downregulation of Numb by musashi 2 promotes the development of blast crisis in leukemia [10]. These observations have led to the suggestion that Numb is a tumor suppressor.
Glioblastoma contains a related hierarchy of CD133+/hi and CD133−/lo cancer cells resembling neural stem cells [11, 12]. Like neural stem cells, many glioblastoma stem-like cells (GSCs) are self-renewing. However, CD133+/hi GSCs display increased tumorigenicity and increased resistance to therapy when compared with CD133−/lo GSCs from the same tumors [13]. Accordingly, CD133 expression correlates with increased tumor aggressiveness and decreased survival in glioblastoma and in other cancers [14–16]. Single GSCs can give rise to a mixed population of CD133+/hi and CD133−/lo cells, suggesting that they undergo asymmetric division. Indeed, a recent study indicated that GSCs undergo both asymmetric and symmetric divisions, suggesting a potential role for Numb in regulating glioblastoma growth [17]. However, another recent study suggests that Numb has no effect on glioblastoma cell growth or differentiation [18]. Furthermore, studies indicate that Notch signaling is essential for GSC maintenance and tumorigenicity [19, 20]. Thus, it is unclear whether Numb suppresses glioblastoma growth by antagonizing Notch signaling, whether it establishes the differentiation hierarchy of GSCs as it does for normal neural stem/progenitor cells, or whether it plays no role in glioblastoma.
Here, we show that the predominant Numb isoforms in glioblastoma, Numb4 and a novel isoform (Numb4 delta 7 [Numb4d7]), are asymmetrically localized during GSC divisions and promote a self-renewing CD133+/hi phenotype. Low Numb favors the production of more rapidly proliferating CD133−/lo GSCs that retain self-renewal capacity. Numb4 and Numb4d7 exert similar effects on GSC differentiation, but they have opposing effects on GSC proliferation, Notch signaling and AKT activation. Mechanistically, these two Numb isoforms differentially interact with Skp1-Cullin-F-box (SCF) ubiquitin ligases to exert opposite effects on Notch levels and tumor growth. Both Numb isoforms decrease EGFR activity, and this contributes to maintenance of the CD133-hi stem-like cell phenotype.
Results
Numb Is Alternatively Spliced in Glioblastoma
Immunostaining of human glioblastoma specimens revealed an altered pattern of Numb immunoreactivity in approximately 80% of the specimens (n = 13) when compared with nontumor brain (Fig. 1A). Whereas Numb immunoreactivity in nontumor brain was primarily present in neurons, Numb staining was more intense and was localized primarily to tumor cells in some glioblastomas. In contrast, approximately 20% of glioblastomas displayed very low levels of Numb immunoreactivity. Immunostaining of Grade II and Grade III oligodendrogliomas and astrocytomas also revealed increased Numb immunoreactivity in tumor cells in a subgroup of specimens when compared with nontumor brain (Supporting Information Fig. S1).
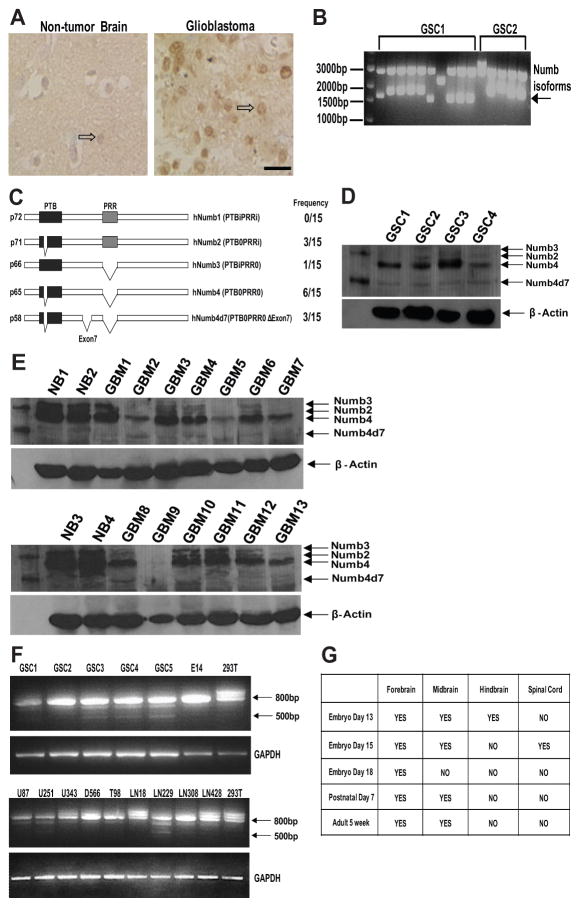
Figure 1. Numb isoform expression in glioblastoma.
(A): Numb immunostaining of paraffin-embedded human nontumor brain or glioblastoma specimens. Open arrows point to a Numb-immunoreactive neuron in the nontumor brain specimen and a Numb-immunoreactive tumor cell in the glioblastoma specimen. (B): Reverse transcription (RT)-PCR of Numb isoforms expressed in two primary human GSC lines. (C): Domain structure and frequency of occurrence of Numb isoforms cloned from four primary human GSC lines. (D): Western blot analysis of Numb isoforms in four primary GSCs. (E): Western blot analysis of Numb isoforms in GBM and human NB specimens. (F): RT-PCR analysis of Numb isoform expression in five primary human GSC cell lines, nine established glioblastoma cell lines, 293T cells, and E14 cells. Five hundred base pair fragment represents Numb4d7 mRNA. (G): RT-PCR analysis of presence or absence of Numb4d7 mRNA during mouse neural development. Abbreviations: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GBM, glioblastoma; GSCs, glioblastoma stem-like cells; NB, nontumor brain; Numb4d7, Numb4 delta 7; PRR, proline-rich region; PTB, phosphotyrosine-binding; PCR, polymerase chain reaction.
Alternative splicing yields several isoforms of Numb that are developmentally regulated and that differentially affect proliferation and differentiation [21]. Numb2 and Numb4 primarily regulate differentiation, while Numb1 and Numb3 promote the proliferation of neural cells. To determine which isoforms of Numb are expressed in glioblastoma, we used reverse transcription (RT)-PCR (polymerase chain reaction) to clone and sequence several Numb transcripts from two primary GSC lines (GSC1 and GSC2; Fig. 1B). To determine their frequency of occurrence, we randomly selected 15 clones for direct sequencing (Fig. 1C). The most frequent Numb isoform among the 15 clones selected for sequencing was Numb4 (40%), followed by Numb2 (20%) and a novel form of Numb4 that lacks exon 7 (20%), which we have designated as Numb4d7 (Supporting Information Fig. S2). One clone encoding a novel Numb4 isoform that lacked exons 7 and 8 was also detected. Numb3 was encoded by one of the 15 sequenced clones, and Numb1 was not detected. Western blot using protein derived from GSC1, GSC2, and two additional primary human GSC cell lines (GSC3 and GSC4) confirmed the presence of Numb4d7 protein expression in all four primary GSC lines (Fig. 1D).
We overexpressed the cloned Numb variants to facilitate identification of the Numb protein isoforms by Western blot (Supporting Information Fig. S2) and subsequently examined Numb expression in protein lysates derived from 13 surgical human glioblastoma and four nontumor brain specimens (Fig. 1E). Numb4 and Numb4d7 were the most abundant Numb isoforms detected in primary GSCs isolated from surgical glioblastoma specimens and maintained as tumorspheres [12]. Whereas expression of Numb4 and Numb2 protein was decreased in glioblastoma when compared with nontumor brain, Numb4d7 protein expression was increased (Fig. 1E). RT-PCR confirmed the presence of Numb4d7 mRNA in all five primary human GSC lines examined (Fig. 1F). In contrast to primary GSCs, however, only three of nine established human glioblastoma cell lines expressed Numb4d7. Numb4d7 mRNA was not detected in mouse E14 neural stem cells or in HEK293T cells.
We examined Numb4d7 expression during mouse nervous system development using PCR primers designed to detect mouse Numb4d7, followed by direct sequencing of the PCR products. As a source of mRNA, we used a commercially available mouse cDNA panel (Rapid-Scan, OriGene) containing mRNAs isolated from various regions of the mouse nervous system throughout development. Low levels of Numb4d7 mRNA were detected from E13 to adulthood during mouse nervous system development. Interestingly, Numb4d7 mRNA expression was present in a rostrocaudal gradient in the mouse nervous system. Numb4d7 expression was most frequently detected in the forebrain and midbrain, with low or no expression detected in more caudal areas (Fig. 1G).
Numb Is Asymmetrically Localized in Glioblastoma and Specifies Stem-Like Cell Fate
A recent in vitro study indicated that primary human GSCs can divide either symmetrically or asymmetrically [17]. We have examined this phenomenon in vivo, using paraffin-embedded glioblastoma specimens stained for Numb immunoreactivity. Although most cells displayed a symmetric pattern of Numb immunoreactivity, approximately 15% of dividing GSCs displayed asymmetric Numb localization in vivo (Fig. 2A). Costaining for the neural stem cell marker, CD133 [22], revealed frequent colocalization of CD133 and Numb (Fig. 2A). Western blot analysis of protein lysates obtained from nine surgical glioblastoma specimens revealed a strong correlation between high CD133 expression and high Numb expression (Fig. 2B).
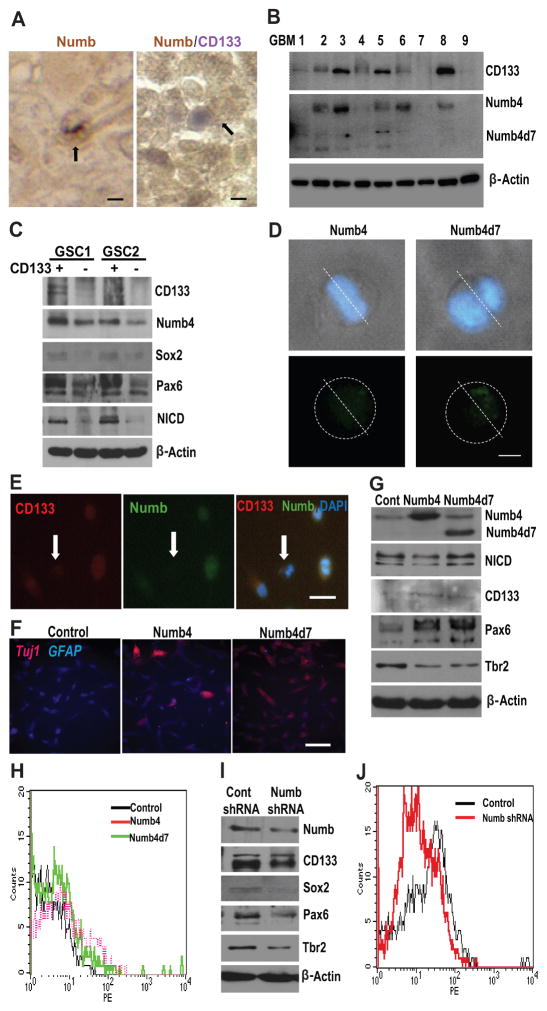
Figure 2. Numb is asymmetrically localized and specifies cell fate in glioblastoma.
(A): (left panel) Paraffin section of glioblastoma specimen stained for Numb immunoreactivity. Arrow indicates asymmetric distribution of Numb immunoreactivity in a single GSC during mitosis. (right panel) Colocalization of Numb and CD133 immunoreactivity in paraffin section of glioblastoma. Arrow indicates asymmetric colocalization of Numb and CD133 immunoreactivity in a glioblastoma cell pair. Scale is approximately 10 μm. (B): Correlation between Numb and CD133 protein expression in nine surgical glioblastoma specimens. (C): Correlation between Numb, CD133, Sox2, Pax6, and cleaved Notch (NICD) in two primary human GSC lines sorted into CD133-hi (CD133+) and CD133-lo (CD133−) cell populations. (D): Light microscopy images illustrating asymmetric localization of Numb4-GFP or Numb4d7-GFP in dividing primary human GSCs. Nuclei are counterstained with DAPI. Scale is approximately 5 μm. (E): Colocalization of CD133 and Numb in primary cultured GSCs. Arrow identifies CD133−/lo/Numb-lo GSC. Scale is approximately 20 μm. (F): Effect of Numb4 or Numb4d7 overexpression on Tuj1 and GFAP immunoreactivity after serum-induced differentiation of primary GSCs. Scale is approximately 50 μm. (G): Effect of Numb4 and Numb4d7 overexpression on NICD, CD133, Pax6, and Tbr2 expression in GSCs. (H): FACS analysis of CD133-expression on the surface of cultured human GSCs after overexpression of Numb4, Numb4d7, or a control vector. (I): Effect of Numb knockdown on CD133, Sox2, Pax6, and Tbr2 expression in primary human GSCs. (J): Fluorescence-activated cell sorting analysis of CD133 surface expression in GSCs after overexpression of Numb shRNA or a control vector. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; GFAP, glial fibrillary acidic protein; GSCs, glioblastoma stem-like cells; NICD, Notch intracellular domain; Numb4d7, Numb4 delta 7.
Alternative splicing yields several isoforms of Numb that are developmentally regulated and that differentially affect proliferation and differentiation [21]. Numb2 and Numb4 primarily regulate differentiation, while Numb1 and Numb3 promote the proliferation of neural cells. To determine which isoforms of Numb are expressed in glioblastoma, we used reverse transcription (RT)-PCR (polymerase chain reaction) to clone and sequence several Numb transcripts from two primary GSC lines (GSC1 and GSC2; Fig. 1B). To determine their frequency of occurrence, we randomly selected 15 clones for direct sequencing (Fig. 1C). The most frequent Numb isoform among the 15 clones selected for sequencing was Numb4 (40%), followed by Numb2 (20%) and a novel form of Numb4 that lacks exon 7 (20%), which we have designated as Numb4d7 (Supporting Information Fig. S2). One clone encoding a novel Numb4 isoform that lacked exons 7 and 8 was also detected. Numb3 was encoded by one of the 15 sequenced clones, and Numb1 was not detected. Western blot using protein derived from GSC1, GSC2, and two additional primary human GSC cell lines (GSC3 and GSC4) confirmed the presence of Numb4d7 protein expression in all four primary GSC lines (Fig. 1D).
We overexpressed the cloned Numb variants to facilitate identification of the Numb protein isoforms by Western blot (Supporting Information Fig. S2) and subsequently examined Numb expression in protein lysates derived from 13 surgical human glioblastoma and four nontumor brain specimens (Fig. 1E). Numb4 and Numb4d7 were the most abundant Numb isoforms detected in primary GSCs isolated from surgical glioblastoma specimens and maintained as tumorspheres [12]. Whereas expression of Numb4 and Numb2 protein was decreased in glioblastoma when compared with nontumor brain, Numb4d7 protein expression was increased (Fig. 1E). RT-PCR confirmed the presence of Numb4d7 mRNA in all five primary human GSC lines examined (Fig. 1F). In contrast to primary GSCs, however, only three of nine established human glioblastoma cell lines expressed Numb4d7. Numb4d7 mRNA was not detected in mouse E14 neural stem cells or in HEK293T cells.
We examined Numb4d7 expression during mouse nervous system development using PCR primers designed to detect mouse Numb4d7, followed by direct sequencing of the PCR products. As a source of mRNA, we used a commercially available mouse cDNA panel (Rapid-Scan, OriGene) containing mRNAs isolated from various regions of the mouse nervous system throughout development. Low levels of Numb4d7 mRNA were detected from E13 to adulthood during mouse nervous system development. Interestingly, Numb4d7 mRNA expression was present in a rostrocaudal gradient in the mouse nervous system. Numb4d7 expression was most frequently detected in the forebrain and midbrain, with low or no expression detected in more caudal areas (Fig. 1G).
To investigate the relationship between CD133 and Numb expression further, we used immunoaffinity columns to separate cultured primary human GSCs into CD133+/hi and CD133−/lo cell populations. Western blots using protein lysates derived from these cell populations revealed increased Numb expression in the CD133+/hi cell population when compared with CD133−/lo cells from the same tumor (Fig. 2C). In addition, increased expression of the radial glial stem cell markers Sox2 and Pax6 [23], as well as increased levels of cleaved Notch (Notch intracellular domain [NICD]), were associated with high Numb levels in CD133+/hi GSCs.
To determine whether asymmetric localization of Numb contributes to the differential expression of CD133 in GSCs, we first transfected primary human GSCs using expression vectors for Numb4-GFP or Numb4d7-GFP fusion proteins. Both Numb4-GFP and Numb4d7-GFP were asymmetrically localized in approximately 32% (n = 19) of dividing GSCs under these conditions (Fig. 2D). Immunocytochemistry using dissociated primary human GSCs confirmed increased Numb immunoreactivity in CD133+/hi cells when compared with CD133−/lo cells (Fig. 2E).
When primary human GSCs (GSC1) overexpressing Numb4 or Numb4d7 were placed into serum-containing medium to induce differentiation, we observed increased expression of the neuronal differentiation marker, TuJ1/BetaIII tubulin, and decreased expression of the astrocyte marker, glial fibrillary acidic protein (GFAP) (Fig. 2F). Furthermore, overexpression of Numb4 and Numb4d7 in GSCs increased expression of the radial glial markers CD133 and Pax6 and decreased expression of Tbr2, a marker of neurogenic intermediate progenitor cells [24] (Fig. 2G). Fluorescence-activated cell sorting (FACS) analysis indicated that overexpression of Numb4 and Numb4d7 increased the fraction of primary human GSCs that expressed high surface levels of CD133 (Fig. 2H). Taken together, these data indicate that Numb4 and Numb4d7 are asymmetrically localized during a fraction of GSC divisions and promote adoption of a CD133+/hi radial glial-like phenotype.
To examine the effect of Numb overexpression on GSC self-renewal, we cultured primary glioblastoma cells overexpressing green fluorescent protein (GFP), Numb4-GFP, or Numb4d7-GFP as tumorspheres in serum-free medium for more than five passages, and then used FACS to sort them as single cells into 96-well plates. All three cell groups continued to form tumorspheres with similar frequency, indicating that Numb overexpression does not prevent GSC self-renewal. Interestingly, tumorspheres formed by cells overexpressing Numb4-GFP tended to be smaller than those formed by GFP-expressing control cells, while spheres formed by Numb4d7-GFP-expressing GSCs were larger and less tightly organized than controls (Supporting Information Fig. S3).
We next transduced primary GSCs with a lentivirus containing a Numb shRNA or a scrambled control shRNA. This approach afforded a 50% knockdown of endogenous Numb protein levels. Numb knockdown decreased CD133, Sox2, Pax6, and Tbr2 expression (Fig. 2I). Collectively, this pattern of marker expression suggested that decreased Numb expression promotes a gliogenic progenitor-like phenotype in human GSCs. Importantly, FACS analysis indicated that Numb knockdown decreased the fraction of GSCs with strong surface expression of CD133 (Fig. 2J).
After several passages, single GSCs expressing Numb shRNA or control shRNA were sorted by FACS into 96-well plates and maintained in serum-free medium. Numb knockdown did not significantly alter the frequency of tumorsphere formation when compared with control GSCs (Supporting Information Fig. S3; p = .074, t test). Taken together, these data indicate that asymmetric localization of Numb4 and Numb4d7 establishes a related hierarchy of radial glial-like and gliogenic intermediate progenitor-like GSCs that display self-renewal capacity.
Endogenous Numb Constrains Growth in Human GSCs
Notch is essential for the maintenance of neural stem cells and CD133+/hi GSCs [19]. However, Numb reportedly antagonizes Notch signaling [3], and we observed that Numb is preferentially expressed in CD133+/hi GSCs. To investigate this apparent paradox, we examined the effect of overexpressing Numb4 and Numb4d7 on GSC growth. Numb4 overexpression caused a small but significant decrease in the proliferation of primary GSCs in vitro (Fig. 3A; p < .05, t test; Supporting Information Fig. S4). In contrast, Numb4d7 robustly increased the proliferation of these cells (Fig. 3A; p < .04, t test; Supporting Information Fig. S4). MTT cell growth assays revealed increased growth of Numb4d7-expressing GSCs when compared with control cells (Fig. 3B, p = .004, t test). GSCs overexpressing Numb4 initially grew at a slower pace but grew more quickly at later time points (Fig. 3B, p = .009, t test). When human U87 glioblastoma cells overexpressing Numb4, Numb4d7, or a control vector were transplanted subcutaneously into nude mice, cells overexpressing Numb4d7 formed significantly larger tumors (Fig. 3C, p = .034, t test), while Numb4 overexpression had no effect on subcutaneous tumor growth. When primary human GSCs overexpressing Numb4, Numb4d7, or a control vector were transplanted into the brains of nude mice, a nonsignificant trend toward shorter survival was observed for mice that received Numb4d7-expressing cells when compared with control mice (Supporting Information Fig. S4; p = .16, log-rank test). Intracranial transplantation of GSCs overexpressing Numb4 did not alter survival (p = .90, log-rank test). Thus, overexpression of Numb in primary human GSCs (which express significant amounts of endogenous Numb at baseline) did not inhibit tumor growth, in agreement with previous reports [18].
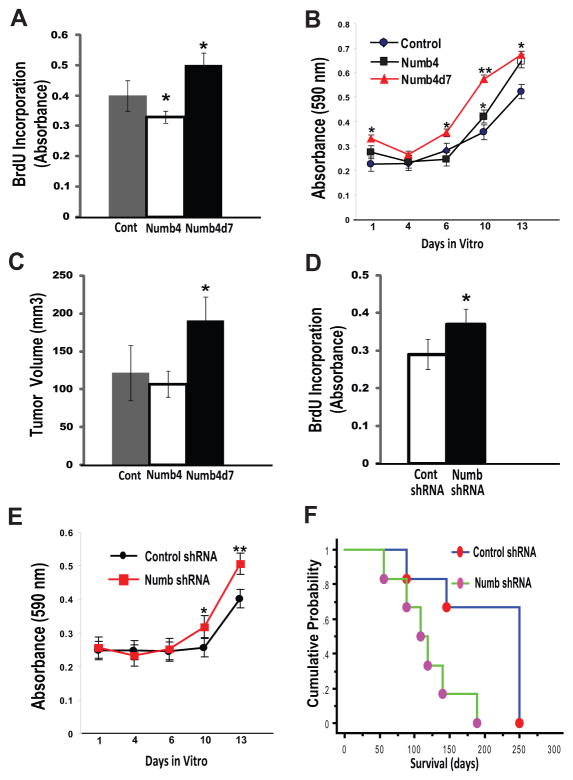
Figure 3. Numb isoforms differentially regulate glioblastoma growth.
(A): BrdU incorporation into DNA of glioblastoma stem-like cells (GSCs) overexpressing Numb4, Numb4d7, or a control vector. Data shown are mean ± SEM. *, p < .05, t test. (B): MTT cell growth assay for GSCs overexpressing Numb4, Numb4d7, or a control vector. *, p < .05; **, p < .01 compared to control, t test. (C): Tumor growth after subcutaneous transplantation of 1 × 106 human U87 glioblastoma cells in nude mice. Data shown are mean ± SEM. *, p < .04 compared to Numb4, t test. (D): BrdU incorporation into GSCs after Numb knockdown using shRNA. Data shown are mean ± SEM. *, p < .05, t test. (E): MTT cell growth assay for GSCs overexpressing Numb shRNA or a control vector. Data shown are mean ± SEM. *, p < .05; **, p < .01 compared to control, t test. (F): Mouse survival after intracranial injection of 5 × 105 primary human GSCs overexpressing Numb shRNA or a control vector. p = .016, log-rank test. Abbreviations: BrdU, Bromodeoxyuridine; Numb4d7, Numb4 delta 7.
To more clearly determine the role of Numb in GSC growth, we examined the effect of knockdown of endogenous Numb on GSC growth. Numb knockdown using shRNA significantly increased primary human GSC proliferation (Fig. 3D; p < .05, t test) and increased GSC growth in vitro (Fig. 3E, p = .036, t test). Moreover, intracranial transplantation of human GSCs overexpressing a Numb shRNA vector significantly decreased survival in nude mice (Fig. 3F; p = .016, log-rank test). Histochemical analysis of the brains of mice transplanted with GSCs expressing Numb shRNA revealed increased tumor cell density and an increased fraction of cells immunoreactive for the proliferation marker, Ki67 (Supporting Information Fig. S4). Taken together, these data demonstrate a role for Numb in regulating the growth and differentiation of primary human GSCs in vitro and in vivo.
Numb4d7 Increases Notch Signaling
Consistent with previous reports [3], we observed that Numb4 overexpression decreased levels of cleaved Notch (NICD) in GSCs and established glioblastoma cell lines and HEK293T cells. Surprisingly, however, Numb4d7 increased NICD levels in these same cells (Fig. 4A). Numb4 also decreased activity of the Notch pathway transcription factor, Hes1 (Fig. 4B; p = .0054, t test), while Numb4d7 increased it (p = .0019, t test).
Figure 4. Numb isoforms differentially regulate Notch and AKT.
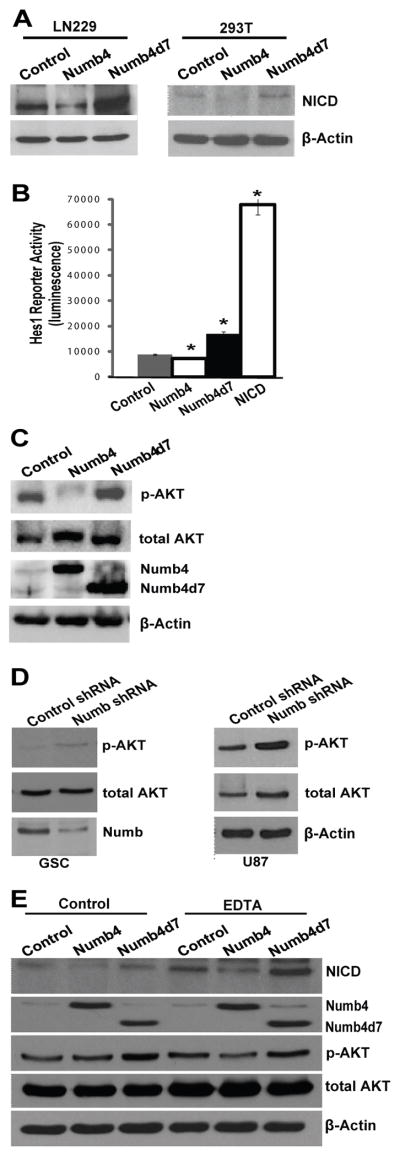
(A): Effect of Numb4 or Numb4d7 overexpression on cleaved Notch (NICD) expression in HEK293T cells and in human LN229 glioblastoma cells. (B): Hes1 reporter assay in HEK293T cells after overexpression of Numb4, Numb4d7, NICD, or a control vector. Data shown are mean ± SEM. *, p < .05, t test. (C): Effect of overexpression of Numb4, Numb4d7, or a control vector on AKT activation in primary human glioblastoma stem-like cells (GSCs). (D): Effect of Numb shRNA on AKT activation in primary human GSCs or in human U87 glioblastoma cells. (E): Effect of overexpression of Numb4, Numb4d7, or a control vector on cleaved Notch (NICD) and AKT activation in primary GSCs. EDTA was used to trigger Notch cleavage. Abbreviations: NICD, Notch intracellular domain; Numb4d7, Numb4 delta 7.
In immune cells and in GSCs, Notch activates AKT [19, 25]. In primary GSCs, we observed that Numb4 decreased AKT activation, while Numb4d7 increased it (Fig. 4C). Furthermore, knockdown of endogenous Numb using shRNA increased AKT activation in primary GSCs and in human U87 glioblastoma cells (Fig. 4D). The effect of Numb4 and Numb4d7 on Notch and AKT was observed after enforced Notch activation using EDTA, indicating that this phenomenon does not require ligand-mediated Notch cleavage (Fig. 4E). This observation is consistent with reports that Numb promotes NICD degradation downstream of Notch cleavage [3]. Taken together, these findings suggest that the differential effects of Numb4 and Numb4d7 on GSC proliferation derive from differences in their effects on Notch and AKT signaling, and they suggest the existence of a Numb-dependent mechanism governing GSC differentiation that does not require Notch inhibition.
Numb Regulates SCF Ubiquitin Ligase Assembly and Activity
Numb4d7 differs from Numb4 in that it lacks exon 7, although both isoforms retain the PTB and PRR domains. The PTB domain is critical for Numb-mediated Notch degradation [3]. We observed that Numb4-GFP and Numb4d7-GFP bound Notch equally in immunoprecipitation experiments (Fig. 5A). However, we also observed that Numb4-GFP was localized to the cytoplasm, while Numb4d7 was located in both the cytoplasm and the nucleus of primary human GSCs and 293T cells (Supporting Information Fig. S5). This observation suggested that Numb4 and Numb4d7 differ with respect to their interactions with other proteins. To identify such differentially interacting proteins, we overexpressed Numb4-GFP or Numb4d7-GFP in HEK293T cells, immunoprecipitated the overexpressed proteins, and separated them by Coomassie Blue gel electrophoresis. Bands that differed between Numb4 and Numb4d7 were then excised and analyzed by mass spectrometry. One of the proteins interacting more strongly with Numb4 than Numb4d7 was cullin-associated and neddylation-dissociated 1 (CAND1). CAND1 binds Cullin1 (CUL1) and prevents its binding to substrate adaptors and its neddylation, a modification necessary for full activation of SCF ubiquitination complexes [26]. The F-box protein, Fbw7, binds Notch and promotes its ubiquitination and degradation, indicating that SCF complexes mediate Notch degradation [27]. Numb has not previously been reported to regulate SCF complex activity or to interact with CAND1. However, the differential interaction of Numb4 and Numb4d7 with CAND1, combined with the known role of Fbw7 in regulating Notch levels, led us to select CAND1 for further study.
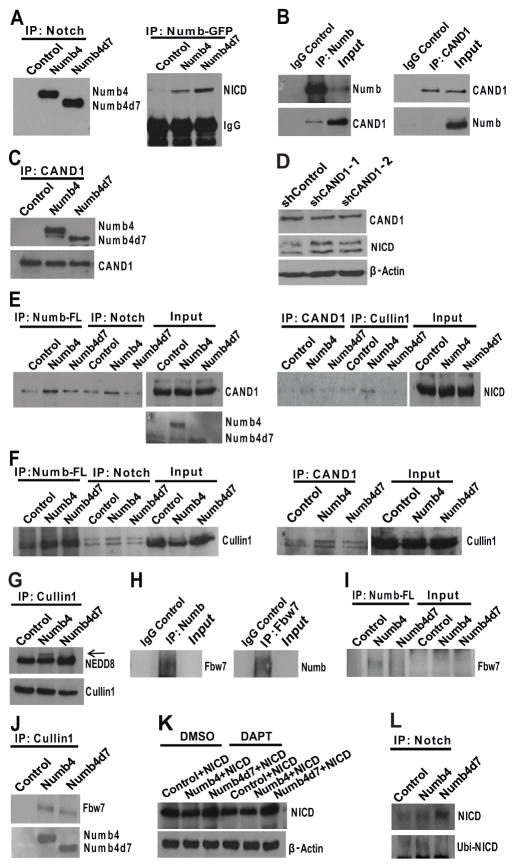
Figure 5. Numb isoforms differentially interact with CAND1 and promote Skp1-Cullin-F-box assembly.
(A): (left panel) Immunoprecipitation of Notch followed by Western blot for Numb using lysates from primary human glioblastoma stem-like cells (GSCs) overexpressing Numb4-GFP, Numb4d7-GFP, or a control vector. Control cells were transfected with a GFP control vector. (right panel) Immunoprecipitation of Numb-GFP using a GFP antibody, followed by Western blot for NICD using lysates from GSCs overexpressing Numb4-GFP, Numb4d7-GFP, or a GFP control vector. (B): (left panel) Immunoprecipitation of endogenous Numb followed by Western blot for Numb and CAND1 using lysates from primary human GSCs. An isotype control antibody was used as a control. (C): Immunoprecipitation of endogenous CAND1 followed by Western blot for CAND1 and Numb using lysates from primary human GSCs. (D): Effect of CAND1 shRNA or a scrambled control shRNA on NICD expression in HEK293T cells. (E): (left panel) Immunoprecipitation of Numb-FLAG or Notch followed by Western blot for CAND1 using lysates from GSCs overexpressing Numb4-FLAG, Numb4d7-FLAG, or an empty control vector. (right panel) Immunoprecipitation of CAND1 and Cullin followed by Western blot for NICD using lysates from GSCs overexpressing Numb4, Numb4d7, or an empty control vector. (F): Immunoprecipitation of CAND1, Numb-FLAG, and Notch followed by Western blot for Cullin1 using lysates from GSCs overexpressing Numb4-FLAG, Numb4d7-FLAG, or a control vector. (G): Immunoprecipitation of Cullin1 followed by Western blot for Cullin1 and NEDD8 using lysates from GSCs overexpressing Numb4, Numb4d7, or a control vector. (H): (left panel) Immunoprecipitation of endogenous Numb followed by Western blot for Fbw7. (right panel) Immunoprecipitation of endogenous Fbw7 followed by Western blot for Numb. (I): Immunoprecipitation of Numb-FLAG followed by Western blot for Fbw7 using lysates from GSCs overexpressing Numb4-FLAG, Numb4d7-FLAG, or an empty control vector. (J): Immunoprecipitation of Cullin1 followed by Western blot for Fbw7 or Numb isoforms using lysates from GSCs overexpressing Numb4-FLAG, Numb4d7-FLAG, or an empty control vector. (K): Western blot for NICD expression using lysates from HEK293T cells overexpressing NICD in combination with Numb4-GFP, Numb4d7-GFP, or a control vector. Cells were treated with DMSO vehicle or DAPT (5 μM) to inhibit endogenous Notch cleavage. (L): Immunoprecipitation of Notch, followed by Western blot for NICD and ubiquitin using lysates from primary human GSCs overexpressing Numb4-GFP, Numb4d7-GFP, or a control vector. Abbreviations: CAND1, cullin-associated and neddylation-dissociated 1; DAPT, N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester; DMSO, dimethylsulfoxide; GFP, green fluorescent protein; NICD, Notch intracellular domain; Numb4d7, Numb4 delta 7.
Immunoprecipitation of endogenous Numb resulted in coimmunoprecipitation of endogenous CAND1 (Fig. 5B). However, reverse immunoprecipitation of endogenous CAND1 did not result in coimmunoprecipitation of endogenous Numb. This finding suggests that only a small fraction of cellular CAND1 interacts with Numb under physiological conditions or, alternatively, that the CAND1-Numb interaction requires the participation of other proteins. When Numb4-GFP or Numb4d7-GFP was overexpressed, immunoprecipitation of CAND1 pulled down Numb4 more effectively than Numb4d7 (Fig. 5C). Although CAND1 binds CUL1 and prevents SCF complex assembly, it is nevertheless required for full SCF activity [28]. Indeed, we observed that knockdown of CAND1 expression in HEK293T cells increased NICD levels, demonstrating a role for CAND1 in Notch degradation (Fig. 5D).
The increased association of CAND1 with Numb4 versus Numb4d7 was confirmed in reverse immunoprecipitation experiments, in which immunoprecipitation of Numb4-FLAG pulled down CAND1 more effectively than immunoprecipitation of Numb4d7-FLAG (Fig. 5E). Importantly, Numb4-FLAG (and to a lesser extent Numb4d7-FLAG) increased the association between CAND1 and Notch (Fig. 5E). To examine whether Numb, Notch, and CAND1 interact as part of the SCF complex, we examined whether Numb associates with CUL1. CUL1 coimmunoprecipitated with both Numb4-FLAG and Numb4d7-FLAG (Fig. 5F). However, only Numb4-FLAG (but not Numb4d7-FLAG) increased the association of NICD with CUL1 or the association of CAND1 with CUL1.
Inspection of Western blots from immunoprecipitation experiments (Fig. 5F) suggested that Numb4 increased the amount of neddylated CUL1 associated with NICD and CAND1. To investigate this phenomenon further, we immunoprecipitated CUL1 and performed Western blots for CUL1 and NEDD8. We observed an increase in neddylated CUL1 in the presence of Numb4 when compared with Numb4d7 (Fig. 5G). Because neddylation promotes SCF activity [29], these findings suggest that Numb4 (but not Numb4d7) promotes assembly and activation of the SCF complex.
The F-box protein, Fbw7, is a substrate adaptor for Notch and promotes its ubiquitination as part of the SCFFbw7 complex [27]. We observed that endogenous Fbw7 coimmunoprecipitates with endogenous Numb and, conversely, endogenous Numb coimmunoprecipitates with endogenous Fbw7 (Fig. 5H). Importantly, Fbw7 coimmunoprecipitated with Numb4-FLAG to a greater extent than Numb4d7-FLAG (Fig. 5I). In addition, Numb4-FLAG (and to a lesser extent Numb4d7-FLAG) increased the association between Fbw7 and CUL1 (Fig. 5J). These data suggest a model in which Numb4 promotes assembly of the SCFFbw7 complex to increase Notch ubiquitination and degradation more effectively than Numb4d7.
We sought to test this model further. When NICD was overexpressed by transient transfection in the presence of a gamma secretase inhibitor (to prevent endogenous Notch cleavage), coexpression of Numb4d7-GFP increased NICD levels, suggesting that Numb4d7 activates Notch signaling downstream of Notch cleavage (Fig. 5K). We next overexpressed Numb4-GFP or Numb4d7-GFP in GSCs, immunoprecipitated Notch, and performed Western blots for NICD and ubiquitin. Numb4 (but not Numb4d7) increased NICD ubiquitination (Fig. 5L).
Based upon these findings, we propose a model in which Numb4 interacts with NICD, Fbw7, and CUL1 to promote SCF assembly and activation. Numb4d7 also interacts with NICD and CUL1 but is less effective in binding Fbw7 and promoting SCF assembly. In this model, Numb4 (but not Numb4d7) also recruits CAND1 to augment SCFFbw7-CAND1 cycling, thereby optimizing Notch degradation (Supporting Information Fig. S6).
Numb Decreases EGFR Expression to Regulate GSC Differentiation
Our finding that Numb4 and Numb4d7 have similar effects on GSC differentiation but opposite effects on Notch signaling suggests the existence of a Notch-independent mechanism that regulates GSC differentiation. Numb has been reported to regulate the sonic hedgehog pathway by downregulating the Gli1 transcription factor [7]. However, we found that overexpression of Numb4 or Numb4d7 had no effect on Gli1 expression levels in primary human GSCs (Supporting Information Fig. S7), in agreement with a previous report [18].
Numb also binds Hdm2 and stabilizes p53 [6]. We confirmed binding of endogenous Numb and Hdm2 and found that Hdm2 coimmunoprecipitated with both Numb4-FLAG and Numb4d7-FLAG (Supporting Information Fig. S7). siRNA-mediated knockdown of endogenous Numb decreased p53 levels after DNA damage caused by the topoisomerase I inhibitor, camptothecin (50 μM), and overexpression of either Numb4 or Numb4d7 increased p53 levels (Supporting Information Fig. S7). However, RT-PCR revealed inactivating mutations in the p53 binding domain in both of the primary GSC lines used in this study. Moreover, a majority of glioblastomas harbor defects in the p53 pathway [30], making it unlikely that p53 stabilization is responsible for the observed effects of Numb on GSC differentiation.
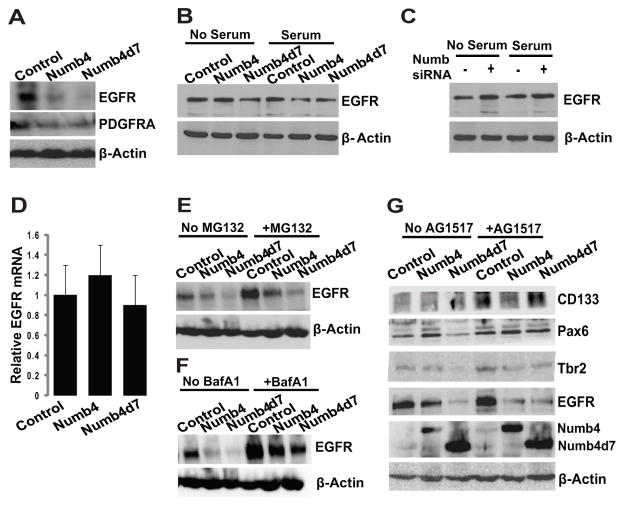
A recent study identified the EGFR as a critical determinant of the transition between neural stem cells and intermediate progenitors [2]. This study suggested that the effect of the EGFR is mediated through transcriptional upregulation of Numb which, in turn, inhibits Notch signaling. Although this model is Notch dependent, it nevertheless implicates the EGFR as a key player in neural stem cell differentiation. Other studies suggest that the EGFR regulates the switch between symmetric and asymmetric division in the developing brain, although the mechanism underlying this effect has not been fully established [31, 32]. We therefore examined the effect of Numb on EGFR expression in GSCs. Overexpression of either Numb4 or Numb4d7 decreased EGFR expression in primary GSCs (Fig. 6A). This effect was also observed in the LN229 glioblastoma cell line (which expresses low endogenous levels of Numb) and was potentiated in the presence of serum (Fig. 6B). Likewise, knockdown of Numb in U87 glioblastoma cells (which express high endogenous levels of Numb) increased EGFR expression (Fig. 6C).
Figure 6. Effect of Numb isoforms on EGFR expression.
(A): Effect of Numb4, Numb4d7, or a control vector on EGFR and PDGFRA expression in primary human glioblastoma stem-like cells (GSCs). (B): Effect of Numb4, Numb4d7, or a control vector on EGFR expression in human LN229 glioblastoma cells in the presence or absence of serum. (C): Effect of Numb knockdown on EGFR expression in human U87 glioblastoma cells in the presence or absence of serum. (D): Real-time PCR illustrating effect of Numb isoforms on EGFR mRNA expression in primary human GSCs. (E): Effect of Numb isoforms on EGFR expression in human GSCs in the presence or absence of the proteasome inhibitor, MG132. (F): Effect of Numb isoforms on EGFR expression in human GSCs in the presence or absence of the lysosome inhibitor, bafilomycin A1. (G): Effect of Numb isoforms on neural differentiation marker expression in GSCs in the presence or absence of the EGFR inhibitor, AG1517. Abbreviations: EGFR, epidermal growth factor receptor; Numb4d7, Numb4 delta 7; PCR, polymerase chain reaction.
Real-time PCR of EGFR mRNA in GSCs overexpressing Numb4 or Numb4d7 failed to show significant differences, suggesting a post-transcriptional mechanism of action (Fig. 6D). A proteasome inhibitor (MG132, 10 μM; Fig. 6E) or a lysosome inhibitor (bafilomycin A1, 100 nM; Fig. 6F) partially antagonized the effect of Numb on EGFR, suggesting that the decrease was due to Numb-mediated endocytosis. We reasoned that if Numb-dependent downregulation of EGFR expression regulates GSC differentiation, then exogenous inhibition of EGFR activity should also regulate GSC differentiation. Consistent with this hypothesis, exposure of GSCs to the small molecule EGFR inhibitor, AG1517, increased expression of CD133 and Pax6 (Fig. 6G). Taken together, these data implicate a Numb-EGFR pathway in the specification of GSC fate.
Numb Expression Correlates with Glioblastoma Subclass and Survival
Using array comparative genomic hybridization data for 197 glioblastoma specimens obtained from The Cancer Genome Atlas (TCGA) Data Portal [30] as well as published glioblastoma subclass designations for those specimens [33], we found that homozygous NUMB deletions occurred primarily in a small subset of proneural glioblastomas (Fig. 7A). This finding suggests that defects in asymmetric division may contribute to tumorigenesis in this subset. Gene expression analysis using these same 197 specimens revealed low, intermediate, and high Numb mRNA expression in the proneural, classic, and mesenchymal glioblastoma subclasses, respectively (Fig. 7B). In addition, survival analysis indicated that increased Numb mRNA expression correlated with decreased patient survival (Fig. 7C; p = .024, log-rank test). This result was confirmed using an independent dataset containing mRNA microarray expression data for 24 surgical glioblastoma specimens from our own institution (Fig. 7C; p = .021, log-rank test). Because the proneural and mesenchymal glioblastoma subclasses are associated with either long or short patient survival, respectively [33, 34], the relatively high expression of Numb mRNA in mesenchymal glioblastomas helps to explain the correlation observed between increased Numb mRNA and decreased survival in glioblastoma patients.
Figure 7. Numb expression correlates with glioblastoma subclass and patient survival.
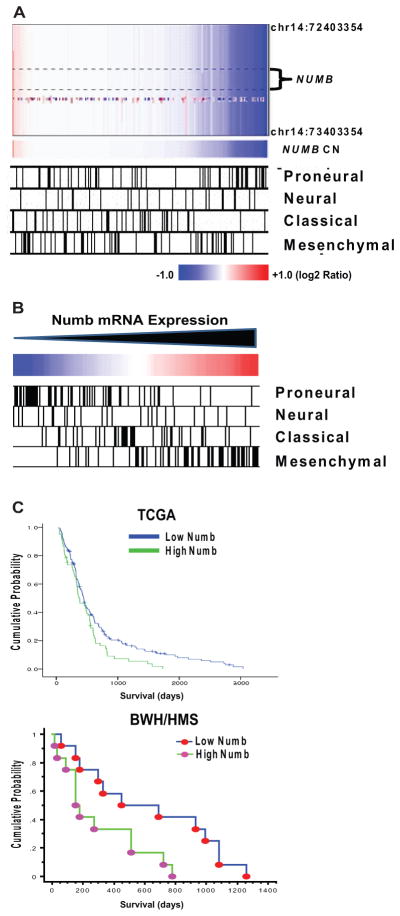
(A): TCGA array CGH data from 197 glioblastomas illustrating relationship between NUMB copy number and glioblastoma subclass. Subclass designations for TCGA glioblastoma specimens are from [33]. (B): mRNA expression data from 197 glioblastomas illustrating relationship between Numb expression and glioblastoma subclass. Subclass designations for tumor specimens are the same as in (A). (C): (upper panel) Cox regression analysis of Numb mRNA and survival for 197 glioblastoma patients (data from TCGA for glioblastoma). p = .024, log-rank test. (lower panel) Kaplan–Meier analysis of Numb mRNA and survival in 24 glioblastoma specimens from our institution (BWH/HMS). Specimens were divided into two equal groups based upon Numb mRNA expression. p = .021, log-rank test. Abbreviation: TCGA, The Cancer Genome Atlas.
Discussion
The differentiated state of tumor cells in glioblastoma and other cancers affects their growth and response to therapy [11, 10, 35, 36]. In glioblastoma, expression of CD133 correlates with growth rate and the response to radiation and chemotherapy [37–40]. Our findings reveal a role for Numb in establishing the hierarchy of differentiated glioblastoma cell types. A model summarizing the role of Numb in regulating glioblastoma growth and differentiation is shown in Supporting Information Figure S8.
We show here that Numb is asymmetrically localized during GSC divisions and regulates the differentiation and growth rate of these cells. High Numb expression increases the number of CD133-hi GSCs while simultaneously decreasing their proliferation. These changes are accompanied by increased expression of radial glia markers such as Pax6 and Sox2. Conversely, low Numb expression leads to the adoption of a highly proliferative CD133-lo/Sox2-lo/Pax6-lo/Tbr2-lo phenotype that is reminiscent of a transit amplifying glial progenitor. Importantly, both Numb-hi/CD133-hi and Numb-lo/CD133-lo cells retain self-renewal capacity. This observation is in agreement with previous reports indicating the existence of a related hierarchy of glioblastoma cells in which both CD133-hi and CD133-lo cells display self-renewal and tumor-initiating capacity [11, 41].
In this study, we show that Numb4d7 is upregulated in glioblastoma and increases Notch signaling, AKT activation and GSC growth while preserving asymmetric cell division. Recent studies have reported the existence of Numb isoforms lacking exon 10 that are upregulated in cancer and that are less antagonistic to Notch signaling than previously identified isoforms [42]. We observed that the frequency and extent of Numb4d7 upregulation are greater in primary human GSCs and in glioblastoma specimens than in glioblastoma cell lines or in nontumor brain specimens, with the ratio of Numb4d7 to Numb4 approaching 1:2 in some tumors. These findings suggest that Numb4d7 opposes the growth-inhibitory effect of Numb4.
Previous studies indicated that Numb regulates Notch degradation by activating the HECT-domain E3 ubiquitin ligase, Itch [3]. However, another major pathway regulating Notch degradation involves Fbw7, an F-box protein component of the SCF family of ubiquitin ligases [27]. The gene encoding Fbw7 is a bona fide tumor suppressor gene, and loss of Fbw7 function leads to tumor formation via increased Notch signaling [27].
Here, we provide the first evidence that Numb interacts with Fbw7, CUL1, and CAND1, all components of the SCFFbw7 complex. Although Numb4 and Numb4d7 have opposite effects on Notch signaling, they interact similarly with NICD. They differ, however, in their ability to promote SCFFbw7 assembly and to interact with Fbw7 and CAND1, suggesting that these are key regulatory steps in Numb-dependent SCF regulation. Numb4 promotes assembly and activation of the SCFFbw7 complex, and it also increases the association of CAND1 with the SCF complex. Studies suggest that CAND1 not only inhibits SCF assembly but also promotes optimal SCF activity [28]. Moreover, only a small fraction of CUL1 is bound by CAND1 at any one time [43], raising the question of how CAND1 can interact with only a small proportion of CUL1 molecules and yet effectively regulate SCF activity. In the case of the SCFFbw7 complex, we propose that Numb4 recruits CAND1 specifically to those SCF complexes where Numb4 is complexed with NICD, Fbw7, and CUL1 (Supporting Information Fig. S6). After substrate ubiquitination is complete, this association facilitates CAND1-mediated disassembly of the SCF complex, thereby increasing substrate/adaptor recycling. Numb4d7 binds NICD but interacts less effectively with Fbw7 and CAND1. Consequently, it fails to promote SCF assembly or activation. By competing with Numb4 for binding to NICD, Numb4d7 downregulates SCFFbw7 activity and increases NICD levels. Upregulation of Numb4d7 in glioblastoma thus provides a mechanism for increasing cancer stem-like cell diversity while maintaining cancer cell proliferation.
Surprisingly, both Numb4 and Numb4d7 promoted the expression of the CD133-hi/Sox2-hi/Pax6-hi phenotype in GSCs, despite having opposite effects on Notch signaling. Thus, it appears that Numb can affect GSC differentiation through mechanisms that do not require Notch inhibition. Because Numb4 and Numb4d7 are both asymmetrically localized, cosegregation or asymmetric regulation of other cell fate determinants with Numb may explain this phenomenon. Several reports have identified a role for the EGFR in regulating the transition between symmetric and asymmetric division in neural stem cells and in determining the transition between neural stem cells and intermediate progenitors [2, 31, 32]. The proposed mechanism involves Notch inhibition via EGFR-dependent upregulation of Numb. However, our findings suggest that Numb also maintains neural stem cells by suppressing EGFR expression via endocytic trafficking [5].
In leukemia and breast cancer, Numb inhibits growth by stabilizing p53 and inhibiting Notch [6, 10, 35]. In glioblastoma, Numb knockdown increased GSC proliferation, indicating that the net effect of Numb expression in glioblastoma is to constrain growth. This notion is further supported by our finding of an overall decrease in Numb protein expression in glioblastoma when compared with nontumor brain. This relative decrease in Numb expression may contribute to the preponderance of symmetric divisions observed in GSCs, as has recently been reported for GSCs and for oligodendrogliomas [17, 32]. Homozygous NUMB deletions and low Numb mRNA expression occurred primarily in a subpopulation of proneural glioblastomas, suggesting that Numb-related defects in asymmetric division play a role in tumorigenesis in this glioblastoma subgroup. Interestingly, a majority of secondary glioblastomas fall within the proneural subclass [30], and secondary glioblastomas contain fewer CD133-hi GSCs than primary glioblastomas [38].
Numb knockdown increased Notch levels and tumor growth, consistent with our finding that the predominant form of Numb (Numb4) in most primary glioblastomas inhibits proliferation. These findings raise questions regarding the functional significance of Numb4d7, which increases Notch and GSC proliferation. Our data suggest that Numb4d7 maintains GSC fate specification while ameliorating the inhibitory effect of Numb4 on tumor growth. Interestingly, a minority of glioblastomas express Numb4 and Numb4d7 in nearly equal amounts (Fig. 1E). It will be of interest to determine the net effect of Numb expression on growth in such tumors.
Conclusions
Increased numbers of CD133-hi cells correlate with increased tumor aggressiveness and decreased survival in glioblastoma and other cancers [14–16]. Among glioblastomas, we observed that increased Numb mRNA correlates with the mesenchymal phenotype and decreased patient survival. The fact that Numb-hi/CD133-hi cells divide asymmetrically to produce a cellular hierarchy comprising rapidly proliferating Numb-lo/CD133-lo progeny as well as more slowly proliferating Numb-hi/CD133-hi GSCs that are resistant to therapy [37–40] may help to explain why patients harboring mesenchymal glioblastomas with high Numb expression have decreased survival.
We report here that Numb expression is decreased overall in glioblastoma when compared with nontumor brain, and that Numb knockdown accelerates glioblastoma growth. These findings appear to be at odds with clinical data indicating a correlation between increased Numb mRNA expression and decreased survival in glioblastoma, and with the observation that Numb promotes adoption of a CD133+/hi phenotype in GSCs. We observed that the CD133−/lo GSCs generated by Numb knockdown in this study retained self-renewal capacity that was comparable to that of the CD133+/hi cells, a finding that is consistent with previous reports [11, 38]. Importantly, CD133−/lo GSCs can be divided into two groups, one with tumorigenicity similar to that of CD133+/hi cells and the other with much lower tumorigenic potential [11]. Using Numb shRNA or siRNA, we were only able to achieve a 50% reduction in Numb protein expression. Collectively, our data indicate that the CD133−/lo cells generated as a result of this moderate degree of Numb depletion corresponded to the intermediate group of CD133−/lo cells with tumorigenicity similar to that of CD133+/hi cells. Studies in normal neural stem cells indicate that complete knockout of Numb and its homolog, Numb-like, depletes the neural stem cell pool [1]. It will thus be of interest to determine whether a greater degree of Numb depletion leads to GSCs with decreased tumorigenicity, as suggested by published reports of a subpopulation of CD133−/lo cells with markedly decreased tumor-forming ability [11], and by the predominance of less aggressive proneural tumors among glioblastomas with homozygous Numb deletions.
Materials and Methods
Tumor Specimens and Cell Lines
Frozen human tissue specimens of World Health Organization Grade II, III, and IV astrocytomas and nontumor brain samples were obtained from the BWH Brain Tumor Tissue Bank under the auspices of a protocol approved by the Brigham and Women’s Hospital IRB. Histology was confirmed by hematoxylin and eosin. Human glioma-derived stem-like cells (GSCs) were isolated from five surgical glioblastoma specimens using a CD133 affinity column (Miltenyi, Cambridge, MA, http://www.miltenyibiotec.com) and maintained as tumorspheres in serum-free medium containing EGF (20 ng/ml) and basic fibroblast growth factor (bFGF) (20 ng/ml) [12]. Established human glioblastoma cell lines (U87, U251, U343, T98, LN18, LN229, LN308, and LN428) were obtained from the American Type Culture Collection (Manassas, VA, http://www.atcc.org) or were a gift (D566, D. Bigner, Duke University). Established glioblastoma cell lines were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% serum and penicillin/streptomycin at 37°C in a 5% CO2 atmosphere. Mouse E14 neural stem cells were obtained from Stem Cell Technologies (Vancouver, BC, http://www.stemcell.com).
Animal Studies
All animal procedures were performed in the animal facility at Brigham and Women’s Hospital/Harvard Medical School in accordance with federal, local, and institutional guidelines under an IACUC protocol approved by the Harvard Medical School Subcommittee on Animals. Adult male athymic CD1 nude mice (Charles River, Cambridge, MA, http://www.criver.com) were used. Intracranial transplantation of 5 × 105 cells per animal or subcutaneous transplantation of 1 × 106 cells per flank was performed as described [44].
Copy Number and mRNA Expression Analysis
NUMB array CGH and mRNA expression and survival data for 197 glioblastoma samples were obtained from the TCGA website (http://cancergenome.nih.gov/dataportal/data/about/). To eliminate cases of perioperative death unrelated to glioblastoma, only samples from patients who survived at least 30 days after diagnosis were used (n = 192). Segmentation of copy number data was performed using GISTIC software. Analysis of NUMB mRNA expression among glioblastoma subclasses was performed using subclass annotations for 197 TCGA samples, the unified NUMB probeset, and consensus clustering as described [33]. Survival analysis was performed using NUMB mRNA expression data (Affymetrix probeset 207545_s_at) for 24 glioblastoma specimens obtained from our institution. Samples were divided into two equal groups based upon NUMB mRNA expression. Statistical significance was determined using the log-rank test.
RT-PCR and Subcloning
Total RNA was isolated from cultured cells or from surgical glioblastoma specimens. Human NUMB cDNAs were cloned by RT-PCR. FLAG-tagged NUMB cDNAs were generated using the primers 5′-ACTAGTATGGACTACAAAGACGATGACGACAAGAACAAATTACGGCAAAG-3′ and 5′-GTCGACTTAAAGTTCAATTTCAAACGTCTTC-3′. The PCR products were cloned into a pGEM-Teasy vector (Promega, Madison, WI, http://www.promega.com) and sequenced. Numb isoforms were also subcloned into lentiviral IRES-EGFP or pEGFPC2 vectors. Numb shRNA and a control shRNA vector were a gift from Dr. S. Pece [6]. Numb siRNA and a control siRNA were purchased from Invitrogen (Grand Island, NY, http://www.invitrogen.com). CAND1 shRNA was purchased from Open Biosystems (Lafayette, CO, https://www.openbiosystems.com).
RT-PCR primers for specific NUMB isoform cDNA fragments were generated: 5′-GTTGACCAGACGATAGAGAAAG-3′ (sense, nucleotides 606–627) and 5′-GAAGGTAGGAGATTGTGGTGCCAC-3′ (antisense, nucleotides 1,379–1,355). The predicted cDNA fragments with or without exon 7 were 800 bp and 500 bp, respectively. Primers for GAPDH were: 5′-GTCAACGGATTTGGTCTGTATT-3′ (sense) and 5′-AGTCTTCTGGGTGGCAGTGAT-3′ (antisense).
Semiquantitative RT-PCR Analysis
A mouse cDNA panel (Rapid Scan) derived from mRNA obtained from various regions of the mouse nervous system throughout development was purchased from Origene (Rockville, MD, http://www.origene.com). Primers to detect mouse Numb4 and Numb4d7 mRNA were: 5′-CTAAGGACCTCATAGTTGACCAGAC-3′ (sense, nucleotides 305–329) and 5′-CTGGAGAGGCAGCACCAGAAG-3′ (antisense, nucleotides 1,401–1,422). The expected cDNA fragments with or without exon 7 were 1,100 bp and 800 bp, respectively. RT-PCR was then performed, and PCR products were sequenced directly.
Western Blots and Immunoprecipitation
Antibodies used were: anti-Numb, anti-Fbw7, anti-Tbr2, and anti-EGFR (Abcam, Cambridge, MA, http://www.abcam.com); anti- CUL1 (Invitrogen); anti-CD133 (Miltenyi Biotec); anti-cleaved Notch (NICD), anti-SOX2, and anti-Hdm2; anti-CAND1 and anti-GFP (SantaCruz Biotechnology, Santa Cruz, CA, http://www.scbt.com); anti-NEDD8, anti-phospho-Akt (Ser473), and anti-Akt (Cell Signaling Technology, Beverly, MA, http://www.cellsignal.com); anti-PAX6 (Covance, Princeton, NJ, http://www.covance.com); anti-Tuj1 (Stem Cell Technologies); anti-GFAP (BD Biosciences, Bedford, MA, http://www.bdbiosciences.com); anti-β-actin and anti-FLAG (Sigma, St. Louis, MO, http://www.sigmaaldrich.com).
Western blots were performed as described [45]. For immunoprecipitation experiments, whole cell lysates from cultured cells were immunoprecipitated with 1 μg of antibody prebound to 30 μl of protein A-Sepharose 4B (GE Healthcare, Pittsburgh, PA, http://www.gelifesciences.com). Appropriate isotype antibodies were used as controls. The beads were washed and boiled in SDS buffer, and the eluates were processed for Western blot as described.
CD133 and Numb Immunohistochemistry and Immunocytochemistry
A tissue microarray containing 63 paraffin-embedded human glioma and nontumor brain samples (Cybrdi, Rockville, MD, http://www.cybrdi.com) was stained for Numb immunoreactivity using the peroxidase method. Additionally, paraffin-embedded glioblastoma samples obtained from the BWH Brain Tumor Tissue Bank were costained using anti-CD133 and anti-Numb antibodies, and secondary antibodies conjugated with peroxidase and alkaline phosphatase. Control sections were stained using the secondary antibodies alone.
Primary cultured human GSCs were transiently transfected with Numb4-GFP or Numb4d7-GFP expression vectors. The cells were fixed, and the nuclei were counterstained with DAPI. Images of dividing cells containing low levels of Numb-GFP fusion protein expression were obtained under epifluorescence.
MTT [3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide] Cell Growth Assay and Bromodeoxyuridine Cell Proliferation Assays
For MTT growth assays, 1 × 103 cells were plated in a 96-well plate, and MTT was detected at 590 nm serially for 13 days according to the manufacturer’s protocol (Roche Applied Sciences, Indianapolis, IN, http://www.roche-applied-science.com). For bromodeoxyuridine (BrdU) proliferation assays, 5 × 103 GSCs were plated on poly(L-ornithine) and maintained in serum-free medium with EGF (20 ng/ml) and FGF (20 ng/ml). The cells were then incubated in BrdU (10 μM) at 37°C for 1 hour, fixed with cold methanol for 10 minutes, washed, and incubated with anti-BrdU antibody followed by a fluorescent secondary antibody for visualization. For quantification, BrdU was measured by ELISA assay. Eight wells were assayed for each condition, and experiments were repeated in triplicate. Statistical significance was determined using the t test.
Hes1 Luciferase Reporter Assay
HEK293T cells were cotransfected with a luciferase Hes1 reporter vector (50 ng) or a control vector lacking Hes1 binding sites (50 ng), a β-galactosidase vector to facilitate normalization (pCMVbeta-gal, 50 ng), and expression vectors for the genes of interest. The total amount of DNA in each transfection was kept constant by the addition of an appropriate amount of empty control vector. Luciferase activity was assayed using a luminometer. For normalization, β-galactosidase activity was measured by ELISA at 420 nm in the same lysates. Samples were assayed in triplicate. Statistical significance was determined by t test.
FACS
GSCs overexpressing Numb4-GFP and Numb4d7-GFP, Numb shRNA-IRES-GFP, or appropriate GFP control vectors were dissociated and filtered to yield single cells. Cells were labeled using a CD133-PE antibody, and FACS analysis of the GFP-expressing cells was performed to measure CD133 expression using a FACScan Flow Cytometer.
Supplementary Material
Acknowledgments
This work was supported by an American Brain Tumor Association Fellowship to X.J. and by a Brain Science Foundation Research Award, a Harvard Stem Cell Institute grant, a Hagerty Fund Research Award, and an NIH Director’s New Innovator Award (DP2OD002319) to M.D.J.
Footnotes
Author contributions: X.J.: data collection and assembly, data analysis and interpretation, and manuscript writing; T.K., Y.J., and P.P.: genomics and biostatistical analysis; H.X., W.H., H.Y., and S.S.: data collection; R.C.: data interpretation; M.J.: conception and design, GBM provision, data collection and assembly, data analysis and interpretation, statistical analysis, manuscript writing, and final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest
References
- 1.Petersen PH, Zou K, Krauss S, et al. Continuing role for mouse Numb and Numbl in maintaining progenitor cells during cortical neurogenesis. Nat Neurosci. 2004;7:803–811. doi: 10.1038/nn1289. [DOI] [PubMed] [Google Scholar]
- 2.Aguirre A, Rubio ME, Gallo V. Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal. Nature. 2010;467:323–327. doi: 10.1038/nature09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem. 2003;278:23196–23203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- 4.Roncarati R, Sestan N, Scheinfeld MH, et al. The gamma-secretase-generated intracellular domain of beta-amyloid precursor protein binds Numb and inhibits Notch signaling. Proc Natl Acad Sci USA. 2002;99:7102–7107. doi: 10.1073/pnas.102192599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGill MA, Dho SE, Weinmaster G, et al. Numb regulates post-endocytic trafficking and degradation of Notch1. J Biol Chem. 2009;284:26427–26438. doi: 10.1074/jbc.M109.014845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colaluca IN, Tosoni D, Nuciforo P, et al. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- 7.Di Marcotullio L, Ferretti E, Greco A, et al. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat Cell Biol. 2006;8:1415–1423. doi: 10.1038/ncb1510. [DOI] [PubMed] [Google Scholar]
- 8.Abbott LA, Natzle JE. Epithelial polarity and cell separation in the neoplastic l(1)dlg-1 mutant of Drosophila. Mech Dev. 1992;37:43–56. doi: 10.1016/0925-4773(92)90014-b. [DOI] [PubMed] [Google Scholar]
- 9.Klezovitch O, Fernandez TE, Tapscott SJ, et al. Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. 2004;18:559–571. doi: 10.1101/gad.1178004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito T, Kwon HY, Zimdahl B, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen R, Nishimura MC, Bumbaca SM, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17:362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 12.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 13.Fatoo A, Nanaszko MJ, Allen BB, et al. Understanding the role of tumor stem cells in glioblastoma multiforme: A review article. J Neurooncol. 2011;103:397–408. doi: 10.1007/s11060-010-0406-3. [DOI] [PubMed] [Google Scholar]
- 14.Colman H, Zhang L, Sulman EP, et al. A multigene predictor of outcome in glioblastoma. Neuro Oncol. 2010;12:49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kappadakunnel M, Eskin A, Dong J, et al. Stem cell associated gene expression in glioblastoma multiforme: Relationship to survival and the subventricular zone. J Neurooncol. 2010;96:359–367. doi: 10.1007/s11060-009-9983-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan X, Ma L, Yi D, et al. A CD133-related gene expression signature identifies an aggressive glioblastoma subtype with excessive mutations. Proc Natl Acad Sci USA. 2011;108:1591–1596. doi: 10.1073/pnas.1018696108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lathia JD, Hitomi M, Gallagher J, et al. Distribution of CD133 reveals glioma stem cells self-renew through symmetric and asymmetric cell divisions. Cell Death Dis. 2011;2:e200. doi: 10.1038/cddis.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Euskirchen P, Skaftnesmo KO, Huszthy PC, et al. Numb does not impair growth and differentiation status of experimental gliomas. Exp Cell Res. 2011;317:2864–2873. doi: 10.1016/j.yexcr.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Fan X, Khaki L, Zhu TS, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purow BW, Haque RM, Noel MW, et al. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 2005;65:2353–2363. doi: 10.1158/0008-5472.CAN-04-1890. [DOI] [PubMed] [Google Scholar]
- 21.Verdi JM, Bashirullah A, Goldhawk DE, et al. Distinct human NUMB isoforms regulate differentiation vs. proliferation in the neuronal lineage. Proc Natl Acad Sci USA. 1999;96:10472–10476. doi: 10.1073/pnas.96.18.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corti S, Nizzardo M, Nardini M, et al. Isolation and characterization of murine neural stem/progenitor cells based on Prominin-1 expression. Exp Neurol. 2007;205:547–562. doi: 10.1016/j.expneurol.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 23.Tonchev AB, Yamashima T, Sawamoto K, et al. Transcription factor protein expression patterns by neural or neuronal progenitor cells of adult monkey subventricular zone. Neuroscience. 2006;139:1355–1367. doi: 10.1016/j.neuroscience.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 24.Englund C, Fink A, Lau C, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sade H, Krishna S, Sarin A. The anti-apoptotic effect of Notch-1 requires p56lck-dependent, Akt/PKB-mediated signaling in T cells. J Biol Chem. 2004;279:2937–2944. doi: 10.1074/jbc.M309924200. [DOI] [PubMed] [Google Scholar]
- 26.Hotton SK, Callis J. Regulation of cullin RING ligases. Annu Rev Plant Biol. 2008;59:467–489. doi: 10.1146/annurev.arplant.58.032806.104011. [DOI] [PubMed] [Google Scholar]
- 27.Welcker M, Clurman BE. FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 28.Lo SC, Hannink M. CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1. Mol Cell Biol. 2006;26:1235–1244. doi: 10.1128/MCB.26.4.1235-1244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bornstein G, Ganoth D, Hershko A. Regulation of neddylation and deneddylation of Cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate. Proc Natl Acad Sci USA. 2006;103:11515–11520. doi: 10.1073/pnas.0603921103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egger B, Gold KS, Brand AH. Regulating the balance between symmetric and asymmetric stem cell division in the developing brain. Fly. 2011;5:237–241. doi: 10.4161/fly.5.3.15640. [DOI] [PubMed] [Google Scholar]
- 32.Sugiarto S, Persson AI, Munoz EG, et al. Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer Cell. 2011;20:328–340. doi: 10.1016/j.ccr.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 35.Pece S, Confalonieri S, Romano RP, et al. NUMB-ing down cancer by more than just a NOTCH. Biochim Biophys Acta. 2011;1815:26–43. doi: 10.1016/j.bbcan.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Rennstam K, McMichael N, Berglund P, et al. Numb protein expression correlates with a basal-like phenotype and cancer stem cell markers in primary breast cancer. Breast Cancer Res Treat. 2010;122:315–324. doi: 10.1007/s10549-009-0568-x. [DOI] [PubMed] [Google Scholar]
- 37.Beier CP, Beier D. CD133 negative cancer stem cells in glioblastoma. Front Biosci (Elite Ed) 2011;3:701–710. doi: 10.2741/e280. [DOI] [PubMed] [Google Scholar]
- 38.Beier D, Hau P, Proescholdt M, et al. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 39.Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thon N, Damianoff K, Hegermann J, et al. Presence of pluripotent CD133+ cells correlates with malignancy of gliomas. Mol Cell Neurosci. 2010;43:51–59. doi: 10.1016/j.mcn.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 41.Sun Y, Kong W, Falk A, et al. CD133 (Prominin) negative human neural stem cells are clonogenic and tripotent. PLoS One. 2009;4:e5498. doi: 10.1371/journal.pone.0005498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karaczyn A, Bani-Yaghoub M, Tremblay R, et al. Two novel human NUMB isoforms provide a potential link between development and cancer. Neural Dev. 2010;5:31. doi: 10.1186/1749-8104-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennett EJ, Rush J, Gygi SP, et al. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell. 2010;143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang HW, Menon LG, Black PM, et al. SNAI2/Slug promotes growth and invasion in human gliomas. BMC Cancer. 2010;10:301. doi: 10.1186/1471-2407-10-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Y, Jiang X, Schoch BS, et al. Aberrant splicing of cyclin-dependent kinase-associated protein phosphatase KAP increases proliferation and migration in glioblastoma. Cancer Res. 2007;67:130–138. doi: 10.1158/0008-5472.CAN-06-2478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.