Abstract
Bovine abortion of unknown infectious etiology still remains a major economic problem. Thus, we investigated whether Brucella spp., Listeria monocytogenes, Salmonella spp., Campylobacter spp. and Coxiella burnetii are associated with abortion and/or stillbirth in Tunisian dairy cattle. Using a pan-Chlamydiales PCR, we also investigated the role of Chlamydiaceae, Waddlia chondrophila, Parachlamydia acanthamoebae and other members of the Chlamydiales order in this setting. Veterinary samples taken from mid to late-term abortions from twenty dairy herds were tested. From a total of 150 abortion cases collected, infectious agents were detected by PCR in 73 (48.66%) cases, 13 (8.66%) of which represented co-infections with two infectious agents. Detected pathogens include Brucella spp (31.3%), Chlamydiaceae (4.66%), Waddlia chondrophila (8%), Parachlamydia acanthamoebae (5.33%), Listeria monocytogenes (4.66%) and Salmonella spp. (3.33%). In contrast, Campylobacter spp. and Coxiella burnetii DNA were not detected among the investigated veterinary samples. This demonstrates that different bacterial agents may cause bovine abortion in Tunisia. This is the first report suggesting the role of Parachlamydia acanthamoebae in bovine abortion in Africa. Further studies with a larger number of samples are necessary to confirm whether this emerging pathogen is directly linked to abortion in cattle.
Introduction
Abortion among dairy cows is one of the major causes of economic losses in the cattle industry. Abortions may be idiopathic or the result of metabolic or hormonal abnormalities, nutritional deficiencies, trauma, toxicities, or infectious agents. The latter represents the leading etiology of reproductive disorders [1], [2] A variety of infectious agents have been reported to cause bovine abortion throughout the world. The major bacterial agents that have been implicated in bovine abortion during mid- to late-gestation are Brucella spp., Chlamydia spp., Salmonella spp., Campylobacter spp., Listeria monocytogenes and Coxiella burnetii [3]–[5]. Bovine brucellosis was distributed worldwide, and its importance is related not only to the economic losses in animal production but also to a significant risk for human health. In cattle, brucellosis is caused by Brucella abortus, a facultative intracellular Gram negative coccobacillus. The abortion generally occurs from 6 to 9 months of gestation. It is frequently followed by fetal membrane retention and endometritis, which may be the cause of infertility in subsequent pregnancies [6], [7]. Even in the absence of abortion, excretion of the infectious organism occurs, for instance via placenta, foetal fluids, vaginal discharge, and milk. In addition to brucellosis, chlamydiosis and Q fever should be considered among the most common zoonotic diseases and are distributed all around the world. They are respectively caused by Chlamydia and C. burnetii, two strictly intracellular Gram negative bacteria. In cattle, C. burnetii infection is associated with abortions and stillbirths even if the infection is often asymptomatic. Inversely, chlamydial infections in cattle cause a variety of syndromes such as conjunctivitis, polyarthritis, encephalomyelitis, mastitis, infertility, abortion and other urogenital tract infections [8], [9]. C. abortus, C. pecorum and W. chondrophila are additional etiologies of chlamydial abortions in cattle [10]–[12]. Moreover, there is increasing evidences supporting the role of another Chlamydia-related bacteria, P. acanthamoebae, in abortion in both cattle [13]–[15] and humans [16]. However, no information is available regarding the presence of this bacterium in Africa.
Listeriosis, salmonellosis and campylobacteriosis are also serious zoonotic diseases. Bovine listeriosis is caused by L. monocytogenes and the organism is excreted in feces, urine and milk. Abortions are usually sporadic but may affect 10–20% of a herd. Abortions occur most commonly during the last trimester of pregnancy. The aborted fetus is often autolyzed. Bovine salmonellosis is caused predominantly by S. enterica serotypes Typhimurium and Dublin [17], [18]. Occasionally, Salmonella spp. cause abortion storms. The cows are usually sick and the fetuses and placentas are autolyzed and emphysematous. Salmonella can be isolated from the fetal tissues, vaginal fluids, feces and milk. Several Campylobacter species can be associated with abortion in cattle. Bovine genital campylobacteriosis is a venereal disease caused by Campylobacter fetus subsp. venerealis that can be found in the genital tract of cattle, in which it may cause genital tract infection and sporadic abortions [19].
Bacteriological isolation by culture on blood agar is usually used for the diagnosis of bovine brucellosis, but it is difficult, time consuming, hazardous, and sometimes inconclusive [20], [21]. Routine diagnosis of Q fever is often made by the use of serological tests [22], which have the disadvantage of indicating post-exposure rather than ongoing infection. Diagnosis of chlamydial infections in animals still represents a considerable challenge [23]. Isolation in cell culture remains difficult, time consuming, and depends on the presence of sufficient numbers of viable bacteria. The reliability of standard diagnostic procedures for Campylobacter fetus, which are based on phenotypic methods, is difficult because of specific nutritional and atmospheric requirements [24]. Furthermore, all these traditional culture and serology-based methods can be relatively insensitive depending on the quality and timing of sampling. Rapid detection of abortigenic agents at the early stage of an outbreak using molecular methods contributes to minimizing the spread of infection, the outbreak burden and increasing treatment efficiency. Therefore, real-time PCR have been increasingly used as a diagnostic tool for etiologic diagnosis of abortion in cattle [3], [15], [25], as a complement or replacement of time consuming traditional diagnostic methods such as bacterial culture [26].
Thus, the present study aimed (i) to investigate the role of some abortigenic agents in cattle from different geographical regions of Sfax in Tunisia (ii) to define the role of P. acanthomoebae as a new abortigenic agent in African cattle and (iii) to detect a possible co-infection of members of the Chlamydiales order with other abortigenic agents.
Materials and Methods
Animals and samples
Twenty dairy herds from different regions of Sfax (Tunisia) that experienced reproductive disorders (mainly abortions) from October 2010 to May 2012 were included in this study. Informations on individual animal such as age, sex and abortion history were recorded separately on sample data sheets. Herd sizes ranged from 20 to 1500 cows, knowing that all cows studied in this work are the descendents of pure Holsteins from the national production. All these dairy cows were vaccinated against foot and mouth disease (FMD) during national immunization campaigns. The majority of included cows were kept on limited pastures or tethered on a pasture. Seventy percent of the herds possessed small ruminants, and only twenty percent reported having bought new cattle. In dairy herds, nutrient requirements may not be the same depending on the animal's age and stage of production. Forages, which refer especially to hay or straw, are the most common type of feed used. Barley is an example of cereal grain that is extensively used in these herds. In all herds, samples from (i) cows with clinical signs (cases) and (ii) cows with normal pregnancies and normal parturitions (controls) were taken. A total of 214 animals were sampled: 150 cases and 64 controls. They were bled on the same day or up to 8 months after abortion. A total of 214 blood, 214 vaginal swabs and 214 milk samples were collected by local veterinarians and sent to the laboratory. Practically, blood samples of about 5 ml were aseptically collected using plain tubes from cows through jugular venipuncture. Serum samples were separated within 12 h of collection and transported to the laboratory using an ice box where they are stored at −20°C until tested. Vaginal swab samples were collected from each cow, using sterile swabs after vulva disinfection with chlorhexidine solution. All swabs were put into tubes containing 1 ml of 2-sucrose phosphate medium (2-SP). They were kept at 4°C during transportation in ice-pack containers, and then stored at −80°C until use. All milk samples were stored at −80°C until tested.
DNA extraction and real-time PCR
The vaginal swab samples collected in 2-SP medium were thawed; 1 ml was transferred to a new microtube, and then centrifuged at 13,000 g for 20 min. The pellet was resuspended in 200 µl sterile water. The total volume was extracted by Quick-gDNA MiniPrep D3006 Kit (Zymo Research, Irvine, CA, USA) as recommended by the manufacturer. Extracted DNA was re-suspended in 50 µl of elution buffer and stored at −20°C until subsequent analysis.
For milk samples, DNA was extracted by using 1 ml aliquot which was centrifuged at 6,000× g for 10 min. The clear whey portion was suctioned out with a transfer pipette and discarded. The remaining milk solids and butterfat were resuspended in 200 µl sterile water and used for DNA extraction using Quick-gDNA MiniPrep D3006 Kit (Zymo Research, Irvine, CA, USA) as recommended by the manufacturer. Extracted DNA was re-suspended in 50 µl of elution buffer and stored at −20°C until subsequent analysis. All samples were tested in duplicate.
Brucella spp. PCR
The real-time PCR assay was performed on a CFX96™ real-time PCR cycler (Biorad, Hercules, CA, USA). Real-time amplifications were carried out in a total reaction volume of 20 µl containing 10 µl iTaq Supermix with ROX (Bio-Rad, Reinach, Switzerland), 0.3 µM of each primer, 0.2 µM of probe and 5 µl of purified DNA to a final volume of 20 µl using nuclease-free water. Primers and probe sequences are provided in Table 1. The optimal PCR efficacy was obtained using a cycling profile that included an initial denaturation step at 95°C for 3 min, then 45 cycles of 15 s at 95°C and 1 min at 60°C. In all experiments, each PCR run included a negative extraction control (sterile water) and a negative PCR control containing 5 µl Diethylpyrocarbonate (DEPC) treated H2O instead of DNA extract, to detect possible contamination of DNA. Samples were run in duplicate.
Table 1. Sequences of primers and probes used in the study.
| Name | Sequence (5′ to 3′) | Product size (bp) | Reference | |
| Brucella spp. | IS711-F | GCTTGAAGCTTGCGGACAGT | 63 | This study |
| IS711-R | GGCCTACCGCTGCGAAT | |||
| IS711-P | HEX-AAGCCAACACCCGGCCATTATGGT-BHQ1 | |||
| Chlamydiale order | panCh16F2 | CCGCCAACACTGGGACT | 207 to 215 | Lienard et al., 2011 |
| panCh16R2 | GGAGTTAGCCGGTGCTTCTTTAC | |||
| panCh16S | FAM-CTACGGGAGGCTGCAGTCGAGAATC-BHQ1 | |||
| Salmonella spp. | invA-F | AGACGACTGGTACTGATTGATAAT | 243 | This study |
| invA-R | ACAGTGCTCGTTTACGACCTGAAT | |||
| L. monocytogenes | hly-F | CATGGCACCACCAGCATCT | 63 | This study |
| Hly-R | ATCCGCGGTGTTTCTTTTCGA | |||
| C. burnetii | IS1111-F | GCGTCATAATGCGCCAACATA | 201 | This study |
| IS1111-R | CGCAGCCCACCTTAAGACTG | |||
| Campylobacter spp. | Camp16SF | CACGTGTCACAATGGCATAT | 108 | This study |
| Camp16SR | GGCTTCATGCTCTCGAGTT |
Chlamydiales-specific real-time PCR
A pan-Chlamydiales PCR that may amplify all members of the Chlamydiales order including Chlamydiaceae, Waddliaceae and Parachlamydiaceae was performed as described previously by Lienard et al. [27]. The CFX96™ real-time PCR (Biorad) was used to perform the amplification and the detection of DNA with a cycling program of 3 min at 95°C, followed by 50 cycles of 15 s at 95°C, 15 s at 67°C, and 15 s at 72°C. Samples were tested at least in duplicates and were considered negative if no amplification was observed during all 50 cycles. Samples positive for members of the Chlamydiaceae family were further tested by corresponding commercial kits.
Commercial standard kits for the quantification of C. abortus and C. pecorum
The PrimerDesign genesig Kits for C. abortus and C. pecorum Genomes are designed for the in vitro quantification of these two bacteria genomes. The two kits are targeting the ompA gene, allowing specific in vitro quantification of these species and detecting no other Chlamydia. A highly conserved sequence within the ompA gene has previously been shown to be a good target sequence in other clinical real time PCR based studies [28]. The primers and probe sequences in this kit have 100% homology with a broad range of clinically-relevant reference sequences.
Listeria monocytogenes PCR
Real-time PCR was performed on the CFX96™ real-time PCR cycler (Biorad). Each reaction contained 10 µl SsoAdvancedTM SYBR1 Green Supermix (Biorad), 0.6 µM of each primer (Table 1), 2 µl template DNA and nuclease free H2O to a final volume of 20 µl. The cycling parameters consisted of 3 min incubation at 95°C followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. A melting curve analysis was performed using the following cycling parameters: 60°C for 30 s, and 5°C temperature changes to the end temperature of 95°C. In all experiments, each PCR run included a negative extraction control (sterile water) and a negative PCR control; containing 2 µl DEPC treated H2O instead of DNA extract, to detect possible contamination of DNA. Samples were run in duplicate.
Salmonella spp. PCR
The real-time PCR was performed on the CFX96™ real-time PCR cycler (Biorad). Amplification reactions were carried at a final volume of 25 µl containing 0.2 µM of each primer (Table 1), 12.5 µl of 2× SYBR Permix Ex Taq™ Tli RNaseH Plus (TaKaRa) and 1 µl of genomic DNA. PCR amplification was conducted by incubating the samples at 95°C for 30 s, followed by 40 cycles of 5 s at 95°C and 30 s at 60°C. A melting curve analysis was performed using the following cycling parameters: 60°C for 30 s, and 5°C temperature changes to the end temperature of 95°C. In all experiments, each PCR run included a negative extraction control (sterile water) and a negative PCR control; containing 1 µl DEPC treated H2O instead of DNA extract, to detect possible contamination of DNA. Samples were run in duplicate.
Coxiella burnetii PCR
The real-time PCR assay was performed on a CFX96™ real-time PCR cycler (Biorad). The optimal concentration of primers was assessed by testing different concentrations (0.05, 0.15, 0.2, 0.3, and 0.5 µM) and by defining the one that gave the highest recorded fluorescence and the lowest threshold cycle (Ct), defined as the point at which the fluorescence crosses the threshold. Each reaction was run in duplicate in a mastermix containing 10 µl of SsoAdvanced SYBR Green Supermix (Biorad), 0.2 µM of each primer (Table 1), and 1 µl of purified DNA to a final volume of 20 µl. The thermal cycling conditions were assessed by testing different annealing temperatures (between 54°C and 60°C) during different times (5, 10, 20, 30 and 60 s.). The optimal qPCR efficacy was obtained using cycling profile included an initial denaturation step at 95°C for 3 min, then 40 cycles of 10 s at 95°C and 10 s at 60°C. A melting curve analysis was performed using the following cycling parameters: 60°C for 30 s, and 5°C temperature changes to the end temperature of 95°C. In all experiments, each PCR run included a negative extraction control (sterile water) and a negative PCR control (containing 1 µl DEPC treated H2O instead of DNA extract, to detect possible contamination of DNA). Samples were run in duplicate.
Campylobacter spp. PCR
The real-time PCR assay was performed on a CFX96™ real-time PCR cycler (Biorad). Each reaction was run in duplicate in a mastermix containing 12.5 µl of 2× SYBR Permix Ex Taq Tli RNaseH Plus (TaKaRa), 0.2 µM of each primer (Table 1), and 1 µl of purified DNA to a final volume of 25 µl using nuclease-free water. The thermal cycling conditions were assessed testing different annealing temperatures (between 54°C and 60°C) during different times (5, 10, 20, 30 and 60 s.). The optimal qPCR efficacy was obtained using cycling profile included an initial denaturation step at 95°C for 30 s, then 40 cycles of 5 s at 95°C and 30 s at 60°C. A melting curve analysis was performed using the following cycling parameters: 60°C for 30 s, and 5°C temperature changes to the end temperature of 95°C. The generated melt peak represented the specific amplified product. In all experiments, each PCR run included a negative extraction control (sterile water) and a negative PCR control (containing 1 µl DEPC treated H2O instead of DNA extract, to detect possible contamination of DNA).
Rose Bengal plate test
All serum samples collected were screened using the Rose Bengal plate test (RBPT), according to the procedures described by Alton et al. [29]. B. abortus antigen (obtained from the Institut Pourquier, Montpellier, France) was used to screen sera for the presence of antibodies to Brucella spp. In brief, 30 µl of serum was mixed with an equal volume of antigen suspension on a glass plate and agitated. After four minutes of rocking, any visible agglutination was considered as positive. The degree of agglutination was graded on an ordinal scale from 0 (no agglutination) to 3 (coarse clumping), with corresponding RBPT scores of 0, 1, 2 and 3. All doubtful reactions were recorded as negative or zero scores.
Detection of the amplification product and sequencing
Ten µl of each PCR product were electrophoresed in agarose gel, stained with ethidium bromide and observed under UV illumination. The fragments at correct size were excised and further purified for sequencing. Briefly, DNA was extracted using the QIAquick Gel extraction Kit (Qiagen, Courtaboeuf, France), was then subjected to cycle sequencing using the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) and processed by the ABI 3100 Genetic Analyzer. The obtained sequences were compared with the sequences available in GenBank by using the BLAST server from the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST).
Statistical analysis
The association of abortion with positive results for different agents by real-time PCR and frequencies of positive results between vaginal and milk samples were assessed using the Fisher's exact test. The correlation between real-time PCR and the serological test (RBPT) for Brucella cases was assessed using Kruskal-Wallis test (nonparametric test). In all tests, a p value<0.05 was considered to be statistically significant. We decided to use a one-sided p value as all the evidence points to the directionality of this relationship only going one way. All tests were conducted using the Graph-Pad Prism 5 statistical package (Graph-Pad Software, San Diego, CA, USA).
Ethical considerations
Samples were collected by clinical veterinarians as part of the usual screening scheme on farms and Tunisian ethical guidelines and animal welfare regulations were strictly respected. All herd owners had given an informed consent prior to the study. All samples were collected as part of routine care.
Results
Detection of Brucella spp
The results of this study showed that 47 out of 150 investigated samples (31.3%) were positive for Brucella using the RBPT (Table 2). As many as 37 sera showed a weak reaction (1+/2+) whereas 10 sera showed a strong reaction (3+). Only 5 sera were detected as positive among the 64 control sera with a strong reaction (3+) (OR = 5.38, 95% CI [2.02–14.29], p<0.0001) (Table 2; Table 3). The results of the RBPT were confirmed by the real-time PCR performed on vaginal discharges and on milk samples. As many as 46 vaginal swabs out of 150 (30.6%) were positives (Table 3; Fig. S1A). For milk, only 21/150 samples (14%) were found to be positives (OR = 2.71, 95% CI [1.52–4.84], p value = 0.0004) (Table 3; Fig. S2A). All control samples remained negative by real-time PCR. Samples showing Ct values above 43 were considered as negative. Interestingly, the mean Ct-value of samples negative for Brucella was significantly lower than that of samples with a strong reactivity, which is lower than that of samples with a weak reactivity (p<0.0001) (Fig. 1). In conventional gel PCR, all positive samples by real-time PCR produced amplicon of 63 bp when resolved on an agarose gel (Figs. S1B, S2A). All of them were identified as B. abortus based on species-specific PCR [30]. To confirm that positive samples were due to the presence of B. abortus DNA, PCR products were sequenced and analyzed using the BLAST web interface (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The amplicons of these positive samples were found to be 100% identical to the B. abortus gene in the database. Details of cases positive for B. abortus are given in Table 2.
Table 2. Details of cases positive for Brucella abortus.
| Number of positive cases | Rose Bengal plate test (RBPT) | Real-time PCR | ||
| Vaginal swab samples | Milk samples | |||
| Aborted cows | 21 | + | + | + |
| 25 | + | + | − | |
| 1 | + | − | − | |
| Control cows | 5 | + | − | − |
+ positive/ − negative.
Table 3. The association of abortion with positive results for different agents and frequencies of positive results between vaginal and milk samples.
| Tests | Cases (n = 150) | Controls (n = 64) | Odds Ratio 95% CI | P value |
| RBPT | 47 (31.3%) | 5 (7.81%) | OR 5.38 (2.02–14.29) | <0.0001 |
| Brucella spp. PCR | 46 (30.6%) | 0 | OR 57.4 (3.47–948.3) | <0.0001 |
| Chlamydiales PCR | 27 (18%) | 0 | OR 28.7 (1.72–478.9) | <0.0001 |
| L. monocytogenes PCR | 7 (4.6%) | 0 | OR 6.74 (0.37–119.9) | 0.0796 |
| Salmonella spp. PCR | 5 (3.3%) | 0 | OR 4.87 (0.26–89.5) | 0.1658 |
The association of abortion with positive results for different agents by real-time PCR and frequencies of positive results between vaginal and milk samples were assessed using the Fisher's exact test. A p value<0.05 was taken as a level of significance. Cases: cows with clinical signs, controls: cows with normal pregnancies and normal parturitions, OR: odds ratio, CI: confidence interval.
Figure 1. Correlation between real-time PCR and the RBPT results.
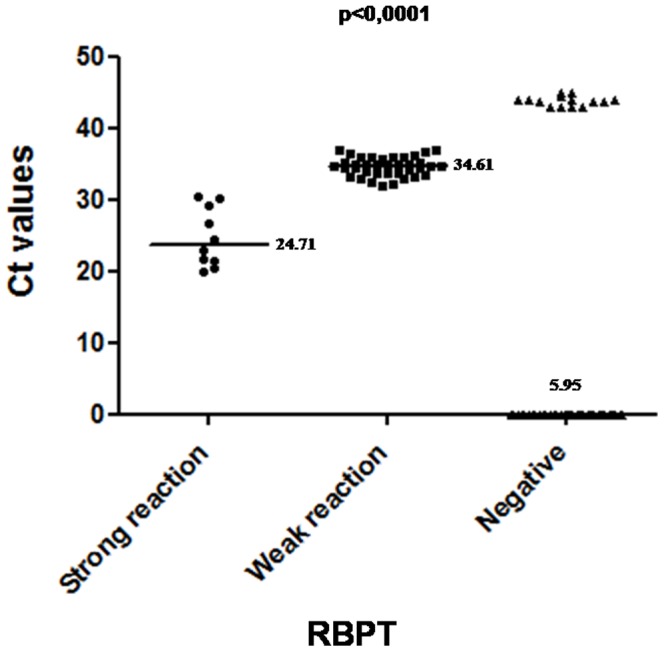
The mean Ct-value of samples negative for Brucella (5.95) was significantly lower than that of samples with a strong reaction results (24.71), which is lower than that of samples with a weak reaction (34.61) (p<0,0001).
Detection of Chlamydiaceae, W. chondrophila, P. acanthamoebae
Chlamydial DNA was detected in 27 (18%) vaginal swab samples, all obtained from the 150 cows that suffered from an abortion (OR = 28.7, 95% CI [1.72–478.9], p<0.0001). Briefly, these cows exhibited symptoms of reproductive disorders like abortion and other symptoms like fever, appetite loss, weakness and diarrhea. No milk sample was positive (OR = 67, 95% CI [4.04–1111], p<0.0001). In conventional gel PCR, the 27 vaginal swab samples positive by real-time PCR produced as expected amplicons of 207 bp when resolved on agarose gel 1.5% (Fig. S3A, S3B). PCR products were sequenced and analyzed using the BLAST web interface (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Among 27 sequences obtained, 12 (8%) belonged to the Waddliaceae family, 8 (5.3%) belonged to the Parachlamydiaceae family and 7 (4.6%) belonged to the Chlamydiaceae family (Fig. S3A, S3B). All sequences corresponding to a Waddliaceae species demonstrated ≥99% similarity with W. chondrophila. All of them had already been identified by the SYBR Green real-time PCR we used in our previous work [12] to detect the presence of W. chondrophila. Since both PCR are targeting two different DNA regions, these results may clearly not result from PCR contamination with amplicons. All the sequences belonging to Parachlamydiaceae family demonstrated ≥99% of similarity with P. acanthamoebae. Four among the 7 sequences belonging to the Chlamydiacaeae family demonstrated ≥99% sequence similarity with C. abortus, whereas the remaining 3 sequences exhibited ≥99% sequence similarity with C. pecorum. These results were confirmed by two commercial kits (PrimerDesign genesig) specific for the quantification of C. abortus and C. pecorum. All control samples remained negative by real-time PCR. Details of cases positive for P. acanthamoebae, C. abortus, C. pecorum and W. chondrophila are given in Table 4.
Table 4. Sequencing results of vaginal swab samples positive with the pan-Chlamydiales PCR.
| Animal no. | Age (years) | Other information | Other etiology | Ct values (mean values) | % 16S rRNA gene homology with most similar GenBank sequence (corresponding family) |
| 1 | 2 | Abortion, fever, loss of appetite, diarrhea | Brucella abortus | 28.34 | 99% Parachlamydia acanthamoebae |
| 2 | 2 | Abortion, fever, diarrhea | 34.91 | 99% Parachlamydia acanthamoebae | |
| 3 | 3 | Abortion, fever, weakness, diarrhea | 34.91 | 99% Parachlamydia acanthamoebae | |
| 4 | 2 | Abortion, fever, diarrhea | Brucella abortus | 35.72 | 99% Parachlamydia acanthamoebae |
| 5 | 2 | Abortion | 31.8 | 99% Parachlamydia acanthamoebae | |
| 6 | 2 | Abortion | Brucella abortus | 34.52 | 99% Parachlamydia acanthamoebae |
| 7 | 2 | Abortion, fever, diarrhea | 35.22 | 99% Parachlamydia acanthamoebae | |
| 8 | 2 | Abortion, fever, diarrhea | 35.64 | 99% Parachlamydia acanthamoebae | |
| 9 | 4 | Abortion, fatigue | Brucella abortus | 19.97 | 99% Chlamydia abortus |
| 10 | 3 | Abortion, respiratory disease | Brucella abortus | 22.91 | 99% Chlamydia abortus |
| 11 | 4 | Abortion | 25.4 | 99% Chlamydia abotus | |
| 12 | 2 | Abortion | Brucella abortus | 25.64 | 99% Chlamydia abortus |
| 13 | 2 | Abortion, diarrhea, fatigue | 27.79 | 99% Chlamydia pecorum | |
| 14 | 3 | Abortion | 31.66 | 99% Chlamydia pecorum | |
| 15 | 2 | Abortion, loss appetite | 34.52 | 99% Chlamydia pecorum | |
| 16 * | 2 | Abortion | Listeria monocytogenes | 32.88 | 99% Waddlia chandrophila |
| 17 * | 2 | Abortion, fatigue | 35.49 | 99% Waddlia chandrophila | |
| 18 * | 2 | Abortion, fever, loss appetite, weakness, diarrhea | 29.72 | 99% Waddlia chandrophila | |
| 19 * | 3 | Abortion | 34.26 | 99% Waddlia chandrophila | |
| 20 * | 3 | Abortion, respiratory diseases | 33.24 | 99% Waddlia chandrophila | |
| 21 * | 2 | Abortion, diarrhea | 31.33 | 99% Waddlia chandrophila | |
| 22 * | 3 | Abortion | 34.1 | 99% Waddlia chandrophila | |
| 23 * | 3 | Abortion | Listeria monocytogenes | 36.1 | 99% Waddlia chandrophila |
| 24 * | 4 | Abortion, fever, loss appetite | 31.97 | 99% Waddlia chandrophila | |
| 25 * | 2 | Abortion | 34.62 | 99% Waddlia chandrophila | |
| 26 * | 2 | Abortion | 33.83 | 99% Waddlia chandrophila | |
| 27 * | 4 | Abortion, fever, diarrhea | 32.84 | 99% Waddlia chandrophila |
*These results were previously published by Barkallah et al. (2013).
Detection of L. monocytogenes
L. monocytogenes DNA was detected in 7/150 (4.66%) vaginal swab samples all obtained from the 150 cows that suffered of an abortion (OR = 6.74, 95% CI [0.37–119.9], p value = 0.08) (Table 3). Only 3/150 (2%) milk samples were found to be positive (Table 3). These cases originated from different herds in different regions of Sfax. All control samples remained negative by real-time PCR. The real-time PCR melt curve data identified one peak with a melting temperature of 87°C for all samples (Fig. S4A, S4B). In conventional gel PCR, all samples positive by real-time PCR produced amplicon of 63 bp when resolved on an agarose gel 3% (Fig. S4A, S4B). To confirm that positive samples were due to the presence of L. monocytogenes DNA, PCR products were sequenced and analyzed using the BLAST web interface (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The amplicons of these positive samples were 100% identical to the L. monocytogenes hlyQ gene in the databases. All control samples remained negative by real-time PCR.
Detection of Salmonella spp
Salmonella DNA was detected in 5 (3.33%) milk samples all obtained from the 150 cows that suffered of an abortion (OR = 4.87, 95% CI [0.26–89.5], p value = 0.1658) (Table 3). No salmonella DNA was detected in vaginal swab samples (OR = 0.08, 95% CI [0–1.6], p value = 0.0302) (Table 3). These cases were collected from the same herd located in the southern region of Sfax. The real-time PCR melt curve data identified one peak with a melting temperature of 76°C for all samples (Fig. S5A). In conventional gel PCR, all positive samples by real-time PCR produced amplicon of 243 bp when resolved on an agarose gel 1.5% (Fig. S5B). To confirm that positive samples were due to the presence of Salmonella spp. DNA, PCR products were sequenced and analyzed using the BLAST web interface (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The amplicons of these positive samples were 100% identical to Salmonella enterica serovar Typhimurium in the databases.
Detection of C. burnetii and Campylobacter spp
C. burnetii was absent in all bovine samples taken from the 150 cows that have aborted and from the 64 control cows tested by the real-time PCR. Similarly, using the Campylobacter spp. real-time PCR, none of the bovine samples taken from the 150 cows that have aborted and from the 64 control cows, were positive.
Co-infections
From a total of 150 abortion cases collected, infectious agents were detected in 73 (48.66%) cases, 13 (8.66%) of which represented co-infections with two infectious agents. These observations may suggest that co-infections commonly exist in cattle herds. Table 5 shows the number of positive samples for each pair of pathogens.
Table 5. The number of samples positive for each pair of pathogens.
| B. abortus | C. abortus | C. pecorum | W. chondrophila | P. acanthamoebae | L. monocytogenes | Salmonella Typhimurium | Campylobacter spp. | C. burnetii | |
| B. abortus | 47 | 3 | 0 | 0 | 3 | 0 | 5 | 0 | 0 |
| C. abortus | 3 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C. pecorum | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| W. chondrophila | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 |
| P. acanthamebea | 3 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 |
| L. monocytogenes | 0 | 0 | 0 | 2 | 0 | 7 | 0 | 0 | 0 |
| Salmonella Typhimurium | 5 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 |
| Campylobacter spp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C. burnetii | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Discussion
In many developing countries, studies conducted to estimate the true prevalence of abortigenic agents were only occasional and limited to certain farms or regions. Consequently, the control programs are not very effective, even for established agents of abortion in cattle. Moreover, the etiology remains unknown in many cases of bovine abortion. In this study, we thus focused on new or not well-defined emerging agents in Tunisian cattle. Vaginal swab samples were used in many previous studies for the detection and isolation of many abortive bacteria [28], [31]–[35], because this type of sampling (i) is simple, fast and it is not necessary to have access to the placenta and (ii) limits the external contamination, which is often the case in the collection of the cotyledons if the placenta hit the ground [36]. The choice of milk specimens is based on the fact that milk is the most frequent shedding route in chronically infected cows [37]–[39] and the transmission of infection to humans is mainly due to the consumption of raw milk and dairy products [39]–[43].
This work studies the etiological agents of some infectious causes of bovine abortion during mid- to late-gestation in Tunisian dairy herds. The high percentage of positive cases for B. abortus detected in the present study both through RBPT (31.3%) and PCR (30.6%) was unexpected. Quantitatively, the current results are relatively similar to other studies based on the detection of Brucella in African veterinary samples by serology tests [44]–[46] and by the IS711 real-time PCR [47]. There is a strong correlation between the DNA copies (Ct values) obtained using the real-time PCR on vaginal swab samples and the reactions intensity obtained by the RBPT (p<0.0001) as previously reported by Abdalla and Hamid [47] on Sudanese bovine samples. Using RBPT, five controls were diagnosed as positive versus none by real-time PCR. This difference between the two tests was not significant (p value = 0.058). Incorrect timing or different site of sampling may explain observed false negative results. RBPT may also give false positive results because (i) the antibody response to vaccination cannot be differentiated from the one observed after field infection and (ii) cross-reactivity seen with other bacteria are a well-known problem in serological diagnosis of brucellosis. The real-time PCR set up in this study detected more cases of bovine brucellosis in vaginal swab samples (30.6%) than in milk (14%). Our results suggest that Brucella may be more readily detected in vaginal samples than in milk (30.6% vs 14%, p value = 0.0004) (Table 3), a statistically significant finding. Reports on the ability to detect Brucella spp. in milk samples from infected animals by PCR can be somewhat conflicting when compared to standard serological [48] and bacteriological methods. Thus, the results of our study confirm that milk is not the best sample to detect Brucella DNA. No correlation was found between B. abortus positivity and age, herd size and breeding system. Considering abortion as the only clinical sign, there was an association between the occurrence of abortion and positive results by the RBPT (31.3% vs 7.81%, p<0.0001) and real-time PCR (30.6% vs 0%, P<0.0001) (Table 3), a statistically significant and clinically important finding. The percentage of Brucella contamination showed that brucellosis is endemic in cows from different regions of Sfax, despite the extensive vaccination programs implemented as one of the main control measures.
Chlamydiosis seem also to play an important role in bovine abortion in Tunisia, since twenty-seven samples out of 150 (18%) were found to be positive by real-time PCR. The combination of real-time PCR and two commercial diagnostic kits identified 7 positive cases for C. abortus (n = 4) and C. pecorum (n = 3). Using the specific real-time PCR, three samples were shown to be infected by both C. abortus and B. abortus. The association of these two known abortigenic agents was not described yet. In these cases of mixed infection, it remains unclear if both or only one is the etiological agent of bovine abortion. The 3 cases positive for C. pecorum were negatives for all other agents investigated in this study. C. pecorum is known as the causing agent of conjunctivitis, arthritis, pulmonary inflammation and fertility disorders in Tunisian ruminants and could also be detected in ovine placenta possibly leading to abortion [49]. On the other hand, C. pecorum occurs in asymptomatic gastrointestinal infections of ruminants. Thus, a transmission of infection from sheep to cattle cannot be excluded especially in areas where there is a contact between the two animal species. Chlamydia-related organisms seem to play an important role in bovine abortion since 20 samples were positive (13.3%). Similar results were given by Swiss studies based on the detection of Chlamydia-related organisms in veterinary samples using another PCR [50]. The involvement of W. chondrophila in abortion in Tunisia was previously confirmed in a previous work [12]. In the present study, 8 (5.33%) bovine vaginal samples were also positive for P. acanthamoebae in the subset of 150 samples. The results for all 8 positive samples were confirmed by the P. acanthamoebae PCR previously described by Casson et al. [51]. This is the first description of this Chlamydia-related organism in bovine abortion in African cattle. All positive samples were collected from cows belonging to a same herd including 1500 dairy cows. The percentage of abortion in this herd was 12% during 2011 knowing that all cows in this herd were vaccinated against Brucella, Bovine Viral Diarrhoea (BVD) and Infectious Bovine Rhinotrachetis (IBR) viruses. These 8 samples were however negatives for C. abortus, C. pecorum, B. melitensis, C. burnetii, L. monocytogenes, Salmonella spp., Campylobacter spp., BVD and IBR Viruses. The co-infection of P. acanthamoebae and B. abortus (three cases) has not been described yet. Again it remains unclear if both or only one agent caused bovine abortion. For the five remaining cases, no other agents have been detected and no other conditions explaining abortion were documented. Thus, our data suggest that P. acanthamoebae is a possible factor in some cases of abortion. This is consistent with a Swiss work that identified P. acanthamoebae in placenta of aborted bovines by PCR, immunohistochemistry and electron microscopy [13]. Based on the present work, Chlamydia and other Chlamydia-related bacteria do not appear to be excreted in the milk of cows. This is in concordance with the reports of other authors stating that shedding of the C. abortus in milk is possible but uncommon [52]. It is likely that the excretion of Chlamydia organisms was more strongly manifested with vaginal discharge than with milk (18% vs 0%, p<0.0001), a statistically significant and clinically important finding.
L. monocytogenes is another agent that may be involved in bovine abortion. This is the first time that bovine abortion samples from Tunisia were investigated for L. monocytogenes. Besides the 2 cases of mixed infection with L. moncytogenes and W. chondrophila already described above [12], 5 new cases positive for L. monocytogenes were found, in which no other agents have been detected. There was no association between the occurrence of abortion and positive results for this bacterium by real-time PCR (4.6% vs 0%, p value = 0.08). In addition, our results suggest that Listeria may be readily detectable in vaginal samples and also in milk (2.6% vs 2%, p value = 0.5). Infection with L. monocytogenes is transmissible to humans through milk and milk products [53], in which case it can cause systemic and often fatal diseases. There are other agents such as Salmonella that may be involved in cattle abortion and can be transmitted to humans through the consumption of milk. In this study, S. enterica Typhimurium was detected only in 5 milk samples taken from 5 cows that suffered an abortion. In these 5 cases, B. abortus DNA was also detected by a specific real-time PCR in vaginal and milk samples. When mixed infection occurs, it remains unclear if both or only one agent caused bovine abortion. If we consider abortion as the only clinical sign, there is no association between the occurrence of abortion and positive cases for Salmonella by real-time PCR (3.3% vs 0%, p value = 0.1658). In many previous studies, it was revealed that abortion was a quite unusual clinical finding of S. enterica Typhimurium infection in cattle because it hasn't a higher potential for systemic dissemination in body [54]. S. enterica Typhimurium is often associated with enteritis that usually affects young calves, resulting in marked acute diarrhea [55]. All these data suggest that B. abortus may be the most probable agent of abortion in these 5 cows belonging to the same herd located in the south of Sfax. Unlike to other agents cited above, Campylobacter spp. and C. burnetii were not detected in the limited number of cases studied. This is in contrast with previous studies in ruminants and human beings that supported a role of C. burnetii in different mammals. In fact, the seroprevalence of Q fever in sheep flocks in different Tunisian regions was 40% and C. burnetii was the abortigenic agent in 17% of sheep and goats [56]. C. burnetii does not seem to have a crucial role in bovine abortion in contrast to the small ruminants. Previous studies already showed negligible percentages for the involvement of Campylobacter spp. in bovine abortion [34], [57]. More samples should be collected to survey the infections caused by these two bacteria in dairy herds.
This study highlighted the real prevalence of brucellosis and the presence of diseases that are not subject to routine control diagnosis, such as chlamydiosis, salmonellosis and listeriosis. It also demonstrated the importance of Chlamydia and Chlamydia-related organisms as major causes of abortion. Strong evidence suggests that P. acanthamoebae should be considered as a new abortigenic agent in Africa. As well, this study underlined the notion of co-infection, which emphasizes the great interest of a differential diagnosis of different abortion causes.
Supporting Information
Brucella spp. real-time PCR, direct amplification from bovine vaginal samples.
(TIF)
Brucella spp. real-time PCR, direct amplification from bovine milk samples.
(TIF)
Pan- Chlamydiales real-time PCR, direct amplification from bovine vaginal samples.
(TIF)
Listeria monocytogenes real-time PCR melt curve data and 3% agarose gel images for determining primer specificity and product size.
(TIF)
Salmonella spp. real-time PCR melt curve data and 1.5% agarose gel image for determining primer specificity and product size.
(TIF)
Acknowledgments
The authors are grateful to Pr. Néji Gharsallah fron the Faculty of Sciences of Sfax (University of Sfax) for his fruitful discussions.
Funding Statement
This work received financial support from “Ministère de l'enseignement supérieur et de la recherche scientifique” and the ISESCO Organization. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Givens MD (2006) A clinical, evidence-based approach to infectious causes of infertility in beef cattle. Theriogenology 66: 648–654. [DOI] [PubMed] [Google Scholar]
- 2.Ortega-Mora LM, Gottstein B, Conraths FJ, Buxton D (2007) Ruminants: Guidelines for Diagnosis and Control in Farm Protozoal Abortion. CAB International, UK. [Google Scholar]
- 3. Tramuta C, Lacerenza D, Zoppi S, Goria M, Dondo A, et al. (2011) Development of a set of multiplex standard polymerase chain reaction assays for the identification of infectious agents from aborted bovine clinical samples. J Vet Diagn Invest 23 (4) 657–664. [DOI] [PubMed] [Google Scholar]
- 4. Yang N, Cui X, Qian W, Yu S, Liu Qs (2012) Survey of nine abortifacient infectious agents in aborted bovine fetuses from dairy farms in Beijing, China, by PCR. Acta Vet Hung 60 (1) 83–92. [DOI] [PubMed] [Google Scholar]
- 5. Boukary AR, Saegerman C, Abatih E, Fretin D, Alambédji Bada R, et al. (2013) Seroprevalence and potential risk factors for Brucella spp. Infection in traditional cattle, sheep and goats reared in urban, periurban and rural areas of niger. PLoS One 8 (12) e83175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radostits M, Gay C, Hinchcliff W, Constable D (2007) Veterinary Medicine, A text book of the diseases of cattle, horses, sheep, pigs and goats. 10th ed. Grafos, S.A. Arte Sobre Papel, Spain. [Google Scholar]
- 7. Ahmad MA, Adbelsalam QT, Mustafa MA, Mohammed MA (2009) Seroprevalence and risk factors for bovine brucellosis. Jordan Journal of Veterinary Science 10: 61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Storz J, Kaltenboeck B (1993) The Chlamydiales. In: Woldehiwet, Z., Ristic, M. (Eds.), Rickettsial and Chlamydial Diseases of Domestic Animals. Pergamon Press, Oxford, UK, pp. 363–393. [Google Scholar]
- 9. Holliman A, Daniel RG, Parr JG, Griffiths PC, Bevan BJ, et al. (1994) Chlamydiosis and abortion in a dairy herd. Vet Rec 134: 500–502. [DOI] [PubMed] [Google Scholar]
- 10. Livngstone M, Longbottom D (2006) What is the prevalence and economic impact of chlamydial infections in cattle? The need to validate and harmonise existing methods of detection. Vet J 172 (1) 3–5. [DOI] [PubMed] [Google Scholar]
- 11. Baud D, Goy G, Osterheld MC, Borel N, Vial Y, et al. (2011) Waddlia chondrophila: from bovine abortion to human miscarriage. Clin Infect Dis 52 (12) 1469–1471. [DOI] [PubMed] [Google Scholar]
- 12. Barkallah M, Fendri I, Dhieb A, Gharbi Y, Greub G, et al. (2013) First detection of Waddlia chondrophila in Africa using SYBR Green real-time PCR on veterinary samples. Vet Microbiol 164 (1–2) 101–107. [DOI] [PubMed] [Google Scholar]
- 13. Borel N, Ruhl S, Casson N, Kaiser C, Pospischil A, et al. (2007) Parachlamydia spp. and related Chlamydia-like organisms and bovine abortion. Emerg Infect Dis 13 (12) 1904–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blumer S, Greub G, Waldvogel A, Hässig M, Thoma R, et al. (2011) Waddlia, Parachlamydia and Chlamydiaceae in bovine abortion. Vet Microbiol 152 (3–4) 385–393. [DOI] [PubMed] [Google Scholar]
- 15. Wheelhouse N, Howie F, Gidlow J, Greub G, Dagleish M, et al. (2012) Involvement of Parachlamydia in bovine abortions in Scotland. Vet J 193 (2) 586–588. [DOI] [PubMed] [Google Scholar]
- 16. Baud D, Regan L, Greub G (2008) Emerging role of Chlamydia and Chlamydia-like organisms in adverse pregnancy outcomes. Curr Opin Infect Dis 21 (1) 70–76. [DOI] [PubMed] [Google Scholar]
- 17. Richardson A (1975) Salmonellosis in cattle. Vet Rec 96 (15) 329–331. [DOI] [PubMed] [Google Scholar]
- 18. Rings DM (1985) Salmonellosis in calves. Veterinary Clinics of North America Food Animal Practice 1: 529–539. [DOI] [PubMed] [Google Scholar]
- 19. Mshelia GD, Amin JD, Egwu GO, Yavari CA, Murray RD, et al. (2010) Detection of antibodies specific to Campylobacter fetus subsp. venerealis inthe vaginal mucus of Nigerian breeding cows. Vet Ital 46 (3) 337–344. [PubMed] [Google Scholar]
- 20. Nielsen KH, Duncan JR (1990) Detection of Brucella cells and cell components. Animal brucellosis 5: 97–120 CRC Press, Boca Raton, FL cells and cell components. [Google Scholar]
- 21.Kirkbride CA (1990) Laboratory diagnosis of abortion caused by various species of bacteria. Laboratory diagnosis of livestock abortion, ed. Kirkbride CA 17–19. Iowa State University Press, Ames, IA. [Google Scholar]
- 22. Peter O, Dupuis M, Peacock MG, Burgdorfer W (1987) Comparison of enzyme-linked immunosorbent assay and complement fixation and indirect fluorescent-antibody test for detection of Coxiella burnetii antibody. J Clin Microbiol 25: 1063–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Storz J (1988) Overview of animal diseases induced by chlamydial infections. Microbiology of Chlamydia, ed. Barron AL, 167–192. CRC Press, Boca Raton, FL. [Google Scholar]
- 24. On SL, Holmes B (1991) Reproducibility of tolerance tests that are useful in the identification of campylobacteria. J Clin Microbiol 29: 1785–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Iraola G, Hernández M, Calleros L, Paolicchi F, Silveyra S, et al. (2012) Application of a multiplex PCR assay for Campylobacter fetus detection and subspecies differentiation in uncultured samples of aborted bovine fetuses. J Vet Sci 13 (4) 371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anderson ML (2007) Infectious causes of bovine abortion during mid- to late-gestation. Theriogenology 68: 474–486. [DOI] [PubMed] [Google Scholar]
- 27. Lienard J, Croxatto A, Aeby S, Jaton K, Posfay-Barbe K, et al. (2011) Development of a new Chlamydiales-specific real-time PCR and its application to respiratory clinical samples. J Clin Microbiol 49 (7) 2637–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pantchev A, Sting R, Bauerfeind R, Tyczka J, Sachse K (2010) Detection of all Chlamydophila and Chlamydia spp. of veterinary interest using species-specific real-time PCR assays. Comp Immunol Microbiol Infect Dis 33 (6) 473–484. [DOI] [PubMed] [Google Scholar]
- 29.Alton GG, Jones LM, Pietz DE (1975) Laboratory techniques in brucellosis. World Health Organization (WHO) monogram series No. 55. WHO, Geneva, 1–163. [PubMed] [Google Scholar]
- 30. Hinic V, Brodard I, Thomann A, Cvetnic Z, Makaya PV, et al. (2008) Novel identification and differentiation of Brucella melitensis, B. abortus, B. suis, B. ovis, B. canis, and B. neotomae suitable for both conventional and real-time PCR systems. J Microbiol Methods 75 (2) 375–378. [DOI] [PubMed] [Google Scholar]
- 31. Petit T, Spergser J, Aurich J, Rosengarten R (2008) Prevalence of Chlamydiaceae and Mollicutes onthe genital mucosa and serological findings in dairy cattle. Vet Microbiol 18: 127 (3–4) 325–333. [DOI] [PubMed] [Google Scholar]
- 32. Gutierrez J, Williams EJ, O'Donovan J, Brady C, Proctor AF, et al. (2011) Monitoring clinical outcomes, pathological changes and shedding of Chlamydophila abortus following experimental challenge of periparturient ewes utilizing the natural route of infection. Vet Microbiol 147 (1–2) 119–126. [DOI] [PubMed] [Google Scholar]
- 33. Natale A, Bucci G, Capello K, Barberio A, Tavella A, et al. (2012) Old and new diagnostic approaches for Q fever diagnosis: correlation among serological (CFT, ELISA) and molecular analyses. Comp Immunol Microbiol Infect Dis 35 (4) 375–379. [DOI] [PubMed] [Google Scholar]
- 34. Mshelia GD, Amin JD, Egwu GO, Woldehiwet Z, Murray RD (2012) The prevalence of bovine venereal campylobacteriosis in cattle herds in the Lake Chad basin of Nigeria. Trop Anim Health Prod 44 (7) 1487–1489. [DOI] [PubMed] [Google Scholar]
- 35. Poudel A, Elsasser TH, Rahman KhS, Chowdhury EU, Kaltenboeck B (2012) Asymptomatic endemic Chlamydia pecorum infections reduce growth rates in calves by up to 48 percent. PLoS One 7 (9) e44961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rekiki A (2004) Chlamydiose abortive en Tunisie : Tests de Diagnostic, caracterisation moleculaire, phylogenetique de souches de Chlamydophila et étude de la protection du vaccin vivant 1B, Th. : Sciences de la Vie et de la Sante : Tours, U. F. R. 233.
- 37. Rodolakis A, Berri M, Héchard C, Caudron C, Souriau A, et al. (2007) Comparaison of Coxiella burnetii shedding in milk of dairy bovine, caprine, and ovine herds. J Dairy Sci 90 (12) 5352–5360. [DOI] [PubMed] [Google Scholar]
- 38. Guatteo R, Joly A, Beaudeau F (2012) Shedding and serological patterns of dairy cows following abortions associated with Coxiella burnetti DNA detection. Vet Microbiol 155 (2–4) 430–433. [DOI] [PubMed] [Google Scholar]
- 39. Ning P, Guo K, Xu L, Xu R, Zhang C, et al. (2012) Evaluation of Brucella infection of cows by PCR detection of Brucella DNA in raw milk. J Dairy Sci 95: 4863–4867. [DOI] [PubMed] [Google Scholar]
- 40. Fishbein DB, Raoult D (1999) A cluster of Coxiella burnetii infections associated with the exposure to vaccinated goats and their unpasteurised dairy products. Amer J of Trop Med 247: 35 40. [DOI] [PubMed] [Google Scholar]
- 41. Centers for Disease Control and Prevention (CDC) (2008a) Outbreak of multidrug-resistant salmonella enterica serotype Newport infections associated with consumption of unpasteurized Mexican-style aged cheese-Illinois, March 2006–April 2007. Morb Mort Wkly Rep 57: 432–435. [PubMed] [Google Scholar]
- 42. Centers for Disease Control and Prevention (CDC) (2008b) Outbreak of Listeria monocytogenes infections associated with pasteurized milk from a local dairy-Massachusetts, 2007. Morb Mort Wkly Rep 57: 1097–1100. [PubMed] [Google Scholar]
- 43. Alter T, Bereswill S, Glünder G, Haag LM, Hänel I, et al. (2011) Campylobacteriosis of man: livestock as reservoir for Campylobacter species. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 54 (6) 728–734. [DOI] [PubMed] [Google Scholar]
- 44. Aggad H, Boukraa L (2006) Prevalence of bovine and human brucellosis in western Algeria: comparison of screening tests. East Mediterr Health J 12 (1–2) 119–128. [PubMed] [Google Scholar]
- 45. Makita K, Fèvre EM, Waiswa C, Eisler MC, Thrusfield M, et al. (2011) Herd prevalence of bovine brucellosis and analysis of risk factors in cattle in urban and peri-urban areas of the Kampala economic zone, Uganda. BMC Vet Res 7: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dean AS, Bonfoh B, Kulo AE, Boukaya GA, Amidou M, et al. (2013) Epidemiology of brucellosis and Q fever in linked human and animal populations in northern togo. PLoS One 8 (8) 71501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Abdalla A, Hamid ME (2012) Comparaison of conventional and non-conventional techniques for diagnosis of bovine brucellosis in Sudan. Trop Anim Health Prod 44 (6) 1151–1155. [DOI] [PubMed] [Google Scholar]
- 48. Romero C, Pardo M, Grillo MJ, Diaz R, Blasco JM, et al. (1995) Evaluation of PCR and indirect enzyme-linked immunosorbent assay on milk samples for diagnosis of brucellosis in dairy cattle. J Clin Microbiol 33 (12) 3198–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Berri M, Rekiki A, Boumedine KS, Rodolakis A (2009) Simultaneous differential detection of Chlamydophila abortus, Chlamydophila pecorum and Coxiella burnetii from aborted ruminant's clinical samples using multiplex PCR. BMC Microbiol 9: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Borel N, Thoma R, Spaeni P, Weilenmann R, Teankum K, et al. (2006) Chlamydia-related abortions in Cattle from Graubunden, Switzerland. Vet Pathol 43: 702–708. [DOI] [PubMed] [Google Scholar]
- 51. Casson N, Posfay-Barbe KM, Gervaix A, Greub G (2008) New diagnostic real-time PCR for specific detection of Parachlamydia acanthamoebae DNA in clinical samples. J Clin Microbiol 46 (4) 1491–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Thomas R, Davison HC, Wilsmore AJ (1990) Use of the IDEIA ELISA to detect Chlamydia psittaci (ovis) in material from aborted fetal membranes and milk from ewes affected by ovine enzootic abortion. Br Vet J 146 (4) 364–367. [DOI] [PubMed] [Google Scholar]
- 53. Okwumabua O, O'Connor M, Shull E, Strelow K, Hamacher M, et al. (2005) Characterization of Listeria monocytogenes isolates from food animal clinical cases: PFGE patter similarity to strains from human listeriosis cases. FEMS Microbiol Lett 249 (2) 275–281. [DOI] [PubMed] [Google Scholar]
- 54. Carrique-Mas JJ, Willmington JA, Papadopoulou C, Watson EN, Davies RH (2010) Salmonella infection in cattle in Great Britain, 2003 to 2008. Vet Rec 167 (15) 560–565. [DOI] [PubMed] [Google Scholar]
- 55. Rankin JD, Taylor RJ (1966) The estimation of doses of Salmonella typhimurium suitable for the experimental production of disease in calves. Vet Rec 78 (21) 706–707. [DOI] [PubMed] [Google Scholar]
- 56. Rekiki A, Thabti F, Dlissi I, Russo P, Sanchis R, et al. (2005) Enquête sérologique sur les principales causes d'avortements infectieux chez les petits ruminants en Tunisie. Revue Méd Vet 156 (7) 395–401. [Google Scholar]
- 57. Njiro SM, Kidanemariam AG, Tsotetsi AM, Katsande TC, Mnisi M, et al. (2011) A study of some infectious causes of reproductive disorders in cattle owned by resource-poor farmers in Gauteng Province, South Africa. J S Afr Vet Assoc 82 (4) 213–218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Brucella spp. real-time PCR, direct amplification from bovine vaginal samples.
(TIF)
Brucella spp. real-time PCR, direct amplification from bovine milk samples.
(TIF)
Pan- Chlamydiales real-time PCR, direct amplification from bovine vaginal samples.
(TIF)
Listeria monocytogenes real-time PCR melt curve data and 3% agarose gel images for determining primer specificity and product size.
(TIF)
Salmonella spp. real-time PCR melt curve data and 1.5% agarose gel image for determining primer specificity and product size.
(TIF)


