Abstract
Denitrification is an important pathway for nitrogen removal from aquatic systems and this could benefit water quality. However, little is known about the denitrifier community composition and key steps of denitrification in the freshwater environments, and whether different bacteria have a role in multiple processes of denitrification reduction. In this study, quantitative PCR, quantitative RT-PCR, clone library and 454 pyrosequencing were used together to investigate the bacterial and denitrifier community in a subtropical deep reservoir during the strongly stratified period. Our results indicated that the narG gene recorded the highest abundance among the denitrifying genes (2.76×109 copies L−1 for DNA and 4.19×108 copies L−1 for RNA), and the lowest value was nosZ gene (7.56×105 copies L−1 for DNA and undetected for RNA). The RNA: DNA ratios indicated that narG gene was the most active denitrifying gene in the oxygen minimum zone of Dongzhen Reservoir. Further, α-, β- and γ- Proteobacteria were the overwhelmingly dominant classes of denitrifier communities. Each functional gene had its own dominant groups which were different at the genus level: the narG gene was dominated by Albidiferax, while nirS gene was dominated by Dechloromonas. The main OTU of nirK gene was Rhodopseudomonas palustris, but for norB and nosZ genes, they were Bacillus and Bradyrhizobium, respectively. These results contribute to the understanding of linkages between denitrifier community, function and how they work together to complete the denitrification process. Studies on denitrifier community and activity may be useful in managing stratified reservoirs for the ecosystem services and aiding in constructing nitrogen budgets.
Introduction
Nitrogen (N), the fourth most abundant element (after oxygen, carbon and hydrogen) in microorganisms, is essential for the synthesis of nucleic acids and proteins [1]. It is commonly found as amine or amide groups in organic matter but is readily oxidized or reduced and thus, has an additional significance in aquatic systems as both an electron acceptor and a donor for energy metabolism [2]. All living organisms require a mass of nitrogen. Depending on the life form, nitrogen constitutes approximately 14% of the weight of a microbial cell [3]. Even though nitrogen is the overwhelmingly abundant element in the atmosphere, nitrogen gas (N2) is virtually inert. Most organisms cannot fix nitrogen but rather obtain their nitrogen directly as NH4 + (or organic nitrogen) from the environment or from the reduction of NO3 − to NH4 + through assimilatory nitrate reduction [4]. However, anthropogenic influences on the biogeochemistry of nitrogen for instance, combustion of fossil fuels, production of nitrogen fertilizers, cultivation of nitrogen-fixing legumes, and other actions have resulted in major changes in the earth's nitrogen cycle [5]. Whilst the production and industrial use of artificial nitrogen fertilizers worldwide has enabled humankind to greatly increase food production, consequently it has also led to a host of environmental problems, especially potential eutrophication of freshwater and coastal ecosystems [6].
Denitrification is a dissimilatory process in which oxidized nitrogen is used as an alternative electron acceptor for energy production when oxygen is limiting [7]. This process in the ocean consists of four reaction steps in which denitrifiers respire nitrate (NO3 −) sequentially to nitrite (NO2 −), nitric and nitrous oxides (NO, N2O) and finally to N2 [8]. It plays an important role in nitrogen transformation and removal and is responsible for the return of fixed nitrogen back to the atmosphere [9]. Alexander et al [10] pointed out that more than 75% of the anthropogenic nitrogen (N) entering watersheds were lost along landscape flow paths before reaching the oceans by heterotrophic denitrification. Thus, heterotrophic denitrification is an important microbial process in terms of water quality [11]. However, denitrification is also a large source of nitrous oxide (N2O) emissions in terrestrial and aquatic ecosystems, because the N2O is a potent greenhouse gas that contributes to climate change and stratospheric ozone destruction [12]. Thus only when the nitrate reduction process proceeds down to the production of N2 will it benefit the water-atmosphere systems.
Lakes and reservoirs are now being recognized as an important location for the removal of nitrogen from the lacustrine environment due to long water residence times and high biological activity in the oxygen minimum zone (OMZ) [13]. The oxygen concentrations in the OMZ are low enough to induce anaerobic metabolism and 20% to 40% of the total loss of nitrogen is estimated to occur in these zones [14]. In fact, denitrification is a facultative anaerobic microbial process [15]. The denitrifiers are known to belong to more than 50 genera of phylogenetic bacteria, including members of the Proteobacteria, Firmicutes, Actinobacteria, Bacteroides, and Planctomyces [16]. As many denitrifying organisms are unable to produce the entire suite of enzymes to complete the denitrification reduction processes, other organisms within the community are required to cooperate to complete the process [17]. Molecular techniques targeting functional genes that code for the enzymes in nitrate reduction processes have been established as molecular markers [18]. Over the past decades, narG (encoding the membrane-bound nitrate reductase), nirS (encoding the cytochrome cd1-containing nitrite reductase), nirK (encoding the Cu-containing nitrite reductase), norB (encoding nitric oxide reductase) and nosZ (encoding the nitrous oxide reductase) genes had been widely used to describe denitrifier communities [19]–[22].
In the present study, the distributions of denitrifier communities were investigated in a typical subtropical deep reservoir, where strong thermal stratification and a clear anaerobic layer marked the autumn season in the bottom water column. Clone library, quantitative PCR, quantitative RT-PCR and 454 pyrosequencing were used together to investigate the bacterial and denitrifier abundance, activity and composition in the freshwater OMZ. We aimed to explore the main steps of denitrification in a freshwater environment, and identify whether different bacteria have different roles in the denitrification reduction processes. This research aims to improve understanding of the linkages between denitrifier community structure, function and how they work together to complete the denitrification process.
Materials and Methods
Ethics statement
No specific permissions were required for these activities. Informed consent was obtained from all participants and this field study did not involve endangered or protected species.
Study sites, sample collection and nucleic acid extraction
Dongzhen Reservoir (25°28′–25°30′N, 118°54′–118°59′E) is a seasonally stratified reservoir located in southeast China. Samples were collected within the main lacustrine zone close to the dam in October 2011 when water was fully stratified. We characterized the water temperature, dissolved oxygen (DO), chlorophyll a (Chl-a), pH, and electrical conductivity (EC) of the water column at 1-m intervals using a multi-parameter water quality analyzer (Hydrolab DS5, Hach Co., USA). The water oxygen minimum zone was identified in the benthic zone (24–36 m) where the oxygen concentration was as low as 0.2 mg/L. Total nitrogen (TN) was determined using a Shimadzu TOC-VCPH analyzer (Shimadzu, Japan). Total phosphorus (TP) was analyzed by spectrophotometry after digestion. Ammonium nitrogen (NH4-N), nitrite and nitrate nitrogen (NOx-N) and phosphate phosphorus (PO4-P) were measured with a Lachat QC8500 Flow Injection Analyzer (Lachat Instruments, USA). Three 1 litre replicate water samples were filtered immediately through a 0.22-μm polycarbonate membrane (47 mm diameter, Millipore, USA). The membranes were frozen at −80°C prior to further molecular analyses. All sampling and instrument casts were made from a station over the deepest area of the reservoir (36 m). Total DNA was extracted directly from the membranes using the FastDNA spin kit (Bio101, USA) according to the manufacturer's instructions. Purified DNA was dissolved in 50 μl ddH2O and stored at −20°C until use. Total RNA was extracted from the membranes using the E.Z.N.A. total RNA kit II (Omega Bio-Tek, USA) following the manufacturer's instructions. After the extraction procedure, RNA was transcribed into complementary DNA using the Takara OneStep RT-PCR kit Version 2.0 following the manufacturer's instructions. Reverse transcription was performed as a 15 min reaction at 37°C terminated by 5 sec incubation at 95°C.
Clone library and sequence analysis
The 16 S rRNA gene and five denitrifying genes (narG, nirS, nirK, norB and nosZ) were amplified from extracted DNA using the primers following Table 1 [23]–[26]. We pooled the three replicates of PCR products from each of the denitrifying functional genes and 16 S rRNA gene for clone libraries analysis. Agarose gel electrophoresis of the 50 μl PCR products were performed prior to purification (QIAquick Gel Extraction Kit, Qiagen). Purified PCR products were ligated into the pMD18-vector (Takara, Japan) and transformed into Escherichia coli DH5α competent cells (Takara, Japan). Positive clones were grown in Luria-Bertan (LB) medium overnight at 37°C. Plasmids containing target gene fragments were identified by agarose gel electrophoresis and sequenced using an automatic capillary sequencer (ABI3730, USA).
Table 1. PCR Primers used in this study.
| Target gene | Primer | Primer sequence | Annealing temperature (°C) | Amplicon size (bp) | Reference |
| Clone library | |||||
| 16 S rRNA | 341F | CCTACGGGNGGCWGCAG | 53 | 465 | [23] |
| 805R | GACTACHVGGGTATCTAATCC | ||||
| NarG | 1960F | TA(CT)GT(GC)GGGCAGGA(AG)AAACTG | 55 | 691 | [24] |
| 2650R | TTYTCRTACCABGTBGC | ||||
| NirS | cd3aF | GT(C/G) AAC GT(C/G) AAG GA(A/G) AC(C/G) GG | 60 | 426 | [25] |
| R3cd | GA(C/G) TTC GG(A/G) TG(C/G) GTC TTG A | ||||
| NirK | F1aCu | ATCATGGT(C\G)CTGCCGCG | 53 | 473 | [25] |
| R3Cu | GCCTCGATCAG(A/G)TTGTGGTT | ||||
| NorB | 2F | GACAARHWVTAYTGGTGGT | 52 | 428 | [26] |
| 7R | CCRTGGSTRWARWARTTSAC | ||||
| NosZ | F | CG(C/T)TGTTC(A/C)TCGACAGCCAG | 58 | 454 | [25] |
| 1622R | CG(G/C)ACCTT(G/C)TTGCC(C/G)T(T/C)GCG | ||||
| Quantitative PCR | |||||
| 16S rRNA | 341F | CCTACGGGNGGCWGCAG | 60 | 175 | [27] |
| 515R | ATTCCG CGG CTG GCA | ||||
| NarG | 1960m2F | TA(CT)GT(GC)GGGCAGGA(AG)AAACTG | 60 | 100 | [27] |
| 2050m2R | CGTAGAAGAAGCTGGTGCTGTT | ||||
| NirS | 2F | TACCACCC(C/G)GA(A/G)CCGCGCGT | 60 | 165 | [19] |
| 3R | GCCGCCGTC(A/G)TG(A/C/G)AGGAA | ||||
| NirK | 876F | ATYGGCGGVAYGGCGA | 60 | 165 | [28] |
| 1040R | GCCTCGATCAGRTTRTGGTT | ||||
| NorB | 2F | GACAARHWVTAYTGGTGGT | 60 | 394 | [26] |
| 6R | TGCAKSARRCCCCABACBCC | ||||
| NosZ | 1F | WCSYTGTTCMTCGACAGCCAG | 60 | 260 | [29] |
| 1R | ATGTCGATCARCTGVKCRTTYTC | ||||
Quantitative PCR and RT-PCR
The successfully sequenced plasmids from clone libraries were extracted using the MiniPrep kit (Qiagen, Germany) and the plasmid concentrations were determined by spectrophotometry using a BioPhotometer (Eppendorf, Germany). Standard primer sets were prepared from linearized plasmid serial dilutions containing between 102 and 1010 of 16 S rRNA gene and denitrifying gene copies calculated directly from the concentration of extracted plasmid. Standard curves were generated by plotting the threshold cycle values versus log10 of the gene copy numbers. The amplification efficiency (E) was estimated using the slope of the standard curve through the following formula: E = (10−1/slope)−1. The efficiency of the PCR was between 95% and 105% in this study. The relation efficiencies of the standard curve with r2 were ≥0.99. The quantitative PCR and quantitative RT-PCR assays were carried out in a volume of 20 μl including 10 μl SYBR Premix Ex Taq, 0.4 μl of ROX Reference Dye II, 0.5 μM of each primer [19], [26], [27], [28], [29], 2 μl of total DNA or cDNA, and 6.8 μl RNase-free water. The thermocycling steps of real-time PCR were run according to the manufacturer's instructions (Takara, Japan). All the measurements were performed and verified in triplicate. Real-time PCR with standard curves was used as the absolute quantification to calculate the concentrations of 16 S rRNA gene and denitrifying functional genes. After evaluation of the analysis parameters, the relation efficiencies for the standard curves were: r2 = 0.992 for 16 S rRNA, 0.990 for narG, 0.993 for nirS, 0.994 for nirK, and 0.992 for nosZ, respectively. The efficiency of the PCR amplification was 103.9% (16 S rRNA), 102.4% (narG), 102.0% (nirS), 101.8% (nirK) and 101.3% (nosZ), respectively. As a useful approach to estimate the gene activity of the bacterial and denitrifier community, we also compared the RNA to DNA ratios of 16 S rRNA and denitrifying genes.
454 pyrosequencing
In order to independently verify our clone library results, 454 pyrosequencing was used to facilitate an in-depth investigation of the bacterial community composition. PCR was performed using 454 sequencing adaptor-linked primers flanking the 16 S rRNA gene V3–V5 region [30]. The 16 S rRNA genes were PCR amplified using broad range bacterial primers. The primers: 357F (CCTACGGGAGGCAGCAG) and 926R (CCGTCAATTCMTTTRAGT) were complemented with 454 adapters and sample specific barcodes. The 50-μl PCR mixture contained 1 μl of the primer set (10 μm each), 0.125 μl (5 U/μl) of Ex Taq DNA polymerase (Takara Bio, Otsu, Japan), 2.5 μl of Ex Taq buffer (20 mM MgCl2), 2 μl of deoxyribonucleotide triphosphate mixture (2.5 mM each, Takara Bio, Otsu, Japan) and 50 ng of DNA template under the following running conditions: initial denaturation at 94°C for 4 min, 25 cycles of 30 s at 94°C, 45 s at 50°C, 1 min at 72°C, and a final elongation step for 8 min at 72°C. PCR products were confirmed using agarose gel electrophoresis and these were subsequently isolated from the gel and purified using a GeneJET gel extraction kit (Thermo Fisher Scientific). Sequencing reactions were performed by utilizing a Roche 454 FLX instrument (Roche, Indianapolis, IN) with Titanium reagents at Personal Biotechnology Company (Shanghai, China). Sequences were processed by using MOTHUR v.1.20.1 [31]. Briefly, any sequences with a length <200 or >1000, mean quality <25, ambiguous bases >1, homopolymer length >6, maximum primer mismatch >0 were removed from further analysis. Sequence reads were clustered to give similarity-based OTUs using a cluster database at high identity with tolerance (cd-hit) at minimum sequence identity set of 97% [32]. The main phylum level data of bacterial communities were used to compare with the clone library analysis results.
Data analysis
For the clone library, the operational taxonomic unit (OTU) accumulation curve, abundance-based coverage estimator (ACE) and Chao1 estimator were calculated in MOTHUR v.1.20.1 [31]. The sequences with similarities greater than 97% were grouped in one OTU. Each sequence was compared with sequences available in GenBank databases using BLAST. Nucleotide Blast was used for the 16 S rRNA gene analysis and for denitrifying genes to search the translated nucleotide database using the translated denitrification nucleotide. The closest relatives were identified for phylogenetic analysis. The identities of our denitrifying fragments ranged from 76% to 99% (average identity of 88.7%) at the nucleotide level. Denitrifier abundances at both genus and phylum levels were measured with 85% and 65% sequence similarity cutoffs [33]. The sequences were realigned and manually edited with the ClustalX aligner and the phylogenetic analyses were performed with the MEGA5.0 software package using the neighbor-joining and maximum-likelihood method. The Jukes-Cantor model numbers at the branches show the bootstrap percentages (above 50% only) after 1000 replications of bootstrap sampling [34].
Nucleotide sequence accession number
All sequence data from this study have been deposited in the public NCBI database (http://www.ncbi.nlm.nih.gov/) under the accession number KF978754-KF978818 and SRR1069588.
Results
Gene abundance, diversity and activity
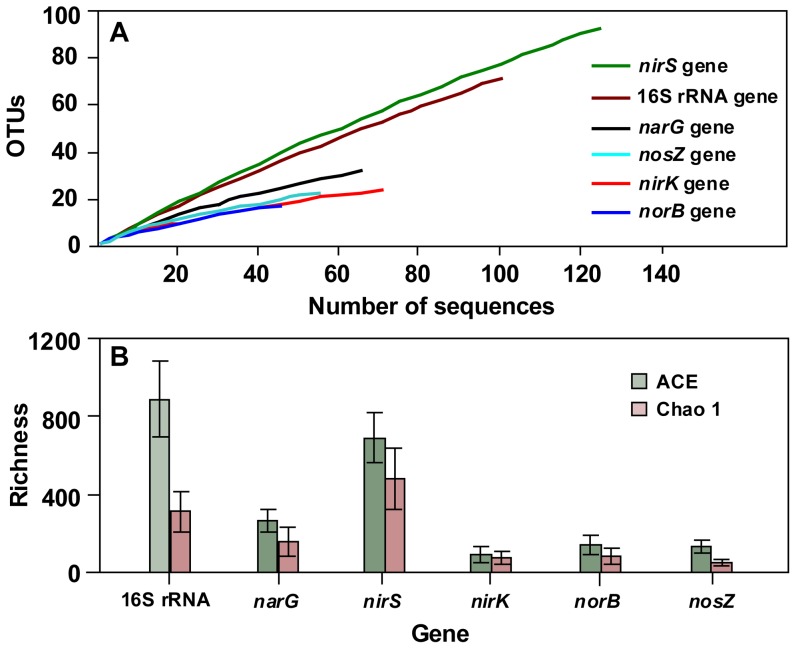
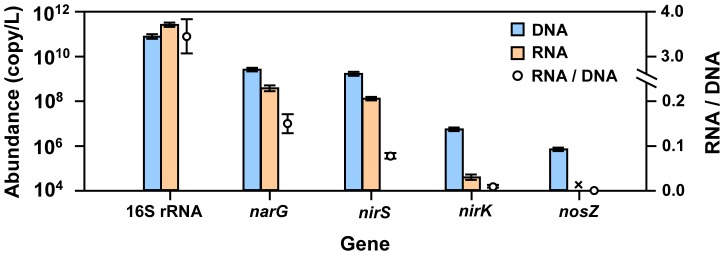
Six clone libraries including 16 S rRNA, narG, nirS, nirK, norB and nosZ genes were successfully constructed (Figure 1). Based on the 99 (16 S rRNA), 64 (narG), 124 (nirS), 69 (nirK), 43 (norB) and 53 (nosZ) sequenced clones, 71, 31, 93, 24, 17 and 22 OTUs were obtained for each gene at the 97% similarity level, respectively (Figure 2A). The order of the denitrifying genetic richness (number of estimated genotypes based on ACE and Chao1) was as follows: nirS > narG > nosZ > norB > nirK for ACE and nirS > narG > nirK > norB > nosZ for Chao1 (Figure 2B). A standard curve was used as a reference to calculate the concentrations of environmental DNA/RNA samples. The narG, nirS, nirK, and nosZ gene copy numbers were lower than the 16 S rRNA gene at both DNA and RNA levels (Figure 3). The norB gene was not detected in our samples. The narG gene recorded the highest abundance among the denitrifying genes in OMZ, and its mean value was 2.76 × 109 copies L−1 for DNA and 4.19 × 108 copies L−1 for RNA, respectively. The lowest value of denitrifying gene was nosZ, its mean value was 7.56× 105 copies L−1 for DNA and undetected for RNA. The order of the denitrifying gene abundance and activity was as follows: narG > nirS > nirK > nosZ. The ratio of ribosomal RNA to DNA can be a good indicator of cellular activity, RNA to DNA ratios (RNA: DNA) were calculated for each gene. The highest RNA: DNA ratios were in the 16 S rRNA (3.452±0.381), followed by decreasing activity in the narG (0.150±0.021), nirS (0.078±0.006), and nirK (0.007±0.001). This indicated that narG gene was the most active denitrifying gene in the oxygen minimum zone of Dongzhen Reservoir.
Figure 1. Scheme for nitrogen transformation from NO3 − to N2 by denitrification.
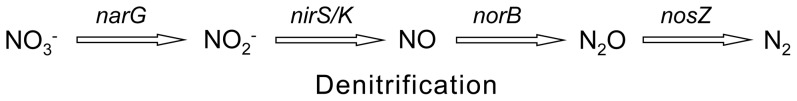
Genes encoding enzymes that mediate the denitrification steps include those for nitrate reductase (narG), nitrite reductase (nirS/nirK), nitric oxide reductase (norB) and nitrous oxide reductase (nosZ).
Figure 2. (A) Rarefaction curves of OTUs, which were defined at 97% sequence similarity for the 16 S rRNA gene and denitrifying gene sequences.
(B) Richness estimates (ACE, Chao1) for six different clone libraries with MOTHUR at 97% similarity level.
Figure 3. Number of 16S rRNA and denitrifying gene copies per litre water and RNA: DNA ratio of five genes in Dongzhen Reservoir.
Error bars indicate standard errors of the three replicates samples with three triplicate qPCR reactions. “x” indicate undetected data from quantitative RT-PCR.
Denitrifying community
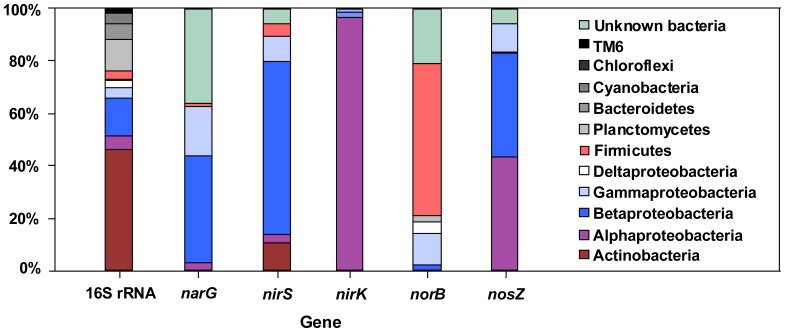
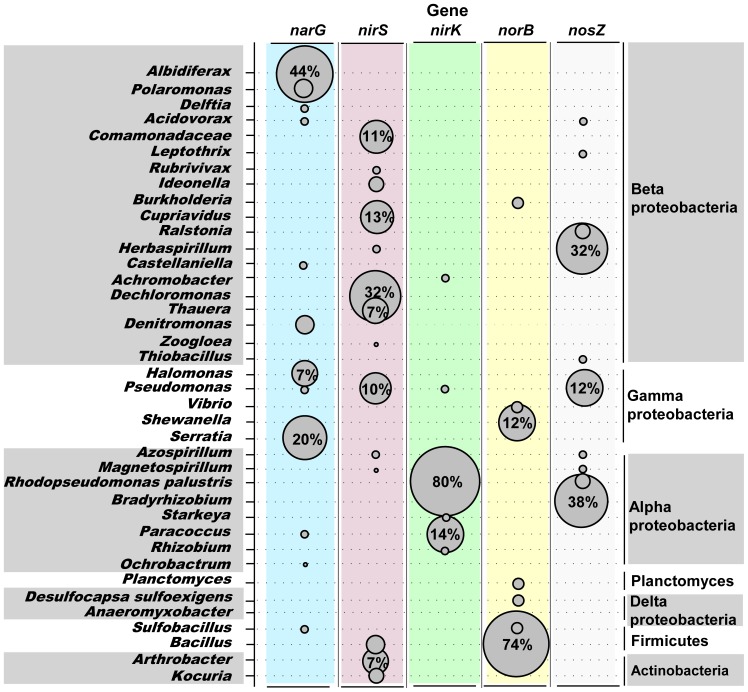
Gene encoding for the membrane-bound nitrate reductase (narG) was distributed among taxonomically diverse bacteria, from α-, β-, γ- Proteobacteria and Firmicutes, and the most represented narG gene group was the β- Proteobacteria (40.6%). The amplified nirS fragments were distributed from α-, β-, γ- Proteobacteria, Actinobacteria and Firmicutes. About 66.1% of the clone library sequences were from the β- Proteobacteria. In contrast to the high richness of nirS, the Cu-containing enzyme (nirK) had 24 OTUs and α- Proteobacteria was the overwhelmingly dominant group which occupied 96.9% of the total sequences. The norB gene was found to be present in the bacteria of β-, γ-, δ- Proteobacteria, Planctomycetes and Firmicutes, and most likely due to the bacterial group of Firmicutes (occupied 58.1% of the total norB gene sequences). For the nosZ gene most bacterial taxa were distributed from α-, β-, γ- Proteobacteria. Strong support groups were observed for α- and β- Proteobacteria (43.2% and 39.6% respectively) (Figure 4). In order to conduct more detailed analysis in the denitrifying community composition, we organized all of the sequencing reads and assigned to the genus and species level. The denitrifier communities were found to be different at the genus/species level. For example, the narG gene was dominated by Albidiferax but the nirS gene was dominated by Dechloromonas. The main OTU of nirK gene was Rhodopseudomonas palustris, but for norB and nosZ genes, they were Bacillus and Bradyrhizobium, respectively. In total, 452 sequences were compared with the GenBank databases one by one, and there were very little two enzymes have been found to coexist in the same denitrifying organism. The same bacterial genus seldom had multi denitrifying genes except only a few genera such as Pseudomonas, Herbaspirillum, Paracoccus (Figure 5).
Figure 4. Relative abundance of different bacterial groups from 16 S rRNA gene and five denitrifying genes based on the clone library analysis.
Figure 5. Relative abundance of different denitrifying genes at the genus (or species) level in Dongzhen Reservoir.
Shown are OTUs assigned to the highest taxonomic level possible using a BLASTN in GenBank database. The circle size corresponds to the relative average abundance OTUs for each denitrifying gene.
Comparison of the microbial community composition obtained using pyrosequencing and clone library
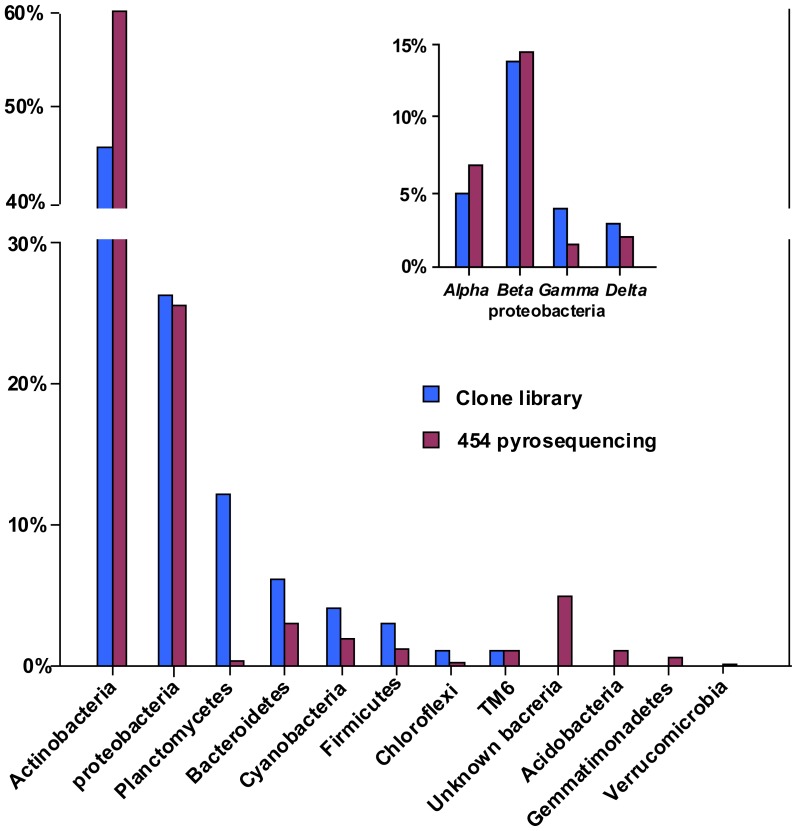
At the phylogenetic phylum level, the predominant bacteria indicated by 454 pyrosequencing and clone library methods were roughly the same proportion (Figure 6). The two methods were both indicated that Actinobacteria was the overwhelmingly dominant bacterial phylum in Dongzhen Reservoir, and α-, β-, δ- and γ- Proteobacteria, Bacteroidetes, Cyanobacteria, Firmicutes and Chloroflexi were found with similar patterns. However, the pyrosequencing detected fewer sequences from the phylum Planctomycetes but more sequences from a broader range for the low abundance bacterial groups (for example, Acidobacteria, Gemmatimonadetes and Verrucomicrobia) compared with the clone library approach (Figure S1 and Figure S2).
Figure 6. Bacterial community compositions at phylum level and Proteobacteria at class level revealed by clone library and 454 pyrosequencing.
Discussion
Nitrate and nitrite reduction as the dominant denitrification process
Denitrification often occurs in specific habitats where oxygen is limited, such as the oxic-anoxic interface of benthic sediments, or in the water column at the edge of suboxic or anoxic water masses in OMZs [35]. Denitrification has been proved to be an important pathway to reduce nitrate loads in aquatic ecosystems [1], [36]. Previous research has shown that different environmental conditions may favor organisms with different key denitrifying genes [2]. For instance, in estuarine wetland sites, the nitrate (narG) and nitrite (nirS) functional genes were relatively dominant during cold and warm seasons, respectively [37]. Further, in rice paddy field soil, the community structures based on nirS and nirK sequences were different among the sampling times and nirK was more abundant than nirS [38]. In the marine ecosystem, N is often a limiting resource and the relative importance of denitrifier communities differs along the environmental N gradient [15]. Our results indicated that in the freshwater OMZ, all five denitrifying genes were present and the narG and nirS genes were identified to be the dominant and diverse denitrification reduction genes. The RNA: DNA ratio of the narG gene was higher than those of three other denitrifying genes, suggesting that narG gene was the most active denitrifying gene in terms of nitrogen loss in the reservoir's OMZ. Further research is warranted to establish whether this is a universal phenomenon in other freshwater ecosystems. Normally, denitrification occurs when three conditions are satisfied: i) N sources for denitrification are available; ii) oxygen concentrations are reduced; iii) electron donors are available [39]. In the present study, the reservoir's OMZ met these conditions. The concentrations of nitrate (NO3 −) and nitrite (NO2 −) (Table 2) in our sampling sites were suitable for encoding microbial narG and nirS genes. Abundance of organic substances that can be used as electron donors for the heterotrophic denitrifiers exists in the reservoir's OMZ. Interestingly, the nosZ gene was detected from extracted DNA with an average copy number of 7.56 × 105 L−1, but it was not detected at the RNA level. A possible explanation could be that the gaseous products of denitrification are the major biological pathway for N loss, then the gaseous intermediates (NO and N2O) would escape from the water to the atmosphere. Finally the nitrous oxide reduction process lacks N2O input as the electron acceptors in the OMZs [2]. The relative activity of the enzymes involved in denitrification may sometimes be affected by denitrifier composition, but in other cases nutrient limitation and environmental factors may be the dominant determinants of activity [15].
Table 2. Environmental variables in the oxygen minimum zone of Dongzhen Reservoir in October 2011.
| Environmental variables | |
| Temperature (°C) | 15.07±0.01 |
| DO (mg L−1) | 0.20±0.00 |
| Chlorophyll a (μg L−1) | 0.53±0.02 |
| EC (mS m−1) | 63.95±4.65 |
| pH | 6.765±0.045 |
| TN (mg L−1) | 8.641±0.197 |
| NH4-N (mg L−1) | 0.410±0.106 |
| NOx-N (mg L−1) | 0.883±0.027 |
| TP (mg L−1) | 0.037±0.002 |
| PO4-P (mg L−1) | 0.026±0.001 |
Values are mean ± SE (n = 3).
Taxonomic diversity of denitrifiers
Our results clearly indicated that each denitrifying gene had its own dominant taxonomic group. The increasing numbers of sequenced bacterial genomes allow a comparison of the composition of denitrifying genes between different taxonomical groups [40]. For the narG gene, the dominant group showed highest similarity to the Albidiferax of β- Proteobacterium, which was originally isolated from sediment and is capable of reducing nitrate to nitrite [41]. Earlier studies of narG diversity in soil environments had previously identified sequences related to those from the Actinobacteria or Proteobacterial classes (α-, β- or γ- Proteobacteria) [42], which were consistent with the results of this study. Both nirS and nirK genes are two functionally-equivalent nitrite reductase enzymes which mediate reduction of nitrite to nitric oxide [2], [43]. However, previous research has shown that denitrifiers possess either nirS or nirK, and no strain is known to harbor both genes and enzymes so far [44], [45]. Our results revealed that the nirS and the nirK genes were different at the class level, nirS was dominated by β- Proteobacteria and nirK was overwhelmingly dominated by α- Proteobacteria (Figure 4 and Figure 5). The norB gene which encodes nitric oxide reductase catalyzes the reduction of NO to N2O, which represents an unusual reaction in biology, the formation of an N-N bond [20]. Our primer sets developed to detect the classes of norB genes and phylogenetic analysis identified nitrifier norB homologues as distinct from other denitrifier sequences. The majority of known denitrifiers which harbor norB genes are α- and β- subdivisions of the Proteobacteria [26], but the dominant group in the present study was Firmicutes. This phenomenon may be linked with environmental specificity [40]. The nosZ cluster has only been identified in the α-, β- and γ- Proteobacteria and no putative nitrous oxide gene cluster has been identified in Gram-positive or Archaebacteria [40]. In summary, our results indicated that the Proteobacteria were identified as the predominant denitrifier in the reservoir OMZ. These denitrifying bacteria were connected by their ability to grow chemolithotrophically at the expense of reduced nitrogen compounds. The distribution of abundant Proteobacterial denitrifiers in the water column may improve the self-purification ability of eutrophic reservoirs.
Conclusions and implications for reservoir management
Denitrification is a stepwise reduction process involving four reaction steps: nitrate reduction, nitrite reduction, nitric oxide reduction, and nitrous oxide reduction [4], [24]. These key steps of the denitrification pathway are catalyzed by nitrite and nitrous oxide reductase [46]. In this study, we have shown a comprehensive view of the denitrifier abundance and composition in an oxygen minimum zone (OMZ) of a subtropical typical stratified reservoir. Proteobacteria were the overwhelmingly dominant phylum in the denitrifier community. The expression of five denitrifying functional genes had nine orders of magnitude of difference. Both narG and nirS gene groups were the dominant and active components of the OMZ denitrifier community with potential direct ties to the denitrification pathway. Previous researches have focused on denitrifying functional genes in marine and soil ecosystems [36]. Our research will enhance the understanding of the denitrification processes in freshwater ecosystems, in particular subtropical deep reservoirs. Normally, phosphorus is the most limiting nutrient in freshwater reservoirs [47], while nitrogen limitation is common in estuaries and oceans [15]. Denitrification is an immensely important process in removing nitrates and nitrites from inland waters before entering the coastal environments and driving eutrophication in those ecosystems. Thus understanding the denitrification in reservoirs along with pathways for nitrogen removal may benefit for water quality. Further research into denitrifier abundance, activity and community structure in freshwater OMZs is needed in order to formulate effective strategies for the water quality protections and improvements of drinking water sources.
Supporting Information
Neighbor-joining phylogenetic tree of 16 S rRNA gene sequences showing the positions of OTUs and taxonomic distribution.
(TIF)
Maximum-likelihood phylogenetic tree of 16 S rRNA gene sequences showing the positions of OTUs and taxonomic distribution.
(TIF)
Acknowledgments
We thank Dmitri Mauquoy and Angela Creevy for improving the English of the manuscript. We also thank Lina Wang for sequences submission and Lin Liu for data analysis assistance.
Funding Statement
This research was supported by the National High-Tech Research and Development Program of China (2012AA062607), the International Science and Technology Cooperation Program of China (2011DFB91710), the Natural Science Foundation for Distinguished Young Scholars of Fujian Province (2012J06009), the National Natural Science Foundation of China (31172114, 31370471). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Conley DJ, Paerl HW, Howarth RW, Boesch DF, Seitzinger SP, et al. (2009) Controlling eutrophication: nitrogen and phosphorus. Science 323: 1014–1015. [DOI] [PubMed] [Google Scholar]
- 2. Zehr JP, Kudela RM (2011) Nitrogen cycle of the open ocean: from genes to ecosystems. Ann Rev Mar Sci 3: 197–225. [DOI] [PubMed] [Google Scholar]
- 3. Vidali M (2001) Bioremediation. An overview. Pure Appl Chem 73: 1163–1172. [Google Scholar]
- 4. Canfield DE, Glazer AN, Falkowski PG (2010) The evolution and future of earth's nitrogen cycle. Science 330: 192–196. [DOI] [PubMed] [Google Scholar]
- 5. Vitousek PM, Aber JD, Howarth RW, Likens GE, Matson PA, et al. (1997) Human alteration of the global nitrogen cycle: sources and consequences. Ecol Appl 7: 737–750. [Google Scholar]
- 6. Gruber N, Galloway JN (2008) An earth-system perspective of the global nitrogen cycle. Nature 451: 293–296. [DOI] [PubMed] [Google Scholar]
- 7. Mounier E, Hallet S, Chèneby D, Benizri E, Gruet Y, et al. (2004) Influence of maize mucilage on the diversity and activity of the denitrifying community. Environ Microbiol 6: 301–312. [DOI] [PubMed] [Google Scholar]
- 8. Ward BB, Devol AH, Rich JJ, Chang BX, Bulow SE, et al. (2009) Denitrification as the dominant nitrogen loss process in the Arabian Sea. Nature 461: 78–81. [DOI] [PubMed] [Google Scholar]
- 9. Piña-Ochoa E, Høgslund S, Geslin E, Cedhagen T, Revsbech NP, et al. (2010) Widespread occurrence of nitrate storage and denitrification among Foraminifera and Gromiida. Proc Natl Acad Sci USA 107: 1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alexander RB, Smith RA, Schwarz GE (2000) Effect of stream channel size on the delivery of nitrogen to the Gulf of Mexico. Nature 403: 758–761. [DOI] [PubMed] [Google Scholar]
- 11. Burgin AJ, Hamilton SK, Jones SE, Lennon JT (2012) Denitrification by sulfur-oxidizing bacteria in a eutrophic lake. Aquat Microb Ecol 66: 283–293. [Google Scholar]
- 12. Beaulieu JJ, Tank JL, Hamilton SK, Wollheim WM, Hall RO, et al. (2011) Nitrous oxide emission from denitrification in stream and river networks. Proc Natl Acad Sci USA 108: 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saunders D, Kalff J (2001) Nitrogen retention in wetlands, lakes and rivers. Hydrobiologia 443: 205–212. [Google Scholar]
- 14.Gruber N (2004) The dynamics of the marine nitrogen cycle and its influence on atmospheric CO2 variations. In: Follows M, Oguz T, editors. The Ocean Carbon Cycle and Climate, NATO ASI series. Kluwer Academic: Dordrecht. pp. 97–148.
- 15. Wallenstein MD, Myrold DD, Firestone M, Voytek M (2006) Environmental controls on denitrifying communities and denitrification rates: insights from molecular methods. Ecol Appl 16: 2143–2152. [DOI] [PubMed] [Google Scholar]
- 16. Zumft WG (1997) Cell biology and molecular basis of denitrification. Microbiol Mol Biol R 61: 533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zumft WG (1993) The biological role of nitric oxide in bacteria. Arch Microbiol 160: 253–264. [DOI] [PubMed] [Google Scholar]
- 18. Smith CJ, Nedwell DB, Dong LF, Osborn AM (2007) Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments. Appl Environ Microb 73: 3612–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Braker G, Fesefeldt A, Witzel KP (1998) Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Environ Microb 64: 3769–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Braker G, Tiedje JM (2003) Nitric oxide reductase (norB) genes from pure cultures and environmental samples. Appl Environ Microb 69: 3476–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scala DJ, Kerkhof LJ (1998) Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiol Lett 162: 61–68. [DOI] [PubMed] [Google Scholar]
- 22. Cheneby D, Hallet S, Mondon M, Martin-Laurent F, Germon J, et al. (2003) Genetic characterization of the nitrate reducing community based on narG nucleotide sequence analysis. Microb Ecol 46: 113–121. [DOI] [PubMed] [Google Scholar]
- 23. Herlemann DP, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, et al. (2011) Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. The ISME J 5: 1571–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Philippot L, Piutti S, Martin-Laurent F, Hallet S, Germon JC (2002) Molecular analysis of the nitrate-reducing community from unplanted and maize-planted soils. Appl Environ Microb 68: 6121–6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Throbäck IN, Enwall K, Jarvis Å, Hallin S (2004) Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol Lett 49: 401–417. [DOI] [PubMed] [Google Scholar]
- 26. Casciotti KL, Ward BB (2005) Phylogenetic analysis of nitric oxide reductase gene homologues from aerobic ammonia-oxidizing bacteria. FEMS Microbiol Lett 52: 197–205. [DOI] [PubMed] [Google Scholar]
- 27. López-Gutiérrez JC, Henry S, Hallet S, Martin-Laurent F, Catroux G, et al. (2004) Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J Microbiol Meth 57: 399–407. [DOI] [PubMed] [Google Scholar]
- 28. Henry S, Baudoin E, López-Gutiérrez JC, Martin-Laurent F, Brauman A, et al. (2004) Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J Microbiol Meth 59: 327–335. [DOI] [PubMed] [Google Scholar]
- 29. Henry S, Bru D, Stres B, Hallet S, Philippot L (2006) Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16 S rRNA, narG, nirK, and nosZ genes in soils. Appl Environ Microb 72: 5181–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sim K, Cox MJ, Wopereis H, Martin R, Knol J, et al. (2012) Improved detection of bifidobacteria with optimised 16 S rRNA gene based pyrosequencing. PLOS ONE 7: e32543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, et al. (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. [DOI] [PubMed] [Google Scholar]
- 33. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, et al. (2011) Enterotypes of the human gut microbiome. Nature 473: 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Naqvi S, Voss M, Montoya J (2008) Recent advances in the biogeochemistry of nitrogen in the ocean. Biogeosciences 5: 1033–1041. [Google Scholar]
- 36. Burgin AJ, Hamilton SK (2007) Have we overemphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Front Ecol Environ 5: 89–96. [Google Scholar]
- 37. Chon K, Chang JS, Lee E, Lee J, Ryu J, et al. (2011) Abundance of denitrifying genes coding for nitrate (narG), nitrite (nirS), and nitrous oxide (nosZ) reductases in estuarine versus wastewater effluent-fed constructed wetlands. Ecol Eng 37: 64–69. [Google Scholar]
- 38. Yoshida M, Ishii S, Otsuka S, Senoo K (2009) Temporal shifts in diversity and quantity of nirS and nirK in a rice paddy field soil. Soil Biol Biochem 41: 2044–2051. [Google Scholar]
- 39. Seitzinger S, Harrison JA, Böhlke J, Bouwman A, Lowrance R, et al. (2006) Denitrification across landscapes and waterscapes: a synthesis. Ecol Appl 16: 2064–2090. [DOI] [PubMed] [Google Scholar]
- 40. Philippot L (2002) Denitrifying genes in bacterial and archaeal genomes. BBA-Gen Struct Expr 1577: 355–376. [DOI] [PubMed] [Google Scholar]
- 41. Finneran KT, Johnsen CV, Lovley DR (2003) Rhodoferax ferrireducens sp. nov., a psychrotolerant, facultatively anaerobic bacterium that oxidizes acetate with the reduction of Fe (III). Int J Syst Evol Micr 53: 669–673. [DOI] [PubMed] [Google Scholar]
- 42. Deiglmayr K, Philippot L, Tscherko D, Kandeler E (2006) Microbial succession of nitrate-reducing bacteria in the rhizosphere of Poa alpina across a glacier foreland in the Central Alps. Environ Microb 8: 1600–1612. [DOI] [PubMed] [Google Scholar]
- 43. Philippot L, Hallin S, Schloter M (2007) Ecology of denitrifying prokaryotes in agricultural soil. Adv Agron 96: 249–305. [Google Scholar]
- 44. Yoshida M, Ishii S, Otsuka S, Senoo K (2009) nirK-harboring denitrifiers are more responsive to denitrification-inducing conditions in rice paddy soil than nirS-harboring bacteria. Microbes Environ 25: 45–48. [DOI] [PubMed] [Google Scholar]
- 45. Angeloni NL, Jankowski KJ, Tuchman NC, Kelly JJ (2006) Effects of an invasive cattail species (Typha × glauca) on sediment nitrogen and microbial community composition in a freshwater wetland. FEMS Microbiol Lett 263: 86–92. [DOI] [PubMed] [Google Scholar]
- 46. Kandeler E, Deiglmayr K, Tscherko D, Bru D, Philippot L (2006) Abundance of narG, nirS, nirK, and nosZ genes of denitrifying bacteria during primary successions of a glacier foreland. Appl Environ Microb 72: 5957–5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang J, Yu XQ, Liu LM, Zhang WJ, Guo PY (2012) Algae community and trophic state of subtropical reservoirs in southeast Fujian, China. Environ Sci Pollut Res 19: 1432–1442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Neighbor-joining phylogenetic tree of 16 S rRNA gene sequences showing the positions of OTUs and taxonomic distribution.
(TIF)
Maximum-likelihood phylogenetic tree of 16 S rRNA gene sequences showing the positions of OTUs and taxonomic distribution.
(TIF)