Abstract
MALDI-TOF MS has been shown capable of rapidly and accurately characterizing bacteria. Highly reproducible spectra are required to ensure reliable characterization. Prior work has shown that spectra acquired manually can have higher reproducibility than those acquired automatically. For this reason, the objective of this study was to optimize automated data acquisition to yield spectra with reproducibility comparable to those acquired manually. Fractional factorial design was used to design experiments for robust optimization of settings, in which values of five parameters (peak selection mass range, signal to noise ratio (S:N), base peak intensity, minimum resolution and number of shots summed) commonly used to facilitate automated data acquisition were varied. Pseudomonas aeruginosa was used as a model bacterium in the designed experiments, and spectra were acquired using an intact cell sample preparation method. Optimum automated data acquisition settings (i.e., those settings yielding the highest reproducibility of replicate mass spectra) were obtained based on statistical analysis of spectra of P. aeruginosa. Finally, spectrum quality and reproducibility obtained from non-optimized and optimized automated data acquisition settings were compared for P. aeruginosa, as well as for two other bacteria, Klebsiella pneumoniae and Serratia marcescens. Results indicated that reproducibility increased from 90% to 97% (p-value 0.002) for P. aeruginosa when more shots were summed and, interestingly, decreased from 95% to 92% (p-value
0.002) for P. aeruginosa when more shots were summed and, interestingly, decreased from 95% to 92% (p-value  0.013) with increased threshold minimum resolution. With regard to spectrum quality, highly reproducible spectra were more likely to have high spectrum quality as measured by several quality metrics, except for base peak resolution. Interaction plots suggest that, in cases of low threshold minimum resolution, high reproducibility can be achieved with fewer shots. Optimization yielded more reproducible spectra than non-optimized settings for all three bacteria.
0.013) with increased threshold minimum resolution. With regard to spectrum quality, highly reproducible spectra were more likely to have high spectrum quality as measured by several quality metrics, except for base peak resolution. Interaction plots suggest that, in cases of low threshold minimum resolution, high reproducibility can be achieved with fewer shots. Optimization yielded more reproducible spectra than non-optimized settings for all three bacteria.
Introduction
Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) has emerged as a rapid and accurate technology to characterize bacteria at the genus and species levels [1]–[7]. Such characterization is based on unique mass spectra associated with different bacteria and obtained by analysis of whole cells or cellular extracts [2], [8]. Peaks represent biological molecules, typically proteins, that originate from cell surfaces, intracellular membranes, and ribosomes [9], [10], and thus are unique and constitute fingerprints. Mass spectra can be acquired either manually [11], [12] or automatically [12], [13], but automated data acquisition can enhance the high-throughput nature of this approach. Due to the rapidity and efficacy of this technique, there has been keen interest in application of MALDI-based approaches to characterize bacteria at the strain and subspecies levels [10], [14]–[18]. Strain level characterization is challenging because strains within a single species are often extremely similar and yield mass spectra with only subtle differences [18], [19]. Several studies have shown that spectra with poor quality and/or low reproducibility may confound bacterial identification and lead to misclassifications [8], [18]–[20]. Consequently, strain level identification is effective only when the reproducibility (i.e., similarity) of replicate spectra of individual strains exceeds that of spectra of unique strains of interest [21].
Because high quality and highly reproducible mass spectra are required to ensure reliable strain level characterization [22], [23], many efforts have been made to optimize data collection conditions to improve spectrum quality and reproducibility. Optimization strategies are generally divided into two categories: optimization of pre-analytical procedures and optimization of post-processing procedures. Many pre-analytical parameters have been reported to influence spectrum quality and reproducibility, including culture age [24], growth medium [25], matrix [26], solvent composition [27], and sample preparation and deposition method [18], [28]. Several of these parameters have been optimized using common univariate approaches in which one variable is changed at a time, and a series of experiments is conducted to determine the optimal condition for each parameter [17], [29]–[31]. Though often effective, optimization based on univariate approaches presents a number of limitations. First, optimization may be not universal, because few studies have tested the resulting optimal condition beyond the species or strains that undergo optimization. Second, univariate-based optimization procedures are time-consuming and labor intensive. Another drawback of univariate approaches is that it is difficult to estimate interaction effects of parameters on reproducibility. With regard to optimization of post-processing criteria (e.g. to optimize Bruker MALDI Biotyper score cutoffs to improve the percentage of bacteria correctly identified [32]), spectrum quality and reproducibility are generally not quantified.
We previously reported that automated data acquisition yielded less reproducible spectra than manual data acquisition [11]. To automate MALDI data acquisition, users specify threshold values of several parameters (e.g., base peak intensity, minimum resolution, signal to noise ratio (S:N), etc.) necessary for the automation algorithm to acquire spectra. We hypothesized that the lower reproducibility associated with automated data acquisition may be due to non-optimized values of data acquisition parameters [11]. In fact, it has been noted previously that automated acquisition settings of the MALDI-TOF mass spectrometer needed to be optimized for better performance of fingerprint-based approaches [2]. Further, effects of data acquisition parameters on reproducibility and quality have not been thoroughly investigated. For each of these reasons, we sought to optimize automated data acquisition. Preliminary work in our lab using univariate approaches did not markedly enhance spectrum reproducibility [11]. As a result, we chose a multivariable method, factorial design of experiments, to characterize and optimize automated data acquisition. Several studies have shown that this approach to statistical design of experiments is an efficient way to provide richer information and extract the maximum amount of information from the most economic number of experiments [33]–[36].
The specific objective of this study was to optimize automated data acquisition to yield spectra with reproducibility comparable to those obtained manually. Pseudomonas aeruginosa, which was shown previously to yield spectra of significantly lower quality and reproducibility when data were acquired via automation than when acquired manually [11], was used as a model bacterium in the designed experiments. Finally, the optimal combination of automated data acquisition parameters we obtained using P. aeruginosa was also tested with Klebsiella pneumoniae and Serratia marcescens. Both of these bacteria are Gram-negative bacteria that showed much lower reproducibility when data were acquired via automation than when acquired manually [11].
Materials and Methods
Bacteria and reagents
Pseudomonas aeruginosa (ATCC 27853), Klebsiella pneumoniae (ATCC 132), and Serratia marcescens (ATCC 13880) were purchased from Carolina Biological Supply Company (Burlington, NC, USA) and stored as freezer stock cultures (nutrient broth:glycerol, 87.5∶12.5) at −80 °C.
Sinapinic acid was purchased from Sigma-Aldrich (St. Louis, MO, USA). Trifluoroacetic acid (TFA) and acetonitrile were purchased from ACROS (Fair Lawn, NJ, USA). MALDI calibrants were purchased from Sigma-Aldrich (St. Louis, MO, USA). Nutrient agar and nutrient broth were purchased from Carolina Biological Supply Company (Burlington, NC, USA).
Parameter selection
Automated data acquisition is typically achieved using a software algorithm which requires the user to specify various parameters. These parameters control the laser power, peak evaluation, mass spectra accumulation, and laser movement on sample. Effects of these parameters on spectrum quality and reproducibility have not been thoroughly studied. Moreover, the values of some of these parameters have not been specified in the literature. For each of these reasons, we evaluated five parameters which are frequently reported in the literature [37]–[41]. The five factors which are adjusted through the Bruker FlexControl software (version 3.0; Bruker Daltonics) during automated data acquisition included: A) peak selection mass range; B) base peak S:N; C) base peak intensity; D) base peak minimum resolution; and E) number of shots summed (Table 1). Peak selection mass range defines the mass range for peak evaluation during automated data acquisition. The base peak is the highest peak observed in the peak selection mass range during automated data acquisition. The value of S:N, intensity and minimum resolution for the base peak must exceed the user-defined levels of these parameters; otherwise, the entire spectrum will be excluded during automated data acquisition.
Table 1. Factors and levels used in the fractional factorial experimental design.
| Level a | ||||
| Name | Factor | −1 | 0 | +1 |
| Peak selection mass range (kDa)b | A | 1 | 4.5 | 8 |
| S:N | B | 1 | 2 | 3 |
| Base peak intensity | C | 100 | 200 | 300 |
| Minimum resolution | D | 100 | 250 | 400 |
| Number of shots summed | E | 100 | 300 | 500 |
−1 represents the low level; 0 represents the center point; +1 represents the high level.
Peak selection mass range represents a continuous mass range. The mid-point of the mass range was 10 kDa. The interval of the mass range was represented by an absolute value. Specifically, the peak selection value for level −1, 0 and +1 were 9 to 11 kDa, 5.5 to 14.5 kDa and 2 to 18 kDa, respectively.
Fractional factorial experimental design
Factorial experimental designs are those which involve two or more factors of interest and where all possible combinations of the factor levels are tested. In this study, spectrum reproducibility was the response. Five independent factors (automated data acquisition parameters) were varied (Table 1). All five factors were numeric type (i.e., values represented by numbers) in the designed experiments.
Factorial-based designs can be separated into two main groups which are full factorial design and fractional factorial design [42]. Different types of factorial design yield different numbers of experiments. For example, if three factors are investigated and each factor has four levels which are to be tested, a full factorial design would require 64 (43) experimental runs. As the number of factors and/or the number of levels of each factor increases, the experimental design becomes prohibitively large [36]. A full factorial design is generally used when the number of factors is small, for example, less than four [43]. If k factors could be set at two levels each, then a 2k factorial design can be implemented. For three factors, each at two levels, a 23 factorial design would require only 8 experimental runs. These designs are often used as screening designs to aid in identifying important factors and interactions [36]. Although using two levels of each factor is efficient, the experimental design can still become quite large as k increases. Therefore, a fractional factorial design denoted as 2k -p (k: the number of factors; p: the fraction index) was used in this study based on assumptions that higher-order interactions are negligible. Higher-order interactions are those which involve three or more factors. In this study, we focused on the interactions involving two factors. Because of the sparsity of effects principle [36], we assumed that the higher-order interactions were negligible and did not need to be estimated. As a result, a 25-1 design was used. This yielded 16 experimental runs. It is important to note the 16 runs are not chosen at random or haphazardly. The 16-run orthogonal design is selected in order to eliminate confounding between main effects and minimize confounding between two-factor interactions [36].
The two levels for each factor were selected as they represent commonly used values [2], [18], [20], [44] and prior work in our lab [11]. Specifically, in the literature, many mass ranges have been used for peak selection, for example, 2 to 20 kDa [11], [18], [40], [41], [45], 2 to 6 kDa [37], 3 to 20 kDa [38], and 7 to 10 kDa [2]. Base peak S:N is usually set as 2 or 3 in literature [2], [11], [38]. Base peak intensity can vary from 100 to 600 [2], [11]. Few studies have specified base peak minimum resolution. In our previous work, this value was set at 400 [3], [11], [18]. The number of shots summed in different studies often varies from 100 to 1000 shots [2], [13], [38]–[41], [46]. Based on these reported values, two levels of each factor were selected and are shown in Table 1. The low level is designated as -1 and the high level is designated at +1 for coding purposes (Table 1).
In addition to the high and low levels of each factor, we also included center points (designated as 0) (Table 1) to assess whether the response changed linearly as the factor moved from its low to high level or if curvature in the response was present. Center points are those experiments where all five factors are set at their center value (Table 1). In this study, three center points were added, resulting in a total of 19 experiments (Table 2). The design of experiment software used in this study was Minitab Statistical Software (version 16) (Minitab Inc, PA, USA).
Table 2. Experimental design matrix of the fractional factorial designs and the resulting reproducibility of mass spectra.
| Factor | ||||||
| Experiment | Peak selection range (kDa) | S:N | Threshold intensity | Minimum resolution | Number of shots summed | Interreplicate imilarity (%)a |
| 1 | 1 | 1 | 300 | 400 | 500 | 95.5±3.4 |
| 2 | 8 | 3 | 100 | 400 | 100 | 86.6±7.9 |
| 3 | 8 | 1 | 300 | 100 | 500 | 97.0±1.4 |
| 4 | 8 | 3 | 300 | 100 | 100 | 92.1±3.4 |
| 5 | 1 | 3 | 300 | 400 | 100 | 87.6±6.0 |
| 6 | 1 | 3 | 300 | 100 | 500 | 97.7±0.9 |
| 7 | 1 | 1 | 100 | 100 | 500 | 98.0±0.9 |
| 8 | 1 | 3 | 100 | 100 | 100 | 92.4±2.3 |
| 9 | 8 | 1 | 100 | 400 | 500 | 96.3±1.5 |
| 10 | 8 | 3 | 300 | 400 | 500 | 97.9±1.1 |
| 11 | 4.5 | 2 | 200 | 250 | 300 | 95.2±3.4 |
| 12 | 8 | 3 | 100 | 100 | 500 | 96.5±2.3 |
| 13 | 8 | 1 | 300 | 400 | 100 | 91.5±4.4 |
| 14 | 1 | 3 | 100 | 400 | 500 | 96.3±2.3 |
| 15 | 1 | 1 | 100 | 400 | 100 | 87.1±7.7 |
| 16 | 4.5 | 2 | 200 | 250 | 300 | 94.3±2.7 |
| 17 | 4.5 | 2 | 200 | 250 | 300 | 95.4±1.8 |
| 18 | 1 | 1 | 300 | 100 | 100 | 92.9±5.3 |
| 19 | 8 | 1 | 100 | 100 | 100 | 92.8±3.0 |
Values reported are the average correlation coefficients of 10 replicates ± the standard deviations of the correlation coefficient.
Sample preparation
A nutrient agar plate was streaked from freezer stock and incubated at 37 °C for 24 hours. A single colony was inoculated into 5 ml sterile nutrient broth, and the broth was incubated at 37°C for 24 hours on an orbital shaker at 200 rpm. Samples were prepared as previously described [11]. Briefly, 1 ml of culture (O.D. 600 = 0.8) was centrifuged at 14,000 × g for 5 minutes. After removal of the supernatant, the cell pellet was resuspended in 1 ml of sterile double-distilled water (ddH2O) (Millipore Corp.; Bedford, MA, USA) and centrifuged again at 14,000 × g for 5 min. The supernatant was decanted and the resulting cell pellet was resuspended in 100 μl of sterile ddH2O. Sinapinic acid matrix solution was prepared as previously described [11]. Equal volumes of cell suspension and matrix solution were mixed. Aliquots (2 μl) of this mixture were spotted onto a MSP 96 ground steel target plate (Bruker Daltonics; Billerica, MA, USA) and allowed to air dry.
MALDI-TOF MS analysis
MALDI-TOF MS analyses were performed using a Bruker Microflex LRF MALDI-TOF mass spectrometer (Bruker Daltonics; Billerica, MA, USA) equipped with a nitrogen laser (λ = 337 nm) under the control of FlexControl software (version 3.0; Bruker Daltonics). Mass spectra were automatically collected in positive linear mode with varying combinations of automated data acquisition parameter values (Table 2). Ion source 1 voltage was set to 20 kV, ion source 2 voltage was set to 18.15 kV, and the lens voltage was set to 9.05 kV. Other parameters were set as described previously [11]. Mass calibration was performed using standard calibrants: ACTH (1–17) 2094.427 Da, ACTH (18–39) 2466.681 Da, Insulin oxidized B 3494.6513 Da, Insulin 5734.518 Da, Cytochrome C 12360.974 Da and Myoglobin 16952.306 Da (Sigma-Aldrich, St. Louis, MO, USA).
Raw spectra were post-processed and peaks were picked using FlexAnalysis software (version 3.0; Bruker Daltonics). Masses from 2 to 20 kDa were used for spectrum evaluation and post-processing. Minimum peak resolution was set at 400 Da. The minimum S:N threshold was set at 2, while the minimum peak intensity threshold was set at 100. Baseline subtraction was performed using the TopHat algorithm [47].
Quantification of spectrum quality and reproducibility
Measures of spectrum quality included base peak intensity, base peak resolution, base peak S:N, number of peaks, and mass range. To quantify reproducibility, peak lists generated by FlexAnalysis were imported into BioNumerics software (version 6.1; Applied Maths; Sint-Martens-Latem, Belgium) using a custom script created by the manufacturers of the software for this application. Similarity coefficients of replicate spectra were calculated using the Pearson product-moment correlation coefficient [48].
Statistical analysis
Each of the 19 runs from the designed experiments (Table 2) consisted of 5 technical replicates of P. aeruginosa. All 19 experiments were carried out on the same day in a randomized order and distribution on the MALDI target, resulting in 95 mass spectra. These spectra constituted one dataset. In total, two datasets were obtained on two consecutive days. Both datasets were subjected to analysis of reproducibility, spectrum quality, main effects, and interactions of factors. Specifically, reproducibility and spectrum quality of each designed experiment were reported using the averaged values of 10 replicates of P. aeruginosa from the two datasets. Main effects and interactions of factors on reproducibility were analyzed based on analysis of variance (ANOVA) and t-tests using a 5% level of significance [36] (Minitab Inc, PA, USA).
Optimization
Most optimization efforts using univariate approaches have not evaluated optimized experimental conditions beyond the species or strains that undergo optimization. We hypothesized that the optimized settings may improve spectrum quality and reproducibility of spectra from bacteria other than P. aeruginosa. Therefore, two other gram negative bacteria, Klebsiella pneumoniae and Serratia marcescens, both of which showed low reproducibility when using non-optimized settings [11], were also analyzed via MALDI using optimized settings. For either optimized or non-optimized settings, 20 spectra were acquired representing 20 technical replicates for each bacterium. The reproducibility and quality of spectra from each bacterium before and after optimization were reported using the averaged values of the corresponding 20 mass spectra. Differences in spectrum quality and reproducibility before and after optimization were identified using t-tests with a 5% level of significance (Minitab Inc, PA, USA).
Results and Discussion
Design matrix and reproducibility
The highest reproducibility achieved for P. aeruginosa using optimized automated data acquisition was 98.0% (Table 2), which is higher than the previously reported value (88.3%) (p-value 0.001) for non-optimized automated data acquisition, and was comparable to the reproducibility (96.1%) obtained manually [11]. The corresponding experimental settings for this high reproducibility were: peak selection mass range = 9 to 11 kDa, S:N = 1, base peak intensity = 100, base peak minimum resolution = 100, and number of shots summed = 500. In contrast, low reproducibility was also observed in these 19 experiments, ranging from 86% to 88%, which was comparable to the previously reported value (88.3%) for non-optimized automated data acquisition [11]. These results show clearly that the values of parameters used in the automated data acquisition procedure influence reproducibility.
0.001) for non-optimized automated data acquisition, and was comparable to the reproducibility (96.1%) obtained manually [11]. The corresponding experimental settings for this high reproducibility were: peak selection mass range = 9 to 11 kDa, S:N = 1, base peak intensity = 100, base peak minimum resolution = 100, and number of shots summed = 500. In contrast, low reproducibility was also observed in these 19 experiments, ranging from 86% to 88%, which was comparable to the previously reported value (88.3%) for non-optimized automated data acquisition [11]. These results show clearly that the values of parameters used in the automated data acquisition procedure influence reproducibility.
Spectrum reproducibility and quality
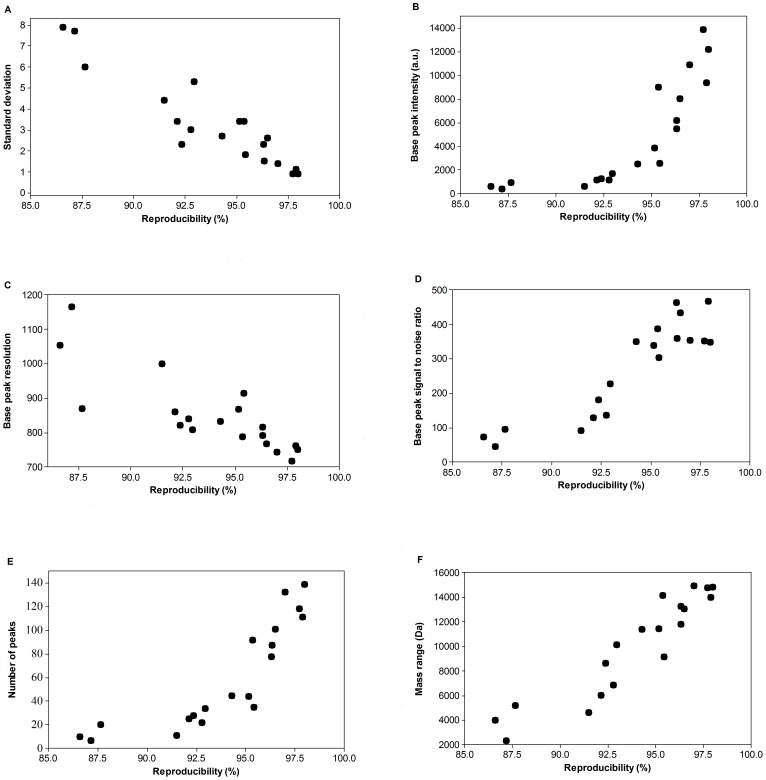
To further investigate spectrum quality and reproducibility obtained using different automated data acquisition settings, we assessed metrics of spectrum quality as a function of reproducibility for all 19 experiments. Specifically, we examined spectra exhibiting varying levels of reproducibility with regard to the standard deviation of the reproducibility and their spectrum quality, including base peak intensity, base peak resolution, base peak S:N, number of peaks, and mass range (Fig. 1).
Figure 1. Characterization of spectra exhibiting varying levels reproducibility.
Metrics of quality include standard deviation (A), base peak intensity (B), base peak resolution (C), base peak S:N (D), number of peaks (E), and mass range (F).
As expected, our analysis revealed that spectra with higher reproducibility tended to have lower standard deviations (Fig. 1A). With regard to spectrum quality, spectra with higher reproducibility tended to have higher base peak intensities (Fig. 1B), higher base peak S:N (Fig. 1D), greater numbers of peaks (Fig. 1E), and broader mass ranges (Fig. 1F). These results indicate that highly reproducible spectra are associated with high spectrum quality.
Interestingly, we observed a counterintuitive relationship between base peak resolution and reproducibility. Highly reproducible spectra tended to have lower resolution base peaks than spectra with lower reproducibility (Fig. 1C). While base peak resolution is an important parameter to assess spectrum quality (high resolution is typically desired), our results suggest that spectra with high reproducibility more commonly had lower base peak resolutions. To investigate the possibility that our result was based on anomalous spectra, we manually and rigorously examined each spectrum to ensure each spectrum contained at least 5 peaks which had intensities higher than 100 arbitrary units. These results suggest that efforts to increase base peak resolution when optimizing MALDI-TOF settings may not necessarily increase spectrum reproducibility. Our results further suggest that a conventional standard for assessing spectrum quality, base peak resolution, may have more limited applicability to microbial characterization via MALDI than to more traditional applications of mass spectrometry (e.g., protein identification). Accordingly, future attempts to optimize automated data acquisition should not place undue emphasis on base peak resolution.
Effects of automated data acquisition parameters on reproducibility
Statistical analysis was used to identify main effects and two-factor interaction effects of automated data acquisition parameters on reproducibility. The estimated effect for any factor or interaction is the difference between the average response at the high level of that factor or interaction and its low level. For example, the estimated effect of factor A would be (if 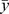 represents the response of interest):
represents the response of interest):  . The plus and minus superscripts represent values of the responses at the high and low levels, respectively. If this difference is large (in absolute value) then factor A would be considered statistically significant. The analysis of variance results are displayed in Table 3. Factors and interactions that had a p-value less than 0.05 were considered significant. Based on the p-values, threshold peak resolution (D) (p-value
. The plus and minus superscripts represent values of the responses at the high and low levels, respectively. If this difference is large (in absolute value) then factor A would be considered statistically significant. The analysis of variance results are displayed in Table 3. Factors and interactions that had a p-value less than 0.05 were considered significant. Based on the p-values, threshold peak resolution (D) (p-value 0.013) and number of shots summed (E) (p-value
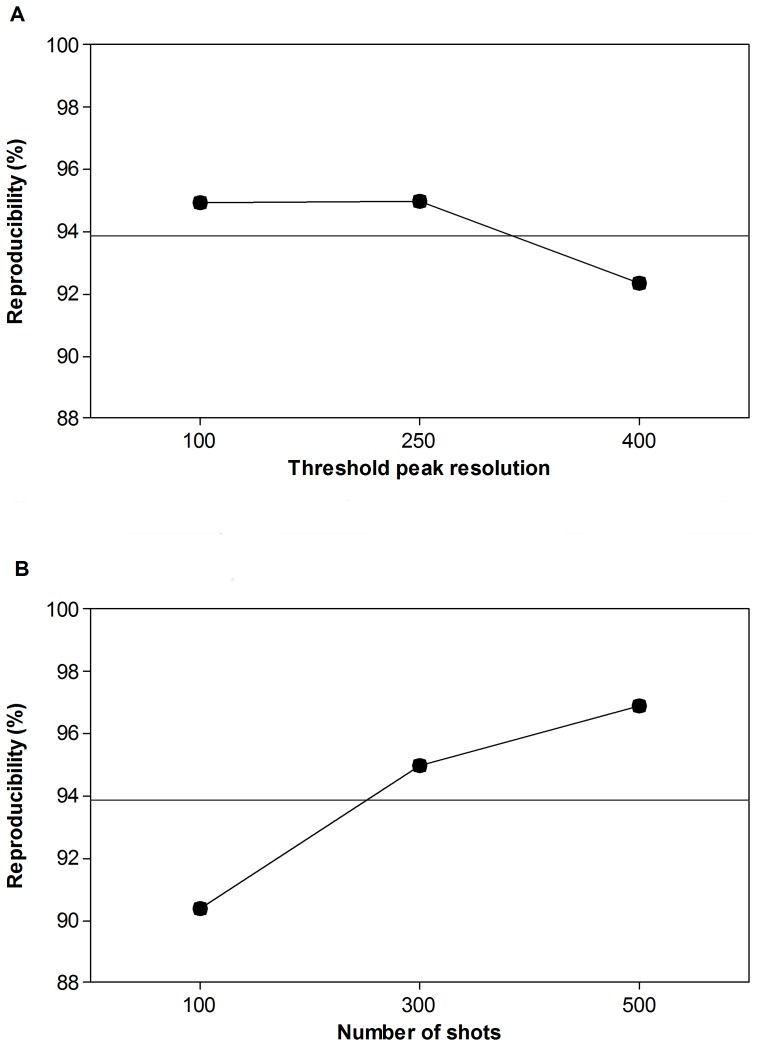
0.013) and number of shots summed (E) (p-value 0.002) were found to be significant. The main effects are shown in Figure 2. The mean value of reproducibility obtained with the high level of threshold resolution decreased in comparison with that obtained with the low level of threshold resolution (Fig. 2A). It is also illustrated in Figure 2 that the effect of number of shots on reproducibility shows a positive trend, in which spectrum reproducibility increased with the number of shots summed (Fig. 2B).
0.002) were found to be significant. The main effects are shown in Figure 2. The mean value of reproducibility obtained with the high level of threshold resolution decreased in comparison with that obtained with the low level of threshold resolution (Fig. 2A). It is also illustrated in Figure 2 that the effect of number of shots on reproducibility shows a positive trend, in which spectrum reproducibility increased with the number of shots summed (Fig. 2B).
Table 3. Analysis of variance for reproducibility.
| Sourcea | DF | Seq SS | Adj SS | Adj MS | F | P |
| Main effects | 5 | 200.105 | 200.105 | 40.021 | 112.51 | 0.009 |
| A | 1 | 0.643 | 0.643 | 0.643 | 1.81 | 0.311 |
| B | 1 | 0.951 | 0.951 | 0.951 | 2.67 | 0.244 |
| C | 1 | 2.380 | 2.380 | 2.380 | 6.69 | 0.123 |
| D | 1 | 26.631 | 26.631 | 26.631 | 74.87 | 0.013 |
| E | 1 | 169.501 | 169.501 | 169.501 | 476.52 | 0.002 |
| Two-way interactions | 10 | 28.278 | 28.278 | 2.828 | 7.95 | 0.117 |
| A * B | 1 | 1.596 | 1.596 | 1.596 | 4.49 | 0.168 |
| A * C | 1 | 2.593 | 2.593 | 2.593 | 7.29 | 0.114 |
| A * D | 1 | 4.488 | 4.488 | 4.488 | 12.62 | 0.071 |
| A * E | 1 | 0.398 | 0.398 | 0.398 | 1.12 | 0.401 |
| B * C | 1 | 0.072 | 0.072 | 0.072 | 0.20 | 0.696 |
| B * D | 1 | 0.001 | 0.001 | 0.001 | 0.00 | 0.965 |
| B * E | 1 | 3.430 | 3.430 | 3.430 | 9.64 | 0.090 |
| C * D | 1 | 2.162 | 2.162 | 2.162 | 6.08 | 0.133 |
| C * E | 1 | 1.253 | 1.253 | 1.253 | 3.52 | 0.201 |
| D * E | 1 | 12.285 | 12.285 | 12.285 | 34.54 | 0.028 |
| Curvature | 1 | 4.399 | 4.399 | 4.399 | 12.37 | 0.072 |
| Residual error | 2 | 0.711 | 0.711 | 0.356 | ||
| Pure error | 2 | 0.711 | 0.711 | 0.356 | ||
| Total | 18 | 233.494 |
A, B, C, D and E represent peak selection mass range, S:N, base peak intensity, minimum resolution, and number of shots summed, respectively.
Figure 2. Main effects plots.
Effects of threshold minimum resolution (A) and number of shots summed (B) on spectrum reproducibility during characterization of P. aeruginosa using automated data acquisition with MALDI-TOF MS.
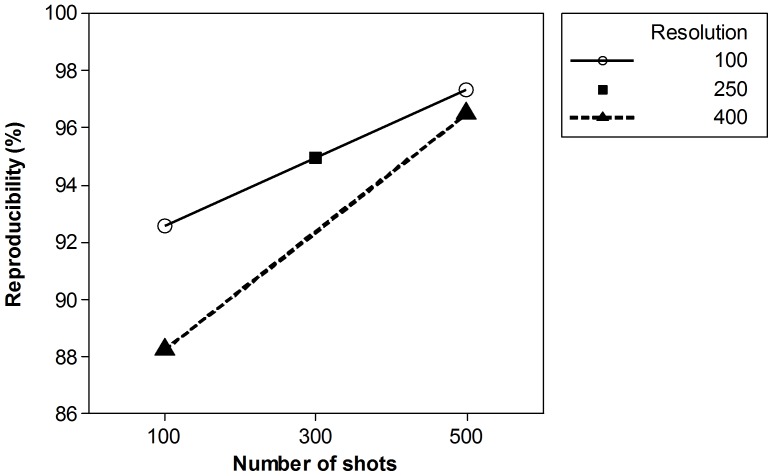
An interaction between minimum resolution and number of shots summed (D*E) (p-value  0.028) was observed (Fig. 3) indicating that the number of shots summed is more important in the case of higher threshold resolution (e.g., 400). In contrast, when using a lower threshold resolution, for example 100, fewer shots appeared to yield reproducibility comparable to that obtained using more shots and a higher threshold resolution (Fig. 3). This finding is intriguing, because it suggests that fewer shots may be used to obtain spectra of reproducibility comparable to that obtained with many more shots. Reducing shot number has the potential to reduce the time required for analysis. This might be particularly valuable information in a clinical microbiology lab setting in which the number of samples processed per day is very high.
0.028) was observed (Fig. 3) indicating that the number of shots summed is more important in the case of higher threshold resolution (e.g., 400). In contrast, when using a lower threshold resolution, for example 100, fewer shots appeared to yield reproducibility comparable to that obtained using more shots and a higher threshold resolution (Fig. 3). This finding is intriguing, because it suggests that fewer shots may be used to obtain spectra of reproducibility comparable to that obtained with many more shots. Reducing shot number has the potential to reduce the time required for analysis. This might be particularly valuable information in a clinical microbiology lab setting in which the number of samples processed per day is very high.
Figure 3. Interaction of threshold minimum resolution and number of shots summed.
Effects on spectrum reproducibility during characterization of P. aeruginosa using automated data acquisition with MALDI-TOF MS.
A prediction equation (Eq. 1) was fitted for P. aeruginosa to predict reproducibility for each experimental run, where ŷ is predicted reproducibility (%), D is threshold minimum resolution and E is number of shots summed.
| (Eq. 1) |
Based on the interaction plot (Fig. 3), setting the number of shots at 500 and resolution at 100 yielded an overall higher average reproducibility than any other combination of the two factors. A response optimization algorithm was also used to find best combinations of threshold minimum resolution and number of shots summed for high reproducibility. This showed the same settings as the interaction plot suggested (data not shown). As a result, we input the threshold minimum resolution at its low level and number of shots summed at its high level, which were −1 and +1, respectively, into the fitted equation (Eq.2). As shown in Eq. 2, the predicted reproducibility for P. aeruginosa was 97.3%.
| (Eq. 2) |
Effects of optimization on automated data acquisition
Finally, we compared spectrum quality and reproducibility using non-optimized and optimized automated data acquisition settings. The non-optimized settings were previously described [11], in which peak selection ranged from 2 to 20 kDa; S:N was 2; base peak intensity was 100; minimum resolution was 400 and number of shots summed was 300. The optimized settings were those used in Eq. 2 as described above.
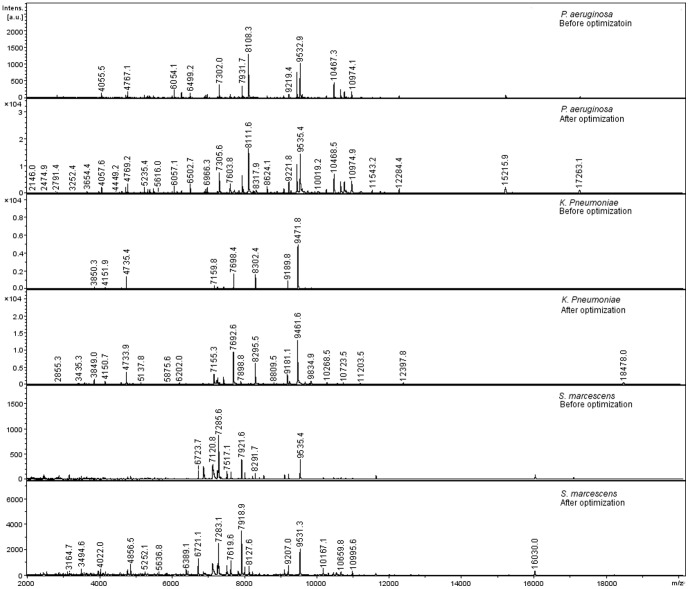
Representative mass spectra obtained before and after optimization are shown in Figure 4, and corresponding spectrum quality and reproducibility metrics are summarized in Table 4. Generally, base peak intensity, number of peaks, and mass range increased when optimized data acquisition settings were used for all three bacteria (Table 4; Fig. 4). No difference was observed for S:N between non-optimized and optimized settings. With regard to base peak resolution, spectra obtained using optimized settings had a lower base peak resolution than those obtained using non-optimized settings for all three bacteria.
Figure 4. Representative mass spectra acquired via non-optimized and optimized automation settings.
Mass spectra of P. aeruginosa, K. pneumoniae and S. marcescens were acquired via non-optimized and optimized settings using automated data acquisition with MALDI-TOF MS.
Table 4. Effect of optimization on spectrum quality and reproducibility.
| Spectrum qualitya | |||||||
| Bacteria | Condition | Base peak intensity | Base peak resolution | Base peak S:N | Number of peaks | Mass range (Da) | Reproducibility (%)b |
| P. aeruginosa | Before optimization | 2143.5± 1623.5* | 869.2±134.1** | 224.6±127.8* | 40.6±28.9* | 9102.6±4040.6 * | 90.4±5.5* |
| After optimization | 13751.8±4907.3** | 718.5±71.6* | 259.0±96.7* | 93.6±35.2** | 14649.4±650.4** | 97.2±1.2** | |
| p value Pr >| t | | 0.000 | 0.000 | 0.344 | 0.000 | 0.000 | 0.000 | |
| K. pneumoniae | Before optimization | 3859.7±1641.1* | 920.9±83.3** | 585.1±206.0* | 27.6±24.9* | 6188.0±1498.7* | 93.6±5.1* |
| After optimization | 21431.4±3387.6** | 776.1±59.0* | 660.6±153.6* | 152.1±28.7** | 15734.8±400.1** | 97.5±1.8** | |
| p value Pr >| t | | 0.000 | 0.000 | 0.197 | 0.000 | 0.000 | 0.000 | |
| S.marcescens | Before optimization | 1543.9±919.2* | 957.7±232.7** | 121.1±37.1* | 38.8±22.1* | 7126.2±4091.3* | 84.6±9.9* |
| After optimization | 8333.4±2137.1** | 750.3±51.6* | 100.7±27.2* | 30.3±18.7* | 12087.2±1410.4** | 93.9±2.9** | |
| p value Pr >| t | | 0.000 | 0.000 | 0.056 | 0.199 | 0.000 | 0.000 | |
Values reported are the means of 20 replicates ± the standard deviations of the mean. Data were analyzed by t test (α = 0.05).
Values reported are the average correlation coefficients of 20 replicates ± the standard deviations of the correlation coefficient.
Before and after optimization values for each bacterium followed by different numbers of asterisks are significantly different.
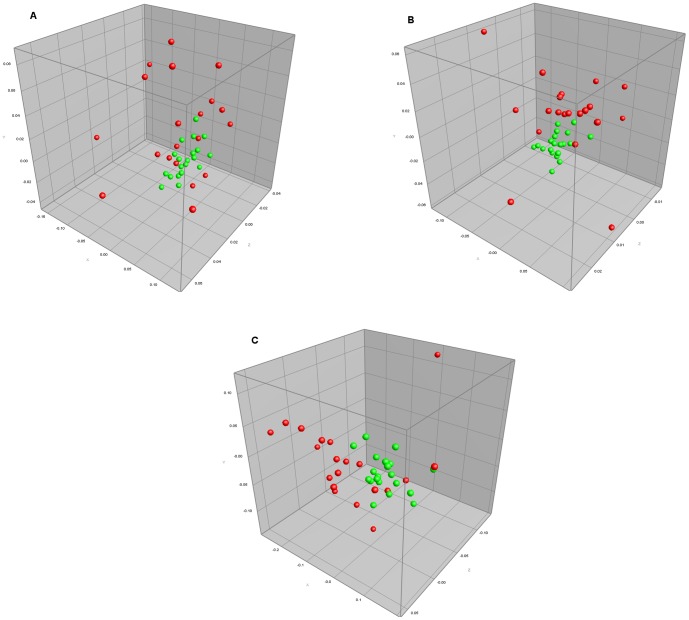
Optimization increased spectrum reproducibility (Table 4). For example, reproducibility for P. aeruginosa, K. pneumoniae and S. marcescens before optimization was 90.4±5.5%, 93.6±5.1% and 84.6±9.9%, respectively, while after optimization, reproducibility was 97.2±1.2%, 97.5±1.8% and 93.9±2.9%, respectively (Table 4). Multidimensional scaling (MDS) was used to visualize effects of optimization on reproducibility (Fig. 5). For all three bacteria, replicate spectra (20 replicates for each bacterium) acquired using optimized settings grouped more closely than replicate spectra (20 replicates for each bacterium) acquired without optimization.
Figure 5. Multidimensional scaling (MDS) representation of spectrum reproducibility.
MDS representing reproducibility associated with spectra of P. aeruginosa (A), K. pneumoniae (B) and S. marcescens (C) acquired via non-optimized (red) and optimized (green) settings.
The reproducibility (97.2%) of P. aeruginosa using optimized settings was strikingly similar to that predicted using the fitted equation (97.3%). Values of peak selection mass range, S:N and threshold peak intensity can have multiple selections. Other selections of these three parameters with constant values of threshold minimum resolution (100) and number of shots summed (500) also yielded spectra with reproducibility comparable to predicted values (data not shown).
We further compared the reproducibility obtained using optimized automated data acquisition settings with reproducibility previously reported which was obtained from spectra acquired manually [11]. They were comparable for all three bacteria. Specifically, reproducibility reported previously for manual data acquisition was 96% to 97% for P. aeruginosa, 95% to 96% for K. pneumoniae and 93% to 96% for S. marcescens [11]. For automated data acquisition using optimized settings, the reproducibility was approximately 97% for P. aeruginosa, 98% for K. pneumoniae and 94% for S. marcescens (Table 4).
The optimized settings were effective in increasing spectrum reproducibility for bacteria beyond the one that served as the model for optimization, suggesting that these settings, to some extent, are effective in improving the reproducibility of spectra for a range of bacteria. However, our model and equation are based on data acquired using P. aeruginosa, a Gram-negative bacterium. With regard to other bacteria, particularly Gram-positive bacteria, the relevance of settings obtained here may have limited utility, and coefficients of models may need to be adjusted. Accordingly, it may be necessary to run designed experiments for specific strains to obtain unique optimum settings. Conversely, such optimization may not always be necessary. For example, Mellmann et al. 2009 [49] reported high reproducibility using parameters for automated data acquisition that had not been rigorously optimized for the bacteria characterized in that work.
Conclusions
A fractional factorial design was applied to optimize five data acquisition parameters (peak selection mass range, S:N, threshold peak intensity, threshold minimum resolution and number of shots summed) and one response (reproducibility of replicate spectra). Both threshold minimum resolution and number of shots summed affected reproducibility, and an interaction was observed between these two data acquisition parameters. In the case of low threshold minimum resolution, high reproducibility could be achieved with fewer shots. After optimization, reproducibility of replicate spectra approached/exceeded those obtained manually for P. aeruginosa, K. pneumoniae and S. marcescens, suggesting that the main effects and interaction found in this study may be applicable to a broad range of bacteria. To our knowledge, this is the first report of use of designed-experiments to optimize automated data acquisition during MALDI-TOF fingerprint-based experiments.
Funding Statement
This work was supported by the New College of Interdisciplinary Arts & Sciences at Arizona State University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hsieh SY, Tseng CL, Lee YS, Kuo AJ, Sun CF, et al. (2008) Highly efficient classification and identification of human pathogenic bacteria by MALDI-TOF MS. Mol Cell Proteomics 7: 448–456. [DOI] [PubMed] [Google Scholar]
- 2. Sauer S, Kliem M (2010) Mass spectrometry tools for the classification and identification of bacteria. Nat Rev Microbiol 8: 74–82. [DOI] [PubMed] [Google Scholar]
- 3. Giebel RA, Fredenberg W, Sandrin TR (2008) Characterization of environmental isolates of Enterococcus spp. by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Water Res 42: 931–940. [DOI] [PubMed] [Google Scholar]
- 4. Barbuddhe SB, Maier T, Schwarz G, Kostrzewa M, Hof H, et al. (2008) Rapid identification and typing of Listeria species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl Environ Microbiol 74: 5402–5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang J, Zhou N, Xu B, Hao HJ, Kang L, et al. (2012) Identification and cluster analysis of Streptococcus pyogenes by MALDI-TOF mass spectrometry. PLoS One 7: e47152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benagli C, Demarta A, Caminada A, Ziegler D, Petrini O, et al. (2012) A rapid MALDI-TOF MS identification database at genospecies level for clinical and environmental Aeromonas strains. PLoS One 7: e48441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ziegler D, Mariotti A, Pfluger V, Saad M, Vogel G, et al. (2012) In situ identification of plant-invasive bacteria with MALDI-TOF mass spectrometry. PLoS One 7: e37189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Edwards-Jones V, Claydon MA, Evason DJ, Walker J, Fox AJ, et al. (2000) Rapid discrimination between methicillin-sensitive and methicillin-resistant Staphylococcus aureus by intact cell mass spectrometry. J Med Microbiol 49: 295–300. [DOI] [PubMed] [Google Scholar]
- 9. Evason DJ, Claydon MA, Gordon DB (2001) Exploring the limits of bacterial identification by intact cell-mass spectrometry. J Am Soc Mass Spectrom 12: 49–54. [DOI] [PubMed] [Google Scholar]
- 10. Giebel R, Worden C, Rust SM, Kleinheinz GT, Robbins M, et al. (2010) Microbial fingerprinting using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS): applications and challenges. Adv Appl Microbiol, Vol 71 71: 149–184. [DOI] [PubMed] [Google Scholar]
- 11. Schumaker S, Borror CM, Sandrin TR (2012) Automating data acquisition affects mass spectrum quality and reproducibility during bacterial profiling using an intact cell sample preparation method with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 26: 243–253. [DOI] [PubMed] [Google Scholar]
- 12. Khot PD, Couturier MR, Wilson A, Croft A, Fisher MA (2012) Optimization of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry Analysis for Bacterial Identification. J Clin Microbiol 50: 3845–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eddabra R, Prevost G, Scheftel JM (2012) Rapid discrimination of environmental Vibrio by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Microbiol Res 167: 226–230. [DOI] [PubMed] [Google Scholar]
- 14. Haag AM, Taylor SN, Johnston KH, Cole RB (1998) Rapid identification and speciation of Haemophilus bacteria by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Mass Spectrom 33: 750–756. [DOI] [PubMed] [Google Scholar]
- 15. Feldsine PT, Lienau AH, Leung SC, Mui LA, Humbert F, et al. (2003) Detection of Salmonella in fresh cheese, poultry products, and dried egg products by the ISO 6579 Salmonella culture procedure and the AOAC official method: Collaborative study. J AOAC Int 86: 275–295. [PubMed] [Google Scholar]
- 16. Stephan R, Cernela N, Ziegler D, Pfluger V, Tonolla M, et al. (2011) Rapid species specific identification and subtyping of Yersinia enterocolitica by MALDI-TOF Mass spectrometry. J Microbiol Methods 87: 150–153. [DOI] [PubMed] [Google Scholar]
- 17. Jackson KA, Edwards-Jones V, Sutton CW, Fox AJ (2005) Optimisation of intact cell MALDI method for fingerprinting of methicillin-resistant Staphylococcus aureus . J Microbiol Methods 62: 273–284. [DOI] [PubMed] [Google Scholar]
- 18. Goldstein JE, Zhang L, Borror CM, Rago JV, Sandrin TR (2013) Culture conditions and sample preparation methods affect spectrum quality and reproducibility during profiling of Staphylococcus aureus with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Lett Appl Microbiol 57: 144–150. [DOI] [PubMed] [Google Scholar]
- 19. Sandrin TR, Goldstein JE, Schumaker S (2013) MALDI TOF MS profiling of bacteria at the strain level: A review. Mass Spectrom Rev 32: 188–217. [DOI] [PubMed] [Google Scholar]
- 20. Kern CC, Usbeck JC, Vogel RF, Behr J (2013) Optimization of matrix-assisted-laser-desorption-ionization-time-of-flight mass spectrometry for the identification of bacterial contaminants in beverages J Microbiol Methods. 93: 185–191. [DOI] [PubMed] [Google Scholar]
- 21. Rupf S, Breitung K, Schellenberger W, Merte K, Kneist S, et al. (2005) Differentiation of mutans streptococci by intact cell matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Oral Microbiol Immun 20: 267–273. [DOI] [PubMed] [Google Scholar]
- 22. Tani A, Sahin N, Matsuyama Y, Enomoto T, Nishimura N, et al. (2012) High-throughput identification and screening of novel Methylobacterium species using whole-cell MALDI-TOF/MS analysis. PLoS One 7: e40784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wolters M, Rohde H, Maier T, Belmar-Campos C, Franke G, et al. (2011) MALDI-TOF MS fingerprinting allows for discrimination of major methicillin-resistant Staphylococcus aureus lineages. Int J Med Microbiol 301: 64–68. [DOI] [PubMed] [Google Scholar]
- 24. Ruelle V, El Moualij B, Zorzi W, Ledent P, De Pauw E (2004) Rapid identification of environmental bacterial strains by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 18: 2013–2019. [DOI] [PubMed] [Google Scholar]
- 25. Walker J, Fox AJ, Edwards-Jones V, Gordon DB (2002) Intact cell mass spectrometry (ICMS) used to type methicillin-resistant Staphylococcus aureus: media effects and inter-laboratory reproducibility. J Microbiol Methods 48: 117–126. [DOI] [PubMed] [Google Scholar]
- 26. Sedo O, Sedlacek I, Zdrahal Z (2011) Sample preparation methods for MALDI-MS profiling of bacteria. Mass Spectrom Rev 30: 417–434. [DOI] [PubMed] [Google Scholar]
- 27. Liu HH, Du ZM, Wang J, Yang RF (2007) Universal sample preparation method for characterization of bacteria by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl Environ Microbiol 73: 1899–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Toh-Boyo GM, Wulff SS, Basile F (2012) Comparison of Sample Preparation Methods and Evaluation of Intra- and Intersample Reproducibility in Bacteria MALDI-MS Profiling. Anal Chem 84: 9971–9980. [DOI] [PubMed] [Google Scholar]
- 29. Sedo O, Vorac A, Zdrahal Z (2011) Optimization of mass spectral features in MALDI-TOF MS profiling of Acinetobacter species. Syst Appl Microbiol 34: 30–34. [DOI] [PubMed] [Google Scholar]
- 30. Goyer M, Lucchi G, Ducoroy P, Vagner O, Bonnin A, et al. (2012) Optimization of the preanalytical steps of matrix-assisted laser desorption ionization-time of flight mass spectrometry identification provides a flexible and efficient tool for identification of clinical yeast isolates in medical laboratories. J Clin Microbiol 50: 3066–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vargha M, Takats Z, Konopka A, Nakatsu CH (2006) Optimization of MALDI-TOF MS for strain level differentiation of Arthrobacter isolates. J Microbiol Methods 66: 399–409. [DOI] [PubMed] [Google Scholar]
- 32. Ford BA, Burnham CAD (2013) Optimization of routine identification of clinically relevant gram-negative bacteria by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry and the Bruker Biotyper. J Clin Microbiol 51: 1412–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Avila A, Sanchez EI, Gutierrez MI (2005) Optimal experimental design applied to the dehydrochlorination of poly(vinyl chloride). Chemometrics and Intelligent Laboratory Systems 77: 247–250. [Google Scholar]
- 34. Kincl M, Turk S, Vrecer F (2005) Application of experimental design methodology in development and optimization of drug release method. Int J Pharm 291: 39–49. [DOI] [PubMed] [Google Scholar]
- 35. Zhang X, Wang RJ, Yang XX, Yu JG (2007) Central composite experimental design applied to the catalytic aromatization of isophorone to 3,5-xylenol. Chemometr Intell Lab Syst 89: 45–50. [Google Scholar]
- 36.Montgomery DC (2013) Design and analysis of experiments. Hoboken, NJ: John Wiley & Sons, Inc. xvii, 730 p. [Google Scholar]
- 37. Araujo PW, Brereton RG (1996) Experimental design.1. Screening. Trac-Trends in Anal Chem 15: 26–31. [Google Scholar]
- 38. Stojanovic BJ (2013) Factorial-based designs in liquid chromatography. Chromatographia 76: 227–240. [Google Scholar]
- 39. Uhlik O, Strejcek M, Junkova P, Sanda M, Hroudova M, et al. (2011) Matrix-assisted laser desorption ionization (MALDI)-time of flight mass spectrometry- and MALDI Biotyper-based identification of cultured biphenyl-metabolizing bacteria from contaminated horseradish rhizosphere soil. Appl Environ Microbiol 77: 6858–6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Salaun S, Kervarec N, Potin P, Haras D, Piotto M, et al. (2010) Whole-cell spectroscopy is a convenient tool to assist molecular identification of cultivatable marine bacteria and to investigate their adaptive metabolism. Talanta 80: 1758–1770. [DOI] [PubMed] [Google Scholar]
- 41. Stets MI, Pinto AS, Huergo LF, de Souza EM, Guimaraes VF, et al. (2013) Rapid identification of bacterial isolates from wheat roots by high resolution whole cell MALDI-TOF MS analysis. J Biotechnol 165: 167–174. [DOI] [PubMed] [Google Scholar]
- 42. Dieckmann R, Graeber I, Kaesler I, Szewzyk U, von Dohren H (2005) Rapid screening and dereplication of bacterial isolates from marine sponges of the Sula Ridge by Intact-Cell-MALDI-TOF mass spectrometry (ICM-MS). Appl Microbiol Biotechnol 67: 539–548. [DOI] [PubMed] [Google Scholar]
- 43. Ichiki Y, Ishizawa N, Tamura H, Teramoto K, Sato H, et al. (2008) Environmental distribution and novel high-throughput screening of APEO-degrading bacteria using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-MS). J Pestic Sci 33: 122–127. [Google Scholar]
- 44. Stafsnes MH, Dybwad M, Brunsvik A, Bruheim P (2013) Large scale MALDI-TOF MS based taxa identification to identify novel pigment producers in a marine bacterial culture collection. Anton Leeuw Int J G 103: 603–615. [DOI] [PubMed] [Google Scholar]
- 45. Thevenon F, Regier N, Benagli C, Tonolla M, Adatte T, et al. (2012) Characterization of fecal indicator bacteria in sediments cores from the largest freshwater lake of Western Europe (Lake Geneva, Switzerland). Ecotoxicol Environ Saf 78: 50–56. [DOI] [PubMed] [Google Scholar]
- 46. Koubek J, Uhlik O, Jecna K, Junkova P, Vrkoslavova J, et al. (2012) Whole-cell MALDI-TOF: rapid screening method in environmental microbiology. Int Biodeterior Biodegradation 69: 82–86. [Google Scholar]
- 47. Statham PJ (1977) Deconvolution and background subtraction by least-squares fitting with prefiltering of spectra. Anal Chem 49: 2149–2154. [Google Scholar]
- 48.Johnson RA, Wichern DW (2007) Applied multivariate statistical analysis (6th Edition). Upper Saddle River, NJ: Pearson Education, Inc. 800 p. [Google Scholar]
- 49. Mellmann A, Bimet F, Bizet C, Borovskaya AD, Deake RR, et al. (2009) High interlaboratory reproducibility of matrix-assisted laser desorption ionization-time of flight mass spectrometry-based species identification of nonfermenting bacteria. J Clin Microbiol 47: 3732–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]