Significance
Autophagy allows the lysosomal degradation of intracellular material. It is a tightly regulated process controlled by the tumor-suppressor gene p53, among others. Here we report a unique regulator of autophagy, BAT3, which modulates the intracellular localization of the enzyme p300, an enzyme that adds some acetyl residues on targets proteins (acetylation) to modulate their activity. To stimulate autophagy, BAT3 allows the acetylation of p53 by p300 in the nucleus, but limits the p300-dependent acetylation of ATG7, a protein specific for autophagy, in the cytosol. Thus, BAT3 acts on both the cytosol and the nucleus to tightly modulate autophagy.
Keywords: degradation, signalisation, nucleo-cytoplasmic shuttling
Abstract
Autophagy is regulated by posttranslational modifications, including acetylation. Here we show that HLA-B–associated transcript 3 (BAT3) is essential for basal and starvation-induced autophagy in embryonic day 18.5 BAT3−/− mouse embryos and in mouse embryonic fibroblasts (MEFs) through the modulation of p300-dependent acetylation of p53 and ATG7. Specifically, BAT3 increases p53 acetylation and proautophagic p53 target gene expression, while limiting p300-dependent acetylation of ATG7, a mechanism known to inhibit autophagy. In the absence of BAT3 or when BAT3 is located exclusively in the cytosol, autophagy is abrogated, ATG7 is hyperacetylated, p53 acetylation is abolished, and p300 accumulates in the cytosol, indicating that BAT3 regulates the nuclear localization of p300. In addition, the interaction between BAT3 and p300 is stronger in the cytosol than in the nucleus and, during starvation, the level of p300 decreases in the cytosol but increases in the nucleus only in the presence of BAT3. We conclude that BAT3 tightly controls autophagy by modulating p300 intracellular localization, affecting the accessibility of p300 to its substrates, p53 and ATG7.
Autophagy allows the lysosomal degradation of intracellular macromolecules and organelles after their sequestration in a vacuole known as the autophagosome (1). Basal autophagy is a cytoplasmic quality control mechanism that limits the production of reactive oxygen species and genomic instability. Autophagy is also induced to improve cell survival under stress.
Signaling pathways involved in the regulation of autophagy have been widely studied (2). Autophagy is modulated at two levels: (i) the molecular machinery involved in autophagosome biogenesis, dependent on specific genes known as Atg (AuTophaGy) genes, and (ii) the upstream signaling pathways (e.g., PI3K, MAPK, mTOR) that act on ATG proteins. Posttranslational modifications are crucial for the regulation of autophagy. The first example came from Y. Oshumi’s laboratory with the discovery of the conjugation systems for the ATG5-ATG12 and ATG8-phosphatidylethanolamine complexes in yeast (3). Phosphorylation is probably the most thoroughly investigated posttranslational event in autophagy. It appears that modulation of acetylation also affects Atg gene expression or activity; for instance, acetylation of the Unc-51-like kinase 1 (ULK1) (mammalian homolog of ATG1) by the acetylase Tat interacting protein 60 kDa (TIP60) induces autophagy after growth factor deprivation (4). In yeast, ESA1-dependent acetylation of ATG3 is essential for its interaction with ATG8 and ATG8 lipidation (5). Conversely, acetylation of ATG5, ATG7, microtubule associated protein 1 light chain 3 (LC3), and ATG12 by the acetyltransferase p300 inhibits autophagy (6), whereas their deacetylation by Sirtuins 1 (SIRT1) stimulates autophagy (7).
HLA-B–associated transcript 3 (BAT3) is a nucleo-cytoplasmic shuttling protein that contains, among other, a nuclear export signal (NES) and a nuclear localization signal (NLS). BAT3 has nuclear and cytosolic functions. In the cytosol, BAT3 has roles in quality control (8) and apoptosis (9–12). In the nucleus, thanks to its NLS domain (13), BAT3 modulates the methylase activity of SET1A and DOT1L, with roles in transcription and DNA damage response, respectively (14, 15). BAT3 is also involved in p53 activation by modulating its p300-dependent acetylation during apoptosis (16).
Here we show that BAT3 is essential for basal and starvation-induced autophagy in embryonic day (E) 18.5 mouse embryos and mouse embryonic fibroblasts (MEFs) through the regulation of p300 intracellular localization. In the presence of BAT3, p300 is mainly nuclear and stimulates p53 acetylation during starvation while maintaining a low level of ATG7 acetylation. In contrast, in the absence of BAT3, p300 accumulates in the cytosol, ATG7 is hyperacetylated, and p53 acetylation during starvation is abolished, leading to inhibition of autophagy. We propose a unique function for BAT3 in the regulation of autophagy through modulation of p300 intracellular localization and the subsequent acetylation of p53 and ATGs.
Results
BAT3 Is Essential for Basal Autophagy in Mice.
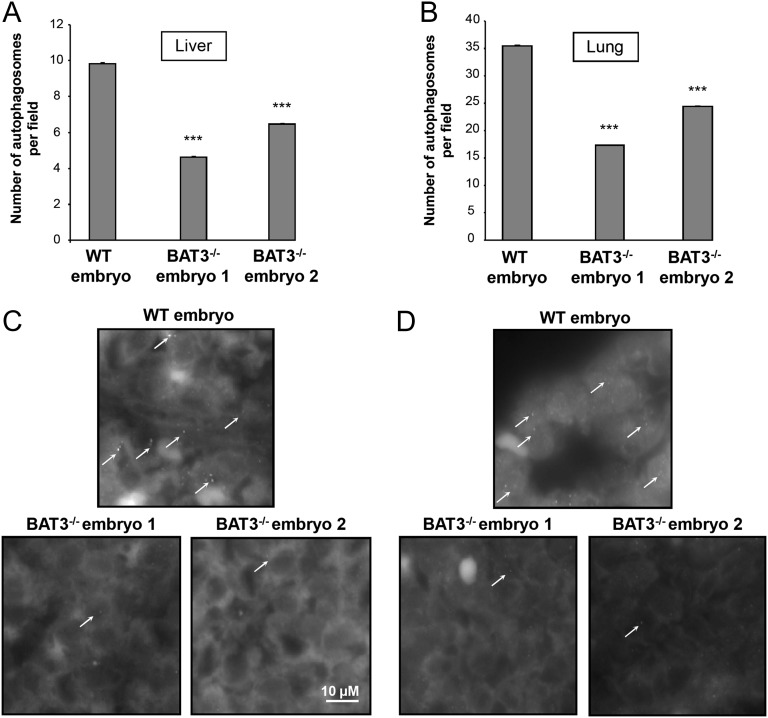
Homozygous deletion of BAT3 in mice is lethal, leading to major cell death and proliferation defects (17). Staining with an anti-LC3 antibody (a marker of autophagosomes) showed that BAT3 is essential for basal autophagy in E18.5 embryos. Indeed, the number of LC3-positive dots was significantly lower in the liver and the lung of BAT3−/− embryos compared with WT embryos (Fig. 1).
Fig. 1.
Autophagy is impaired in liver and lungs of E18.5 BAT3−/− mouse embryos. (A and B) Light-microscopy quantification of autophagy in liver (A) and lung (B) sections of WT and BAT3−/− E18.5 mouse embryos immunostained with an anti-LC3B antibody. Results are mean (SD) numbers of autophagosomes per field (approximately 200 fields counted per organ) in liver and lungs. ***P ≤ 0.001 vs. WT embryo. (C and D) Representative images of LC3 staining in liver (C) and lungs (D). Arrows indicate LC3 dots.
BAT3 Is Essential for Basal and Starvation-Induced Autophagy in MEFs.
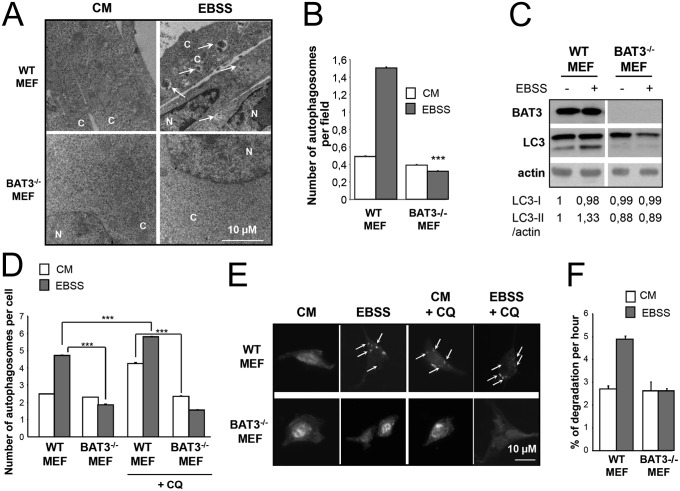
We next studied the role of BAT3 in autophagy in WT and BAT3−/− MEFs. By using transmission electron microscopy, we observed that starvation increased the number of autophagosomes in WT MEFs, and that the number of autophagosome was significantly lower in Earle’s balanced salt solution (EBSS)-treated BAT3−/− MEFs compared with WT MEFs (Fig. 2 A and B). We also tested the maturation of the autophagosome marker LC3-I (16 kDa, cytosolic staining) into LC3-II (18 kDa, punctuated staining). LC3 maturation was considerably reduced in BAT3−/− MEFs grown in complete medium (CM) or switched to EBSS, suggesting that BAT3 is required for basal and starvation-induced autophagy (Fig. 2C). The significantly lower number of GFP-LC3 dots per cell in BAT3−/− MEFs compared with WT MEFs confirms that BAT3 is essential for autophagy (Fig. 2 D and E). In addition, autophagic rates in BAT3−/− MEFs are similar to those in ATG7−/− MEFs, further supporting an essential role of BAT3 in autophagy (Fig. S1). We obtained similar results with other BAT3−/− MEF clones (Fig. S2).
Fig. 2.
BAT3 is essential for autophagy in MEFs. (A) Representative ultrastructural images of WT and BAT3−/− MEFs in CM or EBSS for 2 h. Arrows indicate autophagosomes. C, cytosol; N, nucleus. (B) Quantification of the number of autophagosomes per field as shown in A. ***P ≤ 0.001 vs. WT MEFs in EBSS. (C) WT and BAT3−/− MEFs were cultured in CM (−) or EBSS for 2 h (+), then immunoblotted as indicated. Densitometric analysis of the LC3-I and LC3-II /actin ratio was arbitrarily set at 1 for WT MEFs in CM and represents the mean (SD) of five independent experiments. (D and E) Quantification of the autophagosomes per cell (D) and representative images (E) of cells transfected with the peGFP-LC3 plasmid and incubated in CM or EBSS for 2 h and then with or without 20 µM chloroquine (CQ) for 6 h. Results are the mean (SD) of three independent experiments. ***P ≤ 0.001. (F) Measurement of long-lived protein degradation in WT and BAT3−/− MEFs cultured in CM or EBSS for 2 h.
To confirm that the induction of autophagy observed in WT MEFs during starvation was related to stimulation of the autophagic pathway rather than to increases in both the formation of autophagosomes and their consumption by the lysosome, we measured the autophagic flux by two independent assays: a GFP-LC3 assay using chloroquine, an inhibitor of lysosomal acidification, and the measurement of long-lived protein degradation. Chloroquine treatment led to the accumulation of GFP-LC3 dots only in WT MEFs and not in BAT3-deficient cells, showing a blockage of autophagosome formation in the absence of BAT3 (Fig. 2 D and E). In BAT3−/− MEFs, the absence of BAT3 abrogated protein degradation during starvation compared with WT MEFs (Fig. 2F). Taken together, these data suggest that BAT3 is essential for the early steps of autophagy.
BAT3 Is Responsible for p53 Acetylation by p300 During Starvation.
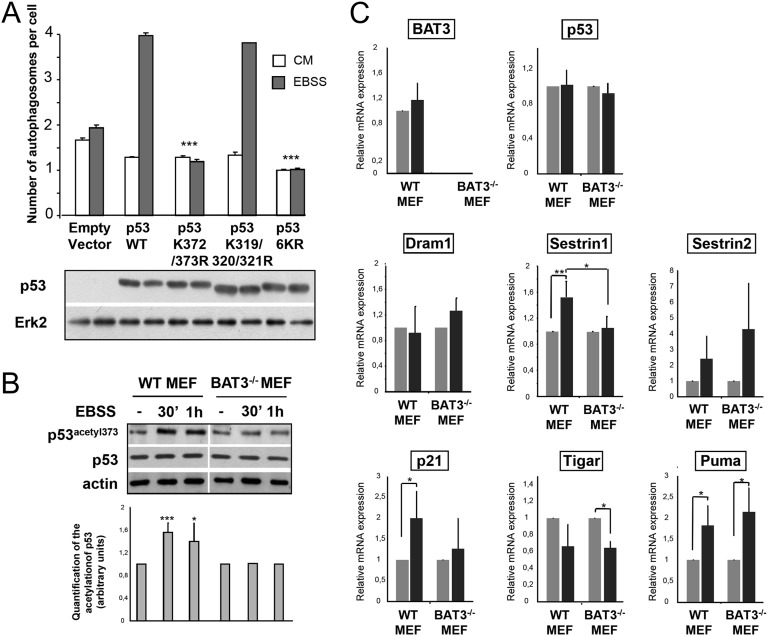
We next investigated how BAT3 modulates autophagy. BAT3 is essential for the p300-mediated acetylation of p53 on lysine 373 during genotoxic stress (16). To investigate whether p53 acetylation is involved in autophagy, we transfected p53-deficient H1299 cells with peGFP-LC3 and empty vector or with constructs encoding WT p53; the p53K372/373R mutant, in which lysines 372 and 373 cannot be acetylated by p300; the p53K319/320/321R mutant, in which lysines 319, 320, and 321 targeted by P300/CBP-associated factor (PCAF) cannot be acetylated; or the p53K6R mutant, in which all of the important lysines in the C terminus are replaced by arginines (Fig. 3A, Lower). We observed an increase in the number of GFP-LC3 dots per cell after transfection of WT p53 compared with empty vector in H1299 cells starved for 2 h (Fig. 3A, Upper). We obtained similar results with the p53K319/320/321R mutant. Conversely, transfection of the p53K372/373R or p536KR mutant totally abolished the starvation-induced autophagy observed in cells transfected with WT p53 (Fig. 3A, Upper). These results strongly suggest that p53 acetylation on lysines 372 and 373, which are known to be acetylated by p300, is essential for starvation-induced autophagy.
Fig. 3.
BAT3 is essential for p53 activity during autophagy. (A) (Upper) Quantification of autophagy in H1299 cells cotransfected with GFP-LC3 and the indicated plasmids and grown in CM or EBSS for 2 h. Results are the mean (SD) of three independent experiments. ***P ≤ 0.001 versus cells with WT p53. (Lower) Western blot analysis of p53 and ERK2 expression. (B) (Upper) Western blot analysis of p53 acetylation at lysine 373 in WT and BAT3−/− MEFs using antibodies against p53 acetylated at lysine 373 and actin. Cells were grown in CM (−) or switched to EBSS for the indicated time. (Lower) Densitometric quantification for five independent experiments ***P ≤ 0.001; *P ≤ 0.05 vs. starved WT MEFs. (C) Relative mRNA expression in WT and BAT3−/− MEFs cultured in EBSS or CM for 4 h (with expression value set at 1 for the CM condition). Results are representative of three independent experiments.
We next investigated the role of BAT3 in p53 acetylation during autophagy using an antibody that recognizes p53 acetylated at lysine 373. Starvation for 30 min and 1 h significantly increased p53 acetylation at lysine 373, a site acetylated by p300 (by 60% and 40%, respectively) in WT MEFs but not in BAT3−/− MEFs (Fig. 3B), indicating that BAT3 is essential for p53 acetylation during starvation. As a control, we confirmed that p53 acetylation at lysine 320, a site targeted by PCAF, was not modified during starvation (Fig. S3). Because p53 enhances the expression of proautophagic genes, we checked whether p53 dependent-stimulation of autophagy was accompanied by increased expression of p53 target genes during starvation. Indeed, in WT MEFs, the expression of SESTRIN1, p21, and Puma was significantly greater during starvation than in CM (Fig. 3C). The qPCR data for BAT3−/− MEFs revealed that BAT3 is required for SESTRIN1 up-regulation. We conclude that BAT3 is essential for p53 proautophagic activity by modulating p53 acetylation by p300, as well as the expression of proautophagic target genes.
BAT3 Inhibits p300-Dependent Acetylation of ATG7.
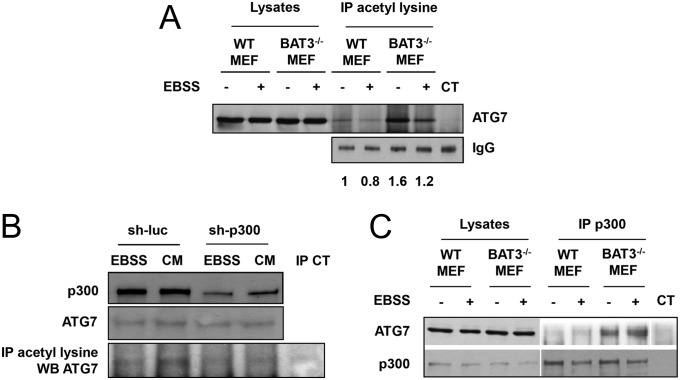
P300 inhibits autophagy by stimulating the p300-mediated acetylation of specific ATG proteins, such as ATG5, ATG7, ATG12 and LC3 (6). We first investigated whether BAT3 affects the acetylation of endogenous ATG7. Immunoprecipitation using an anti-acetylated lysine antibody followed by immunoblotting with an anti-ATG7 antibody demonstrated greater ATG7 acetylation in WT MEFs grown in CM (−) compared with those grown in EBSS, as expected (Fig. 4A). ATG7 acetylation was markedly increased in CM and EBSS in BAT3−/− MEFs compared with WT MEFs. This result may explain the autophagy inhibition observed in basal conditions in both BAT3−/− mice and MEFs (Figs. 1 and 2). In the absence of BAT3, we also observed an increase in ATG5 and LC3-I acetylation with no affect on ATG12 acetylation (Fig. S4).
Fig. 4.
BAT3 inhibits ATG7 acetylation and its interaction with p300. (A) Western blot analysis of ATG7 expression in protein lysates or after immunoprecipitation (IP) using an anti-acetylated lysine antibody. WT and BAT3−/− MEFs were grown in CM (−) or EBSS (+) for 2 h. Control immunoprecipitation (CT) was performed using rabbit IgG (Upper), and loading was checked using an anti-rabbit antibody (Lower). Results are representative of three independent experiments. (B) Coimmunoprecipitation of ATG7 and p300 using an anti-p300 antibody in cells grown in CM (−) or EBSS (+) for 2 h. The controls are the same as in A. Results are representative of three independent experiments. (C) Western blot analysis of ATG7 expression and acetylation in BAT3−/− MEFs in the presence of sh-p300 or sh-luciferase (sh-luc) cultured in CM or EBSS for 2 h as in A.
To confirm the role of p300 in the BAT3 dependent acetylation of ATG7, we decreased p300 expression using a shRNA in BAT3−/− MEFs and observed a decrease in ATG7 acetylation in CM (Fig. 4B). The remaining level of ATG7 acetylation may be attributed to the incomplete inhibition of p300 expression. We also checked whether BAT3 modulates the interaction between endogenous p300 and ATG7 by coimmunoprecipitation and found greater interaction in BAT3−/− MEFs compared with WT MEFs (Fig. 4C). We obtained similar results using another BAT3−/− MEF clone (Fig. S5).
BAT3 Intracellular Localization Is Important for Autophagy.
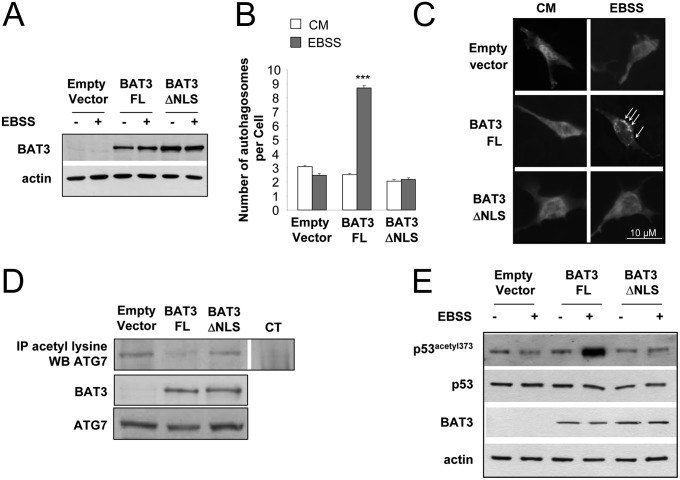
Our data indicate that BAT3 stimulates autophagy via two opposite mechanisms: stimulation of p53 acetylation during starvation and inhibition of ATG7 acetylation in basal conditions and during starvation. ATG proteins are located in the cytosol, whereas active p53 is located in the nucleus. Because BAT3 is a shuttling nuclear-cytosolic protein, we investigated the role of BAT3 localization by cotransfecting BAT3−/− MEFs with peGFP-LC3 and BAT3-FL, BAT3ΔNLS mutant located exclusively in the cytosol after deletion of the NLS, or empty vector (13) (Fig. 5A). Ectopic expression of BAT3-FL restored autophagy in BAT3−/− MEFs, whereas BAT3ΔNLS did not (Fig. 5 B and C). Moreover, ATG7 acetylation was increased in BAT3ΔNLS-expressing cells compared with BAT3 FL-expressing cells (Fig. 5D), whereas p300-dependent acetylation of p53 was reduced in cells expressing empty vector or BAT3ΔNLS compared with cells expressing BAT3-FL (Fig. 5E). Thus, the absence or cytosolic localization of BAT3 results in hyperacetylation of ATG7, repression of p53 acetylation during starvation, and inhibition of autophagy. This finding highlights the importance of the nuclear localization of BAT3 for its proautophagic activity.
Fig. 5.
The BAT3∆NLS mutant does not restore autophagy in BAT3−/− MEFs. (A) Western blot analysis of BAT3 and actin expression in BAT3−/− MEFs transfected with empty vector, BAT3-FL, or BAT3ΔNLS. (B) Quantification of the autophagosomes per cell in BAT3−/− MEFs cotransfected with peGFP-LC3 and the indicated plasmid. Results are the mean (SD) of three independent experiments. ***P ≤ 0.001 vs. empty vector. (C) Representative images of B. (D) Immunoprecipitation (IP) of acetylated lysine residues followed by Western blot analysis using an anti-ATG7 antibody in BAT3−/− MEFs transfected with empty vector, BAT3-FL, or BAT3ΔNLS. Control immunoprecipitation (CT) was carried out using rabbit IgG. Expression of BAT3 and ATG7 was assessed by Western blot analysis. Results are representative of three independent experiments. (E) Western blot analysis of p53 acetylation at lysine 373 in BAT3−/− MEFs stably transfected with the indicated vector and incubated in CM or EBSS for 30 min. Expression of p53, BAT3 and actin is shown as well. Results are representative of three independent experiments.
BAT3 Regulates p300 Cellular Localization.
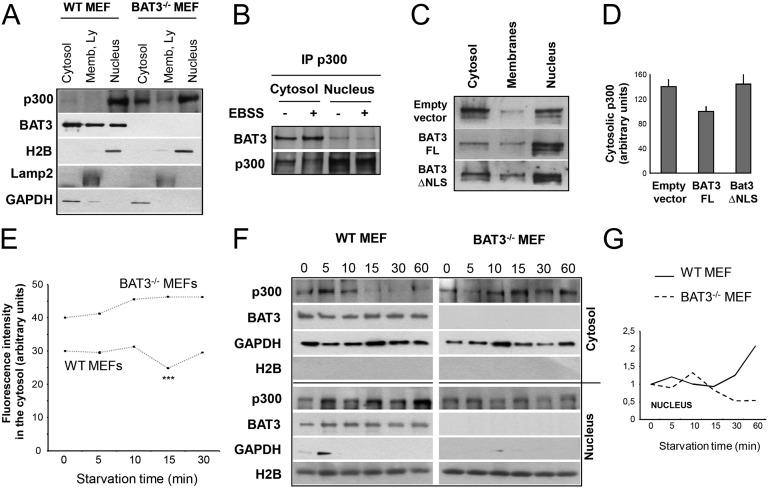
Because BAT3 is involved in the nuclear localization of p21 during the cell cycle (18), we analyzed whether BAT3 has a role in p300 intracellular localization by fractionating WT and BAT3−/− MEFs. We found that in WT MEFs, p300 was located almost entirely in the nucleus, whereas BAT3 was present in both the nucleus and cytosol. Conversely, in BAT3−/− MEFs, the p300 was present in significantly higher amounts in the cytosol (Fig. 6A). This finding was confirmed by quantifying the cytosol fluorescence in WT and BAT3−/− MEFs after staining with an anti-p300 antibody (Fig. S6 A and B).
Fig. 6.
BAT3 modulates p300 intracellular localization. (A) Western blot analysis of the subcellular fractionation in WT and BAT3−/− MEFs using anti-p300, anti-BAT3, anti-GAPDH (cytosol fraction), anti-LAMP2 (membrane fraction), and anti-H2B (soluble nuclear fraction) antibodies. Results are representative of five independent experiments. (B) Coimmunoprecipitation of BAT3 and p300 in cytosolic and nuclear extracts of WT MEFs in CM or EBSS for 2 h. Results are representative of three independent experiments. (C) Western blot analysis of the subcellular localization of p300 in BAT3−/− MEFs stably expressing empty vector, BAT3-FL, or BAT3ΔNLS. (D) Densitometric quantification of cytosolic p300 assessed in C and arbitrarily set at 100 for BAT3−/− MEFs expressing BAT3-FL. (E) Quantification of the fluorescence intensity in the cytosol of WT and BAT3−/− MEFs after immunostaining of p300 and during starvation kinetics as indicated below the x-axis. Results are the mean (SD) cytosol fluorescence intensity (in arbitrary units) of 100 cells from three independent experiments. ***P ≤ 0.001 vs. WT MEFs in CM. (F) Western blot analysis of the subcellular localization of p300 and BAT3 in the cytosol and the nucleus of WT and BAT3−/− MEFs during starvation kinetics. Controls are as in A. (G) Densitometric quantification of the p300 level in the nucleus assessed by Western blot analysis in F. The unit was arbitrarily set at 1 for WT and BAT3−/− MEFs in CM.
We next investigated the interaction between endogenous BAT3 and p300 in the cytosol and nucleus of WT MEFs by coimmunoprecipitation. Although p300 was located mainly in the nucleus, the BAT3–p300 interaction was stronger in the cytosol than in the nucleus (Fig. 6B). In addition, by cell fractionation, we found that ectopic expression of the BAT3ΔNLS mutant in BAT3−/− MEFs induced a cytosolic accumulation of p300 such as in the absence of BAT3 (Fig. 6 C and D), suggesting that BAT3 is responsible for the nuclear shuttling of p300. Thus, we asked whether BAT3 modifies p300 intracellular localization during starvation by quantifying the cytosolic fluorescence signal in WT and BAT3−/− MEFs stained with an anti-p300 antibody at different time points after switching to EBSS. As observed previously (Fig. 6 A, C, and D and Fig. S6 A and B), p300 accumulated in the cytosol of BAT3−/− MEFs (Fig. 6E). In addition, short-term starvation (15 min) induced a significant decrease in the p300 cytosolic pool in WT MEFs, but not in BAT3−/− MEFs (Fig. 6E). By cell fractionation, we also observed a relocation of p300 into the nucleus after 15 min of starvation (Fig. 6 F and G).
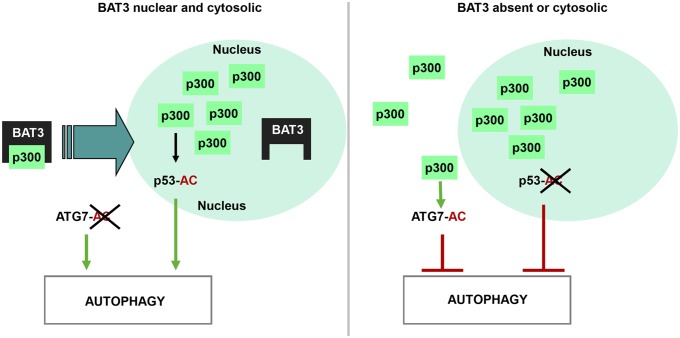
To conclude, we propose a model of BAT3 role in autophagy (Fig. 7). In starved WT MEFs, BAT3 allows relocation of p300 in the nucleus to induce autophagy by targeting p53 and inhibiting ATG7 acetylation. In the absence of BAT3, or when BAT3 is exclusively cytosolic, the starvation-induced p300 nuclear shuttling is inhibited, resulting in accumulation of p300 in the cytosol, where it acetylates ATG7, thereby inhibiting autophagy. We speculate that BAT3 is in charge of shuttling a small pool of p300 between the nucleus and cytoplasm. This p300 pool inhibits autophagy in the cytosol, whereas it stimulates autophagy after its relocation in the nucleus during starvation.
Fig. 7.
Model of autophagy regulation by BAT3. BAT3 regulates autophagy by modulating p300 localization and p300-dependent acetylation of p53 and ATG7. BAT3, allows the shuttling of p300 into the nucleus, where it targets p53 to stimulate autophagy. In contrast, when BAT3 is absent or exclusively cytosolic, p300 nuclear shuttling is inhibited, leading to its accumulation in the cytosol and to ATG7 hyperacetylation.
Discussion
Here we report that BAT3 regulates autophagy through its capacity to modulate the p300-dependent acetylation of p53 and ATG7. We first showed that BAT3 is involved in the p300-dependent acetylation of p53 on its lysine 373 during starvation and in the subsequent transcriptional activity of p53, allowing SESTRIN1 up-regulation. The role of p53 in autophagy remains a matter of debate. This role depends on its intracellular localization, among other factors. Cytoplasmic p53 was initially shown to inhibit autophagy, whereas nuclear p53 stimulates this process (19). It has been reported that the acetylation of p53 on its lysine 120 by TIP60 and sumoylation on its lysine 386 leads to the cytoplasmic accumulation of p53 and enhancement of autophagy, although this is controversial (20). This finding supports the importance of stimuli and subsequent modifications of p53 for the modulation of autophagy. In HCT116 colon cancer cells, a 6-h starvation stimulates p53 acetylation at lysine 382, a site deacetylated by SIRT1 (21). We found that p53 acetylation at lysine 373 occurs mainly 30 min after starvation. The duration of starvation is a critical parameter in the regulation of autophagy, and long-term starvation may reactivate mTor to inhibit autophagy and preserve lysosomal homeostasis (22).
Our findings also demonstrate that BAT3 is involved in the regulation of p300-dependent acetylation of ATG7, the E1 enzyme involved in the ATG5-ATG12 and ATG8-phoshatidylethanolamine conjugation systems. The mechanism by which ATG7 inhibits autophagy after acetylation and the lysine targeted by p300 remain unknown, but little data are available for other ATG proteins. In yeast, the acetylation of ATG3 by ESA1P (yeast homolog of the acetyltransferase TIP60) on lysines 19, 48, and 183 is essential for LC3 lipidation during starvation (5). In mouse cells, the kinase GSK3, activated by serum starvation, phosphorylates TIP60, increasing its affinity for ULK1 (mammalian ortholog of ATG1), which is then acetylated at lysines 162 and 606, leading to autophagy (4). Interestingly, TIP60, like p300, is located mainly within the nucleus, but also has cytoplasmic activities involving ATG proteins. The deacetylase SIRT1 is also essential for autophagy through two mechanisms: (i) deacetylation of ATG5, ATG7, and LC3, allowing autophagy induction, and (ii) deacetylation of the transcription factor forkhead box O3a (FOXO3a), leading to increased expression of BNIP3 (7, 23).
Interestingly, both a deacetylase activator (resveratrol) and an acetylase inhibitor (spermidine) induce autophagy. The acetylproteome of cells treated with these drugs demonstrates that a general cytosolic deacetylation and nuclear acetylation occur during autophagy (24). These findings agree with ours showing that BAT3 inhibits the acetylation of cytosolic ATG7 and stimulates the acetylation of nuclear p53 to induce autophagy.
The relative importance of the p300-p53 and p300-ATG7 pathways in autophagy is of interest. In H1299, the role of p53 appears to be predominant. However, ATG7 acetylation is essential, because it is the last mandatory step of autophagy, and p300 down-regulation stimulates autophagy by inhibiting ATG7 acetylation (6). Interestingly, ATG7 has been shown to regulate p53 activity during starvation (25).
BAT3 stimulates autophagy through opposing effects on p300-dependent acetylation of p53 and ATG7. Because BAT3 is a nucleocytoplasmic protein, we propose that it modulates autophagy differentially through the regulation of p300 localization. Indeed, in the absence of BAT3, the cytoplasmic accumulation of p300 may favor its interaction with ATG7. In addition, we observed a relocation of p300 from the cytosol to the nucleus during starvation. BAT3 is also involved in p21 nuclear translocation during G2/M phases, followed by its degradation during the transition between the G1 and S phases (18); however, the mechanism responsible for BAT3 nuclear shuttling remains elusive. A recent study showed that BAT3 variants are generated by alternative splicing of exon11B, leading to a different pattern of expression and intracellular localization. Thus, BAT3 may affect autophagy, depending on its variant expression (26).
In conclusion, our study demonstrates that BAT3 is essential for autophagy through the regulation of p300 intracellular localization, thereby controlling access to its proautophagic substrates.
Materials and Methods
Cells and Plasmids.
WT and BAT3−/− MEFs were obtained as described previously (17). Cells were cultured in DMEM supplemented with 10% FCS (CM) or in depleted medium (EBSS) (Life Technologies) for the indicated times. Plasmids encoding human WT and p53 mutants (pcDNA p53 WT, pcDNA p53K372/373R, pcDNA p53K319/320/321R, and pcDNA p53K6R) were obtained as described previously (27). Plasmids encoding full-length (FL) and mutant BAT3 (PCI-HA-BAT3-FL and PCI-HA-BAT3∆NLS) were obtained as described previously (13).
Autophagy Measurement.
WT and BAT3−/− E18.5 mouse embryo tissue sections were obtained as described previously (17) and stained with an anti-LC3B antibody (28). Electron microscopy sections were analyzed by counting the number of autophagosome per field. The maturation of LC3 was evaluated by immunoblot analysis. The GFP-LC3 assay was performed after transfection of rat peGFP-LC3, kindly provided by T. Yoshimori, Osaka University, Osaka, Japan (29). For the measurement of long-lived protein degradation, cells were incubated with l-[14C]valine. After a cold valine chase, degradation was measured as acid-soluble radioactivity recovered from both cells and medium as previously described (30).
Immunoprecipitation.
The acetylation of ATG proteins was measured by Western blot analysis after immunoprecipitation with an antibody directed against acetyl-lysine (Cell Signaling Technology). Endogenous p300 was immunoprecipitated with a rabbit polyclonal anti-p300 antibody (N15; 1:1,000), followed by detection of BAT3 or ATG7 by immunoblotting.
Cell Fractionation.
Cell fractionation was performed using a Pierce Subcellular Protein Fractionation Kit (Thermo Scientific).
Real-Time qPCR.
qPCR was performed with the primers described in SI Materials and Methods.
Staining of Endogenous p300.
WT and BAT3−/− MEFs were submitted to starvation, then immunostained with an anti-p300 antibody (N15; 1:200).
Statistical Analysis.
Quantified data were analyzed using the Student t test. A P value of < 0.001, < 0.01, or < 0.05 versus control was considered to indicate significance and was denoted by ***, **, or *. Data are reported as mean ± SD, with error bars indicating SD.
Additional information on autophagy assays, immunoprecipitation, retroviral infection, and qPCR primers is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank P. Pourquier for his inestimable help in reading the manuscript; the RHEM histology platform of Montpellier, especially F. Bernex, for their help with immunostaining of endogenous LC3 in vivo; the Centre de ressources en Imagerie Cellulaire, C. Cazevieille, and C. Tillier for their technical assistance and interpretation of ultrastructural data; T. Yoshimori for providing the peGFP-LC3 constructs; L. Le Cam and M. Lacroix for their help with the gateway cloning system and retroviral infection; and P. Montcourrier for assistance with the microscope. This work is supported by the Institut National de la Santé et de la Recherche Médicale, by the ARC foundation (C.G., P.C., L.K.L., E.L.-C., and S.P.), La Ligue Régionale contre Le Cancer (Comités du Gard et de l'Hérault; S.P.), Cancéropole Grand Sud-Ouest (S.P.), and the CHEMORES consortium (Tumour Chemotherapy Resistance EU FP6, to E.L.-C.). S.S. has fellowships from the Fond National de la Recherche Luxembourg (FNR Luxembourg) and La Ligue Nationale contre le Cancer.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313618111/-/DCSupplemental.
References
- 1.Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22(1):R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 2.Mehrpour M, Esclatine A, Beau I, Codogno P. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010;20(7):748–762. doi: 10.1038/cr.2010.82. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N, et al. A protein conjugation system essential for autophagy. Nature. 1998;395(6700):395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 4.Lin SY, et al. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science. 2012;336(6080):477–481. doi: 10.1126/science.1217032. [DOI] [PubMed] [Google Scholar]
- 5.Yi C, et al. Function and molecular mechanism of acetylation in autophagy regulation. Science. 2012;336(6080):474–477. doi: 10.1126/science.1216990. [DOI] [PubMed] [Google Scholar]
- 6.Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem. 2009;284(10):6322–6328. doi: 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee IH, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA. 2008;105(9):3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawahara H, Minami R, Yokota N. BAG6/BAT3: Emerging roles in quality control for nascent polypeptides. J Biochem. 2013;153(2):147–160. doi: 10.1093/jb/mvs149. [DOI] [PubMed] [Google Scholar]
- 9.Thress K, Henzel W, Shillinglaw W, Kornbluth S. Scythe: A novel reaper-binding apoptotic regulator. EMBO J. 1998;17(21):6135–6143. doi: 10.1093/emboj/17.21.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desmots F, Russell HR, Michel D, McKinnon PJ. Scythe regulates apoptosis-inducing factor stability during endoplasmic reticulum stress-induced apoptosis. J Biol Chem. 2008;283(6):3264–3271. doi: 10.1074/jbc.M706419200. [DOI] [PubMed] [Google Scholar]
- 11.Preta G, Fadeel B. AIF and Scythe (Bat3) regulate phosphatidylserine exposure and macrophage clearance of cells undergoing Fas (APO-1)-mediated apoptosis. PLoS ONE. 2012;7(10):e47328. doi: 10.1371/journal.pone.0047328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minami R, Shimada M, Yokosawa H, Kawahara H. Scythe regulates apoptosis through modulating ubiquitin-mediated proteolysis of the Xenopus elongation factor XEF1AO. Biochem J. 2007;405(3):495–501. doi: 10.1042/BJ20061886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manchen ST, Hubberstey AV. Human Scythe contains a functional nuclear localization sequence and remains in the nucleus during staurosporine-induced apoptosis. Biochem Biophys Res Commun. 2001;287(5):1075–1082. doi: 10.1006/bbrc.2001.5701. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen P, et al. BAT3 and SET1A form a complex with CTCFL/BORIS to modulate H3K4 histone dimethylation and gene expression. Mol Cell Biol. 2008;28(21):6720–6729. doi: 10.1128/MCB.00568-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wakeman TP, Wang Q, Feng J, Wang XF. Bat3 facilitates H3K79 dimethylation by DOT1L and promotes DNA damage-induced 53BP1 foci at G1/G2 cell-cycle phases. EMBO J. 2012;31(9):2169–2181. doi: 10.1038/emboj.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasaki T, et al. HLA-B-associated transcript 3 (Bat3)/Scythe is essential for p300-mediated acetylation of p53. Genes Dev. 2007;21(7):848–861. doi: 10.1101/gad.1534107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desmots F, Russell HR, Lee Y, Boyd K, McKinnon PJ. The reaper-binding protein scythe modulates apoptosis and proliferation during mammalian development. Mol Cell Biol. 2005;25(23):10329–10337. doi: 10.1128/MCB.25.23.10329-10337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yong ST, Wang XF. A novel, non-apoptotic role for Scythe/BAT3: A functional switch between the pro- and anti-proliferative roles of p21 during the cell cycle. PLoS ONE. 2012;7(6):e38085. doi: 10.1371/journal.pone.0038085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tasdemir E, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10(6):676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naidu SR, Lakhter AJ, Androphy EJ. PIASy-mediated Tip60 sumoylation regulates p53-induced autophagy. Cell Cycle. 2012;11(14):2717–2728. doi: 10.4161/cc.21091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morselli E, et al. p53 inhibits autophagy by interacting with the human ortholog of yeast Atg17, RB1CC1/FIP200. Cell Cycle. 2011;10(16):2763–2769. doi: 10.4161/cc.10.16.16868. [DOI] [PubMed] [Google Scholar]
- 22.Yu L, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465(7300):942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kume S, et al. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest. 2010;120(4):1043–1055. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morselli E, et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol. 2011;192(4):615–629. doi: 10.1083/jcb.201008167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee IH, et al. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 2012;336(6078):225–228. doi: 10.1126/science.1218395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kämper N, et al. A novel BAT3 sequence generated by alternative RNA splicing of exon 11B displays cell type-specific expression and impacts on subcellular localization. PLoS ONE. 2012;7(4):e35972. doi: 10.1371/journal.pone.0035972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Cam L, et al. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell. 2006;127(4):775–788. doi: 10.1016/j.cell.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 28.Ladoire S, et al. Immunohistochemical detection of cytoplasmic LC3 puncta in human cancer specimens. Autophagy. 2012;8(8):1175–1184. doi: 10.4161/auto.20353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bauvy C, Meijer AJ, Codogno P. Assaying of autophagic degradation. Methods enzymol. 2009;452:47–61. doi: 10.1016/S0076-6879(08)03604-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.