Significance
Iron is essential for eukaryotic cell survival but toxic at higher concentrations. In yeast, iron levels are tightly regulated by the transcriptional activators Aft1 and Aft2 (activators of ferrous transport), which activate iron-uptake genes when iron levels are low. We report the first crystal structure of DNA-bound Aft2 and show that Aft2 senses cellular iron levels via direct [2Fe-2S]-cluster binding, which promotes Aft2 dimerization and deactivation of the regulated genes. We further demonstrate that Aft2 acquires a [2Fe-2S] cluster from glutaredoxin-3 and Fe repressor of activation-2, two [2Fe-2S]-binding proteins with homologs in higher eukaryotes. This study unveils the molecular mechanism of the Aft family of iron-regulatory proteins and emphasizes the importance of Fe-S clusters in cellular iron sensing in eukaryotes.
Keywords: iron signaling, iron–sulfur cluster, yeast, Fra2, Grx3
Abstract
The paralogous iron-responsive transcription factors Aft1 and Aft2 (activators of ferrous transport) regulate iron homeostasis in Saccharomyces cerevisiae by activating expression of iron-uptake and -transport genes when intracellular iron is low. We present the previously unidentified crystal structure of Aft2 bound to DNA that reveals the mechanism of DNA recognition via specific interactions of the iron-responsive element with a Zn2+-containing WRKY-GCM1 domain in Aft2. We also show that two Aft2 monomers bind a [2Fe-2S] cluster (or Fe2+) through a Cys-Asp-Cys motif, leading to dimerization of Aft2 and decreased DNA-binding affinity. Furthermore, we demonstrate that the [2Fe-2S]-bridged heterodimer formed between glutaredoxin-3 and the BolA-like protein Fe repressor of activation-2 transfers a [2Fe-2S] cluster to Aft2 that facilitates Aft2 dimerization. Previous in vivo findings strongly support the [2Fe-2S] cluster-induced dimerization model; however, given the available evidence, Fe2+-induced Aft2 dimerization cannot be completely ruled out as an alternative Aft2 inhibition mechanism. Taken together, these data provide insight into the molecular mechanism for iron-dependent transcriptional regulation of Aft2 and highlight the key role of Fe-S clusters as cellular iron signals.
As a redox-active cofactor for a wide variety of enzymes engaged in essential functions, iron is crucial for cell survival. Despite its abundance, iron has limited bioavailability because of its tendency to form insoluble ferric hydroxides in aerobic and neutral pH environments. Most organisms have developed efficient iron-uptake strategies, but excess iron is toxic because of its role in generation of reactive oxygen species and interference in other metal-trafficking pathways. As such, intracellular iron in most organisms is tightly controlled (1). In the model eukaryote Saccharomyces cerevisiae, iron homeostasis is primarily regulated by the paralogous transcriptional activators Aft1 and Aft2 (activators of ferrous transport) (2–4). Aft1/2 activate the transcription of over 25 genes, designated the iron regulon, when the cellular iron concentration is low. The protein products of the iron regulon genes function in one of three general categories: iron uptake, intracellular sequestration, or metabolic utilization (5–7). Aft1 and Aft2 have overlapping, as well as separate, gene targets that possess a conserved iron-responsive element (Fe-RE) in their promoters consisting of a core sequence CACCC (4, 8–10). Variations in the nucleotide sequence flanking this core element, particularly on the 5′ side, allow for preferential binding of Aft1 or Aft2. The ultimate importance of Aft1/2 in regulating the cellular response to iron deprivation is highlighted by phenotypic analysis: growth of aft1∆ cells is inhibited in iron-deficient medium and the defect is exacerbated in aft1∆aft2∆ strains (2–4). A gain-of-function mutant allele, AFT1-1up or AFT2-1up, harboring a single cysteine to phenylalanine substitution (C291F in Aft1 and C187F in Aft2) causes iron-independent activation of the iron regulon, suggesting that this cysteine is critical for inhibition of Aft1/2 in response to iron (2–4).
The molecular mechanism by which Aft1 and Aft2 sense and respond to the intracellular iron level is unclear, but much research has provided pieces of the puzzle. Aft1 DNA-binding activity and nuclear localization is influenced by the cellular iron status: when iron is low, Aft1 accumulates in the nucleus and activates the iron regulon, whereas, upon iron sufficiency, Aft1 dissociates from DNA and is shuttled to the cytosol by the nuclear exporter Msn5 in an iron-dependent manner (8, 11, 12). Interestingly, iron sufficiency also induces multimerization of Aft1 in vivo (13). Genetic evidence indicates that Aft2 responds to changes in intracellular iron metabolism via a similar mechanism to Aft1. Iron-dependent inhibition of both Aft1 and Aft2 activity is regulated by a cytosolic signaling pathway comprised of the Cys-Gly-Phe-Ser (CGFS) monothiol glutaredoxins Grx3 and Grx4, the BolA-like protein Fe repressor of activation-2 (Fra2), and the aminopeptidase P-like protein Fe repressor of activation-1 (Fra1), which relays an inhibitory signal that is dependent on the synthesis of mitochondrial Fe-S clusters (12, 14–17). Aft1 and Aft2 share a conserved CDC motif that is essential for in vivo iron signaling, as well as Aft1/2 multimerization and interaction with Grx3/4. Amino acid substitution of either Cys residue in this motif leads to constitutive Aft1/2 nuclear localization and activation of the iron regulon (2, 13, 17).
Grx3/4 interacts with itself and with Fra2 in vivo and in vitro, forming homo- or heterodimers bridged by a [2Fe-2S] cluster (14, 18, 19). Similar to other members of the monothiol Grx family, the ligands for the [2Fe-2S] cluster in Grx3/4 come from the CGFS motif in Grx3/4 and glutathione (GSH) (20–22), whereas in the Fra2-Grx3/4 heterodimer, ligands are provided by a cysteine from Grx3/4, a histidine from Fra2, and GSH (18, 23, 24). The Fe-S–binding ligands in both Grx3/4 and Fra2 are essential for their ability to inhibit Aft1/2 in response to iron availability in vivo, indicating that Fe-S cluster binding is critical to their function.
To elucidate the molecular mechanism for iron-dependent transcriptional inhibition of Aft1/2, we aimed to uncover structural and biochemical information about Aft2 (and, by extension, Aft1) and the iron-sensing mechanism. Herein, we report the previously unidentified X-ray crystal structure of the DNA-binding domain of Aft2 bound to its target DNA. We show that Aft2 is a direct metal-binding protein that can bind a [2Fe-2S] cluster or a single Fe2+ ion, resulting in dimerization. In addition, we provide evidence that Aft2 specifically interacts with [2Fe-2S] Fra2-Grx3 heterodimer in vitro to transfer the intact [2Fe-2S] cluster from Fra2-Grx3 to Aft2, leading to Aft2 dimerization. Our results indicate that dimerization of Aft2, in turn, decreases its DNA-binding affinity, which leads to deactivation of the iron regulon. Collectively, these data suggest a regulation model for Aft1/2 that is in agreement with in vivo observations.
Results
The amino acid sequences of Aft1 and Aft2 share 26% overall identity, with the highest identity (39%) in the N-terminal domain (Fig. 1B). Therefore, a truncated form of Aft2 was constructed that consists of residues 1–204 and harbors the DNA-binding domain and iron-responsive CDC motif (referred to as Aft2 hereafter). The construct overexpressed well in Escherichia coli and yielded protein for subsequent structural and biochemical studies (SI Appendix, Fig. S1).
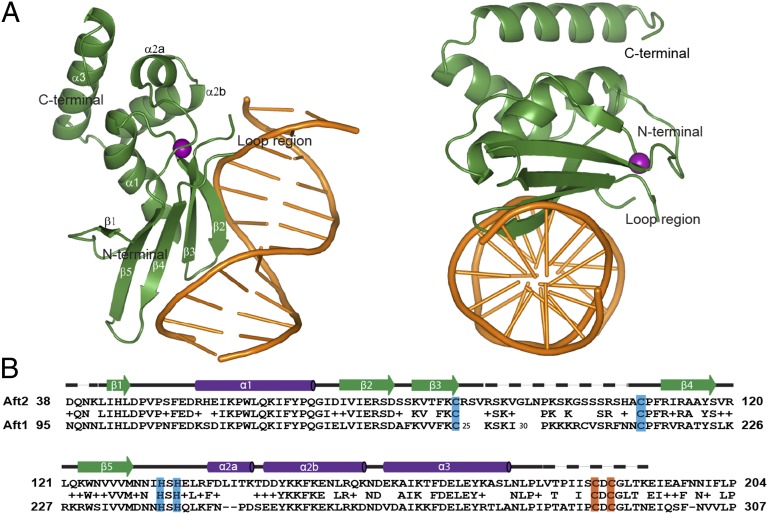
Fig. 1.
(A) Structure of the Aft2-DNA complex. DNA is colored orange, Aft2 green, and a zinc(II) ion purple. (B) Sequence alignment of the homologous N termini of Aft2 and Aft1. Identical residues are lettered and similar residues are marked “+.” The four residues coordinating zinc(II) are highlighted in blue and the iron-sensing cysteines in orange. Secondary structural elements as determined by the crystal structure are indicated above the sequence: green arrows for β-strands and purple tubes for α-helices. Solid black lines represent loops, and dashed lines disordered residues not modeled in the structure.
Aft2 Has a Structural WRKY-GCM1 Zn2+-Binding Domain for DNA Recognition.
Aft2 crystallized as a complex with an oligodeoxynucleotide derived from the promoter of FET3, a gene in the iron regulon encoding a multicopper oxidase required for iron uptake. Both Aft1 and Aft2 have been shown to bind to the FET3 promoter in vitro and in vivo, and this promoter sequence, 5′-AAGTGCACCCATT-3′, contains the consensus Fe-RE sequence of CACCC (shown in bold) (2–4, 9). The Aft2-DNA complex crystallized in the space group P21. The structure was solved using single-wavelength anomalous dispersion data from the bound zinc ion and was refined to a resolution of 2.2 Å (SI Appendix, Table S1). Aft2 binds to DNA as a monomer with a 1:1 protein:Fe-RE sequence stoichiometry (SI Appendix, Fig. S2). There are two copies of the Aft2-DNA complex in the asymmetric unit, and they are highly similar.
The structure of Aft2 consists of both α-helices and β-strands ordered as follows: β1-α1-β2-β3-β4-β5-α2a-α2b-α3 (Fig. 1A). Overall, the protein contains two structural domains, with the five β-strands arranged in a β-sheet forming one domain and the three α-helices packed against one side of the β-sheet forming the other domain. As predicted in a bioinformatics analysis (25), Aft2 possesses a WRKY-GCM1 fold (SI Appendix, Fig. S3), consisting of four β-strands (β2–β5) tethered together by a zinc(II) ion (Fig. 2A). The zinc ligands are provided by Cys86, Cys109, His133, and His135. Cys86 is at the end of β3, Cys109 is at the beginning of β4, and the two histidines are at the end of β5. Within the WRKY-GCM1 core fold, Aft2 has an insert of 22 residues between the two cysteines, most of which could not be included in the structural model because of disorder. Aft1 has an insert of 71 residues in the same position, and the difference in the size of this insert is the biggest difference between the two paralogs in their N-terminal domains (Fig. 1B and SI Appendix, Fig. S3).
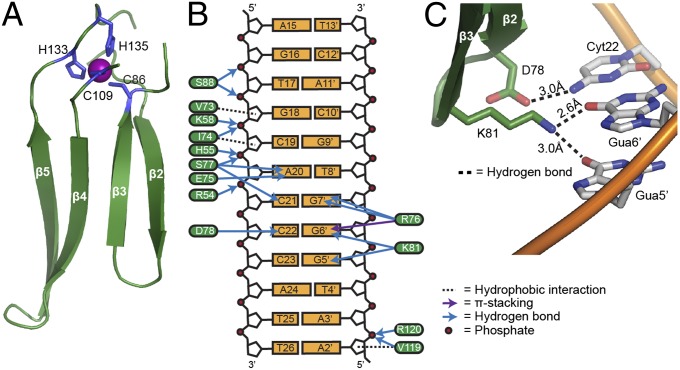
Fig. 2.
Aft2 interactions with DNA. (A) Structure of the WRKY-GCM1 domain in Aft2. The zinc(II) ion is shown as a purple sphere and the metal-coordinating residues as blue sticks. The proper folding of this domain is essential to the integrity of the protein. (B) Schematic representation of Aft2-DNA contacts. (C) Close-up of protein residues within the WRKY-GCM1 domain making specific contacts with bases in the Fe-RE.
Additional analysis of the metal content of purified Aft2 was performed with inductively coupled plasma MS (ICP-MS), which confirmed that Aft2 binds one equivalent of zinc when expressed in complete media (SI Appendix, Fig. S4 A and B). However, when Aft2 was expressed in minimal media or when one of the zinc-coordinating amino acids was substituted for a noncoordinating amino acid, no soluble protein was obtained. Addition of zinc to the minimal growth media restored Aft2 solubility, suggesting that the zinc site is structural and crucial for proper folding of the protein. The zinc ion can be replaced by cobalt, and the UV-visible absorption spectroscopic features of Co2+-loaded Aft2 were characteristic of a two histidine and two cysteine tetrahedral coordination environment to cobalt(II) (26) (SI Appendix, Fig. S4C). Zn2+ extended X-ray absorption fine structure (EXAFS) analysis of as-purified Aft2 was also performed to confirm the Zn2+-coordination site in solution. The data are best modeled with 2 N at 2.09 Å and 2 S at 2.33 Å, in agreement with the crystal structure (SI Appendix, Fig. S5).
Aft2 Specifically Recognizes the Iron-Responsive Element of DNA.
Contacts with DNA are mediated primarily by the WRKY-GCM1 domain of Aft2 (Fig. 2B). The β-sheet inserts into the major groove nearly perpendicular to the duplex DNA axis, allowing 10 residues in the domain to make contact with DNA. The side chains of Val73, Ile74, and Val119 form hydrophobic interactions with the deoxyriboses of Gua18, Cyt19, and Ade2′, respectively. Ser88 makes polar contacts with the phosphates of Thy17 and Gua18, one with its side chain and one with its backbone amide. Additional hydrogen bonds to backbone phosphates are made between the backbone amide of Ile74 and Cyt19, the side chain of Ser77 and Ade20, the backbone amide of Val119 and Ade3′, and the backbone amide of Arg120 and Ade3′. Base-specific contacts are mediated by residues within strands β2 and β3, including Glu75, Arg76, Ser77, Asp78, and Lys81. The carbonyl oxygen of Glu75 forms a hydrogen bond with N6 of Ade20. The guanidinium group of Arg76 makes hydrogen bonds with both N6 and O7 of Gua7′. Ser77 forms a hydrogen bond with N7 of Ade20 through its backbone amide and another with N4 of Cyt21 through its carbonyl oxygen. The OD2 atom of Asp78 forms a hydrogen bond with N4 of Cyt22; and finally, NZ of Lys81 bonds with O6 of both Gua5′ and Gua6′ (Fig. 2C). Outside of the WRKY-GCM1 domain, the side chains of residues Lys58, His55, and Arg54 of helix α1 form hydrogen bonds with the backbone phosphates of Cyt19, Ade20, and Cyt21, respectively.
Strand β2 and the β-hairpin formed by strands β2 and β3 are the most important recognition elements of Aft2. Hydrogen bonds between protein residues and DNA bases provide direct recognition of bases within the consensus Fe-RE sequence. Changes to nucleotides within the duplex Fe-RE would cause a mismatch between hydrogen-bond donors and acceptors, create van der Waals clashes, or reduce the number of hydrogen bonds made, thereby weakening or preventing sequence-selective recognition by Aft2. For example, the O6 atoms of Gua5′ and Gua6′ accept a hydrogen bond from NZ of Lys81; if guanine were a cytosine or adenine, two hydrogen-bond donors would align, and the 5′ methyl group of a thymine would sterically clash with Lys81 (Fig. 2C). Therefore, we provide structural evidence for how Aft2 specifically recognizes the Fe-RE–containing promoter sequences.
Aft2 Can Bind a [2Fe-2S] Cluster or a Fe2+ Ion.
A major question surrounding the mechanism of Aft1/2 regulation is how Aft1 and Aft2 sense iron. Previous studies suggest that Aft1 and Aft2 activate the iron regulon in response to mitochondrial Fe-S cluster synthesis because disruptions of the Fe-S cluster biogenesis and signaling pathways result in increased activation (12, 14, 17, 27, 28). Substitution of either cysteine in the CDC motif (C291 and C293 in Aft1 and C187 and C189 in Aft2) makes Aft1/2 insensitive to iron levels, a finding that suggests direct metal binding (2, 4). According to the structure of Aft2, the CDC motif is connected to the end of the α3 helix and is disordered in the structure in the absence of iron binding. This structural feature suggests the involvement of more than one monomer in binding to either an iron(II) ion or an Fe-S cluster. To determine whether Aft2 can bind iron or an Fe-S cluster, we loaded recombinant Aft2 with each species in an anaerobic environment. Analysis of the iron and sulfur content revealed that two monomers of Aft2 are indeed able to bind a single iron ion (Aft2:Fe ratio of 1:0.5) or a [2Fe-2S] cluster (Aft2:Fe:S ratio of 1:1:1) (SI Appendix, Fig. S6).
Binding of Fe-S Cluster or Fe2+-Induced Aft2 Dimerization.
Aft1 has been shown to interact with itself in an iron-dependent manner in vivo, and this interaction is disrupted when either cysteine in the CDC motif is replaced by another amino acid (13). To determine whether Aft2 can interact with itself in vitro and to investigate the metal-binding mechanism, we used analytical ultracentrifugation and small-angle X-ray scattering (SAXS) in the presence and absence of different metal species.
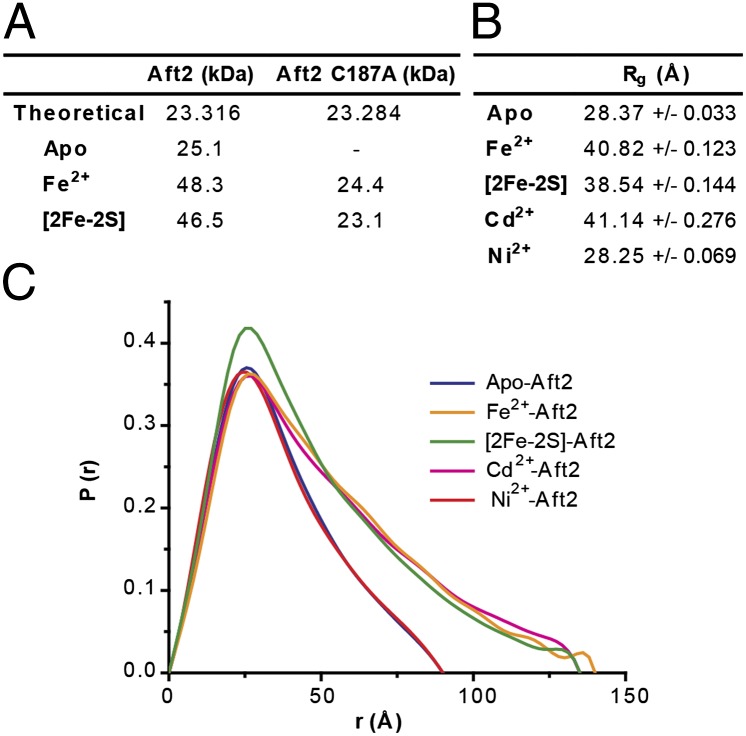
Aft2 is monomeric in solution under the conditions of ultracentrifugation analysis (Fig. 3A). Loading Aft2 with either Fe2+ or a [2Fe-2S] cluster changed the sedimentation velocity from that of the monomeric species, and the fitting to the Lamm equation gave a single peak with the molecular mass of a dimer both for Fe2+-Aft2 and [2Fe-2S]-Aft2 (Fig. 3A and SI Appendix, Fig. S7). On the other hand, the mutant Aft2 C187A was consistently a monomer and did not dimerize in the presence of Fe2+ or a [2Fe-2S] cluster. Similar results were observed with SAXS analysis (Fig. 3 B and C). The radius of gyration, a measure of the overall molecular size, of each Aft2 species was determined from the electron-pair distribution function. The radius of gyration of apo Aft2, measured at 28.37 ± 0.03 Å, is significantly smaller than that of Fe2+-Aft2 or [2Fe-2S]-Aft2, at 40.82 ± 0.12 Å and 38.54 ± 0.14 Å, respectively. In addition, the extended shapes of the electron-pair distribution function of Fe2+-Aft2 and [2Fe-2S]-Aft2 suggest an elongated structure in contrast with Aft2 alone. Both nickel and cadmium also were tested with SAXS: Ni2+-Aft2 has a similar radius of gyration and shape in the electron-pair distribution function as apo Aft2, but Cd2+-Aft2 is similar to Fe2+-Aft2 and [2Fe-2S]-Aft2. Nickel(II) prefers a square planar binding geometry, which is different from the tetrahedral coordination favored by cadmium(II) and iron(II), as well as binding to a [2Fe-2S] cluster. All of these data support a metal-recognition mechanism through dimerization of Aft2 and direct metal binding with two CDC motifs from the dimer.
Fig. 3.
Dimerization of Aft2 in the presence of Fe2+ or [2Fe-2S] cluster. (A) Molecular mass of metal-loaded Aft2 determined by analytical ultracentrifugation. (B) Radii of gyration of metal-loaded Aft2 determined by SAXS. (C) Normalized electron-pair distribution function profiles of Aft2 loaded with various metals and metal species.
With evidence of direct metal binding and metal-induced dimerization, we investigated whether the presence of metal could disrupt binding of Aft2 to DNA. In an electrophoretic mobility-shift assay, the Aft2-DNA complex was disrupted in the presence of Fe2+ and higher concentrations of Cd2+ but not a variety of other metal cations, including Ni2+ (SI Appendix, Fig. S8). This result indicates that the metal-dependent dimerization of Aft2 decreases its affinity for DNA and could be part of the gene deactivation mechanism.
Aft2 Specifically Interacts with [2Fe-2S]-Fra2-Grx3.
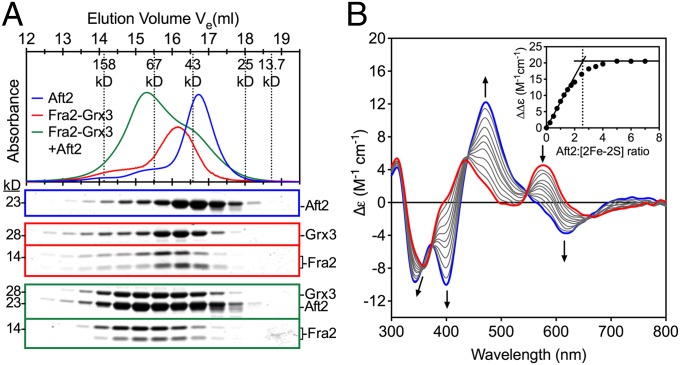
Because the results above demonstrate that Aft2 binds a [2Fe-2S] cluster in vitro, we sought to determine how Aft2 may acquire this Fe-S cluster in the cell. The [2Fe-2S]-binding glutaredoxins Grx3 and Grx4 interact with and inhibit Aft1 (and presumably Aft2) in vivo (14–16) and thus may deliver the Fe-S cluster to Aft1/2 as previously proposed (12, 18, 23). Fra2 also interacts with Grx3 and Grx4 in vivo and plays an unknown role in Aft1/2 inhibition in response to iron. It is not clear whether the interaction between Aft1/2 and Grx3/4 is dependent on Fra2 and/or formation of the [2Fe-2S]-Fra2-Grx3/4 complex. To address this issue, we mixed apo and [2Fe-2S]-loaded Fra2-Grx3 heterodimer and [2Fe-2S]-loaded Grx3 homodimer with purified Aft2 anaerobically and assessed complex formation by analytical gel-filtration chromatography. When Aft2 monomer and [2Fe-2S]-Fra2-Grx3 heterodimer are mixed together (Fig. 4A), all three proteins elute at a higher molecular mass than the individual proteins, indicating formation of a complex and/or multimerization. In contrast, when apo-Grx3, apo-Fra2, or [2Fe-2S]-Grx3 homodimer are mixed with Aft2, the proteins elute separately (SI Appendix, Fig. S9). Thus, these results suggest that Aft2 specifically interacts with the [2Fe-2S]-Fra2-Grx3 heterodimer. To determine whether the interaction between [2Fe-2S]-Fra2-Grx3 and Aft2 also influences the coordination environment of the [2Fe-2S] cluster, we titrated [2Fe-2S]-Fra2-Grx3 with increasing concentrations of Aft2. We observed a dramatic change in the UV-visible CD spectrum (Fig. 4B) within 3 min of Aft2 addition, suggesting a change in the cluster ligation and/or chirality of the cluster environment. The saturation binding curve suggests that the stoichiometry of the interaction is ∼2.5 with a plateau at ∼5:1 Aft2:[2Fe-2S] cluster (Fig. 4B, Inset). In contrast, addition of Aft2 to [2Fe-2S]-Grx3 homodimer did not cause any significant change in the CD spectrum, suggesting that Aft2 does not interact with the Fe-S cluster in [2Fe-2S]-Grx3 homodimer (SI Appendix, Fig. S10A).
Fig. 4.
Interaction of Aft2 with [2Fe-2S]-Fra2-Grx3. (A, Upper) Gel-filtration chromatograms of Aft2 (blue), [2Fe-2S]-Fra2-Grx3 (red), and Aft2 plus [2Fe-2S]-Fra2-Grx3 (green). The elution positions of molecular mass standards used for column calibration are shown as dotted lines. (A, Lower) SDS/PAGE analysis of the fractions collected. Note: Fra2 typically runs as two bands on the gel (18). (B) Titration of [2Fe-2S]-Fra2-Grx3 (red line) with 0.25- to 5-fold excess Aft2 (black lines) monitored by UV-visible CD spectroscopy. Arrows indicate the direction of intensity changes with increasing Aft2 added. Blue line is 5:1 [Aft2]:[2Fe-2S] ratio. (Inset) Difference in CD intensity between 400 and 472 nm with increasing Aft2:[2Fe-2S] ratios. The dotted line highlights the 2.5:1 binding stoichiometry between Aft2 and [2Fe-2S]-Fra2-Grx3.
Fra2-Grx3 Transfers a [2Fe-2S] Cluster to Aft2.
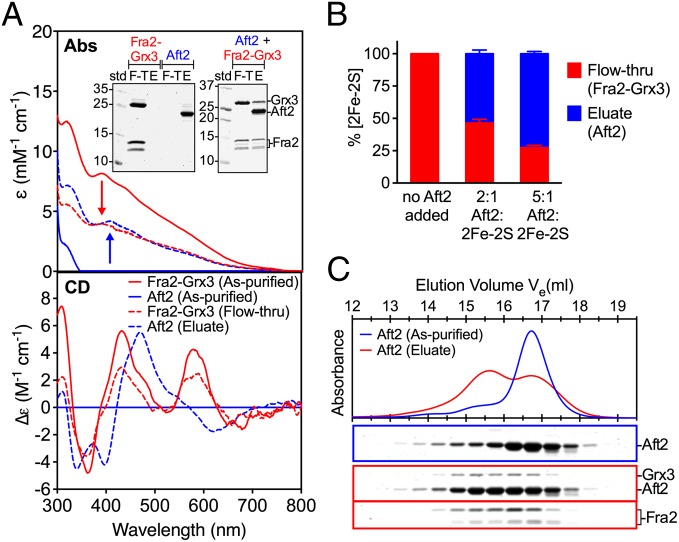
The coelution of Aft2 with Fra2 and Grx3 at a higher molecular mass (Fig. 4A) does not necessarily imply a stable interaction but may instead reflect oligomeric or conformational changes in both Aft2 and Fra2-Grx3 induced by a transient interaction. Given the clear changes in the Fe-S cluster coordination environment shown in Fig. 4B, we tested whether Fra2-Grx3 transfers the [2Fe-2S] cluster to Aft2, facilitating Aft2 dimerization. Heparin-affinity chromatography was used to separate Aft2 from Fra2-Grx3 following the interaction. The flow-through and eluate were assessed by UV-visible absorption and CD spectroscopy (Fig. 5A) and SDS/PAGE (Fig. 5A, Inset). In addition, the Fe and S content of the flow-through and eluate were measured. As shown in the Inset in Fig. 5A, Aft2 alone binds to the heparin column, but Fra2-Grx3 does not. In contrast, for the Aft2/Fra2/Grx3 reaction mixture, ∼65–70% Fra2-Grx3 comes off in the flow-through, whereas all of Aft2 is found in the eluate, together with ∼30–35% Fra2-Grx3. After separation, the [2Fe-2S] cluster content of Fra2-Grx3 drops with a concomitant increase in the [2Fe-2S] cluster binding to Aft2 in the eluate (Fig. 5B), with a maximum of ∼72% of the Fe-S cluster transferred to Aft2 at a 5:1 ratio of Aft2:[2Fe-2S]-Fra2-Grx3. The UV-visible absorption and CD spectra of the fractions also demonstrate that Aft2 is involved in the cluster coordination, because spectra of the flow-through sample are distinct from the Aft2-containing eluate (Fig. 5A). On the other hand, when Aft2 is mixed with [2Fe-2S]-Grx3 there is only 15.8 ± 1.7% Fe-S cluster and a negligible level of Grx3 identified in the eluate (SI Appendix, Fig. S10B), indicating the weak interaction between Aft2 and [2Fe-2S]-Grx3 homodimer and a low level of cluster transfer to Aft2. Overall, we conclude that Fra2 acts as a mediator to facilitate the interaction between Aft2 and Grx3 and transfer of the [2Fe-2S] cluster.
Fig. 5.
[2Fe-2S] cluster transfer from Fra2-Grx3 to Aft2. (A) UV-visible absorption (Upper) and CD (Lower) spectra of heparin column flow-through (red dotted line) and eluate (blue dotted line), compared with as-purified [2Fe-2S]-Fra2-Grx3 (red line) and Aft2 (blue line). ε and Δε values are normalized to Fra2-Grx3 heterodimer or Aft2 homodimer concentrations. (Inset) SDS/PAGE analysis of heparin-affinity chromatography fractions of Aft2 or [2Fe-2S]-Fra2-Grx3 alone or a 2:1 mixture of Aft2 to [2Fe-2S]-Fra2-Grx3. E, eluate; F-T, flow-through. (B) Fe-S cluster quantification in F-T and E after separation by heparin-affinity chromatography. (C, Upper) Gel-filtration chromatograms of as-purified Aft2 and eluate of [2Fe-2S] Fra2-Grx3 plus Aft2 after heparin separation. (C, Lower) SDS/PAGE analysis of the fractions collected.
Next, the oligomeric state of Aft2 after the cluster transfer was assessed by analytical gel-filtration chromatography. As shown in Fig. 5C, Aft2 prepared from the Aft2/Fra2/Grx3 reaction mixture ([Aft2]:[2Fe-2S] ratio, 2:1) shifts from monomer to dimer after Fe-S cluster transfer. In contrast, a very small increase in dimer formation was observed for Aft2 prepared from the Aft2 and [2Fe-2S]-Grx3 homodimer reaction mixture (SI Appendix, Fig. S10C).
ICP-MS analyses on Aft2 before and after Fe-S cluster transfer from Fra2-Grx3 verified that Fe-S cluster binding does not displace the structural Zn (before: ∼0.96 Zn/Aft2 monomer; after: ∼0.92 Zn/Aft2 monomer). To confirm that Fra2-Grx3 can directly transfer an Fe-S cluster to Aft2 and to rule out cluster degradation and reassembly on Aft2, we verified that: (i) [2Fe-2S] Fra2-Grx3 is resistant to EDTA at a 1:1 [EDTA]:[Fe] ratio, at which over 85% cluster is still intact (SI Appendix, Fig. S11A); and (ii) the addition of EDTA at a 1:1 ratio with Fe2+ in the reaction has little effect on the Aft2-Fra2-Grx3 transfer reaction (SI Appendix, Fig. S11B). These results show that the presence of EDTA does not alter the Fe-S cluster transfer process, ruling out the possibility that disassembly of the Fe-S cluster on Fra2-Grx3 is a prerequisite for Fe-S cluster assembly on Aft2.
Finally, we compared the DNA-binding affinity of as-purified, monomeric Aft2 and [2Fe-2S]-Aft2 prepared via cluster transfer from [2Fe-2S]-Fra2-Grx3 (SI Appendix, Fig. S12). The results show that [2Fe-2S] transfer to Aft2 causes at least a 3.3-fold reduction in DNA-binding affinity; the DNA dissociation constant of monomeric Aft2 is ∼26.1 ± 3.3 nM, which changed to ∼85.2 ± 2.1 nM for Aft2 with 0.5–0.6 [2Fe-2S] cluster bound per dimer. The difference is expected to be larger with fully loaded Aft2. Thus, both Fe2+- and Fe-S–induced dimerization of Aft2 interferes with its DNA-binding ability. Furthermore, we confirmed that substitution of one or both Cys in the CDC motif with Ala does not significantly alter the DNA-binding affinity (SI Appendix, Fig. S12C), which is consistent with the monomeric structure in vitro (Fig. 3A) and constitutive activity of these mutant forms in vivo (2, 4).
Discussion
In the current study, we present the biochemical and structural characterization of a major eukaryotic transcriptional regulator of iron homeostasis. Aft1 and Aft2 are 39% identical in their N-terminal DNA-binding domains. We have cloned, expressed, and purified a truncation of Aft2 that contains this homologous portion and includes the domains responsible for DNA binding and iron sensitivity.
The crystal structure of Aft2 bound to DNA reveals the molecular basis for selective binding of Aft2 to the consensus Fe-RE sequence, CACCC, found in the promoter regions of genes in the iron regulon. Aft2 possesses a WRKY-GCM1 fold that is responsible for many of the protein–DNA contacts. Protein residues within strands β2 and β3 and the β-hairpin connecting them establish base-specific contacts that underlie the direct recognition of the Fe-RE. All of the Aft2 residues involved in direct base contacts are conserved in Aft1, indicating that Aft1 recognizes the Fe-RE in the same manner as Aft2.
Both Aft1 and Aft2 belong to a class of proteins that has a WRKY-GCM1 domain with an insert between strands β2 and β3 of the four-strand fold (β3 and β4 in Aft1/2) (25). Aft2 has 22 residues in this insert and Aft1 has 71 residues; the difference in the size of this insert is the major difference between the two paralogs in their DNA-binding and metal-sensing domains. The inserts of both proteins are rich in basic amino acids, which suggest that they might interact with DNA. This insert in Aft2 is disordered in the crystal structure and could not be included in the model. However, based on the modeled portions of where the disordered insert must connect, the C terminus of β3 and N terminus of β4, the insert could contact the major groove at the 5′ end of the Fe-RE. (Another Aft2 monomer blocks access to that region of DNA by binding nonspecifically.) Variation in nucleotide sequence on the 5′ end of the Fe-RE allows for preferential binding of Aft1 or Aft2, and we propose that this insert might provide for such discrimination.
We have demonstrated that Aft2 has two metal-binding sites, one for a structural zinc and one for iron. Zinc is bound in a tetrahedral geometry by Cys86, Cys109, His133, and His135 in the DNA-binding domain, as shown in the crystal structure and supported by Zn2+-EXAFS and UV-visible spectroscopy of the Co2+-substituted protein. Substitutions in any of the Zn-coordinating ligands prevent proper folding of the protein, indicating that the zinc site is structural. Our data suggest that the second site is formed by cysteine residues in the CDC motif, C187 and C189, from two Aft2 monomers. Analytical ultracentrifugation and SAXS analyses reveal that Aft2 dimerizes in the presence of Fe2+ or a [2Fe-2S] cluster, leading us to conclude that four cysteines provide the ligands to coordinate iron.
Although Aft2 can bind both Fe2+ and a [2Fe-2S] cluster in vitro, previously published in vivo studies strongly favor [2Fe-2S] binding as the physiological iron-sensing event. The dependence of Aft1/2 inhibition on Fe-S cluster biogenesis has been revealed by numerous studies. The iron regulon is constitutively active in yeast strains with defects in mitochondrial Fe-S cluster biogenesis despite high cytosolic iron levels (14, 28, 29). Disrupting the mitochondrial Fe-S cluster export machinery and deletion of cytosolic proteins Grx3/4 or Fra2 also leads to constitutive activation of Aft1/2 target genes (14–16, 28, 29). In addition, the in vivo interaction between Aft1 and Grx3/4 and iron-induced dissociation of Aft1 from its target DNA are dependent on the mitochondrial Fe-S cluster biogenesis and export machineries (12). We have shown that the [2Fe-2S] cluster-bridged Fra2-Grx3 heterodimer transfers a [2Fe-2S] cluster to Aft2, providing a plausible mechanism for acquisition of an Fe-S cluster by Aft2 in vivo (Fig. 6). Fe-S binding by Aft2, in turn, decreases its DNA-binding affinity. The rapid Fe-S cluster transfer reaction between Fra2-Grx3 and Aft2 clearly distinguishes [2Fe-2S]-Fra2-Grx3 heterodimer from [2Fe-2S]-Grx3 homodimer and emphasizes the functional role of Fra2 in mediating the interaction between Aft2 and Grx3. However, in fra2∆ mutants, Aft1/2-regulated genes are still partially repressed under iron sufficiency, suggesting that Grx3/4 alone may be able to deliver Fe-S clusters to Aft1/2 in vivo, although somewhat less efficiently than Fra2-Grx3/4 (14).
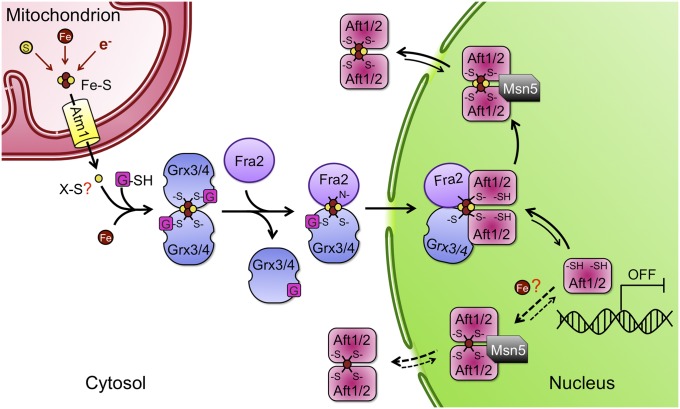
Fig. 6.
Proposed model for iron regulation via Aft1 and Aft2 under iron replete conditions. During conditions of iron sufficiency, Fe–S clusters are synthesized in mitochondria via integration of iron, sulfur, and redox control pathways. An unknown substrate produced by the mitochondrial Fe-S cluster biogenesis machinery is exported to the cytosol by the transporter Atm1. GSH is also required for export of this signal. Grx3 and Grx4, which form GSH-ligated, Fe-S–bridged homodimers, are proposed to form heterodimers with Fra2 to relay this signal to Aft1 and Aft2. Interaction of Grx3/4 with Aft1 promotes dissociation of the transcriptional activator from its target DNA and export to the cytosol, leading to deactivation of Aft1/2-regulated genes. The exportin Msn5 facilitates iron-dependent export of both Aft1 and Aft2. We also observed Fe2+-dependent dimerization of Aft2 in vitro; however, the biological relevance still needs to be investigated in vivo (dotted lines).
In addition to [2Fe-2S]-induced dimerization of Aft2, we present in vitro evidence that Aft2 dimerizes via Fe2+ binding to the CDC motifs. Because the expression of Aft1/2-regulated genes is unresponsive to changes in bioavailable cytosolic iron when mitochondrial Fe-S cluster biogenesis is fully functional (28, 29), this argues against direct binding of Fe2+ to Aft1 and Aft2 as the primary regulation mechanism. However, it is possible that under certain conditions Fe2+ binding may be an alternate dimerization mechanism. For example, the iron regulon is activated in grx3grx4aft1 null cells in response to iron deprivation despite the absence of Grx3/4, suggesting that Aft2 could respond through other pathways (16). Similarly, the expression of Aft1/2-regulated genes in cells with defective mitochondrial iron-sulfur cluster biogenesis is enhanced upon iron depletion (14), implying that high cytosolic iron in these cells may partially inhibit Aft1/2 activity via direct Fe2+ binding. However, the mitochondrial Fe-S cluster assembly pathway is still partially functional in these strains, and thus further iron limitation may deplete Fe-S clusters bound to proteins in a relatively labile fashion, such as found in Grx3/4 and Aft2 homodimers. Ultimately, our results provide the molecular details that support the previously proposed mechanism for iron-dependent inhibition of Aft1/2 activity (12, 13, 23, 24), namely that dimerization of Aft1/2 by coordination of a [2Fe-2S] cluster (or Fe2+) disrupts DNA binding and promotes cytoplasmic localization, resulting in the inactivation of the iron regulon (Fig. 6).
Materials and Methods
Aft2 Expression, Crystallization, Fe2+, and [2Fe-2S] Cluster Loading, Ultracentrifugation and SAXS Analysis, [2Fe-2S] Cluster Transfer from Fra2-Grx3 Heterodimer to Aft2.
Aft2(1–204) was cloned into pET30a, and the protein was expressed in BL21star(DE3) at 16 °C overnight. Aft2 was purified with either a HiTrap Heparin or SFF column, followed by a Superdex 200 column. Aft2(38–193) crystallized as a complex with a 12-bp oligonucleotide in the presence of 0.1 M Bis⋅Tris [pH 6.6], 23% PEG 2000 monomethyl ether, and 0.2 M MgCl2 at ambient temperature after 5 d using the hanging-drop vapor diffusion method. Fe2+ and [2Fe-2S] cluster were loaded into Aft2 inside an anaerobic chamber (O2, <5ppm) and then analyzed for iron and sulfur content. Ultracentrifugation was done at 60,000 rpm (AN 60 Ti rotor, Beckman Optima XL-A) for 15 h for complete sedimentation, and sedimentation profiles were fit to the Lamm equation. SAXS measurements were performed with ∼100 µM protein and electron-pair distribution curves and radii of gyration were calculated by GNOM. For transfer experiments, [2Fe-2S]-Fra2-Grx3 heterodimer was incubated with 2 equivalents of Aft2 and the cluster transfer was characterized by UV-visible and CD spectroscopy and SDS/PAGE.
Other Procedures.
Detailed procedures are available in SI Appendix.
Supplementary Material
Acknowledgments
We thank all beamline staff for their assistance. We thank Prof. T. Sosnick and Dr. E. Solomaha for assistance with the small-angle X-ray scattering and ultracentrifugation. GM/CA at the Advanced Photon Source (APS) at the Argonne National Laboratory has been funded, in whole or in part, by National Cancer Institute Grant Y1-CO-1020 and National Institute of General Medical Sciences (NIGMS) Grant Y1-GM-1104. The Biophysics Collaborative Access Team was supported by National Center for Research Resources (NCRR) Grant 2P41RR008630-17 and NIGMS Grant 9 P41 GM103622-17 from the National Institutes of Health (NIH). Use of the APS was supported by the US Department of Energy (DOE), Basic Energy Sciences, Office of Science, under Contract DE-AC02-06CH11357. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource (SSRL), a national user facility operated by Stanford University on behalf of the DOE, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the DOE, Office of Biological and Environmental Research, and by the NIH, NCRR, Biomedical Technology Program. This work was financially supported by National Science Foundation (NSF) Grant CHE-1213598 (to C.H.), NIH Grant GM100069 (to C.E.O.), a NSF Graduate Research Fellowship (to C.B.P.), and a Henry Dreyfus Teacher-Scholar Award from the Camille and Henry Dreyfus Foundation (to P.J.R.-G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4LMG).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318869111/-/DCSupplemental.
References
- 1.Kwok E, Kosman D. Topics in Current Genetics. Berlin, Heidelberg: Springer; 2005. Iron in yeast: Mechanisms involved in homeostasis; pp. 59–99. [Google Scholar]
- 2.Yamaguchi-Iwai Y, Dancis A, Klausner RD. AFT1: A mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 1995;14(6):1231–1239. doi: 10.1002/j.1460-2075.1995.tb07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaiseau PL, Lesuisse E, Camadro JM. Aft2p, a novel iron-regulated transcription activator that modulates, with Aft1p, intracellular iron use and resistance to oxidative stress in yeast. J Biol Chem. 2001;276(36):34221–34226. doi: 10.1074/jbc.M104987200. [DOI] [PubMed] [Google Scholar]
- 4.Rutherford JC, Jaron S, Ray E, Brown PO, Winge DR. A second iron-regulatory system in yeast independent of Aft1p. Proc Natl Acad Sci USA. 2001;98(25):14322–14327. doi: 10.1073/pnas.261381198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Philpott CC, Protchenko O. Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7(1):20–27. doi: 10.1128/EC.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rutherford JC, Bird AJ. Metal-responsive transcription factors that regulate iron, zinc, and copper homeostasis in eukaryotic cells. Eukaryot Cell. 2004;3(1):1–13. doi: 10.1128/EC.3.1.1-13.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casas C, et al. The AFT1 transcriptional factor is differentially required for expression of high-affinity iron uptake genes in Saccharomyces cerevisiae. Yeast. 1997;13(7):621–637. doi: 10.1002/(SICI)1097-0061(19970615)13:7<621::AID-YEA121>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi-Iwai Y, Stearman R, Dancis A, Klausner RD. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 1996;15(13):3377–3384. [PMC free article] [PubMed] [Google Scholar]
- 9.Rutherford JC, Jaron S, Winge DR. Aft1p and Aft2p mediate iron-responsive gene expression in yeast through related promoter elements. J Biol Chem. 2003;278(30):27636–27643. doi: 10.1074/jbc.M300076200. [DOI] [PubMed] [Google Scholar]
- 10.Courel M, Lallet S, Camadro JM, Blaiseau PL. Direct activation of genes involved in intracellular iron use by the yeast iron-responsive transcription factor Aft2 without its paralog Aft1. Mol Cell Biol. 2005;25(15):6760–6771. doi: 10.1128/MCB.25.15.6760-6771.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamaguchi-Iwai Y, Ueta R, Fukunaka A, Sasaki R. Subcellular localization of Aft1 transcription factor responds to iron status in Saccharomyces cerevisiae. J Biol Chem. 2002;277(21):18914–18918. doi: 10.1074/jbc.M200949200. [DOI] [PubMed] [Google Scholar]
- 12.Ueta R, Fujiwara N, Iwai K, Yamaguchi-Iwai Y. Iron-induced dissociation of the Aft1p transcriptional regulator from target gene promoters is an initial event in iron-dependent gene suppression. Mol Cell Biol. 2012;32(24):4998–5008. doi: 10.1128/MCB.00726-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ueta R, Fujiwara N, Iwai K, Yamaguchi-Iwai Y. Mechanism underlying the iron-dependent nuclear export of the iron-responsive transcription factor Aft1p in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18(8):2980–2990. doi: 10.1091/mbc.E06-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumánovics A, et al. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J Biol Chem. 2008;283(16):10276–10286. doi: 10.1074/jbc.M801160200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ojeda L, et al. Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J Biol Chem. 2006;281(26):17661–17669. doi: 10.1074/jbc.M602165200. [DOI] [PubMed] [Google Scholar]
- 16.Pujol-Carrion N, Belli G, Herrero E, Nogues A, de la Torre-Ruiz MA. Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J Cell Sci. 2006;119(Pt 21):4554–4564. doi: 10.1242/jcs.03229. [DOI] [PubMed] [Google Scholar]
- 17.Rutherford JC, et al. Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron-sulfur protein biogenesis. J Biol Chem. 2005;280(11):10135–10140. doi: 10.1074/jbc.M413731200. [DOI] [PubMed] [Google Scholar]
- 18.Li H, et al. The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry. 2009;48(40):9569–9581. doi: 10.1021/bi901182w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mühlenhoff U, et al. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 2010;12(4):373–385. doi: 10.1016/j.cmet.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bandyopadhyay S, Chandramouli K, Johnson MK. Iron-sulfur cluster biosynthesis. Biochem Soc Trans. 2008;36(Pt 6):1112–1119. doi: 10.1042/BST0361112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Picciocchi A, Saguez C, Boussac A, Cassier-Chauvat C, Chauvat F. CGFS-type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster. Biochemistry. 2007;46(51):15018–15026. doi: 10.1021/bi7013272. [DOI] [PubMed] [Google Scholar]
- 22.Iwema T, et al. Structural basis for delivery of the intact [Fe2S2] cluster by monothiol glutaredoxin. Biochemistry. 2009;48(26):6041–6043. doi: 10.1021/bi900440m. [DOI] [PubMed] [Google Scholar]
- 23.Li H, et al. Histidine 103 in Fra2 is an iron-sulfur cluster ligand in the [2Fe-2S] Fra2-Grx3 complex and is required for in vivo iron signaling in yeast. J Biol Chem. 2011;286(1):867–876. doi: 10.1074/jbc.M110.184176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Outten CE. Monothiol CGFS glutaredoxins and BolA-like proteins: [2Fe-2S] binding partners in iron homeostasis. Biochemistry. 2012;51(22):4377–4389. doi: 10.1021/bi300393z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babu MM, Iyer LM, Balaji S, Aravind L. The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res. 2006;34(22):6505–6520. doi: 10.1093/nar/gkl888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krizek BA, Merkle DL, Berg JM. Ligand variation and metal ion binding specificity in zinc finger peptides. Inorg Chem. 1993;32:937–940. [Google Scholar]
- 27.Foury F, Talibi D. Mitochondrial control of iron homeostasis. A genome wide analysis of gene expression in a yeast frataxin-deficient strain. J Biol Chem. 2001;276(11):7762–7768. doi: 10.1074/jbc.M005804200. [DOI] [PubMed] [Google Scholar]
- 28.Chen OS, et al. Transcription of the yeast iron regulon does not respond directly to iron but rather to iron-sulfur cluster biosynthesis. J Biol Chem. 2004;279(28):29513–29518. doi: 10.1074/jbc.M403209200. [DOI] [PubMed] [Google Scholar]
- 29.Li L, et al. A role for iron-sulfur clusters in the regulation of transcription factor Yap5-dependent high iron transcriptional responses in yeast. J Biol Chem. 2012;287(42):35709–35721. doi: 10.1074/jbc.M112.395533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.