Significance
Members of the 2-oxoglutarate (2OG)-dependent oxygenase superfamily catalyze a range of important biological oxidations. Structurally informed bioinformatic predictions suggest that the human genome encodes as yet unassigned members of the superfamily. We describe work demonstrating that 2OG and Fe(II)-dependent oxygenase domain-containing protein 1 (OGFOD1) is a protein hydroxylase that modifies the small ribosomal subunit protein RPS23 at a conserved prolyl residue in the ribosome-decoding center and that suppression or deletion of OGFOD1 is associated with the activation of translational stress pathways. Together with studies of homologous genes in flies and yeast described in accompanying manuscripts, the work identifies a unique function for 2OG oxygenase-catalyzed hydroxylation in ribosome biology.
Keywords: translational control, ribosome, 2-oxoglutarate oxygenase, hypoxia
Abstract
2-Oxoglutarate (2OG) and Fe(II)-dependent oxygenase domain-containing protein 1 (OGFOD1) is predicted to be a conserved 2OG oxygenase, the catalytic domain of which is related to hypoxia-inducible factor prolyl hydroxylases. OGFOD1 homologs in yeast are implicated in diverse cellular functions ranging from oxygen-dependent regulation of sterol response genes (Ofd1, Schizosaccharomyces pombe) to translation termination/mRNA polyadenylation (Tpa1p, Saccharomyces cerevisiae). However, neither the biochemical activity of OGFOD1 nor the identity of its substrate has been defined. Here we show that OGFOD1 is a prolyl hydroxylase that catalyzes the posttranslational hydroxylation of a highly conserved residue (Pro-62) in the small ribosomal protein S23 (RPS23). Unusually OGFOD1 retained a high affinity for, and forms a stable complex with, the hydroxylated RPS23 substrate. Knockdown or inactivation of OGFOD1 caused a cell type-dependent induction of stress granules, translational arrest, and growth impairment in a manner complemented by wild-type but not inactive OGFOD1. The work identifies a human prolyl hydroxylase with a role in translational regulation.
The human genome encodes ∼60 2-oxoglutarate (2OG)-dependent oxygenases that catalyze diverse biological oxidations including hydroxylation of small molecules and proteins, and demethylation of histones and DNA/RNA (1). The identification of two types of 2OG oxygenase that regulate the transcriptional response to hypoxia by prolyl and asparaginyl hydroxylations in hypoxia-inducible factor (HIF), has led to the proposal that the hydroxylation of intracellular proteins may be involved in other signaling mechanisms (2). We have recently assigned 2OG oxygenases related to the HIF asparaginyl hydroxylase, factor-inhibiting HIF, as histidinyl and argininyl hydroxylases that catalyze hydroxylation of eukaryotic and prokaryotic ribosomes, respectively (3).
2OG and Fe(II)-dependent oxygenase domain-containing protein 1 (OGFOD1) is a highly conserved 2OG oxygenase in eukaryotes. In the fission yeast, Schizosaccharomyces pombe, the homolog of OGFOD1, Ofd1, mediates oxygen-sensitive degradation of the N-terminal region of the transcription factor Sre1 [a homolog of sterol-response element-binding protein (SREBP)] after cleavage from the endoplasmic reticulum membrane, so contributing to oxygen-dependent regulation of the sterol response (4). The oxygen-sensitive SREBP pathway is not conserved in Saccharomyces cerevisiae (5), despite a conserved homolog of OGFOD1 [termination and polyadenylation 1 (Tpa1p)] in this species, suggesting Tpa1p has other roles. Indeed, TPA1 was identified in a screen for genes promoting stop codon readthrough (6). Tpa1p activity is linked to termination efficiency, mRNA polyadenylation, and mRNA stability. Structures of Tpa1p reveal the 2OG oxygenase characteristic double-stranded β-helix (DSBH) fold domain with typical Fe(II) and 2OG binding residues, but also indicate, as predicted for OGFOD1, the presence of a second DSBH fold domain in the C terminus, likely without a direct role in oxygenase catalysis (7, 8). Analysis of the active site of Tpa1p suggests it is a protein hydroxylase with distant similarity to the mammalian HIF prolyl hydroxylase domain (PHD)-containing family of enzymes (7, 8). Substitution of predicted active site residues in Ofd1 or Tpa1p abrogates the relevant function (4, 7), implying function is dependent on oxidation of an unassigned substrate.
Few reports describe the cellular role of mammalian OGFOD1. However, human OGFOD1 has recently been shown to be incorporated into stress granules upon arsenite-induced stress and associate with heme-regulated kinase and eIF2α in regulating eIF2α phosphorylation (9). Separately, it was reported that cells deficient in OGFOD1 manifest a survival advantage upon exposure to ischemic stress (10).
We report cellular and biochemical analyses demonstrating that OGFOD1 is a prolyl hydroxylase acting on a ribosomal protein. We show that OGFOD1 knockdown can result in translational arrest and stress granule formation. Using biochemical assays involving peptide screening, affinity purification coupled to MS, and purification of ribosomal proteins, we define RPS23 as an OGFOD1 substrate. Consistent with effects on translational control, the proline target site (Pro-62) occupies a critical position in the decoding center. Our results reveal the existence of a unique type of protein hydroxylation that is involved in the regulation of cellular growth.
Results
OGFOD1 Knockdown Inhibits Proliferation in a Range of Cell Lines and Can Induce Stress Granule Formation and Translation Arrest.
To investigate roles for OGFOD1 in cell proliferation we performed RNAi-mediated knockdown of OGFOD1 in human cell lines using pooled siRNA, and assessed responses by tetrazolium colorimetric [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS)] assay. OGFOD1 knockdown had a marked effect on the proliferation of HCT116, HEK293, MDA-MB-231, SW620 and U2OS cells. However, HeLa and NCI-H727 cells were relatively insensitive to OGFOD1 siRNA despite efficient OGFOD1 depletion (Fig. 1A and Fig. S1A). Similar results were obtained with single siRNA sequences and concordant data were collected with an alternate proliferation assay (Fig. S1 B and C). In contrast to the siRNA positive control (siDeath), OGFOD1 siRNA did not induce cell death and did not increase apoptosis as assessed by activated caspase-3 and cleaved PARP. To confirm the OGFOD1-dependent phenotype we stably expressed doxycycline (dox)-regulable OGFOD1 shRNA in U2OS and HeLa cells. Two U2OS-derived clonal lines that demonstrated substantial OGFOD1 knockdown were tested in an MTS assay. These clones expressed reduced OGFOD1 protein levels compared with the nontargeting control shRNA (CON) in the uninduced state, presumably as a result of leaky shRNA production. Whereas in dox-free culture the growth rates of control and OGFOD1 shRNA clones were similar, upon dox induction OGFOD1 shRNA clones displayed a marked reduction in proliferation that correlated with efficacy of OGFOD1 depletion (Fig. 1B). HeLa shRNA clones did not display a growth defect, consistent with siRNA results (Fig. S2A). Cumulatively, these data reveal a role for OGFOD1 in context-dependent regulation of cellular proliferation.
Fig. 1.
OGFOD1 knockdown inhibits proliferation in a range of cell lines and induces stress granules and translational stress in U2OS cells. (A) Effect of OGFOD1 siRNA on proliferation of cell lines. Proliferation was assessed by MTS assay upon transfection with a nontargeting siRNA pool (siCON) or equivalent targeting OGFOD1 (siOGFOD1). Data are presented as the ratio of absorbance (A490) at endpoint relative to day 1. siDeath served as a transfection control. Efficacy of OGFOD1 knockdown was confirmed by immunoblotting. Data (n = 3) are expressed as the mean; error bars ± SD. **P < 0.01; n.s., not significant. (B) shRNA-mediated knockdown of OGFOD1 inhibits proliferation of U2OS cells. U2OS clones encoding dox-inducible shRNAs against OGFOD1 (OGF) (clones 7 and 12) or control (CON) (clone 1) were assayed in the presence or absence of dox. MTS data are expressed as the A490 ratio at endpoint over start. OGFOD1 knockdown was confirmed by immunoblotting. Data are n = 6 and expressed as the mean; error bars ± SD. **P < 0.01. (C) shRNA-mediated knockdown of OGFOD1 induces stress granule formation in U2OS cells. U2OS shRNA clones were cultured in the presence (48 h) or absence of dox before arsenite stress (1 mM, 30 min). G3BP1 stress granules were detected by indirect immunofluorescence (green). Nuclei were stained with DAPI. Representative confocal images of dox-treated cells are depicted. Quantitation data (percent age of cells with G3BP1 foci) are from three independent experiments; values represent the mean; error bars ± SD. *P < 0.05. (D) Temporal induction of p–eIF2α following knockdown of OGFOD1. Immunoblot of U2OS shRNA clones treated with dox for the indicated time. (E) OGFOD1 knockdown inhibits protein synthesis rate. shRNA clones were labeled with [35S]-methionine for the indicated time. 35S incorporation was monitored by scintillation counting. Data (n = 4) represent the mean; error bars ± SD. **P < 0.01. (F) OGFOD1 knockdown induces expression of stress response targets, ATF4 and CHOP. Immunoblot detection of indicated proteins in U2OS shRNA cells treated with dox for 48 h.
OGFOD1 has been implicated in translational stress responses; specifically, stress granule resolution via modulation of eIF2α phosphorylation (9). We considered whether perturbations in this pathway might underlie the proliferative defect in U2OS cells following OGFOD1 knockdown. In cells manifesting an OGFOD1-dependent proliferative defect, we observed diffuse cytoplasmic staining of the stress granule marker G3BP1, which in ∼40% of cells concentrated into large foci upon OGFOD1 depletion, similar to that observed upon addition of arsenite (Fig. 1C). In contrast, neither the proliferative impairment nor the associated stress granule phenotype was observed in dox-regulable HeLa shRNA clones following depletion of OGFOD1 (Fig. S2), supporting the contextual specificity of the siRNA effect and suggesting that the observed phenotypes are, at least in part, linked.
In many settings, stress granule formation is associated with activation of inhibitory controls on protein synthesis. Indeed, we detected a temporal increase in phospho–eIF2α (p–eIF2α) with OGFOD1 depletion (Fig. 1D), which manifests as a ∼50% reduction in protein synthesis (Fig. 1E). Induction of p–eIF2α was associated with increased expression of ATF4 transcription factor and its target CHOP (Fig. 1F). Other predicted consequences of the eIF2α–ATF4 pathway activation were observed with increased LC3B lipidation in OGFOD1-depleted U2OS cells (Fig. S3), consistent with the onset of autophagy (11). Interestingly, despite efficient OGFOD1 knockdown, induction of p–eIF2α was not observed in HeLa cells, in keeping with the resistance of these cells to OGFOD1 siRNA-induced stress granule formation (Fig. S2).
OGFOD1 Catalysis Is Required for Rescue of Defective Growth and Suppression of Stress Granule Formation.
RNAi-mediated knockdown of Ogfod1 in SV40T immortalized mouse embryonic fibroblasts (MEFs) had no effect on cell growth, but following transformation with K-rasG12V MEFs manifest clear Ogfod1-dependent effects on proliferation (Fig. S1D). To determine whether these effects are observed in cells in which Ogfod1 is genetically inactivated, we generated immortalized and ras-transformed MEFs from embryos harboring floxed Ogfod1 (Ogfod1Fl/Fl) alleles. Pools of immortalized Ogfod1Fl/Fl MEFs and their transformed derivatives were adenovirally transduced with either control or cre recombinase, to promote loxP recombination of exon 4 and induce premature termination of Ogfod1 upstream of the catalytic site. Consistent with siRNA treatments, when wild-type (WT) (Ogfod1fl/fl) and Ogfod1−/− (KO) cells are directly compared it can be seen that inactivation of Ogfod1 had a significant effect on the proliferation of transformed MEFs (Fig. 2A). To investigate the role of OGFOD1 catalysis in proliferation, cDNAs encoding empty vector (EV), WT human OGFOD1 (WT rescue), or a mutant targeting an active site residue of OGFOD1 (D157A, MUT rescue) were expressed in transformed Ogfod1Fl/Fl MEFs, before cre-mediated excision of the floxed Ogfod1 allele. Cells reexpressing WT OGFOD1 protein proliferated more rapidly than cells expressing an EV or the D157A mutant, when assayed either in monolayer culture (Fig. 2B) or in a spheroid culture model (Fig. 2C).
Fig. 2.
OGFOD1 enzyme function rescues growth and protects against arsenite-induced stress granule formation. (A) Ras-transformed Ogfod1−/− MEFs display reduced proliferation rates relative to WT counterparts. Immortalized (SV40T) and ras-transformed (K-rasG12V) Ogfod1fl/fl MEFs were transduced with control (WT) or cre-recombinase (KO) to delete Ogfod1 alleles. Proliferation rates were determined by MTS assay and expressed as A490 ratio at endpoint relative to day 1. Data (n = 4) represent the mean; error bars ± SD. *P < 0.05; **P < 0.01. (B) Introduction of WT human OGFOD1 into transformed KO MEFs confers a proliferative advantage over hydroxylase mutant and null lines as determined by CyQUANT assay. Proliferation rates of transformed MEFs reexpressing either an EV, WT human OGFOD1, or D157A mutant (MUT) OGFOD1. Data are presented as the ratio of relative luminescence (RLU) at endpoint versus day 1. Data (n = 4) represent the mean; error bars ± SD. **P < 0.01; n.s., not significant. (C) Reexpression of WT OGFOD1 in transformed Ogfod1−/− MEFs confers a proliferative advantage over the D157A catalytic mutant and null lines in a spheroid culture model. Data are n = 36 spheroids per time point and expressed as mean values; error bars ± SD. **P < 0.01; n.s., not significant. (D) Low expression of p–eIF2α in unstressed transformed Ogfod1−/− MEFs. Immunoblot analysis of eiF2α and p–eIF2α in ras-transformed rescue MEFs. +, arsenite control for p–eIF2α immunoblot. (E) WT OGFOD1 suppresses arsenite-induced G3BP1 stress granule formation. Stress granules were detected and quantified by indirect immunofluorescence of G3BP1 (red) in transformed rescue MEFs. Nuclei were stained with DAPI. Cells were treated with arsenite (1 mM, 30 min) and representative confocal microscopy images are presented. Quantitation data (percent age of cells with G3BP1 foci) are from three independent experiments and represent mean values; error bars ± SD. **P < 0.01; n.s., not significant.
We next analyzed formation of G3BP1 stress granules in the same set of transformed Ogfod1−/− MEFs rescued with OGFOD1. Under basal conditions (in which OGFOD1-dependent effects on growth were observed) neither significant stress granule formation nor significant p–eIF2α induction was observed in Ogfod1−/− (EV) MEFs (Fig. 2D). However, upon exposure of the transformed MEFs to arsenite, we observed measurable stress granule formation in EV cells that was reversed in cells reexpressing WT, but not mutant OGFOD1 (Fig. 2E). Together, these data support the existence of context-dependent role(s) for OGFOD1 in cell growth and stress granule formation that are dependent on catalytic function.
Identification of the Small Ribosomal Subunit Protein RPS23 as an OGFOD1-Interacting Protein.
We next sought to define interaction partners of OGFOD1 using proteomics. Because the yeast homolog, Ofd1, regulates the stability of Sre1 (4), OGFOD1 complexes were purified from both control and proteasome inhibitor-treated HEK293 cell lysates in which FLAG-tagged OGFOD1 was stably expressed. SDS/PAGE revealed a major species of ∼80 kDa consistent with the molecular weight of OGFOD1, and two less-abundant species (∼100 and ∼16 kDa) that were detected with similar intensity in control and MG132-treated cells (Fig. 3A). Tandem MS assigned the 80-kDa species as OGFOD1, whereas the 16-kDa species was assigned as the small ribosomal subunit protein subunit, RPS23. Peptide-level quantitation indicated that the 100-kDa species predominantly contained equimolar amounts of OGFOD1 and RPS23, suggesting that these proteins form a high-affinity complex that is stable under SDS/PAGE conditions. To confirm the physiological existence of these complexes, interactions between endogenous OGFOD1 and RPS23 were assayed in HEK293 cells. Immunoblotting anti-OGFOD1 immunoprecipitates with an antibody to RPS23 revealed a similar profile, with RPS23 migrating at 16 kDa and as a 100-kDa OGFOD1–RPS23 complex (Fig. 3B).
Fig. 3.
Identification of the small ribosomal protein RPS23 as an interaction partner of OGFOD1. (A) Assignment of RPS23 as an OGFOD1-interacting protein. Coomassie/SDS/PAGE of FLAG immunoprecipitate (IP) from HEK293 cells expressing 3xFLAG–OGFOD1 or EV (CON). Species migrating at ∼100, ∼80, and ∼16 kDa were observed in OGFOD1 immunoprecipitates that were not altered by proteasomal inhibition (25 μM MG132, 8 h). MS/MS assigned the 80-kDa species as OGFOD1, the 16-kDa species as RPS23, and the 100-kDa species (denoted by “*”) as an OGFOD1–RPS23 complex. (B) Determination of interaction between endogenous OGFOD1 and RPS23. Anti-OGFOD1 antibody, and not control (CON) (rabbit IgG), coprecipitates RPS23 at its native molecular weight (16 kDa; arrow) and as a complexed form with OGFOD1 (100 kDa denoted by “*”) as determined by anti-OGFOD1 and anti-RPS23 immunoblot (IB) analysis. (C) Delineation of the RPS23 interaction domain of OGFOD1. FLAG IP from HEK293 cells expressing the indicated FLAG–OGFOD1 species. Formation of the 100-kDa RPS23–OGFOD1 complex is indicated by “*”. The arrow denotes RPS23 coprecipitating with OGFOD1 at 16 kDa (IB: RPS23). (D) Hydroxylase inhibitors block OGFOD1–RPS23 complex formation. HEK293 cells were transfected with 3xFLAG–OGFOD1 and exposed to DIP (100 μM, 12 h), DMOG (1 mM, 12 h) or DMSO vehicle control (No Tx). The position of the 100-kDa RPS23–OGFOD1 species upon FLAG immunoblotting is indicated by “*.” Controls for hydroxylase inhibition and protein loading were provided by HIF1α and p84 immunoblot, respectively.
OGFOD1 encodes a 542-residue protein consisting of a putative N-terminal oxygenase domain (134–239) and a C-terminal domain that does not contain a predicted catalytic site (262–538). To investigate the contribution of the N- and C-terminal domains to the RPS23 interaction, and the role of predicted catalytic (iron coordinating) residues, we transfected HEK293 cells with plasmids encoding FLAG-OGFOD1 variants. Robust associations were only observed with WT and near full-length (24-End) OGFOD1 (Fig. 3C). Interestingly, the D157A catalytic variant did not form the 100-kDa OGFOD1–RPS23 complex although a weak association with “native” RPS23 was preserved; a finding that was replicated in preparative FLAG-D157A pulldowns. To explore the dependence of these interactions on OGFOD1 catalysis, we expressed WT OGFOD1 in cells exposed to broad-spectrum 2OG oxygenase inhibitors. Formation of the 100-kDa species was inhibited by the Fe(II) chelator 2,2-dipyridyl (DIP) and the pan-hydroxylase inhibitor dimethyloxalylglycine (DMOG) (Fig. 3D).
Pro-62 of RPS23 Is Hydroxylated by OGFOD1.
Based on these data, we postulated that RPS23 is an OGFOD1 substrate. To investigate, we synthesized a tiled array of 20-mer peptides spanning the RPS23 protein and reacted individual peptides with recombinant OGFOD1 and analyzed the products by MS. Overlapping peptides encompassing residues 51–70 and 61–80 of RPS23 display a +16 Da mass increment, consistent with hydroxylation (Fig. S4A). Tandem MS analysis assigns the site of OGFOD1-catalyzed hydroxylation to Pro-62 of RPS23 (Fig. S4B). Supporting the assignment, a P62A mutant peptide was not modified by OGFOD1 (Fig. S4C).
Next we sought to determine the hydroxylation status of the RPS23 species copurifying with OGFOD1 by liquid chromatography mass spectrometry (LC-MS) (Fig. 3A). Surprisingly, we observed that both the ∼100- and ∼16-kDa RPS23 species were hydroxylated (Fig. S5). Together with earlier observations (Fig. 3C), these findings suggest that hydroxylation is necessary for the formation of the 100-kDa OGFOD1–RPS23 complex.
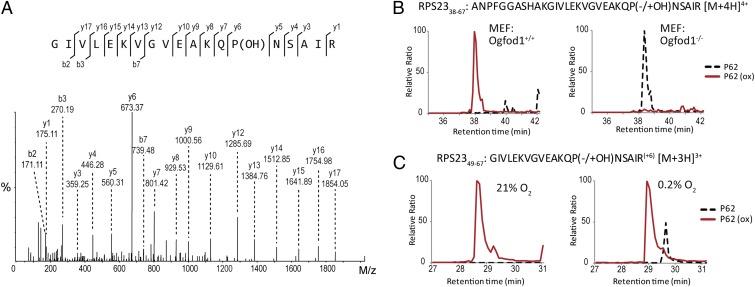
To determine whether the modification is present in RPS23 that is incorporated into ribosomes, we purified ribosomes from different mammalian sources. Tandem MS revealed that RPS23 hydroxylation was widespread; Pro-62 hydroxylation being detected in all samples examined to date (see Fig. 4A for MS/MS from HEK293 cells). Moreover, the modification was near complete; quantification of purified ribosomes by either intact protein MS or peptide-based MS indicated that >95% of ribosomal RPS23 was hydroxylated in normal tissue (mouse liver) and across a panel of cell lines (Fig. S6). OGFOD1 siRNA suppressed RPS23 hydroxylation to a similar extent in HeLa and U2OS cells indicating that OGFOD1 is nonredundant in both cell types and that the resistance of HeLa cells to the siRNA-associated growth and stress granule phenotypes is not due to the absence of an effect on RPS23 hydroxylation (Fig. S6). RPS23 isolated from Ogfod1−/− MEFs was not hydroxylated, confirming that RPS23 hydroxylation is a nonredundant function of Ogfod1 (Fig. 4B and Fig. S7A). Analysis of the rescue MEFs demonstrates that reintroduction of WT human OGFOD1, but not the D157A mutant, restores RPS23 hydroxylation (Fig. S7B), supporting the proposal that the observed effects on cell growth and stress granule formation relate to OGFOD1 catalysis (Fig. 2).
Fig. 4.
Pro-62 of RPS23 is hydroxylated by OGFOD1. (A) MS/MS assignment of Pro-62 hydroxylation in RPS23 isolated from HEK293 cells. RPS23 was subject to Arg-C proteolysis and MS/MS analysis. A mass identical to a hydroxyproline-containing fragment is observed in the GIVLEKVGVEAKQPNSAIR peptide ([M+3H]3+ = m/z 675.38) at the y6 ion that assigns hydroxylation to Pro-62. (B) Hydroxylation of RPS23 is Ogfod1-dependent. Extracted ion chromatograms (EICs) of m/z 762.18 and m/z 766.18 corresponding to unhydroxylated (dashed) and hydroxylated (red) forms of the Pro-62 containing RPS2338–67 peptide isolated from WT (Left) or KO (Right) OGFOD1 MEFs. (C) Incomplete suppression of RPS23 hydroxylation in 0.2% hypoxia. Media supplemented with isotopically labeled arginine (R+6) was added to U2OS cells before hypoxic (0.2% O2) (Right) or control (21% O2) (Left) exposure. Labeled peptides encompassing Pro-62 were quantified by LC-MS; EICs of m/z 672.06 and m/z 677.38 corresponding to unhydroxylated (dashed) and hydroxylated (red) forms of the GIVLEKVGVEAKQPNSAIR+6 ([M+3H]3+) peptide.
To investigate the effect of hypoxia on RPS23 hydroxylation, cells were exposed to different levels of hypoxia before MS analysis. Quantitation of RPS23 hydroxylation across a range of oxygen tensions revealed that the hydroxylation was relatively refractory to hypoxic stress. In U2OS cells exposed to severe hypoxia (0.2%, 24 h), 80% of newly synthesized RPS23 peptides that were isolated remained hydroxylated (Fig. 4C). Limiting nutrient supply (i.e., reduced serum and/or glutamine deprivation) did not constrain RPS23 hydroxylation in cells (Fig. S8A). In contrast, DMOG markedly inhibited RPS23 hydroxylation (Fig. S8B). Taken together, our analyses demonstrate that OGFOD1 is a bona fide prolyl hydroxylase that catalyzes mono-hydroxylation of Pro-62 in RPS23. Hydroxylation is prevalent across a range of cell lines and tissues and on protein incorporated into the assembled ribosome is near complete.
Studies on OGFOD1 Modulation of Translational Termination Efficiency.
Studies in prokaryotes and yeasts have implicated homologs of mammalian RPS23 in the maintenance of translational fidelity (12, 13). Because the target proline residue lies in a conserved loop that projects into the decoding center (14), we hypothesized that it may be required for efficient translation termination in mammalian cells as has been reported for Tpa1p in S. cerevisiae (6, 15), and that failure of this process might underlie aspects of the OGFOD1-associated translational stress phenotype. Thus, we expressed reporter vectors encoding different termination signals in the previously characterized Ogfod1−/− and reexpressing MEFs, and in the U2OS shRNA cell lines (16) (Fig. 5A). In the first instance we tested the termination signal from Tobacco mosaic virus (TMV) that promotes high-level stop codon readthrough in mammals and simple eukaryotes (17). Using this reporter we surprisingly observed an approximately two- to threefold increase in termination efficiency in Ogfod1−/− MEFs (Fig. 5B). These effects were restored by reexpression of WT human OGFOD1, but not the catalytically inactive D157A mutant, consistent with the effects being mediated by RPS23 hydroxylation. To determine whether variability in the response to different termination signals could reconcile our findings with the effects of Tpa1p in S. cerevisiae, a reporter that had previously been shown to direct increased readthrough in TPA1-defective yeast was tested (6); again we found no evidence for increased readthrough, and either no consistent effect, or a modest increase in the termination efficiency on the weaker TGA stop codon reporter (Fig. S9A). OGFOD1 knockdown via shRNA evokes a similar response (Fig. S9B). Although our data does not exclude the possibility that inactivation of OGFOD1 may promote readthrough of specific mammalian mRNAs and/or readthrough in other contexts, it does not support generalized readthrough or decoding defects as an explanation for the translational stress and growth phenotypes that were observed in association with altered OGFOD1 status.
Fig. 5.
OGFOD1 modulates translation termination efficiency. (A) Schematic of readthrough reporter encoding renilla luciferase (RLuc) and firefly luciferase (FLuc) reporters separated by an in-frame nonsense codon (as described by ref. 17). (B) OGFOD1 modulates termination efficiency of nonsense codons in the leaky TMV context. Data (n = 3) are expressed as mean values; error bars ± SD and are representative of three independent experiments.
Discussion
Our results assign mammalian OGFOD1 as a prolyl hydroxylase that targets the small ribosomal subunit protein RPS23. The work demonstrates a physiological role for OGFOD1 in the regulation of protein translation and cellular growth. In most cell lines, we observed that reduced, or absent, expression of OGFOD1 is associated with marked reduction in growth and the appearance of a translational stress phenotype. The latter was characterized by increased levels of p–eIF2α, a reduction in translation rate and the appearance of stress granules. Together, these findings indicate that OGFOD1 is a ribosomal oxygenase that plays a role in the regulation of translation. The target prolyl residue in RPS23 is conserved across evolution and in accompanying papers we demonstrate that OGFOD1 homologs (Sudestada1/Tpa1p/Ofd1) catalyze RPS23 hydroxylation in Drosophila and yeasts (15, 18).
Our findings implicating OGFOD1 in protein translation and stress granule formation are in part consistent with a report by Wehner et al., who identified OGFOD1 in stress granules and described the more slowly migrating OGFOD1 species that we show to be OGFOD1 complexed to hydroxylated RPS23 (9). However, there are differences: Wehner et al. examined the role of OGFOD1 on arsenite-induced translational stress in HeLa cells and reported that OGFOD1 knockdown reduced p–eIF2α and improved recovery from arsenite stress. These observations differ from ours: in HeLa cells we did not observe an effect of OGFOD1 intervention on p–eIF2α either in unstressed cells or under similar conditions of arsenite stress. However, the effects reported by Wehner et al. were relatively modest in relation to the large induction of p–eIF2α we observed in other cell lines. Because ribosomal stresses can induce p53 pathways (19), we considered whether the cell context specificity of the effects could be related to p53, but observed no correlation between the effects of OGFOD1 knockdown on proliferation and p53 status (Fig. S1A). The reasons for the cellular differences we observed in response to reduction or ablation of OGFOD1 are unclear, but may include different stress pathway activation mechanisms and/or the integrity of other checkpoint systems.
Unusual for 2OG oxygenases, we observed that OGFOD1 binds tightly to its product, hydroxylated RPS23, forming a SDS/PAGE resistant complex. Inhibition of OGFOD1 catalysis, by small molecules or substitution of residues that coordinate the catalytic iron, prevents complex formation. That OGFOD1 forms a tight complex with its hydroxylated product suggests stoichiometric complexation of RPS23 by OGFOD1 may be of functional significance. In the case of the related prolyl hydroxylase PHD2, hydroxylation substantially reduces binding to the enzyme–Fe(II)–2OG complex, but not the enzyme–Fe(II) complex (20). Thus, in the presence of sufficient 2OG, hydroxylation promotes release of HIF from PHD2. We have not yet been able to analyze the metal and small-molecule content of the OGFOD1–RPS23 complex. The observation that RPS23 is efficiently hydroxylated in cells suggests that 2OG levels are not limiting and infer an alternative explanation for the tight interaction that is likely unprecedented.
Structural studies indicate that Pro-62 forms the apex of a loop that projects into the codon–anticodon interface of the tRNA acceptor site (14). Assignment of hydroxylation at this position potentially rationalizes the finding that genetic inactivation of the OGFOD1 ortholog in S. cerevisiae, TPA1, is associated with increased readthrough across stop codons (6, 7, 15). We investigated whether the impaired growth and translational stress phenotype is related to coding inaccuracy; monitoring stop codon readthrough using transfected reporter genes. However, we found no clear evidence of increased readthrough. In cells expressing shRNA to OGFOD1 and in transformed Ogfod1−/− MEFs we observed increases in termination fidelity. In contrast, in an accompanying paper we observed clear bidirectional effects on stop codon readthrough in yeast (15); it may be that in mammalian cells other factors mask these effects or that they occur in other contexts. However, our findings indicate that it is unlikely that the translational stress phenotype we observe arises as a consequence of a general increase in readthrough or miscoding, although it is possible that coding accuracy at specific site(s) is responsible for signaling the translational stress phenotype.
The companion paper by Katz et al. reports the phenotypic consequences of silencing the Drosophila OGFOD1 ortholog, Sudestada1 (18). Striking similarities were observed in the associated knockdown phenotypes, with increased levels of translational stress and similar effects on cell growth control. However, there were differences, possibly relating to greater complexity in the pathways activating translational stress in mammalian cells: at least four kinase pathways are known to activate eiF2α as opposed to two (PERK and GCN2) in flies. We did not observe a canonical unfolded protein response (UPR) in mammalian cells; neither XBP1 mRNA cleavage, up-regulation of Bip1 transcript, nor induction of phospho-PERK was detected upon OGFOD1 silencing. Thus, UPR activation does not appear to be a general component of the OGFOD1-associated stress response in mammals.
In mammalian cells, effects on growth and translational stress were cell-type specific. It is likely that changes in OGFOD1 protein expression, as opposed to reduction below a threshold level, are responsible for effects on cell behavior. Dox-regulable U2OS shRNA clones manifesting chronic OGFOD1 knockdown (as a consequence of leaky expression) and transformed MEFs with biallelic inactivation of Ogfod1 display smaller changes in growth than those observed after acute knockdown. These dynamic effects argue against a signaling role of OGFOD1 being mediated directly via the hydroxylation status of the ribosome, where constituent proteins are stable postassembly. An alternative hypothesis is that RPS23 hydroxylation status has effects before ribosome assembly. MS analysis of RPS23 hydroxylation under conditions of incomplete hydroxylation (achieved by OGFOD1 knockdown) did not reveal any differences in RPS23 hydroxylation within the 40S and 80S fractions, suggesting that hydroxylation is not affecting 80S assembly or translation initiation directly, or acting as a “quality control” step at these stages (Fig. S10A). To date we have not observed effects of OGFOD1 status on ribosomal biogenesis as manifest by altered 40S:60S ratios assayed by polysome profiling. However, the absence of significant copurification of OGFOD1 with other ribosomal proteins, the existence of stable RPS23–OGFOD1 complexes and the localization of OGFOD1 in the nucleus (and not the nucleolus) are suggestive of an early preribosomal or extraribosomal signal (Fig. S10B).
Because OGFOD1 is primarily a nuclear protein, one possibility is that hydroxylation promotes complexation with RPS23 to connect ribosomal biogenesis with transcriptional regulation. The OGFOD1 ortholog in S. pombe, Ofd1, plays a key role in the oxygen-dependent control of the sterol response via proteolytic control of the Sre1 transcription factor (4). In animals, transcriptional regulation of the sterol response is not mediated in this way (5). Nevertheless, the unusual catalytic dependent formation of a nuclear complex containing a ribosomal protein suggests a link between ribosomal biogenesis and transcription that merits further exploration.
Materials and Methods
Cell Culture, Cloning, Enzyme Preparation/Hydroxylation Assays, and Mass Spectrometry.
Associated techniques and reagents are described in SI Materials and Methods.
Immunofluorescence.
Immunofluorescence used 4% (vol/vol) paraformaldehyde fixative, 0.5% Triton X-100 permeabilization and anti-OGFOD1 (Sigma Prestige) and anti-G3BP1 (BD Biosciences) antibodies. Secondary detection used Alexa-fluor (488/555; Invitrogen) conjugate. Samples were mounted in Vectashield with DAPI (Vector Laboratories) and images were acquired using a Zeiss LSM510 MetaHead confocal microscope.
Immunoblotting and Immunoprecipitation.
Lysates were immunoblotted using standard protocols and immunoprecipitations were as described (3). Antibodies are described in SI Materials and Methods.
Protein Synthesis Rates by 35S Methionine Incorporation.
Cells were incubated with 100 μCi/mL [35S]-methionine/cysteine for up to 1 h. Lysates were clarified and the incorporation of radiolabel was monitored on trichloroacetic acid filters by scintillation counting (3).
Dual-Luciferase Assays.
Cells were transfected with p2luci (16) variants and luciferase activity was determined using Dual-Luciferase assay (Promega). Recoding frequency is derived from the ratio of FLuc:RLuc for the nonsense reporter divided by the ratio of FLuc:RLuc of the analogous sense reporter, multiplied by 100. Assays were in triplicate and percent recoding values are expressed as mean ± SD.
Statistical Analysis.
Quantitative results are expressed as mean values ± SD. Statistically significant differences were obtained using either unpaired t test or one-way ANOVA with appropriate post hoc analysis (Dunnett’s or Tukey’s). *P < 0.05 and **P < 0.01 were considered to be significant.
Supplementary Material
Acknowledgments
We acknowledge the Transgenic Core (Wellcome Trust Centre for Human Genetics) and P. Jat, K. Kranc, and J. Atkins for provision of reagents. P.L.-Y. received a China Scholarship Council Award. A.W. received a European Molecular Biology Organization long-term fellowship. This work was supported by the Biotechnology and Biological Sciences Research Council, the Wellcome Trust, and the Ludwig Institute for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314482111/-/DCSupplemental.
References
- 1.Loenarz C, Schofield CJ. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem Sci. 2011;36(1):7–18. doi: 10.1016/j.tibs.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Cockman ME, Webb JD, Kramer HB, Kessler BM, Ratcliffe PJ. Proteomics-based identification of novel factor inhibiting hypoxia-inducible factor (FIH) substrates indicates widespread asparaginyl hydroxylation of ankyrin repeat domain-containing proteins. Mol Cell Proteomics. 2009;8(3):535–546. doi: 10.1074/mcp.M800340-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ge W, et al. Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. Nat Chem Biol. 2012;8(12):960–962. doi: 10.1038/nchembio.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes BT, Espenshade PJ. Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. EMBO J. 2008;27(10):1491–1501. doi: 10.1038/emboj.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bien CM, Espenshade PJ. Sterol regulatory element binding proteins in fungi: Hypoxic transcription factors linked to pathogenesis. Eukaryot Cell. 2010;9(3):352–359. doi: 10.1128/EC.00358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keeling KM, Salas-Marco J, Osherovich LZ, Bedwell DM. Tpa1p is part of an mRNP complex that influences translation termination, mRNA deadenylation, and mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26(14):5237–5248. doi: 10.1128/MCB.02448-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henri J, et al. Structural and functional insights into Saccharomyces cerevisiae Tpa1, a putative prolylhydroxylase influencing translation termination and transcription. J Biol Chem. 2010;285(40):30767–30778. doi: 10.1074/jbc.M110.106864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim HS, et al. Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex. Nucleic Acids Res. 2010;38(6):2099–2110. doi: 10.1093/nar/gkp1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wehner KA, Schütz S, Sarnow P. OGFOD1, a novel modulator of eukaryotic translation initiation factor 2alpha phosphorylation and the cellular response to stress. Mol Cell Biol. 2010;30(8):2006–2016. doi: 10.1128/MCB.01350-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saito K, Adachi N, Koyama H, Matsushita M. OGFOD1, a member of the 2-oxoglutarate and iron dependent dioxygenase family, functions in ischemic signaling. FEBS Lett. 2010;584(15):3340–3347. doi: 10.1016/j.febslet.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 11.B’chir W, et al. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41(16):7683–7699. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anthony RA, Liebman SW. Alterations in ribosomal protein RPS28 can diversely affect translational accuracy in Saccharomyces cerevisiae. Genetics. 1995;140(4):1247–1258. doi: 10.1093/genetics/140.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma D, Cukras AR, Rogers EJ, Southworth DR, Green R. Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. J Mol Biol. 2007;374(4):1065–1076. doi: 10.1016/j.jmb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandramouli P, et al. Structure of the mammalian 80S ribosome at 8.7 A resolution. Structure. 2008;16(4):535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loenarz C, et al. Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc Natl Acad Sci USA. 2014;111:4019–4024. doi: 10.1073/pnas.1311750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4(4):479–486. [PMC free article] [PubMed] [Google Scholar]
- 17.Stahl G, Bidou L, Rousset JP, Cassan M. Versatile vectors to study recoding: Conservation of rules between yeast and mammalian cells. Nucleic Acids Res. 1995;23(9):1557–1560. doi: 10.1093/nar/23.9.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Katz MJ, et al. (2014) Sudestada1, a Drosophila ribosomal prolyl-hydroxylase required for protein translation, cell homeostasis, and organ growth. Proc Natl Acad Sci USA 111:4025–4030. [DOI] [PMC free article] [PubMed]
- 19.Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: Probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010;29(30):4253–4260. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- 20.Flashman E, et al. Kinetic rationale for selectivity toward N- and C-terminal oxygen-dependent degradation domain substrates mediated by a loop region of hypoxia-inducible factor prolyl hydroxylases. J Biol Chem. 2008;283(7):3808–3815. doi: 10.1074/jbc.M707411200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.