Significance
Transcription factors (TFs) and their coregulators constitute two components of transcriptional regulatory complexes. TFs are thought to mediate genomic site selection, whereas coregulators appear to modulate the assembly/disassembly of the regulatory complex, which in turn specifies mechanisms of regulation. Here we show that coregulator hydrogen peroxide-inducible clone-5 (Hic-5) acts gene selectively in both modes. On some glucocorticoid receptor (GR) target genes it contributes to the assembly of transcription complexes. However, on other GR target genes it acts before genome occupancy by GR and thereby influences the set of sites occupied by GR. Because genomic occupancy by GR and other TFs varies in different cell types and regulatory contexts, Hic-5 represents an alternative mechanism for regulating DNA binding by TFs, complementing the influence of chromatin and pioneer factors.
Keywords: nuclear receptor, steroid receptor, enhancer
Abstract
Ligand activation and DNA-binding dictate the outcome of glucocorticoid receptor (GR)-mediated transcriptional regulation by inducing diverse receptor conformations that interact differentially with coregulators. GR recruits many coregulators via the well-characterized AF2 interaction surface in the GR ligand-binding domain, but Lin11, Isl-1, Mec-3 (LIM) domain coregulator Hic-5 (TGFB1I1) binds to the relatively uncharacterized tau2 activation domain in the hinge region of GR. Requirement of hydrogen peroxide-inducible clone-5 (Hic-5) for glucocorticoid-regulated gene expression was defined by Hic-5 depletion and global gene-expression analysis. Hic-5 depletion selectively affected both activation and repression of GR target genes, and Hic-5 served as an on/off switch for glucocorticoid regulation of many genes. For some hormone-induced genes, Hic-5 facilitated recruitment of Mediator complex. In contrast, many genes were not regulated by glucocorticoid until Hic-5 was depleted. On these genes Hic-5 prevented GR occupancy and chromatin remodeling and thereby inhibited their hormone-dependent regulation. Transcription factor binding to genomic sites is highly variable among different cell types; Hic-5 represents an alternative mechanism for regulating transcription factor-binding site selection that could apply both within a given cell type and among different cell types. Thus, Hic-5 is a versatile coregulator that acts by multiple gene-specific mechanisms that influence genomic occupancy of GR as well transcription complex assembly.
Glucocorticoid receptor (GR, NR3C1) regulates diverse physiological programs via gene activation and repression. The selection of target genes, directionality of regulation (up or down), and magnitude of the response are determined by a complex interplay of several factors: the activating ligand and DNA binding sequence that together modulate GR conformation and the local “regulatory environment” of each gene. The latter comprises chromatin conformation and other bound transcription factors (TFs). Together, these environmental determinants influence the recruitment of and requirement for specific coregulators that regulate transcription complex assembly. Current evidence suggests that scores of coregulators cooperate in the transcriptional regulation of each gene, with each coregulator contributing a specific molecular function, e.g., remodeling of chromatin or transcription complex assembly/disassembly (1). Moreover, coregulators act gene specifically, using different activation and repression domains to perform different actions on different genes, as specified by the bound GR (2–6). Protein allostery and the combinatorial nature of transcriptional regulation are responsible for the gene specificity of receptor and coregulator function (7, 8), but the specific steps facilitated by a coregulator in a given context are mostly unknown. Although deciphering the exact mechanisms driving context-dependent specificity of gene regulation will require system-wide studies that explore the influence of all interacting components of a regulatory network, the identification of such molecular mechanisms also will demand analyses at the individual gene level.
Combinatorial regulation by nuclear receptors is determined by interaction of the coregulator with various surfaces of the receptor (1, 9). Multiple functional surfaces have been implied by biochemical, molecular, genetic, and structural studies (8, 10), but only one of these surfaces, termed “AF2,” has been defined in detail (9). Another such functional surface is the hinge region of GR, which serves as a flexible linker between the DNA-binding domain (DBD) and the ligand-binding domain (LBD). The hinge contains a nuclear localization signal (11) and an acetylation motif that has been implicated in transcriptional regulation (12). Mutations that alter certain residues in the hinge region of steroid receptors can alter the specificity of response element occupancy (13). GR has a transcriptional activation function called “tau2,” mapped by deletion analysis to 29 amino acids (527–556 in human GR) that span the junction between the hinge and LBD (14). Hinge regions of other nuclear receptors are also important for transcriptional activation (15–17). Only a few coregulators that interact with the hinge region of nuclear receptors have been identified, including HEXIM1, JDP2, TAFII30, SNURF, ASC-1, BAF57, and hydrogen peroxide-inducible clone-5 (Hic-5, TGFB1I1) (18–24). Hic-5 is the only protein known to bind the relatively uncharacterized GR tau2 (24).
Lin11, Isl-1, Mec-3 (LIM) domain coregulator Hic-5 is a member of the paxillin family with multiple cellular locations and functions. In the cytosol, it acts as a scaffold at focal adhesions and controls important cellular processes such as proliferation, invasion, and apoptosis (25–27). In the nucleus, we and others have shown that it functions as a nuclear receptor coactivator in transient reporter gene assays (24, 28). Several physiological and pathological programs are known to depend on Hic-5 through its action with nuclear receptors [e.g., regulation of epithelial fat cell differentiation by PPARγ (29), endometriosis by modulation of PR (30), and prostate tumorigenesis by AR (31)]. Hic-5 also coregulates other TFs, e.g., SMADs and Sp1 (32, 33).
The two defining characteristics of transcriptional regulatory complexes—specific genomic occupancy and coassembly of multiple effector components to generate regulatory mechanisms—have long been considered to be functional roles resident separately in TFs and coregulators, respectively. Assuming that GR carries out genomic site selection, in this study we investigated the scope, specificity, and mechanism of Hic-5 coregulator function by combining experimental analyses of genome-wide expression, factor occupancy, chromatin structure, and gene-specific determination of regulatory mechanisms. Our results challenge the paradigm that genomic site selection by TFs is independent of coregulators.
Results
Hic-5 Depletion Affects both Activation and Repression of GR Target Genes.
After Hic-5 was depleted from U2OS-GR human osteosarcoma cells (which stably express GRα, the major GR isoform) using transfected siRNA, we performed gene-expression profiling of cells treated with the GR ligand dexamethasone (Dex) or ethanol (vehicle) for various lengths of time. Cells transfected with nonspecific siRNA were used as control. Each of two nonoverlapping siRNAs directed against the Hic-5 coding sequence efficiently reduced endogenous Hic-5 protein and mRNA (Fig. S1 A and B). Hic-5 depletion persisted through 24 h of Dex treatment (Fig. S1C). Genome-wide transcript levels were measured using the Illumina bead microarrays (Fig. S1D).
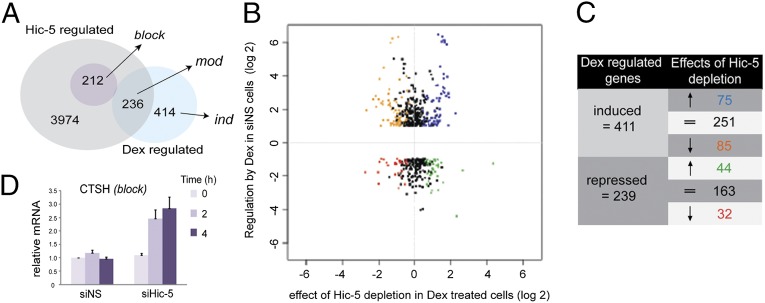
The Dex-regulated subset of genes included 650 genes in Hic-5+ cells that were either positively or negatively regulated by Dex at 4-h treatment (Fig. 1A and Dataset S1, sheet 1). Comparing the differences in transcript levels at 4 h of Dex treatment in Hic-5+ and Hic-5–depleted samples, we found 4,422 genes with significant changes in mRNA levels caused by Hic-5 depletion (Fig. S1E, compare d and b). The intersection of this subset with the Dex-regulated subset included 236 genes that were both Dex regulated and Hic-5 regulated (Fig. 1A, Dataset S1, sheet 2). This subset, the Hic-5–modulated class (mod), represented 36% of all Dex-regulated genes, indicating a broad but selective role for Hic-5 on GR-mediated transcription. The 414 Dex-regulated genes that were unaffected by Hic-5 depletion were designated the “Hic-5–independent” class (ind) (Fig. 1A and Dataset S1, sheet 3), although it is likely that some ind genes were differentially regulated but did not yield a statistically significant result.
Fig. 1.
Differential effects of Hic-5 depletion on transcriptional regulation by GR. (A) Venn diagram showing effects of Hic-5 depletion and Dex treatment on global gene expression. The Hic-5–regulated set indicates the number of genes with mRNA levels that differ significantly (q < 0.02) after 4 h of Dex treatment for cells treated with Hic-5–directed siRNA#2 (siHic-5) versus nonspecific siRNA (siNS). The Dex-regulated set shows the number of genes with mRNA levels that differ at least twofold and q < 0.02 between siNS-transfected cells treated with ethanol or Dex for 4 h. The overlap shows genes belonging to the Dex-regulated set that also are significantly affected by Hic-5 depletion (Hic-5–modulated, mod). The Dex-regulated genes outside the overlap are Hic-5–independent (ind), and the small circle in the Hic-5–regulated set indicates Hic-5–blocked genes (block). (B) Scatter plot showing the effect of Dex versus the effect of Hic-5 depletion on the mRNA level for genes in the Dex-regulated set. The y-axis shows the log2 of the fold change when comparing 4-h Dex–treated versus ethanol–treated Hic-5+ cells; the x-axis shows the log2 of the fold change when comparing Hic-5–depleted cells versus Hic-5+ cells at 4 h Dex treatment. Black dots indicate ind genes. Colored dots (coordinated with C) indicate mod genes. (C) Number of mod genes in each color-coded subset from B. (D) RT-qPCR validation of a candidate block gene (CTSH). mRNA expression values were normalized to Gapdh mRNA levels for each sample and are represented as mean ± SEM of three biological replicates.
To analyze further the effect of Hic-5 on Dex regulation of mod genes, we generated a scatter plot with effects of Dex in Hic-5+ cells on the y-axis and the corresponding effects of Hic-5 depletion in cells treated with Dex for 4 h on the x-axis (Fig. 1 B and C). The black dots represent ind genes. The colored dots in the different quadrants represent mod genes and include (i) genes up-regulated by Dex with the induction inhibited by Hic-5 depletion (orange, 85 genes); (ii) genes up-regulated by Dex with the induction further enhanced by Hic-5 depletion (blue, 75 genes); (iii) genes repressed by Dex with the repression reversed by Hic-5 depletion (green, 44 genes); and (iv) genes repressed by Dex with the repression further augmented by Hic-5 depletion (red, 32 genes). Four mod genes that were substantially affected by Hic-5 depletion in the microarray data and represented positive and negative effects of Hic-5 on Dex-regulated genes were validated by reverse transcription coupled with quantitative PCR (RT-qPCR) along with two ind genes (Fig. S2 A and B). WNT5A was induced by Dex in the presence of endogenous Hic-5, but Hic-5 depletion essentially eliminated induction by Dex (Fig. S2A), indicating Hic-5′s coactivator function on WNT5A. On the other hand, CX3CL1 expression is repressed by Dex when Hic-5 is present but is repressed less efficiently after Hic-5 depletion (Fig. S2B). Thus, Hic-5 can function as either a coactivator or a corepressor. In addition, Hic-5 also opposed the action of Dex and GR on other genes; e.g., Dex-induced expression of RGS2 was further enhanced by Hic-5 depletion, and KLF10 repression by Dex was stronger when Hic-5 was depleted (Fig. S2 A and B). We also validated the ind genes CRYGC and CCRN4L, whose regulation by Dex was not affected by Hic-5 depletion (Fig. S2 A and B).
We also found another distinct subset of genes that normally was unresponsive or weakly responsive to Dex in the presence of Hic-5 but became robustly Dex regulated upon Hic-5 depletion (Fig. 1A, Fig. S2C, and Dataset S1, sheet 4). This subset of 212 genes was categorized as the “Hic-5–blocked” class (block) and included genes that become Dex induced (Fig. S2C, Upper Right Quadrant, darker blue, 115 genes) or Dex repressed (Fig. S2C, Lower Right Quadrant, darker green, 97 genes) only upon Hic-5 depletion. RT-qPCR analysis validated CTSH as an example of a block gene (Fig. 1D). Almost half (107 genes) of the mod genes (e.g., RGS2 and KLF10) resemble block genes, because their regulation by Dex was augmented by Hic-5 depletion. However, mod genes already were regulated at least twofold (q < 0.02) by Dex before Hic-5 depletion, whereas block genes did not pass the imposed twofold change and q < 0.02 cutoffs until Hic-5 was depleted. Altogether, 319 genes became considerably more Dex regulated after Hic-5 depletion.
We used Gene Ontology (GO) analysis to test whether we might find a striking distinction between Hic-5–dependent (combined mod and block genes) and ind genes; such a distinction might strongly infer Hic-5 function. Although two of the top categories of Hic-5–dependent genes also were strongly represented among the ind genes, the two classes of Dex-responsive genes were largely distinct (Fig. S3). The Hic-5–dependent group included response to hypoxia, regulation of cell migration, adhesion, differentiation, and apoptosis, but no single class was so dominant as to infer a single primary function of Hic-5 in the context of GR action (Dataset S2, sheets 1 and 2).
Hic-5 Exerts Its Effects at the Level of Transcription.
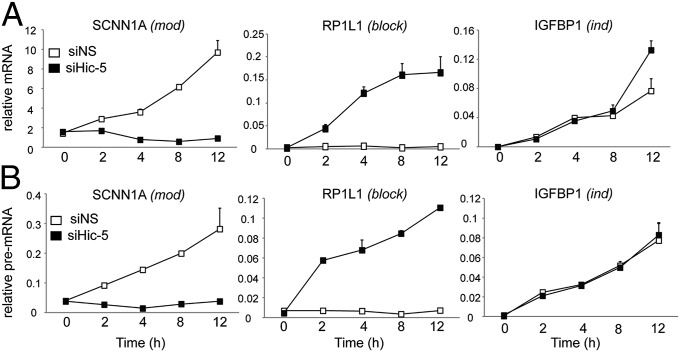
To characterize Hic-5 action further, we tracked mRNA expression profiles of SCNN1A (mod), IGFBP1 (ind), and RP1L1 (block) upon Dex treatment in the presence and absence of Hic-5, focusing on genes that were induced upon Dex treatment. Induction of SCNN1A by Dex in Hic-5+ cells was completely inhibited by Hic-5 depletion (Fig. 2A). In contrast, RP1L1 was not regulated by Dex in the presence of Hic-5, but Hic-5 depletion allowed robust induction by Dex. Dex regulation of IGFBP1 was unchanged by Hic-5 depletion (Fig. 2A). Because mRNA accumulation is affected by several factors in addition to synthesis (e.g., splicing, transport, and turnover), we measured the levels of newly synthesized primary transcripts. The pre-mRNA expression profiles of the three genes closely resembled the mRNA patterns (Fig. 2B), indicating that Hic-5 acts to regulate pre-mRNA synthesis. To increase confidence that effects of Hic-5 depletion on transcription were not mediated by off-target effects of the Hic-5 siRNA#2, we used an additional siRNA construct (siHic-5#1), which also efficiently depleted Hic-5 levels (Fig. S1 A and B), and found that it reproduced the effects of siHic-5#2 on SCNN1A, RP1L1, and IGFBP1 (Fig. S4). Thus, Hic-5 served as an on/off switch for Dex regulation of some genes.
Fig. 2.
Hic-5 affects Dex-regulated gene expression at the level of transcription. Cells were transfected with siNS and siHic-5 for 2 d and then were exposed to 100 nM Dex for 2, 4, 8, or 12 h. Relative levels of mRNA (A) and pre-mRNA (B), determined by RT-qPCR, are shown. Treatment with an equivalent amount of ethanol for 4 h was used for the 0-h time point. Relative RNA values were normalized to Gapdh mRNA levels for each sample and are mean ± SEM of three biological replicates.
Hic-5 Facilitates Dex-Induced Coregulator Recruitment to mod Genes.
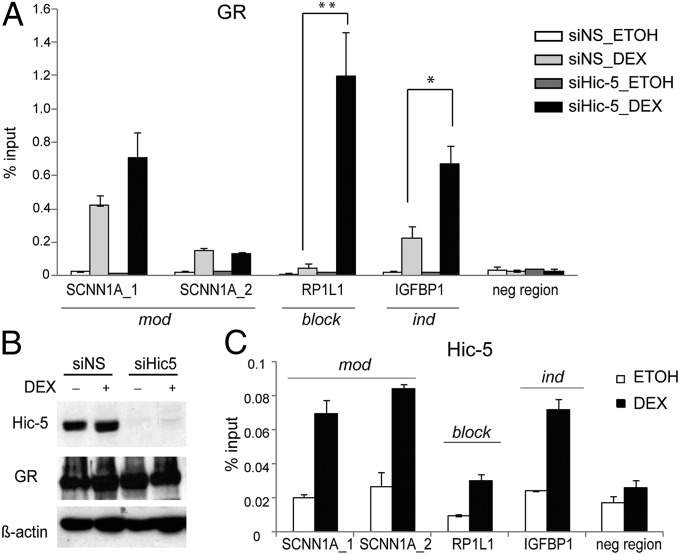
To explore the mechanism by which Hic-5 contributes to Dex-induced expression of mod genes, we evaluated whether GR occupancy on the presumed glucocorticoid (GC) response elements (GRE) of those genes was altered by Hic-5 depletion. Information on locations of GR binding regions (GBRs) in U2OS-GR cells was acquired by deep-sequencing analysis of ChIP-enriched DNA obtained with a GR-specific antibody, and the Genomic Regions Enrichment of Annotations Tool (34) was used to assign peaks to genes. Selected peaks were validated subsequently by ChIP-qPCR analysis (Fig. S5). The two GBRs identified near SCNN1A (mod) had similar levels of Dex-induced occupancy by GR when Hic-5 was present or absent (Fig. 3A), indicating that loss of transcriptional activation upon Hic-5 depletion (Fig. 2A) was not caused by loss of GR binding but instead involved Hic-5 actions subsequent to GR binding. Two other mod genes, FKBP5 and SGK1 (Fig. S6A), also showed Dex-induced GR occupancy despite Hic-5 depletion (Fig. S6B). In fact, Hic-5 depletion increased Dex-induced GR occupancy at the GBRs of most genes tested, including ind genes IGFBP1, MSX2, and CRYGC (Fig. 3A and Fig. S6B). Because Hic-5 depletion did not affect ind genes, we failed to detect a simple correlation between the level of GR occupancy and the level of transcriptional activity for the mod and ind genes.
Fig. 3.
GR and Hic-5 occupancy of GR target genes. (A) GR occupancy. Cells were transfected with siNS or siHic-5 and treated with Dex or ethanol for 1 h. DNA from immunoprecipitated chromatin was analyzed by qPCR using the indicated primers and normalized to input chromatin. *P ≤ 0.05, **P < 0.01 calculated using a paired t test for results from three independent experiments. Abbreviations are as in Fig. 1; neg region, negative control region of DNA. (B) Immunoblot showing effects of Hic-5 depletion on Hic-5 and GR protein levels in cells treated with Dex or ethanol for 1 h, with β-actin protein as internal control. (C) Hic-5 occupancy. ChIP was performed in cells treated with Dex or ethanol for 1 h. Hic-5 occupancy at the indicated GBR is expressed as the mean and range of variation for two technical replicates from one experiment and is representative of at least three independent experiments.
Hic-5 conceivably could facilitate transcriptional activation by direct action at the GR target gene or indirectly by regulating transcription of a primary gene whose product regulates the GR target gene. Because direct action would require Hic-5 occupancy at the gene, we conducted ChIP analysis using a Hic-5-specific antibody. Hic-5 was recruited in a Dex-dependent manner to the GBRs of SCNN1A and FKBP5 (mod) and to the GBRs of MSX2 and IGFBP1 (ind) (Fig. 3C and Fig. S6C), as is consistent with direct action of Hic-5 on GR target genes. The recruitment of Hic-5 to ind genes, which do not require Hic-5 for their Dex-induced expression, likely reflects the combinatoriality and context dependence of metazoan transcriptional regulation, in which the actions of a given coregulator (e.g., recruitment of a general transcription factor) may produce a regulatory function (e.g., increased initiation rate) in some cellular or physiologic settings but not in others. We confirmed by coimmunoprecipitation that endogenous Hic-5 interacts with stably over-expressed GR in U2OS-GRα cells (Fig. S6D). Interestingly, the interaction was Dex independent. We also were able to detect an interaction when the two proteins were transiently expressed in Cos-7 cells (Fig. S6 E and F). The GR–Hic-5 interaction presumably is responsible for Hic-5 recruitment to GR target genes and provides a potential avenue for direct Hic-5 action on these genes.
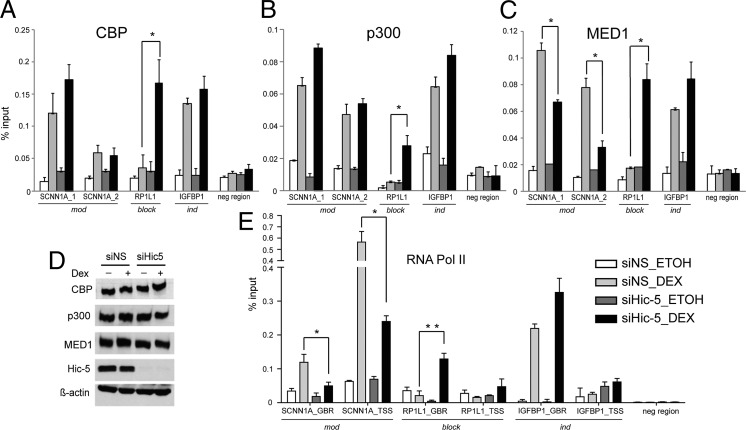
Next, we used quantitative ChIP to analyze the recruitment of coregulators that control important steps in transcription complex assembly. CREB-binding protein (CBP) and p300, two protein/histone acetyltransferases, were recruited efficiently to the SCNN1A GBRs in response to Dex whether Hic-5 was present or depleted (Fig. 4 A and B). In contrast, Dex-induced occupancy of Mediator complex subunit 1 (MED1) on SCNN1A was reduced when Hic-5 was depleted (Fig. 4C). CBP, p300, and MED1 protein levels were not altered by Hic-5 depletion (Fig. 4D). Similar observations were made on FKBP5 (mod) (Fig. S7 A–C). In agreement with the known role of MED1 as a bridge between enhancers and promoters of genes by directly contacting the basal transcriptional machinery and RNA polymerase II (Pol II) (35), we found that Hic-5 depletion caused reduced Dex-induced RNA Pol II occupancy of the SCNN1A transcription start site (TSS) and GBR (Fig. 4E). Similar results were observed at the TSS of FKBP5 (Fig. S7D). In contrast to the mod genes, recruitment of MED1 and RNA Pol II to ind genes IGFBP1 and MSX2 was not diminished by Hic-5 depletion and instead was increased on many ind genes (Fig. 4 C and E and Fig. S7 C and D); although the increase was consistent, the differences were not significant over multiple experiments. The fact that ind gene expression did not increase upon Hic-5 depletion (despite increased occupancy by GR, coregulators, and RNA Pol II) indicates that the occupancy levels of these proteins in the presence of Hic-5 were not rate limiting for optimal Dex-induced expression of ind genes.
Fig. 4.
Hic-5 either facilitates or blocks transcription complex assembly. ChIP assays evaluated CBP (A), p300 (B), MED1 (C), and RNA Pol II (E) recruitment in cells transfected with the indicated siRNA and treated with ethanol or Dex for 1 h. DNA from immunoprecipitated chromatin was analyzed by qPCR using primers specific for the indicated GBR in A–C and for the indicated GBR or TSS in E. Data shown are the mean of two PCR reactions performed on the same DNA samples from a single experiment, normalized to input DNA, and are representative of three independent experiments. Error bars indicate the range of variation of two technical replicates. *P ≤ 0.05 calculated using a paired t test for three independent experiments. Abbreviations are as in Fig. 3A. (D) Immunoblot showing effects of Hic-5 depletion on CBP, p300, and MED1 protein levels after treatment with ethanol or Dex for 1 h.
Hic-5 Inhibits Dex-Induced GR Binding to block Genes.
To investigate the mechanism by which Hic-5 prevents GC regulation of block genes, we evaluated GR occupancy of selected block GBRs. The GBR associated with the RP1L1 gene was weakly occupied by Dex-activated GR in the presence of Hic-5 (Fig. 3A). However, Hic-5 depletion caused a dramatic enhancement of GR occupancy at this site (Fig. 3A), which was not caused by alterations in cellular GR protein levels (Fig. 3B). Hic-5 occupancy at the RP1L1 GBR was weaker than in mod and ind genes (Fig. 3C), presumably because of weak GR binding at that site. Similar results for GR and Hic-5 occupancy were observed at the GBR of another block gene, HOXD1 (Fig. S6 B and C). Although Hic-5 depletion caused moderate enhancement of GR binding to several mod and ind genes, the dramatic change in GR occupancy at block genes after Hic-5 depletion indicates that restricted GR binding may be the underlying mechanism by which Hic-5 prevents efficient transcription of those genes. We examined steps in transcription complex assembly that occur after GR binding, and found that Hic-5 depletion caused dramatic enhancement of Dex-induced occupancy of most of the transcription complex components tested (coregulators CBP, p300, and MED1 and RNA Pol II) at the GBRs of the block genes RP1L1 and HOXD1 (Fig. 4 and Fig. S7). Notably even though weak Dex-induced GR binding was seen at block genes when Hic-5 was present (Fig. 3A and Fig. S6B), there was little or no Dex-induced binding of CBP, p300, MED1, or RNA Pol II in the presence of Hic-5. Collectively, for block genes, Hic-5 strongly inhibited GR binding and precluded transcription complex assembly.
Hic-5 Regulates Chromatin Remodeling of block Genes.
To explore further how Hic-5 restricts GR binding and coregulator recruitment to block genes, we examined chromatin remodeling at GBRs of selected genes using Formaldehyde Assisted Isolation of Regulatory Elements (FAIRE) followed by qPCR. For mod genes (SCNN1A, FKBP5, and SGK1) and ind genes (IGFBP1, MSX2, and CRYGC), treatment with Dex increased the FAIRE signals at the GBRs in Hic-5+ cells, indicating opening of the chromatin conformation. Hic-5 depletion moderately increased the Dex-induced FAIRE signals at these genes (Fig. 5 and Fig. S7E). In sharp contrast, Dex caused little or no increase in open chromatin, as seen by low FAIRE signals, at GBRs of block genes (RP1L1 and HOXD1) when Hic-5 was present. However, Hic-5 depletion caused robust induction of FAIRE signals by Dex at these regions (Fig. 5 and Fig. S7E). These results suggest that the weak GR binding and coregulator recruitment at block genes may reflect Hic-5 inhibition of GR-mediated chromatin remodeling.
Fig. 5.
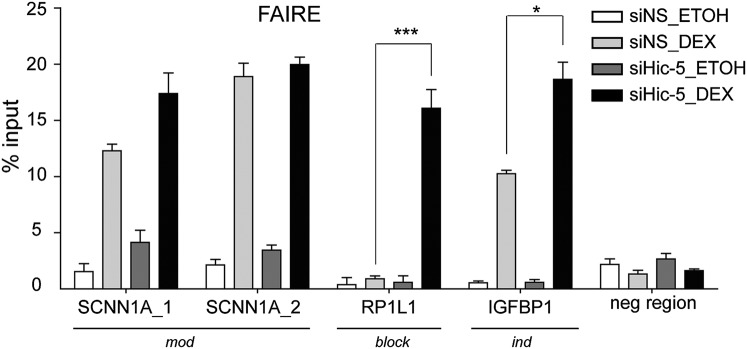
Hic-5 regulation of Dex-induced chromatin remodeling. Chromatin remodeling was measured by FAIRE coupled with qPCR at GBRs of the indicated genes in cells transfected with siNS or siHic-5 and then treated with Dex or ethanol for 1 h. The graph depicts the percentage of input DNA recovered in the soluble fraction after chromatin sonication, expressed as mean ± SD of the FAIRE signal for three technical replicates from a single experiment and is representative of at least three independent experiments.*P ≤ 0.05 and ***P < 0.001 calculated using a paired t test for three independent experiments.
Discussion
Gene-Specific Roles of Hic-5 in Dex-Regulated Transcription.
In this study, we provide insights into the nature and mechanism of Hic-5 coregulator function in global GC-regulated transcription. We defined distinct subsets of genes for which GR function was influenced by Hic-5. Hic-5 plays diverse roles characterized by several distinct principles. First, Hic-5 action was highly gene specific, because it was not required for regulation of all genes by Dex. Second, Hic-5 depletion had dramatic effects on Dex-regulated transcription of some genes, in which Hic-5 served as an all-or-none switch, so that Dex-induced transcriptional regulation by GR was completely dependent on the presence of Hic-5 for some genes and on the absence of Hic-5 for other genes. In contrast, depletion studies with many other coregulators have indicated that they serve as modulators rather than as all-or-none determinants of steroid hormone-regulated gene expression (2, 36–39). Third, Hic-5 was able to function as a coactivator and as a corepressor. Fourth, Hic-5 supported the regulatory actions of Dex and GR on some genes but opposed GR actions on other genes. Finally, and perhaps most noteworthy, Hic-5 functioned either before or after the establishment of genomic occupancy by GR. The highly gene-selective actions of Hic-5 indicate that Hic-5 controls specific physiological pathways regulated by GR. Indeed, a tissue-specific pattern of Hic-5 expression has been reported, with high levels in smooth muscle tissues and myoepithelial cells compared with undetectable levels in epithelial cells from the stomach, colon, liver, skin, and breast (40), suggesting that Hic-5 may influence cell type-specific gene regulation by GR.
Selective Modulation of Regulatory Complex Assembly by Hic-5.
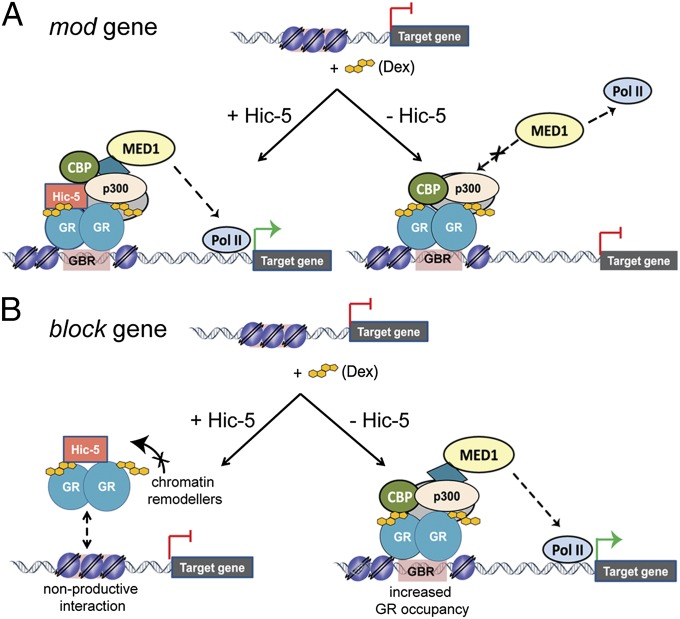
In this study, we were able to distinguish two mechanistic classes of Dex-responsive genes regulated by Hic-5, mod and block genes. In our analysis of the mod class we focused on genes that require Hic-5 for Dex-induced transcription. We found that depletion of Hic-5 did not affect Dex-induced GR binding, chromatin remodeling, or recruitment of CBP and p300 to the GBRs; rather, it reduced MED1 and RNA Pol II recruitment, revealing the mechanism of Hic-5 action on these genes (Fig. 6A). Mediator complex (of which MED1 is a subunit) facilitates RNA Pol II recruitment to promoters (35), explaining why the Dex induction of mod genes was inhibited by Hic-5 depletion. The observation that efficient MED1 recruitment is regulated by Hic-5 supports the view that Hic-5 serves as a molecular scaffold, possibly through its LIM domains, which are known protein-interaction motifs, to stabilize higher-order coregulator complexes (41).
Fig. 6.
Proposed models for Hic-5 action on Dex-induced GR target genes. (A) For mod genes that require Hic-5 for Dex-induced expression, Hic-5 facilitates Dex-induced recruitment of the Mediator complex (MED1), which recruits RNA Pol II (Lower Left). In the absence of Hic-5 (Lower Right), GR binding, nucleosome repositioning to create open chromatin at the GBR, and recruitment of p300 and CBP are still induced by Dex, but MED1 and RNA Pol II are not recruited efficiently. (B) For block genes that become Dex-inducible after Hic-5 depletion, induction by Dex in the presence of Hic-5 induces weak GR binding, and GR recruits Hic-5, which prevents recruitment and/or action of chromatin-remodeling coregulators, resulting in a repressive chromatin conformation at GBRs. Hic-5 depletion allows more robust Dex-induced GR recruitment, nucleosome repositioning, and transcription complex assembly.
Selective Modulation of GR-Binding Site Selection by Hic-5.
The block genes represent the second mechanistic class of Dex-responsive, Hic-5–regulated genes and revealed an unexpected action of a coregulator on genomic site selection by GR. Our FAIRE results indicate that Hic-5 precludes Dex-induced chromatin remodeling, whereas depletion of Hic-5 results in robust Dex-induced chromatin remodeling and GR occupancy at block genes that become Dex inducible after Hic-5 depletion. A parsimonious model for the block subclass is that Hic-5 might impede the establishment of robust GR occupancy at block GREs by preventing GR-dependent chromatin remodeling (Fig. 6B), although other models also are feasible. The demonstrated GR and Hic-5 interaction (Fig. S6 D–F) and the Dex-induced occupancy of Hic-5 on the GBRs associated with the genes examined in this study (Fig. 3C) are consistent with Hic-5 action on block genes through its interaction with GR; however, we cannot rule out the possibility that action by Hic-5 before Dex treatment by a GR-independent mechanism might create an unfavorable chromatin environment that precludes robust Dex-induced GR occupancy. Differences in the local chromatin environment of mod, ind, and block GBRs could account for the different modes of regulation (42).
The regulation by Hic-5 of GR binding to a specific gene set is striking and unexpected. As a coregulator that limits genomic occupancy of a TF selectively, Hic-5 provides a unique opportunity to understand a mechanism that strongly influences TF-binding site selection. Because Hic-5 expression is strongly tissue specific (40), and GBRs differ in different cell types (43), understanding how Hic-5 regulates GR binding to genomic sites and the specific characteristics that define block genes will provide powerful insights into the mechanisms that regulate differential TF binding in cells.
In summary, Dex responsiveness of a substantial proportion of GR target genes in U2OS-GR cells is influenced by Hic-5. Furthermore, Hic-5 completely blocks ligand-mediated regulation of some genes. The diverse mechanisms by which Hic-5 selectively regulates target genes are likely caused by the variable regulatory context of genes, which determines the requirement for and the actions of Hic-5. A comparison of chromatin features in the different Hic-5–regulated gene classes would provide clues to the mechanisms. Because Hic-5 binds to the GR tau2 domain, which is adjacent to the GR DBD, a detailed structural and functional comparison of the interaction of Hic-5 with GR bound to DNA representing the various Hic-5–regulated gene classes could provide important insights into how GR transcriptional specificity is imparted. The selectivity of Hic-5 function on GR target genes also might provide future opportunities for therapeutic intervention by targeting Hic-5 or the GR tau2 region to modulate the set of genes regulated by GR.
Materials and Methods
Extended descriptions of the experimental procedures and details of reagents are provided in SI Materials and Methods.
RNAi.
A clonal line of U2OS cells stably expressing rat GRα was grown as described (44). Cells were plated in medium supplemented with 5% (vol/vol) FBS and transfected with siRNA using Lipofectamine RNAiMAX (Invitrogen). For Hic-5 depletion, siRNA#2 was used unless otherwise indicated. After 2 d (24 h time point) or 3 d (for the ethanol, 2 and 4 h Dex time points) cells were treated with 100 nM Dex. Samples from all treatment groups were harvested at the same time.
Gene-Expression Microarray Analysis.
Total RNA samples were used for Illumina HT12v4 bead chip analysis by standard Illumina procedures. Bioinformatics analysis is described in SI Materials and Methods. The Gene Expression Omnibus accession number for the microarray data is GSE46448.
ChIP Assay.
ChIP was performed as described (6). Primers for GBRs were designed using the data deposited in the National Center for Biotechnology Information’s Sequence Read Archive via accession number SRP020242.
FAIRE.
The protocol for FAIRE was described previously (45). The resulting DNA was analyzed by qPCR amplification.
Supplementary Material
Acknowledgments
We thank Kwang Won Jeong for experimental advice and Dan Gerke, Kelly Chang, and Mengrao Zhang for expert technical assistance. This work was supported by National Institutes of Health (NIH) Grants DK043093 (to M.R.S.) and CA020535 (to K.R.Y.) and by Cancer Center Support Grant P30CA014089 from the National Cancer Institute. R.C. and D.-Y.W. were supported in part by NIH Training Grants T32 CA009320 and T32 GM067587, respectively.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data presented in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE46448).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400522111/-/DCSupplemental.
References
- 1.Wolf IM, Heitzer MD, Grubisha M, DeFranco DB. Coactivators and nuclear receptor transactivation. J Cell Biochem. 2008;104(5):1580–1586. doi: 10.1002/jcb.21755. [DOI] [PubMed] [Google Scholar]
- 2.Bittencourt D, et al. G9a functions as a molecular scaffold for assembly of transcriptional coactivators on a subset of glucocorticoid receptor target genes. Proc Natl Acad Sci USA. 2012;109(48):19673–19678. doi: 10.1073/pnas.1211803109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rogatsky I, Luecke HF, Leitman DC, Yamamoto KR. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc Natl Acad Sci USA. 2002;99(26):16701–16706. doi: 10.1073/pnas.262671599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chinenov Y, Sacta MA, Cruz AR, Rogatsky I. GRIP1-associated SET-domain methyltransferase in glucocorticoid receptor target gene expression. Proc Natl Acad Sci USA. 2008;105(51):20185–20190. doi: 10.1073/pnas.0810863105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang CK, Kim JH, Stallcup MR. Role of the N-terminal activation domain of the coiled-coil coactivator in mediating transcriptional activation by beta-catenin. Mol Endocrinol. 2006;20(12):3251–3262. doi: 10.1210/me.2006-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Won Jeong K, Chodankar R, Purcell DJ, Bittencourt D, Stallcup MR. Gene-specific patterns of coregulator requirements by estrogen receptor-α in breast cancer cells. Mol Endocrinol. 2012;26(6):955–966. doi: 10.1210/me.2012-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392(6679):885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- 8.Watson LC, et al. The glucocorticoid receptor dimer interface allosterically transmits sequence-specific DNA signals. Nat Struct Mol Biol. 2013;20(7):876–883. doi: 10.1038/nsmb.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darimont BD, et al. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12(21):3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogatsky I, et al. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc Natl Acad Sci USA. 2003;100(24):13845–13850. doi: 10.1073/pnas.2336092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Picard D, Yamamoto KR. Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J. 1987;6(11):3333–3340. doi: 10.1002/j.1460-2075.1987.tb02654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kino T, Chrousos GP. Acetylation-mediated epigenetic regulation of glucocorticoid receptor activity: Circadian rhythm-associated alterations of glucocorticoid actions in target tissues. Mol Cell Endocrinol. 2011;336(1-2):23–30. doi: 10.1016/j.mce.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoenmakers E, et al. Differential DNA binding by the androgen and glucocorticoid receptors involves the second Zn-finger and a C-terminal extension of the DNA-binding domains. Biochem J. 1999;341(Pt 3):515–521. [PMC free article] [PubMed] [Google Scholar]
- 14.Milhon J, et al. Identification of amino acids in the tau 2-region of the mouse glucocorticoid receptor that contribute to hormone binding and transcriptional activation. Mol Endocrinol. 1997;11(12):1795–1805. doi: 10.1210/mend.11.12.0018. [DOI] [PubMed] [Google Scholar]
- 15.Clinckemalie L, Vanderschueren D, Boonen S, Claessens F. The hinge region in androgen receptor control. Mol Cell Endocrinol. 2012;358(1):1–8. doi: 10.1016/j.mce.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 16.Daniel AR, et al. The progesterone receptor hinge region regulates the kinetics of transcriptional responses through acetylation, phosphorylation, and nuclear retention. Mol Endocrinol. 2010;24(11):2126–2138. doi: 10.1210/me.2010-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaffer PL, McDonnell DP, Gewirth DT. Characterization of transcriptional activation and DNA-binding functions in the hinge region of the vitamin D receptor. Biochemistry. 2005;44(7):2678–2685. doi: 10.1021/bi0477182. [DOI] [PubMed] [Google Scholar]
- 18.Yoshikawa N, et al. Role of the hinge region of glucocorticoid receptor for HEXIM1-mediated transcriptional repression. Biochem Biophys Res Commun. 2008;371(1):44–49. doi: 10.1016/j.bbrc.2008.03.155. [DOI] [PubMed] [Google Scholar]
- 19.Hill KK, Roemer SC, Jones DN, Churchill ME, Edwards DP. A progesterone receptor co-activator (JDP2) mediates activity through interaction with residues in the carboxyl-terminal extension of the DNA binding domain. J Biol Chem. 2009;284(36):24415–24424. doi: 10.1074/jbc.M109.003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacq X, et al. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell. 1994;79(1):107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 21.Moilanen AM, et al. Identification of a novel RING finger protein as a coregulator in steroid receptor-mediated gene transcription. Mol Cell Biol. 1998;18(9):5128–5139. doi: 10.1128/mcb.18.9.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Link KA, et al. Targeting the BAF57 SWI/SNF subunit in prostate cancer: A novel platform to control androgen receptor activity. Cancer Res. 2008;68(12):4551–4558. doi: 10.1158/0008-5472.CAN-07-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee YS, et al. Activating signal cointegrator 1 is highly expressed in murine testicular Leydig cells and enhances the ligand-dependent transactivation of androgen receptor. Biol Reprod. 2002;67(5):1580–1587. doi: 10.1095/biolreprod.102.006155. [DOI] [PubMed] [Google Scholar]
- 24.Yang L, Guerrero J, Hong H, DeFranco DB, Stallcup MR. Interaction of the tau2 transcriptional activation domain of glucocorticoid receptor with a novel steroid receptor coactivator, Hic-5, which localizes to both focal adhesions and the nuclear matrix. Mol Biol Cell. 2000;11(6):2007–2018. doi: 10.1091/mbc.11.6.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noguchi F, Inui S, Nakajima T, Itami S. Hic-5 affects proliferation, migration and invasion of B16 murine melanoma cells. Pigment Cell Melanoma Res. 2012;25(6):773–782. doi: 10.1111/pcmr.12005. [DOI] [PubMed] [Google Scholar]
- 26.Inui S, Noguchi F, Nishiyama A, Itami S. Multipotential functions of Hic-5 in growth, differentiation, migration and adhesion of human keratinocytes. J Dermatol Sci. 2012;68(3):197–199. doi: 10.1016/j.jdermsci.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 27.Deakin NO, Turner CE. Distinct roles for paxillin and Hic-5 in regulating breast cancer cell morphology, invasion, and metastasis. Mol Biol Cell. 2011;22(3):327–341. doi: 10.1091/mbc.e10-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujimoto N, et al. Cloning and characterization of androgen receptor coactivator, ARA55, in human prostate. J Biol Chem. 1999;274(12):8316–8321. doi: 10.1074/jbc.274.12.8316. [DOI] [PubMed] [Google Scholar]
- 29.Drori S, et al. Hic-5 regulates an epithelial program mediated by PPARgamma. Genes Dev. 2005;19(3):362–375. doi: 10.1101/gad.1240705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aghajanova L, Velarde MC, Giudice LC. The progesterone receptor coactivator Hic-5 is involved in the pathophysiology of endometriosis. Endocrinology. 2009;150(8):3863–3870. doi: 10.1210/en.2009-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, et al. Epithelial Hic-5/ARA55 expression contributes to prostate tumorigenesis and castrate responsiveness. Oncogene. 2011;30(2):167–177. doi: 10.1038/onc.2010.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Song K, Krebs TL, Yang J, Danielpour D. Smad7 is inactivated through a direct physical interaction with the LIM protein Hic-5/ARA55. Oncogene. 2008;27(54):6791–6805. doi: 10.1038/onc.2008.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shibanuma M, Kim-Kaneyama JR, Sato S, Nose K. A LIM protein, Hic-5, functions as a potential coactivator for Sp1. J Cell Biochem. 2004;91(3):633–645. doi: 10.1002/jcb.10754. [DOI] [PubMed] [Google Scholar]
- 34.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kornberg RD. The molecular basis of eucaryotic transcription. Cell Death Differ. 2007;14(12):1989–1997. doi: 10.1038/sj.cdd.4402251. [DOI] [PubMed] [Google Scholar]
- 36.Chen W, Rogatsky I, Garabedian MJ. MED14 and MED1 differentially regulate target-specific gene activation by the glucocorticoid receptor. Mol Endocrinol. 2006;20(3):560–572. doi: 10.1210/me.2005-0318. [DOI] [PubMed] [Google Scholar]
- 37.Jeong KW, et al. Recognition of enhancer element-specific histone methylation by TIP60 in transcriptional activation. Nat Struct Mol Biol. 2011;18(12):1358–1365. doi: 10.1038/nsmb.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim JH, et al. CCAR1, a key regulator of mediator complex recruitment to nuclear receptor transcription complexes. Mol Cell. 2008;31(4):510–519. doi: 10.1016/j.molcel.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chinenov Y, et al. Role of transcriptional coregulator GRIP1 in the anti-inflammatory actions of glucocorticoids. Proc Natl Acad Sci USA. 2012;109(29):11776–11781. doi: 10.1073/pnas.1206059109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuminamochi T, et al. Expression of the LIM proteins paxillin and Hic-5 in human tissues. J Histochem Cytochem. 2003;51(4):513–521. doi: 10.1177/002215540305100413. [DOI] [PubMed] [Google Scholar]
- 41.Heitzer MD, DeFranco DB. Mechanism of action of Hic-5/androgen receptor activator 55, a LIM domain-containing nuclear receptor coactivator. Mol Endocrinol. 2006;20(1):56–64. doi: 10.1210/me.2005-0065. [DOI] [PubMed] [Google Scholar]
- 42.George CL, Lightman SL, Biddie SC. Transcription factor interactions in genomic nuclear receptor function. Epigenomics. 2011;3(4):471–485. doi: 10.2217/epi.11.66. [DOI] [PubMed] [Google Scholar]
- 43.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43(3):264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rogatsky I, Trowbridge JM, Garabedian MJ (1997) Glucocorticoid receptor-mediated cell cycle arrest is achieved through distinct cell-specific transcriptional regulatory mechanisms. Mol Cell Biol 17(6):3181–3193. [DOI] [PMC free article] [PubMed]
- 45.Simon JM, Giresi PG, Davis IJ, Lieb JD. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat Protoc. 2012;7(2):256–267. doi: 10.1038/nprot.2011.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.