Significance
Natural killer (NK) cells are potent tumor-cell killers, but exposure to transforming growth factor beta-1 (TGF-β) abrogates their effectiveness. Here, we show that this suppression is a result of TGF-β induction of microRNA (miR)-183, which binds and represses DNAX activating protein 12 kDa (DAP12), a signal adaptor for lytic function in NK cells. Because introduction of miR-183 alone or its functional blockade in the presence of TGF-β reduced or restored DAP12 levels in NK cells, we define miR-183 as a key factor in TGF-β–mediated immunosuppression. Since DAP12 is required for signaling through multiple NK cytotoxicity receptors and TGF-β is overexpressed by diverse solid malignancies, our data may have significant importance in the development of NK-based cancer immunotherapies.
Keywords: posttranscriptional silencing, immune suppression, non-small cell lung cancer
Abstract
Transforming growth factor β1 (TGF-β), enriched in the tumor microenvironment and broadly immunosuppressive, inhibits natural killer (NK) cell function by yet-unknown mechanisms. Here we show that TGF-β–treated human NK cells exhibit reduced tumor cytolysis and abrogated perforin polarization to the immune synapse. This result was accompanied by loss of surface expression of activating killer Ig-like receptor 2DS4 and NKp44, despite intact cytoplasmic stores of these receptors. Instead, TGF-β depleted DNAX activating protein 12 kDa (DAP12), which is critical for surface NK receptor stabilization and downstream signal transduction. Mechanistic analysis revealed that TGF-β induced microRNA (miR)-183 to repress DAP12 transcription/translation. This pathway was confirmed with luciferase reporter constructs bearing the DAP12 3′ untranslated region as well as in human NK cells by use of sense and antisense miR-183. Moreover, we documented reduced DAP12 expression in tumor-associated NK cells in lung cancer patients, illustrating this pathway to be consistently perturbed in the human tumor microenvironment.
Natural killer (NK) cells express stochastic patterns of germ-line–encoded cytotoxicity receptors, most of which lack signaling domains and require association with adaptor proteins like DNAX activating protein 12 kDa (DAP12) to facilitate surface stabilization and confer activating signals. Upon ligand binding, receptor-associated DAP12 is tyrosine phosphorylated by Src kinases and proceeds through the phosphatidylinositide 3-kinase/extracellular signal-regulated kinase activation cascade to mobilize lytic granules that ultimately kill target cells (1). DAP12 is the exclusive signaling adaptor of all allelic variants of the activating killer Ig-like receptor family (aKIR; allelic variants are known as KIRxDSx), one or more of which is expressed by all human NK cells. Although ligands have not yet been explicitly demonstrated for all activating KIRs, most bind HLA alleles (2). Additionally, DAP12 associates with the C-type lectins, natural killer group (NKG)2C and NKG2E, which bind the nonclassical major histocompatibility (MHC) I allele, HLA-E (3). Thus, DAP12 is required for any NK cell-mediated tumor killing via MHC I ligand interactions. DAP12 is also used by NKp44, a non-MHC-restricted natural cytotoxicity receptor (NCR) induced in cytokine-activated NK cells (4). Therefore, DAP12 plays a crucial role in NK effector responses, and its dysregulation could critically impact NK cell function.
Currently, lung cancer is the leading cause of cancer-related death in the United States, due in part to the failure of protective immunity against malignant cells (5). NK cells constitute nearly 10% of resident lymphocytes in the lungs, making lungs the most NK-rich nonlymphoid tissue (6). NK cells are poised to kill neoplastic cells; however, tumor cells evade NK cell surveillance by creating an immunosuppressive environment via factors including transforming growth factor β1 (TGF-β) (7). TGF-β is broadly immunosuppressive, because both NK cells and cytotoxic T lymphocytes exposed to TGF-β are unable to kill tumor cells in humans or mice (8). Consequently, elevated serum TGF-β levels are observed in metastatic stages of many cancers and correlate with poor prognoses (9). In vivo depletion of TGF-β or blockade of TGF-β signaling can restore the NK cell-mediated antitumoral response (10). Reports indicate that TGF-β suppresses NK IFN-γ and CD16-mediated cytotoxicity through SMAD-dependent regulation (11); however, mechanistic analysis of DAP12-associated KIR and NCR cytotoxicity has not been conducted.
microRNAs (miRs) are ∼22-nucleotide noncoding RNAs that repress gene expression by binding complementary sequences within the 3′ untranslated region (UTR) of targeted mRNAs. This event triggers recruitment of the RNA-silencing complex (RISC) that leads to mRNA degradation or translational arrest (12). miRs are powerful gene regulators involved in diverse cellular processes, including carcinogenesis (13), immune development, and response to infection (12). Whether TGF-β alters NK cell function via miRs is unknown. Here, we show that tumor-derived TGF-β induces miR-183 in NK cells to suppress DAP12 expression. By targeting DAP12, miR-183 efficiently silences NK cells, effectively depleting the stimulatory signaling capacity necessary for lysis through associated receptors. We verified that DAP12 down-regulation occurs in the human tumor microenvironment because tumor-infiltrating NK cells consistently expressed lower DAP12 levels than peritumoral NK cells across all known lung cancer subtypes. Because DAP12 is the common adaptor for many NK cell-activating receptors, this miR-183–dependent pathway drives pan NK cell immunosuppression in the TGF-β–rich tumor microenvironment.
Results
TGF-β Alters NK Cell Phenotype and Cytolytic Function.
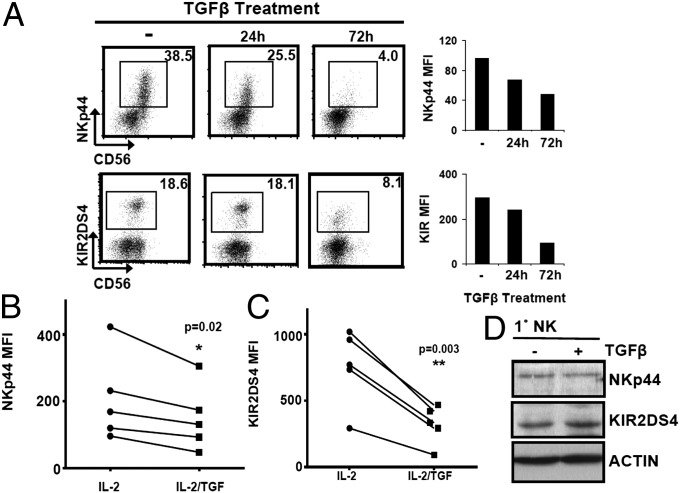
Activating KIR can mediate a strong anticancer effect, as demonstrated by aKIR-dependent prevention of leukemia relapse and recognition of melanoma and leukemia antigens (14, 15). NKp44 drives non-MHC-restricted tumor cell lysis and is inducible by IL-2 in primary human NK cells (4). Because of evidence for their anticancer roles, we elected to study regulation of KIR2DS4 and NKp44 in response to TGF-β. Because IL-2 is required for human NK cell survival and maintenance in culture, all TGF-β stimulation assays were conducted in the presence of IL-2. Flow-cytometric analysis indicated that TGF-β effectively reduced the percentage of NKp44+ cells from 38.5% to 4.0% at 72 h. Moreover, the surface density, as measured by mean fluorescence intensity (MFI), of NKp44 was reduced accordingly (Fig. 1A). Similarly, TGF-β suppressed the surface density and percentage of KIR2DS4+ primary NK cells (Fig. 1A). Further confirmation of NKp44 and KIR2DS4 surface depletion was obtained from NK cells of five healthy donors treated with IL-2 or IL-2/TGF-β for 72 h (Fig. 1 B and C). Because receptor surface expression may not reflect total protein stores, we measured total NKp44 and KIR2DS4 levels in TGF-β–treated and untreated NK cells by immunoblot. Surprisingly, cytoplasmic levels of NKp44 and KIR2DS4 were equivalent irrespective of TGF-β treatment (Fig. 1D), indicating that TGF-β does not disrupt NKp44 or KIR2DS4 protein abundance but alters surface membrane translocation.
Fig. 1.
TGF-β deregulates NK receptor expression. (A) Flow-cytometric analysis of NKp44 and KIR2DS4 in primary NK cells treated with TGF-β. Bar graphs depict MFI of gated cells for each time point. (B and C) MFI of NKp44 (B) or KIR2DS4 (C) of NK cells from five healthy donors after 72 h of IL-2 or IL-2/TGF-β treatment. P values were generated by Student t test. (D) Immunoblot of NKp44 or KIR2DS4 in primary NK cells after 72 h of IL-2 or IL-2/TGF-β treatment. β-actin served as a loading control. Results are representative of three experiments.
NK cells kill targets through a variety of DAP12-associated or DNAX-activating protein of 10 kDa (DAP10)-associated receptors. DAP12-dependent cytolysis may be particularly important for NK cell reactivity against lung tumors because normal and neoplastic lung tissues lack ligands for NKG2D (the major DAP10-associated cytotoxicity receptor) (16). In support of this hypothesis, meta-analysis of a lung adenocarcinoma (ADC) cohort (17) verified that the NKG2D ligands, MHC (HLA) class I chain-related gene A (MICA), MICB, UL16 binding protein 1 (ULBP1), and ULBP2, were absent, or expressed at very low levels (median signal intensity <1.5) in normal and malignant lung tissue (Fig. S1A). Similarly, this trend was detected in large cell carcinoma (LCC), squamous cell carcinoma (SQU), carcinoid tumors (CAR), and small cell lung cancer (SCLC) cohorts (18, 19). Thus, NK cells likely recognize lung tumor cells not through NKG2D/DAP10 but via DAP12-associated receptors. Because of this caveat, we focused solely on DAP12-mediated tumor lysis and used tumor target cell lines that express HLA but little to no MICA/B (namely, Raji lymphoma and H1299 and A549 ADC cell lines) (Fig. S1B).
To analyze the effect of TGF-β on DAP12-mediated NK cell tumor lysis, we tested primary NK and NK92 cells with or without TGF-β treatment in the presence of IL-2. We also compared IL-2 with IL-15 for maintenance of NK cell survival and function, because both cytokines are candidates for clinical trials to engender robust NK cell responses against cancer (20). TGF-β was equally capable of suppressing cytolysis of all three tumor targets (Fig. S2A), illustrating that the DAP12-dependent killing is suppressed despite pretreatment with activating cytokines, an important caveat for NK cell therapy of cancer. Analysis of perforin polarization and immune synapse formation in NK-92 cells interacting with Raji tumor cells indicated that TGF-β blocked perforin mobilization to the immune synapse, as shown in representative images (Fig. S2B). For accuracy, the average distance of the total perforin granules to the tumor contact site in each NK cell/tumor conjugate was calculated, and quantification of 50 conjugates from each treatment group confirmed that perforin granules were less mobile in TGF-β–treated cells (Fig. S2C). Importantly, this phenomenon occurs despite equivalent levels of total perforin protein and mRNA (Fig. S2 D and E). Collectively, these data suggest that TGF-β specifically disrupts DAP12-associated granule movement in NK cells.
Tumor-Derived TGF-β Suppresses NK Cells via Depletion of DAP12.
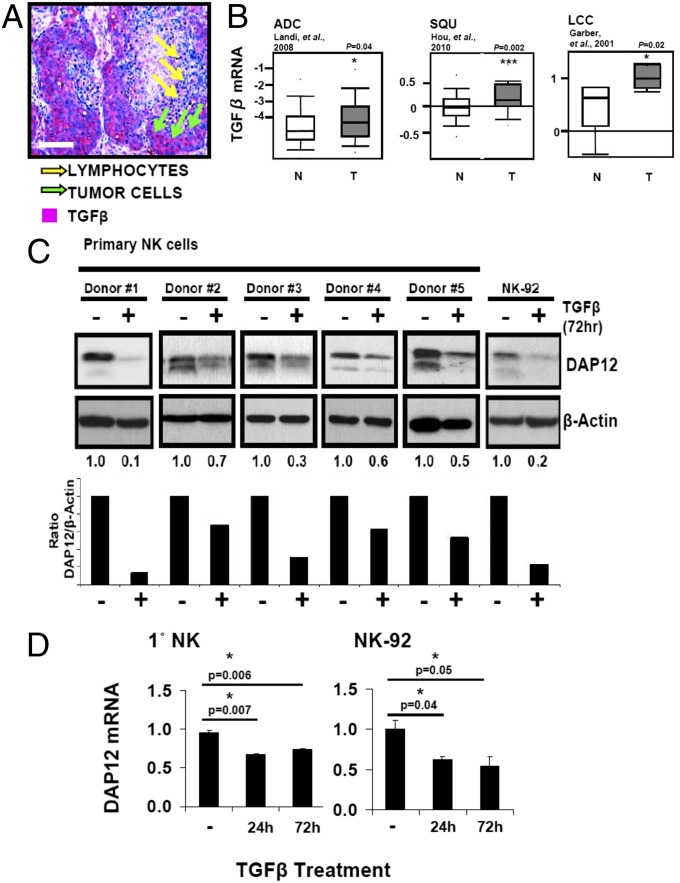
The tumor microenvironment is shaped by TGF-β (8), which is expressed at high levels in non-small-cell lung cancer (NSCLC) (21) and other cancers. We confirmed TGF-β enrichment in human NSCLC tissues, as shown in a representative immunohistochemical evaluation (Fig. 2A), and in analysis of microarrays of ADC (17), SQU (19), and LCC (ref. 22; Fig. 2B). Because we observed that TGF-β reduced surface KIR2DS4 and NKp44 receptor density without loss of cytoplasmic stores (Fig. 1), we reasoned that DAP12, required to stabilize membrane receptor expression, might be the direct target of TGF-β. Therefore, primary NK and NK92 cells were cultured in IL-2 with or without TGF-β for 3 d before immunoblot analysis of DAP12 expression. Strikingly, TGF-β drastically reduced DAP12 levels in both primary purified NK cells from five healthy donors and NK92 (Fig. 2C). Quantitative PCR (qPCR) demonstrated that DAP12 mRNA levels were also reduced as early as 24 h after TGF-β treatment (Fig. 2D). These data suggest that TGF-β specifically disrupts DAP12 mRNA and protein expression in human NK cells, consequently leading to decreased receptor density, failed synapse formation, and impaired cytolysis.
Fig. 2.
TGF-β depletes DAP12. (A) Formalin-fixed paraffin-embedded (FFPE) human lung biopsies were stained with anti–TGF-β (red) and hematoxylin (blue). Representative SQU tumor is shown. (Scale bar, 100 μm.) (B) Meta-analysis of TGF-β mRNA in ADC (Left; ref. 17), SQU (Center; ref. 19), and LCC (Right; ref. 22) (T) and nonmalignant (N) tissue. (C, Upper) Primary NK (five donors) or NK92 cells were treated with IL-2 (−) or IL-2 /TGF-β (+) for 72 h and analyzed by immunoblot. (Lower) Densitometry values (DAP12 normalized to β-actin). (D) qPCR analysis of DAP12 in primary NK (Left) and NK92 (Right) cells treated with IL-2 or IL-2/TGF-β for indicated times. Error bars, SEM. P values were generated by paired Student’s’ t test. Results are representative of three experiments.
TGF-β–Induced miR-183 Targets DAP12.
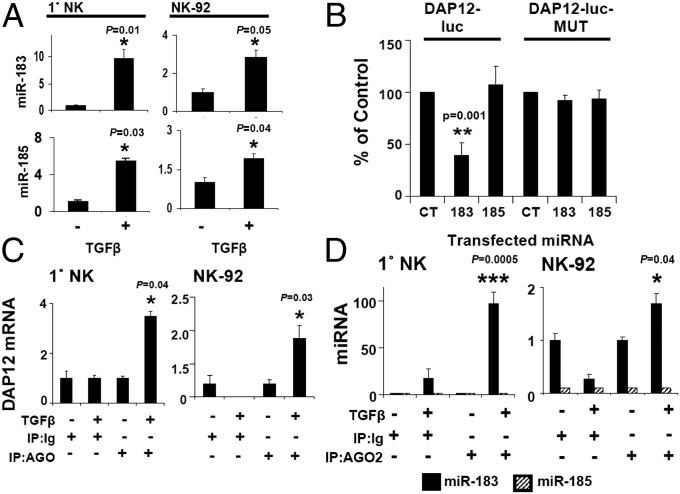
In primary NK cells, DAP12 protein levels were reduced by ∼64% (Fig. 2C; mean of five donors), whereas mRNA was reduced by ∼30% (Fig. 2D; 72 h). Similarly, in NK92, DAP12 was reduced by ∼80% with only a ∼40% reduction in mRNA levels (Fig. 2 C and D). These observations led us to consider miRs for DAP12 regulation because miRs can repress target mRNAs either by mRNA degradation or by translational arrest, and miR-mediated reduction of protein can occur in the absence of reduced mRNA levels (23, 24). Bioinformatic analysis of the 161-nucleotide DAP12 3′ UTR (Fig. S3A) (miRANDA and TargetScan) revealed miR-183 and -185 binding sites (Fig. S3B). Notably, these miR sites were absent in the DAP10 3′ UTR (Fig. S3A). Interestingly, no predicted miR-183 or -185 sites were present within the 3′ UTRs of any known NK activating receptors, including KIR2DS4 and NKp44. Further analysis indicated that TGF-β–treated NK cells expressed more mature miR-183 and -185 transcripts than untreated cells (Fig. 3A). To validate the putative miR-183 and -185 binding sites, we generated luciferase reporter constructs containing the DAP12 3′ UTR (DAP12-luc) (Fig. S4). Cotransfection of HeLa cells with DAP12-luc and miR-183 reduced luciferase expression by 60% compared with DAP12-luc/scrambled control cotransfection, whereas miR-185 cotransfection failed to reduce luciferase expression (Fig. 3B). The seed sequence, 6–8 nucleotides on the 3′ UTR of the target mRNA complimentary to the miR, is required for effective repression (25). Mutation of the miR-183 seed sequence in DAP12-luc (Fig. S4) eliminated luciferase repression by mir-183 without affecting the miR-185 response (Fig. 3B), suggesting that only miR-183 negatively regulates DAP12.
Fig. 3.
miR-183 targets the DAP12 3′ UTR. (A) qPCR analysis of miR-183 and -185 in IL-2 (−) or IL-2/TGF-β (+)-treated primary NK (Left) and NK92 (Right) cells. (B) HeLa cells were transfected with DAP12-luc (250 ng), renilla luciferase (5 ng), and miR-183, miR-185, or control (CT) oligonucleotides (25 nM). Twenty-four hours later, transfectants were lysed for quantification of firefly luciferase activity. (C and D) qPCR analysis of DAP12 (C) or miR-183 and -185 (D) isolated from AGO2 immunoprecipitates from primary NK (Left) and NK92 (Right) cells. Results are presented as the percentage of input RNA. P values were generated by paired Student’s t test. Results are representative of three experiments.
miRs must associate with the RISC, which contains RNA-binding proteins including Argonaute 2 (AGO2), to function (25). To validate miR-183 interaction with endogenous DAP12 and RISC machinery, we immunoprecipitated AGO2 and probed for associated DAP12 mRNA and miRs in IL-2 and IL-2/TGF-β–treated primary NK and NK92 cells (Fig. S5A). AGO2 was expressed at equivalent levels despite TGF-β treatment (Fig. S5B), yet AGO2-associated DAP12 mRNA was detected only in TGF-β–treated cells (Fig. 3C). Furthermore, miR-183, but not miR-185, was detected in AGO2 immunoprecipitates of TGF-β–treated cells (Fig. 3D), providing evidence that TGF-β specifically triggers miR-183/DAP12 localization to the RISC in NK cells.
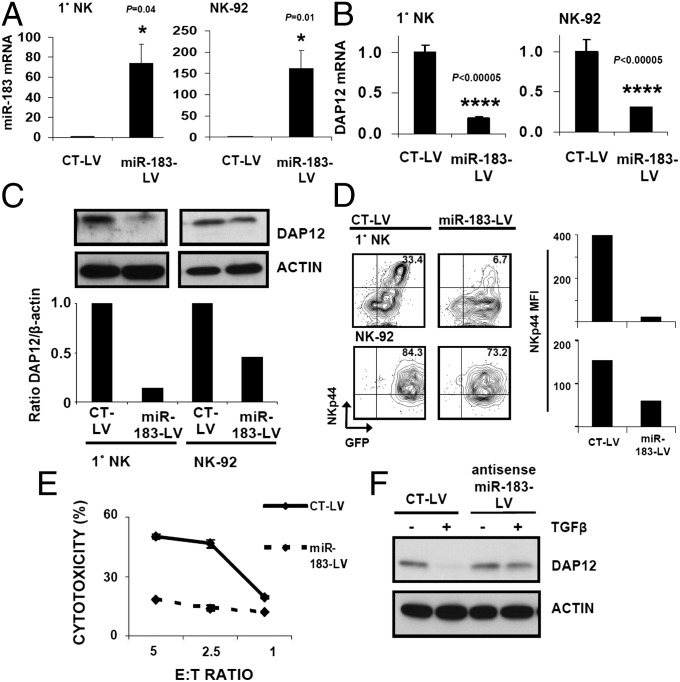
We next verified that the TGF-β–mediated DAP12 reduction relies on miR-183 by transfection of miR-183 or -185 into U937 myeloid cells. Only miR-183 overexpression effectively reduced endogenous DAP12 protein in these cells (Fig. S6). Similarly, lentiviral overexpression of miR-183 (miR-183–LV) (Fig. 4A) in primary NK and NK92 cells repressed DAP12 mRNA (Fig. 4B) and protein levels (Fig. 4C). Importantly, DAP10 mRNA and perforin mRNA levels were not reduced by miR-183 overexpression, confirming the lack of miR-183 binding sites within their 3′ UTRs (Fig. S7). DAP10 mRNA was not detected in NK92 and is not depicted. Notably, primary NK and NK92 cells infected with miR-183–LV exhibited markedly lower NKp44 surface levels (Fig. 4D), and miR-183–LV-infected NK92 cells were less able to kill Raji target cells (Fig. 4E). Conversely, lentiviral expression of antisense miR-183 in NK92 cells abrogated the ability of TGF-β to reduce DAP12 (Fig. 4F). Collectively, these data demonstrate that TGF-β inhibits NK cells by induction of miR-183, which depletes DAP12.
Fig. 4.
miR-183 is sufficient to deplete DAP12 and modulate NK function. (A–E) Lentiviral expression constructs containing scrambled control (CT-LV) or miR-183 (miR-183–LV) were used for NK cell infection. (A and B) Infected (GFP+) primary NK (Left) or NK92 (Right) cells were sorted to >95% purity, followed by qPCR analysis of miR-183 (A) or DAP12 (B). Error bars, SEM. P values were generated by paired Student’s t test. (C, Upper) Immunoblot of DAP12 in CT–LV-infected or miR-183–LV-infected cells. (Lower) Densitometry of DAP12 normalized to β-actin. (D) Flow cytometry of NKp44 in CT–LV-infected (Left) or miR-183–LV-infected (Center) cells. (Right) MFI of NKp44 in GFP+ cells. (E) GFP+ CT–LV-infected or miR-183–LV-infected NK92 were plated at indicated ratios with 51Cr-labeled Raji cells; cytotoxicity was determined by 51Cr release. (F) NK92 cells were infected with CT-LV or antisense–miR-183 (antisense–miR-183–LV). Infected (GFP+) cells were sorted to >95% purity, followed by culture in TGF-β for 72 h and immunoblot analysis. Data shown are representative of three experiments.
Loss of DAP12 in Tumor-Infiltrating Lymphocytes Is a Common Feature of Lung Cancer.
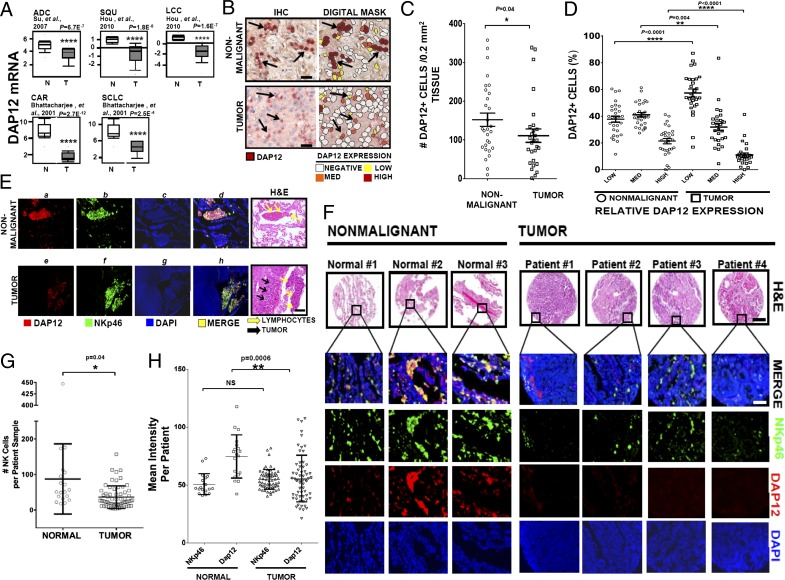
Given that DAP12 depletion abrogates NK cell tumoricidal function, we examined its expression in human lung cancer. Meta-analysis of three large lung cancer studies cataloged in Oncomine revealed a marked reduction in DAP12 mRNA levels. This result occurred not only in the three most common NSCLC subtypes, ADC (26), SQU (19) and LCC (22), but also in SCLC and CAR (19) (Fig. 5A). Additional gene expression studies of lung cancers recorded in Oncomine yielded similar results (17). Thus, the loss of DAP12 is ubiquitous in lung cancer.
Fig. 5.
Reduction of DAP12+ cells in lung tumor biopsies. (A) Meta-analysis of DAP12 mRNA expression in human ADC (Left Upper; ref. 26), SQU (Center Upper; ref. 19), LCC (Right Upper; ref. 19), SCLC (Right Lower; ref. 18), and CAR (Left Lower; ref. 18) (T) vs. normal (N) tissue from gene expression microarrays. (B) Analysis of DAP12+ cells in lung tumors. Twenty-nine FFPE human lung biopsies were stained with anti-DAP12 (red) and hematoxylin. Lymphocyte-restricted DAP12 expression levels in nonmalignant and tumor regions were quantified by an imaging algorithm. A nonmalignant and overt tumor region (Left) with coordinating digital mask (Right) illustrating negative, low, medium, or high DAP12 expression. Arrows depict DAP12+ cells. (Scale bars, 25 μm.) (C and D) Sum of DAP12+ cells (C) and percent of DAP12-low, intermediate, or high cells (D) in representative fields of nonmalignant and tumor regions. (E) Twenty-nine human lung biopsies (E) or a human lung ADC tissue microarray (TMA) (F) were dually stained with anti-DAP12 and -NKp46 and mounted in DAPI medium. (E) An adjacent nonmalignant and tumor region from one patient. (Scale bar, 100 μm.) (F) Representative images from three normal and four tumor tissues. H&E-stained serial sections depict morphology. (Scale bars, H&E, 100 μm; fluorescent images, 50 μm.) (G) Total number of NKp46+ cells per tissue core of the TMA. (H) MFI of NK cell-restricted DAP12 and NKp46 protein. Each marker depicts the average stain intensity per cell for a single tissue core of the TMA. Error bars, SEM. P values were generated by unpaired Student’s’ t test.
Because lung tissue does not express DAP12, this differential is likely due to dysregulation in hematopoietic cells. To clarify this distinction, we stained for DAP12 protein in 29 human lung tumor biopsies. Histopathological classifications and tumor grades are detailed in Table S1. We observed that DAP12+ lymphocytes extensively populated the nonmalignant periphery (Fig. 5B, Left), whereas fewer of them infiltrated the tumor (151 ± 17.17/0.2 mm2 in nonmalignant tissue vs. 110.89 ± 17.29/0.2 mm2 in tumor) (Fig. 5C). We used an imaging algorithm to quantify the level of DAP12 expression per cell as negative, low, intermediate, or high (Fig. 5B, Right). By using this algorithm, parenchymal and tumor cells were consistently DAP12-negative (Fig. 5B). Strikingly, the percentages of DAP12-high and -intermediate lymphocytes were profoundly decreased within the tumor compared with the nonmalignant periphery (Fig. 5D); conversely, the percentage of DAP12-low lymphocytes was increased.
DAP12 Expression Is Reduced in Intratumoral NKp46+ NK Cells.
We next focused on NK-restricted DAP12 by using NKp46, a stable receptor expressed early in differentiation (27, 28), as a marker in the lung biopsies described above. Numerous NKp46+ cells (Fig. 5 E, b) expressing DAP12 (Fig. 5 E, a) as depicted by colocalization (Fig. 5 E, d) populated the nonmalignant regions. In contrast, NKp46+ cells proximal to tumor cells expressed severely diminished DAP12 (Fig. 5 E, e, f, and h). We confirmed our results in a larger human lung ADC tissue microarray (TMA) containing 19 normal and 63 tumor samples, with 3 normal and 4 tumor samples depicted in Fig. 5F. Tissue characteristics are detailed in Experimental Procedures. Again, significantly fewer NKp46+ NK cells infiltrated the tumors (88 ± 22 in nonmalignant tissue, n = 19; 36.68 ± 4.04 in tumor, n = 63) (Fig. 5G). When we assessed the levels of NKp46 and DAP12, we found that NKp46 density per NK cell was equivalent in normal and malignant tissue (normal NKp46 stain intensity = 50.70 ± 2.08; intratumoral NKp46 stain intensity = 54.85 ± 1.08), similar to a recent report (28). However, the DAP12 level was significantly reduced in intratumoral NK cells (normal DAP12 stain intensity = 74.68 ± 4.40; intratumoral DAP12 stain intensity = 55.53 ± 2.56) (Fig. 5H). This differential modulation of DAP12, but not NKp46, expression is a critical observation, allowing us to confidently monitor NK cells. Collectively, our results reveal that the reduction in DAP12 is common to all lung cancers and is a result of both decreased NK cell infiltration and severely diminished DAP12 expression in tumor-infiltrating NK cells.
Discussion
Here we describe a previously unidentified set of interactions between TGF-β, miR-183, and DAP12, a vital component of the NK cell stimulatory signaling pathway. By meta-analysis of published datasets and immunostaining patient tissues, we verified DAP12 dysregulation to be common to diverse lung cancers and restricted to infiltrating NK cells (Fig. 5). Of relevance, meta-analysis of gene expression studies revealed that DAP12 depletion (Fig. 5A) inversely correlated with TGF-β up-regulation (Fig. 2B). In support of this causal relationship, purified TGF-β reduced DAP12 expression in NK cells (Fig. 2C). Bioinformatic analysis revealed a putative miR-183 binding site in the DAP12 3′ UTR consisting of 8 consecutive nucleotides complementary to miR-183 (Fig. S3). Indeed, exposure of NK cells to TGF-β induced miR-183 transcription as well as transcription of miR-185 (Fig. 3A), which also exhibits sequence complimentarity to the DAP12 3′ UTR. Using luciferase reporter assays, we showed that miR-183, but not miR-185, bound and negatively repressed DAP12, illustrating the specificity of miR-183 (Fig. 3B). Analysis of the processing machinery revealed miR-183 and DAP12 transcripts localized with AGO2-containing RISC only upon TGF-β treatment (Fig. 3 C and D). Interestingly, we did not detect miR-185 associated with AGO2 (Fig. 3D) despite its up-regulation in TGF-β–treated cells. It is possible that miR-185 requires cooperation with other yet-unidentified miRs to associate with the RISC and repress targets, as has been demonstrated with other miRs (29). Alternatively, the binding of miR-183 could cause steric hindrance and prevent miR-185 from binding and associating with RISC because the two sites are proximal on the short DAP12 3′ UTR (30). Introduction of miR-183 alone into myeloid (Fig. S6) or NK cells (Fig. 4C) was sufficient to deplete endogenous DAP12 protein stores, whereas antisense blockade of miR-183 in the presence of TGF-β restored DAP12 levels to those of untreated cells (Fig. 4F). The significance of this work lies in the following three seminal discoveries: (i) TGF-β disrupts NK cells downstream of cytotoxicity receptors via depletion of the shared signaling protein, DAP12; (ii) depletion of DAP12 confers a drastically reduced cytolytic potential to NK cells; and (iii) TGF-β uses miR-183 for DAP12 elimination. This study constitutes a report of the previously unidentified miR control of DAP12, a key stimulatory signal adaptor linked to numerous NK receptors. Furthermore, this work adds to the body of previously reported mechanisms by which TGF-β suppresses NK cell function, including SMAD2–, SMAD3–, and SMAD4–dependent suppression of IFN-γ and T-BET (11, 31).
It is well known that DAP12 pairs with NKp44 and aKIR receptors. The ligands for NKp44 include viral proteins (32) and the recently discovered NKp44L, a novel isoform of the mixed-lineage leukemia-5 protein, which is expressed on transformed cells (33); thus, this receptor may be particularly important for the anticancer response. The reduction of NKp44 and KIR2DS4 surface levels (Fig. 1) as a result of TGF-β/miR-183/DAP12 interactions likely contributes to the reported ineffective phenotype of peri- and intratumoral NK cells (6). These changes in cytotoxicity receptor surface expression almost certainly impact NK cell responses, because increased receptor density and multimerization of receptors enhances the sensitivity to their ligands and lowers the threshold for activation (34). Surface density is potentially even more important for KIR2DS4, which binds its ligand HLA–Cw4 with relatively low affinity (15). Indeed, TGF-β–treated NK cells exhibit a drastically impaired cytolysis (Fig. S2A), accompanied by abrogated perforin polarization (Fig. S2 B and C). Thus, DAP12 depletion via TGF-β–induced miR-183 profoundly affects the NK cell response to cancer cells. Of note, TGF-β also represses DAP10 (35). However, the DAP10 3′ UTR lacks miR-183 binding sites (Fig. S4A), and mRNA levels are not affected by miR-183 overexpression (Fig. S7), suggesting that TGF-β may deactivate these two major NK adaptors via separate mechanisms.
DAP12 is broadly expressed in hematopoietic cells of myeloid and lymphocytic lineages. In myeloid cells, DAP12 pairs with several receptors, including myeloid DAP12-associating lectin, triggering receptor expressed on myeloid cells 1, and signal-regulatory protein β1 (36). Ligation of these receptors results in secretion of proinflammatory cytokines and chemokines. Additionally, T cells that have undergone multiple rounds of proliferation can express DAP12 and aKIR. These aKIR+/DAP12+ cells produce IFN-γ and kill target cells independently of TCR stimulation (37). Thus, expression of DAP12 in hematopoietic cells promotes a proinflammatory environment beneficial for tumor rejection. Importantly, TGF-β suppresses myeloid cell function by reducing levels of the FCR-γ–associated γ subunit, which is required for surface stabilization and function of FCR-γ receptors (38); whether DAP12-dependent functions in non-NK cells are affected by TGF-β remains to be investigated.
miR-183 is cotranscribed with miR-92 and -182. This cluster is overexpressed in multiple cancers, including NSCLC (39), where it regulates diverse mediators of tumor survival and function, including targeting the tumor suppressor EGR1 (40). Thus, miR-183 within the tumor microenvironment may act as a double-edged sword by promoting tumor survival and suppressing NK cell function. Significantly, TGF-β specifically depleted DAP12 and not cytoplasmic stores of associated receptors (Fig. 1D), suggesting that blocking miR-183 could restore cytotoxicity through DAP12 stabilization and rescued receptor surface expression. This postulation is illustrated by a recent study showing that only introduction of DAP12 allows for KIR2DS1, -2, and -4 surface expression in NKL cells (41). Clinically, blockade of TGF-β itself is not optimal because it has critical functions in lung homeostasis, including the maintenance of tolerance to airway antigens via regulatory T cells (42). Targeting of the TGF-β/miR-183/DAP12 pathway via miR-183 blockade to restore immunity against cancer may thus provide a new strategy to treat cancer. Unfortunately, generation of an animal model for observation of TGF-β/miR-183/DAP12 interactions is not feasible, because, although miR-183 is conserved in the mouse, the murine DAP12 3′ UTR sequence has no homology to the human counterpart and does not contain miR-183 binding sites. Nevertheless, the most significant relevance for targeting miR-183 to restore DAP12 clearly comes from our studies of human lung cancer samples and in external databases (Fig. 5), which document the loss of DAP12 in association with disease. As yet, DAP12, but not any of the associated NK receptors, is the only known target of miR-183; however, further analysis is needed to determine whether other components important for NK and myeloid cell function might also be controlled by miR-183. Importantly, we provide further mechanistic explanation of the widely observed phenomenon of TGF-β–mediated NK suppression and suggest that, because of its dual prooncogenic and immunosuppressive nature, specific targeting of miR-183 may provide both chemotherapeutic and positive immunomodulatory effects in lung cancer.
Experimental Procedures
Cell Culture and Reagents.
All cell culture, reagents, antibodies for flow cytometry/immunoblot, and suppliers are listed in SI Experimental Procedures.
Molecular Biology.
qPCR was performed to detect DAP12, DAP10, perforin, and miRs. AGO2–RNA immunoprecipitation was performed to detect DAP12 and miR-183 within the RISC.
The DAP12 3′ UTR was cloned into a luciferase reporter vector (Promega) to generate DAP12-Luc or miR-183 seed-site mutant (DAP12-luc-M). For reporter assays, HeLa cells were transfected with luciferase constructs, renilla luciferase, and premiR-precursors followed by quantification of luciferase activity (Dual-Luciferase Reporter Assay; Promega).
NK Cell Lentiviral Infection.
Concentrated viral stocks carrying HIV-based lentiviral expression constructs (miR-183, antisense miR-183, or control) were used to infect primary NK or NK92 cells (multiplicity of infection 20). Infected cells were sorted (>95% GFP) prior to experimentation.
Immunohistochemistry and Fluorescent Microscopy.
FFPE lung biopsies of 29 patients were stained for DAP12 (Santa Cruz; FL-113) or TGFβ (LSBio; LS-B4772) and developed for immunohistochemistry. These biopsies and a human tissue microarray were dually stained for DAP12 and NKp46 (LSbio; LS-B2105), and the staining intensities were independently quantified and averaged by an imaging algorithm.
Supplementary Material
Acknowledgments
We thank Michelle Maurin for assistance with molecular cloning. The following shared resources were critical for the completion of this work: Flow Cytometry, Analytical Microscopy, Molecular Genomics, and Tissue Core Facilities. This work was supported by the Manuel and Adeline Garcia Endowed Chair (to J.Y.D.), Lung SPORE Grant P50 CA119997 (to E.B.H.), and the T32 Training Grant CA115308 (to S.S.D. and E.A.E).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319269111/-/DCSupplemental.
References
- 1.Djeu JY, Jiang K, Wei S. A view to a kill: Signals triggering cytotoxicity. Clin Cancer Res. 2002;8(3):636–640. [PubMed] [Google Scholar]
- 2.Graef T, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med. 2009;206(11):2557–2572. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yokoyama WM, Plougastel BF. Immune functions encoded by the natural killer gene complex. Nat Rev Immunol. 2003;3(4):304–316. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- 4.Vitale M, et al. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. J Exp Med. 1998;187(12):2065–2072. doi: 10.1084/jem.187.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 6.Carrega P, et al. Natural killer cells infiltrating human nonsmall-cell lung cancer are enriched in CD56 bright CD16(-) cells and display an impaired capability to kill tumor cells. Cancer. 2008;112(4):863–875. doi: 10.1002/cncr.23239. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Flavell RA, Sanjabi S, Wrzesinski SH, Licona-Limón P. The polarization of immune cells in the tumour environment by TGFbeta. Nat Rev Immunol. 2010;10(8):554–567. doi: 10.1038/nri2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikushima H, Miyazono K. TGFbeta signalling: A complex web in cancer progression. Nat Rev Cancer. 2010;10(6):415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 10.Friese MA, et al. RNA interference targeting transforming growth factor-beta enhances NKG2D-mediated antiglioma immune response, inhibits glioma cell migration and invasiveness, and abrogates tumorigenicity in vivo. Cancer Res. 2004;64(20):7596–7603. doi: 10.1158/0008-5472.CAN-04-1627. [DOI] [PubMed] [Google Scholar]
- 11.Trotta R, et al. TGF-beta utilizes SMAD3 to inhibit CD16-mediated IFN-gamma production and antibody-dependent cellular cytotoxicity in human NK cells. J Immunol. 2008;181(6):3784–3792. doi: 10.4049/jimmunol.181.6.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: New regulators of immune cell development and function. Nat Immunol. 2008;9(8):839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 13.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giebel S, et al. Association of KIR2DS4 and its variant KIR1D with leukemia. Leukemia. 2008;22(11):2129–2130, discussion 2130–2131. doi: 10.1038/leu.2008.108. [DOI] [PubMed] [Google Scholar]
- 15.Katz G, et al. MHC class I-independent recognition of NK-activating receptor KIR2DS4. J Immunol. 2004;173(3):1819–1825. doi: 10.4049/jimmunol.173.3.1819. [DOI] [PubMed] [Google Scholar]
- 16.Busche A, Goldmann T, Naumann U, Steinle A, Brandau S. Natural killer cell-mediated rejection of experimental human lung cancer by genetic overexpression of major histocompatibility complex class I chain-related gene A. Hum Gene Ther. 2006;17(2):135–146. doi: 10.1089/hum.2006.17.135. [DOI] [PubMed] [Google Scholar]
- 17.Landi MT, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS ONE. 2008;3(2):e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharjee A, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98(24):13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou J, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS ONE. 2010;5(4):e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geller MA, et al. A phase II study of allogeneic natural killer cell therapy to treat patients with recurrent ovarian and breast cancer. Cytotherapy. 2011;13(1):98–107. doi: 10.3109/14653249.2010.515582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saji H, et al. Significance of expression of TGF-beta in pulmonary metastasis in non-small cell lung cancer tissues. Ann Thorac Cardiovasc Surg. 2003;9(5):295–300. [PubMed] [Google Scholar]
- 22.Garber ME, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98(24):13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12(2):99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 24.Pillai RS, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309(5740):1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 25.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su LJ, et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narni-Mancinelli E, et al. Tuning of natural killer cell reactivity by NKp46 and Helios calibrates T cell responses. Science. 2012;335(6066):344–348. doi: 10.1126/science.1215621. [DOI] [PubMed] [Google Scholar]
- 28.Platonova S, et al. Profound coordinated alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res. 2011;71(16):5412–5422. doi: 10.1158/0008-5472.CAN-10-4179. [DOI] [PubMed] [Google Scholar]
- 29.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13(9):849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 30.Saetrom P, et al. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007;35(7):2333–2342. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu J, et al. Pro- and antiinflammatory cytokine signaling: Reciprocal antagonism regulates interferon-gamma production by human natural killer cells. Immunity. 2006;24(5):575–590. doi: 10.1016/j.immuni.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 32.Arnon TI, et al. Recognition of viral hemagglutinins by NKp44 but not by NKp30. Eur J Immunol. 2001;31(9):2680–2689. doi: 10.1002/1521-4141(200109)31:9<2680::aid-immu2680>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 33.Baychelier F, et al. Identification of a cellular ligand for the natural cytotoxicity receptor NKp44. Blood. 2013;122(17):2935–2942. doi: 10.1182/blood-2013-03-489054. [DOI] [PubMed] [Google Scholar]
- 34.Turnbull IR, Colonna M. Activating and inhibitory functions of DAP12. Nat Rev Immunol. 2007;7(2):155–161. doi: 10.1038/nri2014. [DOI] [PubMed] [Google Scholar]
- 35.Park YP, et al. Complex regulation of human NKG2D-DAP10 cell surface expression: Opposing roles of the γc cytokines and TGF-β1. Blood. 2011;118(11):3019–3027. doi: 10.1182/blood-2011-04-346825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lanier LL. DAP10- and DAP12-associated receptors in innate immunity. Immunol Rev. 2009;227(1):150–160. doi: 10.1111/j.1600-065X.2008.00720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snyder MR, Nakajima T, Leibson PJ, Weyand CM, Goronzy JJ. Stimulatory killer Ig-like receptors modulate T cell activation through DAP12-dependent and DAP12-independent mechanisms. J Immunol. 2004;173(6):3725–3731. doi: 10.4049/jimmunol.173.6.3725. [DOI] [PubMed] [Google Scholar]
- 38.Tridandapani S, et al. TGF-beta 1 suppresses [correction of supresses] myeloid Fc gamma receptor function by regulating the expression and function of the common gamma-subunit. J Immunol. 2003;170(9):4572–4577. doi: 10.4049/jimmunol.170.9.4572. [DOI] [PubMed] [Google Scholar]
- 39.Lin Q, et al. A cluster of specified microRNAs in peripheral blood as biomarkers for metastatic non-small-cell lung cancer by stem-loop RT-PCR. J Cancer Res Clin Oncol. 2012;138(1):85–93. doi: 10.1007/s00432-011-1068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 2010;70(23):9570–9580. doi: 10.1158/0008-5472.CAN-10-2074. [DOI] [PubMed] [Google Scholar]
- 41.Mulrooney TJ, Posch PE, Hurley CK. DAP12 impacts trafficking and surface stability of killer immunoglobulin-like receptors on natural killer cells. J Leukoc Biol. 2013;94(2):301–313. doi: 10.1189/jlb.0213093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soroosh P, et al. Lung-resident tissue macrophages generate Foxp3+ regulatory T cells and promote airway tolerance. J Exp Med. 2013;210(4):775–788. doi: 10.1084/jem.20121849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.