Significance
Molecular hydrogen is present in trace concentrations in the Earth’s lower atmosphere and is rapidly turned over through a biogeochemical cycle. Microbial soil processes are responsible for the majority of net uptake of H2 from the atmosphere, but the enzymes involved have remained elusive. In this communication, we use genetic dissection and GC measurements to show that the soil actinobacterium Mycobacterium smegmatis can oxidize trace concentrations of H2. This process depends on two oxygen-dependent, membrane-associated [NiFe] hydrogenases with picomolar thresholds. Activity is most pronounced during carbon limitation, suggesting that mycobacteria oxidize a dependable trace gas to survive starvation. We propose that these enzymes and their homologs in other actinobacteria and soil phyla constitute the principal sink of global atmospheric H2.
Keywords: atmospheric chemistry, biogeochemical cycles, enzyme kinetics, mycobacteria
Abstract
In the Earth’s lower atmosphere, H2 is maintained at trace concentrations (0.53 ppmv/0.40 nM) and rapidly turned over (lifetime ≤ 2.1 y−1). It is thought that soil microbes, likely actinomycetes, serve as the main global sink for tropospheric H2. However, no study has ever unambiguously proven that a hydrogenase can oxidize this trace gas. In this work, we demonstrate, by using genetic dissection and sensitive GC measurements, that the soil actinomycete Mycobacterium smegmatis mc2155 constitutively oxidizes subtropospheric concentrations of H2. We show that two membrane-associated, oxygen-dependent [NiFe] hydrogenases mediate this process. Hydrogenase-1 (Hyd1) (MSMEG_2262-2263) is well-adapted to rapidly oxidize H2 at a range of concentrations [Vmax(app) = 12 nmol⋅g⋅dw−1⋅min−1; Km(app) = 180 nM; threshold = 130 pM in the Δhyd23 (Hyd1 only) strain], whereas Hyd2 (MSMEG_2719-2720) catalyzes a slower-acting, higher-affinity process [Vmax(app) = 2.5 nmol⋅g⋅dw−1⋅min−1; Km(app) = 50 nM; threshold = 50 pM in the Δhyd13 (Hyd2 only) strain]. These observations strongly support previous studies that have linked group 5 [NiFe] hydrogenases (e.g., Hyd2) to the oxidation of tropospheric H2 in soil ecosystems. We further reveal that group 2a [NiFe] hydrogenases (e.g., Hyd1) can contribute to this process. Hydrogenase expression and activity increases in carbon-limited cells, suggesting that scavenging of trace H2 helps to sustain dormancy. Distinct physiological roles for Hyd1 and Hyd2 during the adaptation to this condition are proposed. Soil organisms harboring high-affinity hydrogenases may be especially competitive, given that they harness a highly dependable fuel source in otherwise unstable environments.
Although hydrogen is the universe’s most fundamental and abundant element, H2 is present at only trace concentrations in the Earth’s atmosphere. The biogeochemical cycle of molecular hydrogen results in rapid turnover of tropospheric H2 (lifetime = 1.4–2.1 y−1) (1) and maintains its levels at low steady-state concentrations (0.53 ppmv/0.40 nM) (2). It is well-established that geochemical and anthropogenic processes are responsible for the majority of net H2 production (3, 4). The role of biological processes is more complex. Because of intra- and interspecies hydrogen cycling, most microbial H2-oxidizing and H2-producing processes are balanced and hence make minor or null net contributions to the global hydrogen budget (5). It has nevertheless been known for several decades that soil ecosystems can oxidize tropospheric H2 in an apparent enzymatic process (6–8). This ecologically essential process is estimated to be accountable for 80% of the net loss of tropospheric H2 (3). However, the enzymes responsible remain to be resolved.
GC and tritium exchange measurements in soil samples have demonstrated that H2 oxidation by soil is a biphasic process, containing both low-affinity (Km > 1,000 nM) and high-affinity (Km = 10–70 nM) activities (9, 10). These activities are thought to be catalyzed by [NiFe] hydrogenases, of which there are five defined phylogenetically and functionally distinct groups (11, 12). The majority of characterized hydrogen-oxidizing soil bacteria (e.g., Paracoccus denitrificans, Ralstonia eutropha) have a low affinity for H2 (Km ≥ 1 μM, threshold ≥ 1 nM) and hence are probably responsible for the former process; their uptake hydrogenases (principally group 1 [NiFe] hydrogenases) serve to recycle the relatively high levels of H2 produced by biological processes (8, 13, 14). However, it has recently been shown that certain soil-dwelling actinomycetes are capable of scavenging H2 at tropospheric concentrations (12, 15). The first isolated high-affinity H2-oxidizer, Streptomyces sp. PCB7, can oxidize H2 with Michaelis–Menten kinetics with an apparent Km ≤30 nM and a threshold concentration ≤0.1 nM (15–17). An earlier study demonstrated that Mycobacterium smegmatis can oxidize low concentrations of H2, but did not determine the affinity or threshold of this activity (18).
It is proposed that group 5 [NiFe] hydrogenases are responsible for high-affinity H2 oxidation in streptomycetes (12, 17). The genes encoding this newly discovered hydrogenase lineage (hhyLS) are highly conserved and taxonomically restricted; among sequenced genomes, this hydrogenase is only widespread in environmental actinomycetes, including Streptomyces, Mycobacterium, Rhodococcus, and Frankia species (Fig. S1B) (17, 19). However, the evidence that these enzymes are responsible for the observed activity is correlative and somewhat conflicting. High-affinity hydrogenase activity has never been unambiguously determined, e.g., through purifying proteins or making genetic deletions. In addition, many organisms harboring hhySL genes only have low to medium affinities for H2 (e.g., Streptomyces scabiei, Streptomyces griseoflavus) (17). Furthermore, the recently purified group 5 [NiFe] hydrogenase from the β-proteobacterium R. eutropha is a low-affinity, low-activity enzyme (20).
Our laboratory has used M. smegmatis as a model organism to study hydrogen metabolism in soil actinomycetes. This saprophytic, nonsporulating bacterium is known for its ability to survive in nonreplicating states when carbon and oxygen sources become limiting (21, 22). The organism expresses three phylogenetically distinct [NiFe] hydrogenases, Hydrogenase-1 (Hyd1; MSMEG_2262-2263), Hydrogenase-2 (Hyd2; MSMEG_2719-2720), and Hydrogenase-3 (Hyd3; MSMEG_3928-3932) (19, 23). The group 2a [NiFe] hydrogenase Hyd1 and the group 5 [NiFe] hydrogenase Hyd2 are known to oxidize H2 (23). They are expressed at low levels during exponential growth and are significantly up-regulated during carbon or oxygen limitation (19, 23). By contrast, the group 3b enzyme Hyd3 (MSMEG-3928-3932) is predicted to couple NADPH oxidation to H2 evolution and is induced in oxygen-limitation by the dormancy response regulator DosR (23). Homologs of all three hydrogenases are distributed in other actinomycetes (Fig. S1). In this study, to confirm the determinants of high-affinity H2 oxidation, we compared the kinetics of H2 uptake by M. smegmatis mc2155 and strains harboring markerless deletions of its hydrogenases. We definitively confirm that Hyd1 and Hyd2 can oxidize tropospheric H2 and have distinct but overlapping characteristics.
Results
M. smegmatis mc2155 Constitutively Oxidizes Tropospheric H2.
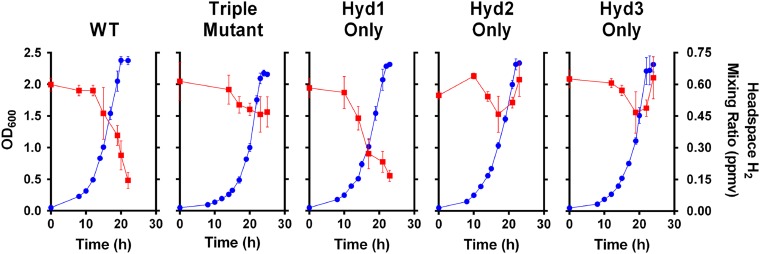
H2 oxidation was measured in exponentially growing and carbon-limited cultures of M. smegmatis mc2155 in sealed serum vials. In both conditions, the wild-type (WT) strain oxidized the majority of the H2 in the headspace (Fig. 1 and Fig. S2). Activity only occurred when the cultures were aerated and agitated, suggesting that O2 serves as the terminal electron acceptor. Consistent with [NiFe] hydrogenases being responsible for this activity, oxidation was fourfold faster when exogenous NiSO4 was introduced in the media (Fig. S2). Oxidation continued when subtropospheric concentrations of H2 were available. During exponential growth in a headspace of ambient air, the WT strain consumed the majority of the H2 available (initial mixing ratio ∼ 0.60 ppmv; final mixing ratio ∼ 0.14 ppmv). This activity was dependent on Hyd1 and did not occur in strains harboring markerless deletions of this enzyme (Δhyd1, Δhyd12, Δhyd13, and Δhyd123; Fig. 1).
Fig. 1.
H2 oxidation during exponential growth of M. smegmatis mc2155 and derivatives in sealed serum vials. H2 mixing ratios were measured upon inoculation of culture and throughout growth. All strains entered stationary phase at OD ∼ 2.5 (∼1.5 × 108 cfu⋅mL−1) after consuming the majority of the oxygen available in the headspace and hence becoming oxygen-limited. Blue circles represent OD600 of the cultures. Red squares represent the H2 mixing ratio in the headspace. Error bars are SDs from three biological replicates.
Hyd1 and Hyd2 Oxidize Tropospheric H2 in Kinetically Distinct Activities.
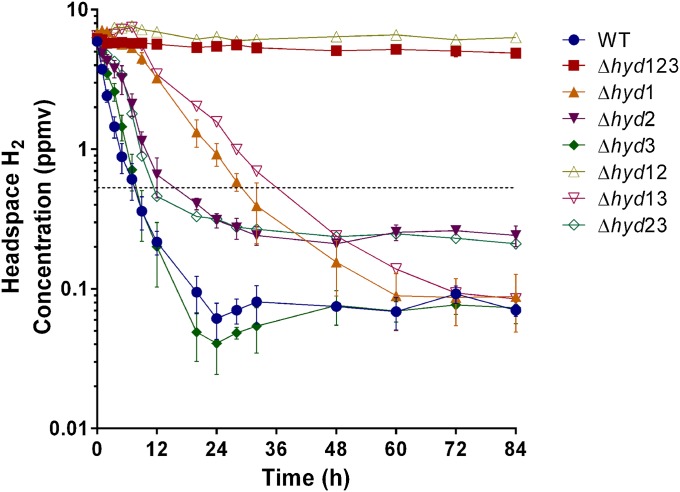
Hydrogenase activity was subsequently measured in nonreplicating carbon-limited cultures of the M. smegmatis strains over an extended time course. Consistent with the increased expression of Hyd1 and Hyd2 during carbon limitation (23), Hyd1 was more active in this condition and Hyd2 activity could be resolved (Fig. 2). The WT strain mc2155 oxidized the available headspace H2 (initial mixing ratio ∼ 6.3 ppmv) in a first-order kinetic process (Fig. 2 and Fig. S3). This activity was mainly attributable to the group 2a [NiFe] hydrogenase Hyd1; the Δhyd23 (Hyd1 only) strain rapidly oxidized H2 to subtropospheric concentrations (final mixing ratio ∼ 0.21 ppmv). However, high-affinity activity corresponding to the group 5 [NiFe] hydrogenase Hyd2 was also observed; the Δhyd13 (Hyd2 only) strain slowly oxidized the headspace H2 and functioned at even lower concentrations than the Δhyd23 strain (final mixing ratio ∼ 0.07 ppmv). The strain lacking Hyd3 (Δhyd3) behaved similarly to WT, reinforcing that this enzyme is not expressed in aerobic cultures. In the strain lacking any hydrogenases (Δhyd123), no significant changes in headspace H2 concentration could be observed.
Fig. 2.
H2 oxidation by carbon-limited cultures of M. smegmatis mc2155 and derivatives. All strains were inoculated to a density of 3 × 107 cfu⋅mL−1 (OD600 = 0.4). H2 mixing ratios were measured upon inoculation of culture and at regular intervals over 84 h. The mixing ratios are displayed on a logarithmic scale. Error bars represent SDs from three biological replicates. The dashed lines represent the average global mixing ratio of tropospheric H2 (0.53 ppmv).
To further profile the hydrogenases, the kinetic parameters of H2 oxidation were determined in carbon-limited cultures of the WT, Δhyd23 (Hyd1 only), and Δhyd13 (Hyd2 only) strains incubated at low (6.3 ppmv) and high (500, 1,000, and 2,000 ppmv) initial mixing ratios of H2 (Table 1). In both cases, Hyd1 consumed the majority of the H2 present and Hyd2 activity was fivefold lower. Whereas Hyd1 is well-adapted to function at a wide range of H2 concentrations [Km(app) = 184 ± 40 nM; threshold = 133 ± 7 pM], Hyd2 can oxidize lower concentrations of H2 overall [Km(app) = 51 ± 16; threshold = 54 ± 3 pM]. Although it is clear that Hyd1 is the dominant uptake hydrogenase in M. smegmatis, it remains to be fully resolved whether the enzyme is intrinsically faster than Hyd2 or instead more abundant in the cell.
Table 1.
Kinetic parameters of hydrogenase activity measured in whole cells
| Parameter | WT | Hyd1 only | Hyd2 only |
| Vmax(app), nmol g dw−1 min−1 | 16.3 ± 1.5 | 12.3 ± 1.7 | 2.5 ± 0.8 |
| k(app), nmol g dw−1 min−1 | 0.069 ± 0.005 | 0.067 ± 0.013 | 0.019 ± 0.0015 |
| Km(app), nM | 113 ± 11 | 184 ± 40 | 51 ± 16 |
| Threshold, pM | 51 ± 13 | 133 ± 7 | 54 ± 3 |
SDs were calculated from three biologically independent replicates.
Hydrogen Oxidation Is Membrane-Associated.
The localization of the uptake hydrogenases was determined by using zymographic and amperometric assays on cell fractions (Fig. 3). Hyd2 efficiently coupled the oxidation of H2 to the reduction of the artificial electron acceptor/redox dye nitroblue tetrazolium chloride acceptor (Eo′ = −80 mV) (24). This activity was enriched in the membrane fraction (Fig. 3B). Attempts to stain with more electronegative acceptors, such as benzyl viologen [Eo′ = −360 mV (24)], were unsuccessful. Although Hyd1 activity could not be visualized zymographically, this highly active enzyme could be detected by using an H2 microsensor. As previously observed in cyanobacterial group 2a [NiFe] hydrogenases (25), its activity was concentrated in the membrane fractions of the WT and Δhyd23 strains (Fig. 3C). Hyd1 and Hyd2 lack predicted transmembrane helices and signal peptides, and are not transcribed with genes encoding probable membrane proteins (PFAM/TMHMM analysis). They are therefore unlikely to be classical membrane-bound proteins and instead may associate with the cytosolic side of the membrane through amphiphilic helices or protein-protein interactions.
Fig. 3.
Determination of uptake hydrogenase localization. Protein samples of each cell fraction (5 µg) were separated on native polyacrylamide gels. (A) Total protein content was stained by using Coomassie brilliant blue. (B) Uptake hydrogenase activity was detected by using the artificial electron acceptor nitroblue tetrazolium chloride. (C) Cell fractions were added to a reaction chamber saturated with H2 and O2. A H2 microsensor determined the rates of hydrogen consumption (negative values) or production (positive values). Error bars represent SDs from three technical replicates. C, cytosolic fraction; L, crude cell lysate; M, membrane fraction.
Discussion
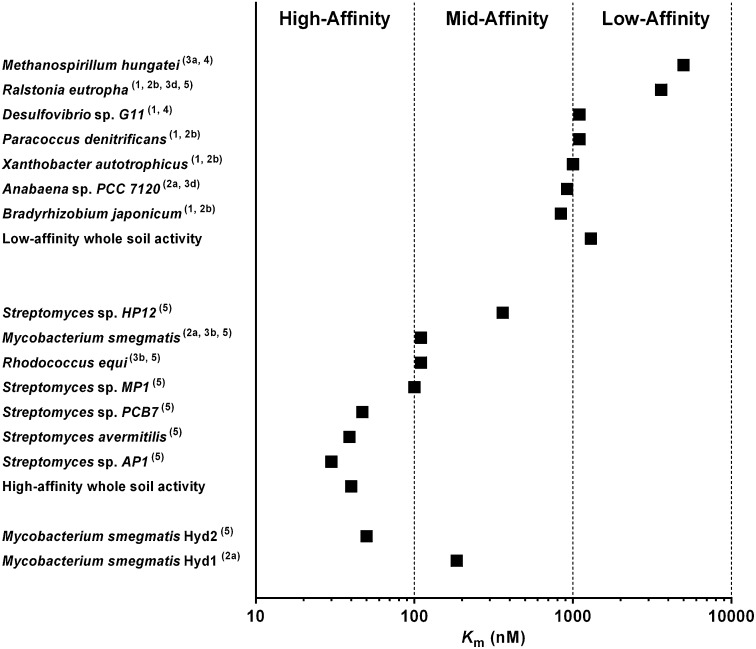
In summary, this work demonstrates that M. smegmatis mc2155 oxidizes tropospheric H2 by using two membrane-associated, oxygen-dependent [NiFe] hydrogenases. We definitively confirm that group 5 [NiFe] hydrogenases can mediate high-affinity H2 oxidation and provide strong support for the hypothesis that these enzymes are the principal sinks for atmospheric H2. The kinetic parameters of whole-cell H2 oxidation by Hyd2, as observed in isolation in the Δhyd13 strain and deleted in the Δhyd2 strain, are very similar to the high-affinity processes observed in whole soil samples (9, 10) and cultured streptomycetes (12, 15) (Fig. 4). Genes encoding group 5 [NiFe] hydrogenases are widespread in soil members of the order Actinomycetales and are also distributed in undercultured soil phyla such as Acidobacteria (Fig. S1B). It is also evident from this study that some group 2a [NiFe] hydrogenases have the capacity to oxidize low concentrations of H2; however, these enzymes are likely to make a smaller contribution to the global hydrogen budget, given they are found in fewer sequenced actinomycetes (Fig. S1A) (23) and primarily serve to recycle nitrogenase-derived H2 in other organisms (e.g., cyanobacteria) (25).
Fig. 4.
The wide spectrum of affinities for H2 observed in hydrogen-oxidizing organisms. The types of [NiFe] hydrogenase encoded by each organism are shown in brackets. Pure cultures of the soil actinomycetes M. smegmatis (this work), R. equi (26), and certain Streptomyces species (12, 15, 17) have high affinities for H2 (Km = 30–130 nM). These activities depend on group 5 [NiFe] hydrogenases and correlate with the high-affinity portion of H2 oxidation in whole soils (10, 16). By contrast, hydrogen-oxidizing soil Proteobacteria such as Bradyrhizobium japonicum (28), P. denitrificans (29), Xanthobacter autotrophicus (14), and R. eutropha (20) have low affinities for H2 (Km = 1–5 µM) similar to that observed for low-affinity portion of H2 oxidation in whole soils (10). H2 affinities of the cyanobacterium Anabaena sp. PCC 7120 (30), sulfate-reducing bacterium Desulfovibrio sp. G11, and methanogen Methanospirillum hungatei (31) are also shown for reference. Many organisms harboring group 5 [NiFe] hydrogenases (e.g., R. eutropha) and group 2a [NiFe] hydrogenases (e.g., Anabaena sp. PCC 7120) still have low affinities for H2.
The ability to oxidize tropospheric H2 may serve as a major selection pressure in soil ecosystems. These environments are characterized by significant interspecies competition, rapid nutrient fluxes, and changing physical conditions. Although present in trace concentrations, tropospheric H2 is ubiquitous, unlimited, and energy-rich; it therefore constitutes a dependable fuel source in an otherwise unstable environment. It is now clear that M. smegmatis grows mixotrophically on H2 and organic carbon sources during exponential growth in pure cultures, in accordance with our previous expression and phenotypic studies (23). However, the expression (23) and activities of Hyd1 and Hyd2 are significantly higher during carbon limitation. Group 5 [NiFe] hydrogenases are also more active during energy limitation in other actinomycetes (e.g., Rhodococcus equi) and sporulation in streptomycetes (e.g., Streptomyces sp. PCB7) (12, 26). In such conditions, H2 may substitute as the primary electron donor and be used to maintain redox balance and a membrane potential. The finding that group 5 [NiFe] hydrogenases can sustain catalytic activity at a wide range of pH values, temperatures, and O2 concentrations further suggests that these enzymes confer robustness and flexibility in response to environmental shifts (20).
Hyd1 and Hyd2 share many characteristics, including similar expression profiles (23), high-affinity oxidation activities, and membrane association. However, it is clear that their physiological roles in the cell are not redundant. Despite continued activity of Hyd1, the growth yield of the Δhyd2 strain was 40% lower than the WT in continuous culture (19). Hyd2 must therefore use tropospheric H2 for an important and unique purpose in the cell, e.g., to maintain the quinone pool or intracellular cofactors in a reduced state. In contrast, Hyd1 is likely to be a respiratory enzyme that sustains a membrane potential by coupling H2 oxidation to O2 reduction through a physical association with the electron transport chain. This enzyme has a high Vmax(app) and is efficient at a range of H2 concentrations. It can metabolize the high concentrations that will occur in certain microenvironments [e.g., in waterlogged or rhizobial soils (27)] or as a result of Hyd3 activity during oxygen limitation. The enzyme also enables M. smegmatis to oxidize tropospheric H2 at significantly faster rates than cultured streptomycetes harboring only group 5 [NiFe] hydrogenases (12).
Hyd1 and Hyd2 are the first hydrogenases to have been unambiguously shown to mediate high-affinity H2 oxidation. Their apparent Kms are more than 10-fold lower than most purified uptake hydrogenases (Fig. 4). However, the features responsible for this adaptation remain to be established; it remains unclear whether the hydrogenases themselves are intrinsically high-affinity enzymes or whether their affinities are modulated by their cellular context (e.g., through their redox partners and interactions with the respiratory chain). Curiously, a wide spectrum of apparent Km values have been observed for group 5 [NiFe] hydrogenases in other organisms (Fig. 4) (12, 26). This is surprising given these enzymes are encoded in relatively conserved operons and share very high amino acid sequence identities (>70%). Our multiple sequence alignments reveal no consistent amino acid substitutions between apparent high-affinity (e.g., M. smegmatis, Streptomyces avermitilis) and low-affinity (e.g., R. eutropha, S. scabiei) members. Hyd1 is also the first group 2a [NiFe] hydrogenase shown to oxidize tropospheric H2. It is notable that this enzyme is cotranscribed with a putative [2Fe-2S] protein (MSMEG_2268), which is likely to serve as the physiological electron acceptor (23). Genes encoding homologous proteins are transcribed with other group 5 [NiFe] hydrogenases (17), including Hyd2 (MSMEG_2718) (23), but are absent from most cyanobacterial genomes. We propose that such proteins may have a relatively high redox potential (>−200 mV), thus ensuring that the oxidation of trace concentrations of H2 is thermodynamically favorable. It is also conceivable that unusual features of the redox metabolism or respiratory chain of M. smegmatis may influence the organism’s affinity for H2. Biochemical and biophysical studies of purified proteins are clearly required to develop an understanding of the high-affinity activities, oxygen tolerance, and redox partners of Hyd1 and Hyd2.
Materials and Methods
Bacterial Strains and Growth Conditions.
All bacterial strains used in this study are listed in Table S1. M. smegmatis mc2155 (32) and derived mutants (19, 23) were maintained on lysogeny broth agar plates (33) supplemented with 0.05% (wt/vol) tyloxapol (Sigma-Aldrich). For broth culture, M. smegmatis was grown at 37 °C in modified Hartmans–de Bont minimal medium (34) supplemented with 25 mM glycerol, 0.05% tyloxapol, and 10 µM NiSO4. Cultures were incubated with agitation (200 rpm) at 37 °C toward carbon limitation (30 mL medium in a 125-mL aerated flask) or oxygen limitation (30 mL medium in a 120-mL rubber-stoppered serum vial). For fractionation studies, cultures were grown into carbon limitation in larger volumes (750 mL medium in a 2,500-mL aerated flask). All cultures were inoculated to an initial optical density of 0.05. Optical densities to assess growth were measured at 600 nm in a Jenway 6300 spectrometer. Cultures were diluted in 0.85% saline solution to bring the OD600 below 0.5 when measured in cuvettes of 1 cm light path length. To count cfus per milliliter, the cultures were serially diluted in PBS solution (pH 7.0) and spread onto agar plates.
GC Measurements.
To measure high-affinity H2 uptake, carbon-limited cultures of M. smegmatis mc2155 and derivatives were injected into serum vials containing 30 mL Hartmans–de Bont medium supplemented with 0.05% tyloxapol to a density of 3 × 107 cfu⋅mL−1 (OD600 = 0.4). Standard H2 gas (1,000 ppmv ± 2%; Messer Schweiz) was added to headspaces to obtain initial mixing ratios of 6.3 ± 0.7 ppmv H2. The change in mixing ratio was measured as a function of time through GC. Aliquots of headspace air (250 µL) were sampled by using a gas-tight syringe (VICI Precision Sampling) and loaded on to a Trace Analytical Reduced Gas Analyzer (SRI Instruments). Serum vials were continuously agitated (120 rpm) to enhance transfer of H2 from the gas phase to the liquid phase. Initial tests showed that H2 consumption was proportional to the amount of cells and thus not limited by gas transfer. For Vmax determinations, the rate of H2 uptake was measured after addition of pure H2 gas (Messer Schweiz) at initial mixing ratios of 500, 1,000, and 2,000 ppmv. Km was estimated by dividing the Vmax by the first-order rate constant k as previously described (35). The kinetics of H2 uptake were also measured in exponentially growing cells (initial mixing ratio = 0.60 ± 0.07 ppmv H2). All experiments were performed by using biological triplicates. To avoid gas buildup, serum vials were sealed with rubber stoppers only after autoclaving. The reproducibility of the measurements was assessed before and during each time course by analysis of standard H2 gas (2.0 ppmv ± 5%, 50 ppmv ± 2%, and 1,000 ppmv ± 2% H2; Messer Schweiz).
Preparation of Cell Fractions.
M. smegmatis mc2155 and derivatives were fractionated into membranes and cytosols. Cultures (750 mL) were pelleted by centrifugation (15 min, 5,000 × g, 4 °C), washed in PBS solution, and resuspended in 25 mL lysis buffer [50 mM 3-(N-morpholino)propanesulfonic acid, pH 7.5, 1 mM PMSF, 15 mM MgCl2, 5 mg⋅mL−1 lysozyme, 1 mg DNase]. The mixture was incubated (60 min, 50 rpm, 37 °C) and passed through a cell disruptor (35,000 psi, four times; Constant Systems). After removal of unbroken cells by centrifugation (10 min, 10,000 × g, 4 °C), the crude cell lysate was ultracentrifuged (45 min, 150,000 × g, 4 °C) to yield cytosols (supernatant) and membranes (pellets). The protein concentrations of the crude cell lysates, cytosols, and membranes were measured by using the BCA method (Pierce) against BSA standards.
Activity Staining on Native Polyacrylamide Gels.
Samples of 5 µg protein from each cell fraction were loaded on to two native 7.5% (wt/vol) polyacrylamide gels prepared as previously described (36). The gels were run simultaneously (25 V, 3 h) in Tris-glycine buffer (pH 8.3). One gel was stained with Coomassie brilliant blue to visualize total protein. The other was stained for hydrogenase activity (H2-nitroblue tetrazolium oxidoreductase activity) in 50 mM potassium phosphate buffer (pH 7.0) supplemented with 0.5 mM nitroblue tetrazolium chloride. The samples were incubated overnight in a hydrogen-rich atmosphere [5% (vol/vol) H2, 10% (vol/vol) CO2, 85% (vol/vol) N2] in an anaerobic chamber (Coy Laboratory Products).
Amperometric Hydrogen Measurements.
Hydrogenase activity of the cell fractions was amperometrically measured as previously described (23). Hydrogen-saturated PBS solution (0.1 mL), oxygen-saturated PBS solution (0.1 mL), and diluted cell fractions (0.7 mL; OD600 = 1.5) were introduced and stirred in a 0.9-mL assay chamber. The H2 concentration was measured over time using a Unisense H2-MR hydrogen microsensor (Unisense) polarized at +800 mV and calibrated according to the manufacturer’s instructions. After linear change in hydrogen concentration was observed, the initial rates were calculated for a period of 20 s and normalized to protein concentration.
Taxonomic Analysis.
The large subunit protein sequences of all predicted group 2a, group 5, and group 3b [NiFe] hydrogenases in sequenced genomes were retrieved from the National Center for Biotechnology Information database. For retrieval, the large subunits of the three [NiFe]-hydrogenases from M. smegmatis were used as reference sequences for the Blast Local Alignment Search Tool (37). The classifications were validated by examining the N-terminal L1 and C-terminal L2 signature motifs of each sequence (38).
Supplementary Material
Acknowledgments
We thank Laura Hesse, Melanie Klose, and Peter Klaus for excellent technical advice and support; Dr. Philippe Constant, Dr. Peter Janssen, Dr. Sergio Morales, Prof. Oliver Lenz, Caspar Schäfer, Prof. Frank Sargent, and Dr. Alison Parkin for helpful discussions; as well as the anonymous reviewers for their constructive feedback. This work was financially supported by a Marsden grant from the Royal Society of New Zealand, an Otago Postgraduate Scholarship from the University of Otago (to C.G.), and a James Cook Fellowship from the Royal Society of New Zealand (to G.M.C.). C.G.’s travel to the Max-Planck Institute for Terrestrial Microbiology was funded by the Professor Sandy Smith Memorial Scholarship, the Maurice and Phyllis Paykel Trust, and the University of Otago.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320586111/-/DCSupplemental.
References
- 1.Ehhalt DH, Rohrer F. The tropospheric cycle of H2: A critical review. Tellus B Chem Phys Meterol. 2009;61(3):500–535. [Google Scholar]
- 2.Novelli PC, et al. Molecular hydrogen in the troposphere: Global distribution and budget. J Geophys Res Atmos. 1999;104(23):30427–30444. [Google Scholar]
- 3.Rhee TS, Brenninkmeijer CAM, Röckmann T. The overwhelming role of soils in the global atmospheric hydrogen cycle. Atmos Chem Phys. 2006;6(6):1611–1625. [Google Scholar]
- 4.Constant P, Poissant L, Villemur R. Tropospheric H(2) budget and the response of its soil uptake under the changing environment. Sci Total Environ. 2009;407(6):1809–1823. doi: 10.1016/j.scitotenv.2008.10.064. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz E, Fritsch J, Friedrich B. H2-Metabolizing Prokaryotes. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thomson F, editors. The Prokaryotes: Prokaryotic Physiology and Biochemistry. 4th Ed. Berlin: Springer; 2013. pp. 119–199. [Google Scholar]
- 6.Conrad R, Seiler W. Contribution of hydrogen production by biological nitrogen fixation to the global hydrogen budget. J Geophys Res. 1980;85(1):5493–5498. [Google Scholar]
- 7.Conrad R, Aragno M, Seiler W. Production and consumption of hydrogen in a eutrophic lake. Appl Environ Microbiol. 1983;45(2):502–510. doi: 10.1128/aem.45.2.502-510.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conrad R. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO) Microbiol Rev. 1996;60(4):609–640. doi: 10.1128/mr.60.4.609-640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuler S, Conrad R. Soils contain two different activities for oxidation of hydrogen. FEMS Microbiol Ecol. 1990;73(1):77–84. [Google Scholar]
- 10.Häring V, Conrad R. Demonstration of two different H2-oxidizing activities in soil using an H2 consumption and a tritium exchange assay. Biol Fertil Soils. 1994;17(2):125–128. [Google Scholar]
- 11.Vignais PM, Billoud B, Meyer J. Classification and phylogeny of hydrogenases. FEMS Microbiol Rev. 2001;25(4):455–501. doi: 10.1111/j.1574-6976.2001.tb00587.x. [DOI] [PubMed] [Google Scholar]
- 12.Constant P, Chowdhury SP, Pratscher J, Conrad R. Streptomycetes contributing to atmospheric molecular hydrogen soil uptake are widespread and encode a putative high-affinity [NiFe]-hydrogenase. Environ Microbiol. 2010;12(3):821–829. doi: 10.1111/j.1462-2920.2009.02130.x. [DOI] [PubMed] [Google Scholar]
- 13.Conrad R, Seiler W. The role of hydrogen bacteria during the decomposition of hydrogen by soil. FEMS Microbiol Lett. 1979;6(3):143–145. [Google Scholar]
- 14.Conrad R, Aragno M, Seiler W. The inability of hydrogen bacteria to utilize atmospheric hydrogen is due to threshold and affinity for hydrogen. FEMS Microbiol Lett. 1983;18(3):207–210. [Google Scholar]
- 15.Constant P, Poissant L, Villemur R. Isolation of Streptomyces sp. PCB7, the first microorganism demonstrating high-affinity uptake of tropospheric H2. ISME J. 2008;2(10):1066–1076. doi: 10.1038/ismej.2008.59. [DOI] [PubMed] [Google Scholar]
- 16.Constant P, Chowdhury SP, Hesse L, Conrad R. Co-localization of atmospheric H2 oxidation activity and high affinity H2-oxidizing bacteria in non-axenic soil and sterile soil amended with Streptomyces sp. PCB7. Soil Biol Biochem. 2011;43(9):1888–1893. [Google Scholar]
- 17.Constant P, Chowdhury SP, Hesse L, Pratscher J, Conrad R. Genome data mining and soil survey for the novel group 5 [NiFe]-hydrogenase to explore the diversity and ecological importance of presumptive high-affinity H(2)-oxidizing bacteria. Appl Environ Microbiol. 2011;77(17):6027–6035. doi: 10.1128/AEM.00673-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King GM. Uptake of carbon monoxide and hydrogen at environmentally relevant concentrations by mycobacteria. Appl Environ Microbiol. 2003;69(12):7266–7272. doi: 10.1128/AEM.69.12.7266-7272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berney M, Cook GM. Unique flexibility in energy metabolism allows mycobacteria to combat starvation and hypoxia. PLoS ONE. 2010;5(1):e8614. doi: 10.1371/journal.pone.0008614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schäfer C, Friedrich B, Lenz O. Novel, oxygen-insensitive group 5 [NiFe]-hydrogenase in Ralstonia eutropha. Appl Environ Microbiol. 2013;79(17):5137–5145. doi: 10.1128/AEM.01576-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartmans S, de Bont JAM, Stackebrandt E. The genus Mycobacterium—nonmedical. In: Dworkin M, Falkw S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The Prokaryotes Third Edition: Archaea. Bacteria: Firmicutes, Actinomycetes. Berlin: Springer; 2006. pp. 889–918. [Google Scholar]
- 22.Smeulders MJ, Keer J, Speight RA, Williams HD. Adaptation of Mycobacterium smegmatis to stationary phase. J Bacteriol. 1999;181(1):270–283. doi: 10.1128/jb.181.1.270-283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berney M, Greening C, Hards K, Collins DM, Cook GM. Three different [NiFe]-hydrogenases confer metabolic plasticity in the obligate aerobe Mycobacterium smegmatis. Environ Microbiol. 2014;16(1):318–330. doi: 10.1111/1462-2920.12320. [DOI] [PubMed] [Google Scholar]
- 24.Pinske C, Jaroschinsky M, Sargent F, Sawers G. Zymographic differentiation of [NiFe]-hydrogenases 1, 2 and 3 of Escherichia coli K-12. BMC Microbiol. 2012;12:134. doi: 10.1186/1471-2180-12-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamagnini P, et al. Cyanobacterial hydrogenases: Diversity, regulation and applications. FEMS Microbiol Rev. 2007;31(6):692–720. doi: 10.1111/j.1574-6976.2007.00085.x. [DOI] [PubMed] [Google Scholar]
- 26.Meredith LK, et al. Consumption of atmospheric hydrogen during the life cycle of soil-dwelling actinobacteria. Environ Microbiol Rep. 2013 doi: 10.1111/1758-2229.12116. [DOI] [PubMed] [Google Scholar]
- 27.Conrad R, Seiler W. Field measurements of hydrogen evolution by nitrogen-fixing legumes. Soil Biol Biochem. 1979;11(6):689–690. [Google Scholar]
- 28.Klüber HD, Conrad R. Ferric iron-reducing Shewanella putrefaciens and N2-fixing Bradyrhizobium japonicum with uptake hydrogenase are unable to oxidize atmospheric H2. FEMS Microbiol Lett. 1993;111(2-3):337–341. [Google Scholar]
- 29.Häring V, Conrad R. Kinetics of H2 oxidation in respiring and denitrifying Paracoccus denitrificans. FEMS Microbiol Lett. 1991;78(2-3):259–264. [Google Scholar]
- 30.Houchins JP, Burris RH. Occurrence and localization of two distinct hydrogenases in the heterocystous cyanobacterium Anabaena sp. strain 7120. J Bacteriol. 1981;146(1):209–214. doi: 10.1128/jb.146.1.209-214.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson JA, Tiedje JM. Competition between sulfate-reducing and methanogenic bacteria for H2 under resting and growing conditions. Arch Microbiol. 1984;137(1):26–32. [Google Scholar]
- 32.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4(11):1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 34.Berney M, Weimar MR, Heikal A, Cook GM. Regulation of proline metabolism in mycobacteria and its role in carbon metabolism under hypoxia. Mol Microbiol. 2012;84(4):664–681. doi: 10.1111/j.1365-2958.2012.08053.x. [DOI] [PubMed] [Google Scholar]
- 35.Conrad R, Goodwin S, Zeikus JG. Hydrogen metabolism in a mildly acidic lake sediment (Knaack Lake) FEMS Microbiol Ecol. 1987;45(4):243–249. [Google Scholar]
- 36.Walker JM. Nondenaturing Polyacrylamide Gel Electrophoresis of Proteins. In: Walker JM, editor. The Protein Protocols Handbook. 2nd Ed. Berlin: Springer; 2002. pp. 57–60. [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Vignais PM, Billoud B. Occurrence, classification, and biological function of hydrogenases: An overview. Chem Rev. 2007;107(10):4206–4272. doi: 10.1021/cr050196r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.