Significance
Identifying disease-modifying therapies for Alzheimer’s disease (AD) has been an insurmountable challenge. To provide a new discovery tool for high-throughput compound screening, we used a simple yeast model that makes toxic amounts of β-amyloid (Aβ), a peptide central to AD pathology. Previous genetic analysis established that Aβ compromises yeast biology in a manner relevant to human AD. We screened 140,000 compounds for reversal of toxicity and identified a class of protective metal-binding compounds related to clioquinol (CQ), a compound that alleviates Aβ toxicity in mouse AD models. Treating yeast with CQ promoted rapid degradation of Aβ oligomers, rescuing cellular processes perturbed by this insidious peptide and restoring viability. Our approach provides a method for identifying compounds that may eventually help treat AD.
Keywords: phenotypic small-molecule screen, metal chelation
Abstract
Alzheimer’s disease (AD) is a common, progressive neurodegenerative disorder without effective disease-modifying therapies. The accumulation of amyloid-β peptide (Aβ) is associated with AD. However, identifying new compounds that antagonize the underlying cellular pathologies caused by Aβ has been hindered by a lack of cellular models amenable to high-throughput chemical screening. To address this gap, we use a robust and scalable yeast model of Aβ toxicity where the Aβ peptide transits through the secretory and endocytic compartments as it does in neurons. The pathogenic Aβ 1–42 peptide forms more oligomers and is more toxic than Aβ 1–40 and genome-wide genetic screens identified genes that are known risk factors for AD. Here, we report an unbiased screen of ∼140,000 compounds for rescue of Aβ toxicity. Of ∼30 hits, several were 8-hydroxyquinolines (8-OHQs). Clioquinol (CQ), an 8-OHQ previously reported to reduce Aβ burden, restore metal homeostasis, and improve cognition in mouse AD models, was also effective and rescued the toxicity of Aβ secreted from glutamatergic neurons in Caenorhabditis elegans. In yeast, CQ dramatically reduced Aβ peptide levels in a copper-dependent manner by increasing degradation, ultimately restoring endocytic function. This mirrored its effects on copper-dependent oligomer formation in vitro, which was also reversed by CQ. This unbiased screen indicates that copper-dependent Aβ oligomer formation contributes to Aβ toxicity within the secretory/endosomal pathways where it can be targeted with selective metal binding compounds. Establishing the ability of the Aβ yeast model to identify disease-relevant compounds supports its further exploitation as a validated early discovery platform.
Alzheimer’s disease (AD) is a common and devastating neurodegenerative disorder that is projected to increase in frequency as our population ages. The lack of disease-modifying therapies requires new approaches to address the underlying mechanisms of cellular dysfunction and identify potential therapeutics.
The amyloid-β peptide (Aβ) plays a major role in AD and ultimately leads to neuronal death and cognitive impairment (1). Aβ peptides of ∼40 aa are generated by the successive cleavage of the amyloid precursor protein (APP) by β- and γ-secretases. Mutations in APP or γ-secretase cause familial AD and bias APP cleavage toward a 42-aa Aβ peptide that predominates in Aβ plaques, is more aggregation prone, and is toxic to neurons (2, 3). Although Aβ plaques are a common, conspicuous feature of pathology in diseased brains, increasing evidence suggests that small oligomers of Aβ are the most toxic species (4, 5).
The conservation of protein homeostasis mechanisms—such as protein trafficking and chaperone networks—among all eukaryotes makes yeast a powerful discovery platform for modeling the cellular toxicities caused by neurodegenerative disease proteins (6). In neurons, the cleavage of APP to generate the Aβ peptide occurs within the secretory and endosomal pathways (7). APP is trafficked through the secretory pathway to the plasma membrane and subsequently internalized and recycled through endosomal vesicles and the trans-Golgi network back to the plasma membrane (7). During this recycling, the Aβ peptide is liberated from APP by β/γ-secretases, thus enabling the peptide to interact with multiple membranous compartments within the cell.
We have taken advantage of the great conservation of the secretory and endocytic pathways between yeast and neurons to study Aβ in a simple, highly tractable model organism—budding yeast. By expressing Aβ as a fusion to an endoplasmic reticulum (ER) targeting signal, we have mimicked in yeast the multicompartmental distribution of Aβ (8). This approach bypasses the need to recapitulate the entire APP processing pathway and generates an Aβ peptide with exactly the same sequence as is found in the human brain. The ER targeting signal directs cotranslational transport of the primary translation product into the ER, where the signal sequence is removed by signal peptidase. The peptide then transits through the secretory pathway and is secreted from the cell. In yeast, the cell wall prevents secreted Αβ from diffusing away, allowing it to interact with the plasma membrane and undergo endocytosis. As in the human nervous system (2), the aggregation-prone 42-aa peptide is more prone to forming oligomeric species than the 40-aa peptide (9) and is more toxic (8).
This model allowed us to take advantage of yeast genetics to perform a completely unbiased screen of the yeast genome for suppressers or enhancers of Aβ toxicity. Of the ∼6,000 genes we tested, we recovered only a handful of modifiers. There are ∼25,000 genes in the human genome and less than 20 (10) have been shown to confer risk for AD. However, several of the yeast genes that alter Aβ toxicity are either direct homologs or interacting partners of human risk factors (8, 10). For example, YAP1802, is the yeast homolog of PICALM, one the most highly validated risk factors for AD (10, 11); INP52, is homologous to SYNJ1, which interacts with the risk factor BIN1 (12, 13); and SLA1 is homologous to SH3KBP1, which interacts with the risk factor CD2AP (14–16). All of these proteins are involved in clathrin-mediated endocytosis in yeast and humans. In addition to ameliorating the toxicity of Aβ in yeast, these proteins reduced Aβ toxicity in both Caenorhabditis elegans and rat cortical neuronal models (8). The recovery of genes that promote clathrin-mediated endocytosis in unbiased genome-wide screens suggested that Aβ poisons this process (8). Indeed, the peptide compromised endocytosis of a transmembrane receptor (Ste3), an activity crucial for neuronal function. Importantly, the mechanism of action of these risk factors had not previously been linked to Aβ. Thus, the yeast model has already provided key insights on the nature of Aβ’s cellular toxicity in the human brain.
In this study, we used our yeast Aβ model to identify small molecules that ameliorate toxicity. In an extremely stringent and unbiased screen of 140,000 compounds, we identified a small number of cytoprotective compounds, including 8-hydroxyquinolines (8-OHQs). Members of this family bind metals and are among the few compounds that have been shown to alleviate Aβ toxicity in mouse models of AD (17, 18), and to show early potential as an AD therapeutic (19). Here, we investigate the mechanism of action for the most efficacious member of this family, clioquinol (CQ).
Results
Screen for Compounds That Rescue Aβ Toxicity in Yeast.
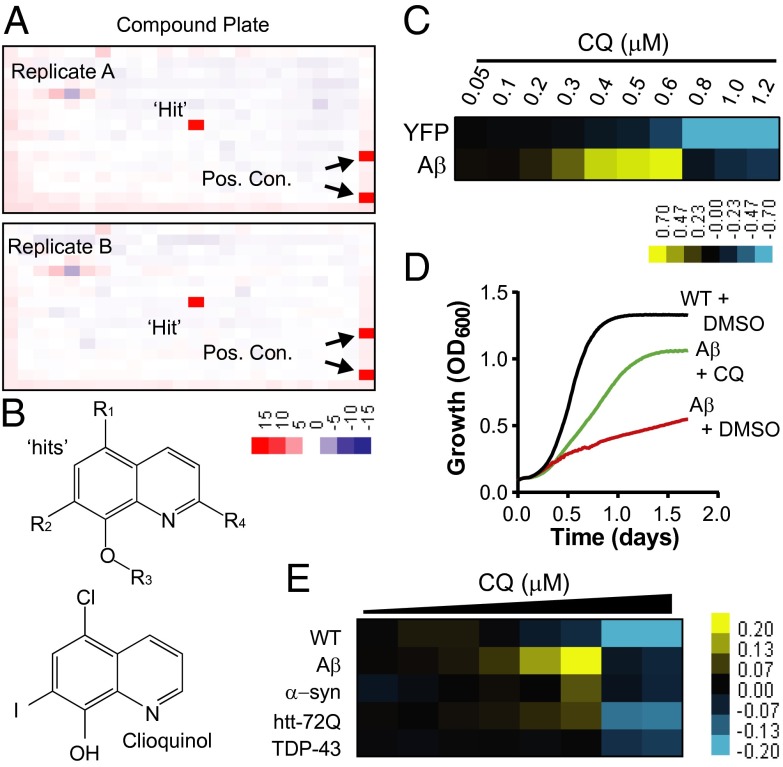
To screen for compounds that ameliorate Aβ toxicity in yeast, we used a strain with a compromised drug efflux system that expresses Aβ under the control of a galactose-inducible promoter (8). This strain exhibited a level of Aβ toxicity that provided sufficient dynamic range and high signal-to-background signal to confidently identify rescuing compounds. Approximately 140,000 compounds were tested in duplicate in 384-well plates using increased growth (optical density) as an end point. The high reproducibility enabled robust detection of “hits” (Fig. 1A). Only ∼30 compounds passed our rigorous scoring criteria (Z score > 3), establishing the high stringency of the screen. Strikingly, one-half of all recovered hits were 8-OHQs (Fig. 1B and Fig. S1).
Fig. 1.
Small-molecule screen identifies 8-OHQ that protect against Aβ toxicity. (A) Representative duplicate 384-well screening plates containing a single “hit” and positive controls. Values are Z scores, where “red” indicates rescue and “blue” indicates toxicity. (B) General structures of 8-OHQ hits and CQ. (C) Dose–response heat map of CQ treatment of both YFP- and Aβ-expressing strains. Data indicate the difference in OD600 between CQ- and DMSO-treated samples. Yellow, rescue. Blue, toxicity. Concentrations are in micromolar. (D) Growth curves of WT yeast and the Aβ strain treated with the maximal effective dose of CQ or DMSO alone. The y axis is OD600, and the x axis is time in days. (E) Dose–response curves of WT and multiple proteotoxicity yeast models treated with CQ in 384-well plates. Data are reported as the difference in OD600 between the CQ-treated and DMSO-treated conditions. Yellow indicates rescue, and blue reflects toxicity. The doses of CQ range from 5 μM down to 0.3 μM CQ with a 30% dilution at each step.
Notably, the effectiveness of this class of compounds against Aβ toxicity has been established in neurons and mouse models. Two closely related 8-OHQs—CQ and PBT2—decrease Aβ accumulation and alleviate cognitive and behavioral symptoms in mouse models of AD (17, 18). 8-OHQs are moderate-strength metal chelators that can extract metals weakly bound to proteins and both redistribute metals within the cell and act as ionophores (17, 18). PBT2 is one of only a few compounds effective in phase II clinical trials with human AD patients (19). CQ rescued our yeast Aβ model in a highly dose-dependent manner (Fig. 1 C and D), but at higher concentrations it inhibited the growth of both Aβ-expressing cells and control strains (Fig. 1C). The increase in optical density of Aβ-expressing cells with CQ was a direct consequence of increased viable colony-forming units and not an artifact of increased cell size (Fig. S2).
Due to our previous findings that different 8-OHQs have distinct activities in different models of proteotoxicity (20), we compared the efficacy of CQ in all of our yeast proteotoxicity models. The Aβ, α-synuclein (i.e., Parkinson disease), TDP-43 (i.e., frontotemporal dementia and ALS), and htt72Q (i.e., Huntington disease) yeast models have comparable levels of toxicity, but in each case the nature of the cellular toxicity is distinct as are the genetic hits obtained from unbiased genome-wide modifier screens (8, 21–23). CQ had significant, yet modest efficacy against α-syn and htt72Q and no effect on TDP-43 (Fig. 1E). However, it rescued the Aβ model more potently than it did any other model, indicating a considerable degree of selectivity.
Although CQ showed some protection in other models, we previously showed that CQ does not rescue the toxicity caused by diverse yeast genes when overexpressed from the same galactose-regulated promoter (20). In addition, CQ was unable to rescue growth defects caused by five different gene deletion mutations and seven toxic compounds with diverse modes of action (Table S1). Given the selectivity of CQ, we focused on investigating its activity in the yeast Aβ toxicity model.
CQ Restores Endosomal Trafficking.
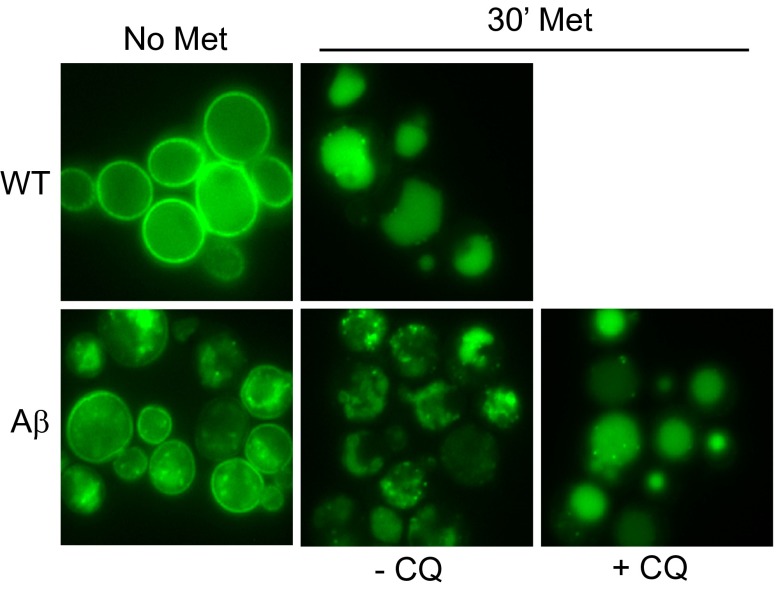
Our recent unbiased screen for suppressors of Aβ toxicity uncovered several proteins involved in clathrin-mediated endocytosis whose homologs are AD risk factors in humans (8). We directly tested the effect of Aβ on clathrin-mediated endocytosis using Ste3, a G-protein–coupled receptor for the mating pheromone. Indeed, steady-state Ste3 endocytosis was severely perturbed by Aβ and rescued by the genetic suppressors involved in this process (8). Here, we extended the analysis with another widely used reporter of endocytic function, a putative 13-pass transmembrane transporter for methionine, Mup1. This reporter provides a facile means of investigating stimulus-dependent trafficking: the endocytosis of Mup1 is rapidly induced by the addition of methionine.
As expected, Mup1-GFP localized primarily to the plasma membrane of WT cells in the absence of methionine (Fig. 2, Upper Left). After 30 min of methionine treatment, Mup1-GFP was endocytosed and exclusively detected in endosomes and the vacuole (Fig. 2, Upper Right). In the absence of methionine, Aβ expression perturbed steady-state trafficking of Mup1, detected by its localization to both the plasma membrane and to intracellular foci (Fig. 2, Lower Left). The addition of methionine promoted Mup1-GFP internalization, but delivery to the vacuole was greatly impeded (Fig. 2, Lower Center). CQ completely restored endosomal transport of Mup1-GFP to the vacuole in the strain expressing Aβ (Fig. 2, Lower Right). Thus, Aβ causes defects in endosomal transport that are manifested downstream of initial transporter internalization and these defects are rescued with CQ treatment.
Fig. 2.
CQ restores endosomal trafficking of Mup1-GFP. Mup1-GFP endocytosis was monitored in WT and Aβ strains (CQ, 0.8 μM). Cultures without methionine (Left) or with 30-min methionine treatment (Right) enabled visualization of stimulated endocytosis of Mup1-GFP from the plasma membrane to the vacuole.
CQ Rescues a C. elegans Model of Aβ Toxicity.
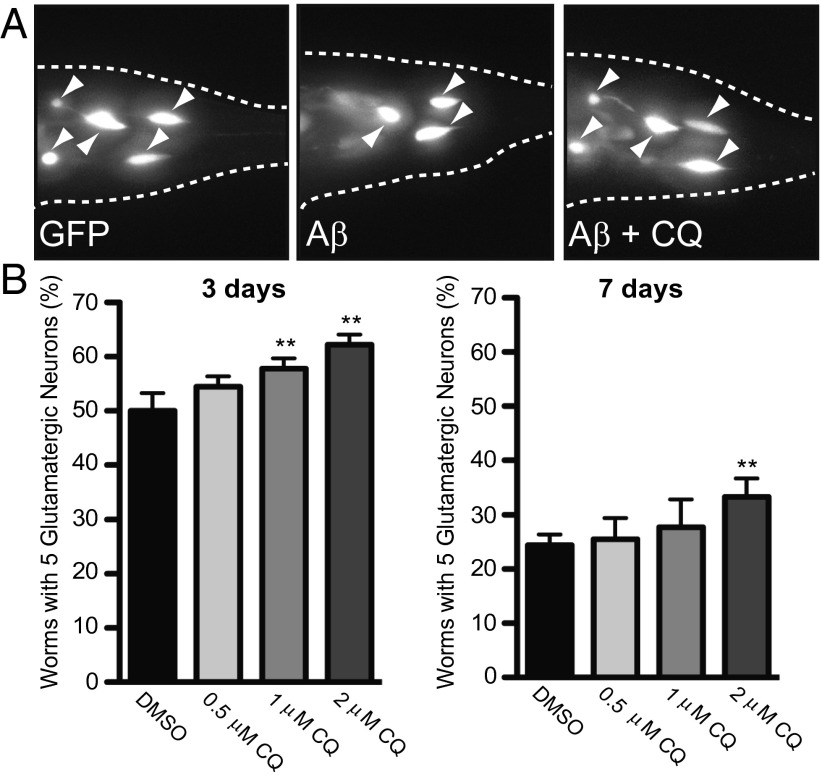
To establish that the ability of CQ to rescue the toxicity of Aβ in the secretory compartment was conserved in neurons, we tested a nematode model of Aβ toxicity. In this model, Aβ is expressed in the secretory pathway of glutamatergic neurons (a neuronal subtype particularly relevant to AD) using the eat-4 promoter and a C. elegans ER signal sequence (8). An advantage of C. elegans is that the transparency of the cuticle allows surviving neurons to be visualized in living animals expressing GFP in the same neurons. Aβ expression produced an age-dependent loss of glutamatergic neurons. Importantly, the endocytosis-related genes we originally identified in our yeast screen, which are also known AD risk factors in humans, suppress Aβ toxicity in this nematode model (8). Expressing Aβ in glutamatergic neurons enables a quantitative measure of neurodegeneration and differs from previous approaches in the nematode where Aβ expressed within the body wall muscle cells caused a motor phenotype (24, 25).
As previously described, the percentage of worms with the WT number of neurons decreased from 3 to 7 d post larval hatching [Fig. 3A) (8)]. Because the cuticle of the worm is relatively impenetrable, we used an established protocol to administer CQ acutely at the earliest larval stage (26). After 24 h, the worms were transferred to normal growth media and allowed to develop and age in the absence of CQ. Even with this single early dosing, CQ increased the percentage of worms with the WT number of neurons. Rescue was apparent at both 3 and 7 d post CQ treatment (Fig. 3 A and B). Thus, as in yeast, the toxicity of secreted forms of Aβ is rescued by CQ in glutamatergic neurons. This encouraged us to take advantage of the yeast model to further investigate CQ’s mechanism of action.
Fig. 3.
CQ rescues worms expressing Aβ in glutamatergic neurons. (A) Fluorescent microscopy images of representative WT or Aβ-expressing worms. The rescue of C. elegans glutamatergic neurons by CQ was monitored with GFP coexpressed in the same cells as Aβ under control of the eat-4 promoter. (B) Quantification of CQ rescue is reported as the percentage of worms with the WT number (5) of anatomically distinct glutamatergic neurons in the hermaphrodite tail at 3 and 7 d after drug treatment. The arrowheads indicate surviving glutamatergic neurons. Data are the average of three independent experiments with 30 worms in each set. Error indicates SD, and the asterisk (*) indicates a value of P < 0.05 according to Student’s t test.
CQ Rescue of Aβ Toxicity Requires Metal Binding.
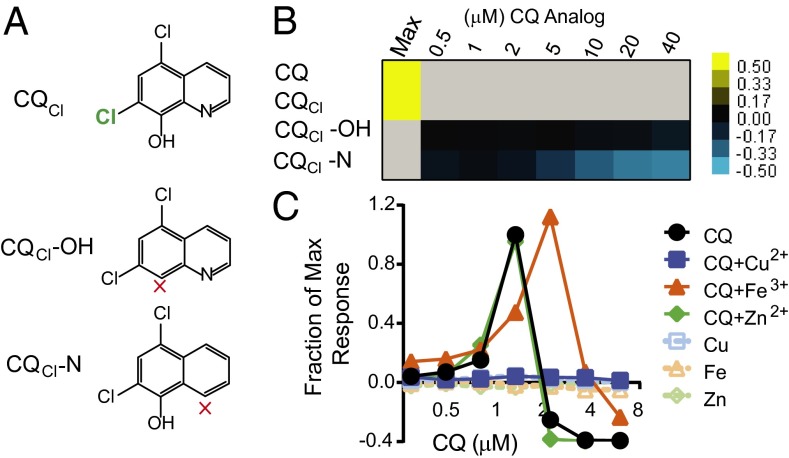
Because 8-OHQs are known metal binding compounds, we first asked whether this functionality was required for the rescue of Aβ toxicity. We tested CQ analogs lacking either the hydroxyl group or the aromatic nitrogen. Together, these groups coordinate the bidentate binding of transition metals ions. Because direct analogs of CQ lacking these groups were not available, we tested analogs of chloroxine (CQCl) (Fig. 4A). This 8-OHQ is identical to CQ except that it has a chlorine atom in place of the iodine atom. It was as effective at alleviating Aβ toxicity in yeast as was CQ (Fig. 4B, “Max”). We then tested CQCl analogs lacking either the hydroxyl group (CQCl-OH) or the nitrogen atom (CQCl-N). Both were inactive against Aβ, indicating CQ efficacy requires bidentate metal binding (Fig. 4B).
Fig. 4.
CQ rescues Aβ through a metal-dependent mechanism. (A) Structures of CQ derivative, chloroxine (CQCl), and analogs lacking either the hydroxyl group (CQCl-OH) or aromatic nitrogen (CQCl-N). (B) Dose–response curves of CQCl analogs. Data are expressed as the difference in OD600 between drug-treated and untreated cultures. Values above graph indicate concentration in micromolar. Yellow indicates rescue; blue indicates toxicity. Only the maximum effective concentration (1 μM, “Max”) is shown for CQ and CQCl. (C) Dose–response curves of Aβ cultures treated with CQ and equimolar copper, iron, or zinc indicate interactions between CQ and copper and iron. Data reflect differences in OD600 between treated and untreated samples normalized to maximal rescue by CQ (micromolar) in the absence of supplemented metals.
Next, we tested the ability of exogenous metals to alter CQ activity. We used copper, iron, and zinc, all of which are known to bind CQ (27). On their own, none of these metals affected Aβ toxicity (Fig. 4C). When coadministered at a concentration that was equimolar with CQ, each metal had completely different effects (Fig. 4C). Copper abolished rescue of Aβ by low concentrations of CQ and also abolished the growth inhibition caused by higher CQ concentrations. Iron shifted the dose–response, increasing the required dose for rescue and reducing the growth inhibitory effects of CQ. Zinc had no effect on CQ efficacy or growth inhibition at higher concentrations. These data suggest that CQ binds copper, and to a lesser extent iron, to alleviate Aβ toxicity.
Effects of Metals and CQ on in Vitro Aβ Assembly.
Metal ions, including copper, iron, and zinc, bind to Aβ both in vitro and within Aβ plaques from the brains of AD patients (28, 29). In in vitro assembly reactions with Aβ, the addition of copper favors the formation of nonfibrillar, amorphous aggregates (29) and oligomers (30, 31) that are more toxic to cells than amyloid fibers of Aβ (18, 30–32). Metal binding compounds, such as CQ and PBT2, can remove metals from Aβ in vitro (17, 18, 33, 34). These compounds also increase the amount of soluble Aβ in the brains of AD mouse models in vivo and stimulate the extraction of Aβ from plaques in brain samples from AD patients (17). Because the predominant form of Aβ 1–42 in yeast is small oligomers (8) and copper appears central to rescue of Aβ toxicity (Fig. 4C), we tested the effects of copper on Aβ oligomerization in vitro, and asked how CQ affects this process.
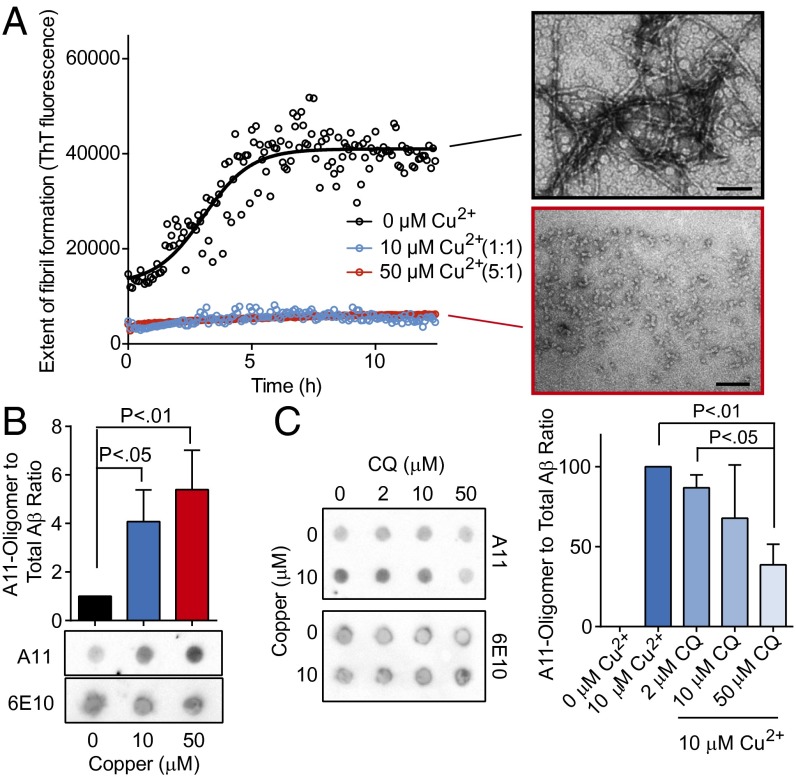
We first tested the effects of copper on Aβ 1–42 fiber formation using ThioflavinT (ThT), which binds β-sheet–rich proteins (i.e., amyloid fibrils). Copper strongly inhibited amyloid fiber formation when present in Aβ assembly reactions (Fig. 5A) (29). To characterize the species of Aβ that accumulate in the presence of copper, we used an antibody known as A11. This antibody, which was raised against a form of Aβ that mimics toxic oligomers, recognizes specific prefibrillar conformers of Aβ that are highly toxic to neurons (35). Aβ assembly reactions were carried out with both equimolar (1:1) and excess (5:1) copper relative to Aβ for 30 min and then subject to dot blot analysis with A11 and another antibody that detects total Aβ(6E10). Copper substantially increased the amount of A11-reactive Aβ (Fig. 5B). CQ had no effect on the formation of A11-reactive Aβ species on its own, but it strongly antagonized the oligomer-potentiating effects of copper (Fig. 5C).
Fig. 5.
Effects of copper and CQ on Aβ oligomers. (A) ThT fluorescence of Aβ assembly reactions in the presence of copper (micromolar) at indicate molar ratios compared with Aβ. EM shows representative fields of Aβ with no or 50 μM Cu2+. (B) Dot blot analysis of Aβ assembly (10 μM) in the presence or absence of both copper and CQ at 1:1 and 5:1 molar ratios. A11 monitors oligomeric Aβ conformations and 6E10 monitors total Aβ. (C) Dot blot (Left) and quantitation (Right) of Aβ assembly reactions in the presence of indicated concentrations of copper and CQ.
CQ Dramatically Reduced Aβ Peptide Levels by Increasing Degradation.
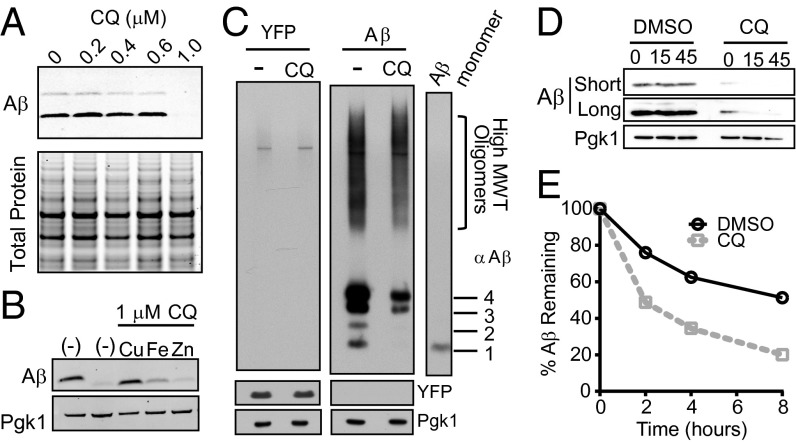
To determine whether CQ has a similar ability to modulate Aβ within the context of the secretory compartment of a living cell, we returned to yeast. First, we examined the effects of CQ on the accumulation of Aβ using denaturing SDS gels. CQ dramatically reduced the accumulation of Aβ, with a sharp dose dependence that mirrored the sharp dose dependence of CQ’s ability to rescue Aβ toxicity (Fig. 6A, Upper). CQ had no effect on total protein levels (Fig. 6A, Lower).
Fig. 6.
CQ promotes Aβ degradation. (A) Denaturing SDS/PAGE and Aβ immunoblot show reduction of Aβ levels in response to CQ treatment compared with total protein (Coomassie). (B) Aβ immunoblots of cells treated with CQ and equimolar Cu2+, Fe3+, and Zn2+. Pgk1 is a loading control. (C) Aβ immunoblot of nondenaturing gel shows a decrease in all forms of Aβ. Aβ oligomeric states are indicated to the Right of the monomer control lane. A YFP control strain is shown on the Left. (D) Immunoblot analysis of Aβ-expressing yeast with DMSO control or CQ after a short CHX time course. Short and long exposures are shown for comparison. (E) The percentage of Aβ remaining after [35S]methionine pulse-labeling Aβ-expressing yeast in presence or absence of CQ. Aβ was immunoprecipitated and quantified at 2-h intervals after pulse labeling.
We next tested whether exogenous metals antagonized the ability of CQ to reduce Aβ levels. Indeed, they did so in a manner similar to their effects on the rescue of Aβ toxicity (Fig. 4C). Copper completely eliminated CQs effects on Aβ accumulation; iron affected it modestly; and zinc had no effect (Fig. 6B). We next asked what species of Aβ were most affected by CQ. Nondenaturing gels, which preserved oligomer structure, demonstrated a decrease in most forms of Aβ when cultures were treated with CQ (Fig. 6C, Right). The levels of a control protein (YFP) expressed from the same promoter did not change (Fig. 6C, Left). Thus, the ability of CQ to rescue Aβ toxicity directly correlated with its ability to prevent the accumulation of oligomeric Aβ in a manner dependent on metal binding.
The decrease in oligomeric Aβ could be due to either its reduced accumulation or accelerated degradation. To distinguish between these two possibilities, we used two methods to assess degradation rates. First, we treated cells with the protein synthesis inhibitor, cycloheximide (CHX), and monitored Aβ levels by immunoblotting. After expressing Aβ for 24 h with either DMSO or CQ, cells were treated with CHX for 15 or 45 min. In CQ-treated cells, Aβ was rapidly degraded within 15 min of inhibiting translation (Fig. 6D). This increased degradation was not observed with a control protein (Pgk1). In contrast, the Aβ peptide that accumulated in cells treated with DMSO did not significantly degrade during this same time frame (Fig. 6D).
Inhibiting translation in this global manner might alter cellular degradation pathways and confound interpretations. We therefore used [35S]methionine pulse labeling and immunoprecipitation. Cells expressing Aβ were transiently labeled with [35S]methionine to allow incorporation of radiolabeled methionine into newly synthesized proteins. After chasing cultures with nonlabeled methionine, we monitored the remaining Aβ by immunoprecipitating the peptide from cell lysates. In agreement with the CHX experiment, CQ specifically increased degradation of Aβ (Fig. 6E and Fig. S3). Together, the two experiments indicate that the stability of Aβ oligomers formed within the secretory pathway is dependent on metals and that, by affecting these oligomers, CQ can directly antagonize the cellular pathologies caused by Aβ.
Discussion
We have used an unbiased phenotypic small-molecule screen of ∼140,000 compounds to identify a clinically relevant class of compounds that prevent the toxic effects of Aβ expressed within the secretory pathway. Although multiple metal-related Aβ pathologies and modes of action for CQ—and PBT2—have been described and postulated (17, 18, 28), our data suggest a mechanism of action in the yeast model that is not ionophore based (18). Rather, CQ promoted the degradation of metal-dependent Aβ oligomers within the secretory and endosomal compartments. Thus, our yeast Aβ model has allowed us to focus on an early aspect of Aβ misfolding and the identification of CQ indicates that these early events are directly modulated by metals. Moreover, the results from this chemical screen indicate that modulating Aβ:metal complexes directly ameliorates the defects caused by Aβ oligomers within the secretory/endosomal pathways, which we previously linked to AD risk alleles through genetic screens (8). This connection further establishes the potential for this class of small molecules to mitigate both early and late aspects of Aβ cellular pathologies.
Endogenous transition metals play an important role in Aβ pathology and their modulation has potential promise in treating AD patients (19, 36, 37). We suggest that the metal-dependent enhanced degradation of Aβ we observe here for CQ rescue of Aβ toxicity plays a role in neuronal systems as well. However, it is important to note that multiple mechanisms likely operate on the longer timescales of Aβ accumulation in mouse models and in human disease. For example, accumulation of transition metals in extracellular Aβ plaques formed late in disease can limit intracellular metal availability (28). CQ can dissolve plaques that have formed in late stages of disease (in both mice and humans) (17) and liberate trapped metals. This then presumably allows the ionophoric capability of CQ and PBT2 to simultaneously restore metal homeostasis and up-regulate proteases that degrade extracellular Aβ (18, 28, 36). Deciphering both early and late aspects of Aβ pathology will be important in understanding cellular pathologies and developing new therapeutic approaches in the treatment of AD.
The nearly complete elimination of Aβ accumulation that is mediated by CQ in yeast far exceeds any effect on protein stability that we have observed for any other compound or genetic suppressor in any of our yeast models of protein-folding pathologies (20, 38). CQ modestly rescues α-syn and htt72Q toxicity [Fig. 1E (20)]. However, it has little effect on the accumulation of these proteins (20). Presumably, this difference in the activities and efficacy of CQ is due to some, as yet unknown, difference in the cellular context and biophysical properties of different metal-binding proteins [of which α-syn is one (39)]. We found that the degradation of Aβ was not affected by the ablation of any single pathway (i.e., autophagy; Fig. S4). This, together with the extreme rate of CQ-mediated Aβ degradation, suggests that multiple mechanisms are at play.
Empowered by significant improvements in DNA sequencing, a considerable effort is now underway to catalog polymorphisms in the human genome and identify those that contribute to pathologies related to protein homeostasis. It is now clear that the total number of human polymorphisms is much greater than previously realized (40), creating an intense need for rapid and tractable model systems to explore genotype–phenotype causality. Our yeast Aβ system meets this need. A previous unbiased genetic screen for modifiers of Aβ toxicity allowed us to link toxicity to recently discovered human AD risk alleles (8) with no previously known relationship to Aβ. Here, the facile yeast system enabled high-throughput compound screening that allowed us to rapidly identify relevant compounds and interrogate their mechanism of action. This ultimately validates our approach as we pursue new compounds with unknown protective mechanisms. Although this initial screen primarily revealed the central importance of metals in cellular Aβ pathology, we anticipate that other compounds with distinct modes of action will be identified. In general, for AD and other protein-misfolding diseases, yeast chemical genetic approaches will help elucidate new targets and “druggable” pathways and aid discovery and validation (38). It is imperative to integrate several orthogonal model systems and many other recent advances (10, 22, 38, 41, 42) to further our understanding of, and eventually our ability to treat, devastating diseases such as AD.
Methods
Constructs and Yeast Strains.
The signal sequence:Aβ (ssAβ) construct has been described (8). The chemical screening strain used here was generated by integrating either GAL1-regulated Aβ or YFP constructs into the trp1 and ura3 loci of a WT strain and deleting the drug efflux pump (pdr5::KanMX) using standard methods (Table S1). The MUP1-GFP Aβ strain was generated by recombining a PCR product of GFP and a downstream HIS3 auxotrophic marker amplified from a chromosomal MUP1-GFP WT yeast strain (38).
Small-Molecule Screen and Compounds.
Small-molecule screening was carried out as described (20). Briefly, late log phase Aβ cultures grown in 2% (wt/vol) raffinose-containing media were diluted (OD600 = 0.03) into galactose-containing media. Compounds (100 nL) were pinned to 384-well plates containing diluted culture and incubated for 40 h at 30 °C. OD600 values were then used to calculate Z scores [(OD600 well – OD600 plate average)/OD600 plate average]. Compounds were purchased as follows: CQ (Sigma-Aldrich); CQCl (Sigma-Aldrich); CQCl-OH (Santa Cruz Biotechnology); CQCl-N (Frontier Scientific Services).
Growth Assays.
Growth assays with compounds were performed by either Bioscreen C or 384-well plates analysis. In both cases, log phase raffinose cultures were diluted (OD600 ∼ 0.02) into galactose-containing media, compounds or DMSO added, and cultures grown at 30 °C with either intermittent shaking (Bioscreen C) or endpoint (384-well) OD600 readings. Data were typically expressed as the difference in OD600 between compound- and DMSO-treated cultures. For all metal experiments, metals were added at the same time as CQ. Specificity experiments were performed in conditions with at least 50% reduction in growth in response to gene deletion or toxic compounds (Table S2). Toxic compound concentrations were selected to inhibit growth of WT yeast by at least 50%.
MUP1-GFP Assay.
The effects of Aβ expression on Mup1-GFP endocytosis were performed as described (38). Cultures induced for ∼16 h with SGalMet in a Bioscreen C with or without CQ (0.8 μM), methionine (50 μg/mL) added for 30 min, and GFP localization then imaged by fluorescence microscopy.
C. elegans CQ Treatment.
The C. elegans strain (UA166) expressing an ssAβ construct in glutamatergic neurons has been described (8). Compound treatment (20) and analysis of glutamatergic neurons (8) have been described.
In Vitro Aβ Oligomer Formation.
Lyophilized, chemically synthesized Aβ(1–42) (American Peptide Company) was dissolved at ∼1 mg/mL in pH 11.5 NaOH and flash frozen. Exact concentration was measured using amino acid analysis (Dana Farber Molecular Biology Core). For ThT assays, Aβ was dissolved in PBS (pH 7.2) to a final concentration of 10 μM and ThT was added to a final concentration of 20 μM. The solution was allowed to aggregate at 30 °C under quiescent conditions and fluorescence at 480 nm was monitored for 12 h using a Tecan Safire plate reader. For dot blots, 300 μL of solutions containing 10 μM Aβ with varying concentrations of Cu and CQ were loaded onto a 0.1-μm nitrocellulose membrane using the Bio-Rad vacuum filter trap device. The membrane was then blocked in 5% (wt/vol) nonfat dry milk in PBS for 1–2 h followed by an overnight incubation in A11 antibody [Millipore; 1:250 in 5% (wt/vol) milk/PBS] and a horseradish peroxidase-conjugated anti-rabbit secondary antibody (Sigma). Quantification of dot blots was performed using ImageJ (NIH). Transmission electron microscopy was performed by incubating Aβ solutions aggregated for ∼12–15 h on a Ni-Formvar grid followed by negative staining by 2% (wt/vol) uranyl acetate.
Effect of CQ on Aβ Levels.
The ssAβ and YFP strains were grown to log phase in raffinose media and then diluted to an OD600 of 0.01 (YFP) or 0.03 (Aβ) in media containing galactose with either 0.6 μM CQ or DMSO and grown in a Bioscreen C plate at 30 °C to an OD600 of ∼0.6 (∼16 h). Lysates were generated and blots probed as previously described (8).
Analysis of Aβ Degradation.
For CHX experiments, Aβ yeast strains were cultured and induced as above with or without CQ. After Aβ induction, CHX was added (35 μg/mL) and aliquots removed at indicated time points. Samples were brought to 40 mM sodium azide, washed with ice-cold water, and Aβ levels analyzed (8).
For [35S]methionine pulse-labeling experiments, cells expressing ssAβ were grown as described above, harvested, washed twice in media lacking methionine, resuspended in the same media (OD600 = 1.0), and incubated at 30 °C for 15 min. l-[35S]Methionine (Perkin-Elmer; specific activity > 1,000 Ci/mmol) was then added (50 μCi/mL) for 20 min. Cultures were diluted into media containing excess cold methionine (450 μg/mL) to an OD600 of ∼0.5. Aliquots were removed, brought to 20 mM Na azide and 0.1 mg/mL BSA and placed on ice. Aβ was immunoprecipitated with the 6E10 antibody (Covance) and Protein A/G Plus Agarose (Pierce). After washing, beads were boiled in sample buffer and supernatant subjected to SDS/PAGE. For total label, Aβ immunoprecipitation total extract was resolved and the entire lane quantified. Imperial blue (Pierce)-stained gels were washed with water, equilibrated with 1 M sodium salicylate in 10% (wt/vol) glycerol, and dried. The gel was exposed to a FujiFilm imaging plate at −20 °C, the plate scanned with a FujiFilm BAS-2500 Bio-Imagining Analyzer, and the data quantitated with Multigage, version 2.2, software.
Supplementary Material
Acknowledgments
We thank the Harvard Institute for Chemistry and Cellular Biology and C. Shamu/A. Daab for help with the small-molecule screen, Nicki Watson and the Keck Imaging Facility for help with EM, Tom O’Halloran and Meera Raja for advice on metal biology, and members of the S.L. laboratory for helpful discussions and comments on the manuscript. This research was funded by a Collaborative Innovation Award from the Howard Hughes Medical Institute (HHMI) (to S.L. and G.A.C.). K.E.S.M. was funded by National Institutes of Health Grant R03 DA032472 and a grant from The Ellison Medical Foundation. D.F.T. was funded by National Research Service Award Fellowship F32NS061419 and research funded by The JPB Foundation and an Edward N. and Della L. Thome Memorial Foundation grant. S.L. is an investigator of the HHMI and gratefully acknowledges its support.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402228111/-/DCSupplemental.
References
- 1.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Iwatsubo T, et al. Visualization of A beta 42(43) and A beta 40 in senile plaques with end-specific A beta monoclonals: Evidence that an initially deposited species is A beta 42(43) Neuron. 1994;13(1):45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- 3.Selkoe DJ, Wolfe MS. Presenilin: Running with scissors in the membrane. Cell. 2007;131(2):215–221. doi: 10.1016/j.cell.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 4.McLean CA, et al. Soluble pool of Abeta amyloid as a determinant of severity of neurodegeneration in Alzheimer’s disease. Ann Neurol. 1999;46(6):860–866. doi: 10.1002/1531-8249(199912)46:6<860::aid-ana8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 5.Hartley DM, et al. Protofibrillar intermediates of amyloid beta-protein induce acute electrophysiological changes and progressive neurotoxicity in cortical neurons. J Neurosci. 1999;19(20):8876–8884. doi: 10.1523/JNEUROSCI.19-20-08876.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khurana V, Lindquist S. Modelling neurodegeneration in Saccharomyces cerevisiae: Why cook with baker’s yeast? Nat Rev Neurosci. 2010;11(6):436–449. doi: 10.1038/nrn2809. [DOI] [PubMed] [Google Scholar]
- 7.Thinakaran G, Koo EH. Amyloid precursor protein trafficking, processing, and function. J Biol Chem. 2008;283(44):29615–29619. doi: 10.1074/jbc.R800019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Treusch S, et al. Functional links between Aβ toxicity, endocytic trafficking, and Alzheimer’s disease risk factors in yeast. Science. 2011;334(6060):1241–1245. doi: 10.1126/science.1213210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarrett JT, Berger EP, Lansbury PT., Jr The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: Implications for the pathogenesis of Alzheimer’s disease. Biochemistry. 1993;32(18):4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 10.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45(12):1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harold D, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seshadri S, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303(18):1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramjaun AR, Micheva KD, Bouchelet I, McPherson PS. Identification and characterization of a nerve terminal-enriched amphiphysin isoform. J Biol Chem. 1997;272(26):16700–16706. doi: 10.1074/jbc.272.26.16700. [DOI] [PubMed] [Google Scholar]
- 14.Hollingworth P, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43(5):429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naj AC, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43(5):436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaidos G, Soni S, Oswald DJ, Toselli PA, Kirsch KH. Structure and function analysis of the CMS/CIN85 protein family identifies actin-bundling properties and heterotypic-complex formation. J Cell Sci. 2007;120(Pt 14):2366–2377. doi: 10.1242/jcs.004333. [DOI] [PubMed] [Google Scholar]
- 17.Cherny RA, et al. Treatment with a copper-zinc chelator markedly and rapidly inhibits beta-amyloid accumulation in Alzheimer’s disease transgenic mice. Neuron. 2001;30(3):665–676. doi: 10.1016/s0896-6273(01)00317-8. [DOI] [PubMed] [Google Scholar]
- 18.Adlard PA, et al. Rapid restoration of cognition in Alzheimer’s transgenic mice with 8-hydroxy quinoline analogs is associated with decreased interstitial Abeta. Neuron. 2008;59(1):43–55. doi: 10.1016/j.neuron.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 19.Lannfelt L, et al. Safety, efficacy, and biomarker findings of PBT2 in targeting Abeta as a modifying therapy for Alzheimer’s disease: A phase IIa, double-blind, randomised, placebo-controlled trial. Lancet Neurol. 2008;7(9):779–786. doi: 10.1016/S1474-4422(08)70167-4. [DOI] [PubMed] [Google Scholar]
- 20.Tardiff DF, Tucci ML, Caldwell KA, Caldwell GA, Lindquist S. Different 8-hydroxyquinolines protect models of TDP-43 protein, α-synuclein, and polyglutamine proteotoxicity through distinct mechanisms. J Biol Chem. 2012;287(6):4107–4120. doi: 10.1074/jbc.M111.308668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper AA, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science. 2006;313(5785):324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung CY, et al. Identification and rescue of α-synuclein toxicity in Parkinson patient-derived neurons. Science. 2013;342(6161):983–987. doi: 10.1126/science.1245296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HJ, et al. Therapeutic modulation of eIF2α phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat Genet. 2014;46(2):152–160. doi: 10.1038/ng.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Link CD. Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans. Proc Natl Acad Sci USA. 1995;92(20):9368–9372. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McColl G, et al. Utility of an improved model of amyloid-beta (Aβ1–42) toxicity in Caenorhabditis elegans for drug screening for Alzheimer’s disease. Mol Neurodegener. 2012;7:57. doi: 10.1186/1750-1326-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su LJ, et al. Compounds from an unbiased chemical screen reverse both ER-to-Golgi trafficking defects and mitochondrial dysfunction in Parkinson’s disease models. Dis Model Mech. 2010;3(3-4):194–208. doi: 10.1242/dmm.004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrada E, et al. Stoichiometry and conditional stability constants of Cu(II) or Zn(II) clioquinol complexes; implications for Alzheimer’s and Huntington’s disease therapy. Neurotoxicology. 2007;28(3):445–449. doi: 10.1016/j.neuro.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Ayton S, Lei P, Bush AI. Metallostasis in Alzheimer’s disease. Free Radic Biol Med. 2013;62:76–89. doi: 10.1016/j.freeradbiomed.2012.10.558. [DOI] [PubMed] [Google Scholar]
- 29.Faller P, Hureau C, Berthoumieu O. Role of metal ions in the self-assembly of the Alzheimer’s amyloid-β peptide. Inorg Chem. 2013;52(21):12193–12206. doi: 10.1021/ic4003059. [DOI] [PubMed] [Google Scholar]
- 30.Smith DP, et al. Concentration dependent Cu2+ induced aggregation and dityrosine formation of the Alzheimer’s disease amyloid-beta peptide. Biochemistry. 2007;46(10):2881–2891. doi: 10.1021/bi0620961. [DOI] [PubMed] [Google Scholar]
- 31.Jin L, Wu WH, Li QY, Zhao YF, Li YM. Copper inducing Aβ42 rather than Aβ40 nanoscale oligomer formation is the key process for Aβ neurotoxicity. Nanoscale. 2011;3(11):4746–4751. doi: 10.1039/c1nr11029b. [DOI] [PubMed] [Google Scholar]
- 32.Smith DP, et al. Copper-mediated amyloid-beta toxicity is associated with an intermolecular histidine bridge. J Biol Chem. 2006;281(22):15145–15154. doi: 10.1074/jbc.M600417200. [DOI] [PubMed] [Google Scholar]
- 33.Raman B, et al. Metal ion-dependent effects of clioquinol on the fibril growth of an amyloid beta peptide. J Biol Chem. 2005;280(16):16157–16162. doi: 10.1074/jbc.M500309200. [DOI] [PubMed] [Google Scholar]
- 34.Atwood CS, et al. Dramatic aggregation of Alzheimer abeta by Cu(II) is induced by conditions representing physiological acidosis. J Biol Chem. 1998;273(21):12817–12826. doi: 10.1074/jbc.273.21.12817. [DOI] [PubMed] [Google Scholar]
- 35.Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300(5618):486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 36.Crouch PJ, Barnham KJ. Therapeutic redistribution of metal ions to treat Alzheimer’s disease. Acc Chem Res. 2012;45(9):1604–1611. doi: 10.1021/ar300074t. [DOI] [PubMed] [Google Scholar]
- 37.Bush AI, et al. Rapid induction of Alzheimer A beta amyloid formation by zinc. Science. 1994;265(5177):1464–1467. doi: 10.1126/science.8073293. [DOI] [PubMed] [Google Scholar]
- 38.Tardiff DF, et al. Yeast reveal a “druggable” Rsp5/Nedd4 network that ameliorates α-synuclein toxicity in neurons. Science. 2013;342(6161):979–983. doi: 10.1126/science.1245321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dudzik CG, Walter ED, Millhauser GL. Coordination features and affinity of the Cu2+ site in the α-synuclein protein of Parkinson’s disease. Biochemistry. 2011;50(11):1771–1777. doi: 10.1021/bi101912q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casals F, Bertranpetit J. Genetics. Human genetic variation, shared and private. Science. 2012;337(6090):39–40. doi: 10.1126/science.1224528. [DOI] [PubMed] [Google Scholar]
- 41.Sandoe J, Eggan K. Opportunities and challenges of pluripotent stem cell neurodegenerative disease models. Nat Neurosci. 2013;16(7):780–789. doi: 10.1038/nn.3425. [DOI] [PubMed] [Google Scholar]
- 42.Weiner MW, et al. The Alzheimer’s Disease Neuroimaging Initiative: A review of papers published since its inception. Alzheimers Dement. 2013;9(5):e111–e194. doi: 10.1016/j.jalz.2013.05.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.