Significance
Giant DNA viruses are visible under a light microscope and their genomes encode more proteins than some bacteria or intracellular parasitic eukaryotes. There are two very distinct types and infect unicellular protists such as Acanthamoeba. On one hand, Megaviridae possess large pseudoicosahedral capsids enclosing a megabase-sized adenine–thymine-rich genome, and on the other, the recently discovered Pandoraviruses exhibit micron-sized amphora-shaped particles and guanine–cytosine-rich genomes of up to 2.8 Mb. While initiating a survey of the Siberian permafrost, we isolated a third type of giant virus combining the Pandoravirus morphology with a gene content more similar to that of icosahedral DNA viruses. This suggests that pandoravirus-like particles may correspond to an unexplored diversity of unconventional DNA virus families.
Keywords: giant DNA virus, late Pleistocene, icosahedral capsid
Abstract
The largest known DNA viruses infect Acanthamoeba and belong to two markedly different families. The Megaviridae exhibit pseudo-icosahedral virions up to 0.7 μm in diameter and adenine–thymine (AT)-rich genomes of up to 1.25 Mb encoding a thousand proteins. Like their Mimivirus prototype discovered 10 y ago, they entirely replicate within cytoplasmic virion factories. In contrast, the recently discovered Pandoraviruses exhibit larger amphora-shaped virions 1 μm in length and guanine–cytosine-rich genomes up to 2.8 Mb long encoding up to 2,500 proteins. Their replication involves the host nucleus. Whereas the Megaviridae share some general features with the previously described icosahedral large DNA viruses, the Pandoraviruses appear unrelated to them. Here we report the discovery of a third type of giant virus combining an even larger pandoravirus-like particle 1.5 μm in length with a surprisingly smaller 600 kb AT-rich genome, a gene content more similar to Iridoviruses and Marseillevirus, and a fully cytoplasmic replication reminiscent of the Megaviridae. This suggests that pandoravirus-like particles may be associated with a variety of virus families more diverse than previously envisioned. This giant virus, named Pithovirus sibericum, was isolated from a >30,000-y-old radiocarbon-dated sample when we initiated a survey of the virome of Siberian permafrost. The revival of such an ancestral amoeba-infecting virus used as a safe indicator of the possible presence of pathogenic DNA viruses, suggests that the thawing of permafrost either from global warming or industrial exploitation of circumpolar regions might not be exempt from future threats to human or animal health.
Ten years ago, the discovery of Acanthamoeba polyphaga Mimivirus revealed the existence of giant DNA viruses with particles large enough to be visible under a light microscope (1, 2). Further sampling of various environments and geographical locations led to the isolation of Mimivirus variants (3, 4) and more distant relatives, two of which have been fully sequenced: Moumouvirus (5) and Megavirus chilensis (6). All of these viruses share the same distinctive particle structure: a unique external fiber layer enclosing a pseudoicosahedral protein capsid of about 0.5 µm in diameter, itself containing lipid membranes surrounding an electron-dense nucleocapsid. They share an adenine–thymine (AT)-rich (>70%) linear DNA genome with sizes up to 1.26 Mb encoding up to 1,120 proteins (6). They all encode a full transcription apparatus allowing them to replicate in the host’s cytoplasm. These common features suggested that all giant viruses were to belong to a single family (Megaviridae) sharing the same particle morphology, genomic features, and replication strategy (7, 8).
This assumption was radically challenged by the discovery of Pandoraviruses (9), which show different and somewhat opposite characteristics. These Acanthamoeba-infecting viruses exhibit much larger amphora-shaped virions 1–1.2 μm in length. Their guanine–cytosine (GC)-rich (>61%) genomes are up to 2.8 Mb long and encode up to 2,500 proteins sharing no resemblance with those of Megaviridae (9). Finally, Pandoravirus particles do not incorporate the transcription machinery that would allow them to entirely replicate in the host’s cytoplasm. Known giant viruses infecting Acanthamoeba were thus thought to belong to two very dissimilar types in terms of particle structure, genome characteristics, and replication strategies. Here we describe a third type of giant virus named “Pithovirus” (from the Greek word pithos designating the kind of large amphora handed over by the gods to the legendary Pandora) propagating in an even larger pandoravirus-like particle, but exhibiting a replication cycle and genomic features reminiscent of those of large icosahedral nucleocytoplasmic DNA viruses. Giant viruses are thus much more diverse than initially assumed.
Results
Particle Morphology.
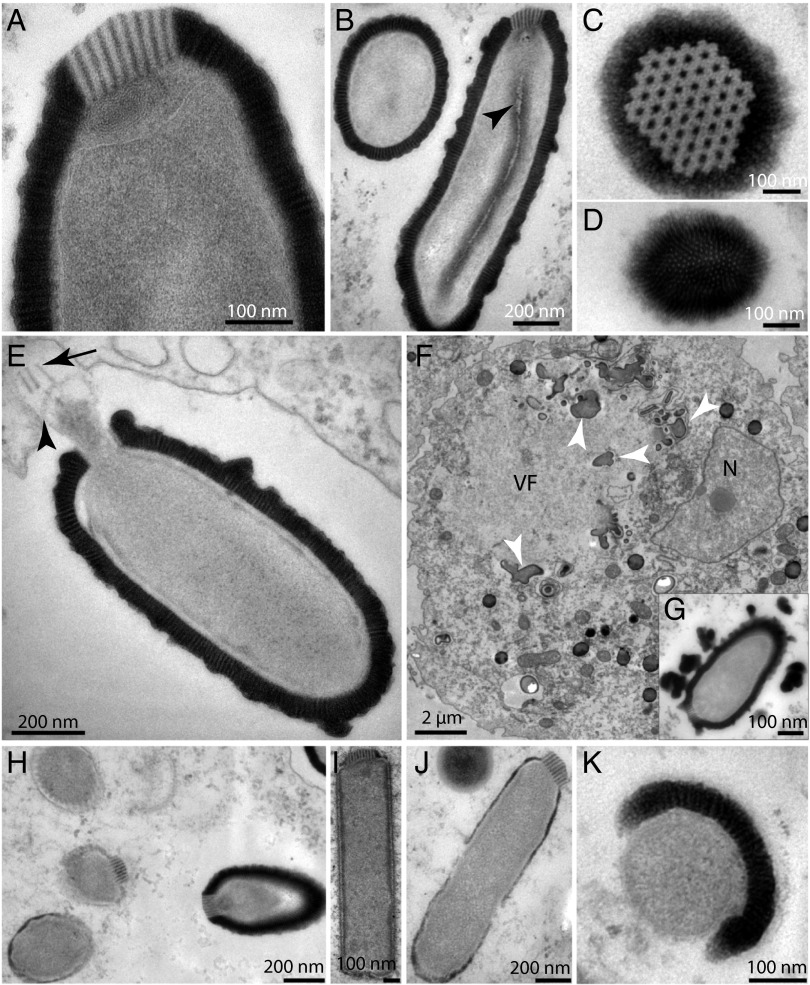
If the notion that DNA may persist over geological timespans (>1 My) is gaining a progressive acceptance, the fact that cellular organisms might survive that long remains a contentious issue (10, 11). Because of its neutral pH and reducing and anaerobic properties, northeast Siberian permafrost is among the most suitable environments to look for long-term surviving microorganisms (12) or even plants (13). Except for few studies targeting the Influenza virus and the Variola virus over historical timespans (14, 15), the possibility that DNA viruses might remain infectious over a much longer time scale has not yet been investigated. We recently initiated such a survey using Acanthamoeba as bait and its giant DNA viruses as safe surrogates for pathogenic viruses. In this context, Pithovirus was initially spotted using light microscopy as ovoid particles (Fig. S1) multiplying in a culture of Acanthamoeba castellanii inoculated with a sample of Siberian permafrost from the Kolyma lowland region (SI Materials and Methods). This sample was aseptically collected from a permafrost layer corresponding to late Pleistocene sediments older than 30,000 y (13). Similar paleosoils are known to contain cysts of Acanthamoeba cells (16). After amplification, the particles were analyzed by transmission electron microscopy confirming that Pithovirus shared the overall morphology of the Pandoraviruses (9) with slightly larger dimensions (∼1.5 µm in length, 500 nm in diameter). The virions exhibit a 60 nm-thick structured envelope made of one layer of parallel stripes (Fig. 1). An internal membrane (Fig. 1A) encases a compartment without discernible substructures except for an electron-dense sphere (50 nm in diameter) seen episodically but in a reproducible fashion (Fig. S1) and a tubular structure parallel to the long axis of the particle (Fig. 1B). At variance with the Pandoraviruses, the apex aperture of the Pithovirus particle appears sealed by a protruding cork (80 nm thick and 160 nm wide) with a hexagonal grid structure (Fig. 1 A and C) reminiscent of the organization of capsomers in icosahedral virions. A coil of rolled-up membrane seems to be connected to this apex structure (Fig. 1A).
Fig. 1.
Electron microscopy imaging of the Pithovirus replication cycle in A. castellanii. (A) Apex of the Pithovirus particle showing its unique cork made of 15 nm-spaced stripes, rolled membranes underneath, and the internal membrane. (B) Two perpendicular views of the Pithovirus particles (cross- and longitudinal sections). The particles are wrapped into a 60 nm-thick envelope made of 10 nm-spaced parallel stripes. A lipid membrane is enclosing a homogeneous interior where a tubular structure is seen episodically, but in a reproducible fashion (arrowhead). (C) Top view of the cork revealing a hexagonal honeycomb-like array. (D) Bottom view of the particle showing the striated organization of the envelope. (E) An opened Pithovirus particle in the host vacuole. Parts of the expelled cork are visible (black arrows) and the internal membrane of the particle (black arrowhead) appears ready to fuse with the vacuole membrane. (F) Maturing virions at a late stage of infection. Structures made of stripes, pieces of cork, and dense material accumulate (white arrowhead) in the periphery of the virion factory (VF). These structures may contain preassembled particle building blocks (Fig. S1). The cell nucleus (N) is visible. (G) Inset highlighting a late stage of virion maturation with globular striated structures accumulating at the virion periphery. (H) Various stages of particle assembly in the same cell. (I) Incompletely assembled rectangular particle lacking its thick envelope. The striated cork is already visible. (J) At a later stage, the particle adopts its final rounded shape while its envelope thickens. (K) Orthogonal view of an immature virion showing the envelope in the process of wrapping the particle.
Replication Cycle.
The Pithovirus replication strategy was documented by following its propagation in axenic Acanthamoeba cultures over an entire multiplication cycle, starting from purified particles. A complete lysis of infected cultures occurred in 10–20 h depending on the initial number of virus particles. As for Pandoraviruses, the replication cycle begins with the phagocytosis of individual particles. First, the Pithovirus particles lose their apical cork allowing the underlying lipid membrane to fuse with the cellular vacuole membrane (Fig. 1E). This creates a channel between the most internal compartment of the virion and the cell cytoplasm (9). In contrast with the Pandoraviruses, the cell nucleus maintains its shape throughout the entire Pithovirus replication cycle (Movie S1). The first visible sign of infection is the formation of an area cleared of cytoplasm subcellular structures, 4–6 h postinfection (Fig. 1F). Numerous vesicles start accumulating in this presumable virion factory (Fig. S1). The process of virion formation is reminiscent of Pandoravirus, the envelope and the interior of the Pithovirus particles being assembled or “knitted” simultaneously (9) (Fig. 1H and Movie S1). First, rectangular-shaped closed particles with their characteristic cork appear at the periphery of the virion factory (Fig. 1I). Later on, their outer wall thickens and the particles take their final ovoid shape, still lacking their thick striated tegument (Fig. 1J). This layer is built subsequently and piecewise, as evidenced in cross section images of immature particles (Fig. 1K) as well as by the fuzzy appearance of the tegument-like envelope in the latest stages of particle maturation (Fig. 1G and Fig. S1). After 6–8 h, particles at various stages of maturation may coexist in the same virion factory.
Besides dense vesicles of unknown composition accumulating inside the infected cells, shapeless blobs made of pieces of striated envelope, pieces of the corks, and diffuse material reminiscent of the mature particles interior (Fig. 1F and Fig. S1) are seen at the periphery of the virion factories. They might be reservoirs of partially organized virion building blocks. Mature particles are found in equal amounts in the cytoplasm or in vacuoles suggesting that they could exit the cell by exocytosis (Movie S1). The replicative cycle ends with the cells releasing hundreds of particles upon lysis. Despite their bacterial-like dimensions no image of particles undergoing binary fission was encountered throughout our comprehensive electron microscopy study, hinting at the viral nature of Pithoviruses before the analysis of their gene content.
Genome Sequencing.
DNA from purified particles was sequenced using the standard Illumina protocol (2× 100 nt paired end reads). The resulting 600 kb of unique sequence data readily allowed the identification of most genes, but could not be assembled in less than 80 nonoverlapping scaffolds. The finished genome was obtained by sequencing a Nextera mate pair library (5–8 kb inserts) using the Illumina MiSeq platform (1,066,320 reads, 2× 250 nt) combined with 77,241 PacBio RS long single-end reads. Unexpectedly, given the similar morphology of their particles, the Pithovirus genome was found to be completely different from the Pandoravirus’s in terms of size, nucleotide composition, topology, and gene content. The Pithovirus genome consists of an AT-rich (64%) dsDNA molecule of a mere 610,033 bp whereas the Pandoraviruses exhibit a GC-rich genome (> 61%) of up to 2.8 Mb. As for the Iridoviridae (17) and the Acanthamoeba-infecting Marseilleviridae (18), the Pithovirus genome sequence either corresponded to a terminally redundant circularly permutated linear DNA molecule or to a closed circle (Fig. S2), in contrast with the linear Pandoravirus genomes flanked by terminal repeats (9). Finally the Pithovirus genome is predicted to encode a mere 467 proteins, much less than the 2,500 predicted proteins of Pandoravirus salinus. The Pithovirus particle appears to be out of proportion with its gene content compared with other DNA viruses such as the Phaeocystis globosa virus packing a similar number of genes into an icosahedral capsid 150 nm in diameter (150-fold less in volume) (19).
Genome Annotation.
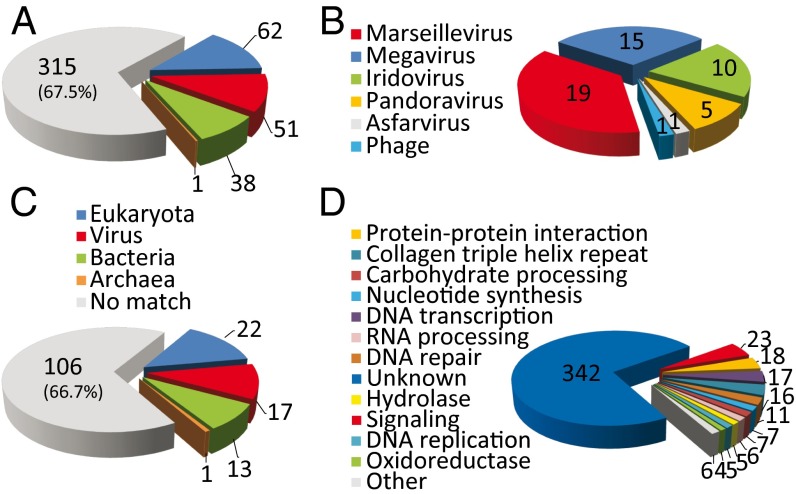
The genome sequence was analyzed using BLAST (National Center for Biotechnology Information, NCBI) and a combination of motif search and protein-fold recognition methods (SI Materials and Methods). As is customary on the discovery of the first member of a previously unknown virus family, the proportion of Pithovirus-predicted proteins with recognizable homologs in the NCBI database was low (152/467 = 32.5%). For comparison, it was 60.6% for Mimivirus (2), 41% for Marseillevirus (18), and only 15.7% for Pandoravirus (9). The best matches are distributed almost equally among DNA viruses, bacteria, and eukaryotes, suggesting the absence of close relatives among previously sequenced organisms (Fig. 2A). A very similar distribution (χ2 = 0.683, P > 0.95) was computed for the 159 predicted protein validated by their detection in the particle proteome (Table S1), confirming that the small fraction of database matches was not due to bioinformatic overpredictions (Fig. 2C).
Fig. 2.
Distributions of the Pithovirus protein closest homologs. (A) All predicted protein sequences against the NCBI NR (non-redundant) database. (B) Distribution of the 51 best-matching viral proteins. (C) Subset of the 159 proteins detected in the particle proteome. (D) Distribution of the predicted protein functions.
Although only one-third of the best database matches corresponded to viral proteins (representing 11% of the total gene content), the absence of genes coding for translation components, ATP-generating enzymes, or related to cell-division confirmed the viral nature of Pithovirus (20). The low level of sequence similarity of these best matches (44% identical residues in average across the highest BLAST scoring segment pairs) (Tables S1 and S2) as well as their dispersion among different DNA virus families argues against Pithovirus being a member of any one of them. On the basis of this distribution (Fig. 2B), the Pithovirus appears globally more similar to Marseilleviridae (19 best hits), then Megaviridae (15 best hits), and then Iridoviridae (10 best hits), all of which are well-established families of icosahedral large DNA viruses. Remarkably, there were only five Pithovirus proteins with their closest homologs in Pandoraviruses. None of these proteins are clearly associated with a functional attribute, except for a remote phosphoglycerate mutase homolog [Pithovirus protein #15 (pv15), 27% identity].
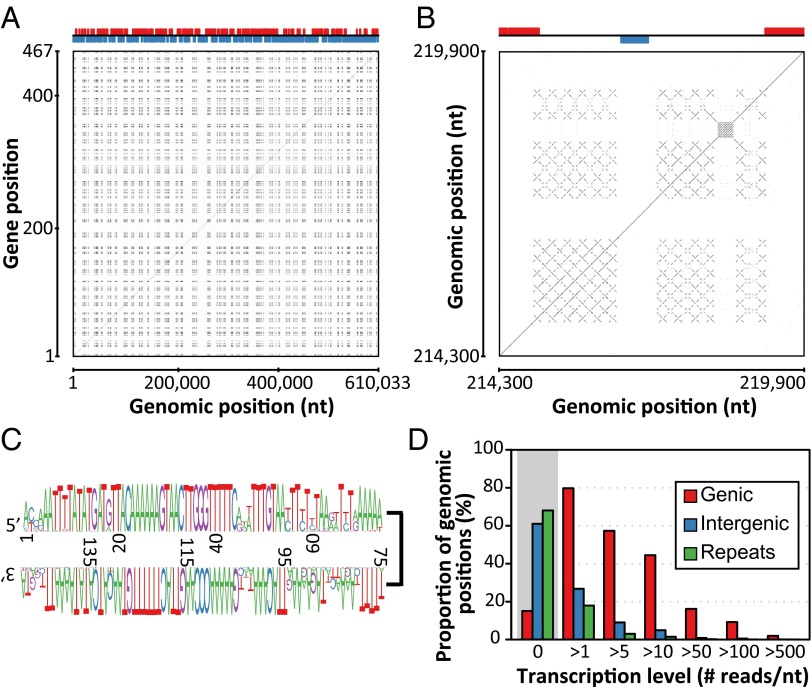
An unusually large fraction of the Pithovirus genome (129 kb, 21.2%) corresponds to multiple regularly interspersed copies of a noncoding repeat, the intricate structure of which produces a unique fractal-like dot-plot pattern (Fig. 3 A and B). These repeats exhibit GC content (23%) much lower than the coding regions (41%), similar to the one of intergenic regions (24%). These repeats are composed of gene-free 2 kb-long tandem arrays of a well-conserved 150-bp palindromic motif (Fig. 3C). This motif is not similar to a previously described mobile element and unrelated to the repeats found in some Iridovirus genomes (21) or Emiliania huxleyi virus (22). The high-repeat content of the Pithovirus genome decreases its coding density to an unusually low 68%. This value is restored to 85.7% (i.e., 1,048 bp per gene), typical of viruses and prokaryotes, when the repeat moiety is not taken into account. The unique Pithovirus genome structure may result from the multiplication of an invasive selfish DNA sequence or from physical constraints reflecting a specific genome organization, mode of replication, packaging, or transcription. The high copy number of these palindromic repeats contrasts with their known instability in cellular genomes where they tend to be rapidly eliminated after several rounds of replication (23, 24).
Fig. 3.
Distribution, structure, and expression of Pithovirus genome repeats. (A) Alignment (dot-plot) of the Pithovirus genome nucleotide sequence against itself. Repeated sequences appear as a black patchwork. The x axis shows genomic position and the y axis shows the gene position. The upper part of the figure shows the distribution of genes on the forward strand (red) and reverse strand (blue). (B) Enlarged view on one of the repeat-containing regions. Each cross is characteristic of a palindromic sequence whereas parallel lines indicate tandem repeated sequences. Notice that each palindromic sequence is itself repeated multiple times. (C) Sequence logo showing the sequence conservation of the palindromic repeats. (D) The transcription level assigned to each genome position is defined as its coverage by RNA-seq reads. The repeat regions are the least expressed.
Among the 152 predicted proteins with a database match, only 125 (26.7% of the 467 predicted proteins) are associated with functional attributes. Most of them are poorly informative such as protein–protein interaction motifs (e.g., zinc-finger, ankyrin domain, leucine-rich, or collagen triple helix repeats), or motifs involved in various signaling/regulatory pathways (e.g., kinase, phosphatase, GTP-binding) (Table S1). Typical of large DNA viruses, the dominant functional categories were DNA transcription (17 genes), DNA repair (11 genes, including an ATP-dependent DNA ligase), nucleotide synthesis (7 enzymes, including a ThyX alternative thymidylate synthase), and DNA replication (5 genes). Other categories include carbohydrate processing, RNA processing, and various hydrolases and oxidoreductases (Fig. 2D). At variance with other giant DNA viruses, no component of the protein translation machinery, including tRNA, is encoded in the Pithovirus genome. The presence of a complete virus-encoded transcription machinery (most of which found in the particle; Table S1) is consistent with the cytoplasmic location of the Pithovirus replication (Fig. 1). In contrast with the extremely high-repeat content of the genome, a single intervening sequence, a group I self-splicing intron, was detected and validated in the DNA-dependent RNA polymerase large subunit gene (RPB1, pv366–368). As borderline database matches were further scrutinized, we detected a remote similarity (E value = 0.002, 21% identical residues) between the pv460 gene product and the divergent major capsid protein (MCP) characteristic of Iridoviruses (e.g., in Megalocytivirus; GenBank accession no. AFE85881.1). The FUGUE server (25) predicted that the pv460 protein would adopt the structural “jelly-roll” fold common to large DNA virus MCPs. However, although the pv460 gene is transcribed, its product was not found in the mature virion. Like the Poxvirus D13 protein (26, 27), the pv460 protein might only play a transient role in the particle morphogenesis.
Phylogenetic Analysis.
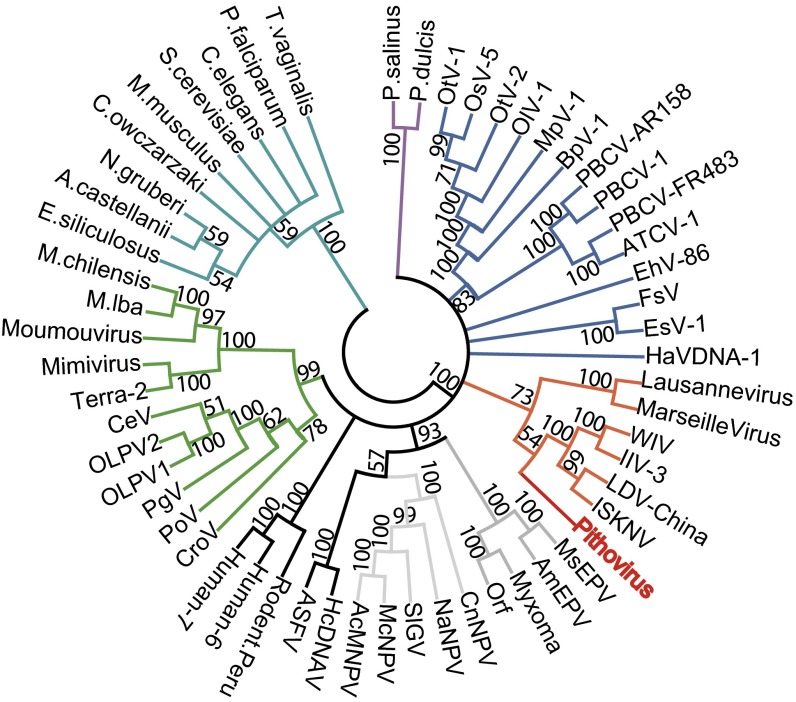
Despite its sizable Pandoravirus-like particle, Pithovirus exhibits a replication cycle and a gene content (Fig. 2 and Table S2) more similar to those of previously described large icosahedral eukaryotic DNA viruses, such as Marseilleviridae and Iridoviridae. This global picture is consistent with the neighbor-joining clustering pattern of the Pithovirus DNA polymerase (Fig. 4). It is further confirmed by the phylogenetic positioning of three other viral core proteins shared with Pandoraviruses using maximum likelihood-based analyses and a cladogram based on the presence/absence of 205 conserved viral genes (Fig. S3). In all cases, Pithovirus was positioned within well-supported clades including Marseilleviruses, Iridoviruses, or both. The tree topologies confirmed the unexpected absence of a close evolutionary relationship between Pithovirus and the look-alike Pandoraviruses.
Fig. 4.
Clustering of viral and eukaryotic DNA polymerases. A multiple alignment of 57 eukaryotic and large virus DNA polymerase sequences (569 ungapped positions) was computed using the default options of the MAFFT server (40). The neighbor-joining tree was built using the JTT substitution model (estimated α = 1.05) and 100 bootstrap resamplings were performed. The tree was rooted at the basis of the eukaryotes and collapsed for bootstrap values <50 before drawing using MEGA5 (41). The Pithovirus DNA polymerase sequence (red) does not cluster with the Pandoraviruses (purple), but falls within a clade clustering the Iridoviruses and Marseilleviruses (orange). Other colors are used to distinguish eukaryotes (turquoise) and viruses from different families: Megaviridae (green), Phycodnaviridae (blue), Herpesviridae (dark gray), Baculoviridae (light gray), Asfar (black), and Poxviridae (gray).
Particle Proteome.
The proteome analysis of purified Pithovirus virions identified 159 different gene products, two-thirds of them corresponding to unknown functions (Fig. 2C and Table S1). The number of proteins making the Pithovirus and Pandoravirus particles (i.e., 210) is thus similar, despite their fivefold difference in gene number (9). This finding refutes the simple idea that large and complex particles should always correlates with large genomes, as observed until now (2, 3, 5, 6, 9). However, despite their overall similar morphology and complexity, the Pithovirus and Pandoravirus particles are made of entirely different sets of proteins, only sharing one pair of homologous proteins (pv384 and ps500) with highly discrepant abundances in their respective proteomes (ranked 8th and 162nd, respectively) (Table S1). In further contrast with the Pandoraviruses, the Pithovirus particles incorporate the complete transcriptional machinery encoded by its genome (Table S1). The availability of a preloaded functional transcription apparatus in the particle is the sine qua non for a fully cytoplasmic replication cycle also characterizing the Megaviridae (8) and the Poxviridae (26, 28). The presence of glycosylated proteins in the Pithovirus particles [also found in Marseillevirus (18) and Mimivirus (29)] is another noticeable difference from Pandoravirus, in which none were found (Fig. S4). In the absence of detectable MCP or core protein homologs, the major structural components of the Pithovirus particle remain to be identified. Potential candidates include the ankyrin domain-containing proteins and collagen triple helix repeat-containing proteins encoded in multiple copies in the genome. None of the ankyrin domain-containing proteins are detected in the proteome whereas 7 of the 12 collagen repeat-containing proteins were readily detected and may participate in the structural scaffold, corresponding to the striated tegument patterns (Fig. 1). Upon semiquantitative analysis (30) (Table S1), the four far most abundant proteins are encoded by genes pv449, pv461, pv93, and pv106. Unfortunately neither their amino acid sequences nor predicted fold provide clues about their functions or evolutionary origin.
Besides most of the enzymes related to nucleotide synthesis or nucleic acid processing, a few others are associated with the particle, such as an adenylosuccinate synthetase (pv90), an ADP ribosyl glycohydrolase (pv118), GTP-binding proteins (pv116, pv127, and pv137) and a GTPase (pv213), a protease (pv133), protein kinases (pv156, pv405, and pv438), hydrolases (pv212 and pv289), oxidoreductases (pv342 and pv467), dehydratase (pv385), and a glycosyltransferase (pv406). These enzymes, as well as 37 A. castellanii proteins, all detected in low abundance (Table S1), might be simple by-standers or play a role at the earliest stages of infection.
Transcript Mapping.
A whole transcriptome shotgun sequencing analysis was performed on Pithovirus-infected cells at various infection times to validate our gene predictions and assess the transcriptional status of the repeated regions. As expected, a vast majority of the transcribed nucleotides fall in the predicted genic regions. Evidence of transcription was provided for 82% of the predicted coding regions (Fig. 3D), irrespectively of the ORF length (Fig. S5). A residual transcription activity was observed in the intergenic regions and at an even lower level in the repeat-containing regions (Fig. 3D). The sequence tags generated at both extremities of the cDNAs during library construction were used to investigate the 5′ and 3′ UTR transcripts structures. The 5′ extremities were unambiguously mapped for 216 transcripts, leading to an estimated short median length of 8 nt for the 5′ UTRs (Fig. S6). Similarly, 3′ extremities were reliably mapped for 130 transcripts, estimating the median length of 3′ UTRs at 16 nt (Fig. S6). These values are markedly shorter than the ones observed for eukaryotic transcripts, leaving little room for regulatory motifs. The precise mapping of the 3′-transcript ends also demonstrated that 70% are overlapped by a nonconserved palindromic nucleotide sequence capable of forming a hairpin structure that may act as a termination/polyadenylation signal as documented for Mimivirus (31) (Fig. S6).
Discussion
Our discovery of the giant Pandoraviruses, exhibiting astonishing gene contents packaged into uniquely amphora-like shaped particles, hinted that an entire world of viruses remained to be unraveled. The simultaneous isolation of two pandoravirus family members (P. salinus and Pandoravirus dulcis) from two remote locations suggested that they were not rare. The lack of gene content similarity between these Pandoraviruses and the newly isolated Pithovirus is now adding another dimension in the exploration of the world of giant viruses. We now have evidence that viruses from entirely different families (from a genomic point of view) may be associated with the same particle morphotype defined by an amphora-like shape, an apex aperture, and a thick electron-dense tegument lined by a lipid membrane enclosing an empty-looking internal compartment. At this point, one may wonder about the diversity of viruses using Pandora box-like particles that remains to be discovered. If this diversity is as large as that of viruses associated with icosahedral capsids, Pandoravirus-like particles might be used to propagate genomes with vastly different sizes and/or molecular structures (e.g., dsDNA, ssDNA, or RNA). The hunt for these viruses is only beginning and speculations about their evolutionary origin should be postponed before many more are found. Unfortunately, there are only three representatives known today (Pithovirus sibericum, P. salinus, and P. dulcis) and they do not share a distinctive set of genes from which to design molecular probes or mine metagenomic data. In the absence of such trademark genes, the identification of additional Pandora box-associated viruses will remain a challenging and serendipitous process.
Beyond their global similarity, a closer inspection of the ultrastructures of the Pithovirus and Pandoravirus particles reveals a number of specific features by which they can be discriminated. These include the spectacular honeycomb-like array of the Pithovirus cork, its striated tegument, and the fuzzy appearance of its surface. Inside their common host, the two viruses also differ by the subcellular structures they generate while replicating. These multiple differences lead us to propose that the uncharacterized KC5/2 endoparasite described more than 10 y ago (32) is a modern relative of the P. sibericum rather than another Pandoravirus (9). KC5/2 exhibits a Pithovirus-like striated envelope surrounding a low-density compartment and a protruding cork (“ostiole” in ref. 32) showing parallel stripes. A roll of membrane underneath the cork is also distinguishable (32) (Fig. 1A). Finally, cells infected by KC5/2 display Pithovirus-like virion factories (“sol cytoplasm” in ref. 32) surrounded by Pithovirus-like particle components such as stripes, pieces of cork, and amorphous dense material.
Conclusion
Using Acanthamoeba as bait, we isolated a large DNA virus from a 30,000 y-old permafrost layer. To our knowledge this is the most ancient eukaryote-infecting DNA virus revived to date. Previous studies reported the detection of genomic signatures of Emiliania huxleyi virus in 7,000-y-old sediments underlying the Black Sea (33) and of tomato mosaic tobamovirus in 140,000-y-old glacial ice in Greenland (34), but did not address their viability. Although Pithovirus is the prototype of a new family, its genome structure and replication cycle are similar to that of other large eukaryotic DNA viruses, many of which are human or animal pathogens. Our results thus further substantiate the possibility that infectious viral pathogens might be released from ancient permafrost layers exposed by thawing, mining, or drilling. Climate change in the Russian Arctic is more evident than in many other regions of the world. Whereas the average global temperature has increased by 0.7 °C during the last 100 y, the average temperatures of the surface layer of Arctic permafrost have increased by 3 °C during the same period. In the 20th century, the permafrost in the Northern Hemisphere has diminished by 7% (35). This no doubt corresponded to a large release of microorganisms from previously frozen soils, an unknown fraction of which was revived upon thawing. Indeed, pathogenic bacteria can survive under low temperatures recurrently causing diseases in circumpolar regions (35). The rich mineral resources and oil reserves of the arctic regions are under increasing pressure for their industrial exploitation (involving mining and drilling). It thus becomes urgent to examine which viruses are expected to be encountered not only near the surface, but in increasingly deeper and more ancient permafrost layers (down to 3 My) (36, 37). Attempting to isolate and revive increasingly ancient amoeba-infecting viruses from permafrost is an inexpensive and safe way to realistically assess the threat posed by pathogenic animal DNA viruses eventually released from soils and sediments frozen in a distant past. Metagenomic analyses will also be used to safely document the eventual presence of known viral pathogens in permafrost samples containing viable amoeba-infecting viruses.
Materials and Methods
Sample Recovery and Radiocarbon Dating.
The P-1084 T permafrost sample was collected in 2000 from the frozen outcrop walls in Chukotka, on the Stanchikovsky Yar, a steep bank 23 m above the level of the Anui River. It had never thawed. See SI Materials and Methods for details.
P. sibericum Isolation and Production.
The buried soil sample was resuspended in Prescott and James medium (38) and used to infect A. castellanii (Douglas) Neff (American Type Culture Collection 30010TM) cells. The wells presenting an infected phenotype were recovered and used to infect four T-75 tissue-culture flasks plated with fresh Acanthamoeba cells. After lysis completion, the virus was purified on a discontinuous sucrose gradient.
Genome Sequencing, Assembly, and Annotation.
Around 10 µg of genomic DNA was recovered from purified particles by combining a phenol/chloroform extraction and purelink genomic DNA extraction mini kit (Life Technologies) protocols. An Illumina 100-bp paired-end library was prepared for sequencing. Another Nextera mate pair library was also prepared and sequenced using 250 bp-length read chemistry on a paired-end flow cell on the Illumina MiSeq (Illumina). SI Materials and Methods contains further details on the bioinformatic genome assembly and annotation procedures.
Transcriptome Study.
A. castellanii -adherent cells (1.75 × 108) were infected by Pithovirus with 30 particles per cell. For each infection time (4, 11, and 19 h pi) three-fourths of the cells were kept aside for RNA extraction, and the rest were pooled to make a fourth point corresponding to the mix of T4-11-19 h pi. RNA was extracted and quantified and full-length cDNA synthesis was performed using the switching mechanism at 5′ end of RNA transcript cDNA library construction kit. The Advantage 2 PCR Kit (Clontech) was used to produce the dscDNA. Illumina 250-bp paired-end libraries were prepared for sequencing. SI Materials and Methods contains experimental details and the bioinformatic procedures used for data analyses.
Proteome Analysis.
Purified particles (108) were resuspended in 100 µL lysis buffer Tris/20 mM CaCl2/1 mM (pH 7.4) and incubated 5 min at 95 °C. Proteins were stacked in the top of a 4–12% (wt/vol) NuPAGE gel (Invitrogen) before R-250 Coomassie blue staining. The gel band was manually excised and cut in pieces before nano–liquid chromatography-tandem MS analyses. The data were processed automatically using Mascot Daemon software (Version 2.3.2; Matrix Science) and the results filtered using the IRMa software (39) (Version 1.31.1). Detailed procedures are presented in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. J.-P. Chauvin, F. Richard, and A. Aouane for their expert assistance on the electronic microscopy platform (Institut de Biologie du Développement de Marseille). Technical assistance was provided by Dr. N. Philippe, V. Schmidt, and M.-A. Galeron. We also thank F. Lopez (Jeol), D. Monville, and S. Searle (Gatan) for the 3D reconstruction shown in Movie S1. This work was partially supported by France Génomique Grant ANR-10-INSB-01-01, by the Provence-Alpes-Côte-d'Azur région (2010 12125), and by the Program of the Presidium of the Russian Academy of Sciences 28 and the Russian Foundation of Basic Research (Projects 12-04-31827 and 12-05-01085).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The Pithovirus genome sequence reported in this paper has been deposited in GenBank database (accession no. KF740664). The MS proteomics data reported in this paper have been deposited in the ProteomeXchange Consortium database, http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD000460, via the Proteomics Identifications database (www.ebi.ac.uk/pride/archive) partner repository [identifier no. PXD000460 (10.6019/PXD000460)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320670111/-/DCSupplemental.
References
- 1.La Scola B, et al. A giant virus in amoebae. Science. 2003;299(5615):2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 2.Raoult D, et al. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306(5700):1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 3.Colson P, et al. Viruses with more than 1,000 genes: Mamavirus, a new Acanthamoeba polyphaga mimivirus strain, and reannotation of Mimivirus genes. Genome Biol Evol. 2011;3:737–742. doi: 10.1093/gbe/evr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desnues C, et al. Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc Natl Acad Sci USA. 2012;109(44):18078–18083. doi: 10.1073/pnas.1208835109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoosuf N, et al. Related giant viruses in distant locations and different habitats: Acanthamoeba polyphaga moumouvirus represents a third lineage of the Mimiviridae that is close to the megavirus lineage. Genome Biol Evol. 2012;4(12):1324–1330. doi: 10.1093/gbe/evs109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arslan D, Legendre M, Seltzer V, Abergel C, Claverie J-M. Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc Natl Acad Sci USA. 2011;108(42):17486–17491. doi: 10.1073/pnas.1110889108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao C, et al. Structural studies of the giant mimivirus. PLoS Biol. 2009;7(4):e92. doi: 10.1371/journal.pbio.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claverie J-M, Abergel C. Mimivirus and its virophage. Annu Rev Genet. 2009;43:49–66. doi: 10.1146/annurev-genet-102108-134255. [DOI] [PubMed] [Google Scholar]
- 9.Philippe N, et al. Pandoraviruses: Amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science. 2013;341(6143):281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 10.Hebsgaard MB, Phillips MJ, Willerslev E. Geologically ancient DNA: Fact or artefact? Trends Microbiol. 2005;13(5):212–220. doi: 10.1016/j.tim.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Lewis K, Epstein S, Godoy VG, Hong S-H. Intact DNA in ancient permafrost. Trends Microbiol. 2008;16(3):92–94. doi: 10.1016/j.tim.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Willerslev E, et al. Long-term persistence of bacterial DNA. Curr Biol. 2004;14(1):R9–R10. doi: 10.1016/j.cub.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 13.Yashina S, et al. Regeneration of whole fertile plants from 30,000-y-old fruit tissue buried in Siberian permafrost. Proc Natl Acad Sci USA. 2012;109(10):4008–4013. doi: 10.1073/pnas.1118386109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tumpey TM, et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310(5745):77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 15.Biagini P, et al. Variola virus in a 300-year-old Siberian mummy. N Engl J Med. 2012;367(21):2057–2059. doi: 10.1056/NEJMc1208124. [DOI] [PubMed] [Google Scholar]
- 16.Podlipaeva IuI, Shmakov LA, Gilichinskiĭ DA, Gudkov AV. [Heat shock protein of HSP70 family revealed in some contemporary freshwater Amoebae and in Acanthamoeba sp. from cysts isolated from permafrost samples] Tsitologiia. 2006;48(8):691–694. [PubMed] [Google Scholar]
- 17.Williams T, Barbosa-Solomieu V, Chinchar VG. A decade of advances in iridovirus research. Adv Virus Res. 2005;65:173–248. doi: 10.1016/S0065-3527(05)65006-3. [DOI] [PubMed] [Google Scholar]
- 18.Colson P, et al. “Marseilleviridae”, a new family of giant viruses infecting amoebae. Arch Virol. 2013;158(4):915–920. doi: 10.1007/s00705-012-1537-y. [DOI] [PubMed] [Google Scholar]
- 19.Santini S, et al. Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes. Proc Natl Acad Sci USA. 2013;110(26):10800–10805. doi: 10.1073/pnas.1303251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Claverie J-M, Abergel C. Open questions about giant viruses. Adv Virus Res. 2013;85:25–56. doi: 10.1016/B978-0-12-408116-1.00002-1. [DOI] [PubMed] [Google Scholar]
- 21.Delhon G, et al. Genome of invertebrate iridescent virus type 3 (mosquito iridescent virus) J Virol. 2006;80(17):8439–8449. doi: 10.1128/JVI.00464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen MJ, Schroeder DC, Wilson WH. Preliminary characterisation of repeat families in the genome of EhV-86, a giant algal virus that infects the marine microalga Emiliania huxleyi. Arch Virol. 2006;151(3):525–535. doi: 10.1007/s00705-005-0647-1. [DOI] [PubMed] [Google Scholar]
- 23.Nasar F, Jankowski C, Nag DK. Long palindromic sequences induce double-strand breaks during meiosis in yeast. Mol Cell Biol. 2000;20(10):3449–3458. doi: 10.1128/mcb.20.10.3449-3458.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bzymek M, Lovett ST. Evidence for two mechanisms of palindrome-stimulated deletion in Escherichia coli: Single-strand annealing and replication slipped mispairing. Genetics. 2001;158(2):527–540. doi: 10.1093/genetics/158.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi J, Blundell TL, Mizuguchi K. FUGUE: Sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol. 2001;310(1):243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 26.Condit RC, Moussatche N, Traktman P. In a nutshell: Structure and assembly of the vaccinia virion. Adv Virus Res. 2006;66:31–124. doi: 10.1016/S0065-3527(06)66002-8. [DOI] [PubMed] [Google Scholar]
- 27.Bahar MW, Graham SC, Stuart DI, Grimes JM. Insights into the evolution of a complex virus from the crystal structure of vaccinia virus D13. Structure. 2011;19(7):1011–1020. doi: 10.1016/j.str.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moss B. Poxvirus DNA replication. Cold Spring Harb Perspect Biol. 2013;5(9):5. doi: 10.1101/cshperspect.a010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renesto P, et al. Mimivirus giant particles incorporate a large fraction of anonymous and unique gene products. J Virol. 2006;80(23):11678–11685. doi: 10.1128/JVI.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishihama Y, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4(9):1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Byrne D, et al. The polyadenylation site of Mimivirus transcripts obeys a stringent ‘hairpin rule’. Genome Res. 2009;19(7):1233–1242. doi: 10.1101/gr.091561.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michel R, Schmid EN, Hoffmann R, Müller K-D. Endoparasite KC5/2 encloses large areas of sol-like cytoplasm within Acanthamoebae. Normal behavior or aberration? Parasitol Res. 2003;91(4):265–266. doi: 10.1007/s00436-003-0944-0. [DOI] [PubMed] [Google Scholar]
- 33.Coolen MJL. 7000 years of Emiliania huxleyi viruses in the Black Sea. Science. 2011;333(6041):451–452. doi: 10.1126/science.1200072. [DOI] [PubMed] [Google Scholar]
- 34.Castello JD, et al. Detection of tomato mosaic tobamovirus RNA in ancient glacial ice. Polar Biol. 1999;22(3):207–212. [Google Scholar]
- 35.Revich B, Tokarevich N, Parkinson AJ. Climate change and zoonotic infections in the Russian Arctic. Int J Circumpolar Health. 2012;71:18792. doi: 10.3402/ijch.v71i0.18792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi T, Reeves RH, Gilichinsky DA, Friedmann EI. Characterization of viable bacteria from Siberian permafrost by 16S rDNA sequencing. Microb Ecol. 1997;33(3):169–179. doi: 10.1007/s002489900019. [DOI] [PubMed] [Google Scholar]
- 37.Gilichinsky DA, et al. Microbial populations in Antarctic permafrost: Biodiversity, state, age, and implication for astrobiology. Astrobiology. 2007;7(2):275–311. doi: 10.1089/ast.2006.0012. [DOI] [PubMed] [Google Scholar]
- 38.Page FC. A New Key to Freshwater and Soil Gymnamoebae. Ambleside, UK: Freshwater Biological Association; 1988. [Google Scholar]
- 39.Dupierris V, Masselon C, Court M, Kieffer-Jaquinod S, Bruley C. A toolbox for validation of mass spectrometry peptides identification and generation of database: IRMa. Bioinformatics. 2009;25(15):1980–1981. doi: 10.1093/bioinformatics/btp301. [DOI] [PubMed] [Google Scholar]
- 40.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9(4):286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 41.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.