Significance
The enzyme activation-induced deaminase (AID) triggers antibody class switch recombination (CSR), a critical mechanism for immune response. CSR is an intrachromosomal rearrangement requiring DNA double strand breaks that are initiated by AID and must be repaired by specific DNA repair pathways. We identify a domain of AID that is required to link the DNA damage step with the subsequent repair during CSR as well as for chromosomal translocations, a collateral effect of CSR. AID influences the recruitment of appropriate end-joining pathways for CSR, preventing aberrant DNA processing that leads to cell death or nonproductive repair and dominant-negative effects. Our results can also explain the basis of an autosomal dominant immunodeficiency caused by C-terminally truncated AID variants.
Keywords: antibody gene diversification, isotype switching, somatic hypermutation
Abstract
Activation-induced deaminase (AID) triggers antibody class switch recombination (CSR) in B cells by initiating DNA double strand breaks that are repaired by nonhomologous end-joining pathways. A role for AID at the repair step is unclear. We show that specific inactivation of the C-terminal AID domain encoded by exon 5 (E5) allows very efficient deamination of the AID target regions but greatly impacts the efficiency and quality of subsequent DNA repair. Specifically eliminating E5 not only precludes CSR but also, causes an atypical, enzymatic activity-dependent dominant-negative effect on CSR. Moreover, the E5 domain is required for the formation of AID-dependent Igh-cMyc chromosomal translocations. DNA breaks at the Igh switch regions induced by AID lacking E5 display defective end joining, failing to recruit DNA damage response factors and undergoing extensive end resection. These defects lead to nonproductive resolutions, such as rearrangements and homologous recombination that can antagonize CSR. Our results can explain the autosomal dominant inheritance of AID variants with truncated E5 in patients with hyper-IgM syndrome 2 and establish that AID, through the E5 domain, provides a link between DNA damage and repair during CSR.
Antibodies change during an immune response by increasing their affinity for cognate antigen and acquiring new biological properties that reside in the constant region of the heavy chain. These changes originate from modifications in the Ig genes. Somatic hypermutation (SHM) introduces single base pair mutations over the Ig variable exon (IgV), changing the antibody affinity (1, 2). Class switch recombination (CSR) exchanges the exons encoding for the constant region of the heavy chain that defines IgM for those exons defining IgG, IgA, or IgE. This process involves the stepwise generation and subsequent repair of DNA double strand breaks (DSBs) (3, 4).
Activation-induced deaminase (AID) initiates both SHM and CSR by deaminating deoxycytidine to deoxyuridine at the Ig loci (2). During CSR, removal of AID-generated deoxyuridine from opposite DNA strands at two distant switch (S) regions by either the uracil DNA-glycosylase (UNG) or components of the mismatch repair pathway initiates DNA processing leading to DSBs. These DSBs at the Igh evoke a DNA damage response and are resolved by either classical nonhomologous end joining (C-NHEJ) requiring the DSBs end-binding heterodimer Ku70/80, the scaffold protein Xrcc4, and Ligase4 (4, 5) or an ill-defined alternative end-joining (A-EJ) pathway (6) for productive CSR. CSR requires the joining of two simultaneous DSBs located far apart and deletion of the intervening chromosomal segment (3). As a side effect, CSR can also produce chromosomal translocations involving the Ig loci (7). The recombination of variable diversity joining (VDJ) gene fragments is also a long-range intrachromosomal joining, but in that case, the initiating recombination-activating gene (RAG)1/2 endonuclease protects the DNA ends and promotes C-NHEJ to prevent aberrant joining (8, 9). No analogous role of AID on DNA repair during CSR has been shown so far, although AID has been suggested to stabilize inter–S-region synapsis (10). The C terminus of AID is necessary for CSR but not SHM for unknown reasons (11, 12). This requirement might reflect a role of this domain in repair, given that C-terminally truncated AID variants still produce DSBs at the S regions in B cells (13–15). However, the fact that AID can be replaced by the yeast endonuclease I-SceI for efficient CSR in engineered mice seems to argue against its need for repair (16). Thus, it is still unclear whether AID contributes to the repair steps of CSR.
AID deficiency causes a hyper-IgM immunodeficiency syndrome (HIGM2) in humans. Most HIGM2 patients carry deleterious mutations in AICDA (the AID gene), which are inherited as autosomal recessive (AR) traits (17, 18). These patients lack SHM and CSR, are susceptible to infections, and develop lymphadenopathies because of germinal center hyperplasia (17, 18). Intriguingly, a small proportion of HIGM2 patients carries only one mutated AICDA allele. There is no explanation as to why these alleles are autosomal dominant (AD), but in every case, the AD allele encodes for an AID protein missing the last 8 or 12 aa (12, 15). Because this region is necessary for CSR and because AD HIGM2 patients show normal SHM, the simplest explanation would be that AD AID variants behave as dominant negatives specifically for CSR, as suggested by the families’ pedigrees (15). We hypothesized that studying this proposed dominant-negative effect could also shed light on the role of AID C terminus and show a role of AID in late steps of CSR.
Results
AID Variants with Enzymatic Activity-Dependent Dominant-Negative Effect on CSR.
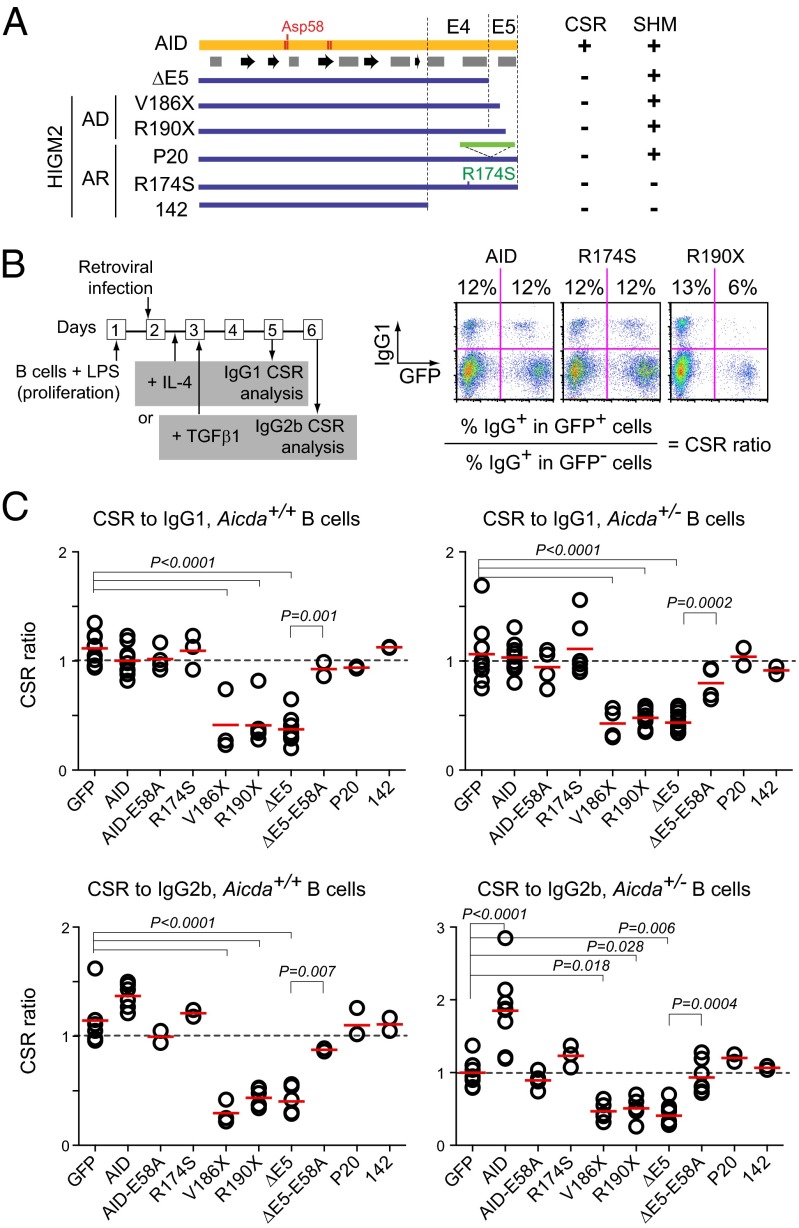
The presumed dominant-negative effect of AD AID variants on CSR has never been shown. Hence, we first tested this possibility for multiple AID variants (Fig. 1A and Fig. S1), including the AD HIGM2 variants R190X and V186X that lack the C-terminal 8 and 12 residues, respectively (15, 19), and the artificial variant ΔE5 that lacks the whole 17 aa encoded by AICDA exon 5 (E5). We also tested the AR variants pD143_L181 > AfsAPVX (mimicked by AID 142), P20 (bearing a 34-residue insertion just before E5), and R174S (a point mutant) (12, 18, 19) (Table S1). The CSR dominant-negative assay (Fig. 1B) consisted of activating naïve B cells from Aicda+/+ (WT) or Aicda+/− mice with LPS and retroviral delivery of the AID variants. Because LPS induces AID and CSR to IgG3, which could give endogenous AID a head start over the transduced variant for switching to IgG3, after retroviral infection, we added cytokines that redirected CSR to other isotypes to favor competition between endogenous and transduced AID. We used IL-4 for CSR to IgG1 and TGF-β1 for CSR to IgG2b. The proportion of switched cells could be compared between the infected (GFP+) and uninfected (GFP−) cells in the same population, with the CSR ratio between GFP+ and GFP− cells providing a measure of the effect of each AID variant on CSR by endogenous AID (Fig. 1C). This normalization corrected for any effect of the infection on the CSR reaction, which was shown by the CSR ratio of ∼1 in cells transduced with control retrovirus (GFP only) (Fig. 1C and Fig. S2). Endogenous AID was not limiting in most of our assay conditions, because transduced AID did not further increase CSR, except in the case of IgG2b in Aicda+/− cells, in which it did (Fig. 1C and Fig. S2). Regardless of the conditions tested, variants R190X and V186X as well as ΔE5 reduced CSR to IgG1 and IgG2b by one-half, indicating a dominant-negative effect (Fig. 1C and Fig. S2). Interestingly, R174S and P20, which have reduced catalytic activity (12, 20), did not interfere with CSR, nor did AID 142 (Fig. 1C and Fig. S2), which is catalytically inactive (Fig. S1 A–E). Indeed, E5 was the only AID domain dispensable for catalytic activity (Fig. S1 A–D). We, therefore, asked whether enzymatic activity was necessary for dominant-negative behavior. Surprisingly, although overexpression of catalytically inactive full-length AID-E58A (bearing Asp58 to Ala) (21) failed to interfere with CSR assay, inactivating ΔE5 through E58A abrogated its dominant-negative effect (Fig. 1C and Fig. S2).
Fig. 1.
AID variants with dominant-negative effect on CSR. (A) Schematics of AID (with predicted secondary structure), AID variants found in AD or AR HIGM2 patients, and artificial variant ΔE5. E4 and E5 denote regions encoded by AICDA exons 4 and 5, respectively. The table indicates CSR and SHM status of homozygous carrier patients. (B) Experimental design of CSR dominant-negative assay with illustrative flow cytometry primary data from WT (Aicda+/+) mouse splenic B cells expressing retrovirally transduced AID- or variant-ires-GFP and stimulated with LPS and IL-4. The proportion of IgG1+ cells was calculated for the GFP− and GFP+ populations (shown above) and then divided to calculate the CSR ratio. (C) Compiled CSR ratios for IgG1 and IgG2b in WT and Aicda+/− B cells. Each symbol represents the CSR ratio for B cells from an individual mouse. A dotted line indicates a ratio of one. Horizontal bars are means. Significant P values from ANOVA with Dunnet posttest are indicated. ΔE5 and ΔE5-E58A were compared by t test.
The E58A mutation did not compromise the structural integrity or DNA binding ability of the protein. First, E58A does not prevent Zn++ coordination (22). Second, it did not inhibit binding to DNA in electrophoretic mobility shift assays (Fig. S1F). Importantly, ΔE5 and ΔE5-E58A bound to ssDNA with comparable affinities, indicating that the dominant-negative behavior did not reflect competition for the DNA substrate. The protein levels of the AID variants did not correlate with their ability to interfere with CSR. Indeed, AID-E58A is expressed at higher levels than the AD or ΔE5 variants but is not dominant negative (Fig. S3A). More importantly, ΔE5 and ΔE5-E58A were similarly expressed in B cells (Fig. S3A). Finally, the AD or ΔE5 variant had no effect on the proliferation of live GFP+ B cells, which was monitored by dilution of a cell division tracking dye (Fig. S3B), also ruling out this possibility to explain the CSR reduction.
We conclude that truncation of AID E5 leads to dominant-negative behavior for CSR by a mechanism that is distinct from molar excess or competition for the DNA substrate by CSR-deficient AID mutants and rather, dependent on enzymatic activity.
Dominant-Negative AID Variants Are Hypermutagenic.
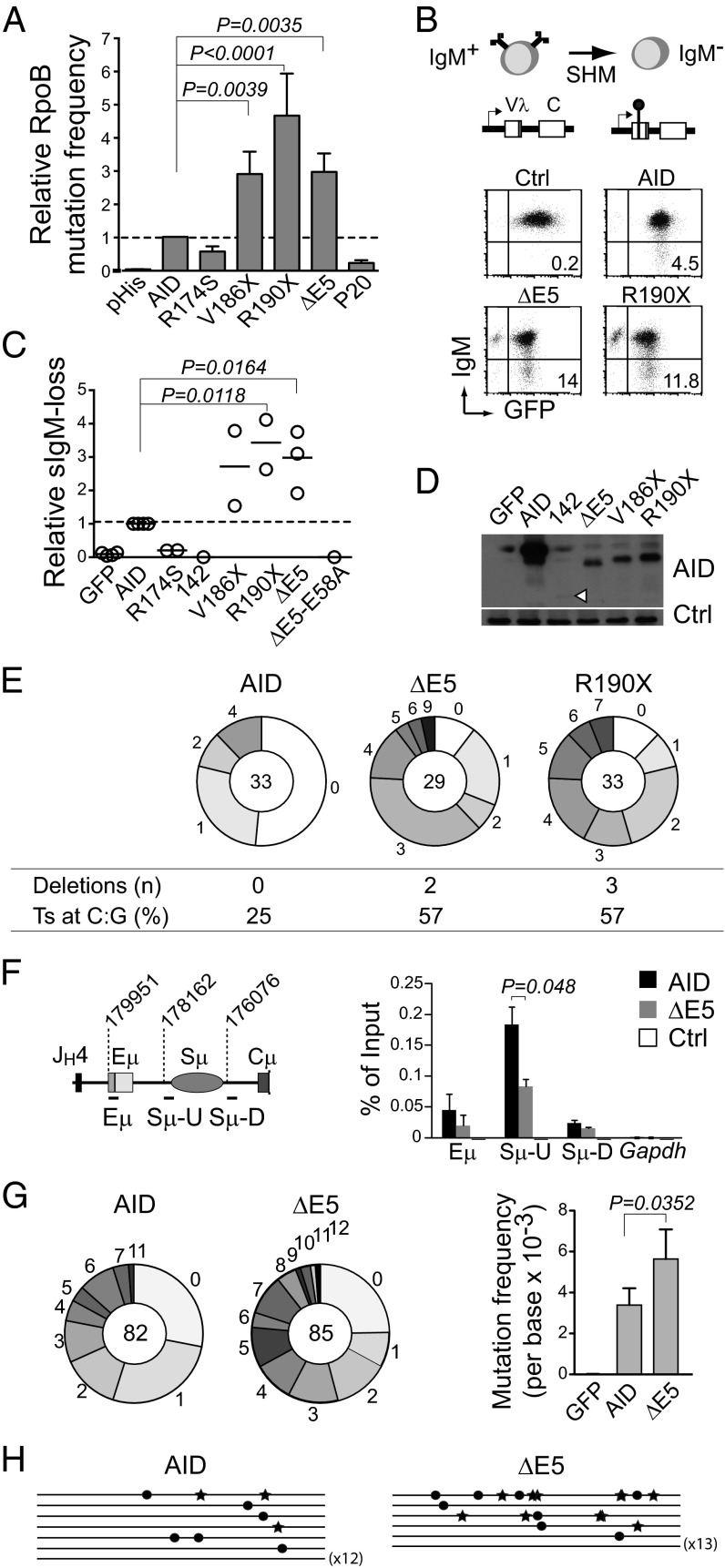
Because the dominant-negative behavior of ΔE5 on CSR required its enzymatic activity, we asked whether truncations in E5 might affect AID mutagenic capacity or quality. Previous reports analyzing these mutants in vitro as recombinant proteins have not been conclusive. Although R190X and V186X showed threefold more activity than AID for deaminating oligonucletides (20), ΔE5 has been reported as threefold higher (23) or slightly lower than the activity of AID (20) using the same assay. Our own measurements on Escherichia coli-purified GST-AID yielded similar estimated Km values for AID and ΔE5 (Km = 41 ± 12 vs. 42 ± 11 nM, respectively). This heterogeneity might suggest limitations in the current biochemical assays for AID. We, therefore, compared the AID variants by a genetic assay, in which the frequency of rifampicin-resistant E. coli cfu, arising from mutations at the rpoB gene, is a sensitive reporter of their DNA deamination activity (24). The ΔE5, R190X, and V186X variants showed approximately threefold higher rpoB mutation frequency than AID, which could not be attributed to increased solubility of the truncated variants (Fig. 2A and Fig. S1 A–E). Altogether, the data indicate that variants bearing E5 truncations are at least equally (and probably more) catalytically active than AID.
Fig. 2.
CSR-deficient and dominant-negative AID variants are hypermutagenic. (A) Relative rpoB mutation frequency of AID variants in E. coli. Means + SDs of the relative medians obtained from three independent experiments normalized to AID are shown. Significant P values by ANOVA with the Holm–Sidak posttest are indicated. P20 and R174S are significantly less active than AID by t test (pHis vs. R174S, P = 0.0002; AID vs. R174S, P = 0.0002; pHis vs. P20, P = 0.0032; AID vs. P20, P < 0.0001). (B) Diagram and representative flow cytometry plots of the surface IgM-loss assay for estimating SHM frequency in IgM+ DT40 ΔψVL Aicda−/− B cells complemented with AID or variant-ires-GFP. (C) Relative IgM-loss capacity of AID variants. Each symbol is the median of ≥12 cultures from one to three experiments performed per variant normalized to their AID control median value. Horizontal lines are means. Statistics are the same as in A. (D) Western blot of AID variants in DT40 B cells. A nonspecific band was used as loading control (Ctrl). (E) Mutation load of the DT40 IgVλ in complemented cells. Pie chart slices represent the proportion of sequences with the indicated number of mutations, with the total number of sequences analyzed at the center. Deletion events and the proportion of transition (Ts) mutations at C:G pairs are indicated. (F) Real-time PCR ChIP analysis of UNG occupancy at the Igh or Gapdh control in mouse Aicda−/− B cells transduced with pMXs empty (Ctrl), AID, or ΔE5-ires-GFP vectors 21 h postinfection. The Igh amplicons and their first nucleotide position are shown on the scheme (according to the National Center for Biotechnology Information contig NG_005838.1). Means + SDs of three independent experiments are plotted. P value by t test. (G) Mutation load (pie charts; plotted as in E) of a 628-bp Sμ fragment (nucleotides 177959–178587 of NG_005838.1) in transduced Aicda−/− Ung−/− B cells. Bars show means + SDs mutation frequencies from four experiments. P value by t test. (H) Sγ1 region sequences from transduced Aicda−/− Ung−/− mouse B cells (1,805-bp fragment, nucleotides 90156–91960 of NG_005838.1). Each line is a sequence, with mutations at G indicated by stars and mutations at C indicated by dots.
We then asked whether the ability to mutate E. coli translated into higher SHM capacity at the Ig genes in B cells. The chicken DT40 B-cell lymphoma line constitutively expresses AID and is a validated system to measure IgV SHM (25). We used the DT40 Aicda−/− ΔΨVλ line, which undergoes SHM on AID complementation (25). SHM can inactivate the Vλ gene and thereby, IgM expression (Fig. 2B). The median fraction of IgM− cells from multiple independent populations is proportional to the SHM frequency (25). IgM loss fluctuation analysis showed approximately threefold higher SHM activity for ΔE5, R190X, and V186X than WT AID, which was confirmed by Vλ sequencing (Fig. 2 C–E). As expected, catalytically compromised variants (142, ΔE5-E58A, and R174S) produced little SHM (Fig. 2C).
SHM in DT40 showed the typical spectrum with >98% mutations at C:G pairs. However, ΔE5 and R190X produced a higher proportion of transition mutations (C/G to T/A) than WT AID (Fig. 2E). It was previously shown that AID lacking the nuclear export signal (AIDΔNES; similar to R190X) mutates the Igh Sμ region with a similar frequency to AID (11); in these data, we noticed an increase in transitions at C:G (25% for AID and 57% for AIDΔNES), reminiscent of our finding at the DT40 IgV. Because transversions at C:G in SHM depend mostly on UNG (26), we measured UNG at the Sμ in mouse Aicda−/− B cells complemented with AID or ΔE5 by ChIP. The AID-dependent UNG occupancy at Sμ was, indeed, reduced by 50% in splenic B cells expressing ΔE5 (Fig. 2F).
Finally, we compared the frequency and quality of deamination by ΔE5 at S regions by complementing B cells from Aicda−/− Ung−/− mice. UNG deficiency prevents CSR and faithful uracil repair to a large extent, thus revealing the deamination footprint of AID (26). We found that ΔE5 deaminated Sμ and Sγ1 with higher frequency than AID, but there were no differences in their footprint or strand preference (Fig. 2 G and H and Fig. S4).
Thus, a common characteristic of the AID variants that are CSR-deficient and dominant negatives is that they target and deaminate the IgV and S regions with higher efficiency than but similar quality to AID, except for a higher proportion of transition mutations at C:G caused by lower UNG recruitment.
AID E5 Prevents Cytotoxicity and DNA Damage Accumulation.
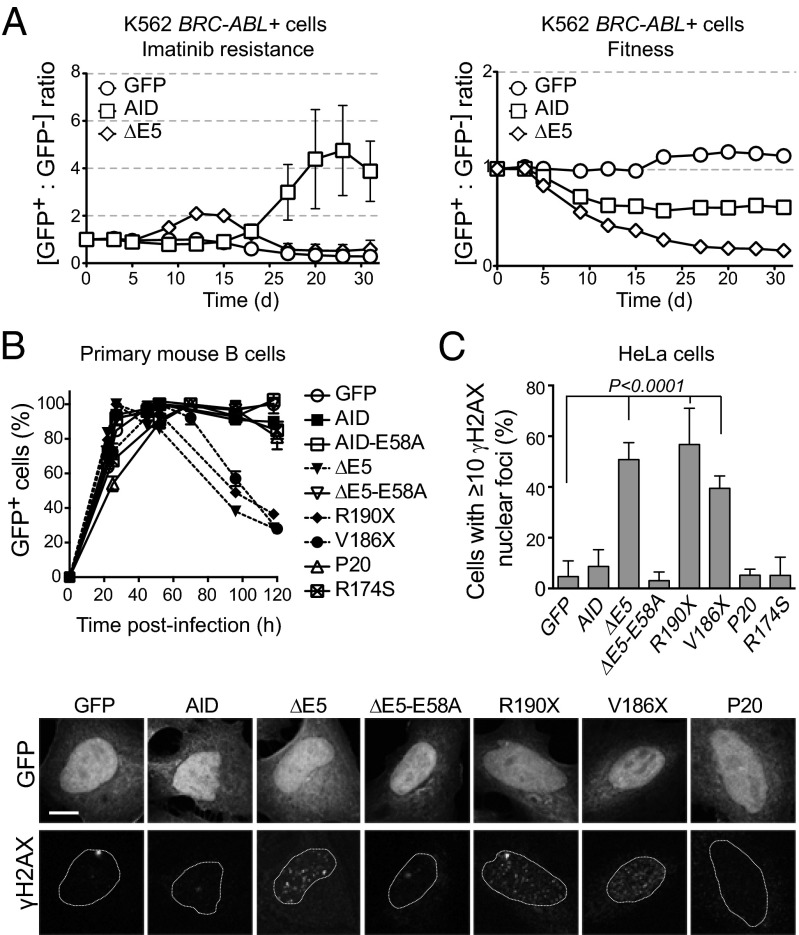
We then asked whether the increased DNA deamination frequency by AD and ΔE5 variants would extend beyond the Ig loci. We first used the BCR-ABL oncogene, an AID target in chronic myelogenous leukemia (27), as a reporter. The BCR-ABL kinase is necessary for cell growth and can be inhibited by imatinib. Expressing AID in the chronic myelogenous leukemia cell line K562 leads to imatinib resistance through BCR-ABL mutations in an AID dose-dependent manner (28). We mixed K562 cells transduced with AID (GFP+) and uninfected cells (GFP−) at a 1:1 ratio and expanded the mixed cultures with imatinib (Fig. S5A). AID expression led to an increase in the GFP+/GFP− cell ratio starting at approximately day 18, whereas ΔE5 caused a much earlier increase in the proportion of GFP+ cells by day 9 (Fig. 3A), consistent with higher mutation frequency. However, ΔE5 cells plateaued at a lower ratio than AID cells and quickly disappeared (Fig. 3A), suggesting an adverse effect on cell expansion. This negative influence was shown by repeating the experiment in the absence of imatinib, which then measures the effect of each variant on cell fitness (Fig. S5A). ΔE5 compromised fitness much more severely than AID, which was shown by the accelerated loss of GFP+ cells over time (Fig. 3A). The latter results explained the expression instability of ΔE5 variants in DT40 B cells, whereas ΔE5-E58A was well-tolerated (Fig. S5B) and could explain the apparently lower infections in retrovirally complemented primary B cells (Fig. S2). We, therefore, analyzed the effect of AID variants on primary B-cell expansion. After retroviral complementation, the proportion of Aicda−/− B cells expressing vector control, AID, AID-E58A, R174S, or P20 peaked at ∼30 h and remained constant for >100 h postinfection. However, cells expressing R190X, V186X, or ΔE5 quickly declined over time, which was fully dependent on enzymatic activity, as shown by ΔE5-E58A that behaved like AID (Fig. 3B).
Fig. 3.
CSR-deficient dominant-negative AID variants are genotoxic. (A) Follow-up of the ratio of K562 BCR-ABL+ cells originally sensitive to imatinib transduced with AID-ires-GFP (GFP+) or untransduced (GFP−). Cells were grown in the presence of imatinib (Left), where the increase in the GFP+/GFP− cells ratio indicates acquisition of imatinib resistance, or without selection (Right), where decrease in the GFP+/GFP− cells ratio reflects the effect of each AID variant on cell fitness. One representative experiment out of three experiments performed is shown. (B) Effect of AID variants on the expansion of retrovirally complemented Aicda−/− splenic B cells evaluated from the relative proportion of GFP+ (complemented) cells in the culture at various times postinfection. Mean + SEM proportion of GFP+ cells in B-cell cultures from two independently infected mice. One of two similar experiments is shown. (C) DNA damage accumulation in HeLa cells transiently expressing AID- or variant-ires-GFP was measured by the presence of γH2AX nuclear foci 48 h posttransfection. Means + SDs proportions of GFP+ cells with ≥10 foci from three to five independent experiments are plotted for each variant with ANOVA and Dunnet posttest P values. Representative confocal images are shown below.
The effect of AD variants on B-cell expansion contrasted with the hyperproliferation described for AID-deficient B cells (29), which leads to germinal center hyperplasia and produces lymphadenopathies in HIGM2 patients (17, 18). However, the analysis of HIGM2 patients data showed that, whereas in 72 AR HIGM2 compiled patients, the incidence of lymphadenopathy was 75%, only 2 of 11 available AD HIGM2 patients displayed clinical lymphadenopathies (18% incidence) (Table 1). Coincidently, AD AID variants compromised B-cell clonal expansion, which is critical in forming germinal centers (Fig. S5C). Notably, the two AD patients with lymphadenopathies (in one patient, giant germinal centers were confirmed in a cervical lymph node biopsy) also lacked SHM (Table 1). The fact that neither the mutated nor the WT allele of AID produces mutations in these two patients suggests an additional defect. Whatever this defect, these patients functionally resemble AID nulls (i.e., CSR- and SHM-deficient) and most likely developed lymphadenopathy, because B-cell proliferation is not hampered by AID activity.
Table 1.
Clinical characteristics of autosomal dominant HIGM2 patients
| Patient | AICDA status | Ig levels (g/L) | CSR in vitro | SHM in vivo | Lymphadenopathies | ||
| IgM | IgG | IgA | |||||
| 1-I-1* | R190X/+ | 4.05† | 1.28† | 0.1† | − | N | − |
| 1-II-2* | R190X/+ | 2.85† | 0.5† | <0.05† | − | N | − |
| 1-II-3* | R190X/+ | 3.02† | 6.33† | <0.05† | − | N | − |
| 1-III-1* | R190X/+ | 1.22 | 7.58 | <0.05† | − | N | − |
| 2-I-1* | R190X/+ | 2.93† | 2.82† | <0.05† | − | N | − |
| 2-II-2* | R190X/+ | 2.34† | 0.21† | <0.05† | − | − | ++ |
| 3-II-1* | R190X/+ | 2.34† | 0.38† | <0.05† | − | N | − |
| 4-I-1 | R190X/+ | 2.03 | 1.30† | <0.05† | − | ND | − |
| 5-I-1‡ | V186X/+ | 2.02 | 5.41† | 1.19 | − | ND | − |
| 5-II-1‡ | V186X/+ | 0.84 | NE | <0.05† | ND | − | ++ |
| 5-II-2‡ | V186X/+ | 1.32 | 4.71† | <0.05† | − | ND | − |
| AR-HIGM2§ | −/− (range) | 0.7–37† | <0.2–1.3† | <0.05–0.2† | − | − | ++ (70%) |
| Reference | +/+ (range) | 0.4–2.3 | 7–16 | 0.7–4 | + | N | − |
Despite the AD and ΔE5 variants negatively impacted on B-cell expansion, they showed no effect on cell proliferation dye dilution (Fig. S3B), and they did not further stimulate B-cell apoptosis compared with AID (Fig. S5D). However, because they were hypermutagenic, they could induce extensive DNA damage, leading to necrotic cell death (30). We assayed the ability of each variant to produce DNA breaks detectable as foci of phosphorylated histone H2AX (γH2AX) by immunofluorescence in transfected HeLa cells. Indeed, cells expressing R190X, V186X, and ΔE5 (but not those cells expressing AID, R174S, P20, or ΔE5-E58A) accumulated abundant γH2AX foci (Fig. 3C).
We conclude that the CSR-deficient and dominant-negative AID variants compromise B-cell expansion and cause or permit the accumulation of DNA damage.
CSR Deficiency and Dominant-Negative Behavior Correlate with Persistent DNA Damage.
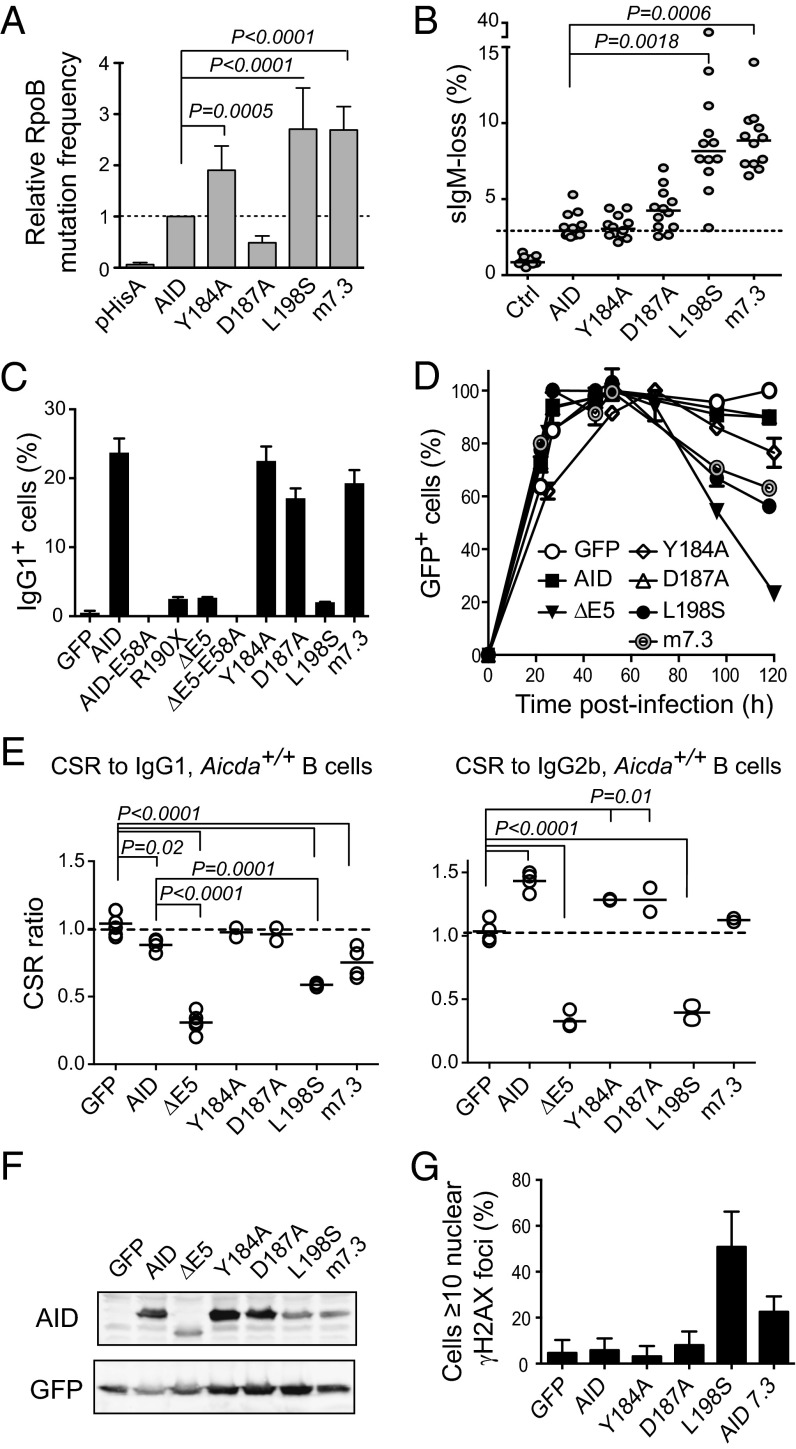
The results above showed that AID variants R190X, V186X, and ΔE5 shared several features. They were hypermutagenic in E. coli and B cells, compromised B-cell expansion, and caused accumulation of DNA damage (Table S1). We wanted to discriminate which of those characteristics correlated with CSR deficiency and dominant-negative behavior. To that end, we compared artificial full-length AID variants having only some of those characteristics (Table S1). We chose the single point mutant L198S, which inactivates the NES (21), and variant m7.3, bearing mutations K10E, T82I, and E156G, which was identified in a screening for AID variants with increased enzymatic activity (31). Both L198S and m7.3 displayed similarly high mutagenic activity in E. coli as well as SHM assays in DT40 cells (Fig. 4 A and B). However, L198S was inactive for CSR, whereas m7.3 was fully proficient (Fig. 4C). Importantly, although L198S and m7.3 similarly impaired primary B-cell expansion (Fig. 4D), only L198S behaved as a dominant negative for CSR (Fig. 4E and Fig. S6). We analyzed two other single residue substitutions within E5. Mutation D187A weakens cytoplasmic retention, leading to increased nuclear accumulation (21), but still showed similar SHM and CSR activities to WT AID (Fig. 4 B and C). This apparent discrepancy could be attributed to its reduced activity, as was shown by the E. coli mutation assay (Fig. 4A), offsetting its higher nuclear abundance. Mutation Y184A eliminates a phosphorylation site of unknown function. Despite Y184A being more active than AID in E. coli mutation assays, it produced normal SHM and CSR levels (Fig. 4 A–F). Neither D187A nor Y184A interfered with CSR (Fig. 4E and Fig. S6). These results clearly segregated hyperactivity and cell growth defects from the CSR dominant-negative effect. However, the unique ability of L198S among the point mutants to interfere with CSR correlated with its capacity to produce a large accumulation of γH2AX foci in transfected cells (Fig. 4G). Neither D187A nor Y184A produced any γH2AX foci accumulation, whereas m7.3 showed only a slight increase, despite being as hypermutagenic as, and presumably producing DNA damage with similar frequency to, L198S (Fig. 4 A, B, and G).
Fig. 4.
Dominant-negative ability of CSR-deficient AID variants correlates with DNA damage accumulation. (A) E. coli rpoB mutation frequency by AID single point mutants or variant m7.3 (bearing mutations K34E, T82,I and E156G). Means + SEMs of the relative medians normalized to AID from two to three independent experiments are plotted with significant P values from one-way ANOVA with the Holm–Sidak posttest. (B) SHM assay by IgM-loss fluctuation analysis in complemented DT40 ΔψVL Aicda−/− B cells. Each symbol is the percent of IgM− cells in an individual population after 2 wk of expansion; horizontal lines are median values for each population, with significant P values by Kruskal–Wallis test with Dunnet posttest comparison indicated. One representative experiment of two experiments is shown. (C) IgG1 CSR activity of AID variants in complemented mouse Aicda−/− B cells 4 d after stimulation with LPS + IL-4. Mean + SEM of two mice for each variant. (D) Effect of AID variants expression on the expansion of Aicda−/− splenic B cells transduced with AID variants-ires-GFP vectors. Mean ± SEM proportion of GFP+ cells over time postinfection in B-cell cultures from two mice. One representative experiment of two experiments is shown. (E) CSR dominant-negative assays for the indicated mutants like in Fig. 1. Each symbol is the CSR ratio of B cells transduced with AID- or variant-ires-GFP to uninfected cells from an individual mouse. Means (lines) and significant P values from one-way ANOVA with Dunnet posttest are indicated. (F) Protein expression level of AID variants by Western blot in packaging cells. (G) DNA damage accumulation in HeLa cells transiently transfected with AID variants assessed by the presence of γH2AX foci at 48 h posttransfection. Mean + SEM proportion of transfected cells showing ≥10 γH2AX nuclear foci from two to three experiments for each variant is plotted.
We conclude that the CSR dominant-negative effect shown by the AD and ΔE5 variants could not be explained solely by their hyperactivity or their effect on B-cell expansion but rather, could be explained by the loss of a specific domain in E5 that was required for CSR as well as prevention of the accumulation of DNA damage.
AID E5 Is Required for Efficient End Joining.
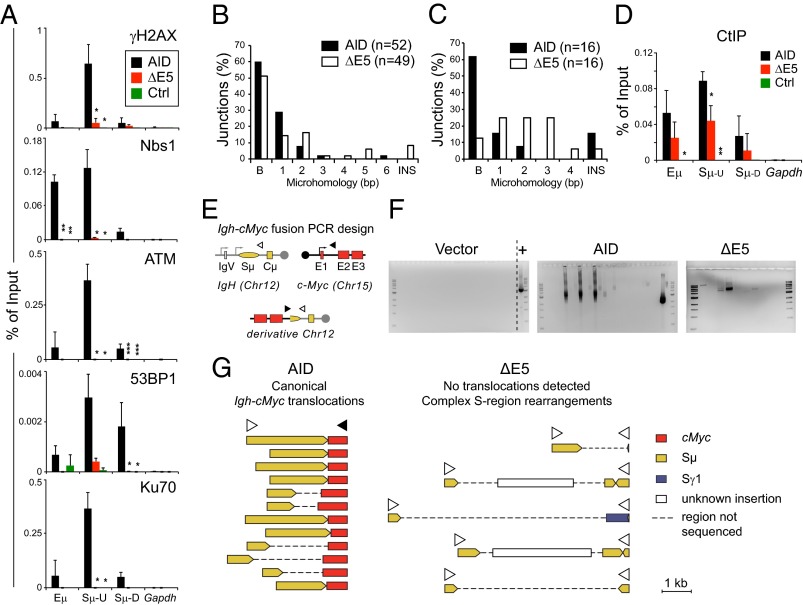
We then investigated the possible function of E5. It has been previously reported that ΔE5 and R190X are capable of initiating as many DSBs as AID at the S regions (13–15). Thus, neither targeting defects nor reduced DSB formation seemed sufficient to explain CSR deficiency and/or the dominant-negative effect by these variants. We, therefore, investigated whether deletion of E5 influenced downstream DNA repair. Early events during NHEJ-mediated CSR are phosphorylation of H2AX and Nijmegen breakage syndrome protein 1 (Nbs1) deposition at the Igh (5), both of which were readily detectable by ChIP at the Sμ in cells expressing AID but absent in those cells expressing ΔE5 (Fig. 5A). Also, ataxia telangiectasia mutated (ATM), which orchestrates much of the DNA repair response during CSR (4), the chromatin modifications reader 53BP1, which is necessary for efficient CSR (32), and Ku70, which is required for CSR by C-NHEJ and perhaps, also a microhomology-mediated pathway (33), were enriched at the Sμ in cells expressing AID but not ΔE5 cells (Fig. 5A).
Fig. 5.
E5 deletion compromises end joining. (A) Real-time PCR ChIP assays for the presence of γH2AX, Nbs1, 53BP1, ATM, and Ku70 at the Igh or Gapdh control in mouse Aicda−/− B cells transduced with pMXs empty (Ctrl), AID, or ΔE5-ires-GFP vectors at 21 h postinfection. Means + SDs of three independent experiments are plotted. Significant differences vs. AID for each amplicon were calculated by ANOVA with Dunnet posttest (*P < 0.05; **P < 0.01; ***P < 0.001). Amplicons in Igh are the same as in Fig. 2F. (B) Analysis of Sμ-Sγ1 junctions amplified from LPS + IL-4–stimulated Aicda−/− B cells complemented with AID or ΔE5 72 h postinfection from three mice. B, blunt (includes 1- or 2-nt insertion); INS, ≥3-nt insertions. (C) Analysis of Sμ-Sα junctions from the CH12 B-cell line depleted of AID with shRNA, complemented, and analyzed as in B. (D) ChIP analysis for CtIP is the same as in A. (E) PCR strategy for detecting Igh-cMyc chromosomal translocation in retrovirally complemented Aicda−/− primary B cells. (F) Representative ethidium bromide-stained agarose gels from PCR assays. Each lane is a PCR performed on DNA extracted from 0.5 to 1 × 106 sorted GFP+ cells expressing each indicated construct. A positive control (+) of the PCR using a cloned translocation is shown. (G) Schematic representation of all independently amplified PCR products from two experiments after cloning and sequencing.
In the absence of C-NHEJ, one or more pathways collectively known as A-EJ can mediate substantial, albeit less efficient, CSR (33). We wanted to determine whether ΔE5 might be shifting CSR resolution from C-NHEJ to A-EJ. The preferential use of A-EJ in CSR is commonly inferred from the increased presence of microhomologies at the switch junctions (6). Sequencing of Sμ-Sγ1 joints from complemented mouse B cells expressing ΔE5 showed longer microhomologies compared with AID (Fig. 5B), with the mean number of overlapping nucleotides at the junctions being 2.3 ± 1.4 bp for ΔE5 vs. 1.5 ± 1.2 bp for AID (P = 0.02, Mann–Whitney test). The effect was not striking, but the homology between Sμ and Sγ1 is low. Indeed, the residual Sμ-Sα junctions in AD HIGM2 patients show unusually long microhomologies (34), and we saw a similar picture in Sμ-Sα junctions of AID-depleted CH12 B cells complemented with ΔE5 and stimulated for CSR to IgA (Fig. 5C), which probably reflects the higher homology between Sμ and Sα. Thus, although microhomology could suggest A-EJ, the unusual Sμ-Sα junctions could also suggest altered processing. Unfortunately, there are no specific markers of A-EJ. However, the factor CtBP-interacting protein (CtIP) facilitates end resection for homologous recombination (HR) (35) and also contributes to the microhomology-mediated subpathway of CSR to IgA in CH12 cells (36). CtIP is dispensable for CSR to IgG1 (37), but it is still recruited to the S region in stimulated mouse B cells (38), where it can promote end resection (37). Therefore, we asked whether CtIP might be increased at the Sμ in B cells expressing ΔE5 as another indication for preferential use of A-EJ. CtIP recruitment to the Sμ in mouse B cells stimulated for CSR to IgG1 was AID-dependent (Fig. 5D). Interestingly, CtIP was, in fact, moderately reduced at the Sμ both in primary (Fig. 5D) and CH12 (Fig. 6) B cells expressing ΔE5.
Fig. 6.
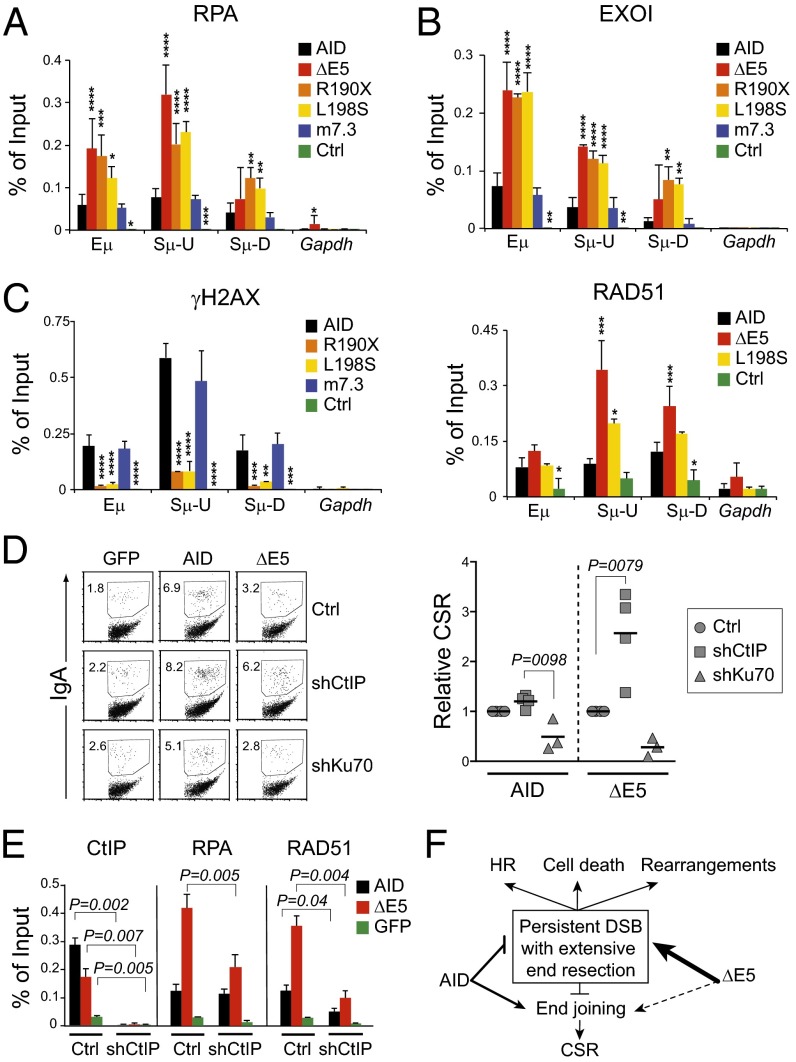
Truncation of E5 leads to end resection at the Sμ. Real-time PCR ChIP assays for (A) RPA, (B) EXOI, and RAD51 and (C) γH2AX at the Igh in cell extracts of Aicda−/− B cells transduced with empty vector (Ctrl) or carrying the indicated AID variants at 21 h postinfection. Amplicons are the same as in Fig. 2F. Means + SDs of three biological replicates for each variant are plotted. Significant differences vs. AID in each amplicon by ANOVA with Dunnet posttest are indicated (****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05). (D) CSR in the complemented CH12 B-cell line. Cells were depleted of AID alone or combined with CtIP or Ku70 by shRNA transfection and then transduced with empty pMXs-ires-GFP (GFP) or encoding human AID or ΔE5. Cells were stimulated, and CSR was measured 24 h later. A representative example of the raw data is shown. The fold change in CSR for AID or ΔE5 when depleting CtIP or Ku70 from three to four independent experiments is plotted. The proportion of IgA+ cells in the GFP+ population was first corrected for the background CSR in the corresponding GFP (vector only) infection. Then, the CSR value of cells expressing AID or ΔE5 in shCtIP (AID- and CtIP-depleted) or shKu70 (AID- and Ku70-depleted) cells was normalized to the CSR value of Ctrl cells (AID-depleted only) expressing AID or ΔE5, respectively. Significant differences by ANOVA with Dunnet posttest are shown for each variant. (E) Real-time PCR ChIP assays for CtIP, RPA, and RAD51 occupancy at the Sμ in the CH12 cells from D. Means + SDs of three biological replicates for each variant and condition are plotted. Significant differences by paired two-tailed t test. (F) Proposed model of the mechanisms operating at the S regions downstream from AID or CSR-deficient and dominant-negative variants (ΔE5).
As an additional functional assay for end joining after AID-mediated damage, we compared the ability of AID and ΔE5 to produce chromosomal translocations in complemented Aicda−/− B cells. These translocations could proceed by either C-NHEJ or A-EJ, although they may preferentially use the A-EJ pathway (6). The Igh-cMyc fusion is a validated reporter of this undesired AID activity and can be detected in B cells by PCR using oligonucleotide pairs, priming one at the Igh and the other one at cMyc (Fig. 5E) (7). AID and ΔE5 both produced PCR bands, which were not present in B cells infected with empty vector (Fig. 5F). However, sequence analysis showed that every band from AID-expressing cells was a canonical Igh-cMyc translocation, whereas each band from ΔE5 cells contained Sμ rearrangements involving inversions, which allowed amplification with the Igh primer at both ends (Fig. 5G).
Together with the very low CSR efficiency shown by ΔE5 mutants (Fig. 4C), these results suggested that E5 was required for productive end joining during CSR as well as chromosomal translocations and that E5 absence might cause altered DNA end processing.
AID E5 Prevents DNA End Resection.
We wanted to better define the nature of the DNA processing defect in the absence of E5. The use of longer microhomologies at the switch joins in AD HIGM2 patients and ΔE5 cells as well as the reduced occupancy of histone-associated factors 53BP1 and γH2AX at the Sμ suggested the possibility of longer than normal end resection. We took advantage of the recent finding that end resection during CSR can be detected by the accumulation of the single strand DNA-binding replication protein RPA A (RPA) (39). ChIP experiments showed a large increase in RPA occupancy at the Sμ of Aicda−/− B cells expressing ΔE5 compared with AID (Fig. 6A). Additional evidence of extensive end resection was provided by the simultaneous increase in the recruitment of EXOI and RAD51 to Sμ in B cells expressing ΔE5 but not in those cells complemented with AID (Fig. 6B). EXOI is an exonuclease involved in mismatch as well as homology-directed repair, and RAD51 is a core component of HR, which decorates long ssDNA stretches in S/G2 (39, 40). Confirming that longer end resection was a common characteristic of CSR-deficient and dominant-negative AID variants, R190X and/or L198S also brought higher levels of RPA and EXOI to the Sμ than AID, whereas the hyperactive variant m7.3, which has an intact E5 and is CSR-proficient, did not (Fig. 6 A and B). Conversely, γH2AX was found at the Sμ of cells expressing AID and m7.3 but not R190X or L198S (Fig. 6C).
We asked whether extended end resection could explain the CSR deficiency of AID variants with truncated E5. We expected that preventing end resection would increase the efficiency of CSR by ΔE5. We chose to target CtIP, because it has a role in HR repair (35, 40) and mediates end resection at the S regions in B cells (37). We used CH12 cells in which we could deplete AID together with CtIP using shRNAs and analyzed CSR to IgA after complementation with either AID or ΔE5 (Fig. S7 A and B). CtIP depletion improved relative switching to IgA by ΔE5 by approximately threefold, although it did not significantly affect the performance of AID (Fig. 6D). As expected, depletion of Ku70 decreased CSR by both variants (Fig. 6D and Fig. S7C). ChIP analyses confirmed CtIP Sμ occupancy in CH12 cells complemented with ΔE5, even if less than in AID cells, which disappeared in CtIP-depleted cells (Fig. 6E and Fig. S7D). As in primary B cells, RPA and RAD51 Sμ occupancy increased in ΔE5 cells, but importantly, this increase was prevented by CtIP depletion, which would be expected from inhibiting end resection (Fig. 6E and Fig. S7D). CtIP depletion did not rescue CSR to the same levels achieved by AID (Fig. S7C), which may be related to the nonredundant role of CtIP in CSR in this particular cell line (36, 38) or could indicate additional roles of E5 (Discussion). In any case, end resection at least in part explained the CSR defect of ΔE5.
We conclude that the E5 domain of AID is required to prevent end resection of the DNA breaks during CSR and thereby promotes end joining. All together, our data suggest a model to explain why E5 truncation eliminates CSR and at the same time, confers dominant-negative behavior, which was observed in AD HIGM2 patients (Fig. 6F).
Discussion
Here, we propose a molecular model to explain the AD HIGM2 syndrome caused by AID variants bearing short C-terminal truncations. Part of the explanation is that a C-terminal domain of AID is required to prevent end resection and promote end joining during CSR, which explains why this region is required for CSR (11, 12). We define the relevant AID domain as the 17 aa exactly encoded by AICDA E5. Indeed, the AD variants R190X and V186X bear truncations in E5, and full deletion of E5 not only eliminates CSR capacity but also, mimics the dominant-negative phenotype of the AD variants, whereas any additional deletion kills enzymatic activity and dominant-negative capacity. Interestingly, E5 is well-conserved down to bony fishes, whereas CSR is first found in amphibians. The fact that fish AID is capable of inducing robust CSR when expressed in mouse B cells (41) suggests that the link of AID to DNA repair reflects some ancestral function that was later exploited for CSR when the Igh locus acquired the translocon configuration with S regions. Promoting DNA repair might be that function, because E5 is also necessary for Igh-cMyc chromosomal translocations.
AID variants R190X, V186X, ΔE5, and L198S (ΔE5 variants) are all CSR-deficient and dominant negatives. We propose that both characteristics are explained by the loss of a motif within E5 that drastically influences DNA processing and repair. E5 also acts as a self-inhibitory domain for deamination, at least in E. coli and B cells. However, the artificial AID variant m7.3 shows that, although exacerbated activity impairs B-cell expansion, it is insufficient to cause CSR deficiency or dominant-negative effect. However, the dominant-negative effect requires intact catalytic activity, which is unusual. Our results would suggest that damage and repair during CSR are closely linked in time, and therefore, failure to engage productive repair soon after deamination by ΔE5 variants not only precludes CSR but in some way, can also compromise CSR by WT AID.
UNG is less abundant at the Ig genes in the wake of ΔE5 variants, leading to a higher proportion of transition mutations at C:G at the Sμ and IgV (11, 13, 15, 42) (this work). However, despite the importance of UNG in generating DSBs for CSR (3), breaks have been found to happen with similar frequency at the Sμ of murine B cells expressing ΔE5 and R190X compared with AID (13, 14) as well as in AICDAR190X/+ vs. AICDA+/+ human B cells (15). Because we see ∼50% reduction in Sμ occupancy by UNG after ΔE5, it may be that UNG is not limiting, which was suggested by the normal CSR Ung haploinsufficient mice and B cells (43). Alternatively, mismatch repair enzymes could compensate for UNG (26), although whether ΔE5 recruits more or less MSH2 to the Sμ is unclear (13, 42). In any case, a partial requirement for E5 to recruit UNG does not prevent DSBs, and it does not explain why this domain is required for CSR. However, we cannot exclude the possibility that the combination of hyperactivity with reduced UNG recruitment might alter the quality of the DNA breaks made by ΔE5 variants, making those breaks more prone to end resection.
The E5 domain dramatically impinges on the processing and repair of DNA breaks at the Igh during CSR. We find little γH2AX, 53BP1, ATM, Nbs1, or Ku70 at the Sμ in primary B cells expressing ΔE5 variants. Similar findings for Ku80, Xrcc4, and DNA-PKcs in CH12 cells were reported while this work was under review (42). Thus, CSR deficiency could be explained, in part, by the lack of recruitment of C-NHEJ core factors. The use of microhomologies at the residual S-S junctions in mouse B cells expressing ΔE5 or in AD HIGM2 patients (34) would suggest preferential use of the A-EJ in these cells. However, B cells lacking core C-NHEJ components switch to IgG1 with considerably higher efficiency (40–50% compared with WT B cells) (6, 33) than B cells overexpressing the ΔE5 variants. Thus, inactivating E5 must reduce the overall efficiency of end joining rather than shifting from C-NHEJ to A-EJ. The inability of ΔE5 to mediate Igh-cMyc translocations would support this proposition.
AID (overexpressed or endogenous) makes DSBs at multiple sites in activated B cells (39, 44) that are not visible as DNA damage foci, except in cells deficient in H2AX, DNA-PKcs, or 53BP1 (39, 45). The reduced recruitment of DNA damage response and end-joining factors downstream from ΔE5 variants is likely to cause persistent DNA damage (4). Accordingly, all CSR-deficient and dominant-negative AID variants, but not the equally hyperactive but CSR-proficient variant m7.3, produce widespread DNA damage foci. This observation seems at odds with the absence of γH2AX at the Sμ. However, it can readily be explained by preferential targeting to the Ig loci. The ΔE5 variants would target the Sμ more frequently than any other genomic site, which is shown by the high deamination frequency at the Sμ, thus resulting in a higher density of breaks. This enhanced frequency, perhaps also impinging on the quality of the DSBs, would increase the probability of extensive end resection, which would displace nucleosomes and reduce γH2AX and histone binding factors. Importantly, a proportion of the genomic breaks made by ΔE5 variants can still engage NHEJ, as shown by their residual CSR capacity. This possibility would also exist at other genomic sites, perhaps more often, because end resection would happen less frequently, and those breaks would trigger a normal DNA damage response and be visible as γH2AX foci. Because CSR breaks are made in G1 (5), the fact that we detect typical S/G2-phase events, like extensive end resection and RAD51 deposition at the Sμ, support our proposition that ΔE5 variants produce DNA breaks that persists in time. Thus, WT cells expressing ΔE5 variants resemble the phenotype of 53BP1- or H2AX-deficient cells, in which AID-induced breaks are resected, producing long ssDNA extensions decorated with RPA and RAD51 in S/G2 (39). Such end resection can explain the increase in microhomologies in the residual CSR, but it would more often prevent CSR by causing crippling rearrangements at the S regions, as we detect in the Igh-cMyc fusion assay, or through error-free HR repair. Indeed, reducing resection by depleting CtIP rescues CSR by ΔE5 in CH12 cells. The effect is substantial but partial, maybe because CtIP is required for maximum CSR levels specifically in CH12 cells (36, 38). Alternatively, AID could also promote end joining by directly recruiting NHEJ. This function of AID would not contradict the fact that I-SceI, which makes homogeneous DSBs, can underpin CSR from engineered S regions (16). Just as 53BP1 is required for normal CSR but dispensable for I-SceI–mediated CSR (46), more complex AID-induced breaks may require AID postcleavage functions. This possibility is reminiscent of the function of RAG in guiding DNA ends to C-NHEJ during VDJ recombination. However, the RAG2 PHD domain inhibits HR as well as A-EJ (9) and prevents chromosomal translocations (47), whereas AID E5 is required for both CSR and translocations, thus suggesting a different mechanism.
Several nonmutually exclusive mechanisms could explain the importance of E5. For example, AID could either recruit scaffold proteins (48) or directly act as a scaffold for DNA end protection or repair factors, with E5 mediating these interactions either autonomously or as part of a larger conformational domain. The interaction of AID with such factors has been reported, although the role of E5 in mediating the interaction is unclear (45, 49, 50). E5 might also be necessary to stabilize the interaction of AID with chromatin. Indeed, others have shown reduced ΔE5 occupancy of the Sμ vs. AID by ChIP (13). Because ΔE5 profusely deaminates S regions, this result probably reflects high turnover rather than impaired association. Unstable AID-chromatin association could hinder the initial steps of the organization of a repair complex or fail to activate some checkpoint, thus allowing the break to persist into S/G2 and/or be resected. A defect in S-S synapsing has been proposed for ΔE5 variants using chromosome conformation capture assays (42). Although we cannot rule out this possibility, end resection could also explain reduced signal in those assays. Finally, as mentioned, by modifying AID activity, E5 could impinge on the quality of DSBs to reduce the chances of end resection.
Whatever the mechanism, our data suggests a model to explain the dominance of the R190X and V186X variants in vivo and the specific deficiency in CSR but not SHM of AD HIGM2. On one side, rearrangements at the Sμ or error-free HR repair could both hinder CSR and probably account for the dominant-negative effect on CSR in vitro. On the other side, alternative explanations for the dominant-negative behavior, such as competition for the substrate, titration of CSR-specific factors, or destabilization of AID multimers by truncated mutants, would not explain the dependence on enzymatic activity. In vivo, the effect would be magnified by compromising B-cell clonal expansion. The cytotoxicity of these hyperactive variants would eliminate many of the B cells attempting CSR during the germinal center reaction, which would also explain the lack of lymphadenopathies in AD patients and perhaps the less severe hyper-IgM compared to AR patients (Table 1). AD AID mutants would affect SHM much less, because DSBs are not necessary intermediates in the mechanism, and therefore, cell death and HR would be much less frequent. Accordingly, SHM is normal in most AD HIGM2 patients (15).
In conclusion, we identify and provide a strong basis to investigate a mechanism by which AID, through its E5 domain, coordinates DNA damage with repair by preventing end resection and promoting end joining to avoid nonproductive or fatal outcomes during CSR.
Materials and Methods
Animals.
C57BL6/J WT (Jackson Laboratories), Aicda−/− (a gift from Tasuku Honjo, Kyoto University, Kyoto), and Aicda−/− Ung−/− mice (Ung−/− mice were a gift from Hans Krokan, Norwegian University of Science and Technology, Trondheim, Norway) were bred at the specific pathogens-free facility of Institut de Recherches Cliniques de Montréal. The Institut de Recherches Cliniques de Montréal animal protection committee (following Canadian Council for Animal Care guidelines) approved animal procedures.
DNA Constructs.
Retroviral vector pMXs AID-ires-GFP has been described (21). AID variant m7.3 was a gift from Michael Neuberger (Medical Research Council, Laboratory of Molecular Biology, Cambridge, United Kingdom), and P20 was a gift from T. Honjo. All AID variants were constructed by PCR amplification with ad hoc oligonucleotides or generated by site-directed mutagenesis by the quick-change protocol using recombinant Thermococcus kodakaraensis KOD1 DNA polymerase (Toyobo Inc.). R190X, ΔE5, and L198S were cloned as BamHI-XhoI fragments and V186X was cloned as a BamHI-EcoRI fragment into pMXs-ires-GFP. Oligonucleotide sequences are available on request.
Cell Culture and Transduction.
CH12 cells, Plat-E, primary B lymphocytes, and K562 cells were cultured in RPMI (Wisent), 1% penicillin/streptomycin (Wisent), and 0.1 mM 2-mercaptoethanol (Bioshop) at 37 °C with 5% (vol/vol) CO2. DT40 Aicda−/− ΔΨVλ cells were supplemented with 1% chicken serum (Wisent). For retroviral infection of DT40, VSV-G, MLV gag-pol, and pMXs AID-ires-GFP vectors (1:1:4 ratio, 2.5 μg DNA total) were transfected into HEK293 cells using Trans-IT LT-1 (Mirus Bio). One milliliter HEK293 supernatant was used 48 h posttransfection to infect 106 DT40 cells with 8 μg/mL polybrene in 24-well plates by spinning at 600 × g for 60 min at 37 °C. Medium was replaced 4 h later. CSR and lentiviral transduction of CH12 cells has been described (38). Lentiviral vector pLKO.1 neomycin bearing an shRNA against mouse AID (GCGAGATGCATTTCGTATGTT) was selected with 250 μg/mL G418. These cells were subsequently transduced with shRNA against Ku70 (AGCTCAGAAGCCCAGCCACTT) or CtIP (ATCCGACAGCAGAACCTTAAG) in pLKO.1 vector and selected in 1 μg/mL puromycin. Protein depletion was confirmed by Western blot, and each population was then transduced using pMXs-human AID or ΔE5-ires-GFP.
CSR and Dominant-Negative Assays in Primary B Cells.
CSR was assayed by complementing naïve splenic B cells from Aicda−/− or Aicda−/− Ung−/− mice through retroviral infection with pMXs AID-ires-GFP vectors using Plat-E ecotropic packaging cells as described (21). For dominant-negative assays, the B cells were purified from either C57BL6/J or Aicda+/− mice, activated with LPS, and infected 22–36 h later. To induce CSR to IgG1, the medium was supplemented with 20 ng/mL mrIL-4 (PreProtech) 4 h postinfection. To induce CSR to IgG2b, 1 ng/mL TGF-β1 was added 24 h postinfection. CSR efficiency in the uninfected (GFP−) and infected (GFP+) subpopulations was measured by flow cytometry using biotinylated anti-IgG1 or -IgG2b (BD) followed by anti–biotin-allophycocyanin (Miltenyi Biotech) and propidium iodide to exclude dead cells.
Western Blots.
The relative expression of untagged truncated and other AID variants was analyzed using a 1:1 mixture of mAbs 52–1 and 39–1 specific for human AID N terminus (a gift from Michael Neuberger). Endogenous mouse AID expression in B cells and depletion in CH12 cells were measured using a rabbit polyclonal raised against mouse AID C-terminal 14 residues (a gift from Jayanta Chaudhuri, Sloan–Kettering Institute, New York). Blots were developed by anti-mouse– or anti-rabbit–AlexaFluor680 and read in an Odyssey CLx apparatus (Li-COR).
Deaminase Activity and DNA Binding Assays.
E. coli mutation assays were performed using the Δung BW310 strain as described (24). AID variants were subcloned as NheI-NotI fragments from pEGFP-N3 into pTrc23-24 (21) to express AID-GFP fusions or as BamHI-XhoI in pTrcHisA (Invitrogen) to express 6XHis-AID fusions after Isopropyl β-D-1-thiogalactopyranoside induction. Mutation frequencies were calculated as the median number of cfu that survived rifampicin selection per 109 ampicillin-resistant cells from at least two experiments with five independent cultures per construct. For biochemical assays, AID variants were cloned into pGEX-5x-3 (GE Healthcare) to express GST-AID fusions and purified as described (51). An end-labeled bubble substrate containing a 7-nt-long single-stranded region with the motif TGC, previously described to be an optimal AID substrate, was used in activity assays and EMSAs (52). For EMSA, 0.015–5 nM substrate was incubated with 0.1–0.25 μg GST-AID in binding buffer (50 mM Tris, pH 7.5, 2 μM MgCl, 50 mM NaCl, 1 mM DTT) in a final volume of 10 μL for 60 min at 37 °C. Samples were electrophoresed at 4 °C on an 8% acrylamide native gel. Enzyme activity was measured using the alkaline cleavage assay as described (52). Alkaline cleavage and EMSA gels were visualized using a PhosphorImager (Bio-Rad). Densitometry was performed using Quantity One 1-D Analysis Software (Bio-Rad). Data were graphed using GraphPad Prism to derive estimated Km and Kd values.
Monitoring Mutation in Eukaryotic Cells.
Ig SHM activity was measured in IgM+ DT40 ΔψVL Aicda−/− cells (a gift from Jean-Marie Buerstedde, Yale University, New Haven, CT) (25) complemented by retroviral infection. For fluctuation analysis of IgM phenotype, 200 GFP+ IgM+ cells were FACS-sorted into 96-well plates and expanded for 3 wk in 24-well plates before analyzing the proportion of GFP+ IgM− cells by flow cytometry and staining with anti-chicken IgM-RPE (Southern Biotech). The IgVλ from DT40 cells was amplified from sorted IgM− cells and analyzed as described (28). S-region mutations were analyzed in Aicda−/− Ung−/− mouse B cells retrovirally complemented with pMX-AID-ires-GFP vectors at 4 d postinfection using KOD1 DNA polymerase to amplify a 607-bp fragment from the Sμ with primers OJ353 (GTAAGGAGGGACCCAGGCTAAG) and OJ354 (CAGTCCAGTGTAGGCAGTAGA) and a 1,805-bp fragment from the Sγ1 with primers OJ300 (CTTCCTACCTTCTCCCCTGAGTCTCAA) and OJ301 (CACCTGGATCAGTTTCTCTGTGACTGC). PCR products were sequenced at Macrogen. The distribution, frequency, and type of mutations were computed after removing clonal mutations from the database. Imatinib resistance assays to select for mutations in the endogenous BCR-ABL1 gene of K562 cells were performed as previously described (28).
DNA Damage Foci.
Transiently transfected HeLa cells were fixed in 3.7% (mass/vol) paraformaldehyde for 10 min, permeabilized with 0.5% Triton-X for 10 min, blocked for 1 h in 5% goat serum, incubated overnight at 4 °C with antiphospho-H2AX (Ser139; 1:1,000; 05–636; Millipore) in blocking buffer, washed, and incubated for 1 h with AlexaFluor 680 goat anti-mouse IgG (1:500; A-21057; Molecular Probes) in blocking buffer. DNA was stained with propidium iodide or DAPI before mounting in Aqua Mount (Thermo Scientific). Multiple random fields were imaged using an LSM700 confocal microscope (Zeiss) at 63×, and the number of foci in transfected (GFP+) cells was blindly scored.
Patient Data.
Clinical, serological, in vitro CSR, and SHM data from HIGM2 patients were compiled from previous publications (15, 17–19). The presence of lymphadenopaties for all patients was determined by physical examination and histological examination in one case.
Chromosomal Translocation Assays.
Aicda−/− splenic B cells stimulated with LPS + IL-4 were transduced two times on consecutive days using pMXs AID or ΔE5-ires-GFP vectors. GFP+ cells were sorted 72 h after the first infection. The fusion between chromosomes 12 and 15 juxtaposing Igh and cMyc was detected by PCR using the Expand Long Template Kit (Roche) following a published protocol (7). PCR products were visualized by ethidium bromide, and the presence of Igh and cMyc was determined by cloning and sequencing each product.
CSR Junction Analysis.
Sμ-Sγ1 joins were amplified from retrovirally complemented splenic B cells using 100 or 450 ng template DNA for AID or ΔE5, respectively, with Expand Long Template PCR system with Buffer 1 (Roche) and oligonucleotides OJ523 (GCTTGAGCCAAAATGAAGTAGACT) and OJ524 (CCCCATCCTGTCACCTATA). Sμ-Sα joins from Ch12 cells were analyzed as described (38). Cycling conditions were 2 min at 94 °C + (10 s at 94 °C, 30 s at 54 °C, and 4 min at 68 °C) × 10 cycles + (10 s at 94 °C, 30 s at 54 °C, and 4 min + 20 s per cycle at 68 °C) × 25 cycles + 7 min at 68 °C. PCR products between 500 and 1,000 bp were cloned in pGEMT-easy (Promega) and sequenced at Macrogen.
ChIP.
Naïve Aicda−/− primary mouse B cells were stimulated with 10 μg/mL LPS and 50 ng/mL mIL-4 and retrovirally transduced 24 h later using pMXs AID variants-ires-GFP vectors or the empty vector as control. Cells were harvested at 18–20 h postinfection when GFP+ cell proportions were 30–50%. ChIP procedures have been described in detail (38). Briefly, cells fixed with 1% formaldehyde were lysed in RIPA buffer and sonicated to generate DNA fragments <500 bp. Lysate fractions of 0.5 mg (2 μg/μL) were precleared with G protein-Sepharose slurry before adding 2–5 μg antibody, incubated overnight at 4 °C, and DNA-purified. Antibodies were from Santa Cruz Biotechnology (anti-CtIP, sc-5970; anti-Ku70, sc-1486; anti-RAD51, sc-8349), Sigma (anti-ATM, A1106; anti-UNG, SAB1406569), Millipore (anti-Nbs1, 04–236; anti-RPA, NA19L; anti-γH2AX, 05–636), and Novus Biologicals (anti-EXO1, NBP1-19709; anti-53BP1, NB100-304). Immunoprecipitated DNA was used as template in real-time PCR reactions containing 1× SYBR Green Mix (Applied Biosystems), 1/10 fraction ChIP-enriched DNA, and 100 nM primers. Primers against C57BL/6J Igh and Gapdh were designed to obtain amplicons Eμ (78 bp), Sμ-U (57 bp), Sμ-D (64 bp), and Gapdh (116 bp). Plates were read in an Applied Biosystems StepOnePlus instrument. Standard curves with different amounts of the input extracts were run in each plate for each individual primer and used to calculate input percentage. The input percentage of the IgG control immunoprecipitation was subtracted from each sample to calculate the values shown in the figures.
Statistics.
Either one-way ANOVA or nonparametric Kruskal–Wallis test, depending on the assumption of normalcy, with appropriate posttests was performed to compare multiple groups. Two-group comparisons were performed by unpaired two-tailed Student t tests. A statistical power of α < 0.5 was considered significant.
Supplementary Material
Acknowledgments
We thank Drs. H. Krokan, T. Honjo, J. M. Buerstedde, J. Chaudhuri, and Michael Neuberger for reagents. Michael’s friendship will be sorely missed. We thank J. Stavnezer and A. Ramiro for advice and D. Durocher, S. Petersen-Mahrt, and D. Muñoz for critical reading. We thank Drs. S. Nonoyama, P. Lane, H. Aarnoud, and S. Kilic for patient care. This work was supported by Canadian Institutes of Health Research (CIHR) Grants MOP130535 (to J.M.D.N.) and MOP111132 (to M.L.), an Institut National de la Santé et de la Recherche Médicale grant (to A.D.), European Union’s 7th Research and Technological Development Framework Programme Grants EURO-PADnet 201549 (to A.D.) and ERC PIDIMMUNE 249816 (to A.D.), and the Stanley J. Glaser Foundation Research Award (to R.E.V.). We acknowledge salary support from the Cole Foundation Fellowship (to S.P.M.), the CIHR New Investigator Award (to M.L.), and the Canada Research Chairs Program (to J.M.D.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320486111/-/DCSupplemental.
References
- 1.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381(6585):751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 2.Peled JU, et al. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 3.Stavnezer J, Guikema JEJ, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniel JA, Nussenzweig A. The AID-induced DNA damage response in chromatin. Mol Cell. 2013;50(3):309–321. doi: 10.1016/j.molcel.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petersen S, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414(6864):660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 7.Ramiro AR, et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118(4):431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117(2):171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 9.Corneo B, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449(7161):483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 10.Wuerffel R, et al. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;27(5):711–722. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barreto V, Reina-San-Martin BR, Ramiro AR, McBride KM, Nussenzweig MC. C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol Cell. 2003;12(2):501–508. doi: 10.1016/s1097-2765(03)00309-5. [DOI] [PubMed] [Google Scholar]
- 12.Ta V-T, et al. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat Immunol. 2003;4(9):843–848. doi: 10.1038/ni964. [DOI] [PubMed] [Google Scholar]
- 13.Ranjit S, et al. AID binds cooperatively with UNG and Msh2-Msh6 to Ig switch regions dependent upon the AID C terminus. J Immunol. 2011;187(5):2464–2475. doi: 10.4049/jimmunol.1101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doi T, et al. The C-terminal region of activation-induced cytidine deaminase is responsible for a recombination function other than DNA cleavage in class switch recombination. Proc Natl Acad Sci USA. 2009;106(8):2758–2763. doi: 10.1073/pnas.0813253106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imai K, et al. Analysis of class switch recombination and somatic hypermutation in patients affected with autosomal dominant hyper-IgM syndrome type 2. Clin Immunol. 2005;115(3):277–285. doi: 10.1016/j.clim.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Zarrin AA, et al. Antibody class switching mediated by yeast endonuclease-generated DNA breaks. Science. 2007;315(5810):377–381. doi: 10.1126/science.1136386. [DOI] [PubMed] [Google Scholar]
- 17.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102(5):565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 18.Quartier P, et al. Clinical, immunologic and genetic analysis of 29 patients with autosomal recessive hyper-IgM syndrome due to Activation-Induced Cytidine Deaminase deficiency. Clin Immunol. 2004;110(1):22–29. doi: 10.1016/j.clim.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Durandy AH, Taubenheim N, Péron S, Fischer A. Pathophysiology of B-cell intrinsic immunoglobulin class switch recombination deficiencies. Adv Immunol. 2007;94:275–306. doi: 10.1016/S0065-2776(06)94009-7. [DOI] [PubMed] [Google Scholar]
- 20.Mu Y, Prochnow C, Pham P, Chen XS, Goodman MF. A structural basis for the biochemical behavior of activation-induced deoxycytidine deaminase class-switch recombination-defective hyper-IgM-2 mutants. J Biol Chem. 2012;287(33):28007–28016. doi: 10.1074/jbc.M112.370189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patenaude A-M, et al. Active nuclear import and cytoplasmic retention of activation-induced deaminase. Nat Struct Mol Biol. 2009;16(5):517–527. doi: 10.1038/nsmb.1598. [DOI] [PubMed] [Google Scholar]
- 22.Uchimura Y, Barton LF, Rada C, Neuberger MS. REG-γ associates with and modulates the abundance of nuclear activation-induced deaminase. J Exp Med. 2011;208(12):2385–2391. doi: 10.1084/jem.20110856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohli RM, et al. A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. J Biol Chem. 2009;284(34):22898–22904. doi: 10.1074/jbc.M109.025536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418(6893):99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 25.Arakawa H, Saribasak H, Buerstedde J-M. Activation-induced cytidine deaminase initiates immunoglobulin gene conversion and hypermutation by a common intermediate. PLoS Biol. 2004;2(7):E179. doi: 10.1371/journal.pbio.0020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell. 2004;16(2):163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Klemm L, et al. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer Cell. 2009;16(3):232–245. doi: 10.1016/j.ccr.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orthwein A, et al. Regulation of activation-induced deaminase stability and antibody gene diversification by Hsp90. J Exp Med. 2010;207(12):2751–2765. doi: 10.1084/jem.20101321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaheen A, et al. AID constrains germinal center size by rendering B cells susceptible to apoptosis. Blood. 2009;114(3):547–554. doi: 10.1182/blood-2009-03-211763. [DOI] [PubMed] [Google Scholar]
- 30.Ouyang L, et al. Programmed cell death pathways in cancer: A review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45(6):487–498. doi: 10.1111/j.1365-2184.2012.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang M, Yang Z, Rada C, Neuberger MS. AID upmutants isolated using a high-throughput screen highlight the immunity/cancer balance limiting DNA deaminase activity. Nat Struct Mol Biol. 2009;16(7):769–776. doi: 10.1038/nsmb.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manis JP, et al. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5(5):481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 33.Boboila C, et al. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J Exp Med. 2010;207(2):417–427. doi: 10.1084/jem.20092449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kracker S, et al. Impaired induction of DNA lesions during immunoglobulin class-switch recombination in humans influences end-joining repair. Proc Natl Acad Sci USA. 2010;107(51):22225–22230. doi: 10.1073/pnas.1012591108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450(7169):509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee-Theilen M, Matthews AJ, Kelly D, Zheng S, Chaudhuri J. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat Struct Mol Biol. 2011;18(1):75–79. doi: 10.1038/nsmb.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bothmer A, et al. Mechanism of DNA resection during intrachromosomal recombination and immunoglobulin class switching. J Exp Med. 2013;210(1):115–123. doi: 10.1084/jem.20121975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cortizas EM, et al. Alternative end-joining and classical nonhomologous end-joining pathways repair different types of double-strand breaks during class-switch recombination. J Immunol. 2013;191(11):5751–5763. doi: 10.4049/jimmunol.1301300. [DOI] [PubMed] [Google Scholar]
- 39.Yamane A, et al. RPA accumulation during class switch recombination represents 5′-3′ DNA-end resection during the S-G2/M phase of the cell cycle. Cell Rep. 2013;3(1):138–147. doi: 10.1016/j.celrep.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11(3):196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barreto VM, et al. AID from bony fish catalyzes class switch recombination. J Exp Med. 2005;202(6):733–738. doi: 10.1084/jem.20051378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabouri S, et al. C-terminal region of activation-induced cytidine deaminase (AID) is required for efficient class switch recombination and gene conversion. Proc Natl Acad Sci USA. 2014 doi: 10.1073/pnas.1324057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zahn A, et al. Separation of function between isotype switching and affinity maturation in vivo during acute immune responses and circulating autoantibodies in UNG-deficient mice. J Immunol. 2013;190(12):5949–5960. doi: 10.4049/jimmunol.1202711. [DOI] [PubMed] [Google Scholar]
- 44.Staszewski O, et al. Activation-induced cytidine deaminase induces reproducible DNA breaks at many non-Ig Loci in activated B cells. Mol Cell. 2011;41(2):232–242. doi: 10.1016/j.molcel.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu X, Geraldes P, Platt JL, Cascalho M. The double-edged sword of activation-induced cytidine deaminase. J Immunol. 2005;174(2):934–941. doi: 10.4049/jimmunol.174.2.934. [DOI] [PubMed] [Google Scholar]
- 46.Bothmer A, et al. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med. 2010;207(4):855–865. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deriano L, et al. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature. 2011;471(7336):119–123. doi: 10.1038/nature09755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Z, et al. 14-3-3 adaptor proteins recruit AID to 5′-AGCT-3′-rich switch regions for class switch recombination. Nat Struct Mol Biol. 2010;17(9):1124–1135. doi: 10.1038/nsmb.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reina-San-Martin BR, Chaudhuri J. In: DNA Deamination and the Immune System: AID in Health and Disease. Fugmann SD, Diaz M, Papavasiliou FN, editors. London: Imperial College Press; 2010. pp. 62–82. [Google Scholar]
- 50.Vuong BQ, et al. A DNA break- and phosphorylation-dependent positive feedback loop promotes immunoglobulin class-switch recombination. Nat Immunol. 2013;14(11):1183–1189. doi: 10.1038/ni.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dancyger AM, et al. Differences in the enzymatic efficiency of human and bony fish AID are mediated by a single residue in the C terminus modulating single-stranded DNA binding. FASEB J. 2012;26(4):1517–1525. doi: 10.1096/fj.11-198135. [DOI] [PubMed] [Google Scholar]
- 52.Larijani M, et al. AID associates with single-stranded DNA with high affinity and a long complex half-life in a sequence-independent manner. Mol Cell Biol. 2007;27(1):20–30. doi: 10.1128/MCB.00824-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.