Significance
Emerging evidence indicates that posttranslational hydroxylation of intracellularly localized proteins is more prevalent than once thought. We identify Drosophila melanogaster sudestada1 (sud1) as a gene that is needed for normal growth in the fly and show that sud1 encodes a prolyl-hydroxylase that catalyzes posttranslational hydroxylation of a conserved residue in the small ribosomal subunit protein RPS23. Knockdown of Sud1 results in growth impairment and reduced RPS23 hydroxylation, which is associated with activation of the unfolded protein response, induction of apoptosis, and increased autophagy. Together with findings in humans and yeast reported in the companion articles, the work reveals a new type of posttranslational ribosome modification that is highly conserved in eukaryotes.
Keywords: fruit fly, ribosome, dioxygenase, proline, tranlational stress
Abstract
Genome sequences predict the presence of many 2-oxoglutarate (2OG)-dependent oxygenases of unknown biochemical and biological functions in Drosophila. Ribosomal protein hydroxylation is emerging as an important 2OG oxygenase catalyzed pathway, but its biological functions are unclear. We report investigations on the function of Sudestada1 (Sud1), a Drosophila ribosomal oxygenase. As with its human and yeast homologs, OGFOD1 and Tpa1p, respectively, we identified Sud1 to catalyze prolyl-hydroxylation of the small ribosomal subunit protein RPS23. Like OGFOD1, Sud1 catalyzes a single prolyl-hydroxylation of RPS23 in contrast to yeast Tpa1p, where Pro-64 dihydroxylation is observed. RNAi-mediated Sud1 knockdown hinders normal growth in different Drosophila tissues. Growth impairment originates from both reduction of cell size and diminution of the number of cells and correlates with impaired translation efficiency and activation of the unfolded protein response in the endoplasmic reticulum. This is accompanied by phosphorylation of eIF2α and concomitant formation of stress granules, as well as promotion of autophagy and apoptosis. These observations, together with those on enzyme homologs described in the companion articles, reveal conserved biochemical and biological roles for a widely distributed ribosomal oxygenase.
Iron [Fe(II)]- and 2-oxoglutarate (2OG)-dependent oxygenases are a superfamily with diverse biochemical and biological functions. During 2OG oxygenase catalysis, substrate oxidation is coupled to decarboxylation of 2OG, yielding succinate and carbon dioxide (1, 2). Structural studies reveal that the catalytic domain of 2OG oxygenases contains a conserved double-stranded β-helix (DSBH) fold presenting an HXD…H facial triad motif that coordinates an Fe(II) cofactor (3, 4). These and other structural features have been used to predict the existence of multiple uncharacterized 2OG oxygenases. In contrast to microorganisms and plants where 2OG oxygenases catalyze a wide variety of oxidative reactions, in animals their biochemical activities appear limited to hydroxylations or demethylations via hydroxylation (1, 5, 6). Despite progress in making biochemical assignments, the physiological roles of most 2OG oxygenases predicted by bioinformatic analysis of animal genomes are unknown. For instance, we have identified ∼50 putative 2OG oxygenases in the Drosophila genome, but only a few are characterized (7, 8).
The function of Fatiga, the single Drosophila homolog of human hypoxia inducible transcription factor (HIF) prolyl-4-hydroxylases (PHDs), has been well studied in the context of oxygen sensing (9). HIF prolyl-hydroxylation plays a central role in the animal hypoxic response via hydroxylation of HIF, a posttranslational modification that signals for HIF-α degradation in a physiologically relevant oxygen-dependent manner (10, 11). Given the tractability of these enzymes as targets for pharmacological modulation by 2OG analogs and related compounds, elucidation of the function of related 2OG oxygenases in biology is an area of current interest (12, 13).
To identify other oxygenase-catalyzed reactions with signaling roles, we have conducted an RNAi-based screen of 2OG oxygenases for phenotypes in Drosophila. We identified CG44254, a highly conserved gene that is distantly related to oxygen sensing PHDs (14), as necessary for normal growth and mRNA translation in the fly. This gene, which we have termed sudestada1 (sud1) after a wind that blows across the southeastern coast of South America, is highly conserved from yeast to humans; homologous genes in Saccharomyces cerevisiae (TPA1) (15, 16), Schizosaccharomyces pombe (Ofd1) (17, 18), and Homo sapiens (OGFOD1) (19, 20) have been implicated in translation termination, oxygen-dependent regulation of the transcription factor Sre1N, and translational stresses responses, respectively.
In independent work, reported in companion articles, we discovered that Tpa1p, Ofd1, and OGFOD1 are protein hydroxylases that catalyze unique di- and monohydroxylations of a conserved prolyl residue in the small ribosomal subunit protein RPS23 (21, 22). Here we describe the biochemical and physiological characterization of Sud1 in Drosophila melanogaster. We show that Sud1 is a dioxygenase that catalyses monohydroxylation of RPS23 in the fly and that its silencing results in growth defects and impairment of mRNA translation, along with the induction of Eukaryotic Initiation Factor 2α (eIF2α) phosphorylation, stress granule formation, autophagy and apoptosis.
Results
Sudestada1 Encodes the Drosophila Tpa1p/OGFOD1 Homolog and Is Required for Normal Growth.
In initial studies, we carried out an RNAi screen in Drosophila to identify 2OG oxygenases that lead to impaired growth after knockdown (Table S1). These studies led to the identification of a potential Ofd1/TPA1/OGFOD1 homolog (CG44254) that we name sudestada1 (sud1). Two transcripts generated by alternative splicing, sud1 and sud2, are reported in databases that curate high-throughput transcriptomic data (http://flybase.org) (Fig. S1A). RT-PCR analyses confirm the expression of both transcripts in larvae (Fig. S1 B and C). Only Sud1 includes a predicted oxygenase domain encompassing residues 171–275 that manifests a substantial degree of identity with Tpa1p/Ofd1/OGFOD1 (Fig. S1 D and E). Sud2 encodes a predicted phosphatidylinositol-glycan biosynthesis class S protein that is apparently unrelated to oxygenases and has not been investigated in this work.
We then analyzed the mRNA expression profile of sud1 through the fly life cycle. Quantitative real-time RT-PCR (qRT-PCR) assays reveal sud1 mRNA expression at all developmental stages, with the highest levels at the first larval instar (Fig. 1A). We then compared expression in third-instar larval tissues and found that sud1 mRNA is highly expressed in the fat body, with significant expression in other organs including the brain, salivary glands, imaginal discs, and gut (Fig. 1B). To study the subcellular localization of Sud1, we generated a Sud1-GFP fusion construct and expressed this using the Gal4-UAS system in different tissues (Fig. 1 C–H); Sud1 localizes predominantly to the nucleus, although lower levels are also detected in the cytoplasm.
Fig. 1.
Expression and subcellular localization of sudestada1. (A) Temporal sud1 expression pattern throughout the fly life cycle, as determined by real-time RT-PCR (qRT-PCR); error bars represent SD. (B) Expression of sud1 mRNA in different organs of third-instar larvae. (C–H) Expression of a UAS-Sud1-GFP fusion construct in wing imaginal discs (C–E) or the fat body (F–H). In both tissues, the protein is mostly nuclear, although some GFP signal can be seen at the cytoplasm (arrows in F).
To investigate Sud1 functions, we first expressed a double-stranded RNA that specifically targets sud1 sequences, without affecting sud2 mRNA levels (Fig. S1 B and C). Given that S. cerevisiae Tpa1p and S. pombe Ofd1 have putative active sites in the N-terminal of their two DSBH domains that possess similarity to PHD2 (15, 23), we tested whether Sud1 plays a role in the HIF-dependent transcriptional response to hypoxia by observing the effects of Sud1 knockdown on HIF/Sima-dependent transcription in the embryonic tracheal system. Embryos expressing sud1 RNAi failed to modulate a HIF-dependent transcriptional reporter, whereas, as expected, embryos that express an RNAi targeting the prolyl-4-hydroxylase gene fatiga displayed strong up-regulation of the same reporter under mild hypoxic conditions, providing a positive control for the assay (Fig. S2 A–D).
Ubiquitous sud1 RNAi expression mediated by an actin-Gal4 driver was lethal at the second larval instar. To enable phenotypic analysis of the effect of Sud1 suppression at later developmental stages, we used more restricted RNAi interventions. Because Sud1 is most expressed in the fat body, we expressed sud1 RNAi in this organ, using the driver pumpless-Gal4 (ppl-Gal4). Restricted silencing allowed development of viable adults. Analysis of the fat body of third-instar larvae of such flies revealed a significant reduction of cell size (Fig. 2 A–C). Because reduction of fat body size can have a systemic effect on body growth (24, 25), we measured pupal size and observed a significant reduction in the volume of individuals expressing sud1 RNAi in the fat body (Fig. 2 D and E). To determine whether Sud1 suppression impairs growth in other fly organs, sud1 RNAi was expressed in the wing imaginal disc. In both the posterior (Fig. 2 F–H) and the dorsal (Fig. S2 E and F) compartments of the disc, sud1 RNAi significantly reduced growth. To verify that the effect of the RNAi is due to Sud1 silencing, the homologous Drosophila willistoni sud1 gene was coexpressed and found to restore the growth defect (Fig. 2H). To test whether Sud1 function is conserved across species, we coexpressed human OGFOD1, together with sud1 RNAi, in the wing posterior compartment. Although incomplete, we again observed restoration of the growth that had been reduced by Sud1 silencing (Fig. 2H), implying that the mammalian and fly dioxygenases are functionally conserved.
Fig. 2.
Sud1 loss of function affects growth. (A–C) Sud1 silencing in the fat body of third-instar larvae provokes cell size reduction. white (control) (A) or sud1 (B) double-stranded RNAs were expressed under control of a pumpless-Gal4 (ppl-Gal4) driver; fat body cells expressing sud1 RNAi are smaller than those of the control as assessed by quantification of the area of cell nuclei (C). n = 3 independent experiments, error bars represent SD, and *P < 0.01 (Student t test). (D and E) As a consequence of ppl-Gal4–driven expression of sud1 RNAi, pupal volume is reduced in comparison with that of a control line expressing a white RNAi. n ≥ 25 in three independent experiments. Error bars represent SD; **P < 0.001 (Student t test). (F–H) Expression of sud1 RNAi in the wing disc posterior compartment results in growth impairment: the area limited by wing veins L4, L5, the posterior cross-vein, and the wing margin (marked in red in F and G) was measured as an indication of the variation of the area of the posterior compartment; (H) area quantification after sud1 RNAi expression, and rescue of the WT wing phenotype after concomitant expression of a Drosophila willistoni sud1 or human OGFOD1 transgenes (DwSud1; OGFOD1); the rescue after expression of human OGFOD1 was partial. n ≥ 30 in three independent experiments. Error bars represent SD (**P < 0.001; one-way ANOVA with Tukey post hoc test). (I–K) Cell size and cell number are both reduced after expression of sud1 dsRNA: wing hair density increases after expression of sud1 RNAi (K), indicating that cell size is reduced. Cell size reduction (15%) accounts only partially for reduction of the area of the wing posterior compartment after sud1 RNAi treatment (21%) (cf. H and K). The remaining area reduction is due to a decreased number of cells in the compartment (6%). n ≥ 10 in three independent experiments. Error bars represent SD (**P < 0.001; Student t test).
We next studied effects on cell size. Because all epidermal cells of the Drosophila wing produce a single cuticular hair, hair density was used to assess cell density and hence to calculate both cell size and cell number in the wing posterior compartment. In comparison with control RNAi, engrailed-Gal4–driven expression of sud1 RNAi induced an increase in cell density (Fig. 2 I–K), indicating that the 21% reduction of the wing posterior compartment (Fig. 2K) arose from both a reduction in cell size (15%) and a reduction in cell number (6%).
Sudestada1 Hydroxylates the Ribosomal Protein RPS23.
It is known that mutations in ribosomal proteins can provoke growth defects in Drosophila, such as those observed after Sud1 knockdown (26, 27). Consistent with this, in refs. 21 and 22, it is shown that the Sud1 homologs in humans and yeast catalyze a posttranslational modification of a protein of the small ribosomal subunit termed RPS23. Whereas human OGFOD1 mediates monohydroxylation of proline 62 of RPS23, yeast Tpa1p/Ofd1 and its green algae homolog catalyze dihydroxylation of the analogous prolyl residue. Sequence alignments reveal that the RPS23 prolyl residue that is subject to Tpa1p/OGFOD1-dependent hydroxylation is conserved in Drosophila (Fig. 3A).
Fig. 3.
Sudestada1 hydroxylates RPS23. (A) Drosophila and human ribosomal protein RPS23 are almost identical. Conserved residues between the two proteins are marked in black; Pro-62 is highlighted in red and marked with an asterisk. (B) Deconvoluted ESI-MS whole protein spectrum of D. melanogaster RPS23 isolated from S2 cell ribosomes purified by sucrose density sedimentation followed by online UPLC mass spectrometry. A mass of 15,901.2 Da is consistent with a +16-Da shift relative to predicted mass of 15,885.4 Da (N-terminal methionine cleaved), indicative of oxidative modification. Note there is no evidence for proline dihydroxylation. (C and D) Hydroxylation of RPS23 is suppressed by sud1 dsRNA. Extracted ion chromatograms of m/z 670.057 and m/z 675.39 corresponding to unhydroxylated (dashed line) and hydroxylated (solid red line) forms of the Pro-62 containing RPS23 peptide GIVLEKVGVEAKQPNSAIR ([M+3H]3+) isolated from S2 cells treated with control (C) or sud1 dsRNA (D) (see Fig S3 A and B for assignment of species). (E) LC-MS/MS analysis of trypsinized GST-RPS23 after coexpression with Sudestada1. MS/MS spectrum of trypsinized RPS23 after coexpression with His6-Sudestada1 in E. coli reveals a peptide, 55-VGVEAKQPNSAIR-67, with a complete series of y-ions demonstrating monohydroxylation (+16 Da) at Pro-62. The b and y fragment ions are indicated (peptide precursor ion: Mr 1,383.760048 Da; calculated 1,383.7470 Da; see Fig. S3E for assignment of species).
We therefore investigated whether Drosophila RPS23 is posttranslationally modified. Ribosomes were purified from Drosophila Schneider2 (S2) cells grown in culture and then subjected to ultra-performance liquid chromatography (UPLC)-coupled intact protein mass spectrometric analysis. A species corresponding to Drosophila RPS23 was observed with a mass +16 Da greater than that predicted from the primary sequence, consistent with addition of a single oxygen atom (Fig. 3B). To investigate the dependence of this modification on Sud1, S2 cells were treated with sud1 RNAi, and LC-MS analyses were performed on Arg-C–digested ribosomal RPS23 preparations. Consistent with the whole-protein MS data (Fig. 3B), a peptidic fragment containing Pro-62 was observed with a mass increment of +16 Da. No unmodified species was observed in material prepared from untreated cells, consistent with RPS23 being monohydroxylated. However, after Sud1 silencing, a second species corresponding to the unhydroxylated parent peptide became apparent (Fig. 3 C and D and Fig. S3 A and B), indicating that Sud1 is necessary for the hydroxylation. To directly investigate whether Sud1 catalyses RPS23 prolyl hydroxylation, we prepared recombinant RPS23 either alone or coexpressed with Sud1 in a His-tagged form in Escherichia coli. A tryptic RPS23 fragment (residues 55–67) revealed a mass shift of +16 Da, which was assigned using MS/MS analyses to Pro-62, implying that Sud1 is the RPS23 prolyl monohydroxylase (Fig. 3E and Fig S3 C–E). In none of the studies on Sud1/RPS23, either on cellular prepared material or when working with purified protein, did we observe evidence of a +32-Da mass shift corresponding to di-hydroxylation of RPS23 as observed for yeast Tpa1p/Ofd1 and green algae (Ostreococcus tauri) otOGFOD1 (21). Taken together, these experiments indicate that Sud1 catalyzes mono-, but not dihydroxylation of RPS23 Pro-62.
Sudestada1 Knockdown Affects Protein Synthesis and Triggers the Unfolded Protein Response.
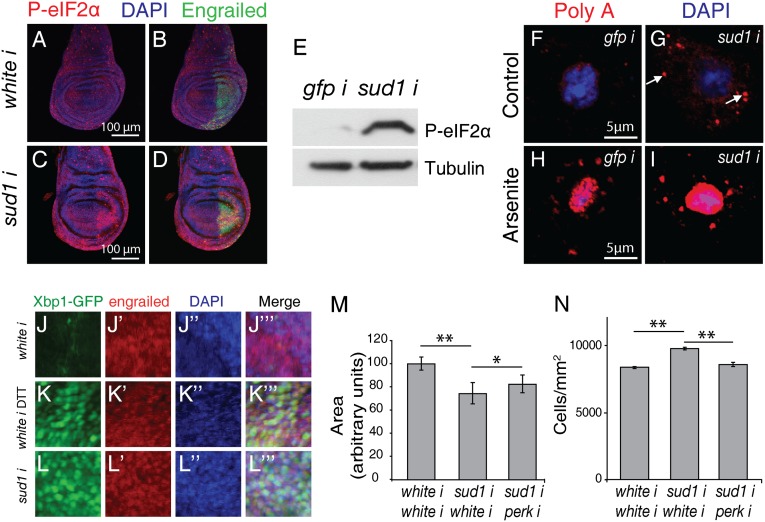
To further investigate the effects of Sud1 knockdown, we analyzed the effects of sud1 RNAi on protein synthesis. Wing imaginal discs, ubiquitously expressing either sud1 or a control dsRNA, were incubated with [14C]-labeled amino acids, and incorporation into protein was measured. A significant decrease of protein synthesis occurred in discs expressing sud1 RNAi in comparison with controls (Fig. S4 A and B). Next, we analyzed the phosphorylation status of the translation initiation factor eIF2α, a key regulatory step in the control of cap-dependent mRNA translation. Sud1 silencing in the posterior wing disc compartment promotes substantial increase in P-eIF2α staining that is clearly limited to the posterior wing disc compartment, whereas in flies expressing control RNAi, P-eIF2α was evenly distributed across the whole disc (Fig. 4 A–D). This effect was specifically dependent on Sud1 silencing, because coexpression of the Drosophila willistoni sud1 transgene largely suppressed the induction of P-eIF2α (Fig. S4 C–F). This effect of sud1 RNAi on eIF2α phosphorylation was further confirmed by immunoblotting of extracts prepared from cultured S2 cells after treatment with sud1 RNAi (Fig. 4E). Because eIF2α phosphorylation induces stress granule formation (28), we analyzed for Sud1-linked formation of stress granules. Extensive stress granule formation was observed in S2 cells treated with sud1 RNAi compared with control RNAi (Fig. 4 F–I and Fig. S4 G and H). Together, these experiments suggest that Sud1 knockdown leads to increased eIF2α phosphorylation, which in turn diminishes protein synthesis and promotes stress granule accumulation.
Fig. 4.
Sudestada1 knockdown affects protein synthesis and leads to activation of the unfolded protein response. (A–D) Phosphorylation of eIF2α is induced at the wing disc posterior compartment after expression of a sud1 RNAi (C and D) but not of a white RNAi (A and B), as revealed by anti–P-eIF2α immunofluorescence (red); anti-Engrailed–positive staining identifies the disc posterior compartment (green) (B and D); DAPI labels cell nuclei (blue). (E) Anti–phospho-eIF2α western blot analysis of S2 cell extracts shows that sud1 but not gfp (control) RNAi treatment promotes eIF2α phosphorylation. (F–I) Sud1 silencing induces stress granules (SGs) formation. S2 cells treated with a sud1 RNAi (G and I) exhibit more stress granules than cells treated with a control gfp RNAi (arrow, F and H), as revealed by polyA FISH. SGs increase after sud1 silencing occurs both in untreated cells (F and G) and in cells exposed to sodium arsenite for 2 h (H and I). (J–L′″) xpb1splicing is activated after sud1 knockdown. An Xbp1-GFP splicing reporter was expressed at the disc posterior compartment through an en-Gal4 driver, and expression was detected with an anti-GFP antibody; a portion of the disc posterior compartment is shown in J–L′″. The reporter is silent in discs expressing a white (control) dsRNA (J) and is activated after DTT treatment (K) or expression of sud1 RNAi (L). In J′, K′, and L′, anti-Engrailed immunostaining confirms the expression of this posterior compartment-specific marker. Wing posterior compartment area (M) and cell size (N) reduction observed after sud1 silencing were partially suppressed by concomitant expression of a perk double-stranded RNA in the same disc compartment. n ≥ 30 wings (M) and n = 10 wings (N) in three independent experiments. Error bars represent SD. One-way ANOVA analysis with Tukey comparison (*P < 0.05, **P < 0.001).
Because eIF2α phosphorylation is triggered by the unfolded protein response (UPR) (29), among other stimuli, we investigated whether the UPR is triggered by Sud1 knockdown. As a marker of UPR induction, we measured splicing of xbp1 mRNA, a transcription factor that induces ER chaperones and that is activated by splicing in response to UPR. We used transgenic flies expressing an Xbp1-GFP reporter that generates an in-frame transcript when splicing of the xbp1 mRNA has occurred (30). This reporter was coexpressed in the wing disc posterior compartment, along with sud1 RNAi or a control RNAi. As expected, xbp1 splicing did not occur with expression of the control RNAi (Fig. 4 J–J′″), but was strongly induced both by exposure of the discs to DTT (DTT is an established inducer of UPR; Fig. 4 K–K′″) and by expression of sud1 RNAi (Fig. 4 L–L′″). Consistent with activation of xbp1 splicing, ubiquitous expression of sud1 RNAi at the first larval instar provoked the up-regulation of the endogenous Xbp1 target gene bip1, as determined by RT-PCR (Fig. S4I). To investigate the extent to which UPR activation and eIF2α phosphorylation account for Sud1-dependent growth impairment, we studied growth of the wing posterior compartment in flies coexpressing the sud1 RNAi simultaneously with an RNAi targeting perk, the Drosophila ortholog of PKR-related ER kinase (PERK). Silencing of PERK partially suppresses the reduction of growth induced by Sud1 knockdown (Fig. 4 M and N), suggesting that UPR activation accounts, at least in part, for the observed growth impairment.
Sudestada1 Knockdown Triggers Autophagy and Apoptosis.
One consequence of UPR activation is induction of autophagy (31). Autophagy affects growth by reducing cell size in various Drosophila tissues (32), so we hypothesized that autophagy contributes to the growth impairment observed with Sud1 silencing. Consistent with this, sud1 RNAi expression in the wing disc posterior compartment provoked induction of autophagy markers in this disc territory, including nucleation of the ATG8-GFP autophagy marker (Fig. 5 A and B); accumulation of the fluorescent lysosomal probe lysotracker (Fig. 5 C and D); and formation of large Lamp1-GFP foci (Fig. 5 E and F). Expression of the D. willistoni sud1 transgene abolished lysosomal dye accumulation at the posterior disc compartment, again indicating that the action of sud1 silencing on autophagy is target specific (Fig. S5 A–C).
Fig. 5.
Autophagy and apoptosis are induced after Sudestada1 knockdown. Expression of a sud1 (B) but not of a white (control) (A) RNAi at the wing disc posterior compartment provokes nucleation of ATG8-GFP expressed at the same compartment, revealing that autophagy was induced. Autophagy induction was confirmed by Lysotracker-positive staining (C and D), as well as by nucleation of Lamp1-GFP (E and F) in cells of the same disc territory. Note in E and F that small Lamp1-GFP foci become bigger after sud1 RNAi treatment. (G–J) Apoptosis is triggered after sud1 silencing: Cells of the posterior wing disc compartment expressing sud1 (H and J) but not those expressing a white (control) RNAi (G and I) become TUNEL positive (the arrows in H show two examples of TUNEL-positive cells). The posterior disc compartment that expresses Engrailed (green staining in I and J) is marked with a dotted line.
Finally, we investigated whether the reduction in the number of cells observed following expression of sud1 RNAi (Fig. 2 I and J) is due to reduced cell proliferation or increased cell death. Phospho-histone3 staining analysis revealed that expression of sud1 dsRNA in the wing disc posterior compartment does not decrease cell proliferation (Fig. S5D), whereas TUNEL staining assays revealed that apoptosis was triggered by sud1 RNAi (Fig. 5 G–J). Reduction of the area of the posterior compartment of the wing following Sud1 silencing was partially suppressed by concomitant expression of the caspase inhibitor p35 (Fig. S5E), confirming that induction of apoptosis accounts in part for growth impairment.
Given that the Target of Rapamicin (TOR) pathway plays a central role in growth regulation in the fat body (32), we analyzed genetic interactions between Sud1 and genes of this pathway in this organ. Interestingly, reduction of function of the TOR pathway led to partial suppression of growth defects provoked by Sud1 silencing (Table S2). These results suggest that slowing down translation, as a consequence of TOR down-regulation (33), alleviates the translational stress provoked by Sud1 knockdown.
Discussion
Ribosomal protein hydroxylation mediated by 2OG oxygenases is emerging as an important evolutionary conserved pathway (34). We analyzed the function of Sudestada1, a Drosophila oxygenase homolog of human OGFOD1, S. cerevisiae Tpa1p, and S. pombe Ofd1. We demonstrated that, like OGFOD1, Sud1 mediates hydroxylation of proline 62 of the small ribosomal subunit protein RPS23. Importantly, Sud1 catalyzes a single prolyl hydroxylation, as observed for human OGFOD1 but contrasting with homologs from lower eukaryotes including Tpa1p in yeast, where RPS23 is di-hydroxylated (21). Thus, RPS23 Pro-62 hydroxylation is a unique and highly conserved ribosomal posttranslational modification, but there appears to be a clear biochemical difference between the extent (i.e., mono- or di-) of RPS23 hydroxylation in animals and lower eukaryotes.
Reduction of Sud1 levels causes growth impairment in various Drosophila tissues, as observed for OGFOD1 in some human-derived cells (22). The growth defects associated with Sud1 silencing correlated with translational stress, characterized by phosphorylation of eIF2α, the formation of stress granules, induction of UPR, and promotion of autophagy and apoptosis. These findings, along with those on the human and yeast homologs described in the accompanying manuscripts (21, 22), reveal a biochemically conserved, but context variable, biological role for Sud1/OGFOD1/Tpa1p in the regulation of growth and stress responses.
In flies, Sud1 suppression induces a strong UPR. Furthermore, partial suppression of the growth defects associated with sud1 RNAi was observed with RNAi directed against perk, a key effector kinase in UPR signaling that targets eIF2α (35). Together, these findings suggest that the UPR is at least in part responsible for activation of stress pathways and impairment of growth related to Sud1. The observed induction of autophagy after sud1 RNAi expression is also consistent with activation of UPR stress pathways. Despite similar effects on growth, eIF2α phosphorylation, and stress granule formation, induction of the UPR was not observed following OGFOD1 inactivation in mammalian cells (see ref. 22), suggesting the operation of additional signaling systems.
It has recently been reported that OGFOD1 is a stress granule component (20). In contrast with our findings in flies, in that report, OGFOD1 knockdown did not induce stress granule formation, suggesting that induction of translational stress is context determined. We observed suppressive effects of sud1 RNAi on growth in both the fat body and wing disc compartments together with consistent effects on the induction of stress pathways in both developing flies and cultured Drosophila cells. By crossing in loss-of-function alleles of the TOR signaling pathway, and presumably reducing the rate of protein synthesis, growth defects provoked by sud1 RNAi were partially suppressed, suggesting that translational stress was alleviated. Together with work in mammalian cells described in the accompanying manuscript (22), these results demonstrate that the activation of stress responses by OGFOD1/Sud1 knockdown occur in a number of settings. Nevertheless, as in mammalian and yeast cells, phenotypic responses to Sud1 suppression vary with context. For instance, the growth suppression was affected by nutritional supplementation, as addition of four times the quantity of yeast to the fly medium largely corrected the impairment in wing growth. Together with the effects in mouse embryonic fibroblasts being enhanced by transformation, it is possible that growth restriction by OGFOD1/Sud1 knockdown is enhanced in settings where there is an imbalance between growth and nutrient supply. We have thus far been unable to determine whether this reflects different effects on RPS23 hydroxylation or the downstream integration of signals on stress pathways.
Although there are differences between the effects of Sud1 and OGFOD1 knockdown in respect of the observed activation of the UPR, our results indicate that aspects of both the biochemical function of OGFOD1/Sud1 as ribosomal oxygenases and cellular functions in translational control and stress are conserved. Nevertheless, in other organisms, notably in the fission yeast S. pombe, the homolog Ofd1 has a defined role as an oxygen sensor in the regulation of nuclear transcription by mediating oxygen-dependent proteolysis of Sre1, the homolog of sterol response element-binding proteins (SREBPs) (17, 18). Whether this response is conserved in flies is unclear. However, our analysis reveals that Sud1, like Tpa1p/OGFOD1, is a predominantly nuclear protein. An interesting possibility that will require exploration in future work is whether Sud1 functions in linking ribosomal signals, either generated at preribosomal stages before cytoplasmic export or from the assembled ribosome, to the regulation of nuclear transcriptional responses.
Currently the precise molecular mechanisms linking the growth/stress phenotypes observed in flies to RPS23 hydroxylation are unclear. This relationship is consistent with other studies in which defects in the production or metabolism of protein/nucleic acid components of the ribosome create cellular stress responses (36, 37). As described in refs. 21 and 22, the site of RPS23 hydroxylation is at the ribosomal decoding site and Tpa1p/OGFOD1 affects translation termination efficiency in yeast and mammalian cells.
In preliminary experiments measuring the effects of Sud1 knockdown on stop codon read through using a transgenic bicistronic reporter in Drosophila, we observed either no change or small increases in read through at different larval stages, indicating that general increases in stop codon readthrough are unlikely to be the sole Sud1-mediated signal activating the UPR. Nevertheless our results do not exclude the possibility that specific coding errors might be the activating signal, and this is the subject of future investigations.
Materials and Methods
Wing Size Measurement.
Wings from 4-d-old females were removed and mounted in a solution containing 1:1 lactic acid/ethanol. Wings were imaged using an Olympus MVX10 stereomicroscope connected to an Olympus DP71 digital camera. The area of the posterior compartment was measured using ImageJ software (National Institutes of Health). To quantify the wing hairs, images were taken using an Olympus BX60 microscope connected to an Olympus DP71 digital camera.
Pupal Size Measurement.
Pupae were photographed using the stereomicroscope as previously described. Pupal width and length were measured using Image J software. Volume was estimated using the following formula: V = πD2(3L − D)/12, where D is the width and L the length of the pupa.
Fat Body Cell Nuclei Measurement.
The area of fat body cell nuclei was measured using the ImageJ Software.
See SI Materials and Methods for additional information.
Supplementary Material
Acknowledgments
We thank Ana Depetris Chauvin for help in statistical analyses, the Vienna Drosophila RNAi Centre (VDRC) and Bloomington Stock Centers, the many colleagues that provided Drosophila lines, the Hybridoma Bank for antibodies, and the Wappner laboratory members for discussion. This work was funded by Wellcome Trust Grant WT087675MA, Biotechnology and Biological Sciences Research Council UK, and Agencia Nacional de Promoción Científica y Tecnológica Grant 2011 N° 0090.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314485111/-/DCSupplemental.
References
- 1.Loenarz C, Schofield CJ. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem Sci. 2011;36(1):7–18. doi: 10.1016/j.tibs.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Ozer A, Bruick RK. Non-heme dioxygenases: Cellular sensors and regulators jelly rolled into one? Nat Chem Biol. 2007;3(3):144–153. doi: 10.1038/nchembio863. [DOI] [PubMed] [Google Scholar]
- 3.McDonough MA, Loenarz C, Chowdhury R, Clifton IJ, Schofield CJ. Structural studies on human 2-oxoglutarate dependent oxygenases. Curr Opin Struct Biol. 2010;20(6):659–672. doi: 10.1016/j.sbi.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Valegård K, et al. Structure of a cephalosporin synthase. Nature. 1998;394(6695):805–809. doi: 10.1038/29575. [DOI] [PubMed] [Google Scholar]
- 5.Tsukada Y. Hydroxylation mediates chromatin demethylation. J Biochem. 2012;151(3):229–246. doi: 10.1093/jb/mvs003. [DOI] [PubMed] [Google Scholar]
- 6.Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat Chem Biol. 2008;4(3):152–156. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- 7.DiTacchio L, et al. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science. 2011;333(6051):1881–1885. doi: 10.1126/science.1206022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herz HM, et al. The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol Cell Biol. 2010;30(10):2485–2497. doi: 10.1128/MCB.01633-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centanin L, Ratcliffe PJ, Wappner P. Reversion of lethality and growth defects in Fatiga oxygen-sensor mutant flies by loss of hypoxia-inducible factor-alpha/Sima. EMBO Rep. 2005;6(11):1070–1075. doi: 10.1038/sj.embor.7400528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaakkola P, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 11.Ivan M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science. 2001;292(5516):464–468. doi: 10.1126/science.1059817. Epub2001Apr. [DOI] [PubMed] [Google Scholar]
- 12.Mecinović J, Loenarz C, Chowdhury R, Schofield CJ. 2-Oxoglutarate analogue inhibitors of prolyl hydroxylase domain 2. Bioorg Med Chem Lett. 2009;19(21):6192–6195. doi: 10.1016/j.bmcl.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Leung IK, et al. Reporter ligand NMR screening method for 2-oxoglutarate oxygenase inhibitors. J Med Chem. 2013;56(2):547–555. doi: 10.1021/jm301583m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDonough MA, et al. Cellular oxygen sensing: Crystal structure of hypoxia-inducible factor prolyl hydroxylase (PHD2) Proc Natl Acad Sci USA. 2006;103(26):9814–9819. doi: 10.1073/pnas.0601283103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henri J, et al. Structural and functional insights into Saccharomyces cerevisiae Tpa1, a putative prolylhydroxylase influencing translation termination and transcription. J Biol Chem. 2010;285(40):30767–30778. doi: 10.1074/jbc.M110.106864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keeling KM, Salas-Marco J, Osherovich LZ, Bedwell DM. Tpa1p is part of an mRNP complex that influences translation termination, mRNA deadenylation, and mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26(14):5237–5248. doi: 10.1128/MCB.02448-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes BT, Espenshade PJ. Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. EMBO J. 2008;27(10):1491–1501. doi: 10.1038/emboj.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee CY, Yeh TL, Hughes BT, Espenshade PJ. Regulation of the Sre1 hypoxic transcription factor by oxygen-dependent control of DNA binding. Mol Cell. 2011;44(2):225–234. doi: 10.1016/j.molcel.2011.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saito K, Adachi N, Koyama H, Matsushita M. OGFOD1, a member of the 2-oxoglutarate and iron dependent dioxygenase family, functions in ischemic signaling. FEBS Lett. 2010;584(15):3340–3347. doi: 10.1016/j.febslet.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 20.Wehner KA, Schütz S, Sarnow P. OGFOD1, a novel modulator of eukaryotic translation initiation factor 2alpha phosphorylation and the cellular response to stress. Mol Cell Biol. 2010;30(8):2006–2016. doi: 10.1128/MCB.01350-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Loenarz C, et al. (2014) Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc Natl Acad Sci USA 111:4019–4024. [DOI] [PMC free article] [PubMed]
- 22. Singleton RS, et al. (2014) OGFOD1 catalyses prolyl hydroxylation of RPS23 and is involved in translation control and stress granule formation. Proc Natl Acad Sci USA 111:4031–4036. [DOI] [PMC free article] [PubMed]
- 23.Kim HS, et al. Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex. Nucleic Acids Res. 2010;38(6):2099–2110. doi: 10.1093/nar/gkp1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delanoue R, Slaidina M, Léopold P. The steroid hormone ecdysone controls systemic growth by repressing dMyc function in Drosophila fat cells. Dev Cell. 2010;18(6):1012–1021. doi: 10.1016/j.devcel.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 25.Colombani J, et al. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114(6):739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- 26.Lambertsson A. The minute genes in Drosophila and their molecular functions. Adv Genet. 1998;38:69–134. doi: 10.1016/s0065-2660(08)60142-x. [DOI] [PubMed] [Google Scholar]
- 27.Lin JI, et al. Drosophila ribosomal protein mutants control tissue growth non-autonomously via effects on the prothoracic gland and ecdysone. PLoS Genet. 2011;7(12):e1002408. doi: 10.1371/journal.pgen.1002408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farny NG, Kedersha NL, Silver PA. Metazoan stress granule assembly is mediated by P-eIF2alpha-dependent and -independent mechanisms. RNA. 2009;15(10):1814–1821. doi: 10.1261/rna.1684009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hetz C. The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 30.Ryoo HD, Domingos PM, Kang MJ, Steller H. Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J. 2007;26(1):242–252. doi: 10.1038/sj.emboj.7601477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40(2):280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott RC, Juhász G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17(1):1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 34.Ge W, et al. Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. Nat Chem Biol. 2012;8(12):960–962. doi: 10.1038/nchembio.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pomar N, et al. Functional characterization of Drosophila melanogaster PERK eukaryotic initiation factor 2alpha (eIF2alpha) kinase. Eur J Biochem. 2003;270(2):293–306. doi: 10.1046/j.1432-1033.2003.03383.x. [DOI] [PubMed] [Google Scholar]
- 36.Boglev Y, et al. Autophagy induction is a Tor- and Tp53-independent cell survival response in a zebrafish model of disrupted ribosome biogenesis. PLoS Genet. 2013;9(2):e1003279. doi: 10.1371/journal.pgen.1003279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112(13):5228–5237. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.