Significance
DNA repair efficiency varies across the genome. This is due, in part, to repair pathways that are linked to transcription. When RNA polymerase stalls at a DNA lesion, repair proteins are recruited in a process called transcription-coupled nucleotide excision repair (TCR). We have examined the substrate requirements for TCR using a system in which transcription stalling is disconnected from the search for the DNA lesion. We show that stalled or paused transcription complexes initiate a damage-detection process that promotes strand-specific repair of lesions over a considerable distance. Our findings help to explain the mechanism by which the repair of active genes is accelerated and suggest that some transcription pause sites may target repair activity to specific regions of the genome.
Keywords: UvrA, DNA translocase, protein roadblock, ATPase
Abstract
Transcription-coupled nucleotide excision repair (TCR) accelerates the removal of noncoding lesions from the template strand of active genes, and hence contributes to genome-wide variations in mutation frequency. Current models for TCR suppose that a lesion must cause RNA polymerase (RNAP) to stall if it is to be a substrate for accelerated repair. We have examined the substrate requirements for TCR using a system in which transcription stalling and damage location can be uncoupled. We show that Mfd-dependent TCR in bacteria involves the formation of a damage search complex that can detect lesions downstream of a stalled RNAP, and that the strand specificity of the accelerated repair pathway is independent of the requirement for a lesion to stall RNAP. We also show that an ops (operon polarity suppressor) transcription pause site, which causes backtracking of RNAP, can promote the repair of downstream lesions when those lesions do not themselves cause the polymerase to stall. Our findings indicate that the transcription-repair coupling factor Mfd, which is an ATP-dependent superfamily 2 helicase that binds to RNAP, continues to translocate along DNA after RNAP has been displaced until a lesion in the template strand is located. The discovery that pause sites can promote the repair of nonstalling lesions suggests that TCR pathways may play a wider role in modulating mutation frequencies in different parts of the genome than has previously been suspected.
Mutations accumulate more quickly in some parts of the genome than in others (1). This is due, in part, to the existence of repair pathways that prioritize the repair of certain regions of DNA. One such pathway is transcription-coupled nucleotide excision repair (TCR), which accelerates the repair of some types of noncoding lesion in the template strand of active genes (2, 3). It has been suggested that localized mutation rates may evolve to allow mutations to occur more frequently in regions of the genome where they are likely to be advantageous and less often in regions where they are likely to be deleterious (4).
TCR is triggered when a lesion causes RNA polymerase (RNAP) to stall, and the need to stall RNAP is thought to underlie the observed strand specificity of the TCR pathway (2). The two strands of DNA are separated as they enter RNAP and take different paths through the enzyme (5). Many types of lesions cause RNAP to pause or stall when they occur in the strand that the enzyme uses as a template for RNA synthesis, but the same lesions are often bypassed without incident when they occur in the nontemplate strand (6, 7). In order for a DNA lesion such as a UV-induced cyclopyrimidine dimer (CPD) to stall RNAP, it must enter the active site of the enzyme (8). The stalled transcription complex, which is highly stable, shields the lesion from the action of repair enzymes (6). Two things must therefore happen if the lesion is to be repaired more quickly than it would be by the transcription-independent global nucleotide excision repair (NER) pathway: The stalled RNAP must be displaced from the site of damage, and the repair proteins must be enabled to act more rapidly than they would do on their own. In bacteria, these two functions can be performed by a single TCR-specific factor: the Mfd protein [also referred to as the transcription-repair coupling factor (TRCF)] (9, 10).
Mfd is an ATP-dependent DNA translocase that binds to stalled transcription complexes and removes RNAP from DNA by pushing RNAP forward (11). Mfd also interacts with the UvrA protein, which is a component of the global NER apparatus (9, 12, 13). During global NER, damage is detected by a search complex made up of a UvrA dimer and two molecules of the repair protein UvrB. Both partners contribute to lesion detection, and it is thought that a single heterotetramer simultaneously examines both strands of the DNA duplex for damage (14, 15). Once a lesion is detected, a single UvrB remains stably bound at the site of damage. This recruits a nuclease, UvrC, which nicks the damaged strand on either side of the lesion. The oligonucleotide containing the lesion is removed by the helicase UvrD, and a repair patch is synthesized by DNA polymerase I and DNA ligase using the nondamaged strand as a template (16). All of the proteins that are required for global NER are also required for TCR, and the steps subsequent to UvrA dissociation are thought to be common to the two pathways (9). Mfd and UvrB both interact with the same surface of UvrA, and the interaction between Mfd and UvrA involves a region of Mfd that is structurally homologous to part of UvrB (12, 13). Mutational analysis of UvrA has shown that the ability of UvrA to distinguish between damaged and undamaged DNA is less important during TCR than during global NER (12), but the mechanism by which the Mfd–UvrA interaction accelerates repair is not understood.
Several different mechanisms might underlie the increased rate of repair during TCR. The simplest model is that interaction with Mfd increases the local concentration of repair proteins in the vicinity of the stalled RNAP. If this is the case, then any secondary lesions found close to a stalled RNAP might also be repaired at an enhanced rate (2). Alternatively, it has been proposed that Mfd-dependent TCR may be restricted to the repair of lesions that enter the RNAP and are located within the ssDNA of the transcription bubble (17). Because strand separation by UvrA is important for the stable binding of UvrB at the site of DNA damage, such a model would help to explain the decreased requirement for damage recognition by UvrA during TCR. Models have been proposed in which UvrA is recruited to Mfd shortly after Mfd binds to RNAP (12) or in which UvrA is not recruited until after Mfd itself has detected the lesion (13). Because Mfd and UvrB interact with UvrA in a similar fashion, they may compete with each other for UvrA, and it is possible that the damage search mechanism of TCR involves a Mfd/UvrA2/UvrB complex in place of the UvrB/UvrA2/UvrB complex involved in global NER. The asymmetry of such a complex may restrict the search for lesions to a single strand.
To help distinguish between these models, we have assessed the ability of the Mfd-dependent TCR pathway to promote the repair of lesions at locations close to, but distinct from, a stalled RNAP. We found that the pathway is not restricted to repair of lesions within the transcription bubble, because it can promote the repair of lesions located well downstream of a stalled RNAP. Only lesions in the template strand were subject to enhanced repair, indicating that the observed strand specificity of TCR does not simply reflect the ability of lesions in the template strand to stall RNAP. We also found that a lesion that does not stall RNAP can be targeted for accelerated repair if it is located in the template strand downstream of a stalled RNAP or a transcriptional pause site that promotes backtracking of RNAP.
Results
Stalled Transcription Complexes Promote Strand-Specific Repair of Distant Lesions.
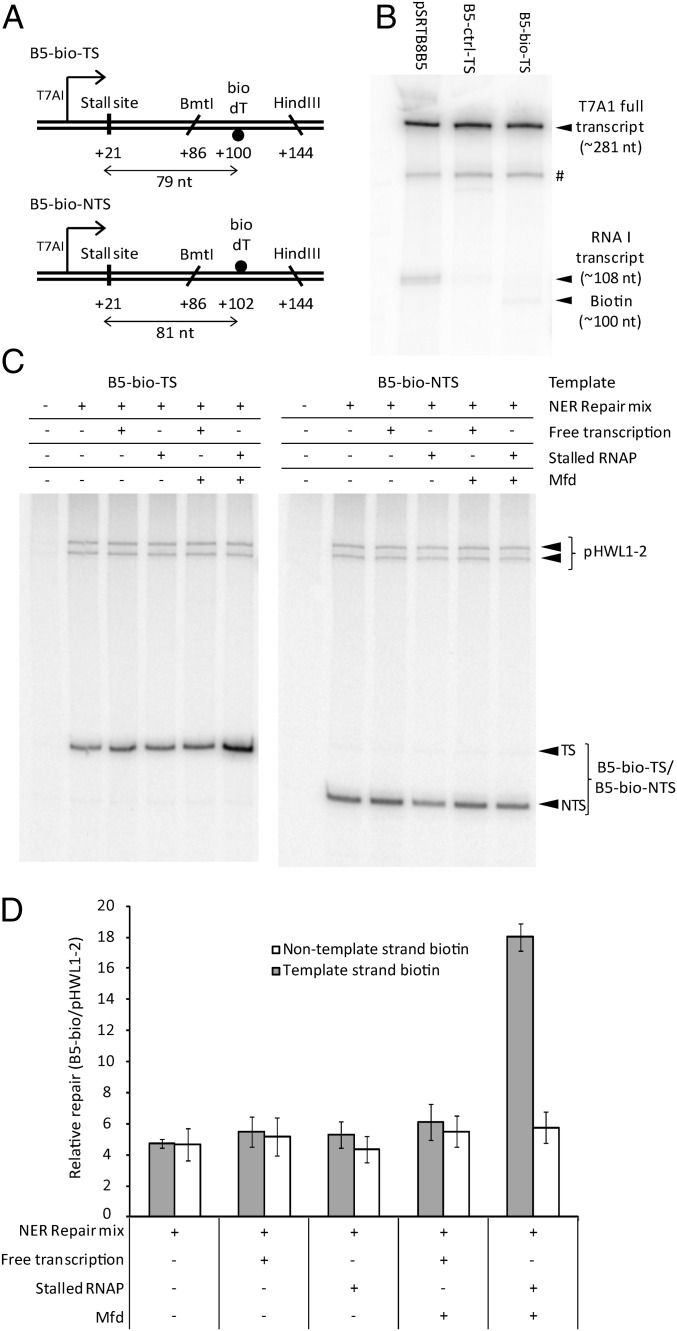
To uncouple the processes of RNAP stalling and lesion detection, we constructed plasmid substrates in which RNAP could be stalled by incorporation of a chain-terminating nucleotide at a specific position upstream of a single defined DNA lesion (Fig. 1A). To minimize the confounding effect of any transcription complexes that might bypass the stall site, we used a biotinylated deoxythymidine residue (bio-dT) as the lesion in our initial experiments. This artificial nucleotide is a substrate for NER (Fig. S1) but has little or no ability to stall RNAP when present on the template strand (Fig. 1B).
Fig. 1.
Effect of stalling RNAP upstream of a bio-dT lesion. (A) DNA substrates. A single bio-dT was present in the template strand (B5-bio-TS) or nontemplate strand (B5-bio-NTS). (B) Effect of a template strand bio-dT lesion on in vitro transcription. pSRTB8B5 is unmodified supercoiled DNA. B5-bio-TS and B5-ctrl-TS are closed circular substrates into which a bio-dT–containing or unmodified control oligonucleotide had been ligated. Reactions contained 20 nM RNAP. The T7A1 transcript terminates 281 bp downstream of the transcription start site. The RNA I transcript is encoded as part of the ColE1 DNA replication origin of the pSR plasmids. The locations of the bio-dT and a product caused by pausing or termination within the A/T-rich TFO binding site (labeled #) are indicated. (C) Patch synthesis assay monitoring repair of bio-dT in the template strand and nontemplate strand. All reactions contained DNA polymerase, DNA ligase, ATP, NADH, and the radiolabeled dNTP mixture required for patch synthesis. “NER Repair mix” indicates that UvrA, UvrB, UvrC, and UvrD were added; “Free transcription” indicates that RNAP was added with GTP, CTP, and UTP; and “Stalled RNAP” indicates that RNAP was added with GTP, CTP, adenylyl (3′-5′) uridine (ApU), and 3′ dUTP. (D) Quantification of the [32P]-dATP incorporated into the bio-dT–containing strand of B5-bio-TS and B5-bio-NTS during patch synthesis assays, normalized to the amount incorporated into the UV-irradiated pHWL1-2 control in each lane. Values are the average of at least three repeats and are shown with SD.
The plasmid substrates contained the strong T7A1 promoter upstream of the lesion. Transcription was halted 21 nt after initiation by replacing UTP with 3′ dUTP, incorporation of which at the 3′ end of the transcript prevents further extension of the RNA chain (Fig. S2). A single bio-dT lesion was placed at +100 on the template strand or at +102 on the nontemplate strand, and the DNA was incubated with combinations of purified transcription and repair proteins. Repair of the lesion was monitored by a patch synthesis assay that measures the incorporation of radiolabeled dATP into repair patches (Fig. 1C). Each reaction also contained a control template, in which repair of UV-induced CPDs within an ∼140-bp nontranscribed region provided a global NER reference within each reaction.
Incubation of the substrates containing single bio-dT lesions with the proteins and cofactors required for global NER resulted in repair patch synthesis (Fig. 1D). Repair of the lesion in the template strand was strongly stimulated by the presence of Mfd when RNAP was stalled upstream of the lesion. No stimulation of repair of the template strand was observed when Mfd was added to reactions in which RNAP was able to transcribe the damaged template strand fully, or in the absence of Mfd. The efficiency with which the nontemplate strand was repaired remained constant regardless of the presence or absence of transcribing RNAP, stalled RNAP, or Mfd.
These results indicate that an RNAP stalled in a damage-independent manner can stimulate repair of a DNA lesion located at least 79 nt downstream. This stimulation is dependent on the presence of the Mfd protein and is strand-specific: Only lesions on the template strand were substrates for this “TCR at a distance” pathway. The absence of any “conventional” TCR of the bio-dT lesion is consistent with the observation that bio-dT causes little or no stalling of RNAP. In reactions that do not cause RNAP to stall at +21, it is likely that RNAP transcribes past the lesion, and so does not recruit Mfd. The observation that freely transcribing RNAP does not trigger TCR of bio-dT supports the conclusion that the stimulation of repair seen when RNAP is stalled at +21 is due to events initiating at the stalled RNAP, rather than any potential bypass of the stall site (and the other sites at which UTP must be incorporated into the transcript between +21 and the lesion) by a small fraction of RNAPs.
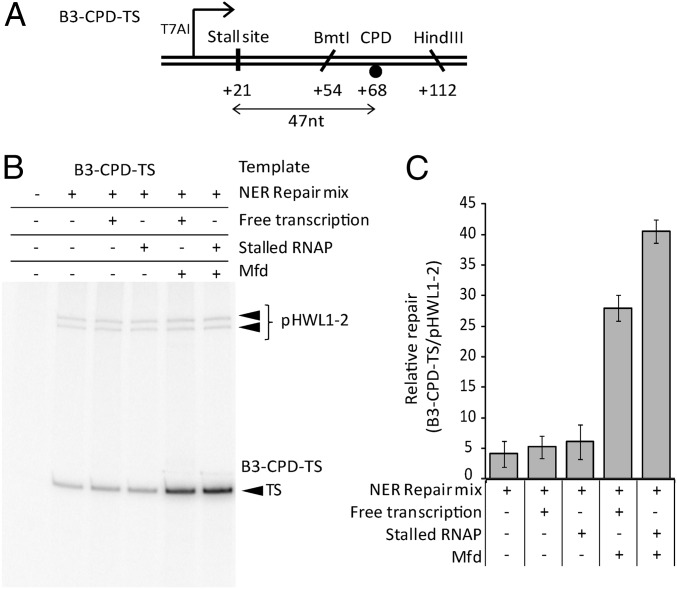
We repeated the assays using a template containing a single CPD located 47 bp downstream of the transcription stall site (Fig. 2 and Fig. S1). As expected, we observed Mfd-dependent stimulation of repair when transcribing RNAPs were able to reach the CPD. We observed a similar level of Mfd-dependent stimulation of repair of the CPD when RNAP was stalled 47 bp upstream of the lesion. We conclude that the Mfd-dependent mechanism for locating lesions downstream of stalled RNAPs can function on a physiologically relevant lesion, as well as on the bio-dT substrate.
Fig. 2.
Effect of stalling an RNAP upstream of a CPD lesion. (A) DNA substrate. A single CPD was present in the template strand. (B) Patch synthesis assay monitoring repair of a single CPD in the template strand. Additions are as described for Fig. 1. (C) Quantification of the [32P]-dATP incorporated into the template strand of B3-CPD-TS during patch synthesis assays, normalized to the amount incorporated into the UV-irradiated pHWL1-2 control in each lane. Values are the average of at least three repeats and are shown with SD.
Downstream Repair Can Be Blocked by a Protein Roadblock.
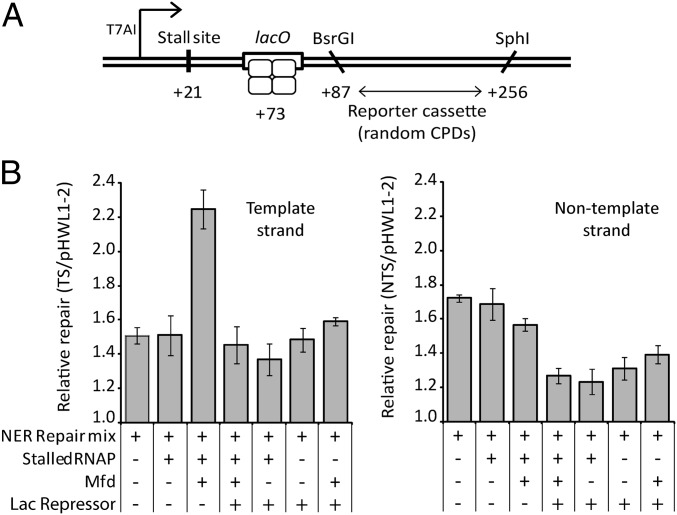
The ability of a stalled RNAP to promote the strand-specific repair of lesions located some distance downstream suggests that one or more proteins recruited to the stall site may move along the DNA until a lesion is encountered. We investigated whether the presence of a protein roadblock between the stall site and the lesions affected this process. We constructed a substrate in which a lac operator was placed between the transcription stall site and a reporter cassette in which we could monitor the repair of UV-induced, randomly distributed lesions in both the template and nontemplate strands (Fig. 3). The absolute levels of repair patch synthesis in this substrate were lower than those observed with substrates containing single defined lesions because the UV fluence used in these experiments generates one to two lesions per plasmid; thus, the majority of substrates did not contain lesions within the reporter region (18) (Fig. 3 and Fig. S3). In agreement with our previous assays, we observed that RNAP stalled at +21 stimulated repair of the template strand when Mfd was present but had little or no effect on the repair of the nontemplate strand. The Mfd-dependent stimulation of repair of the template strand was abolished when Lac repressor protein (which binds to the lac operator) was added to the reaction. This result supports the notion that repair of the downstream lesions is stimulated by the movement of one or more proteins along the DNA from the site at which RNAP is stalled, and indicates that this movement can be blocked by an intervening protein–DNA complex. Curiously, addition of Lac repressor inhibited repair of the nontemplate strand irrespective of the presence or absence of Mfd or RNAP. No such effect was observed on the template strand. UvrAB complexes can slide along DNA when searching for DNA damage during global NER (19), and they may need to approach DNA lesions from the 5′ side (with respect to the damaged strand) (20). The lac operator in these constructs is immediately adjacent to the upstream end of the reporter cassette, and Lac repressor may inhibit global NER of lesions close to the 5′ end of the nontemplate strand because it prevents UvrAB approaching from the 5′ side of those lesions.
Fig. 3.
Effect of a protein roadblock on TCR at a distance. (A) DNA substrate. Plasmid pHWL1-T7A1-2lacO contains a lac operator between the T7A1 promoter and a repair-reporter cassette. DNA lesions were introduced randomly into both strands of the plasmid by exposure to 30 J/m2 of UV light. (B) Quantification of the [32P]-dATP incorporated into the template and nontemplate strands of the pHWL1-T7A1-2lacO reporter cassette during patch synthesis assays normalized to the amount incorporated into the UV-irradiated pHWL1-2 control in each lane. Additions are as described for Fig. 1. Values are the average of at least three repeats and are shown with SD. A sample dataset is shown in Fig. S3.
Role of DNA Translocation by Mfd in Downstream Repair.
When TCR occurs at a lesion that has stalled RNAP, DNA translocation by Mfd pushes RNAP forward and eventually displaces it (11). During this process, Mfd moves along the dsDNA in a 3′ to 5′ direction with respect to the template strand. If Mfd continues to translocate along DNA after it has displaced RNAP, then it would move in the direction needed to encounter lesions located downstream of the site at which the RNAP had stalled. We used a triplex-forming oligonucleotide (TFO) displacement assay to determine whether DNA translocation by Mfd is sufficiently processive to potentially participate in the search for DNA damage downstream of a stalled RNAP.
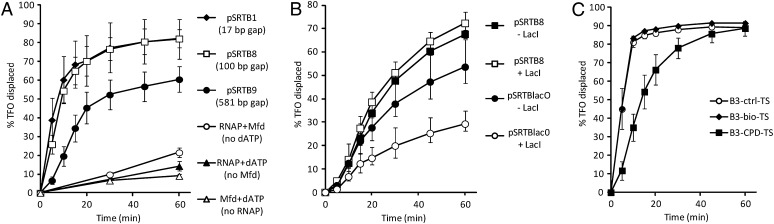
As we have previously shown, Mfd in isolation has little ability to displace a TFO bound in the major groove, because its translocation activity is autoinhibited in the absence of RNAP (21) (Fig. 4A). However, when RNAP is stalled by nucleotide starvation immediately upstream of the TFO, DNA translocation by Mfd results in displacement of the majority of TFO from the DNA. UvrA interacts directly with the triplex-containing DNA substrate, and so its effect on DNA translocation by Mfd could not be readily assessed in this assay. In the pSRTB1 substrate, the distance between the stall site and the proximal end of the TFO is 17 bp, which means that the downstream face of the RNAP is immediately adjacent to the TFO. It is therefore not clear whether the TFO is displaced when the front edge of RNAP is pushed forward by Mfd (in which case Mfd might translocate for only a few base pairs along the DNA) or whether Mfd continues to translocate along the DNA after RNAP has been displaced. We created constructs in which the distance between the RNAP stall site and the TFO binding site was increased to 100 bp or 581 bp (Fig. 4A). We found that Mfd was able to displace a TFO bound 100 bp downstream of a stalled RNAP just as well as it could displace a TFO bound 17 bp downstream. When the TFO was bound 581 bp downstream of the stall site, there was a reduction in the efficiency of displacement, but a substantial proportion of the TFO was displaced by Mfd. We also examined the effect of placing a Lac repressor protein roadblock between the RNAP stall site and the TFO binding site (Fig. 4B). We found that Lac repressor bound to a lac operator located between the stall site and the TFO binding site impaired the ability of Mfd to displace the TFO but did not abolish it. These results indicate that once Mfd has been activated by binding to a stalled RNAP, it is capable of translocating more than 100 bp along the DNA. It is therefore possible that the damage search complex that stimulates repair of lesions in the template strand downstream of stalled RNAPs is driven by the translocation activity of Mfd.
Fig. 4.
DNA translocation by Mfd. (A) Effect of distance on efficiency of translocation by Mfd, measured by TFO-displacement assay. All substrates allow RNAP to be stalled at +21. “Gap” indicates the distance between +21 and the promoter-proximal end of the triplex when the TFO is bound to the substrate. Data labeled pSRTB1, pSRTB8, and pSRTB9 represent experiments in which RNAP was stalled and Mfd and dATP were then added to initiate DNA translocation. Control experiments in which RNAP, Mfd, or dATP was omitted were conducted on pSRTB9. (B) Effect of a Lac repressor roadblock on TFO displacement using substrates that contain (pSRTBlacO) or lack (pSRTB8) a lac operator between the RNAP stall site and the TFO binding site. RNAP was stalled in the presence or absence of Lac repressor, and Mfd and dATP were then added to initiate DNA translocation. (C) Effect of lesions in the template strand between the RNAP stall site and the TFO binding site. The DNA between the stall site and the TFO binding site was either undamaged (B3-ctrl-TS) or contained a single bio-dT (B3-bio-TS) or CPD (B3-CPD-TS) in the template strand. RNAP was stalled, and Mfd and dATP were then added to initiate DNA translocation. Values in A and C are the average of at least three repeats and are shown with SD. Values in B are the average of two or three repeats and are shown with range.
DNA lesions are sometimes detected by their ability to stall DNA helicases (22–24). An attractive model for damage detection by Mfd is that it might stall when it encounters lesions in the template strand. We tested the ability of Mfd to translocate past a single CPD or bio-dT lesion placed on the template strand between an RNAP stall site and a TFO binding site (Fig. 4C). We found that a single CPD on the template strand slowed the Mfd-dependent displacement of TFO, although, ultimately, the same amount of TFO was displaced as on the undamaged template. A single bio-dT on the template strand had no detectable effect on the DNA translocation activity of Mfd, although we cannot exclude the possibility that this lesion causes a short-lived pause that cannot be detected in our assay. These results indicate Mfd is able to translocate past both of the lesions tested, although it seems to pause, or perhaps sometimes dissociate, when it encounters a CPD.
A Transcription Pause Site Can Stimulate Repair of Downstream Lesions.
In addition to acting at RNAPs that stall at DNA lesions, Mfd plays a role in a variety of other transcriptional events. When RNAPs slide backward on DNA, the 3′ end of the nascent transcript moves out of the active site and into the secondary channel of RNAP (25). Mfd reactivates these backtracked complexes by pushing the RNAP forward, allowing the 3′ end of the transcript to reenter the active site (11). In contrast to the situation at permanently stalled RNAPs, once a backtracked RNAP has been pushed forward by Mfd it can resume transcription and is not displaced from the DNA. Certain template sequences, called class II pause sites, promote backtracking (26, 27). Among the best-characterized template sequences are the ops (operon polarity suppressor) pause sites that are involved in the recruitment of the transcription antiterminator protein RfaH to several operons in Escherichia coli (26).
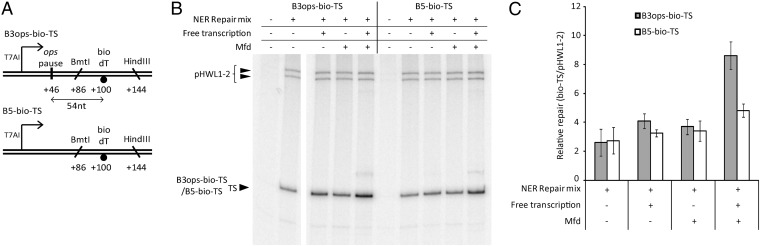
To determine whether Mfd can stimulate repair of lesions located downstream of a backtracked paused RNAP, we constructed templates containing the ops pause site from the E. coli rfaQ operon upstream of a single DNA lesion in the template strand (Fig. 5A). To detect any effects of the paused RNAP, it was necessary to ensure that TCR would not be triggered by RNAP stalling at the lesion after it had passed through the pause site. We therefore used bio-dT, rather than a CPD, as the lesion in these assays. Insertion of the ops site downstream of the T7A1 promoter in our plasmid substrate causes a transient pause in transcript elongation that is not observed with a control substrate that lacks the pause site (Fig. S4). In patch synthesis assays, the ops sequence increased the efficiency of repair of the downstream bio-dT lesion when the substrate was transcribed in the presence of Mfd but had no effect when the substrate was transcribed in the absence of Mfd or when the repair assays were conducted in the absence of transcription (Fig. 5 B and C). This suggests that recruitment of Mfd to RNAP at a class II pause site can stimulate repair of a lesion located in the template strand downstream of the pause site.
Fig. 5.
Effect of pausing RNAP upstream of a nonstalling lesion. (A) DNA substrates. (B) Patch synthesis assay monitoring repair of a bio-dT in the template strand in a substrate containing an ops pause site (B3ops-bio-TS) or a control lacking the ops pause site (B5-bio-TS). Additions are as described for Fig. 1. (C) Quantification of the [32P]-dATP incorporated into the template strand of each substrate during patch synthesis assays normalized to the amount incorporated into the UV-irradiated pHWL1-2 control in each lane. Values are the average of at least three repeats and are shown with SD.
Discussion
Our results show that the bacterial TCR pathway involves the formation of a damage search complex that can detect lesions downstream of a stalled RNAP. Only lesions in the template strand are targeted for preferential repair, indicating that the observed strand bias of TCR reflects an asymmetry in the damage detection process and is not simply a byproduct of the need for a lesion to be in the template strand to stall RNAP. We conclude that Mfd does not act simply to increase the local concentration of UvrA, or to assemble a UvrB/UvrA2/UvrB complex away from the lesion, because these methods of damage detection would be expected to detect lesions in both strands with equal efficiency. We also conclude that there is no requirement for a lesion to be located within the ssDNA of a transcription bubble in order for its repair to be accelerated during Mfd-dependent TCR.
Although Mfd binds transiently to DNA in the absence of other factors, it becomes very stably associated once it has been recruited to a stalled transcription complex (28). In this study, we have shown that after being activated by binding to stalled RNAP, Mfd can translocate several hundred base pairs. It is clear that ATP-dependent translocation by Mfd can continue well beyond the range that would be needed to displace RNAP from a lesion. Mfd initially binds to the DNA immediately upstream of the stalled RNAP (11). It moves along DNA in a direction that is 3′ to 5′ with respect to the template strand, and so it will eventually encounter lesions located within, or downstream of, the stalled RNAP. During conventional TCR, in which a lesion has entered the active site of RNAP and caused transcription to stall, the distance between the site at which Mfd starts to translocate and the point at which it encounters the lesion will be short; the upstream face of RNAP is ∼14 bp from the active site (11). When RNAP stalling is uncoupled from DNA damage detection, as in our assays, it seems that Mfd continues to translocate along the DNA until a lesion is encountered.
DNA lesions are sometimes detected by their ability to stall DNA helicases. For example, it has been shown that stalling of the xeroderma pigmentosum group D helicase in transcription factor II H by CPDs is important for damage detection during NER in eukaryotes (22, 23), and stalling of UvrB helicase activity is thought to play a role in damage detection during NER in prokaryotes (24). Although Mfd is a DNA translocase rather than a strand-separating helicase, we speculated that lesions might be detected by their ability to block the movement of Mfd along DNA. We found that a CPD on the template strand did indeed impede the progress of Mfd, although it was not a complete block. However, the presence of a bio-dT residue on the template strand had no detectable effect on DNA translocation by Mfd, despite the fact that bio-dT can be preferentially repaired by TCR when located downstream of a stalled RNAP. This suggests that stalling of Mfd is not an essential requirement for lesion detection during TCR. An important caveat to this conclusion is that the composition of the damage search complex formed during TCR is not known; it is likely to contain Mfd but may also contain one or more of RNAP, UvrA, and UvrB (28). Our DNA translocation assays contained Mfd and RNAP but did not contain UvrA or other repair proteins, because direct interaction of repair proteins with the template DNA and the DNA lesion makes their use in these assays impractical. If UvrA is part of the complex that translocates along the DNA in search of damage, it may modify the properties of Mfd and render it more susceptible to stalling by DNA lesions. Alternatively, if Mfd does not detect the lesion itself, it might act by “feeding” DNA to repair proteins that are associated with it. A dimer of UvrA associated with Mfd is unlikely to be able to provide the observed strand specificity, because the dimer is symmetrical and is thought to probe both strands of DNA for damage simultaneously (14, 15). The strand specificity could arise from the formation of an Mfd/UvrA2/UvrB complex at some point in the damage recognition process: Such a complex is likely to be able to load UvrB onto only one strand, and so lesions in the other strand would not be processed further.
The ability of RNAP stalled at one location to promote repair at a different location is particularly intriguing in the case of lesions that, like bio-dT, do not block transcription, and so may not trigger TCR directly. Physiologically relevant examples of such lesions include small alkylation products, such as O6-methylguanine, which are detected inefficiently by the global NER machinery (29–31). Although irreversible stalling of RNAP upstream of a lesion is likely to be a rare event in vivo, our finding that a transcription pause site can promote the repair of nonblocking lesions in the adjacent DNA suggests that transcriptional pausing may play an important and previously unsuspected role in genome maintenance. By acting as recruitment sites for Mfd, pause sites could promote repair within specific regions of the genome. The ops pause used in our experiments promotes formation of backtracked transcription complexes, which Mfd can bind to and push forward to reinstate the 3′ end of the transcript in the active site and allow RNAP to resume transcription (11). It will be interesting to discover whether all such transcription-restart events lead to the loading of Mfd onto DNA in a fashion that allows it to start scanning for DNA damage or whether the repair-promoting properties of the pause site arise from a proportion of the transcription complexes that fail to reinitiate and are displaced from the DNA by Mfd.
Pause sites play a variety of roles in the regulation of gene expression, including acting as recruitment sites for transcription elongation factors, coordinating the progress of transcription and translation machinery, and participating in transcription termination (27). Our work shows that some transcription pauses also have the potential to increase localized DNA repair efficiency. Because even small differences in mutation frequency in critical regions may confer evolutionary advantages, the distribution of such pause sites throughout the genome may have arisen, at least in part, to optimize their use as modulators of DNA repair. Further work will be needed to examine the scope and importance of pause-mediated effects on DNA repair and mutation within living cells.
Materials and Methods
Full details of the materials and methods used in this study are presented in SI Materials and Methods.
Proteins.
His-tagged E. coli RNAP was purified as described by Smith and Savery (32). His-tagged E. coli UvrA, UvrB, and UvrC were purified as described by Manelyte et al. (33). Untagged E. coli UvrD was purified as described by Manelyte et al. (33). E. coli DNA polymerase I and DNA ligase were purchased from New England BioLabs. E. coli Lac repressor protein was a gift from P. McGlynn, (University of York, York, UK) (34). Lac repressor concentrations refer to tetramers. All other protein concentrations refer to monomers.
Plasmids.
Substrates were derived from plasmid pSRTB1 (21), which contains a TFO binding site downstream of the T7A1 promoter, or from pHWL1-T7A1-2 (12), which contains a repair-reporter region downstream of the T7A1 promoter. RNAP stalls at +21 of the T7A1 promoter if UTP is replaced with the chain terminator 3′ dUTP. Single-lesion templates were constructed by placing multiple BbvCI recognition sites between the T7A1 promoter and the TFO binding site. Nicking of one or other strands with Nb. BbvCI or Nt. BbvCI generated a region of ssDNA into which complementary oligonucleotides containing DNA modifications were annealed, as described by Luzzietti et al. (35). Details of DNA substrates used for TFO assays and for the generation of plasmids containing single lesions are given in Tables S1–S3.
In Vitro Transcription.
In vitro transcription reactions were carried out under multiround conditions on plasmid DNA. Unless stated otherwise, each reaction contained the following: 1.5 nM template DNA; 1× repair buffer [40 mM Hepes, 100 mM KCl, 8 mM MgCl2, 4% (vol/vol) glycerol, 5 mM DTT, 100 μg/mL BSA]; 200 μM GTP, CTP, and ATP; 10 μM UTP; and 2.5 μCi of [α32P] UTP. Reactions were initiated by the addition of RNAP and were incubated at 37 °C. Reactions were stopped by the addition of an equal volume of formamide stop buffer [95% (vol/vol) formamide, 20 mM EDTA, 0.25% (wt/vol) bromophenol blue, 0.25% (wt/vol) xylene cyanol]. Samples were separated by denaturing PAGE and analyzed using a phosphorimager and ImageQuant software (GE Healthcare Life Sciences).
Patch Synthesis Assay.
Patch synthesis assays were performed essentially as described by Manelyte et al. (12). To reconstitute TCR, circular DNA substrates were incubated with purified DNA polymerase I, DNA ligase, UvrA, UvrB, UvrC, UvrD, Mfd, RNAP, and the nucleotides and cofactors necessary to support transcription, repair protein activity, and synthesis of the repair patch. Repair was monitored by the incorporation of [α32P] dATP into the repair patches. To generate transcripts stalled at +21, UTP was omitted from the reactions and was replaced with ApU (necessary for transcription initiation) and 3′ dUTP. Repair patches in the template and nontemplate strands were distinguished by cutting the substrates with restriction enzymes that generated a 5′ overhang on one side of the lesion and a 3′ overhang on the other. The two released strands, which differed in length, were separated by denaturing PAGE and analyzed as for transcription reactions. To correct for lane-to-lane variation, all reactions contained, as an internal control, pHWL1-2 irradiated with 30 J/m2 of 254-nm UV light. The analysis cassette in pHWL1-2 is not transcribed and is repaired by global NER (12).
TFO-Displacement Assays.
TFO-displacement assays were conducted essentially as described by Smith et al. (21).
Supplementary Material
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council via studentships (to N.M.H. and Y.-I.T.K.) and Research Grant BB/I003142/1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 3905.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322350111/-/DCSupplemental.
References
- 1.Martincorena I, Seshasayee ASN, Luscombe NM. Evidence of non-random mutation rates suggests an evolutionary risk management strategy. Nature. 2012;485(7396):95–98. doi: 10.1038/nature10995. [DOI] [PubMed] [Google Scholar]
- 2.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9(12):958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 3.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342(6245):95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 4.Martincorena I, Luscombe NM. Non-random mutation: The evolution of targeted hypermutation and hypomutation. Bioessays. 2013;35(2):123–130. doi: 10.1002/bies.201200150. [DOI] [PubMed] [Google Scholar]
- 5.Borukhov S, Nudler E. RNA polymerase: The vehicle of transcription. Trends Microbiol. 2008;16(3):126–134. doi: 10.1016/j.tim.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Selby CP, Sancar A. Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J Biol Chem. 1990;265(34):21330–21336. [PubMed] [Google Scholar]
- 7.Saxowsky TT, Doetsch PW. RNA polymerase encounters with DNA damage: Transcription-coupled repair or transcriptional mutagenesis? Chem Rev. 2006;106(2):474–488. doi: 10.1021/cr040466q. [DOI] [PubMed] [Google Scholar]
- 8.Brueckner F, Hennecke U, Carell T, Cramer P. CPD damage recognition by transcribing RNA polymerase II. Science. 2007;315(5813):859–862. doi: 10.1126/science.1135400. [DOI] [PubMed] [Google Scholar]
- 9.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260(5104):53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 10.Deaconescu AM, Artsimovitch I, Grigorieff N. Interplay of DNA repair with transcription: From structures to mechanisms. Trends Biochem Sci. 2012;37(12):543–552. doi: 10.1016/j.tibs.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JS, Marr MT, Roberts JW. E. coli Transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109(6):757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 12.Manelyte L, Kim Y-IT, Smith AJ, Smith RM, Savery NJ. Regulation and rate enhancement during transcription-coupled DNA repair. Mol Cell. 2010;40(5):714–724. doi: 10.1016/j.molcel.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deaconescu AM, Sevostyanova A, Artsimovitch I, Grigorieff N. Nucleotide excision repair (NER) machinery recruitment by the transcription-repair coupling factor involves unmasking of a conserved intramolecular interface. Proc Natl Acad Sci USA. 2012;109(9):3353–3358. doi: 10.1073/pnas.1115105109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pakotiprapha D, Samuels M, Shen K, Hu JH, Jeruzalmi D. Structure and mechanism of the UvrA-UvrB DNA damage sensor. Nat Struct Mol Biol. 2012;19(3):291–298. doi: 10.1038/nsmb.2240. [DOI] [PubMed] [Google Scholar]
- 15.Jaciuk M, Nowak E, Skowronek K, Tańska A, Nowotny M. Structure of UvrA nucleotide excision repair protein in complex with modified DNA. Nat Struct Mol Biol. 2011;18(2):191–197. doi: 10.1038/nsmb.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kisker C, Kuper J, Van Houten B. Prokaryotic nucleotide excision repair. Cold Spring Harb Perspect Biol. 2013;5(3):a012591. doi: 10.1101/cshperspect.a012591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen SE, et al. Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli. Proc Natl Acad Sci USA. 2010;107(35):15517–15522. doi: 10.1073/pnas.1005203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell DL, Jen J, Cleaver JE. Sequence specificity of cyclobutane pyrimidine dimers in DNA treated with solar (ultraviolet B) radiation. Nucleic Acids Res. 1992;20(2):225–229. doi: 10.1093/nar/20.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kad NM, Wang H, Kennedy GG, Warshaw DM, Van Houten B. Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single- molecule imaging of quantum-dot-labeled proteins. Mol Cell. 2010;37(5):702–713. doi: 10.1016/j.molcel.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moolenaar GF, et al. The effect of the DNA flanking the lesion on formation of the UvrB-DNA preincision complex. Mechanism for the UvrA-mediated loading of UvrB onto a DNA damaged site. J Biol Chem. 2000;275(11):8038–8043. doi: 10.1074/jbc.275.11.8038. [DOI] [PubMed] [Google Scholar]
- 21.Smith AJ, Szczelkun MD, Savery NJ. Controlling the motor activity of a transcription-repair coupling factor: autoinhibition and the role of RNA polymerase. Nucleic Acids Res. 2007;35(6):1802–1811. doi: 10.1093/nar/gkm019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugasawa K, Akagi J, Nishi R, Iwai S, Hanaoka F. Two-step recognition of DNA damage for mammalian nucleotide excision repair: Directional binding of the XPC complex and DNA strand scanning. Mol Cell. 2009;36(4):642–653. doi: 10.1016/j.molcel.2009.09.035. [DOI] [PubMed] [Google Scholar]
- 23.Mathieu N, Kaczmarek N, Rüthemann P, Luch A, Naegeli H. DNA quality control by a lesion sensor pocket of the xeroderma pigmentosum group D helicase subunit of TFIIH. Curr Biol. 2013;23(3):204–212. doi: 10.1016/j.cub.2012.12.032. [DOI] [PubMed] [Google Scholar]
- 24.Truglio JJ, et al. Structural basis for DNA recognition and processing by UvrB. Nat Struct Mol Biol. 2006;13(4):360–364. doi: 10.1038/nsmb1072. [DOI] [PubMed] [Google Scholar]
- 25.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149(7):1438–1445. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci USA. 2000;97(13):7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34(Pt 6):1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 28.Howan K, et al. Initiation of transcription-coupled repair characterized at single-molecule resolution. Nature. 2012;490(7420):431–434. doi: 10.1038/nature11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viswanathan A, Doetsch PW. Effects of nonbulky DNA base damages on Escherichia coli RNA polymerase-mediated elongation and promoter clearance. J Biol Chem. 1998;273(33):21276–21281. doi: 10.1074/jbc.273.33.21276. [DOI] [PubMed] [Google Scholar]
- 30.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: The UvrABC system. Chem Rev. 2006;106(2):233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 31.Mazon G, Philippin G, Cadet J, Gasparutto D, Fuchs RP. The alkyltransferase-like ybaZ gene product enhances nucleotide excision repair of O(6)-alkylguanine adducts in E. coli. DNA Repair (Amst) 2009;8(6):697–703. doi: 10.1016/j.dnarep.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 32.Smith AJ, Savery NJ. RNA polymerase mutants defective in the initiation of transcription-coupled DNA repair. Nucleic Acids Res. 2005;33(2):755–764. doi: 10.1093/nar/gki225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manelyte L, et al. The unstructured C-terminal extension of UvrD interacts with UvrB, but is dispensable for nucleotide excision repair. DNA Repair (Amst) 2009;8(11):1300–1310. doi: 10.1016/j.dnarep.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Payne BT, et al. Replication fork blockage by transcription factor-DNA complexes in Escherichia coli. Nucleic Acids Res. 2006;34(18):5194–5202. doi: 10.1093/nar/gkl682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luzzietti N, Knappe S, Richter I, Seidel R. Nicking enzyme-based internal labeling of DNA at multiple loci. Nat Protoc. 2012;7(4):643–653. doi: 10.1038/nprot.2012.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.