Significance
The DNA replication machinery is a multiprotein assembly that is responsible for a rapid and faithful duplication of the genome. Based on biochemical, structural, and genetic studies, we have obtained a very detailed picture of its different enzymatic activities that underlie unwinding of the parental duplex DNA and incorporation of nucleotides into daughter DNA. However, little is known about the dynamic changes in composition of the complex during replication. Here, we use single-molecule fluorescence methods to visualize the dynamics with which individual DNA polymerases, the replication protein responsible for DNA synthesis, associate with and dissociate from the replication machinery. Our results show that the DNA polymerase remains associated with the replication complex for a much shorter time than our previous models suggested.
Keywords: polymerase exchange, single molecule, fluorescence microscopy
Abstract
Replication of DNA plays a central role in transmitting hereditary information from cell to cell. To achieve reliable DNA replication, multiple proteins form a stable complex, known as the replisome, enabling them to act together in a highly coordinated fashion. Over the past decade, the roles of the various proteins within the replisome have been determined. Although many of their interactions have been characterized, it remains poorly understood how replication proteins enter and leave the replisome. In this study, we visualize fluorescently labeled bacteriophage T7 DNA polymerases within the replisome while we simultaneously observe the kinetics of the replication process. This combination of observables allows us to monitor both the activity and dynamics of individual polymerases during coordinated leading- and lagging-strand synthesis. Our data suggest that lagging-strand polymerases are exchanged at a frequency similar to that of Okazaki fragment synthesis and that two or more polymerases are present in the replisome during DNA replication. Our studies imply a highly dynamic picture of the replisome with lagging-strand DNA polymerases residing at the fork for the synthesis of only a few Okazaki fragments. Further, new lagging-strand polymerases are readily recruited from a pool of polymerases that are proximally bound to the replisome and continuously replenished from solution.
The organization of replisomes is highly conserved among various organisms (1), underlining the evolutionary importance of the replication machinery architecture. The bacteriophage T7 replication system offers an attractive model system to study the interplay between replication proteins because its replication machinery is relatively simple; a functional replisome can be reconstituted by just four purified proteins. Three of these proteins are encoded by the phage itself: helicase-primase (gp4), DNA polymerase (gp5), and single-stranded DNA (ssDNA) binding protein (gp2.5). A processivity factor for the gp5 polymerase, thioredoxin (trx), is provided by the host Escherichia coli. T7 gp4 assembles as a hexamer on ssDNA and provides both helicase and primase activities (2–4). The helicase unwinds the parental DNA to provide an ssDNA template for DNA replication, whereas the primase catalyzes synthesis of oligoribonucleotides at specific DNA sequences for use as primers to initiate Okazaki fragment synthesis (5, 6). T7 gp5 forms a complex with trx (7, 8) to increase its processivity from a few nucleotides to ∼1,000 nucleotides per binding event (9). Basic regions of the DNA polymerase, located in the thioredoxin-binding domain (TBD) as well as on the front of the polymerase, loosely interact with the acidic C-terminal tails of the gp4 helicase (10–12), whereas a high affinity interaction with gp4 occurs when the polymerase is in a polymerization mode (13, 14). T7 gp2.5 also has an acidic C-terminal tail that interacts with the same basic residues of the polymerase TBD (10, 15). Beyond coating the ssDNA that is transiently exposed during replication, gp2.5 plays an important role in mediating and timing protein−protein interactions within the replisome (15–17).
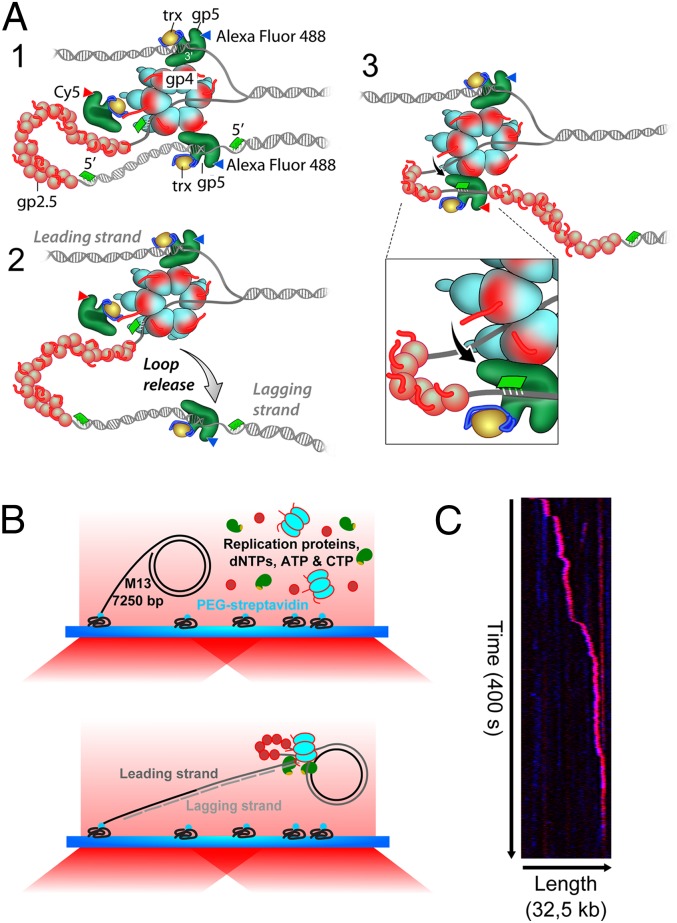
The antiparallel nature of DNA complicates the directional DNA replication by the replisome. In 1975, Alberts et al. (18) proposed a “trombone model” where a tightly bound polymerase replicates the leading strand continuously while the lagging strand is synthesized discontinuously. The Okazaki fragments are located within replication loops and are eventually ligated to form a continuous lagging strand. Upon completion of an Okazaki fragment, the lagging-strand polymerase recycles to a new primer, allowing the polymerase to remain bound to the replication machinery. However, biochemical studies suggest that the association of polymerase to the replisome is more transient. In vivo experiments by Lia et al. showed the exchange of lagging-strand polymerases at the E. coli replisome with cytosolic polymerases (19). In addition, experiments in which actively replicating T7 polymerases are challenged with either a mutant or a fluorescently labeled polymerase show a highly dynamic exchange for both the leading- and lagging-strand polymerase (16, 20, 21). These studies suggest that the two binding modes of the polymerase to the helicase provide a molecular switching mechanism. Relatively weak electrostatic interactions between the TBD or the front basic patch of the polymerase and the C terminus of the helicase ensure the presence of a nonsynthesizing polymerase at the replisome, whereas an alternative and more tight interaction locks actively synthesizing polymerases onto the helicase (10, 12–14). Consequently, the T7 helicase theoretically could bind up to six polymerases corresponding to the six C termini of gp4. These loosely bound, “spare” polymerases can occasionally exchange with the polymerase locked in a replication mode, offering a rapid switching mechanism (21) (Fig. 1A). Also, studies of the E. coli replisome revealed the presence of a third DNA polymerase, in addition to the canonical leading- and lagging-strand polymerases (22, 23).
Fig. 1.
Single-molecule replication experiment. (A) Schematic of hypothesized lagging-strand polymerase exchange. (B) Experimental design. Biotinylated M13 dsDNA molecules are coupled to streptavidin-functionalized coverslips. Addition of the replication proteins and nucleotides initiates DNA replication. The elongated DNA products are flow-stretched, and fluorescently labeled polymerases are observed by TIRF microscopy. (C) Kymograph of the distribution of fluorescently labeled polymerases on the DNA product over time. The fluorescently labeled replisome elongates the DNA in the direction of flow and is visible itself as a bright spot moving away from the surface anchor point.
Even though previous studies elucidated many of the properties of the molecular interactions between the T7 replication proteins in the replisome, important questions on the kinetics and stoichiometry still remain. In particular, direct visualization of the composition of the replication machinery during coordinated leading- and lagging-strand synthesis has not been possible. Observation of the kinetics with which polymerases enter and leave the replisome and the number of polymerases associated with the replication complex is needed to obtain a full picture of the dynamic interplay between the key components of the replisome. Here, we report the visualization of fluorescently labeled T7 DNA polymerases during coordinated replication of a single DNA molecule. To obtain a full description of the dynamic behavior of the polymerases at the replication fork and to identify exchange of polymerases, we perform replication reactions using a mixture of two differently labeled polymerases in equal ratio. The polymerases are labeled with either a blue or a red fluorophore, and we visualize their action simultaneously on hydrodynamically stretched DNA molecules. This experimental design allows us to unravel both the total number of polymerases and their exchange kinetics. The summed intensity of the two colors provides information on the total number of polymerases associated with the replisome, and the fluctuations in the individual colors inform on the kinetics with which they exchange (by the replacement of one color by the second). These experiments show that up to six polymerases can be present within the replisome and that the polymerases are exchanged on the same time scale as the production of Okazaki fragments.
Results
The T7 replication proteins self-assemble into a replisome on a preformed replication fork (16). We have carried out replication reactions on circular double-stranded M13 DNA molecules bearing a replication fork coupled to the surface of a microfluidic flow cell (Fig. 1B) (24, 25). Upon addition of gp4, gp5/trx, gp2.5, and E. coli SSB as well as dNTPs, ATP, and CTP, DNA synthesis was initiated, resulting in rolling-circle DNA replication (see Methods). E. coli SSB was introduced to the replication reactions to more faithfully mimic the physiologically relevant conditions of phage replication in the E. coli host and to minimize nonspecific binding of T7 DNA polymerases to T7 gp2.5-coated ssDNA (Fig. S1). During replication, the newly synthesized leading strand initially becomes part of the circle and is subsequently used as a lagging-strand template. Such a rolling-circle design allows, in principle, for a replication template of indefinite length. By coupling the lagging strand to the surface and by using hydrodynamic flow to extend the continuously growing lagging-strand product, the M13 DNA circle with the replisome moves away from the anchor point of the DNA template. In the experiments described here, we visualized the binding of DNA polymerases to the replisome, located at the most downstream position of the growing DNA chain (Fig. 1C).
To visualize individual gp5/trx, the polymerase was labeled in a 1:1 protein:fluorophore ratio at the amino terminus with either Alexa Fluor 488 or Cy5. The two labeled proteins were used in equal concentrations in the replication reactions. Total internal reflection fluorescence (TIRF) microscopy was used near the critical angle to observe, in real time, the distribution of fluorescence along the entire length of the DNA product. The kymograph in Fig. 1C shows the presence of a bright fluorescent spot containing contributions from both the blue (Alexa Fluor 488) and red (Cy5) polymerases. The polymerases move in the direction of the flow at rates consistent with the known kinetics of the T7 replication reaction. DNA staining after the replication reaction with an intercalating double-stranded DNA dye indicates that this spot resides at the end of the product (Fig. S2), corresponding to the expected location of the replication complex. Formation and release of loops, as has been shown before (26), were not observed during these experiments due to the lower resolution of our assay (∼1,000 bp). As the reaction progressed, the signals from both fluorophores were present continuously, indicating that there are several polymerases bound within the replisome at all times.
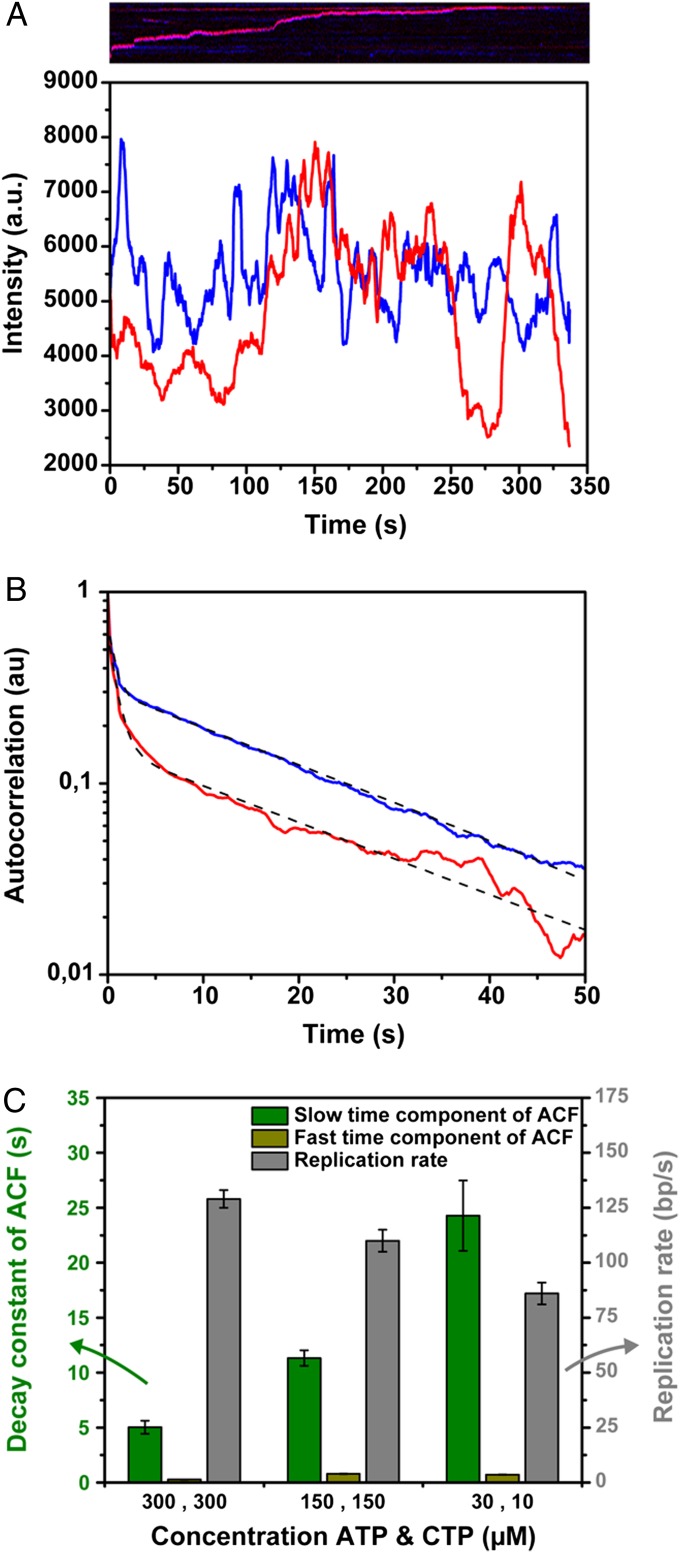
Association and dissociation of polymerases at the replisome were detected by measuring the fluorescence intensity of the replisome spot as a function of time (Fig. 2A). To extract characteristic time scales of fluctuations, we calculated the fluorescence intensity autocorrelation function <I(t)⋅I(t+τ)>, where I denotes fluorescence intensity, t is time, and τ is the time delay (27–29). For all conditions, we observed a double-exponential decay, indicating the presence of two different time scales (Fig. 2B). The faster time scale was consistently observed to be around 1 s (0.8 ± 0.1 s for the conditions used in Fig. 2B), whereas the other was much longer (22.6 ± 0.3 s) and varied with the particular replication reaction conditions used (see below). Because both of the time scales are an order of magnitude lower than the observed lifetime of the fluorophores, bleaching kinetics had a marginal influence on the autocorrelation function (Fig. S3). We interpret the fast time scale as the stochastic binding of polymerases to the carboxy termini of either the gp4 or gp2.5 (10) and the long time scale as a result of the association and dissociation kinetics of single DNA-synthesizing polymerases in the replisome. However, it should be noted that the decay parameters of autocorrelation functions are not suitable to extract information on association and dissociation rate constants separately. Instead, they report on the sum of these properties, henceforth referred to as the exchange time. Due to the equal concentrations of Alexa Fluor 488- and Cy5-labeled polymerases in the DNA replication reactions, half of the polymerase exchange events comprised the exchange of polymerases labeled with the same fluorophore and were thus impossible to observe. Therefore, we halved the polymerase exchange time to correct for exchange of polymerases with the same label. Finally, the autocorrelation functions of both the Alexa Fluor 488- and the Cy5-labeled polymerases for any given reaction condition display the same kinetics, indicating that there is no preferential binding of one of the labeled polymerases at the replication fork. In the subsequent analysis, we therefore average the decay constants of the autocorrelation functions of both fluorophores to determine polymerase exchange times.
Fig. 2.
Autocorrelation function of the fluorescence intensity of the replisome over time. (A) Fluorescence intensity fluctuations of a single replisome over time. The corresponding kymograph is shown above the graph. The blue line represents the Alexa Fluor 488 intensity, whereas the red line shows the Cy5 intensity at the replisome. (B) Averaged normalized autocorrelation function of the fluorescence intensity of 25 replisomes, replicating DNA in the presence of 150 µM ATP and CTP. The autocorrelation function of the Alexa Fluor 488-labeled polymerases is shown in blue, whereas the red curve represents the autocorrelation function of the Cy5-labeled polymerases. The autocorrelation functions are fitted with a dual exponential decay, where we interpret the fast time scale to represent transient electrostatic binding of gp5 to the carboxy termini of either gp4 or gp2.5 (0.92 ± 0.02 s and 0.66 ± 0.02 s for the red and blue curves, respectively), and the slower time scale shows the replacement kinetics of DNA-synthesizing polymerases at the replisome (22.3 ± 0.5 s for the red and 22.9 ± 0.2 s for the blue curve). (C) Bar plot of the slow and fast time scale of the autocorrelation function and the replication rate as a function of ATP and CTP concentration. The characteristic time scales of DNA-synthesizing polymerase exchange (green bars) are constituted of the slower decay constants of the autocorrelation function divided by 2, to correct for exchange events of polymerases with the same dye attached which decrease the exponential decay of the autocorrelation functions by twofold. The grey bars show the DNA replication rate computed by tracking the position of the replisome over time. Both the polymerase exchange times and DNA replication rates were calculated from trajectories of multiple individual replisomes.
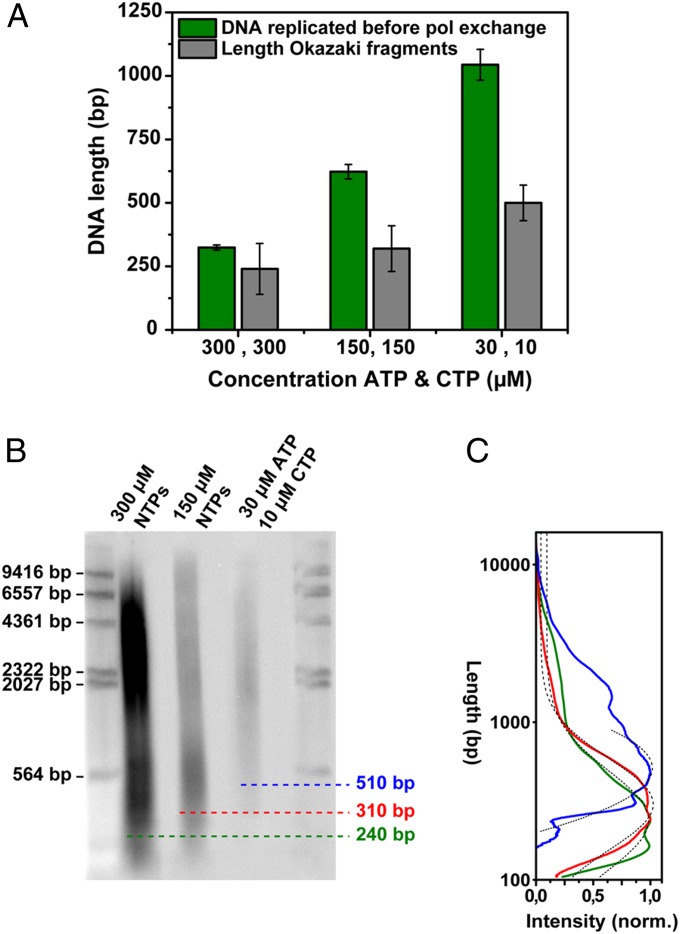
Next, we varied the frequency of primer formation on the lagging strand by changing the concentration of ATP and CTP, the only ribonucleotides required for T7 primer synthesis (30). The priming frequency controls the length of Okazaki fragments (26) and potentially affects the exchange kinetics of polymerases at the fork. Fig. 2C (green bars) shows a strong dependence of the slower time scale of the autocorrelation functions on ATP and CTP concentration, indicating that the polymerase exchange kinetics slow when less ATP and CTP are available. In addition, we tracked the position of the replisome over time to determine the rate of DNA synthesis (Fig. 2C, grey bars). By multiplying the replication rate with the polymerase exchange time, we obtained a measure for the amount of DNA synthesized over the characteristic time scale of polymerase exchange (Fig. 3A, green bars). Separating the replication reaction products on a denaturing gel (Fig. 3B) and determining the average Okazaki fragment length for the different ATP and CTP concentrations (Fig. 3C) allowed us to directly compare Okazaki fragment lengths to the amount of DNA synthesized before polymerases exchange (Fig. 3A). The similarity between these two measures indicate that polymerase exchange occurs on the same time scale as the time required to synthesize at most a few Okazaki fragments.
Fig. 3.
Length of DNA replicated by a single polymerase in comparison with Okazaki fragment length. (A) Bar plot of the average length of DNA replicated on the time scale of polymerase (pol) exchange and the Okazaki fragment length for different ATP and CTP concentrations. (B) Alkaline agarose gel analysis of DNA products. DNA synthesis was carried out in the standard reaction containing minicircular DNA substrates (as described in ref. 32) with varying concentrations of ATP and CTP and with [α-32P]dCTP. After incubation for 15 min, the reaction products were denatured and analyzed by electrophoresis through a 0.8% (wt/vol) alkaline agarose gel. Lanes 1 and 5 contain a HindIII digest of λ DNA to provide length markers. Lanes 2, 3, and 4 represent the lagging-strand products for 300 µM NTPs, 150 µM NTPs, and 30 µM ATP and 10 µM CTP, respectively. (C) Intensity profiles of lanes 2–4 of the denaturing gel. To determine the average product length for a given condition, the intensity profile along a lane was corrected to take into account the higher intensity of larger fragments and subsequently fitted with a single Gaussian. The maximum of the Gaussian peaks are at 240 ± 100 bp, 300 ± 90 bp, and 510 ± 70 bp for reactions with 300 µM ATP and CTP, 150 µM ATP and CTP, and 30 µM ATP with 10 µM CTP, respectively. Correction of the intensity profiles for different amounts of radiolabel present in different fragment lengths results in Okazaki fragment lengths that are shorter (140−580 bp) than previously reported (1,000−6,000 bp) (32), but are more representative of their true distributions.
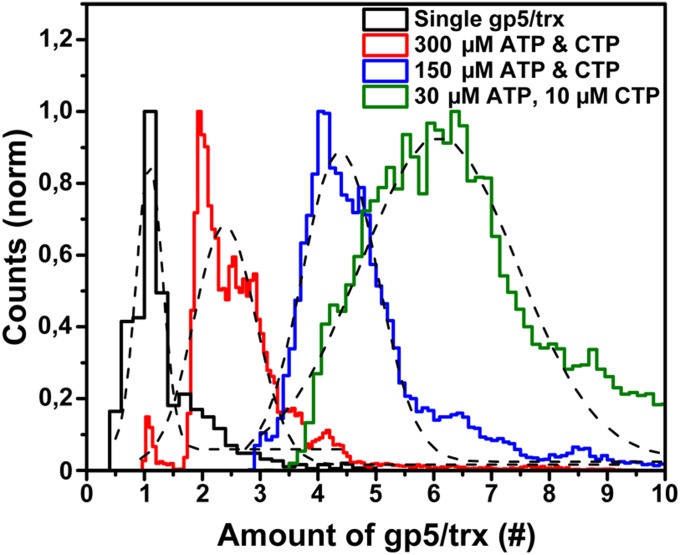
Polymerase exchange events require the presence of readily available polymerases at the replisome in addition to the DNA-synthesizing polymerases. Here, we quantified the fluorescence intensity of the replisome spots to determine the number of polymerases residing at the replication fork. First, we determined the average fluorescence intensity of a single dye-labeled polymerase. Given the challenges associated with controllably and stably binding a single polymerase to the end of a long DNA tether, we chose to perform this calibration by immobilizing individual polymerases in an agarose gel (31) and measuring the intensities of only those proteins that were at the same height above the coverslip, and thus subject to the same excitation intensity, as the polymerases in the replication experiment (see Methods). The narrow nature of this fluorescence intensity distribution suggests that the majority of the polymerases are labeled with a single dye (Fig. 4, black bars), consistent with the spectroscopically obtained ratio of dye to protein of 1:1.
Fig. 4.
Stoichiometry of polymerases at the replication fork. The steady-state binding of polymerases at the replisome for different ATP and CTP concentrations was determined by dividing the fluorescence intensity at the replisome, averaged over 10 s by a sliding average window, by the intensity of a single labeled polymerase. The total fluorescence represents the summation of the number of Alexa Fluor 488- and Cy5-labeled polymerases. Fitting the distributions with Gaussians (black) resulted in maximum values with a SD of 2.4 ± 0.3 gp5/trx, 4.4 ± 0.3 gp5/trx, and 6.1 ± 0.7 gp5/trx for 300 µM ATP and CTP, 150 µM ATP and CTP, and 30 µM ATP with 10 µM CTP, respectively.
The steady-state population of bound polymerases can be quantified by dividing the average fluorescence intensity of the replisomes by the fluorescence intensity of a single polymerase. Fig. 4 shows the stoichiometry of labeled polymerases at the replication machinery as a function of the concentration of ATP and CTP (see also Fig. S4 for the distributions of the individual colored polymerases). A clear correlation can be observed between the ATP and CTP concentration and the number of polymerases bound at the replisome. In particular, at low ribonucleotide concentration, and thus a low priming frequency and a slow polymerase turnover, we found an average of six polymerases to be present at the replication fork (Fig. 4). At high ATP and CTP concentrations, the number of polymerases reduced to two to three. The widths of the distributions mainly stem from the intensity fluctuations caused by the Brownian motions of the replisome in and out of the evanescent excitation field. This artificial broadening explains the apparent existence of a population corresponding to more than six polymerases bound to the replisome. This interpretation is further confirmed by the fact that in none of our experimental conditions was the center of the distribution observed to be higher than six polymerases bound.
The dependency of polymerase copy number on priming frequency is consistent with the notion that at the constant polymerase concentration in our experiments (20 nM), the association rate remains constant, whereas the dissociation rate is modulated by the frequency of primer and Okazaki fragment synthesis. Notably, the number of polymerases in the replisome is two or more for all ribonucleotide concentrations under the experimental conditions used. Thus, besides the leading- and lagging-strand polymerases, additional polymerases can bind to the replisome.
Discussion
Extensive research over the past decades has provided insight into the functioning of the individual proteins involved in DNA replication. However, the architecture of the replisome and its dynamics during replication remain poorly understood. Here, we have visualized the DNA polymerases associated with individual replisomes during coordinated DNA replication. These single-molecule experiments have enabled us to examine the dynamics of polymerase exchange and to determine the number of polymerases under different conditions. An important molecular step controlling the dynamics of the T7 DNA replication machinery is the synthesis of a primer and its handoff to the lagging-strand polymerase (9). This event initiates the synthesis of an Okazaki fragment synthesis and thus effectively controls the timing of the sequence of enzymatic events on the lagging strand. We observe that changing the frequency of primer synthesis directly impacts the dynamics of polymerase exchange. Under conditions where the concentration of DNA polymerase in solution is kept constant, thus giving rise to a constant rate of association of the polymerase with the replisome, a change in exchange dynamics can be interpreted as a change in the rate with which DNA polymerase dissociates from the replisome. A comparison between the observed exchange dynamics and the length of Okazaki fragments with different priming frequencies reveals that the exchange times are comparable to the time needed to synthesize only a few Okazaki fragments. This observation strongly suggests that the majority of the DNA polymerases do not remain associated with the replisome much longer than the time required to synthesize one, or at most a few, Okazaki fragments and are then triggered to be released.
It should be pointed out that the measured exchange dynamics also contain a contribution of the on and off rates of the leading-strand DNA polymerase. Loparo et al. (21) found the binding lifetime of the leading-strand polymerase under similar conditions to be ∼43 s, significantly longer than the exchange times we observe for the total population of polymerases at the fork. In the current studies, the exchange times varied from 5 to 23 s, at high and low priming frequencies, respectively. As a result, the higher frequency of lagging-strand polymerase exchange will dominate the decay constants of the autocorrelation function and will render our measurements less sensitive to the slow component corresponding to exchange of the leading-strand polymerase.
Loparo et al. demonstrated that during leading-strand DNA synthesis, polymerases in addition to the replicating DNA polymerase, can associate with the replisome (21). Supported by earlier biochemical evidence (9, 13), these data suggested that multiple DNA polymerases bind to the acidic C-terminal tail of the gp4 helicase. The question remains how many polymerases are associated with the T7 replisome during coordinated leading- and lagging-strand synthesis. By monitoring the total fluorescence intensity corresponding to labeled polymerases, we show that under all priming conditions, there are two or more polymerases bound to the replisome. At high priming frequencies, we observed the presence of two to three polymerases, and at low frequencies, a total of six polymerases associate with the replisome. The dependence of the number of polymerases present at the fork on priming frequency can be understood by considering the faster dissociation of the polymerases with more frequent priming events. With a constant rate of association, an increase in off rate results in a lower steady-state population residing at the replisome. Conversely, for lower priming frequencies, the low dissociation rate results in a buildup of a large population of DNA polymerases.
The picture that emerges is that of a highly dynamic replisome. Lagging-strand DNA polymerases dissociate frequently from the replisome, conceivably residing at the fork only for the synthesis of one or two Okazaki fragments, while a continuous populating of the gp4 C-terminal binding sites provides a nearby, readily employable, pool of DNA polymerases. The proximity of these DNA polymerases to the priming site results in an extremely high effective local concentration (>100 mM) of DNA polymerases. The result is an efficient recruitment of a polymerase to a newly synthesized primer.
The short exchange times of polymerases seem to be contradictory to the previous observation of leading- and lagging-strand synthesis being highly resistant to dilution and suggesting a very stable complex (32). However, dilution of the replication reaction greatly reduces the association rate of polymerases to the replication fork and thus greatly reduces the number of polymerases bound to the C termini of gp4. In the absence of an excess of polymerases residing on the helicase, there will be no competition for the lagging-strand DNA polymerase to rebind at a new primer and thus synthesize a larger number of Okazaki fragments. Such a mechanism is comparable to that observed for the reuse of beta clamps in the E. coli replication system (25). In this system, there is efficient exchange between beta clamps at the replisome and those in solution in the presence of a solution pool of clamps. An efficient recycling of clamps at the fork occurs in the absence of free clamps. This ability of the replisome to revert to reusing components in the absence of a solution pool may be important to sustain DNA replication under suboptimal replication conditions.
A model in which lagging-strand DNA polymerases remain associated with the replisome for the duration of synthesis of only a few Okazaki fragments does call into question the mechanistic necessity of a replication loop. Originally proposed by Bruce Alberts (18), the replication loop is formed by the lagging-strand DNA polymerase associating with the replisome. This association gives rise to a loop that is created for every new Okazaki fragment, growing from the ssDNA product of helicase unwinding and the dsDNA product of lagging-strand synthesis. One of the attractive features of such a loop model is that the lagging-strand DNA polymerase remains associated with the replisome and thus could support many rounds of Okazaki fragment synthesis. Even though many studies have demonstrated the existence of such loops (26, 33, 34), one can imagine that its formation is an accidental byproduct of the necessity of the DNA polymerase to associate with the replisome to support helicase-coupled synthesis at the leading strand and primer handoff at the lagging strand.
Methods
Single-stranded M13mp18 (New England Biolabs) is biotinylated by annealing a complementary biotinylated oligo to the M13 template. Subsequently, the primed M13 is filled in by adding T7 DNA polymerase (New England Biolabs), dNTPs, and a replication buffer containing MgCl2. Replication proteins are removed from the filled-in DNA products by phenol/chloroform extraction and stored in 10 mM Tris-HCl, 1mM EDTA (TE) buffer (24).
Previously published protocols were used to prepare gp4 (35), gp5/trx (36), gp2.5 (37), and E. coli SSB (38). Polymerase labeling reactions were performed according to the procedure described by Etson et al. (39) with an excess of either Alexa Fluor 488 carboxylic acid succinimidyl ester (Invitrogen) or Cy5 NHS ester (Lumiprobe) for an hour at room temperature. The conjugation reactions were biased to the N-terminal groups by adjusting the pH to 7.6. The N-terminal α-amino group has a pKa of 7.7 ± 0.5, whereas the ε-amino group of a lysine side chain, the other major target of the conjugation reaction, has a pKa of 10.5 ± 1.1 (40). The T7 polymerase contains 1 N-terminal site versus 62 lysine side chains, suggesting labeling to be 10 times more specific for the N-terminal site than for the lysine side chains. Subsequently, the excess of unbound dyes is filtered out by size exclusion spin columns (Micro Bio-Spin 6 Columns, Bio Rad) and the degree of labeling was determined by measuring the absorbance spectrum of the samples and found to be 1.3 ± 0.1 fluorophore per gp5/trx for Alexa Fluor 488-labeled polymerases and 1.0 ± 0.1 dye for Cy5-labeled polymerases.
Our replication reactions contained a protein mixture of 2.5 nM gp4, 20 nM gp5/trx of which 10 nM was Alexa Fluor 488-labeled and 10 nM Cy5-labeled, 180 nM gp2.5, and 100 nM E. coli SSB throughout the whole experiment. The experiments were performed in a buffer containing 40 mM Tris (pH 7.5), 50 mM potassium glutamate, 10 mM MgCl2, 0.1 mg/mL BSA, 0.75 µM DTT, 0.6 mM dNTPs, and various concentrations of ATP and CTP. To increase the lifetime of our fluorophores, we added 1 mM Trolox, 10% (wt/vol) glucose, 0.45 mg/mL glucose oxidase, and 21 µg/mL catalase to our replication reactions. In addition, the combination of 1 mM Trolox with an enzymatic oxygen-scavenging system was shown to eliminate Cy5 blinking and dramatically reduce photobleaching (41). Abovementioned reaction mixtures are continuously in the flow solution to continuously replenish fresh reaction components to the DNA replication reactions.
Fluorescently labeled polymerases were illuminated with a 488-nm and 641-nm laser (Coherent) through a 100× TIRF objective [Olympus, UApoN, N.A. = 1.49 (oil)]. A dual-color inset was used to image both the Alexa Fluor 488 and the Cy5 fluorescent signal simultaneously. Images were captured with an EMCCD camera (Hamamatsu) using Meta Vue imaging software (Molecular Devices) with a typical frame rate of five frames per second.
To identify the fluorescence intensity of a single labeled polymerase, polymerases were dissolved in 6% low-gelling agarose (Sigma Aldrich) containing 10% wt/vol glucose, 0.45 mg/mL glucose oxidase, 21 µg/mL catalase, and 1 mM Trolox. We identified the height of the replisomes above the coverslip during the replication experiments by measuring the height difference in the focal plane of the flow cells coverslip and the replisomes. Subsequently, we imaged the single agarose-trapped polymerases at the same height above the coverslip to reproduce identical excitation conditions as in the replication experiments. The entire field of view was corrected for the illumination profile of the laser and rectified for the background. The fluorescence intensity distributions of the Alexa Fluor 488- as well as the Cy5-labeled polymerases were fitted with a single Gaussian and normalized against their peak fluorescence intensity. The normalized fluorescence intensities of the Alexa Fluor 488- and the Cy5-labeled polymerases were added and normalized against the counts, resulting in the black fluorescence intensity distribution shown in Fig. 4.
To determine the fluorescence intensity of the replisome, the entire field of view was corrected for the illumination profile of the laser, and subsequently a region of interest was selected around the replisome spot and rectified for the background for each frame in the movie. Then, the fluorescence intensity of the region of interest was calculated for each frame of the movie. The location of the replisome spot in time was tracked by fitting a 2D Gaussian to the replisome spot for each frame. Replication rate is calculated by linear fitting the position of the replisome in time.
Okazaki length distributions were determined using a denaturing alkaline agarose gel. Replication reactions are carried out for 30 min at room temperature using a minicircle template with [α-32P]dCTP and different concentrations of ATP and CTP. Subsequently, the reaction is terminated by adding EDTA and running the reaction products over a spin column (PD SpinTrap G-25, GE Healthcare). Then, the reaction products are loaded onto a 0.8% agarose gel, and electrophoresis is performed for 6 h at 4 V/cm. The products are precipitated with trichoroacetic acid. Subsequently, the gel is dried on filter paper overnight to decrease the gel volume for imaging purposes. The gel is imaged on a phosphor storage plate and read out by a Typhoon system (Amersham Biosciences, run in the storage phosphor mode). Intensity profiles of the lanes were background rectified and divided by the DNA length, corresponding to migration distance in the gel. Consequently, the profiles were corrected for the amount of incorporated [α-32P]dCTP per DNA fragment length. Subsequently, the intensity profiles were fitted with single Gaussian distributions.
Supplementary Material
Acknowledgments
The authors would like to thank Prof. N. E. Dixon and Dr. Slobodan Jergic for their kind gift of purified E. coli SSB and S. Moskowitz for illustrations. A.M.v.O. acknowledges funding from the Netherlands Organization for Scientific Research (Vici 680-47-607) and the European Research Council (ERC Starting Grant 281098).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402010111/-/DCSupplemental.
References
- 1.Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 2.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101(6):589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 3.Mendelman LV, Richardson CC. Requirements for primer synthesis by bacteriophage T7 63-kDa gene 4 protein. Roles of template sequence and T7 56-kDa gene 4 protein. J Biol Chem. 1991;266(34):23240–23250. [PubMed] [Google Scholar]
- 4.Kato M, Ito T, Wagner G, Richardson CC, Ellenberger T. Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol Cell. 2003;11(5):1349–1360. doi: 10.1016/s1097-2765(03)00195-3. [DOI] [PubMed] [Google Scholar]
- 5.Fujiyama A, Kohara Y, Okazaki T. Initiation sites for discontinuous DNA synthesis of bacteriophage T7. Proc Natl Acad Sci USA. 1981;78(2):903–907. doi: 10.1073/pnas.78.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frick DN, Richardson CC. Interaction of bacteriophage T7 gene 4 primase with its template recognition site. J Biol Chem. 1999;274(50):35889–35898. doi: 10.1074/jbc.274.50.35889. [DOI] [PubMed] [Google Scholar]
- 7.Doublié S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391(6664):251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 8.Tabor S, Huber HE, Richardson CC. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J Biol Chem. 1987;262(33):16212–16223. [PubMed] [Google Scholar]
- 9.Lee JB, et al. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439(7076):621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 10.Hamdan SM, et al. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc Natl Acad Sci USA. 2005;102(14):5096–5101. doi: 10.1073/pnas.0501637102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SJ, Marintcheva B, Hamdan SM, Richardson CC. The C-terminal residues of bacteriophage T7 gene 4 helicase-primase coordinate helicase and DNA polymerase activities. J Biol Chem. 2006;281(35):25841–25849. doi: 10.1074/jbc.M604602200. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, et al. Helicase-DNA polymerase interaction is critical to initiate leading-strand DNA synthesis. Proc Natl Acad Sci USA. 2011;108(23):9372–9377. doi: 10.1073/pnas.1106678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamdan SM, et al. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol Cell. 2007;27(4):539–549. doi: 10.1016/j.molcel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 14.Kulczyk AW, et al. An interaction between DNA polymerase and helicase is essential for the high processivity of the bacteriophage T7 replisome. J Biol Chem. 2012;287(46):39050–39060. doi: 10.1074/jbc.M112.410647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh S, Hamdan SM, Richardson CC. Two modes of interaction of the single-stranded DNA-binding protein of bacteriophage T7 with the DNA polymerase-thioredoxin complex. J Biol Chem. 2010;285(23):18103–18112. doi: 10.1074/jbc.M110.107656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Chastain PD, 2nd, Kusakabe T, Griffith JD, Richardson CC. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Mol Cell. 1998;1(7):1001–1010. doi: 10.1016/s1097-2765(00)80100-8. [DOI] [PubMed] [Google Scholar]
- 17.He ZG, Rezende LF, Willcox S, Griffith JD, Richardson CC. The carboxyl-terminal domain of bacteriophage T7 single-stranded DNA-binding protein modulates DNA binding and interaction with T7 DNA polymerase. J Biol Chem. 2003;278(32):29538–29545. doi: 10.1074/jbc.M304318200. [DOI] [PubMed] [Google Scholar]
- 18. Alberts BM, et al. (1975) DNA synthesis and its regulation. ICN-UCLA Symposium on Molecular and Cellular Biology, eds Goulian M, Hanawalt P, Fox CF (W.H. Benjamin, Menlo Park, Calif.), Vol 3, pp. 241−269.
- 19.Lia G, Michel B, Allemand JF. Polymerase exchange during Okazaki fragment synthesis observed in living cells. Science. 2012;335(6066):328–331. doi: 10.1126/science.1210400. [DOI] [PubMed] [Google Scholar]
- 20.Johnson DE, Takahashi M, Hamdan SM, Lee SJ, Richardson CC. Exchange of DNA polymerases at the replication fork of bacteriophage T7. Proc Natl Acad Sci USA. 2007;104(13):5312–5317. doi: 10.1073/pnas.0701062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loparo JJ, Kulczyk AW, Richardson CC, van Oijen AM. Simultaneous single-molecule measurements of phage T7 replisome composition and function reveal the mechanism of polymerase exchange. Proc Natl Acad Sci USA. 2011;108(9):3584–3589. doi: 10.1073/pnas.1018824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McInerney P, Johnson A, Katz F, O’Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007;27(4):527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 23.Reyes-Lamothe R, Sherratt DJ, Leake MC. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328(5977):498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanner NA, van Oijen AM. Visualizing DNA replication at the single-molecule level. Methods Enzymol. 2010;475:259–278. doi: 10.1016/S0076-6879(10)75011-4. [DOI] [PubMed] [Google Scholar]
- 25.Tanner NA, et al. E. coli DNA replication in the absence of free β clamps. EMBO J. 2011;30(9):1830–1840. doi: 10.1038/emboj.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamdan SM, Loparo JJ, Takahashi M, Richardson CC, van Oijen AM. Dynamics of DNA replication loops reveal temporal control of lagging-strand synthesis. Nature. 2009;457(7227):336–339. doi: 10.1038/nature07512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madge D, Elson E, Webb WW. Thermodynamic fluctuations in a reacting system—Measurement by fluorescence correlation spectroscopy. Phys Rev Lett. 1972;29(11):705–708. [Google Scholar]
- 28.Hess ST, Huang S, Heikal AA, Webb WW. Biological and chemical applications of fluorescence correlation spectroscopy: A review. Biochemistry. 2002;41(3):697–705. doi: 10.1021/bi0118512. [DOI] [PubMed] [Google Scholar]
- 29.Ries J, Schwille P. Fluorescence correlation spectroscopy. Bioessays. 2012;34(5):361–368. doi: 10.1002/bies.201100111. [DOI] [PubMed] [Google Scholar]
- 30.Tabor S, Richardson CC. Template recognition sequence for RNA primer synthesis by gene 4 protein of bacteriophage T7. Proc Natl Acad Sci USA. 1981;78(1):205–209. doi: 10.1073/pnas.78.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu HP, Xun L, Xie XS. Single-molecule enzymatic dynamics. Science. 1998;282(5395):1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 32.Debyser Z, Tabor S, Richardson CC. Coordination of leading and lagging strand DNA synthesis at the replication fork of bacteriophage T7. Cell. 1994;77(1):157–166. doi: 10.1016/0092-8674(94)90243-7. [DOI] [PubMed] [Google Scholar]
- 33.Chastain PD, 2nd, Makhov AM, Nossal NG, Griffith J. Architecture of the replication complex and DNA loops at the fork generated by the bacteriophage t4 proteins. J Biol Chem. 2003;278(23):21276–21285. doi: 10.1074/jbc.M301573200. [DOI] [PubMed] [Google Scholar]
- 34.Park K, Debyser Z, Tabor S, Richardson CC, Griffith JD. Formation of a DNA loop at the replication fork generated by bacteriophage T7 replication proteins. J Biol Chem. 1998;273(9):5260–5270. doi: 10.1074/jbc.273.9.5260. [DOI] [PubMed] [Google Scholar]
- 35.Notarnicola SM, Park K, Griffith JD, Richardson CC. A domain of the gene 4 helicase/primase of bacteriophage T7 required for the formation of an active hexamer. J Biol Chem. 1995;270(34):20215–20224. doi: 10.1074/jbc.270.34.20215. [DOI] [PubMed] [Google Scholar]
- 36.Tabor S, Richardson CC. Selective oxidation of the exonuclease domain of bacteriophage T7 DNA polymerase. J Biol Chem. 1987;262(32):15330–15333. [PubMed] [Google Scholar]
- 37.Hyland EM, Rezende LF, Richardson CC. The DNA binding domain of the gene 2.5 single-stranded DNA-binding protein of bacteriophage T7. J Biol Chem. 2003;278(9):7247–7256. doi: 10.1074/jbc.M210605200. [DOI] [PubMed] [Google Scholar]
- 38.Mason CE, et al. Escherichia coli single-stranded DNA-binding protein: NanoESI-MS studies of salt-modulated subunit exchange and DNA binding transactions. J Am Soc Mass Spectrom. 2013;24(2):274–285. doi: 10.1007/s13361-012-0552-2. [DOI] [PubMed] [Google Scholar]
- 39.Etson CM, Hamdan SM, Richardson CC, van Oijen AM. Thioredoxin suppresses microscopic hopping of T7 DNA polymerase on duplex DNA. Proc Natl Acad Sci USA. 2010;107(5):1900–1905. doi: 10.1073/pnas.0912664107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grimsley GR, Scholtz JM, Pace CN. A summary of the measured pK values of the ionizable groups in folded proteins. Protein Sci. 2009;18(1):247–251. doi: 10.1002/pro.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Methods. 2006;3(11):891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.