Significance
In response to injury or disease, skeletal muscle has the capacity for regeneration and repair. Muscle regeneration is orchestrated by a population of stem cells called satellite cells that reside between the basal lamina and sarcolemma of muscle fibers. Upon muscle injury, activated satellite cells proliferate and undergo differentiation to recreate functional muscle tissue. In this work, we show that deletion of three members of the MEF2 family of transcription factors, MEF2A, C, and D, in satellite cells prevents muscle regeneration because of a failure of differentiation. Also, we identify a collection of muscle genes regulated by MEF2 in satellite cells. These findings provide a potential molecular inroad into the process of muscle regeneration through modulation of MEF2 activity.
Keywords: myogenesis, myogenic regulatory factor, myotube
Abstract
Regeneration of adult skeletal muscle following injury occurs through the activation of satellite cells, an injury-sensitive muscle stem cell population that proliferates, differentiates, and fuses with injured myofibers. Members of the myocyte enhancer factor 2 (MEF2) family of transcription factors play essential roles in muscle differentiation during embryogenesis, but their potential contributions to adult muscle regeneration have not been systematically explored. To investigate the potential involvement of MEF2 factors in muscle regeneration, we conditionally deleted the Mef2a, c, and d genes, singly and in combination, within satellite cells in mice, using tamoxifen-inducible Cre recombinase under control of the satellite cell-specific Pax7 promoter. We show that deletion of individual Mef2 genes has no effect on muscle regeneration in response to cardiotoxin injury. However, combined deletion of the Mef2a, c, and d genes results in a blockade to regeneration. Satellite cell-derived myoblasts lacking MEF2A, C, and D proliferate normally in culture, but cannot differentiate. The absence of MEF2A, C, and D in satellite cells is associated with aberrant expression of a broad collection of known and unique protein-coding and long noncoding RNA genes. These findings reveal essential and redundant roles of MEF2A, C, and D in satellite cell differentiation and identify a MEF2-dependent transcriptome associated with skeletal muscle regeneration.
Adult skeletal muscle has a remarkable capacity for repair and regeneration in response to injury, aging, and disease (1). Muscle regeneration is a precisely orchestrated process mediated by a population of stem cells, called satellite cells, that reside beneath the basal lamina of myofibers (2–4). In normal skeletal muscle, satellite cells, marked by expression of paired box 7 (Pax7), are maintained in a quiescent state and, upon muscle damage, are activated to reenter the cell cycle (2–4). Activated satellite cells express both Pax7 and the myogenic regulatory factors (MRFs) MyoD and Myf5 and assume a myoblast identity. After extensive proliferation, myoblasts undergo differentiation and fusion with each other or existing myofibers to recreate functional muscle tissue. Satellite cells also undergo asymmetric cell division to replenish the reservoir of quiescent stem cells (5).
Many of the same transcription factors that control embryonic myogenesis are redeployed during adult regenerative myogenesis. Members of the MyoD family of myogenic basic helix–loop–helix proteins, for example, play essential roles in the control of muscle differentiation during embryonic development and adult muscle regeneration (6, 7). The myogenic activity of these transcription factors is enhanced through their interaction with members of the MEF2 family. MEF2 transcription factors lack myogenic activity alone, but they interact with MRFs to synergistically activate muscle-specific genes and the myogenic differentiation program (8). Chromatin immunopreciptation and gene expression analysis revealed that MRFs and MEF2 directly regulate the expression of an extensive array of muscle structural genes, and other transcription factors that propagate and amplify the signals initiated by MRFs (9).
In vertebrates, there are four Mef2 genes, Mef2a, b, c, and d, that display distinct but overlapping temporal and spatial expression patterns in embryonic and adult tissues (10). MEF2 proteins share high sequence homology in their DNA binding and dimerization domains but are divergent in their C-terminal transcriptional activation domains. Global deletion of Mef2a or Mef2d in mice has little or no effect on skeletal muscle development (11). Skeletal muscle-specific deletion of Mef2c results in neonatal lethality due to defects in muscle integrity and failure in sarcomere formation (11, 12). However, the potential involvement of MEF2 proteins in muscle regeneration or adult myogenesis has not been systematically explored. In this regard, a recent study reported that adult Mef2a-null mice showed impaired muscle regeneration in response to injury, concluding that MEF2A is essential for satellite cell activation and regeneration (13). However, because MEF2A is expressed in a range of cell types in addition to skeletal muscle, global deletion of Mef2a cannot definitively distinguish its function in satellite cells versus other cells, such as inflammatory cells, fibroblasts, vascular cells, neurons, and injured myofibers that also influence the regenerative process.
In the present study, we explored the specific functions of MEF2A, C, and D in adult satellite cells and skeletal muscle regeneration. We show that deletion of individual Mef2 genes in satellite cells does not influence muscle regeneration following cardiotoxin injury. However, muscle regeneration is completely abolished when the Mef2a, c, and d genes are deleted in combination in satellite cells. The dramatic regeneration defect associated with the absence of MEF2A, C, and D reflects a failure of satellite cell-derived myoblasts to differentiate and fuse into multinucleated myotubes. Our findings provide unequivocal evidence that MEF2 is essential within satellite cells for activation of the adult myogenic gene program in mice and also reveal a MEF2-dependent transcriptome associated with muscle regeneration.
Results
MEF2 Regulation During Skeletal Muscle Regeneration.
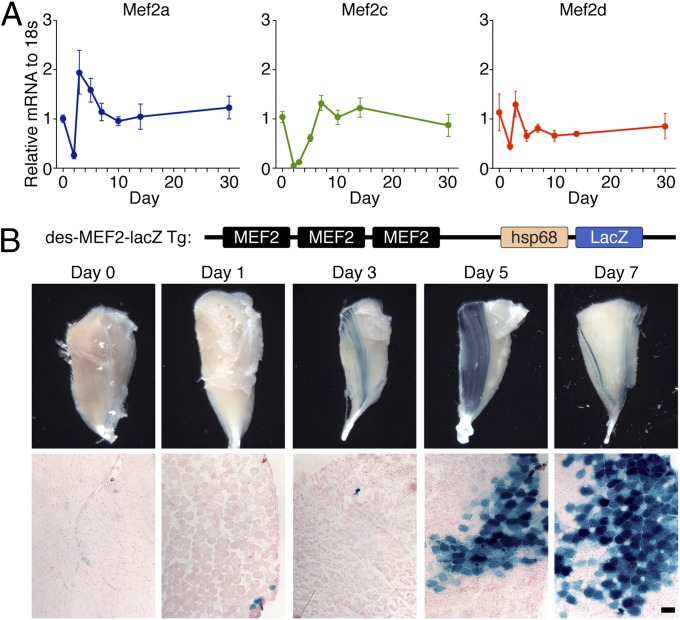
To determine the expression patterns of MEF2 factors during skeletal muscle regeneration, we induced skeletal muscle injury and regeneration in mice by injecting cardiotoxin (CTX) in the tibialis anterior (TA) muscle and analyzed MEF2A, C, and D expression profiles during the course of muscle regeneration. As shown in Fig. 1A, both MEF2A and C were dramatically down-regulated by day 2 after CTX injury, reflecting the loss of muscle tissue caused by CTX injection. On day 3 after injury, MEF2A expression was markedly up-regulated in injured muscle and remained highly expressed throughout the course of regeneration. Expression of MEF2C was gradually restored to normal by 7 d after injury, whereas MEF2D expression was only modestly decreased on day 2 and restored to normal levels by day 3 after injury. These results are consistent with previous reports that MEF2A and C are up-regulated during differentiation of both C2C12 cells and primary myoblasts derived from satellite cells, with MEF2A activation preceding MEF2C by 1 or 2 d (13, 14).
Fig. 1.
Mef2 expression during skeletal muscle regeneration. (A) Real-time RT-PCR showing expression of Mef2a, Mef2c, and Mef2d mRNA during the course of muscle regeneration. TA muscle was subjected to CTX injection and was harvested on indicated days after injury for RNA analysis. For each time point, values are normalized to 18s rRNA, and then normalized to day 0 (before injury), which is set at 1. Data are presented as mean ± SEM. n = 5 for each time point. (B) Expression of lacZ gene in des-MEF2-lacZ transgenic mice following CTX injury. (B, Upper) Whole-mount images of TA muscle before and after injury. (B, Lower) β-galactosidase staining of transverse sections of uninjured (day 0) and injured muscles isolated from des-MEF2-lacZ transgenic mice. Initial activation of expression on day 3 is variable and weak and not detectable in histological sections. (Scale bar: 100 μm.)
We also analyzed transcriptional activity of MEF2 during regeneration, using the des-MEF2-lacZ transgenic mice, which harbor a lacZ reporter gene controlled by three tandem copies of the MEF2 site and flanking sequences from the desmin enhancer (15). This transgene has been shown to serve as a sensitive indicator of MEF2 transcriptional activity in cardiac and skeletal muscle lineages during embryogenesis (15). We subjected the des-MEF2-lacZ mice to CTX injury and analyzed transgene expression by β-galactosidase staining. As shown in Fig. 1B, the des-MEF2-lacZ transgene shows minimal expression in only a few myofibers of adult TA muscle in the absence of injury. Upon CTX injection, the des-MEF2-lacZ transgene began to be expressed at day 3 and became strongly expressed by day 5. We conclude that the transcriptional activity of MEF2 is up-regulated during skeletal muscle regeneration.
Redundant Functions of MEF2A, C, and D During Adult Skeletal Muscle Regeneration.
To determine the potential functions of individual MEF2 factors in adult skeletal muscle regeneration, we generated mice with conditional null alleles of Mef2a, c, and d and bred them to Pax7-CreERT2 mice, which contain a tamoxifen-inducible Cre recombinase-estrogen receptor fusion protein cassette within the Pax7 allele (16). Details of the conditional Mef2 alleles have been described (17–19). Because Pax7 is expressed in both quiescent and activated satellite cells, Pax7-CreERT2 mice allow for inducible Cre-mediated gene deletion specifically in satellite cells (16).
To activate Cre recombinase expression in vivo, we injected 8-wk-old mice of the following genotypes with tamoxifen: Pax7-CreERT2/Mef2aloxp/loxp, Pax7-CreERT2/Mef2cloxp/loxp, and Pax7-CreERT2/Mef2dloxp/loxp. Mef2aloxp/loxp mice lacking the Cre transgene were used as controls (referred to as wild-type; WT) (Fig. S1A).
Mice with conditional deletion of individual Mef2 genes showed neither overt phenotypes nor abnormalities in muscle histology before injury. To evaluate their regenerative potential, we injected CTX into the TA muscle to induce muscle damage and regeneration. Seven days after injury, WT TA muscle was composed primarily of regenerating myofibers, as recognized by the presence of centralized nuclei (Fig. S1B). The regenerative response of all mice lacking individual Mef2 genes was indistinguishable from that of WT mice following CTX injury (Fig. S1B). We also subjected mice lacking Mef2a to BaCl2 injection, as an independent means of muscle injury, and observed efficient regeneration in WT mice and mice lacking the Mef2a gene (Fig. S1C). Quantification of Mef2c and Mef2d mRNA expression in Pax7-CreERT2/Mef2aloxp/loxp muscle showed no compensatory up-regulation of these genes (Fig. S1D).
Deletion of pairs of MEF2 factors such as MEF2A and MEF2C or MEF2A and MEF2D also did not impair regeneration after CTX injury. Moreover, muscle regeneration occurred normally in mice with only a single functional allele of Mef2c or Mef2d in mice of the following genotypes: Pax7-CreERT2/Mef2aloxp/loxp/Mef2cloxp/loxp/Mef2dloxp/+; or Pax7-CreERT2; Mef2aloxp/loxp/Mef2cloxp/+/Mef2d loxp/loxp (Fig. S1E).
Severe Regeneration Defects in MEF2-TKO Mice.
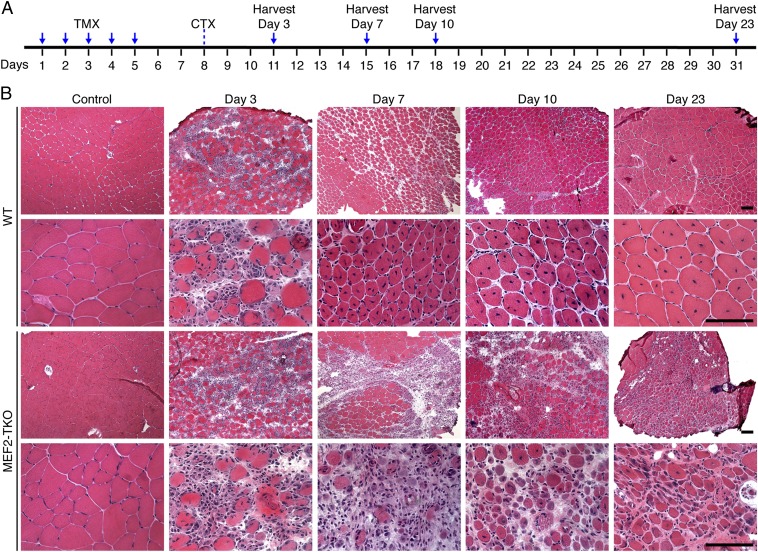
To determine whether MEF2A, MEF2C, and MEF2D play redundant functions in skeletal muscle regeneration, we generated mice carrying satellite cell-specific deletion of all three Mef2 genes, which we refer to as MEF2-triple knockout (MEF2-TKO) mice. We analyzed the regeneration capacity of these mice following the scheme shown in Fig. 2A. MEF2-TKO mice showed normal muscle size and morphology in the absence of injury (Fig. 2B). CTX induced extensive muscle damage and infiltration of inflammatory cells in both WT and MEF2-TKO muscle on day 3 after injury (Fig. 2B). By day 7, WT mice efficiently repaired the damaged muscle and regenerated new myofibers, as evidenced by the presence of myofibers with centralized nuclei (Fig. 2B). In contrast, MEF2-TKO muscles were composed of degenerating myofibers, fibrotic tissues, and inflammatory cells at this time point. Regenerating fibers were rarely seen in MEF2-TKO muscle, and the few that were present were extremely small, compared with those of WT mice (Fig. 2B). Ten days after injury, muscle damage and inflammatory cells in WT mice were largely cleared, and the regenerated myofibers continued to grow and mature, as their sizes became homogenous (Fig. 2B). However, MEF2-TKO muscle was occupied by damaged fibers and inflammatory cells, with only traces of regenerating myofibers. By day 23 after injury, WT muscle had fully regenerated, and muscle architecture was largely restored, with the normal myofiber hypertrophy seen after injury. In contrast, MEF2-TKO mice failed to regenerate and reconstitute muscle structure and instead displayed severe atrophy, compared with WT TA muscle (Fig. 2B).
Fig. 2.
Satellite-cell specific deletion of Mef2a, c and d prevents muscle regeneration upon CTX injury. (A) Schematic of TMX and CTX treatment. (B) TA muscles from WT (Mef2aloxp/loxp; 2c loxp/loxp; 2d loxp/loxp) and MEF2-TKO (Pax7-Cre-ERT2; Mef2aloxp/loxp; 2c loxp/loxp; 2d loxp/loxp) mice were analyzed by H&E staining on days 3, 7, 10, and 23 after injury. (Scale bars: 100 μm.)
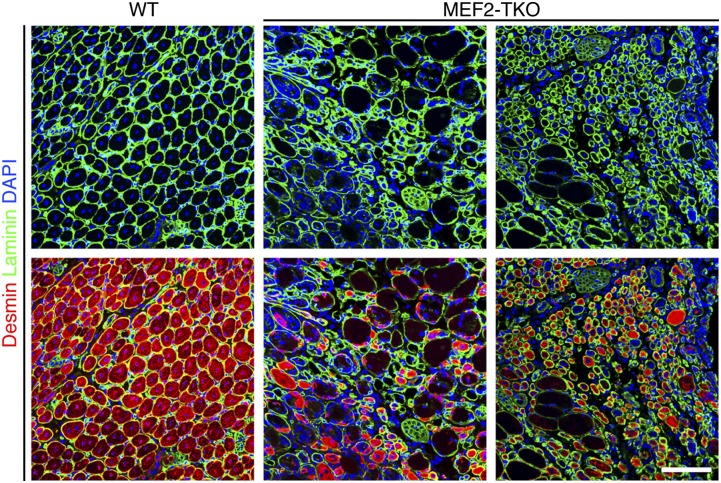
To further visualize the regenerative process, we performed immunostaining for desmin, an intermediate filament protein highly expressed in immature muscle fibers during fetal life and regeneration. As seen in Fig. 3, WT muscle at day 7 after injury showed strong desmin expression. In contrast, desmin expression was dramatically decreased in MEF2-TKO muscle, and desmin-positive cells were much smaller than those of WT muscle (Fig. 3).
Fig. 3.
Severe regeneration defects in MEF2-TKO mice upon CTX injury. Immunostaining for desmin (red) and laminin (green) on WT and MEF2-TKO TA muscles at day 7 after injury showed a dramatic decrease in regenerated myofiber formation. (Scale bar: 100 μm.)
Normal Proliferation and Impaired Differentiation of Satellite Cell-Derived Myoblasts in MEF2-TKO Mice.
Muscle regeneration requires activation and proliferation of satellite cells and differentiation of myogenic progenitors into myotubes. In principle, MEF2 could regulate any or all of these steps in the regenerative process. To further pinpoint the MEF2-dependent steps in muscle regeneration, we isolated activated satellite cells by FACS from WT and MEF2-TKO mice following CTX injury. We found no significant difference in the number of activated satellite cells between WT and MEF2-TKO mice 3 d after CTX injury (Fig. S2A), suggesting that MEF2 does not control the activation or expansion of the satellite cell population in response to injury.
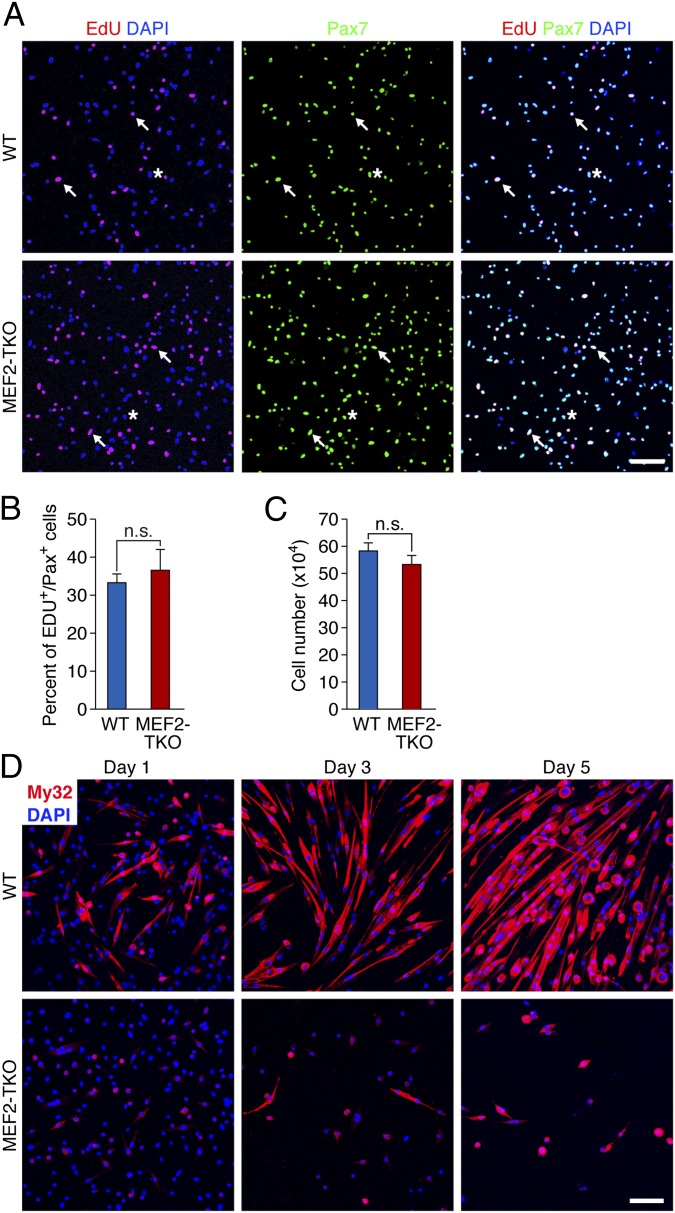
We cultured satellite cell-derived myoblasts from WT and MEF2-TKO mice and further studied their proliferation capacity by several methods. More than 90% of isolated satellite cells expressed Pax7 in both WT and MEF2-TKO mice, confirming the purity of these cells (Fig. S2 B and C). To study the rate of DNA synthesis, we assessed incorporation of 5-Ethynyl-2′-deoxyuridine (EdU) into cells. After 8 h of EdU labeling, approximately 35% of Pax7+ cells from WT and MEF2-TKO mice were positive for EdU, indicative of comparable rates of DNA synthesis (Fig. 4 A and B). EdU+ cells also expressed desmin in both WT and MEF2-TKO samples (Fig. S2D). When cultured at the same density, comparable numbers of WT and MEF2-TKO cells were also obtained after 72 h (Fig. 4C). Together, these results indicate that combined deletion of MEF2A, C, and D in satellite cells does not alter the ability of the cells to become activated in response to injury or to proliferate.
Fig. 4.
MEF2-TKO satellite cell-derived myoblasts proliferate normally, but they fail to differentiate into myotubes. (A) WT and MEF2-TKO myoblasts were labeled with EdU for 8 h and stained for EdU (red), Pax7 (green), and DAPI (blue) to show active DNA synthesis in Pax7+ myoblasts. Arrows indicate EdU+/Pax7+ nuclei. Asterisks indicate EdU−/Pax+ nuclei. (Scale bar: 100 μm.) (B) Quantification of the percentage of EdU+ Pax7+ cells between WT and MEF2-TKO myoblasts. Data are presented as mean ± SEM n.s., not significant. (C) WT and MEF2-TKO myoblasts (1 × 105) were plated, grown for 72 h, and counted. The number represents the average of three independent experiments. Data are presented as mean ± SEM n.s., not significant. (D) WT and MEF2-TKO myoblasts were cultured in differentiation medium for 1, 3, and 5 d and stained for skeletal myosin, using myosin antibody My32 (red) and DAPI (blue). (Scale bar: 100 μm.)
We compared the ability of WT and MEF2-TKO myoblasts to differentiate into myotubes in cell culture. As shown in Fig. 4D, after 24 h in differentiation medium (DM), WT myoblasts express the terminal differentiation marker, skeletal myosin, and start to elongate and fuse. However, very few MEF2-TKO myoblasts expressed myosin (Fig. 4D). After 3 and 5 d in DM, almost all WT cells elongated and fused into multinucleated myotubes with strong myosin expression. In contrast, most MEF2-TKO cells remained spindle shaped and failed to fuse. In addition, extensive cell death was observed in MEF2-TKO cultures, probably because MEF2-TKO myoblasts failed to differentiate (Fig. 4D). These findings indicate that the combined loss of MEF2A, C, and D disrupts terminal differentiation of satellite cell-derived myoblasts into myotubes.
Identification of MEF2-Dependent Genes in Satellite Cells.
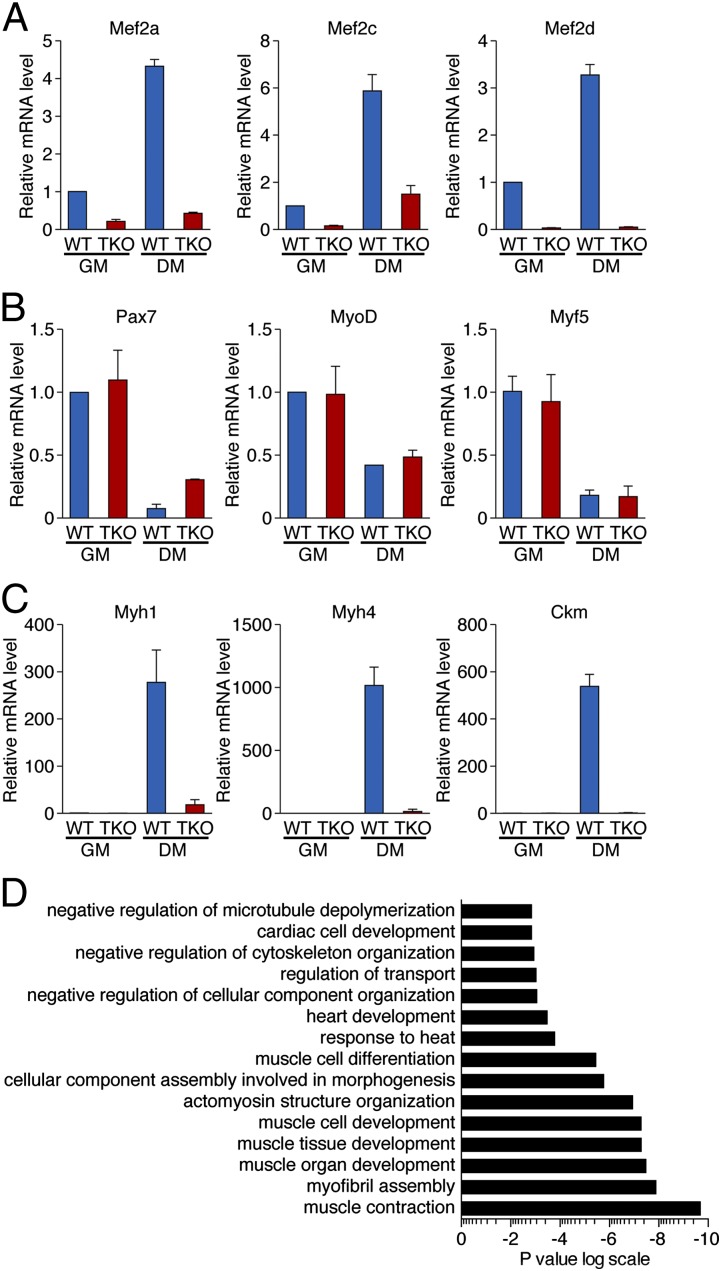
To study the mechanism by which MEF2 controls satellite cell differentiation, we compared gene expression of WT and MEF2-TKO myoblasts in growth medium (GM) and myotubes in DM for 3 d. Consistent with previous reports, all three MEF2 factors were up-regulated during differentiation of WT cells, whereas they were not expressed above background levels in MEF2-TKO cell cultures under differentiation conditions (Fig. 5A), confirming the effective deletion of Mef2a, c, and d in satellite cells using Pax7-CreERT2 mice. Mef2b transcript was not detectable in WT or MEF2-TKO myoblasts or myotubes.
Fig. 5.
Analysis of gene expression in MEF2-TKO myoblasts. (A) Real-time RT-PCR revealed that expression of Mef2a, Mef2c, and Mef2d mRNA is dramatically down-regulated in MEF2-TKO cells. DM, cells were cultured in differentiation medium for 3 d; GM, cells were cultured in growth medium. Data are presented as mean ± SEM. (B) Real-time RT-PCR revealed that expression of Pax7, MyoD, and Myf5 is not changed in MEF2-TKO cells relative to WT cells. Data are presented as mean ± SEM. (C) Real-time RT-PCR revealed that expression of Myh1, Myh4, and Ckm mRNAs is activated in WT cells but not in MEF2-TKO cells upon differentiation. Data are presented as mean ± SEM. (D) Gene ontology analysis was performed with DAVID. Microarray data from WT and MEF2-TKO myotubes were used in the analysis. Significantly (P < 0.05) enriched biological processes are shown. Plotted is the log (P value).
Deletion of Mef2a, c, and d did not significantly alter the expression of Pax7, MyoD, or Myf5, supporting the conclusion that MEF2 does not influence satellite cell specification or proliferation (Fig. 5B). However, MEF2-TKO cells failed to up-regulate Myf6 (Mrf4) during differentiation (Fig. S3B). Consistent with the differentiation defects, expression of terminal differentiation markers such as muscle creatine kinase (Ckm), myosin heavy chain 4 (Myh4), and myosin heavy chain 1 (Myh1) was dramatically decreased in MEF2-TKO myotubes (Fig. 5C).
To further assess the involvement of MEF2 in satellite cell differentiation, we performed microarray analysis on WT and MEF2-TKO cells in DM. There were 376 genes down-regulated by at least twofold and 208 genes up-regulated by at least twofold in MEF2-TKO compared with WT cells. Gene ontogeny analysis of down-regulated transcripts revealed that the most significantly down-regulated genes participate in muscle contraction (Fig. 5D). Other down-regulated genes were involved in myofibril assembly, muscle development, cytoskeleton organization, and transport. It has been shown that MyoD and MEF2 control stress responses during myogenesis (9). Similarly, we observed significant enrichment in genes involved response to heat (Fig. 5D). Down-regulated genes in the muscle contraction category fell into a variety of functional classes, including calcium-binding proteins, cytoskeletal proteins, cell junction and cell adhesion molecules, demonstrating the involvement of MEF2 in many aspects of muscle function (Fig. S3A). Representative genes in each category were confirmed by real-time PCR analysis and are listed in Fig. S3B. The up-regulated genes in MEF2-TKO cells did not fall into distinct biological pathways.
We compared the gene expression profiling data with previously published MEF2D-chromatin immunoprecipitation data in C2C12 cells to identify down-regulated genes that have nearby MEF2-binding sites (20). We found that 29% of the down-regulated genes contained MEF2 binding sites near their promoters. Many of the genes were known MEF2 targets, regulating diverse molecular functions in muscle contraction, calcium signaling, and stress responsiveness. Examples include myomesin 1 and 2, myozenin 1 and 2, Fabp3, Ldb3, and Casq2 (Fig. S3B) (9, 11, 21, 22). Our analysis also identified a group of previously uncharacterized or little characterized genes that are down-regulated in MEF2-TKO cells (Fig. S4). This list includes both protein-coding genes and putative noncoding RNAs, and many genes contain MEF2-binding sites near their promoters, suggesting that they might be direct transcriptional targets of MEF2 (Fig. S4).
Discussion
The results of this study reveal redundant functions of MEF2A, C, and D in the control of adult skeletal muscle regeneration. In response to muscle injury, MEF2 activity is dramatically induced, as shown by expression of the MEF2-dependent reporter. In the absence of MEF2A, C and D, activated satellite cells fail to differentiate or become incorporated in regenerating myofibers, demonstrating the essential functions of MEF2 in differentiation of adult muscle stem cells. The proregenerative activity of these MEF2 isoforms is reflected in their cooperative activation of a broad spectrum of genes required for muscle differentiation and function.
The central role of MEF2 in orchestrating muscle development has been delineated most definitively in Drosophila, where the single Mef2 gene is necessary for myoblast fusion and muscle differentiation (23–25). Despite extensive studies of the role of MEF2 in enhancing myogenesis in vitro, it has not been established whether MEF2 is required for vertebrate skeletal muscle development in vivo, because deletion of individual MEF2 factors has little or no effect on formation of myofibers in mice. In this regard, our finding that combined deletion of MEF2A, C, and D severely impairs regeneration demonstrates that MEF2 proteins are required for adult regenerative myogenesis.
MRFs and MEF2 have overlapping but distinct functions in adult versus embryonic myogenesis. Myf5 expression marks the majority of quiescent satellite cells and all activated satellite cells and myoblasts (5). Myf5 deletion in satellite cells decreases the myoblast proliferation rate and delays the transition from proliferation to differentiation (26, 27). MyoD is expressed in activated satellite cells and myoblasts, and MyoD-null satellite cells fail to differentiate into myotubes (28–30). Interestingly, although both Myf5 and MyoD can each compensate for the loss of the other during embryogenesis (31), they cannot efficiently compensate for each other in the adult context. Surprisingly, myogenin-null myoblasts show normal proliferation and differentiation, suggesting distinct functions of myogenin in adult versus embryonic myogenesis (32). The phenotype of MEF2-TKO mice resembles that of MyoD-null mice, in which terminal differentiation of myoblasts is blocked. Moreover, expression of MRF4 was dramatically decreased in both mice upon differentiation, consistent with the notion that MRF4 up-regulation may be an essential step in adult satellite cell differentiation (28). In addition to defects in myoblast differentiation, MyoD-null myoblasts also exhibited enhanced proliferation, whereas proliferation is normal in MEF2-TKO myoblasts (28, 30). Consistent with this finding, many growth-related genes are dysregulated in MyoD-null, but not MEF2-TKO, myoblasts. These results suggest that whereas MyoD acts upstream of MEF2 to control both proliferation and differentiation of myoblasts, MEF2 functions mainly during differentiation of myoblasts.
Gene expression analysis revealed a large collection of muscle genes that depend on the combined expression of MEF2A, C, and D for expression. A large proportion of these genes encode contractile proteins, calcium-handling proteins, cytoskeleton proteins, and proteins involved in mitochondrial function and metabolism. In addition, numerous uncharacterized protein-coding and long noncoding RNA genes were down-regulated in cultured myotubes from MEF2-TKO mice. It will be of particular interest to learn about the functions of these unique genes in muscle differentiation and regeneration.
Prior studies have implicated MEF2 isoforms in muscle regeneration but the consequences of combined deletion of Mef2a, c, and d have not been investigated in any tissue. Our conclusions regarding the role of MEF2 in muscle regeneration differ from those of Synder et al. who reported that mice with global deletion of Mef2a were defective in skeletal muscle regeneration (13). They further showed that MEF2A regulates transcription of the Gtl2-Dio3 microRNA megacluster that targets sFRP2, an inhibitor of WNT signaling, thus linking MEF2 activity with WNT signaling, in the process of adult regenerative myogenesis (13). In contrast, we show that satellite cell deletion of Mef2a has no observable effect on regeneration. How might these findings be reconciled? Because MEF2A is expressed in a range of cell types, including mature myofibers, fibroblasts, and inflammatory cells, we believe the apparent requirement of MEF2A for regeneration, revealed by global gene deletion, reflects an important role of MEF2A in nonsatellite cells during the regeneration process. In this regard, MEF2A has been shown to participate in inflammatory signaling in macrophages (33), which play an important role in tissue responses to injury, and in fibrotic responses of fibroblasts (34, 35). Knockdown of MEF2A and C by shRNAs in cultured satellite cells was recently found to disrupt differentiation and fusion, suggesting that MEF2D alone was not sufficient for differentiation (14). In contrast, our results show clearly that satellite cells from mice with genetic deletion of Mef2a and c show normal regeneration in vivo and differentiation in vitro. This discrepancy could be attributable to the approaches for inhibition of MEF2 expression used in the studies. Perhaps the functional requirement of MEF2D in regeneration is more pronounced in vivo than in cultured satellite cells in vitro or perhaps compensatory mechanisms in vivo can overcome the absence of MEF2A and C, which does not occur in the context of shRNA knockdown experiments in vitro.
MEF2 proteins are targeted by a variety of signal transduction cascades. MAP kinase signaling, for example, culminates with the phosphorylation of the transcription activation domains of MEF2 proteins, enhancing transcriptional activity (10). Activation of calcium-dependent protein kinase signaling also stimulates MEF2 activity through phosphorylation of class II HDACs and their export from the nucleus, thereby derepressing MEF2 (10). The realization that MEF2 plays a key role in the control of muscle regeneration raises interesting possibilities for augmenting this process through pharmacologic regulation of the signaling pathways that modulate MEF2 activity.
Materials and Methods
Detailed methods for all experiments are available in SI Materials and Methods.
Mice.
All experiments involving animals were approved by the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center. The Pax7-Cre-ERT2 mice were kindly provided by Chen-Ming Fan (Carnegie Institution for Science, Baltimore) (16). Details of the conditional Mef2 alleles have been described (17–19). Tamoxifen injection and cardiotoxin injury were performed on 8-wk-old mice. A detailed description is available in SI Materials and Methods.
Histological Analysis of Skeletal Muscle.
TA muscles were harvested and flash frozen in embedding medium containing a 3:1 mixture of Tissue Freezing Medium (Triangle Biomedical Sciences) and gum tragacanth (Sigma-Aldrich). Frozen sections were cut on a cryotome and stained with H&E as described (36). Immunohistochemistry on frozen sections is described in SI Materials and Methods.
Culture of Satellite Cell-Derived Myolbasts.
CTX was injected into hind limb muscles and activated SCs were isolated 3 d after injection as described (37–39). Culture conditions and EdU labeling of cells are described in SI Materials and Methods.
RT-PCR and Real-Time RT-PCR Analysis.
RNA was treated with Turbo RNase-free DNase (Ambion) before the reverse transcription step. RT-PCR was performed by using random hexamer primers (Invitrogen). Real-time RT-PCR was performed by using TaqMan probes (ABI) or SYBR green probes.
Supplementary Material
Acknowledgments
We thank members of the Olson laboratory for technical help and scientific discussions; Jose Cabrera for graphics; Megan Kong for analysis of microarray data; and Dr. Chen-Ming Fan (Carnegie Institution for Science) for the Pax7-Cre-ERT2 mice. This work was supported by National Institutes of Health (NIH) Grants HL-077439, HL-111665, HL-093039, U01-HL-100401, and DK-099653 and The Welch Foundation Grant 1-0025 (to E.N.O.). N.L. is supported by grants from the American Heart Association. B.R.N. is supported by NIH Training Grant T32-HL-007360.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401732111/-/DCSupplemental.
References
- 1.Carlson BM. The regeneration of skeletal muscle. A review. Am J Anat. 1973;137(2):119–149. doi: 10.1002/aja.1001370202. [DOI] [PubMed] [Google Scholar]
- 2.Brack AS, Rando TA. Tissue-specific stem cells: Lessons from the skeletal muscle satellite cell. Cell Stem Cell. 2012;10(5):504–514. doi: 10.1016/j.stem.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tedesco FS, Dellavalle A, Diaz-Manera J, Messina G, Cossu G. Repairing skeletal muscle: Regenerative potential of skeletal muscle stem cells. J Clin Invest. 2010;120(1):11–19. doi: 10.1172/JCI40373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin H, Price F, Rudnicki MA. Satellite cells and the muscle stem cell niche. Physiol Rev. 2013;93(1):23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129(5):999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol. 2011;12(6):349–361. doi: 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- 7.Tapscott SJ, et al. MyoD1: A nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988;242(4877):405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- 8.Molkentin JD, Olson EN. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci USA. 1996;93(18):9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blais A, et al. An initial blueprint for myogenic differentiation. Genes Dev. 2005;19(5):553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potthoff MJ, Olson EN. MEF2: A central regulator of diverse developmental programs. Development. 2007;134(23):4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 11.Potthoff MJ, et al. Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2c. Mol Cell Biol. 2007;27(23):8143–8151. doi: 10.1128/MCB.01187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potthoff MJ, et al. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J Clin Invest. 2007;117(9):2459–2467. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snyder CM, et al. MEF2A regulates the Gtl2-Dio3 microRNA mega-cluster to modulate WNT signaling in skeletal muscle regeneration. Development. 2013;140(1):31–42. doi: 10.1242/dev.081851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mokalled MH, Johnson AN, Creemers EE, Olson EN. MASTR directs MyoD-dependent satellite cell differentiation during skeletal muscle regeneration. Genes Dev. 2012;26(2):190–202. doi: 10.1101/gad.179663.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naya FJ, Wu C, Richardson JA, Overbeek P, Olson EN. Transcriptional activity of MEF2 during mouse embryogenesis monitored with a MEF2-dependent transgene. Development. 1999;126(10):2045–2052. doi: 10.1242/dev.126.10.2045. [DOI] [PubMed] [Google Scholar]
- 16.Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460(7255):627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akhtar MW, et al. In vivo analysis of MEF2 transcription factors in synapse regulation and neuronal survival. PLoS ONE. 2012;7(4):e34863. doi: 10.1371/journal.pone.0034863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arnold MA, et al. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12(3):377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y, et al. The MEF2D transcription factor mediates stress-dependent cardiac remodeling in mice. J Clin Invest. 2008;118(1):124–132. doi: 10.1172/JCI33255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sebastian S, et al. Tissue-specific splicing of a ubiquitously expressed transcription factor is essential for muscle differentiation. Genes Dev. 2013;27(11):1247–1259. doi: 10.1101/gad.215400.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frey N, Richardson JA, Olson EN. Calsarcins, a novel family of sarcomeric calcineurin-binding proteins. Proc Natl Acad Sci USA. 2000;97(26):14632–14637. doi: 10.1073/pnas.260501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reyes-Juárez JL, Juárez-Rubí R, Rodríguez G, Zarain-Herzberg A. Transcriptional analysis of the human cardiac calsequestrin gene in cardiac and skeletal myocytes. J Biol Chem. 2007;282(49):35554–35563. doi: 10.1074/jbc.M707788200. [DOI] [PubMed] [Google Scholar]
- 23.Bour BA, et al. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 1995;9(6):730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- 24.Lilly B, et al. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267(5198):688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 25.Ranganayakulu G, et al. A series of mutations in the D-MEF2 transcription factor reveal multiple functions in larval and adult myogenesis in Drosophila. Dev Biol. 1995;171(1):169–181. doi: 10.1006/dbio.1995.1269. [DOI] [PubMed] [Google Scholar]
- 26.Gayraud-Morel B, et al. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Dev Biol. 2007;312(1):13–28. doi: 10.1016/j.ydbio.2007.08.059. [DOI] [PubMed] [Google Scholar]
- 27.Ustanina S, Carvajal J, Rigby P, Braun T. The myogenic factor Myf5 supports efficient skeletal muscle regeneration by enabling transient myoblast amplification. Stem Cells. 2007;25(8):2006–2016. doi: 10.1634/stemcells.2006-0736. [DOI] [PubMed] [Google Scholar]
- 28.Cornelison DD, Olwin BB, Rudnicki MA, Wold BJ. MyoD(-/-) satellite cells in single-fiber culture are differentiation defective and MRF4 deficient. Dev Biol. 2000;224(2):122–137. doi: 10.1006/dbio.2000.9682. [DOI] [PubMed] [Google Scholar]
- 29.Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10(10):1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- 30.Sabourin LA, Girgis-Gabardo A, Seale P, Asakura A, Rudnicki MA. Reduced differentiation potential of primary MyoD-/- myogenic cells derived from adult skeletal muscle. J Cell Biol. 1999;144(4):631–643. doi: 10.1083/jcb.144.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudnicki MA, et al. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75(7):1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 32.Meadows E, Cho JH, Flynn JM, Klein WH. Myogenin regulates a distinct genetic program in adult muscle stem cells. Dev Biol. 2008;322(2):406–414. doi: 10.1016/j.ydbio.2008.07.024. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki E, et al. Myocyte enhancer factor 2 mediates vascular inflammation via the p38-dependent pathway. Circ Res. 2004;95(1):42–49. doi: 10.1161/01.RES.0000134631.75684.4A. [DOI] [PubMed] [Google Scholar]
- 34.Mounier R, et al. AMPKα1 regulates macrophage skewing at the time of resolution of inflammation during skeletal muscle regeneration. Cell Metab. 2013;18(2):251–264. doi: 10.1016/j.cmet.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 35.Rigamonti E, et al. Requirement of inducible nitric oxide synthase for skeletal muscle regeneration after acute damage. J Immunol. 2013;190(4):1767–1777. doi: 10.4049/jimmunol.1202903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu N, et al. Mice lacking microRNA 133a develop dynamin 2–dependent centronuclear myopathy. J Clin Invest. 2011;121(8):3258–3268. doi: 10.1172/JCI46267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu N, et al. microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Invest. 2012;122(6):2054–2065. doi: 10.1172/JCI62656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosnakovski D, et al. Prospective isolation of skeletal muscle stem cells with a Pax7 reporter. Stem Cells. 2008;26(12):3194–3204. doi: 10.1634/stemcells.2007-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olguin HC, Olwin BB. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: A potential mechanism for self-renewal. Dev Biol. 2004;275(2):375–388. doi: 10.1016/j.ydbio.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.