Significance
P-glycoprotein exports various hydrophobic chemicals in an ATP-dependent manner, determines their absorption and distribution in the body, and is involved in multidrug resistance (MDR) in tumors. Understanding the mechanism of the multidrug transport is important for designing drugs of good bioavailability and efficient cancer chemotherapy. We determined the high-resolution crystal structures of a eukaryotic P-glycoprotein homolog and revealed the detailed architecture of its transmembrane domains, which contain an exit gate for substrates that opens to the extracellular side and two entrance gates that open to the intramembranous region and the cytosolic side. We propose a motion of the transmembrane domains powered by the association of two nucleotide-binding domains on ATP binding that is different from other transporters.
Keywords: multidrug resistance, ABC transporter, membrane protein, X-ray crystallography, macrocyclic peptide
Abstract
P-glycoprotein is an ATP-binding cassette multidrug transporter that actively transports chemically diverse substrates across the lipid bilayer. The precise molecular mechanism underlying transport is not fully understood. Here, we present crystal structures of a eukaryotic P-glycoprotein homolog, CmABCB1 from Cyanidioschyzon merolae, in two forms: unbound at 2.6-Å resolution and bound to a unique allosteric inhibitor at 2.4-Å resolution. The inhibitor clamps the transmembrane helices from the outside, fixing the CmABCB1 structure in an inward-open conformation similar to the unbound structure, confirming that an outward-opening motion is required for ATP hydrolysis cycle. These structures, along with site-directed mutagenesis and transporter activity measurements, reveal the detailed architecture of the transporter, including a gate that opens to extracellular side and two gates that open to intramembranous region and the cytosolic side. We propose that the motion of the nucleotide-binding domain drives those gating apparatuses via two short intracellular helices, IH1 and IH2, and two transmembrane helices, TM2 and TM5.
Multidrug transporters of the ATP-binding cassette (ABC) superfamily, such as P-glycoprotein (P-gp; MDR1; ABCB1), MRP1 (ABCC1), and ABCG2 (BCRP), transport a large number of structurally unrelated compounds with molecular weights ranging up to several thousand Daltons (1, 2). These transporters not only play important roles in normal physiology by protecting tissues from various toxic xenobiotics and endogenous metabolites but also contribute to multidrug resistance (MDR) in tumors, a major obstacle to effective chemotherapeutic treatment (1, 3–7). Their functional forms consist of a minimum of four core domains: two transmembrane domains (TMDs) that create the translocation pathway for substrates and two nucleotide-binding domains (NBDs) that bind and hydrolyze ATP to power the transport process (8, 9). These four domains can exist either as two separate polypeptides (half-size) or fused together in a single large polypeptide with an internal duplication (full-size). The crystal structures of mouse and nematode P-gps, as well as their bacterial homologs (10–14), have been determined, and they have provided important insights into the relationships between protein structure and the functional and biochemical characteristics of P-gp. However, the detailed architecture of the TMD machinery and the gating mechanism during the transition between the inward- and outward-open states are poorly understood.
Here, we report the structures of a eukaryotic P-gp homolog, unlocked (at 2.6-Å resolution) and locked allosterically with a tailor-made peptide at 2.4-Å resolution. Although CmABCB1 is not a full-length ABC transporter but a half-sized ABC transporter adopting a homodimeric architecture, CmABCB1 showed quite similar functional properties to those of human P-gp (hP-gp). Based on these structures, we propose mechanisms by which the intramembranous entrance gate can take up various substrates from the inner leaflet of the membrane bilayer. Although the entrance and exit gates are assembled from distinct transmembrane helices, and their gating motions are in opposite directions, the mechanisms powered by the dimerization motions of the NBDs enable one gate to close while the other gate simultaneously opens with the aid of two short intracellular helices and two transmembrane helices, which act as a lever arm. This mechanism is totally different from that of solute carrier (SLC) transporters (15–19) and ABC importers (20), in which the intra- and extracellular gating apparatuses are constructed from the same transmembrane helices. Furthermore, a part of the intramembranous entrance gate has another function gating the pathway from the inside of the transporter to the cytosol. The mode of action of the novel inhibitor, which disables the diverging outward motions of the TM helices by clamping them from the outside of the transporter, supports our proposed gating mechanism.
Results and Discussion
Structure Determination.
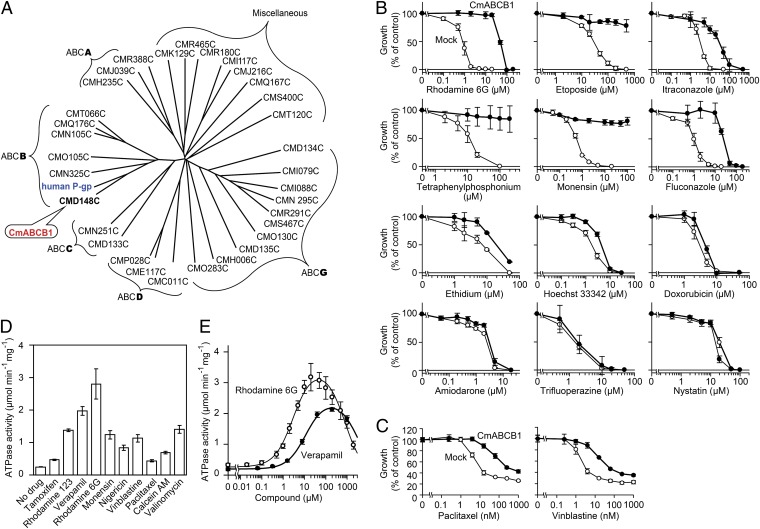
To select a protein for high-resolution structure determination, we searched for ABC transporters closely resembling P-gp in the genome of Cyanidioschyzon merolae (21), a thermophilic unicellular eukaryote. This screen resulted in identification of CMD148C, the protein in the C. merolae genome whose amino acid sequence, multidrug specificity, and kinetics of ATP hydrolysis are most similar to that of hP-gp (SI Results, Fig. 1, Figs. S1 and S2, and Table S1). On the basis of its similarity to hP-gp, we named this protein CmABCB1 and solved its crystal structure at 2.75-Å resolution by multiwavelength anomalous dispersion (MAD) using a mercury derivative (Tables S2 and S3). In the resultant electron-density map, a portion of transmembrane helix 4 (TM4) exhibited very weak density (Fig. S3A). Therefore, we introduced a triple mutation (G277V/A278V/A279V, hereafter referred to as the GAA/VVV mutant) to reduce the flexibility of the helices; as a result, the resolution was improved to 2.6 Å (Fig. S3 B and D and Tables S2 and S3). To further enhance crystallization, we introduced a CmABCB1-binding cyclic peptide, aCAP (anti-CmABCB1 peptide), which we identified from an artificial cyclic peptide library containing more than 1012 unique molecules, using the RaPID system (SI Materials and Methods) (22, 23). The final resolution of the X-ray diffraction was 2.4 Å (Tables S2 and S3).
Fig. 1.
CmABCB1 is an ABC multidrug transporter. (A) Dendrogram of ABC transporters from C. merolae and human P-gp (ABCB1). Among 32 ABC proteins from C. merolae, CMD148C is the most similar to human P-gp and was therefore designated as CmABCB1. This dendrogram is based on multiple sequence alignments of the full amino acid sequences of ABC transporters. Alignments were performed using with ClustalW and visualized using Drawtree. (B) Drug-susceptibility assay in S. cerevisiae AD1-8u− cells. AD1-8u− cells expressing WT CmABCB1 (●) were grown in various concentrations of drugs. For each drug assayed, mock-transfected AD1-8u− cells (○) were also grown as controls. Data are means ± SD (n = 5). (C) Drug-susceptibility assay in HEK293 cells. Mock-transfected HEK293 (○) and CmABCB1-expressing HEK293 cells (●) were incubated with paclitaxel or vinblastine, and viability was measured by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. Data are means ± SD (n = 3). (D) Effect of drugs on CmABCB1 ATPase activity. The ATPase activity was measured in the presence or absence of 50 µM of the indicated drugs with 5 mM ATP at 37 °C. Data are means ± SD (n = 2). (E) Drug concentration dependence of CmABCB1 ATPase activity. The ATPase activity was measured as a function of verapamil or rhodamine 6G concentration with 5 mM ATP at 37 °C. Data are means ± SD (n = 3). The solid line is a fit of the equation to the data as described in SI Materials and Methods (Eq. S2).
Architecture.
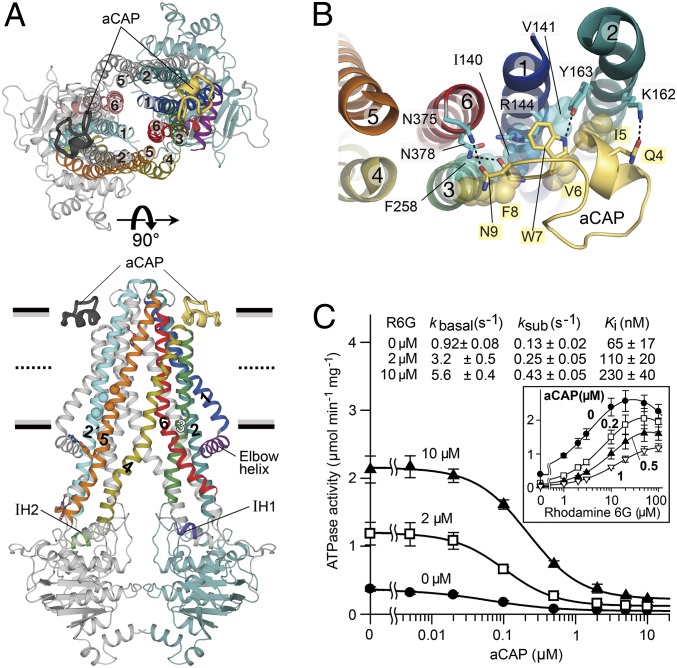
The structure of CmABCB1 reveals an overall inward-open conformation (Fig. 2A and Fig. S4) that is favorable for the uptake of transport substrates. CmABCB1 adopts a homo-dimeric architecture, with each subunit containing an NBD and a cytosolically extruded TMD. The TMD is composed of an N-terminal elbow helix, followed by six α-helices and two short intracellular helices (IH1 between TM2 and TM3, and IH2 between TM4 and TM5). IH1 and IH2 contact the NBD as coupling helices, as shown previously (12) (Fig. 2A). The homo-dimeric TMD region is formed mainly by tight interactions between TM2 and TM5′ (corresponding to TM2 and TM11 in P-gp) or TM5 and TM2′ (corresponding to TM5 and TM8 in P-gp) via Gly-zipper motifs (G179xxxA183 in TM2, and G344xxxG348 in TM5) that allow the two helices to come into close proximity to form a right-handed helix–helix contact while retaining flexibility (secondary structure elements and amino acid residues belonging to the other subunit are indicated with a prime symbol). The two helices are also connected by interhelical hydrogen bonds near the middle of the outer leaflet and at the polar head group of the inner leaflet of the membrane bilayer. Assembly of this helical pair promotes consolidated movement of the helices at the dimer interfaces and acts as a lever arm for extracellular gating, as we will describe below (Fig. 2A and Fig. S5). On the top of CmABCB1, TMs 1, 6, 1′, and 6′ form a right-handed four-helix bundle structure (24) that surrounds the center of the dimer and closes the pathway connecting the inside and outside of the transporter (Fig. 2A).
Fig. 2.
Overall architecture of the CmABCB1–aCAP complex. (A) CmABCB1–aCAP complex structure viewed parallel to the plane of the membrane (Lower) or from the extracellular side (Upper). CmABCB1 and bound aCAPs are depicted in cartoon representation. One subunit is displayed in multiple colors, and the other subunit is shown in gray except for TM2′ (cyan) in the lower panel and TM1′ (light blue) and TM6′ (dark salmon) in the upper panel. Horizontal black bars represent the expected positions of the hydrophilic surfaces of the lipid membrane; gray bars represent the expected positions of the hydrophobic surfaces. Thick dashed lines represent the middle of the membrane bilayer. (B) The binding site of aCAP, viewed from the extracellular side. Dashed lines indicate hydrogen bonds. (C) Inhibitory effect of aCAP on the ATPase activity of CmABCB1. The ATPase activity of WT CmABCB1 was measured as a function of aCAP concentration in the presence or absence of rhodamine 6G at 37 °C; ATP was present at 5 mM. Solid lines are fits of an equation (Eq. S4) to the data, as described in SI Materials and Methods. (Inset) ATPase activity vs. rhodamine 6G concentration at various constant concentrations of aCAP. Solid lines are fits of an equation (Eq. S2) to the data, as described in SI Materials and Methods.
Tailored Macrocyclic Peptide Inhibitor.
A tailored cyclic peptide that we identified, aCAP, acts as a surface clamp by binding TM2 and TM6 together from the outer surface of the CmABCB1 dimer, at the interface between the hydrophobic chains and hydrophilic head groups of the outer leaflet of the membrane bilayer (Fig. 2 A and B). Lys162 and Tyr163 in TM2 form hydrogen bonds with Gln4 and Trp7 in aCAP, respectively, whereas Asn375 and Asn378 in TM6 form hydrogen bonds with Asn9 and the main-chain carbonyl of Phe8 in aCAP, respectively (Fig. 2B). In addition, Phe258 in TM3, and Ile140, Val141, and the alkyl-chain region of the side-chain of Arg144 in TM1 form van der Waals contacts with Phe8, Ile5, Val6, and Trp7 of aCAP, respectively. Consequently, aCAP clamping can restrain the dynamic movement of the CmABCB1 molecule; and therefore the X-ray diffraction ability of the crystals was improved. However, there is little difference between the crystal structures in the presence and absence of aCAP.
aCAP inhibited both the basal and rhodamine 6G–stimulated ATPase activities of CmABCB1. Comparison of the Ki for aCAP and Km for rhodamine 6G revealed that aCAP binds CmABCB1 46-fold more tightly than the substrate (Fig. 2C). Ki values for aCAP increased as a function of rhodamine 6G concentration, suggesting that aCAP binding is weakened by helical packing interaction due to rhodamine 6G docking at the interior binding site of the transporter. These results support the notion that dissociation of the helix-bundle interactions specific to the inward-open conformation is indispensable for transporter function.
Extracellular Gate.
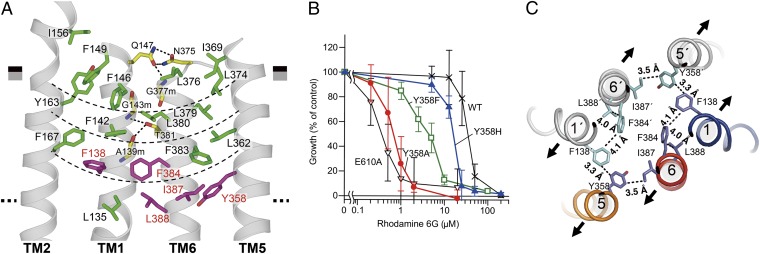
Around the aCAP binding region, TM2, TM1, TM6, and TM5 form a linear helix bundle flanked by a four-layered cluster comprised of interior-facing aromatic and hydrophobic residues such as Phe, Leu, Ile, and Tyr, which contact one another closely by van der Waals and/or hydrophobic interactions (Fig. 3A). The thickness of this cluster is similar to that of the outer leaflet of the lipid bilayer. The TM1–TM6 interactions in the cluster are also mediated by several hydrogen bonds: Gln147 to Asn375 and NH of Gly377, and the main-chain amide of Gly143 and carbonyl of Ala139 to Thr381 (Fig. 3A); these interactions are promoted by the hydrophobic environment created by the four-layered cluster. The linear bundle flanked by the four-layered cluster associates with the corresponding assembly in the other subunit to form the dimer molecule. This architecture stabilizes the inward-open structure, explaining the low basal and high drug-stimulated ATPase activities that are characteristic of P-gps (25–27) and are also observed in CmABCB1 (Fig. 1E). The bulky side chains in the cluster fill the inner space, preventing entry of extracellular fluid into the spacious internal cavity (WT, 7,648 Å3; GAA/VVV mutant, 6,773 Å3), which has been proposed to be the substrate-binding site (10) located in the inner leaflet of the lipid bilayer.
Fig. 3.
Extracellular gate. (A) Close-up view of the four-layered cluster. The boundaries of the four-layered cluster are indicated as thin dashed lines. Residues whose replacement markedly decreased transport activity in the drug-susceptibility assay using rhodamine 6G are colored in purple. Hydrogen bonds are indicated as thick dashed lines. (B) Rhodamine 6G susceptibility of yeast cells expressing CmABCB1 mutants. Yeast cells expressing WT CmABCB1 or mutant proteins were cultured with various concentrations of rhodamine 6G. E610A is an ATPase-deficient mutant (SI Materials and Methods). Cell proliferation was analyzed by measuring the absorbance at 600 nm, and growth (% of control) was plotted against rhodamine 6G concentration. Data are means ± SD (n = 11–32). (C) Key interacting residues at the extracellular gate, viewed from the extracellular side. Contacts within van der Waals and/or hydrophobic interactions are indicated with dashed lines. The expected directions of the initial stages of TM helix motions from the inward-open to outward-open states are indicated as arrows.
Functional Residues Lining the Cavity.
To identify the residues required for the transport function, we performed Ala substitution of the residues in the cluster. Replacement of several residues markedly decreased transport activity in a drug-susceptibility assay using rhodamine 6G (Fig. S6 A and B); these residues, Tyr358, Phe138, Phe384, Ile387, and Leu388, are all located on the ceiling of the cavity (Fig. 3A). Mutants with Ala substitutions also exhibited reduced affinity for rhodamine 6G, as revealed by kinetic analysis of their ATPase activities (Fig. S6C and Table S1). Notably, Ala substitution of Tyr358 caused the greatest loss of both transport activity and affinity. Tyr358 is the only residue bearing a side-chain that can act as a hydrogen-bonding acceptor near the ceiling of the cavity. Many substrates for hP-gp contain hydrogen-bonding donor groups, and Tyr358 may form hydrogen bonds with these moieties (28, 29). To explore this possibility, we replaced Tyr358 with Phe or His. In decreasing order, the effective drug resistances bestowed by the mutant CmABCB1s were as follows: WT > Y358H > Y358F > Y358A (Fig. 3B and Fig. S6B); this result suggests that the hydroxyl group, as well as the steric portion of the side-chain, is important.
These findings imply that a right-handed four-helix bundle consisting of TM1, TM6, TM1′, and TM6′, in association with two other helices, TM5 and TM5′, forms an extracellular gate through spatial coordination of the aromatic and bulky hydrophobic residues Tyr358 (TM5), Phe138 (TM1), Phe384 (TM6), Ile387 (TM6), and Leu388 (TM6) (Fig. 3C). Those residues are arranged such that the distances between them are suitable for van der Waals contacts; in particular, Tyr358 on TM5 is closer to Phe138 on TM1′ and Ile387 on TM6 than those residues are to each another and therefore interacts with each of them more strongly. This arrangement suggests that movement of TM5 results in unlocking and elicits dissociation of the tight interactions between TM1 and TM6 in the inward-open state, thereby opening the extracellular gate. The hydrogen bonding of Tyr358 with bulky hydrophobic substrates is expected to perturb the van der Waals interactions between the aromatic and hydrophobic residues listed above and may trigger the opening of the extracellular gate (Fig. 3C). This extracellular gating mechanism could explain why allosteric inhibition by aCAP is disturbed by substrate binding (Fig. 2C). The corresponding residues of Tyr358 are highly conserved in human and mouse P-gps (Fig. S1), and it is one of the residues that have been proposed to interact with the QZ59 compounds, a family of ATPase modulators, in the internal cavity of mouse P-gp (10).
Intramembranous Gate.
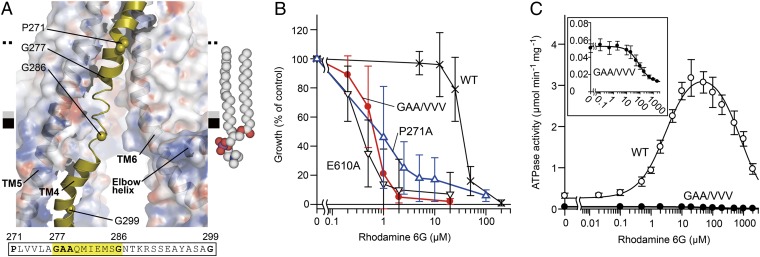
A wide cleft between TM4 and TM6 spans the hydrophobic interface between the TMD and the inner leaflet of the putative lipid bilayer (Fig. 2A). In the WT structure, the region from Gly277 to Gly286 of TM4 near the amphipathic interface in the inner leaflet of the bilayer is flexible (Fig. 4A), as reflected by the fact that this region exhibited very weak electron density (Fig. S3 A and C). This behavior of the TM4 structure was unchanged by aCAP binding, suggesting that flexibility could be an intrinsic property of the transporter. Most substrates of ABC multidrug transporters are hydrophobic and consequently concentrated in the membrane bilayer (1). To take up such substrates, the entrance gate of the transporter needs to open laterally into the lipophilic interior of the membrane bilayer. The width of the cleft between TM4 and TM6 in the membrane bilayer is not sufficient to allow uptake of larger molecules in such a fashion. However, TM4 is unfolded from Gly277 to Gly286 and kinked at Pro271 and Gly299 (Fig. 4A and Fig. S3D), suggesting that because of its flexibility, TM4 enables the entrance gate to open as wide as the distance between TM5 and TM6. This notion is supported by two mutation experiments that we performed on TM4. The GAA/VVV mutant, which restricts TM4 movement (Fig. S3 B and D and Tables S2 and S3), exhibited significantly reduced drug-transport activity for either rhodamine 6G or smaller molecules, tetraphenylphosphonium bromide and fluconazole (Fig. 4B and Fig. S3E), as well as lower basal and substrate-stimulated ATPase activities (Fig. 4C and Table S1). Furthermore, Ala substitution of Pro271, which is a highly conserved residue (Fig. S1) and is located around the midpoint of the bilayer, also decreased transport activity (Fig. 4B and Fig. S3E). Proline is a key residue for hinge bending motion of TM helices (30). Therefore, P271A mutation also could cause perturbation of dynamic motion of TM4. These results suggest that the flexibility or mobility of the TM4 helix is critical for efficient drug binding and transport. Thus, TM4 is a “gatekeeper” of the substrate entrance gate. Consistent with these findings, TM10 of C. elegans P-gp, which corresponds to TM4 of CmABCB1, is also unstructured (11).
Fig. 4.
Intramembranous gate. (A) Close-up view of the disordered region of TM4. The protein (except TM4) is shown as a semitransparent surface colored by electrostatic potential contoured from −10 kT (red) to +10 kT (blue). The amino acid sequence of the unwound region of TM4 is highlighted. A model of a 2-oleoyl-1-palmitoyl-sn-glycero-3-phosphocholine (POPC) molecule in the inner leaflet of the bilayer is shown as spheres. (B) Rhodamine 6G susceptibility of yeast cells expressing CmABCB1 variants. E610A is an ATPase-deficient mutant (SI Materials and Methods). (C) Rhodamine 6G concentration dependence of ATPase activity of WT (○) and GAA/VVV mutant (●) CmABCB1. (Inset) Close-up of the graph for the GAA/VVV mutant.
Structural Comparison Between CmABCB1 and Sav1866.
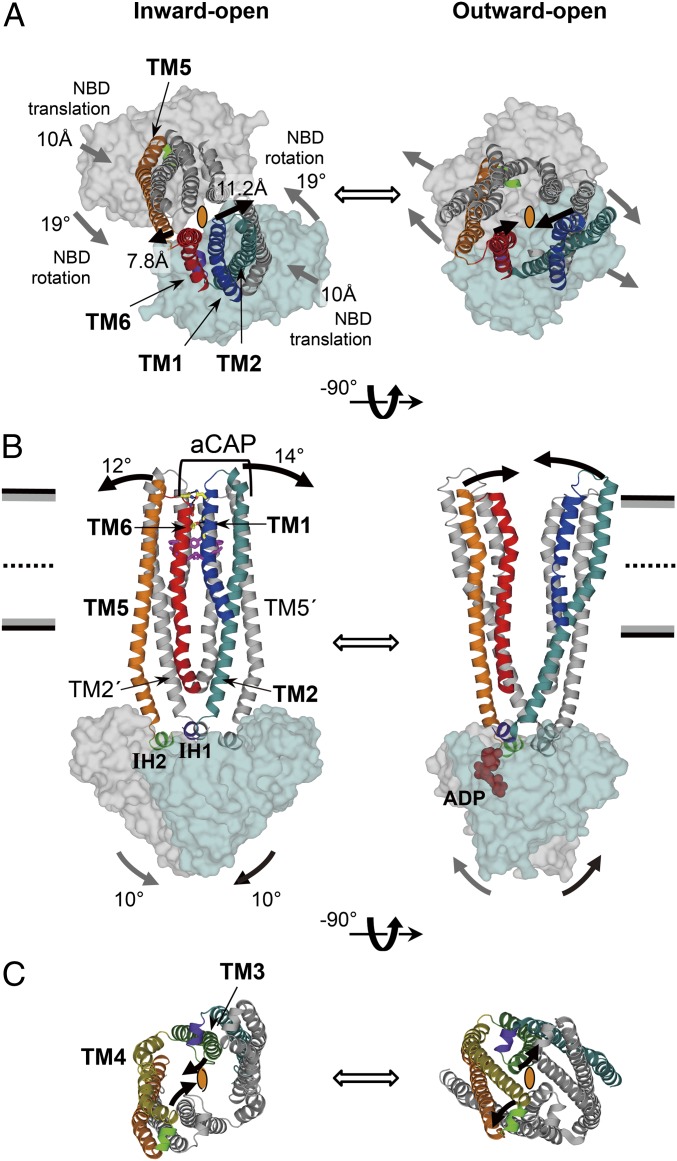
The superposition of the inward-open CmABCB1 and the outward-open Sav1866 (12) reveals that the structural changes of the transmembrane helices of these transporters are induced by a rigid-body motion of the NBDs involving both translation and rotation. Because both structures have homo-dimeric construction and resolutions better than 3.0 Å, their overall structures provide a more reliable structural comparison than other inward- and outward-open pairs of ABC transporter structures. Although the TMDs of these two proteins adopt different conformations, their NBD structures are well aligned, as illustrated by the RMSD value of 2.4 Å for the Cα atoms in a superposition of the two structures (Fig. S7A). Thus, the structural difference between their inward-open and outward-open structures is likely to be caused by rigid-body movements of the NBDs. During dimerization toward the outward-open structure, the NBDs of the inward-open structure rotate by 19° (Fig. 5A and Fig. S7B), tilt by 10° (Fig. 5B), and translate by 10 Å (Fig. 5A and Fig. S7B) with respect to the crystallographic twofold axis at the center of the dimer. Structural comparison also reveals the roles of each TM helix during the structural change from the inward-open to outward-open states (Movie S1). The movements of the NBDs are transmitted to two pairs of TMs (2 and 5′; 2′ and 5) through IH1 and IH2 (Fig. 5B and Fig. S7B). Notably, ATP binding to the NBD may result in formation of a rigid foundation under IH1, preventing trembling of TM2 and facilitating precise transmission of NBD motions to TM2 (Fig. 5B and Fig. S7E). TM2′ and TM5 are tightly associated via hydrogen bonding and Gly-zipper motifs (Fig. S5). The tilting of TM2′ by 14° and TM5 by 12° (Fig. 5B) directly pulls TM1′ and TM6 in the same direction (Fig S7C). Thus, the extracellular terminals of TM1 and TM6 move in opposite directions by 11.2 and 7.8 Å, respectively, thereby dissociating TM1 and TM6 from each other (Fig. 5 A and B and Movie S1). This dissociation of TM1 and TM6 collapses the four-helix bundle structure that closes the pathway from the inner cavity to the extracellular side in the inward-open state, resulting in adoption of the outward-open state (Figs. 2A and 5 A and B). Inhibition by aCAP, which forms bidentate hydrogen bonds to TM2 and TM6, which tightly sandwich TM1, provides experimental evidence that the TM1–TM6 dissociation is indispensable for the transporter activity. As a consequence of the combination of tilt and rotation as well as translation, the direction of outward-gate opening is approximately perpendicular to that of the translational motion of the NBDs (Fig. 5A and Fig. S7C). In the cytosolic region near the NBDs, TMs 3, 4, 3′, and 4′ form another right-handed four-helix bundle structure surrounding the center of the dimer, and this structure closes the pathway from the inside of the transporter to the cytosol (Fig. 5C and Movie S1). During this motion, TM4 is straightened and interacts with TM3 in the cytosol, drawing TM6 away from TM6′ perpendicular to the translational motion of the NBDs (Fig. S7D). This movement of TM6 may also assist in efficient formation of an outward-open state. This perpendicular motion, mediated by the gating mechanisms described above, may permit wide opening of the gates against the lateral pressure exerted by the lipid bilayer.
Fig. 5.
Structural comparison between CmABCB1 and Sav1866. (A–C) Inward-open CmABCB1 (Left) and the outward-open Sav1866 (Right), viewed from the extracellular side (A), parallel to the plane of the membrane (B), and from the cytoplasm (C). In A and C, the crystallographic twofold axis at the center of each homodimer is depicted as an ellipse. TM helices and NBDs are depicted in cartoon and surface representation, respectively. In A and B, TM3 and TM4 are omitted for simplicity. In C, only TMDs except TM1 and TM6 are shown. The expected motions from inward- to outward-open states (or vice versa) are indicated as thick arrows; in A and B, the motions on near and far sides of the molecule are shown in black and gray, respectively.
Transport Mechanism.
The transport mechanism of the eukaryote P-gp homolog ABC multidrug transporter, which we revealed in this study, can be summarized as follows. Transport substrates can enter from the inner-leaflet side of the lipid bilayer. Entry of substrate molecules is regulated by the intramembrane gatekeeper, TM4, whose flexibility enables uptake of substrates with various sizes and structures. On the upper side of the TMDs, the extracellular exit gate consists of the right-handed four-helix bundle (TMs 1, 6, 1′, and 6′), along with two other helices, TM5 and TM5′. These structures are locked by the four-layered cluster of hydrophobic and aromatic residues and hydrogen bonds between TM1 and TM6, which together maintain the structure of ABC transporter in the inward-open state. A substrate molecule attaches to the hydrophobic ceiling of the inner cavity and interacts with a Tyr residue (Tyr358 in CmABCB1), which induces the opening of the extracellular exit gate and accelerates ATPase activity. These structural features and processes are consistent with the “vacuum cleaner” model (31, 32). The movement of these TMs in opposite directions is caused mostly by the tilting of the transmitter helices, TM2 and TM5′ or TM2′ and TM5, which is powered by dimerization of the NBDs. The inhibitor aCAP reinforces the TM1–TM6 interaction and thereby prevents the extracellular gate from opening. This unique inhibitory mechanism strongly suggests that opening of the extracellular gate is required for the substrate-dependent ATP hydrolysis cycle of ABC multidrug transporters.
Materials and Methods
For crystallographic experiments, WT CmABCB1 fused to a tobacco etch virus (TEV) protease–cleavable C-terminal His10 affinity tag was overexpressed in Pichia pastoris SMD1163 cells, whereas mutant CmABCB1 variants with C-terminal FLAG and His6 affinity tags were overexpressed in Saccharomyces cerevisiae AD1-8u− cells. Recombinant proteins were purified by immobilized metal-ion affinity chromatography (IMAC). The C-terminal His10 tag of purified WT CmABCB1 protein was then digested by TEV protease, and the cleaved C-terminal His10 tag and TEV protease were removed by passing the digestion reaction through an IMAC column. The WT CmABCB1 (with the C-terminal His10 removed) and the IMAC purified mutant CmABCB1 variants were further purified by gel filtration after cleavage of the flexible N-terminal region (1–92) by trypsin treatment (Fig. S8). For aCAP–CmABCB1 complex formation, aCAP was mixed with CmABCB1 (2:1 molar ratio). The transporter was concentrated to 10 mg/mL in buffer containing 20 mM Tris⋅HCl (pH 7.0), 150 mM NaCl, and 0.2% (wt/vol) n-decyl-β-d-maltopyranoside. Crystallization was performed by sitting-drop vapor diffusion at 20 °C with a precipitation solution containing 14–16% (wt/vol) polyethylene glycol 2000 monomethyl ether and 100 mM magnesium nitrate. X-ray diffraction data (50–2.4 Å, space group R32) were collected using an MAR225HE detector at beamline BL41XU of SPring-8 and processed using the HKL2000 software suite. The initial phases were derived from multiwavelength anomalous dispersion on a mercury derivative. Each final model was obtained by an iterative process of manual model building and refinement against X-ray diffraction data. Crystallographic data and refinement statistics are shown in Tables S2 and S3. The functional properties of CmABCB1 mutants were examined using ATPase activity assays of the purified preparations and drug susceptibility assays using the CMD148C transformant of S. cerevisiae AD1-8u−, which is susceptible to cytotoxic drugs due to the disruption of seven endogenous ABC transporters. Details of all experimental procedures are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank T. Kuroiwa for the gift of genomic DNA of Cyanidioschyzon merolae, T. Sato for the cloning of CMD148C, and K. Egawa for assistance with fed-batch fermentation. We also thank the members of the beamline staff at SPring-8, especially T. Kumasaka, K. Hasegawa, and N. Shimizu. The X-ray experiments were performed at the BL41XU of SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (Proposals 2012B1348, 2012A1304, 2011B1210, 2011A1289, 2010B1324, 2010A1275, 2009B1515, 2009A1458, and 2008B1537). This research was supported by the Targeted Proteins Research Program (H.K.); the SACLA (SPring-8 Angstrom Compact Free Electron Laser) Research Program (H.K.); Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan Grants-in-Aid for Scientific Research 25251006 and 23118709 (to H.K.), 20228001 and 25221203 (to K.U.), and 24689029 (to A.K.); the Japan New Energy and Industrial Technology Development Organization (Y.K.); Japan Society for the Promotion of Science (JSPS) Specially Promoted Research Grant-in-Aid 21000005 and Platform for Drug Discovery, Informatics, and Structural Life Science from MEXT (to H.S.); and JSPS Post-Doctoral Fellows Grant-in-Aid P11344 (to C.J.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3WME, 3WMF, and 3WMG).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321562111/-/DCSupplemental.
References
- 1.Gottesman MM, Pastan I. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu Rev Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- 2.Lam FC, et al. beta-Amyloid efflux mediated by p-glycoprotein. J Neurochem. 2001;76(4):1121–1128. doi: 10.1046/j.1471-4159.2001.00113.x. [DOI] [PubMed] [Google Scholar]
- 3.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat Rev Cancer. 2002;2(1):48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 4.Szakács G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5(3):219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 5.Schinkel AH, Jonker JW. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family: An overview. Adv Drug Deliv Rev. 2003;55(1):3–29. doi: 10.1016/s0169-409x(02)00169-2. [DOI] [PubMed] [Google Scholar]
- 6.Borst P, Elferink RO. Mammalian ABC transporters in health and disease. Annu Rev Biochem. 2002;71:537–592. doi: 10.1146/annurev.biochem.71.102301.093055. [DOI] [PubMed] [Google Scholar]
- 7.Ling V. Multidrug resistance: Molecular mechanisms and clinical relevance. Cancer Chemother Pharmacol. 1997;40(Suppl):S3–S8. doi: 10.1007/s002800051053. [DOI] [PubMed] [Google Scholar]
- 8.Hyde SC, et al. Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature. 1990;346(6282):362–365. doi: 10.1038/346362a0. [DOI] [PubMed] [Google Scholar]
- 9.Higgins CF. ABC transporters: From microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 10.Aller SG, et al. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323(5922):1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin MS, Oldham ML, Zhang Q, Chen J. Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature. 2012;490(7421):566–569. doi: 10.1038/nature11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443(7108):180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 13.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc Natl Acad Sci USA. 2007;104(48):19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hohl M, Briand C, Grütter MG, Seeger MA. Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation. Nat Struct Mol Biol. 2012;19(4):395–402. doi: 10.1038/nsmb.2267. [DOI] [PubMed] [Google Scholar]
- 15.Forrest LR, Rudnick G. The rocking bundle: A mechanism for ion-coupled solute flux by symmetrical transporters. Physiology (Bethesda) 2009;24:377–386. doi: 10.1152/physiol.00030.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnamurthy H, Gouaux E. X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature. 2012;481(7382):469–474. doi: 10.1038/nature10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301(5633):616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 18.Abramson J, et al. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301(5633):610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 19.Reyes N, Ginter C, Boudker O. Transport mechanism of a bacterial homologue of glutamate transporters. Nature. 2009;462(7275):880–885. doi: 10.1038/nature08616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khare D, Oldham ML, Orelle C, Davidson AL, Chen J. Alternating access in maltose transporter mediated by rigid-body rotations. Mol Cell. 2009;33(4):528–536. doi: 10.1016/j.molcel.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuzaki M, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428(6983):653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- 22.Hipolito CJ, Suga H. Ribosomal production and in vitro selection of natural product-like peptidomimetics: The FIT and RaPID systems. Curr Opin Chem Biol. 2012;16(1-2):196–203. doi: 10.1016/j.cbpa.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Yamagishi Y, et al. Natural product-like macrocyclic N-methyl-peptide inhibitors against a ubiquitin ligase uncovered from a ribosome-expressed de novo library. Chem Biol. 2011;18(12):1562–1570. doi: 10.1016/j.chembiol.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Spencer RH, Rees DC. The alpha-helix and the organization and gating of channels. Annu Rev Biophys Biomol Struct. 2002;31:207–233. doi: 10.1146/annurev.biophys.31.082901.134329. [DOI] [PubMed] [Google Scholar]
- 25.Ambudkar SV, et al. Partial purification and reconstitution of the human multidrug-resistance pump: Characterization of the drug-stimulatable ATP hydrolysis. Proc Natl Acad Sci USA. 1992;89(18):8472–8476. doi: 10.1073/pnas.89.18.8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharom FJ, Yu X, Chu JW, Doige CA. Characterization of the ATPase activity of P-glycoprotein from multidrug-resistant Chinese hamster ovary cells. Biochem J. 1995;308(Pt 2):381–390. doi: 10.1042/bj3080381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato T, et al. Functional role of the linker region in purified human P-glycoprotein. FEBS J. 2009;276(13):3504–3516. doi: 10.1111/j.1742-4658.2009.07072.x. [DOI] [PubMed] [Google Scholar]
- 28.Seelig A. A general pattern for substrate recognition by P-glycoprotein. Eur J Biochem. 1998;251(1-2):252–261. doi: 10.1046/j.1432-1327.1998.2510252.x. [DOI] [PubMed] [Google Scholar]
- 29.Ecker G, Huber M, Schmid D, Chiba P. The importance of a nitrogen atom in modulators of multidrug resistance. Mol Pharmacol. 1999;56(4):791–796. [PubMed] [Google Scholar]
- 30.Barlow DJ, Thornton JM. Helix geometry in proteins. J Mol Biol. 1988;201(3):601–619. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]
- 31.Raviv Y, Pollard HB, Bruggemann EP, Pastan I, Gottesman MM. Photosensitized labeling of a functional multidrug transporter in living drug-resistant tumor cells. J Biol Chem. 1990;265(7):3975–3980. [PubMed] [Google Scholar]
- 32.Higgins CF, Gottesman MM. Is the multidrug transporter a flippase? Trends Biochem Sci. 1992;17(1):18–21. doi: 10.1016/0968-0004(92)90419-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.