A cell’s genome is under constant threat of damage, which if not repaired can lead to mutations or cell death. Common forms of DNA damage found in nature include cyclobutane pyrimidine dimers and 6-4 photoproducts induced by UV-irradiation. These and other helix-distorting lesions are removed by a highly conserved process called nucleotide excision repair (NER) that is found in every kingdom of life (1). NER is initiated in two general ways: by damage recognition proteins that survey the entire genome for damage or lesion-induced transcriptional stalling. This latter pathway, called transcription-coupled repair (TCR), first reported in mammalian cells and then in bacteria, is initiated when RNA polymerase (RNAP) is arrested at a DNA lesion embedded in the transcribed strand (2, 3). However, before DNA repair enzymes obtain access, the stalled RNAP must be pushed away from the lesion by the action of DNA translocases. Thus, the repair “coupling factors,” which recognize the stalled RNAP, must work to both displace the polymerase and simultaneously enlist the repair proteins to remove the damage. In bacteria, two different TCR pathways have emerged involving two different DNA helicases, which help to displace RNAP. The Mfd (mutation frequency decline) protein, also called transcription-repair coupling factor, uses its helicase fold and ATP hydrolysis to literally push RNAP forward (downstream) past the damaged site (Fig. 1A) (reviewed in ref. 4), whereas in a newly discovered alternative pathway UvrD (helicase II) tows the RNAP backward (upstream) with the help of the transcription elongation factor, NusA (5). This second approach more closely resembles what is thought to occur in mammalian cells during TCR (6). As described below, Mfd targets the nucleotide excision repair system to sites of damage through its direct interaction with a stalled RNAP. However, nature has gone even further in devising ways to find and remove potentially RNAP-blocking DNA damage. As described by Haines et al. (7) in PNAS, Nigel Savery’s group at the University of Bristol have found that the Mfd protein that normally accompanies the translocating RNAP can be sent ahead of a blocked RNAP to scout for damage in the transcribed strand and facilitate the recruitment of the bacterial NER machinery.
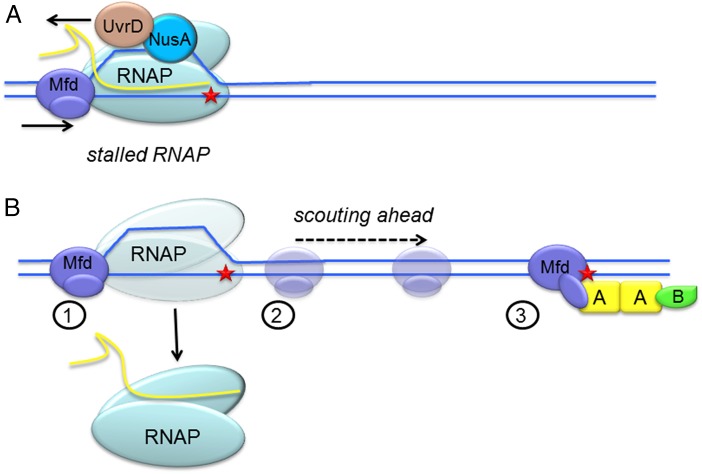
Fig. 1.
(A) RNAP stalled at a DNA lesion can be removed by the action of two different helicases. Mfd can push RNAP downstream past the lesion or UvrD/NusA can tow RNAP upstream of the damage site. Recruitment of UvrAB occurs after RNAP is moved, not shown. (B) 1, Mfd senses RNAP pausing and 2, probably dissociates RNAP from the DNA and begins scouting on the transcribed strand (9); 3, once Mfd has encountered a lesion on the transcribed strand a repair-competent complex is formed. The timing of UvrA and UvrB recruitment at the damaged site remains to be determined.
During the past 15 y structural biologists and biochemists have described in wonderful detail how bacterial NER proteins process and remove DNA damage (reviewed in ref. 1). Once the stalled RNAP has been removed from the damage site, Mfd or UvrD are thought to recruit the UvrAB complex (made up of a UvrA dimer and one or two UvrB molecules) to the site of the damage. Mfd is a multidomain protein that shares a protein fold with UvrB and this motif is responsible for the interaction face with UvrA. Mfd undergoes a large conformational change during its handling of stalled RNAP. This process exposes Mfd’s UvrB homology domain, which is believed to attract UvrA to the damaged site. Single-molecule studies have shown that the UvrAB complex searches for DNA damage using both 3D and 1D sliding on DNA (8). It has been proposed that one monomer of UvrA can interact with Mfd, whereas the other monomer engages UvrB. UvrA first recognizes the lesion-induced conformational change in the DNA and hands off the damage to UvrB for verification. UvrB recruits UvrC, which incises the DNA 3′ and 5′ of the damaged site. The actions of the helicase UvrD and DNA polymerase I are necessary to dissociate UvrB, UvrC, and the damage-containing oligonucleotide. The DNA polymerase fills in the resulting gap and DNA ligase seals the nick to join the repair patch to the contiguous DNA strand (1).
In an ingenious series of experiments, Haines et al. (7) first paused RNAP in the absence of DNA lesions by using the incorporation of dUTP (an RNA chain terminator) instead of UTP, 21 nucleotides after transcription initiation. The researchers then constructed a substrate containing a defined lesion 47 or 81 bases downstream on either the nontranscribed or transcribed strand by incorporating a single biotin-modified deoxythymidine (dT) or a cyclobutane pyrimidine dimer (CPD) into the DNA. In separate experiments they show that a biotin-modified dT is a robust substrate for the UvrABC nuclease system, but that this nucleotide analog does not stall RNAP when located on the transcribed strand. This latter point was important to rule out any effects of RNA polymerase molecules that might escape the pause site. Using an assay comprised of the complete NER system (UvrA, UvrB, UvrC, UvrD, pol I, DNA ligase), as well as RNAP and Mfd, which measures incorporation of [32P]-dATP into repair patches, they found that repair was stimulated only when the lesion was located on the transcribed strand downstream of RNA polymerase. So the question arises, is this enhanced repair the result of a locally higher Mfd concentration or by direct tracking of Mfd downstream of the stalled RNAP? To address this question, the authors placed a lacO operator sequence between the RNAP stall site and a damage cassette consisting of UV-irradiated DNA, reasoning that if Mfd were to track forward away from the stalled RNAP in a 3′→5′ direction on the transcribed strand, then its forward progress would be prevented when the lac0 operator was bound by Lac repressor. This result is exactly what the authors observed, suggesting that Mfd literally tracks along the transcribed strand as it scouts for DNA damage, and that “downstream repair can be blocked by a protein roadblock.” Interestingly, the presence of the Lac repressor road block actually reduced UvrABC-mediated repair by about 50%, independent of RNAP or Mfd. This result strongly suggests that the UvrAB complex uses facilitated diffusion along the DNA to detect damage and blocked access from the 5′ direction, thereby lowering overall repair by about 50%, as expected (8). These data also suggest that the UvrAB motion on DNA occurs through linear diffusion on DNA rather than hopping, which would permit jumping over the Lac repressor roadblock.
The next question is then: how far out ahead of RNAP can the Mfd scouting party go? One way to measure Mfd’s ability to track along the DNA is to make use of its known capacity to displace triplex forming oligonucleotides (TFO). Triplexes are unique three-stranded DNA sequences that are created by Hoogsteen base-pairing of a third strand into the major groove of double-stranded DNA, and are strong blocks to Mfd. By placing a 17-bp TFO either 100 or 581 bp away from the stalled RNAP, Haines et al. (7) found that Mfd can scout ahead of RNAP a full 0.5 kbp from the stalled RNA polymerase. If Mfd facilitates repair at damaged sites by recruiting the UvrAB proteins, it might be expected that Mfd’s translocation ability would be inhibited by damage on the transcribed strand, but not on the nontranscribed strand. The authors show that a single CPD, but not a single biotin-dT “damage site,” impedes the ability of Mfd to displace a TFO. This stalling of the Mfd translocase at a damaged site is reminiscent of the manner by which the damage recognition helicase, XPD (a subunit of TFIIH), in mammalian cells verifies the strand-containing damage. In TFO displacement assays, Mfd stimulated repair at biotin-dT sites, but apparently did not stall at these “damaged sites.” Consequently, it is not clear whether Mfd stalling is essential for its ability to target UvrAB to the damaged site. Ultimately, single-molecule studies using uniquely labeled Mfd, RNAP, and UvrAB proteins may be necessary to determine whether Mfd scouts ahead of the paused RNAP and then remains for a sufficient period at damaged sites to recruit UvrAB.
How frequently does RNAP pause in vivo and could this interval help to facilitate repair? RNAP pause sites in bacteria are best characterized by the ops (operon polarity suppressor), which initiates both pausing and backtracking of RNAP. Mfd has been shown to facilitate reengagement of RNAP transcription at these pause sites. Haines et al. (7) show that the ops sequences can interrupt RNAP and facilitate repair of biotin-dT lesions downstream of these sites. This enhanced repair was only observed when Mfd and RNAP were both present.
Taken together, these experiments strongly support a model of damage surveillance in bacteria in which the pausing of RNAP prompts Mfd to scout downstream for DNA damage on the transcribed strand (Fig. 1B). As Haines et al. (7) suggest, specific sequences that promote RNAP pausing in vivo may result in localized sites of rapid repair and subsequent lower mutations rates. These otherwise beautiful results leave several important questions unanswered. Although not explicitly tested in this work, it has been shown that Mfd displaces RNAP from damaged sites and continues to track on the DNA (9). How long must RNAP pause before it releases Mfd? Does Mfd have to dissociate RNAP to scout ahead for DNA damage? And if so, what conformational change is triggered in RNAP that facilitates Mfd release? Could UvrD working with NusA push RNAP backward, and at the same time release Mfd on a scouting expedition? Finally, how frequently does this process occur in a bacterial cell? Bacterial transcription and translation are highly coupled and transcriptional pausing allows for proper mRNA folding and recruitment of the elongation factors. Mutation frequency decline (MFD) was first described by Evelin Witkin in the 1950s and 1960s (before the discovery of NER in 1964) as a phenomenon in which bacteria grown after exposure to UV under less than ideal conditions (such as reduced protein synthesis) show a reduction in mutations (10, 11). It is tempting to speculate that growth under such conditions promotes increased transcriptional pausing and consequently increases DNA damage scouting by Mfd, with subsequent repair by NER on the transcribed strand. Taken together, these exciting findings by Haines et al. (7) indicate that bacterial cells still have a lot to teach us and that enhanced repair pathways ultimately might be linked to the nutritional status and growth rate of a cell in a more integrated and elegant fashion than previously supposed. Might a similar process occur in mammalian cells, and contribute to the increased longevity associated with caloric restriction?
Acknowledgments
This work is supported by National Institutes of Health Grant 1R01ES019566 (to B.V.H.) and by the Deutsche Forschungsgemeinschaft KI-562/2 and Forschungszentrum FZ-82 (to C.K.).
Footnotes
The authors declare no conflict of interest.
See companion article on page 4037.
References
- 1.Kisker C, Kuper J, Van Houten B. Prokaryotic nucleotide excision repair. Cold Spring Harb Perspect Biol. 2013;5(3):a012591. doi: 10.1101/cshperspect.a012591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342(6245):95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 3.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51(2):241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 4.Ganesan A, Spivak G, Hanawalt PC. Transcription-coupled DNA repair in prokaryotes. Prog Mol Biol Transl Sci. 2012;110:25–40. doi: 10.1016/B978-0-12-387665-2.00002-X. [DOI] [PubMed] [Google Scholar]
- 5.Epshtein V, et al. UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature. 2014;505(7483):372–377. doi: 10.1038/nature12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9(12):958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 7.Haines NM, Kim Y-IT, Smith AJ, Savery NJ. Stalled transciption complexes promote DNA repair at a distance. Proc Natl Acad Sci USA. 2014;111:4037–4042. doi: 10.1073/pnas.1322350111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kad NM, Wang H, Kennedy GG, Warshaw DM, Van Houten B. Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single- molecule imaging of quantum-dot-labeled proteins. Mol Cell. 2010;37(5):702–713. doi: 10.1016/j.molcel.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howan K, et al. Initiation of transcription-coupled repair characterized at single-molecule resolution. Nature. 2012;490(7420):431–434. doi: 10.1038/nature11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witkin EM. Time, temperature, and protein synthesis: A study of ultraviolet-induced mutation in bacteria. Cold Spring Harb Symp Quant Biol. 1956;21:123–140. doi: 10.1101/sqb.1956.021.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Witkin EM. Mutation frequency decline revisited. Bioessays. 1994;16(6):437–444. doi: 10.1002/bies.950160613. [DOI] [PubMed] [Google Scholar]